Abstract
Accumulation of the amyloid β peptide in the cortical and hippocampal regions of the brain is a major pathological feature of Alzheimer disease. Amyloid β peptide is generated from the sequential protease cleavage of the amyloid precursor protein (APP). We reported previously that copper increases the level of APP at the cell surface. Here we report that copper, but not iron or zinc, promotes APP trafficking in cultured polarized epithelial cells and neuronal cells. In SH-SY5Y neuronal cells and primary cortical neurons, copper promoted a redistribution of APP from a perinuclear localization to a wider distribution, including neurites. Importantly, a change in APP localization was not attributed to an up-regulation of APP protein synthesis. Using live cell imaging and endocytosis assays, we found that copper promotes an increase in cell surface APP by increasing its exocytosis and reducing its endocytosis, respectively. This study identifies a novel mechanism by which copper regulates the localization and presumably the function of APP, which is of major significance for understanding the role of APP in copper homeostasis and the role of copper in Alzheimer disease.
Keywords: Alzheimer Disease, Copper, Membrane Proteins, Neurodegeneration, Trafficking, Amyloid Precursor Protein, Amyloid beta Peptide, Metal Dyshomeostasis
Introduction
The neuropathology of Alzheimer disease (AD)3 includes the accumulation of extracellular plaques containing amyloid β peptide (Aβ) in the cortical and hippocampal regions of the brain, intracellular neurofibrillary tangles, and trace metal dyshomeostasis. The trace metals copper, zinc, and iron are significantly enriched in the amyloid plaques of AD patients compared with age-matched subjects (1, 2). Interaction between Aβ and copper or zinc promotes peptide oligomerization and aggregation, eventually leading to further amyloid deposition (3–7). In addition, Aβ is able to catalyze the reduction of Cu(II) and Fe(III), generating reactive oxygen species that contribute to the oxidative stress observed in the AD brain (8–14). Paradoxically, there is evidence of copper deficiency in neighboring cells (15–18), compromising the activity of copper-dependent enzymes, such as cytochrome c-oxidase and copper/zinc-superoxide dismutase (SOD1), which are essential for cellular respiration and as antioxidant defense, respectively (19–21). Restoration of copper balance using ionophores (22), such as clioquionol and PBT-2, has shown promising results in both animal and human trials (23–26). A proposed mechanism of action of these drugs is to bind to extracellular copper and restore intracellular copper levels, hence correcting copper dyshomeostasis. Importantly, these ionophores are capable of reducing amyloid load and attenuate cognitive decline (23–26).
Amyloid precursor protein (APP) is differentially processed by the α-, β-, and γ-secretases in two alternative processing pathways, commonly referred to as the non-amyloidogenic and amyloidogenic pathways (supplemental Fig. S1). In the non-amyloidogenic pathway, APP is initially cleaved by an α-secretase that is predominantly localized to the plasma membrane (27, 28), generating an N-terminal ectodomain (sAPPα) and a C-terminal fragment, C83 (CTFα). Further cleavage of C83 by γ-secretase produces a small non-toxic peptide, p3, and a cytoplasmic polypeptide (APP intracellular domain (AICD)). Alternatively, APP undergoes β-cleavage in BACE-1 (β-site APP-cleaving enzyme)-enriched endosomes (29), producing an N-terminal ectodomain (sAPPβ) and a C-terminal fragment, C99 (CTFβ). Subsequent cleavage of C99 by γ-secretase during endocytic/recycling steps and the secretory pathway generates Aβ and the AICD (30). The α-, β-, and γ- secretases are localized to distinct cellular compartments, and therefore, segregation of APP into particular cellular compartments determines the pathway by which it is processed.
Copper has been shown to modulate APP metabolism. An increase in intracellular copper levels promotes the non-amyloidogenic pathway (31, 32). In addition, copper regulates APP expression, whereby a decrease in intracellular copper down-regulates APP gene expression (33). Conversely, elevated cellular copper levels result in up-regulation of APP gene expression (34). Importantly, APP is proposed to participate in copper homeostasis via a role in the copper efflux pathway (35, 36). Overexpression of APP in cultured cell and animal models leads to decreased cellular copper levels (35, 37, 38). We recently reported that copper promotes an increase in the level of APP at the cell surface in SH-SY5Y human neuroblastoma cells with a reduction in lipid raft-mediated APP processing (39).
In the current study, we investigated the effect of copper on APP cellular localization and the dynamics of changes in its localization. We report that in both neuronal and non-neuronal cell models, APP traffics from the Golgi to intracellular compartments and to the cell surface in response to increases in intracellular copper but not zinc or iron. We provide evidence that this is due to an increase in the rate of APP exocytosis with a concomitant reduction in its rate of endocytosis.
MATERIALS AND METHODS
Antibodies and Reagents
The following antibodies were used in this study: GM-130 (BD Transduction Laboratories), golgin-97 (Invitrogen), CT20 (C-terminal APP antibody; Calbiochem), β-actin (Sigma), W0-2 (40), and 22C11 (41). The antibody CT77 was used to detect the copper transporter ATP7A and was a kind gift from Prof. B. Eipper (Neuroscience and Molecular, Microbial, and Structural Biology Division, University of Connecticut) (42, 43). The APP antibodies W0-2, 22C11, and CT20 all recognize full-length APP. The W0-2 antibody also specifically recognizes the human Aβ sequence and sAPPα, whereas the CT20 antibody specifically recognizes residues 751–770 and will detect C-terminal fragments cleaved by α-, β-, and γ-secretases. In contrast, 22C11 recognizes an epitope site within the N terminus of APP and detects sAPPα and sAPPβ (supplemental Fig. S7). For immunocytochemistry, secondary IgG antibodies conjugated to AlexaFluor® 488 or AlexaFluor® 594 fluorophores (Invitrogen) were used at 1:400 to detect primary antibodies. The nucleus was visualized using DAPI nucleic acid stain (Invitrogen) at a final concentration of 100 ng/ml. Cycloheximide (50 μg/ml; Sigma) was used to inhibit protein synthesis.
Cell Culture
Madin-Darby canine kidney (MDCK) cells (American Type Culture Collection catalog no. CCL-34) were cultured in BME medium (HyClone) supplemented with 2 mm l-glutamine, 1.2 mm NaHCO3, 20 mm HEPES, and 10% fetal calf serum (Bovogen, Victoria, Australia). Human neuroblastoma SH-SY5Y cells (American Type Culture Collection catalog no. CRL-2266) were cultured in DMEM (Invitrogen) containing GLUTAMAXTM-I (Invitrogen) supplemented with 10% fetal calf serum. Primary cortical neurons were isolated from embryonic day 14 mouse embryos as described previously (44). Primary cortical neurons were initially cultured in DMEM with GLUTAMAXTM-I (Invitrogen) containing 7.5% NaHCO3, 5% horse serum, and gentamycin (Invitrogen), which was replaced after 2 h with Neurobasal medium (Invitrogen) containing B27 supplement (Invitrogen), gentamycin, and 200 mm GLUTAMAXTM-I (Invitrogen) for growth and maintenance. All cell lines were cultured at 37 °C and in the presence of 5% CO2.
Generation of MDCK-APP-cherry Stable Cell Line
The pcDNA3.1-APP-cherry expression vector was generated by first subcloning the cherry fluorescent tag (mCherry) at the BamHI/NotI site of the pcDNA3.1 vector (Invitrogen). The cherry tag was a gift from Professor Roger Tsien (University of California). The wild type APP695 cDNA (in the pIRESpuro2 vector), a gift from Robyn Sharples and Assistant Professor Andrew Hill (Bio21 Institute, Melbourne, Australia), was subcloned into the pcDNA3.1-cherry vector at the NheI/HindIII site, N-terminal of the cherry tag. MDCK cells cultured in 6-well plates were transfected with 2.4 μg of plasmid DNA (pcDNA3.1-APP-cherry) using the Lipofectamine 2000TM reagent (Invitrogen) according to manufacturer's instructions. Cells stably expressing APP-cherry were selected and maintained with Geneticin (0.5 mg/ml; Invitrogen) 48 h following transfection. To obtain an enriched population of APP-cherry-expressing cells, transfected MDCK cells were selected by flow cytometry. MDCK cells express low levels of endogenous APP, facilitating investigations using transfected fluorescent tagged APP.
Copper Treatments
For studies on localization of APP, cells were incubated in growth medium containing 10% fetal calf serum in the presence of copper (25–150 μm) for 3 h. The copper chelators used were bathocuproine disulfonate for Cu(I) and d-penicillamine for Cu(II), both of which were used at equal concentrations (150 μm, or 50 μm for overnight treatment). Primary cortical neurons were incubated with copper in serum-free Neurobasal medium (Invitrogen), and therefore lower copper concentrations were used (5–20 μm; 3 h). The copper concentrations chosen were based on previously published data (32, 45, 46) as well as taking into consideration the presence of serum in the growth medium, which contains copper-binding proteins that reduce the amount of bioavailable copper, necessitating the use of higher copper concentrations than in serum-free media (32).
Immunocytochemistry
Cells were grown on 12-mm glass coverslips or Costar Transwells (Corning Glass). For primary cortical neurons, coverslips were precoated for at least 2 h with poly-d-lysine (0.1 mg/ml; Sigma). MDCK cells were cultured on coverslips or Transwells and cultured for ∼7 days to reach polarization prior to treatments and immunocytochemistry. Following copper/chelator treatment, cells were fixed with 4% paraformaldehyde in PBS (pH 7.2; Sigma) and permeabilized with 0.1% Triton X-100, and nonspecific sites were blocked with 1% bovine serum albumin (BSA; Sigma) overnight. Antibodies used for immunocytochemistry were used at the following dilutions: GM130 (1:200), golgin-97 (1:200), W0-2 (1:20), CT20 (1:1,000), ATP7A (1:400). Primary antibodies were detected using secondary IgG antibodies conjugated to AlexaFluor® 488 or AlexaFluor® 594 fluorophores. Rhodamine phalloidin (Invitrogen) was used at a 1:50 dilution. Images were taken using an Olympus FV1000 scanning confocal microscope.
Co-localization Analysis; Pearson's Correlation Coefficient
Pearson's correlation coefficient was used to estimate the level of co-localization between APP and the Golgi marker GM130. Pearson's correlation coefficient (rp) is a statistical analysis of the relationship between fluorescence intensities. An rp value greater than 0.5 indicates a high degree of co-localization, whereas a value of less than 0.5 indicates low levels of co-localization (reviewed in Ref. 47). Pearson's correlation coefficient was calculated for 10 cells/treatment (i.e. copper or copper chelator), and the average was calculated.
Western Blot Analysis
Proteins in cell lysates were separated by SDS-PAGE using a NuPAGE 4–12% BisTris gel (Invitrogen), using the MES running buffer (Invitrogen), followed by protein transfer onto Amersham BiosciencesTM HybondTM-ECL 0.2-μm membrane (GE Healthcare). Following transfer, membranes were boiled in PBS for 45 s and blocked in 5% skim milk in Tris-buffered saline (TBS). For protein detection, membranes were incubated with appropriate primary antibodies diluted in TBS containing 0.1% Tween 20 at the following concentrations: W0-2 (1:40) and β-actin (1:500). Proteins were detected by the use of the corresponding secondary IgG horseradish peroxidase-conjugated antibody (DAKO; 1:5,000). Membrane was developed using an ECLTM Western blotting detection system (GE Healthcare) as per the manufacturer's instructions and visualized using the LAS-3000 Imaging system (Fuji).
Analysis of APP Processing
To analyze the level of secreted APP processing products, including sAPPα, -β, and Aβ, conditioned medium was collected. Protein in 400 μl of conditioned medium was precipitated using 5 volumes of ice-cold 10% trichloroacetic acid, as described previously (48). Protein was pelleted by centrifugation at 10,000 × g for 30 min and resuspended in 50 μl of sample loading buffer (Novex Tricine SDS sample buffer, Invitrogen). The level of sAPPα in conditioned medium from three independent experiments was evaluated by densitometry whereby pixel intensities (arbitrary units) were quantified using the Multi Gauge software (Fuji) and normalized relative to total APP detected in the cell lysate.
Cell Surface Biotinylation
Cell surface biotinylation to isolate cell surface proteins was performed as described previously (45, 49). Briefly, SH-SY5Y cells were treated with either copper (150 μm) or copper chelators (150 μm) for 3 h in the presence of cycloheximide (50 μg/ml) to inhibit protein synthesis. To label cell surface proteins, cells were incubated with 0.5 mg/ml sulfosuccinimidyl-2-[biotinamido]ethyl-1,3-dithiopropionate (Sulfo-NHS-SS-Biotin) for 30 min (Thermo Scientific). Biotinylated proteins were precipitated using streptavidin beads (Pierce), eluted, and analyzed by Western blot using the W0-2 antibody to detect cell surface APP.
Live Cell Imaging
MDCK-APP-cherry cells were cultured on 40-mm coverslips in growth medium until confluent and polarized (approximately 7 days). On the day of the experiment, growth medium was replaced with BME medium without phenol red supplemented with 2% fetal calf serum. Coverslips were then mounted onto a “Bioptics” live cell imaging chamber, and cells were imaged with an Olympus FV1000 laser-scanning confocal microscope equipped with a 60× water immersion objective and fitted with a heated “Bioptics” imaging chamber, perfusion pump, and objective heater. Cells were initially imaged at 2-min intervals for ∼10 min prior to the perfusion of above medium containing 150 μm copper. After cells were immersed in copper-supplemented medium, images were taken at 2-min intervals for ∼25 min.
Densitometry Analysis
The level of APP and APP processing products from three independent experiments was evaluated by densitometry, whereby pixel intensities (arbitrary units) were quantified using the Multi Gauge software (Fuji) and normalized relative to total APP detected in the cell lysate.
Antibody Uptake (Endocytosis) Assays
APP endocytosis assays have been described previously (50, 51). Briefly, SH-SY5Y cells plated on coverslips were washed in ice-cold PCM buffer (phosphate buffered saline supplemented with 1 mm CaCl2, 0.5 mm MgCl2) and incubated on ice with 22C11 antibody (20 μg/ml) in PCM for 20 min. Cells were washed twice with PCM prior to returning them to prewarmed growth medium supplemented with either copper (150 μm) or copper chelators (150 μm) for either 0, 5, 15, or 30 min in a 37 °C incubator. After these time points, unbound antibody was removed by washing cells twice for 2 min with acidic buffer (100 mm glycine, 20 mm magnesium acetate, 50 mm KCl, pH 2.2), followed by two washes in ice-cold PBS. Cells were then fixed with 4% paraformaldehyde in PBS for 15 min and processed as above for standard immunofluorescence analysis. The 22C11 antibody was detected using the mouse AlexaFluor® 488 (1:400) secondary antibody. Cells were also labeled with rhodamine phalloidin (Invitrogen) to visualize the F-actin cytoskeleton and the cell surface.
This experiment was repeated three times. In each experiment, between 40 and 60 cells were scored for undergoing APP endocytosis. The rate of endocytosis was represented as the average percentage of cells undergoing endocytosis per time point and treatment, as described previously (50, 51). Statistical analysis was carried out using Student's t test.
RESULTS
Copper-stimulated Relocalization of APP in Cultured Neuronal and Polarized Epithelial Cells
We previously reported that in the human neuroblastoma SH-SY5Y cell line, an increase in copper concentration stimulated an increase in the level of APP at the cell surface (39). In the present study, we further investigated the influence of copper on APP trafficking in both neuronal and polarized epithelial cell models using confocal fluorescence imaging techniques. Experiments were carried out in normal growth medium, and the concentration of copper used was based on our previous studies (32, 45, 46). Consistent with our previous finding (39), exposure of SH-SY5Y cells to 150 μm CuCl2 in serum containing growth medium also resulted in an increase of APP at the cell surface (as assessed by biotinylation) compared with untreated controls (supplemental Fig. S2). APP levels at the cell surface from three independent cell surface biotinylation experiments were analyzed by densitometry. Here, we report an approximately 2-fold increase in cell surface APP following copper treatment (n = 3; p = 0.009).
To assess if the influence of copper on cellular localization of APP was cell line-specific, in addition to SH-SY5Y cells, we also investigated polarized epithelial cells (MDCK cells) and mouse primary cortical neurons. We investigated APP cellular localization in MDCK cells stably expressing wild type APP695 with a C-terminal cherry fluorescent tag. Our rationale for using MDCK cells as a representative non-neuronal cell line was based on these cells having similar mechanisms for polarized protein sorting to neuronal cells (52), and MDCK cells have been widely used to study APP cellular localization and processing (53). Under copper-deficient conditions, APP-cherry demonstrates a perinuclear localization (Fig. 1). However, exposure to 150 μm copper or greater led to a dispersed localization of APP-cherry throughout the cytoplasm (Fig. 1). APP-cherry also redistributed at lower copper concentrations (25 μm) after a longer period of incubation (12 h) (supplemental Fig. S3). This is consistent with lower rates of copper uptake at lower extracellular copper concentrations.
FIGURE 1.
APP-cherry traffics in response to copper in polarized epithelial cells. MDCK cells stably expressing APP with a C-terminal fluorescent tag (“cherry”) were incubated with copper chelators (bathocuproine disulfonate and d-penicillamine (150 μm; i) or increasing levels of copper (50, 150, or 200 μm; ii–iv) for 3 h. At this time interval and at copper concentrations of 150 μm and above, APP-cherry redistributes from a perinuclear localization to a wider distribution throughout the cell as detected by imaging using confocal microscopy. The nucleus was visualized by using DAPI stain. Scale bar, 5 μm.
In MDCK-APP-cherry cells, a proportion of APP-cherry was observed to co-localize with EEA1-positive endosomal compartments. This co-localization increased in the presence of elevated copper (Fig. 2). Notably, this redistribution of APP was specific to copper, and cells incubated in the presence of iron or zinc did not elicit a detectable change in APP localization (supplemental Fig. S4).
FIGURE 2.
The co-localization of APP and early endosomal marker EEA1 increases following copper treatment. MDCK-APP-cherry were incubated with either copper chelators (−Cu; bathocuproine disulfonate and d-penicillamine) or copper (+Cu; 150 μm), followed by immunolabeling with the W0-2 and EEA1 antibodies to detect APP and early endosomes, respectively. Under copper-deficient conditions (−Cu; top), APP shows a partial co-localization with EEA1, which increases following treatment with copper (+Cu; bottom). The nucleus was visualized by using DAPI stain. Scale bar, 5 μm.
To ensure that the observed copper-responsive change in APP-cherry localization was specific to APP, we used the monoclonal antibody W0-2 to immunolabel APP. There was strong co-localization of APP-cherry with W0-2 under the differential copper conditions tested in this study (supplemental Fig. S5).
Copper-responsive APP Trafficking Is Not Due to Up-regulation of APP Expression
Elevated cellular copper levels up-regulate the transcription of the APP gene (32–34). To determine whether copper-responsive trafficking of APP was due to up-regulation of copper-mediated APP expression within the time frame of these experiments, we evaluated APP protein levels under differential copper conditions in SH-SY5Y cells by Western blot analysis (supplemental Fig. S6A). Results indicated that copper-induced APP trafficking was not a consequence of copper-mediated up-regulation of APP protein synthesis. Furthermore, we showed that in the presence of the protein synthesis inhibitor, cycloheximide (50 μg/ml), copper-responsive redistribution of APP was comparable with that observed in the absence of cycloheximide (supplemental Fig. S6B).
Copper Increases APP Localization to Neurites in Neuronal Cell Models
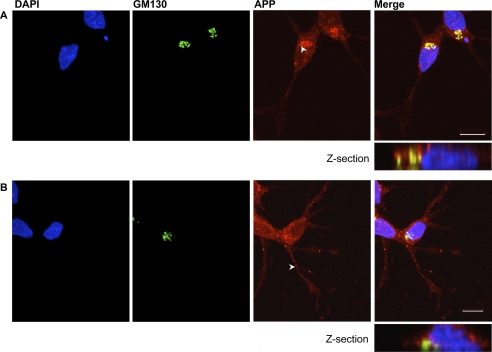
To investigate the effect of copper on APP intracellular distribution in neuronal cells, we evaluated copper-mediated APP trafficking in two neuronal cell models, SH-SY5Y (an established neuronal cell line) and mouse primary cortical neuronal cells, which express relatively high levels of endogenous APP. The APP antibodies used to detect APP include W0-2 (40), 22C11 (41), and CT20 (54) (see supplemental Fig. S7 for a schematic diagram of the epitope sites). These antibodies have different epitope sites and thus were used to detect the various APP processing products. In SH-SY5Y cells, under copper-deficient conditions, there was a high degree of colocalization of APP with Golgi markers GM130 (Fig. 3A) and golgin-97 (data not shown), which was consistent with previous published reports (55–58). Following exposure to elevated copper (150 μm, 3 h), APP redistributed throughout the cell, including in neurites (Fig. 3B). This was accompanied by a decrease in the level of co-localization between APP and GM130 (Fig. 3B). The degree of APP co-localization with GM130 was expressed as Pearson's correlation coefficient (rp) at the Golgi network. Under copper-deficient conditions, we found an average Pearson's correlation coefficient of 0.733 (S.D. = 0.0321), indicative of a relatively high degree of colocalization, which decreased to an average of 0.564 (S.D. = 0.111) following copper treatment, indicating redistribution of APP out of the Golgi compartment. The decrease in rp value following copper treatment was statistically significant (p = 0.0001). We also found similar degrees of co-localization between APP and the Golgi marker golgin-97 following copper and copper chelator treatment (data not shown). It should be noted that copper did not compromise the integrity of the Golgi network, which was evident by the expected localization of the Golgi markers GM130 and golgin-97. This strongly suggested that copper altered the localization of APP rather than affecting Golgi integrity.
FIGURE 3.
APP exits the Golgi in response to elevated intracellular copper in SH-SY5Y cells. SH-SY5Y cells were treated with copper chelators (bathocuproine disulfonate and d-penicillamine; 150 μm) (A) or copper (150 μm) (B) for 3 h. Following fixation steps, cells were immunolabeled with antibodies against APP (W0-2) and GM130, a cis-Golgi marker. The degree of co-localization between GM130 and APP was analyzed using Pearson's correlation coefficient (rp). Under copper-deficient conditions, the average Pearson's correlation coefficient was calculated to be 0.733 (S.D. = 0.0321) in comparison with 0.564 (S.D. = 0.111) following copper treatment. DAPI stain (left panels) was used to visualize the nucleus. The white arrows indicate the localization of APP following chelator or copper incubation. Scale bar, 10 μm.
We previously discovered copper-responsive trafficking of ATP7A, an essential copper-transporting P-type ATPase, from the Golgi to the plasma membrane to mediate cellular copper export (45, 49, 59). To determine whether copper-responsive trafficking of APP utilizes an exocytic pathway similar to that of ATP7A, we compared the cellular localization of APP and ATP7A under differential copper conditions. In the presence of copper chelators, endogenous APP and ATP7A partially co-localized primarily to the Golgi (Fig. 4A). Following copper treatment, both APP and ATP7A demonstrated a wider distribution throughout the cytoplasm (Fig. 4B). However, there was no observable co-localization between APP and ATP7A, suggesting that these proteins traffic via different exocytic pathways.
FIGURE 4.
APP and the copper transporter, ATP7A, exhibit copper-responsive trafficking in SH-SY5Y. SH-SY5Y cells were treated with 150 μm copper chelators (A) or 150 μm copper (B) as described previously. Cells were immunolabeled to detect APP and ATP7A using W0-2 and CT77 antibodies, respectively. DAPI stain (left panels) was used to visualize the nucleus. Under copper-deficient conditions, both APP and ATP7A show perinuclear localization. APP and ATP7A redistribute following copper treatment to differential subcellular compartments. The white arrows indicate the localization of APP or ATP7A following chelator or copper incubation. Scale bar, 10 μm.
Similar to SH-SY5Y cells, endogenous APP in primary cortical neurons exhibited copper-responsive relocalization. Primary cortical neurons used in these experiments were incubated in defined neurobasal medium without serum. Due to the absence of copper-binding proteins present in serum, which reduce “available copper” for cells, lower copper concentrations were used (46). Primary cortical neurons were exposed to a range of copper concentrations (5–20 μm) for 3 h. At copper concentrations of 5 μm and greater, there was a change in the cellular localization of APP from a predominantly perinuclear localization to a wider distribution, specifically toward neurites (Fig. 5A). As expected, ATP7A also showed a copper-responsive redistribution (Fig. 5B).
FIGURE 5.
APP and ATP7A exhibit copper-responsive trafficking in primary cortical neurons. A, primary cortical neurons isolated from embryonic day 14 embryos were exposed to either copper chelators (i) or increasing levels of copper (5, 10, and 20 μm; ii–iv) for 3 h in neurobasal medium (described under “Materials and Methods”). Cells were immunolabeled for APP using the CT20 antibody. Upon the addition of 5 μm copper and above, APP relocalizes from a predominant perinuclear location to neurites, as indicated by the white arrows. B, similarly, in the presence of copper chelators (i), ATP7A demonstrates a perinuclear localization and redistributes to neurites upon the addition of copper (ii; 10 μm). Scale bar, 10 μm.
No Detectable Processing of Endogenous APP in SH-SY5Y Cells after Exposure to Copper
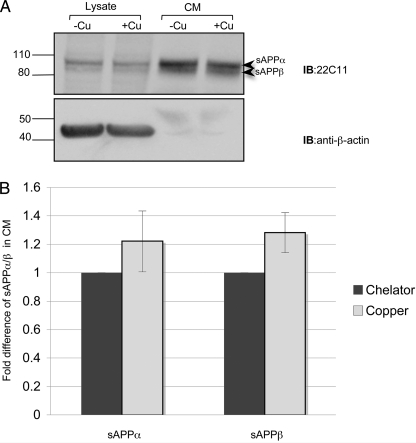
Changes in the cellular localization of APP have been reported to influence APP processing (reviewed in Ref. 60). Borchardt et al. (31) reported an increase in non-amyloidogenic processing, which was evident by an increase in sAPPα and reduced Aβ production following copper treatment. To investigate whether copper-responsive APP trafficking was associated with changes in APP processing, we evaluated the level of APP proteolytic products, including sAPPα/β, CT fragments, and Aβ in SH-SY5Y cells. Under the conditions used for trafficking experiments (i.e. 150 μm copper/copper chelators; 3 h), we did not observe a statistically significant change in the level of sAPPα between treatments (n = 3, p = 0.517; Fig. 6). Similarly, no change in sAPPβ was detected (n = 3, p = 0.152). C-terminal products and Aβ levels were below detection limits. This result suggests that exposure to copper for a short period of time (3 h) promotes full-length APP trafficking without detectable changes in the processing of the endogenous protein.
FIGURE 6.
Copper does not affect the levels of sAPPα and sAPPβ in conditioned medium. A, SH-SY5Y cells were incubated with either copper chelators (−Cu; 150 μm) or copper (+Cu; 150 μm). The 22C11 antibody was used to detect full-length APP in cell lysates and the proteolytic product sAPPα/sAPPβ in the growth medium. The arrows indicate bands corresponding to sAPPα and sAPPβ in conditioned medium. β-Actin was used as a loading control. B, the level of sAPPα and sAPPβ following copper chelator or copper treatment was analyzed by densitometry. There was no statistically significant difference between the levels of sAPPα/β and chelator/copper treatment (n = 3, p > 0.1). IB, immunoblot. Error bars, S.D.
Copper Promotes APP Exocytosis
To investigate the basis of copper-responsive APP relocalization, we studied the dynamics of APP trafficking, following the addition of copper, by live cell imaging using the MDCK cell line stably expressing APP-cherry in a background of low endogenous APP expression in MDCK cells, thus facilitating such analyses. Within 10 min of copper stimulation, we observed a rapid redistribution of APP-cherry away from the Golgi, which exhibited a punctate intracellular distribution (Fig. 7 and supplemental Fig. S8, movie). This result further supported our immunofluorescence data that copper promotes the exocytosis of APP from the Golgi.
FIGURE 7.
Real-time imaging of copper-stimulated APP trafficking. MDCK-APP-cherry cells cultured on 40-mm coverslips were incubated in BME medium without phenol red supplemented with 2% fetal calf serum (described under “Materials and Methods”) and the above medium supplemented with copper (150 μm). Images were captured under basal conditions (00:00) and following the addition of copper at 2-min intervals for ∼25 min using an Olympus FV1000 laser-scanning confocal microscope (see supplemental Fig. S8). Panels represent a series of images acquired at five different time points following the addition of copper. Numbers at the top left-hand corner indicate the minutes and seconds (min:s) following the addition of copper. Scale bar, 5 μm.
The Rate of APP Endocytosis Is Reduced following Copper Treatment
To investigate whether the copper-mediated cell surface retention of APP was due to a reduced rate of APP endocytosis, we studied the rate of APP endocytosis using an antibody uptake assay in SH-SY5Y cells (50, 51). APP at the cell surface was labeled with the 22C11 antibody (which detects an epitope in the extracellular N-terminal domain of APP) prior to exposing cells to growth medium containing either copper (150 μm) or copper chelators (150 μm) for various times. Internalized 22C11 was detected by immunolabeling with Alexa-Fluor 488 mouse secondary antibody (Fig. 8A). To visualize the cell surface, cells were co-labeled with rhodamine phalloidin, a fluorescent probe that labels filamentous actin (Fig. 8A, bottom panels). At t = 0, there was no detectable level of intracellular 22C11-labeled APP. In the absence of copper at t = 15 min, an average of 62% of cells had internalized 22C11-labeled APP from the cell surface, compared with an average of 28% of cells incubated with copper (Fig. 8, A and B). Moreover, at t = 30 min, only an average of 37% of cells exposed to copper showed APP endocytosis compared with an average of 66% of cells incubated with copper chelators. Our results were consistent with previous studies (not involving changes in copper levels) that reported a time-dependent internalization of APP, with approximately one-third (34%) of cells undergoing APP endocytosis at 7 min (61). Moreover, our data indicated that elevated copper significantly reduced the rate of APP endocytosis. Therefore, elevated copper leads to an increase of APP at the cell surface by both promoting its exocytosis from the Golgi and by reducing its rate of endocytosis.
FIGURE 8.
Increase in copper leads to a reduction in the rate of APP internalization from the cell surface. Antibody uptake assays were performed to investigate the rate of APP endocytosis. A, SH-SY5Y cells were incubated with 22C11 (anti-APP ectodomain) for 20 min on ice to label APP at the cell surface. Cells were returned to growth medium containing either copper chelators (−Cu) or copper (+Cu) for various times (5, 15, and 30 min). SH-SY5Y cells were washed, fixed, and processed for immunofluorescence analysis, whereby internalized antibody-APP conjugate was visualized using Alexa Fluor 488 anti-mouse secondary antibody. The bottom image in each panel corresponds to actin staining with rhodamine phalloidin used to visualize the cell membrane. Scale bar, 10 μm. White arrows indicate internalized APP. B, to quantify APP endocytosis, the percentage of cells that demonstrated APP endocytosis was scored per treatment and time point. Values are mean ± S.E. (error bars) (n = 3; 40–60 cells were analyzed per experiment/treatment). Statistical analysis was calculated using Student's t test.
DISCUSSION
In this study, we discovered a novel process whereby changes in copper concentration promote a redistribution of APP from a perinuclear localization to a dispersed cytoplasmic and plasma membrane distribution in cultured neuronal (both primary and established) and polarized epithelial cell models. Using immunofluorescence and live cell imaging methods, we found that an elevation in cellular copper levels promotes the exit of APP from the Golgi to a wider distribution throughout the cytoplasm and to the plasma membrane. In MDCK-APP-cherry cells, copper promotes an increase in APP-cherry localization to EEA1-positive endosomal compartments. In SH-SY5Y cells, endogenous APP showed a partial co-localization with the Golgi marker GM130, which decreased following copper treatment. In addition, APP exhibited an approximately 2-fold increase at the cell surface in response to copper compared with treatment with copper chelators, which is consistent with our previous finding (39) (see also supplemental Fig. S2). Our data suggest that this increase in cell surface APP is the result of a concomitant increase in exocytosis and reduction in endocytosis. Importantly, the observed copper-responsive trafficking was independent of copper-mediated increases in APP protein synthesis.
Copper-induced APP exocytosis and reduced endocytosis is reminiscent of insulin-stimulated trafficking of the glucose transporter GLUT4. GLUT4 exhibits continuous recycling between the PM and several intracellular compartments, with only a small proportion (∼5%) of total GLUT4 protein localized to the PM in the basal state (reviewed in Ref. 62). Stimulation with insulin leads to both a marked increase in GLUT4 exocytosis and a small decrease in endocytosis so that 50% of GLUT4 protein relocalizes to the cell surface, resulting in increased glucose uptake (63–65). Similarly, under steady state conditions, only a small proportion (∼10%) of APP is present at the PM (reviewed in Ref. 66). In this study, we show that, similar to insulin-stimulated GLUT4 trafficking properties, copper-responsive cell surface relocalization of APP results from a concomitant increase in exocytosis and reduced endocytosis. This relates to a potential role for APP in the copper efflux pathway (32, 33, 38).
We previously reported increased copper efflux following the copper-responsive trafficking of ATP7A from the Golgi network to the PM (42, 67). An increase in APP at the PM may be influencing copper efflux as occurs for the copper transporter ATP7A when at the PM or in rapid recycling compartments proximal to the PM (49). Copper-responsive trafficking of APP is therefore consistent with a role for APP in copper efflux pathways (38). Indeed, various studies have reported an interdependent relationship between APP expression and copper levels, implicating an important role for APP in copper homeostasis. For instance, overexpression of APP in primary cortical neurons resulted in decreased intracellular copper levels (38). Increases in APP at the PM may also lead to modulation of signaling processes mediated by APP (68–70).
It has been reported that exposure to copper decreases Aβ levels (31, 32, 39, 71, 72). Under conditions used in this study, we did not observe any detectable changes in APP processing. This could be explained by different expression levels of APP in cell models used in different studies. Previous studies used cell models of AD where WT APP or the familial AD mutant, APPsw (APP K670N/M671L), were overexpressed (31, 32, 39, 71, 72). Overexpression of APPsw promotes its processing by the amyloidogenic pathway, thus generating more Aβ. Consistent with our finding, a previous study using parental SH-SY5Y cells also reported no change in APP C-terminal fragments when cells were exposed to copper (32). There was a 30% increase in Aβ levels in the case of cells overexpressing APP when deprived of copper (32). Overexpression of APP has been reported to stimulate basal and constitutive exocytosis in PC12 cells (73). Hence, studies utilizing cells expressing endogenous levels of APP, as in the current study involving neuronal cells, are likely to be more informative in investigating factors that modulate normal APP function. Our data show that copper promotes a redistribution of APP without any detectable change in processing, suggesting copper-responsive trafficking of full-length APP.
In summary, we provide evidence for a novel process whereby copper regulates APP localization and presumably its function. Future investigations will focus on identifying the physiological function(s) affected by copper-responsive APP relocalization. We recently reported that copper-responsive ATP7A trafficking is regulated by kinase phosphorylation of specific serine residues in ATP7A, implicating a signal transduction pathway(s) (46). Similarly, copper activation of the phosphoinositol 3-kinase pathway (71, 72, 74) can result in altered glycogen synthase kinase-3-mediated phosphorylation of APP, which may then modulate its trafficking. Investigating the complex relationship between copper and APP function will be of considerable importance in understanding copper dyshomeostasis underlying the pathophysiology of Alzheimer disease.
Supplementary Material
Acknowledgments
The cherry tag was provided by Professor Roger Tsien (University of California). The APP695 cDNA was provided by Robyn Sharples and Associate Professor Andrew Hill (Bio21 Institute). The CT77 (ATP7A) antibody was a gift from Prof. B. Eipper (University of Connecticut).
This work was supported by grants from the National Health and Medical Research Council (to J. C. and A. I. B.). The FV1000 Olympus confocal microscope used for this work was co-funded by the Rowden White Foundation.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S8.
- AD
- Alzheimer disease
- APP
- amyloid precursor protein
- Aβ
- amyloid β peptide
- MDCK
- Madin-Darby canine kidney
- PM
- plasma membrane
- EEA1
- early endosomal antigen 1
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- Tricine
- N-[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]glycine.
REFERENCES
- 1. Lovell M. A., Robertson J. D., Teesdale W. J., Campbell J. L., Markesbery W. R. (1998) J. Neurol. Sci. 158, 47–52 [DOI] [PubMed] [Google Scholar]
- 2. Miller L. M., Wang Q., Telivala T. P., Smith R. J., Lanzirotti A., Miklossy J. (2006) J. Struct. Biol. 155, 30–37 [DOI] [PubMed] [Google Scholar]
- 3. Tõugu V., Karafin A., Zovo K., Chung R. S., Howells C., West A. K., Palumaa P. (2009) J. Neurochem. 110, 1784–1795 [DOI] [PubMed] [Google Scholar]
- 4. Ha C., Ryu J., Park C. B. (2007) Biochemistry 46, 6118–6125 [DOI] [PubMed] [Google Scholar]
- 5. Bush A. I., Pettingell W. H., Multhaup G., d Paradis M., Vonsattel J. P., Gusella J. F., Beyreuther K., Masters C. L., Tanzi R. E. (1994) Science 265, 1464–1467 [DOI] [PubMed] [Google Scholar]
- 6. Bush A. I., Multhaup G., Moir R. D., Williamson T. G., Small D. H., Rumble B., Pollwein P., Beyreuther K., Masters C. L. (1993) J. Biol. Chem. 268, 16109–16112 [PubMed] [Google Scholar]
- 7. Atwood C. S., Perry G., Zeng H., Kato Y., Jones W. D., Ling K. Q., Huang X., Moir R. D., Wang D., Sayre L. M., Smith M. A., Chen S. G., Bush A. I. (2004) Biochemistry 43, 560–568 [DOI] [PubMed] [Google Scholar]
- 8. Guilloreau L., Combalbert S., Sournia-Saquet A., Mazarguil H., Faller P. (2007) Chembiochem 8, 1317–1325 [DOI] [PubMed] [Google Scholar]
- 9. Jiang D., Men L., Wang J., Zhang Y., Chickenyen S., Wang Y., Zhou F. (2007) Biochemistry 46, 9270–9282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Jiang D., Li X., Williams R., Patel S., Men L., Wang Y., Zhou F. (2009) Biochemistry 48, 7939–7947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Smith D. G., Cappai R., Barnham K. J. (2007) Biochim. Biophys. Acta 1768, 1976–1990 [DOI] [PubMed] [Google Scholar]
- 12. Huang X., Atwood C. S., Hartshorn M. A., Multhaup G., Goldstein L. E., Scarpa R. C., Cuajungco M. P., Gray D. N., Lim J., Moir R. D., Tanzi R. E., Bush A. I. (1999) Biochemistry 38, 7609–7616 [DOI] [PubMed] [Google Scholar]
- 13. Opazo C., Huang X., Cherny R. A., Moir R. D., Roher A. E., White A. R., Cappai R., Masters C. L., Tanzi R. E., Inestrosa N. C., Bush A. I. (2002) J. Biol. Chem. 277, 40302–40308 [DOI] [PubMed] [Google Scholar]
- 14. Opazo C., Ruiz F. H., Inestrosa N. C. (2000) Biol. Res. 33, 125–131 [DOI] [PubMed] [Google Scholar]
- 15. Deibel M. A., Ehmann W. D., Markesbery W. R. (1996) J. Neurol. Sci. 143, 137–142 [DOI] [PubMed] [Google Scholar]
- 16. Religa D., Strozyk D., Cherny R. A., Volitakis I., Haroutunian V., Winblad B., Naslund J., Bush A. I. (2006) Neurology 67, 69–75 [DOI] [PubMed] [Google Scholar]
- 17. Adlard P. A., Bush A. I. (2006) J. Alzheimers Dis. 10, 145–163 [DOI] [PubMed] [Google Scholar]
- 18. Magaki S., Raghavan R., Mueller C., Oberg K. C., Vinters H. V., Kirsch W. M. (2007) Neurosci. Lett. 418, 72–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. McGeer E. G., McGeer P. L., Harrop R., Akiyama H., Kamo H. (1990) J. Neurosci. Res. 27, 612–619 [DOI] [PubMed] [Google Scholar]
- 20. Bayer T. A., Schäfer S., Simons A., Kemmling A., Kamer T., Tepest R., Eckert A., Schüssel K., Eikenberg O., Sturchler-Pierrat C., Abramowski D., Staufenbiel M., Multhaup G. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 14187–14192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Crouch P. J., Blake R., Duce J. A., Ciccotosto G. D., Li Q. X., Barnham K. J., Curtain C. C., Cherny R. A., Cappai R., Dyrks T., Masters C. L., Trounce I. A. (2005) J. Neurosci. 25, 672–679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ding W. Q., Lind S. E. (2009) IUBMB Life 61, 1013–1018 [DOI] [PubMed] [Google Scholar]
- 23. Cherny R. A., Atwood C. S., Xilinas M. E., Gray D. N., Jones W. D., McLean C. A., Barnham K. J., Volitakis I., Fraser F. W., Kim Y., Huang X., Goldstein L. E., Moir R. D., Lim J. T., Beyreuther K., Zheng H., Tanzi R. E., Masters C. L., Bush A. I. (2001) Neuron 30, 665–676 [DOI] [PubMed] [Google Scholar]
- 24. Ritchie C. W., Bush A. I., Mackinnon A., Macfarlane S., Mastwyk M., MacGregor L., Kiers L., Cherny R., Li Q. X., Tammer A., Carrington D., Mavros C., Volitakis I., Xilinas M., Ames D., Davis S., Beyreuther K., Tanzi R. E., Masters C. L. (2003) Arch. Neurol. 60, 1685–1691 [DOI] [PubMed] [Google Scholar]
- 25. Adlard P. A., Cherny R. A., Finkelstein D. I., Gautier E., Robb E., Cortes M., Volitakis I., Liu X., Smith J. P., Perez K., Laughton K., Li Q. X., Charman S. A., Nicolazzo J. A., Wilkins S., Deleva K., Lynch T., Kok G., Ritchie C. W., Tanzi R. E., Cappai R., Masters C. L., Barnham K. J., Bush A. I. (2008) Neuron 59, 43–55 [DOI] [PubMed] [Google Scholar]
- 26. Lannfelt L., Blennow K., Zetterberg H., Batsman S., Ames D., Harrison J., Masters C. L., Targum S., Bush A. I., Murdoch R., Wilson J., Ritchie C. W. (2008) Lancet Neurol. 7, 779–786 [DOI] [PubMed] [Google Scholar]
- 27. Sisodia S. S. (1992) Proc. Natl. Acad. Sci. U.S.A. 89, 6075–6079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lammich S., Kojro E., Postina R., Gilbert S., Pfeiffer R., Jasionowski M., Haass C., Fahrenholz F. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 3922–3927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ermolieff J., Loy J. A., Koelsch G., Tang J. (2000) Biochemistry 39, 12450–12456 [DOI] [PubMed] [Google Scholar]
- 30. Koo E. H., Squazzo S. L. (1994) J. Biol. Chem. 269, 17386–17389 [PubMed] [Google Scholar]
- 31. Borchardt T., Camakaris J., Cappai R., Masters C. L., Beyreuther K., Multhaup G. (1999) Biochem. J. 344, 461–467 [PMC free article] [PubMed] [Google Scholar]
- 32. Cater M. A., McInnes K. T., Li Q. X., Volitakis I., La Fontaine S., Mercer J. F., Bush A. I. (2008) Biochem. J. 412, 141–152 [DOI] [PubMed] [Google Scholar]
- 33. Bellingham S. A., Lahiri D. K., Maloney B., La Fontaine S., Multhaup G., Camakaris J. (2004) J. Biol. Chem. 279, 20378–20386 [DOI] [PubMed] [Google Scholar]
- 34. Armendariz A. D., Gonzalez M., Loguinov A. V., Vulpe C. D. (2004) Physiol. Genomics 20, 45–54 [DOI] [PubMed] [Google Scholar]
- 35. Maynard C. J., Cappai R., Volitakis I., Cherny R. A., White A. R., Beyreuther K., Masters C. L., Bush A. I., Li Q. X. (2002) J. Biol. Chem. 277, 44670–44676 [DOI] [PubMed] [Google Scholar]
- 36. Treiber C., Simons A., Strauss M., Hafner M., Cappai R., Bayer T. A., Multhaup G. (2004) J. Biol. Chem. 279, 51958–51964 [DOI] [PubMed] [Google Scholar]
- 37. White A. R., Multhaup G., Maher F., Bellingham S., Camakaris J., Zheng H., Bush A. I., Beyreuther K., Masters C. L., Cappai R. (1999) J. Neurosci. 19, 9170–9179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Bellingham S. A., Ciccotosto G. D., Needham B. E., Fodero L. R., White A. R., Masters C. L., Cappai R., Camakaris J. (2004) J. Neurochem. 91, 423–428 [DOI] [PubMed] [Google Scholar]
- 39. Hung Y. H., Robb E. L., Volitakis I., Ho M., Evin G., Li Q. X., Culvenor J. G., Masters C. L., Cherny R. A., Bush A. I. (2009) J. Biol. Chem. 284, 21899–21907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ida N., Hartmann T., Pantel J., Schröder J., Zerfass R., Förstl H., Sandbrink R., Masters C. L., Beyreuther K. (1996) J. Biol. Chem. 271, 22908–22914 [DOI] [PubMed] [Google Scholar]
- 41. Weidemann A., König G., Bunke D., Fischer P., Salbaum J. M., Masters C. L., Beyreuther K. (1989) Cell 57, 115–126 [DOI] [PubMed] [Google Scholar]
- 42. Camakaris J., Petris M. J., Bailey L., Shen P., Lockhart P., Glover T. W., Barcroft C., Patton J., Mercer J. F. (1995) Hum. Mol. Genet. 4, 2117–2123 [DOI] [PubMed] [Google Scholar]
- 43. Nyasae L., Bustos R., Braiterman L., Eipper B., Hubbard A. (2007) Am. J. Physiol. Gastrointest Liver Physiol 292, G1181–G1194 [DOI] [PubMed] [Google Scholar]
- 44. White A. R., Zheng H., Galatis D., Maher F., Hesse L., Multhaup G., Beyreuther K., Masters C. L., Cappai R. (1998) J. Neurosci. 18, 6207–6217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Greenough M., Pase L., Voskoboinik I., Petris M. J., O'Brien A. W., Camakaris J. (2004) Am. J. Physiol. Cell Physiol. 287, C1463–C1471 [DOI] [PubMed] [Google Scholar]
- 46. Veldhuis N. A., Valova V. A., Gaeth A. P., Palstra N., Hannan K. M., Michell B. J., Kelly L. E., Jennings I., Kemp B. E., Pearson R. B., Robinson P. J., Camakaris J. (2009) Int. J. Biochem. Cell Biol. 41, 2403–2412 [DOI] [PubMed] [Google Scholar]
- 47. Bolte S., Cordelières F. P. (2006) J. Microsc. 224, 213–232 [DOI] [PubMed] [Google Scholar]
- 48. Cherny R. A., Legg J. T., McLean C. A., Fairlie D. P., Huang X., Atwood C. S., Beyreuther K., Tanzi R. E., Masters C. L., Bush A. I. (1999) J. Biol. Chem. 274, 23223–23228 [DOI] [PubMed] [Google Scholar]
- 49. Pase L., Voskoboinik I., Greenough M., Camakaris J. (2004) Biochem. J. 378, 1031–1037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Haass C., Lemere C. A., Capell A., Citron M., Seubert P., Schenk D., Lannfelt L., Selkoe D. J. (1995) Nat. Med. 1, 1291–1296 [DOI] [PubMed] [Google Scholar]
- 51. Kaether C., Lammich S., Edbauer D., Ertl M., Rietdorf J., Capell A., Steiner H., Haass C. (2002) J. Cell Biol. 158, 551–561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Winckler B., Mellman I. (1999) Neuron 23, 637–640 [DOI] [PubMed] [Google Scholar]
- 53. Dotti C. G., Simons K. (1990) Cell 62, 63–72 [DOI] [PubMed] [Google Scholar]
- 54. Pinnix I., Musunuru U., Tun H., Sridharan A., Golde T., Eckman C., Ziani-Cherif C., Onstead L., Sambamurti K. (2001) J. Biol. Chem. 276, 481–487 [DOI] [PubMed] [Google Scholar]
- 55. Caporaso G. L., Takei K., Gandy S. E., Matteoli M., Mundigl O., Greengard P., De Camilli P. (1994) J. Neurosci. 14, 3122–3138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Xu H., Sweeney D., Wang R., Thinakaran G., Lo A. C., Sisodia S. S., Greengard P., Gandy S. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 3748–3752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Hartmann T., Bieger S. C., Brühl B., Tienari P. J., Ida N., Allsop D., Roberts G. W., Masters C. L., Dotti C. G., Unsicker K., Beyreuther K. (1997) Nat. Med. 3, 1016–1020 [DOI] [PubMed] [Google Scholar]
- 58. Greenfield J. P., Tsai J., Gouras G. K., Hai B., Thinakaran G., Checler F., Sisodia S. S., Greengard P., Xu H. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 742–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Petris M. J., Mercer J. F., Culvenor J. G., Lockhart P., Gleeson P. A., Camakaris J. (1996) EMBO J. 15, 6084–6095 [PMC free article] [PubMed] [Google Scholar]
- 60. Thinakaran G., Koo E. H. (2008) J. Biol. Chem. 283, 29615–29619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Neumann S., Schöbel S., Jäger S., Trautwein A., Haass C., Pietrzik C. U., Lichtenthaler S. F. (2006) J. Biol. Chem. 281, 7583–7594 [DOI] [PubMed] [Google Scholar]
- 62. Hou J. C., Pessin J. E. (2007) Curr. Opin. Cell Biol. 19, 466–473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Bryant N. J., Govers R., James D. E. (2002) Nat. Rev. Mol. Cell Biol. 3, 267–277 [DOI] [PubMed] [Google Scholar]
- 64. Buhl E. S., Jessen N., Schmitz O., Pedersen S. B., Pedersen O., Holman G. D., Lund S. (2001) Diabetes 50, 12–17 [DOI] [PubMed] [Google Scholar]
- 65. Pessin J. E., Thurmond D. C., Elmendorf J. S., Coker K. J., Okada S. (1999) J. Biol. Chem. 274, 2593–2596 [DOI] [PubMed] [Google Scholar]
- 66. Vetrivel K. S., Thinakaran G. (2006) Neurology 66, Suppl. 1, S69–S73 [DOI] [PubMed] [Google Scholar]
- 67. Petris M. J., Mercer J. F. (1999) Hum. Mol. Genet. 8, 2107–2115 [DOI] [PubMed] [Google Scholar]
- 68. Birkaya B., Aletta J. M. (2005) J. Neurobiol. 63, 49–61 [DOI] [PubMed] [Google Scholar]
- 69. Sabo S. L., Ikin A. F., Buxbaum J. D., Greengard P. (2001) J. Cell Biol. 153, 1403–1414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Sabo S. L., Ikin A. F., Buxbaum J. D., Greengard P. (2003) J. Neurosci. 23, 5407–5415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Crouch P. J., Hung L. W., Adlard P. A., Cortes M., Lal V., Filiz G., Perez K. A., Nurjono M., Caragounis A., Du T., Laughton K., Volitakis I., Bush A. I., Li Q. X., Masters C. L., Cappai R., Cherny R. A., Donnelly P. S., White A. R., Barnham K. J. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 381–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Donnelly P. S., Caragounis A., Du T., Laughton K. M., Volitakis I., Cherny R. A., Sharples R. A., Hill A. F., Li Q. X., Masters C. L., Barnham K. J., White A. R. (2008) J. Biol. Chem. 283, 4568–4577 [DOI] [PubMed] [Google Scholar]
- 73. Lee H. W., Park J. W., Sandagsuren E. U., Kim K. B., Yoo J. J., Chung S. H. (2008) Neurosci. Lett. 436, 245–249 [DOI] [PubMed] [Google Scholar]
- 74. Ostrakhovitch E. A., Lordnejad M. R., Schliess F., Sies H., Klotz L. O. (2002) Arch. Biochem. Biophys. 397, 232–239 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.