Abstract
The 2-Cys peroxiredoxins (Prx) belong to a family of antioxidant enzymes that detoxify reactive oxygen and nitrogen species and are distributed throughout the intracellular and extracellular compartments. However, the presence and role of 2-Cys Prxs in the nucleus have not been studied. This study demonstrates that the PrxII located in the nucleus protects cancer cells from DNA damage-induced cell death. Although the two cytosolic 2-Cys Prxs, PrxI and PrxII, were found in the nucleus, only PrxII knockdown selectively and markedly increased cell death in the cancer cells treated with DNA-damaging agents. The increased death was completely reverted by the nuclearly targeted expression of PrxII in an activity-independent manner. Furthermore, the antioxidant butylated hydroxyanisole did not influence the etoposide-induced cell death. Mechanistically, the knockdown of Prx II expression impaired the DNA repair process by reducing the activation of the JNK/c-Jun pathway. These results suggest that PrxII is likely to be attributed to a tumor survival factor positively regulating JNK-dependent DNA repair with its inhibition possibly sensitizing cancer cells to chemotherapeutic agents.
Keywords: Cancer Therapy, Cell Death, DNA Damage, DNA Repair, JNK, Peroxidase, Peroxiredoxin
Introduction
Reactive oxygen species (ROS)4 are produced by the partial reduction of oxygen as a by-product of metabolism and by exogenous insults, including anti-cancer drugs, ultraviolet light, and γ-irradiation. High levels of endogenous ROS are cytotoxic because they can damage proteins, lipids, and DNA (1). Therefore, they have long been regarded as a possible cause of aging and many chronic diseases like cancer, arteriosclerosis, and neurological disorders (2, 3). In contrast, at relatively low levels, ROS are thought to be the second messenger that plays a key role in signaling events that induce proliferation, differentiation, and apoptosis (4, 5). Accordingly, the cellular ROS should be tightly maintained at noncytotoxic levels and controlled in a regulated manner.
The mammalian cells have a variety of antioxidant systems, which maintain the cellular redox homeostasis and further protect cellular molecules against oxidative damage (6). Although glutathione (GSH) acts as a general redox buffer, several antioxidant enzymes function as local bystanders. Specifically, Cu,Zn-superoxide dismutase and Mn-superoxide dismutase reduce the superoxide anion (O2˙̄) to hydrogen peroxide (H2O2) in cytosol and mitochondria, respectively. Catalase reduces H2O2 to water in peroxisomes. The five isoforms of glutathione peroxidase reduce the peroxides, such as H2O2 and lipid peroxide, to the corresponding alcohol in the presence of a redox recycling system (GSH, GSH reductase, and NADPH) in various compartments, i.e. extracellular, cytosol, and mitochondria (7). The six Prx enzymes reduce the peroxide, such as H2O2, lipid peroxides, and peroxynitrite, to the corresponding alcohol in the presence of another redox recycling system (thioredoxin, thioredoxin reductase, and NADPH) (8, 9). The cellular distribution of Prxs is much more complex than that of other antioxidant enzymes. PrxI and PrxII are found mainly in the cytosol; however, PrxII has been shown to associate with the plasma membrane in the erythrocyte. Furthermore, PrxIII is a mitochondrial peroxide reductase. PrxIV is a secretory peroxidase. PrxV exhibits diverse distribution in the cytosol, peroxisome, and mitochondria. PrxVI, also called 1-Cys Prx, is a cytosolic protein. However, the presence and function of a nuclear antioxidant enzyme have not been determined.
The 2-Cys Prx isoforms of Prx family enzymes, which are widely distributed in the tissues, have been shown to be overexpressed in various types of cancer cells and tumor tissues (10). Because the cancer cells were known to produce large amounts of ROS (11), it is readily appreciated that the overexpression of the antioxidant enzyme Prx could benefit cancer cell survival. For example, PrxI has been shown to suppress JNK activation and in turn cell death in the irradiated lung cancer cells (12). The reduction in the PrxII expression was shown to increase the radiation sensitivity of a head and neck cancer cell line (13). Mitochondrial PrxIII has been shown to sensitize cervical cancer cells to TNF-α-induced cell death (14). However, it has become controversial in other studies, wherein Prx seems to suppress tumor progression. For example, PrxI was shown to interact with c-Myc via Myc Box II domain and inhibit c-Myc-induced cellular transformation (15). PrxI was also found to suppress Ras- or ErbB2-induced transformation by protecting phosphatase and tensin homolog from oxidative inactivation (16). Moreover, PrxI−/− mice developed age-dependent malignant cancers (17, 18). In the case of PrxII, it is noteworthy that there have been no reports showing that PrxII interacts with proteins related to the tumorigenesis. The PrxII-deficient mice exhibited only splenomegaly phenotype due to the major antioxidant role of PrxII in erythrocyte but no cancer-related phenotype (19–21). It is important to determine whether Prx is involved in tumorigenesis or cancer cell survival.
In this study, we found that the two isoforms of 2-Cys Prxs, PrxI and PrxII, are present in the nucleus and demonstrated that the nuclear PrxII prevents the cancer cell death induced by DNA-damaging agents, including topoisomerase inhibitors. More importantly, such protective function of PrxII is only effective in the cancer cells and independent of its peroxidatic activity. Our study also revealed that PrxII is required for the JNK-dependent DNA repair process in the nucleus. Thus, PrxII has turned out to be the first antioxidant enzyme involved in the DNA repair process via regulating JNK activation.
EXPERIMENTAL PROCEDURES
Reagents, RNAi, and Cell Lines
Etoposide, hydroxyurea, doxorubicin, camptothecin, leptomycin B, and butylated hydroxyanisole were purchased from Sigma. KU55933 was from Selleck Chemicals. Benzyloxycarbonyl-VAD-O-Me-fluoromethyl ketone was from R&D Systems. PD98059 and SP600125 were from Calbiochem. Comet assay kit was from Trevigen. Propidium iodide was from Invitrogen. Antibodies against α-tubulin, Lamin B, p-ERK (Thr-202/Tyr-204), p-JNK (Thr-183/Tyr-182), p-p38 (Thr-180/Tyr-182), p-c-Jun (Ser-73), ERK1, JNK1/2, p38, c-Jun, and γ-H2AX were from Cell Signaling Technology. Alexa Fluor 568 goat anti-rabbit and Alexa Fluor 488 goat anti-rabbit were from Molecular Probes. Anti-PrxI, PrxII, c-Myc, catalase, and Prx-SO2/SO3 antibodies were from AbFrontier (Seoul, Korea). Human catalase siRNA SMART-pool and control luciferase siRNA duplex were purchased from Dharmacon. Phospho-JNK peptide (H2N-MMpTPpY-COOH) was synthesized (Peptron, Korea). Sequences of the siRNA duplexes are as follows: human PrxI was 5′-ACTCAACTGCCAAGTGATTUU-3′; human PrxII, 5′-CGCUUGUCUGAGGAUUACGUU-3′ (number 1) and 5′-UCAAAGAGGUGAAGCUGUCUU-3′ (number 2); mouse PrxII, 5′-AAAUCAAGCUUUCGGACUAUU-3′; and human PrxVI, 5′-GGACGTGGCTCCCAACTTTUU-3′. The siRNA oligonucleotide duplexes were synthesized from Dharmacon. Unless otherwise stated, the cancer cells were transfected with the siRNA duplexes for 48 h, using Lipofectamine RNAi MAXTM (Invitrogen) according to the manufacturer's protocol. One of the PrxII siRNA sequences (number 1) was used for designing a small hairpin RNA (shRNA) oligonucleotide pair, which was then subcloned into pSuper.retro.puro plasmid (Oligoengine Co.) with BglII and HindIII sites. A firefly luciferase shRNA was used as control shRNA. HeLa and 293T cells were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum (FBS) at 37 °C in a CO2 incubator. U2OS and HCT116 cells were cultured in McCoy's 5A medium containing 10% FBS. IMR90 human lung fibroblast cells were cultured in Eagle's minimal essential medium containing 10% FBS. The wild-type and PrxII−/− mouse embryonic fibroblasts (MEFs) were prepared as described previously (22).
Subcellular Fractionation
HeLa cells were trypsinized and washed once with ice-cold PBS. Cells were immediately resuspended with detergent-containing nuclei buffer (DNB buffer: 10 mm Hepes (pH 7.5), 10 mm KCl, 2 mm MgCl2, 0.2% Nonidet P-40) containing protease inhibitors and swollen on ice for 10 min. The cells were then gently homogenized by forcing 10 times through 22-gauge needle. The homogenate was centrifuged at 500 × g for 10 min, and the supernatant was taken up as post-nuclear supernatant fraction. The resuspension and homogenization were repeated twice with DNB buffer. From each homogenization step, the supernatants were saved for analysis. The final pellet containing nuclei were resuspended with the same buffer and then disrupted by brief sonication. Along with the supernatants, nuclei fractions were analyzed by immunoblotting.
Immunocytochemistry
HeLa cells were cultured on glass cover slides for 1 day, washed twice with cold PBS, and fixed with pre-warmed 4% paraformaldehyde solution for 30 min. Cells were washed two times with PBS and permeabilized with 0.2% Triton X-100 for 15 min at room temperature. After permeabilization, the cells were then blocked with 2% BSA in PBS (blocking buffer) for 1 h and incubated at 4 °C overnight with the indicated primary antibodies diluted in blocking buffer: anti-p65 (1:300), anti-PrxI (1:300), anti-PrxII (1:300), anti-pJNK (1:100), and anti-γH2AX (1:300). The cells were washed three times with blocking buffer and incubated with Alexa Fluor 568-conjugated secondary antibody. After 30 min, the coverslips were washed three times in a blocking buffer and mounted. The fluorescence images were taken by an LSM510 META confocal laser-scanning microscope (Zeiss). For γ-H2AX staining, the number of foci was counted using Phoretix 2D evolution (Nonlinear Dynamics Ltd.).
Cell Death Assay
The stimulated cells were washed once with PBS, detached by incubating at 37 °C for 2 min in 0.05% trypsin/EDTA, and then gently collected into the 5-ml FACS tubes, where the culture media and PBS wash were collected. The cells were then centrifuged for 3 min and once washed with cold PBS, and the final cell pellets were fixed with 70% ethanol in PBS and stained with propidium iodide (PI) for 5 min on ice. Cell death was assessed by FACS analysis of PI-stained cells in a FACSCalibur system (BD Biosciences). Cell death was determined by the percentage of PI-positive cells. The cell death assays were performed in duplicate.
Measurement of Intracellular ROS
Intracellular ROS generation was assessed with an oxidation-sensitive fluorescent dye 5,6-chloromethyl-2′,7′-dichlorodihydrofluorescein diacetate (Molecular Probes) as described previously (23). The HeLa cells (3 × 105) were cultured on 35-mm dishes and transfected with siRNAs for 24 h. The cells were then deprived of serum for 6 h and stimulated with 20 μm etoposide in phenol red-free media for the indicated periods of time. After stimulation, the cells were quickly rinsed with Krebs-Ringer solution and incubated for 5 min with 5 μm 5,6-chloromethyl-2′,7′-dichlorodihydrofluorescein diacetate. The 2′,7′-dichlorodihydrofluorescein fluorescence was collected for 10 s with an inverted Axiovert200 fluorescence microscope (Zeiss). The relative 2′,7′-dichlorodihydrofluorescein fluorescence was obtained by averaging the fluorescence intensities of the 60–80 cells after background subtraction in each image using ImageQuantTM software (GE Healthcare). The presented data are representative of three experiments with the same result. Note that the detached round cells were omitted from quantification.
Retrovirus Production
The coding sequences for human PrxII wild-type (WT) and activity-dead cysteine double mutant (C51S/C172S) were inserted by PCR cloning into pShooter vectors (Invitrogen), pCMV/myc/cyto© and pCMV/myc/nuc© to construct the plasmids encoding the Myc-tagged PrxII proteins targeted to cytosol and nucleus, respectively. The human PrxII sequence with a Myc tag and/or nuclear localization sequence was then subcloned into pQCXIX (Invitrogen) to produce the retroviral vectors, designated as pQ-PrxII-WT, pQ-PrxIIWT-Nuc, and pQ-PrxIIDN-Nuc. To avoid the ectopic expression being knocked down by PrxII siRNA, the three bases were substituted within the sequence targeted by the PrxII siRNA used in the study. The base substitution was carried out by site-directed mutagenesis using the following primers: sense, 5′-GCT GAC GTG ACC AGA CGC TTA TCC GAA GAT TAC GGC GTG CTG AAA ACA-3′; antisense, 5′-TGT TTT CAG CAC GCC GTA ATC TTC GGA TAA GCG TCT GGT CAC GTC AGC-3′, where the mutated bases are underlined. The final retroviral vector (20 μg) along with the viral packaging vector set (gag-pol and VSVG-encoding plasmid, 20 μg each) were transfected to 293T cells for 24 h by the calcium phosphate method. The cells were then washed twice in phosphate-buffered saline (PBS) and placed in fresh complete medium for virus collection. After 24 h, the culture supernatants containing the retroviruses were collected and filtered using a 0.450-μm filter, and the aliquots were frozen at −70 °C until used. For retroviral infection, the viral aliquots were thawed in warm water bath and mixed with 10 μg/ml Polybrene.
Comet Assay
Neutral comet assays were performed using the Comet assay kit (4250-050-K, Trevigen) according to the manufacturer's protocol. Briefly, the cells were mixed with low melting agarose gel and spread on CometSlide. The cells were lysed by Lysis Solution (catalog no. 4250-050-01) on ice and then equilibrated with neutral electrophoresis buffer. The slides were subjected to electrophoresis and afterward immersed in DNA precipitation solution. DNA was stained with SYBR Green, and the fluorescence image was taken by fluorescence microscopy. The tail moments were measured and averaged from ∼70 randomly selected cells using TriTek CometScore freeware program.
Statistics
Data were analyzed using Student's t test on SigmaPlot 8.0 software, and the p value was derived to assess statistical significance. A p < 0.05 was considered to be significant.
RESULTS
Prx I and II Are Located in the Nucleus
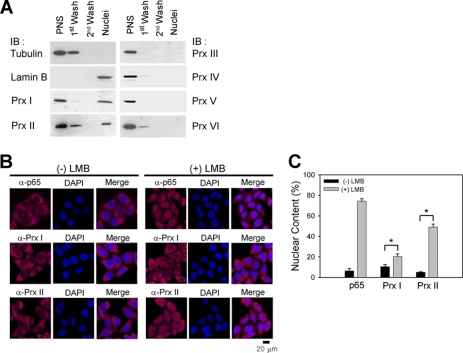
The six human Prxs show broad subcellular distribution, including cytosol, mitochondria, and peroxisome (8). To examine whether any of the Prxs are localized in the nucleus, we used the HeLa cervical carcinoma cells, where the Prxs are highly expressed (24), and prepared the pure nuclear fraction using a hypotonic extraction buffer containing mild detergent (0.2% Nonidet P-40). The last nuclei pellet was enriched with a nuclear protein lamin B but without PrxIII and PrxIV proteins present in the mitochondria and endoplasmic reticulum, respectively (Fig. 1A). This result indicates that several washes with detergent-containing extraction buffer completely eliminated the contamination of other organelles. Nonetheless, the two cytosolic 2-Cys Prxs, PrxI and PrxII, were apparently detected in the nuclei pellet (Fig. 1A). Their nuclear localization was further examined by immunostaining. First, the antisera against PrxI and PrxII were affinity-purified. Second, the specificities of the anti-PrxI and -PrxII antibodies were tested by blocking with the corresponding recombinant antigen proteins (data not shown). A significant amount of immunoreactive signal was detected in the nucleus, about 10% for PrxI and 5% for PrxII of total immunoreactive signals (Fig. 2, B and C). Given that both PrxI and PrxII do not have a featured nuclear localization signal, we hypothesized that their nuclear localization may be achieved by cytosol-nuclear shuttling. To test this, we blocked the shuttling with leptomycin B, an inhibitor of CRM-1-dependent nuclear export pathway (25). As a result, leptomycin B treatment induced the remarkable accumulation of PrxII in the nucleus, although there was less accumulation for PrxI (Fig. 2, B and C). NF-κB p65, which is known to shuttle between cytosol and nucleus (26), was also accumulated by leptomycin B treatment. This result indicates that at least the nuclear localization of PrxII seems more dynamic than that of PrxI.
FIGURE 1.
PrxI and PrxII are located in the nucleus. A, immunoblot (IB) analysis of Prx isoforms in the subcellular fractions of HeLa cells. The α-tubulin and lamin B are cytosolic and nuclear markers, respectively. One representative blot of three experiments is shown. PNS, post-nuclear supernatant. B and C, immunostaining of PrxI and PrxII in HeLa cells treated either with or without leptomycin B (nuclear exportin-1 inhibitor). The NF-κB p65 protein is used as a positive control. Nuclei are labeled with DAPI (blue). One representative set of three experiments is shown. Data in the graph (C) are means ± S.D. of the percent of nuclear immunoreactive fluorescence versus total fluorescence obtained from 25 to 35 cells (n = 3; *, p < 0.005). LMB, leptomycin B.
FIGURE 2.
Knockdown of Prx II enhances DNA damage-induced cell death. A, knockdown of PrxI and PrxII expression by specific siRNAs. An siRNA targeting firefly luciferase is used as a control (C). B–D, enhancement of DNA damage-induced cell death by PrxII, not PrxI, knockdown in HeLa cells. The cells were treated with increasing concentrations of either etoposide (ETO), camptothecin (CPT), or hydroxyurea (HU) for 24 h. E, enhancement of cell death by PrxII knockdown in doxorubicin (DOX)-treated HeLa cells. F and G, enhancement of cell death by PrxII knockdown in HCT116 and U2OS cancer cells treated with 20 μm etoposide for 24 h. H, DNA damage-induced cell death in PrxII-deficient MEFs. I, DNA damage-induced cell death in primary MEFs and human lung fibroblast IMR90 cells with PrxII knockdown. The knockdown of PrxII expression by siRNA is shown. Data in the graph are means ± S.D. of the percent of PI+ cells in the sub-G0 fraction (n = 3; *, p < 0.005; **, p < 0.01, N.S., not significant). N/T, not treated; IB, immunoblot.
PrxII Protects Cancer Cells from DNA Damage-induced Cell Death
We studied the role of the nuclear PrxI and PrxII in the cell death induced by DNA-damaging agents, including topoisomerase inhibitors, a replication blocker, and a DNA intercalating agent. To do so, we knocked down expression of PrxI and PrxII using specific small interfering RNAs (siRNA). The siRNA transfection resulted in the reduction of PrxI and PrxII protein levels in HeLa cells by ∼95% (Fig. 2A). Interestingly, the PrxII knockdown drastically enhanced the death of HeLa cancer cells in response to different DNA-damaging agents, such as etoposide, camptothecin, hydroxyurea, and doxorubicin (Fig. 2, B–E). In contrast, the PrxI knockdown had no influence on the cell death. Similarly, PrxII knockdown also enhanced cell death in HCT116 colon cancer cells and U2OS osteosarcoma cells treated with etoposide (Fig. 2, F and G). Transfection of short hairpin RNA (shRNA) construct and other siRNA duplexes targeting PrxII similarly enhanced the etoposide-induced cell death in HeLa cells (supplemental Fig. S1, A and B), thus confirming the specific effect of PrxII knockdown. Throughout this study, we noticed that PrxII knockdown itself induces a certain amount of basal cell death (around 10% cell death after a 48-h PrxII knockdown) in the absence of DNA damage. The control experiment turned out that the extent of basal cell death is proportional to the duration of PrxII knockdown, suggesting that the PrxII knockdown somehow destabilized cancer cells. Albeit to a lesser extent, a shorter period (24-h transfection) of PrxII knockdown with apparently no basal cell death clearly enhanced the etoposide-induced cell death (supplemental Fig. S1C). Nonetheless, we kept 48 h of transfection of PrxII siRNA for experimental convenience. In addition, knockdown of neither a peroxisomal peroxidase catalase nor other cytosolic Prx isoform called PrxVI influenced the etoposide-induced cell death (supplemental Fig. S1D). These results indicate that PrxII specifically regulates DNA damage-induced cancer cell death. Next, to determine whether protective action of PrxII is applicable to normal cells responding to DNA damage, we used the primary fibroblast cells, such as MEFs and lung fibroblasts. Neither gene knock-out nor transient knockdown of PrxII did enhance cell death of primary fibroblasts in response to DNA damage (Fig. 2, H and I). These results indicate that PrxII is selectively involved in the survival of cancer cells against DNA damage stress.
Nuclearly Targeted PrxII Sufficiently Prevents DNA Damage-induced Death
To further confirm the specific action of PrxII in DNA damage-induced cancer cell death, we carried out a rescue experiment by exogenously expressing PrxII in PrxII siRNA-transfected cells. To avoid targeting of exogenously expressed PrxII by cognate siRNA, the three bases in the region corresponding to siRNA target sequence were substituted without causing amino acid change. Exogenous PrxII, which shows a size shift due to a Myc epitope tag and a nuclear localization signal sequence (NLS), was expressed in a level similar to the endogenous proteins (Fig. 3A), and the NLS-tagged version of PrxII was localized in the nucleus (Fig. 3B). Like endogenous PrxII, only some of the expressed wild-type PrxII (WT-Cyto) was detected in the nucleus. When these cells were treated with etoposide, add-back expression of nuclearly targeted PrxII (WT-NLS), as well as wild-type PrxII (WT-Cyto), rescued the HeLa cells from the augmented death by PrxII knockdown (Fig. 3C). Thus, the result indicates that the nuclear PrxII is sufficient for protecting cancer cells against DNA damage. Given that PrxII is a peroxidase enzyme reducing H2O2 to water (23), we examined whether the peroxidase activity is required. Unexpectedly, both the wild-type and activity-dead mutant (C51S/C172S double mutant) rescued the HeLa cells from the augmented death by PrxII knockdown (Fig. 3D), suggesting that regardless of the peroxidase activity, the protective function of PrxII may rely on the protein-protein interaction.
FIGURE 3.
Nuclearly targeted PrxII protects cancer cell death against DNA damage in a peroxidase-independent manner. A and B, ectopic expression of PrxII in cytosol and nucleus of HeLa cells. HeLa cells were transfected with either control or PrxII siRNA for 24 h. Then PrxII siRNA-transfected cells were infected with the retroviruses encoding PrxII wild type (WT) targeted to cytosol (Cyto) and nucleus (NLS), respectively. The expression was assessed by immunoblotting (IB) (A) and immunostaining (B). Immunostaining of the expressed PrxII proteins were detected with anti-Myc antibody (green). Nuclei are labeled with DAPI (blue). C, reduction of etoposide (ETO)-induced cell death by add-back expression of PrxII WT in cytosol and nuclei. D, reduction of etoposide-induced cell death by add-back expression of PrxII WT and activity-dead mutant (DM) in nuclei. A and D, the extent of endogenous (endo) and exogenous (exo) Myc-tagged PrxII expression shows percent of the band intensities versus one in control cells (mean values from three experiments). Data in the graph are means ± S.D. of results from three independent death assays as done in Fig. 2 (n = 3; *, p < 0.005). N/T, not treated.
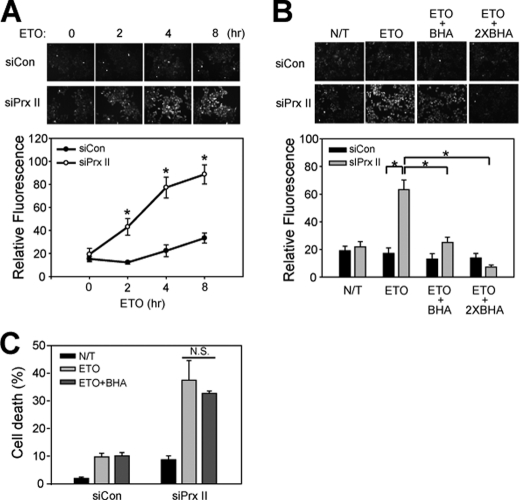
DNA Damage-induced Cell Death Is Independent of ROS Generation
It has been controversial whether ROS production is involved in the DNA damage-induced cell death (27–30). Therefore, we decided to determine whether the ROS production is increased by PrxII knockdown in etoposide-treated cells; if so, it is involved in the cell death. To determine the relationship between ROS and cell death, we first measured the intracellular ROS with an oxidation-sensitive fluorescent dye, 5,6-chloromethyl-2′,7′-dihydrochlorofluorescein diacetate. The intracellular ROS was barely detected in the etoposide-treated control HeLa cells (Fig. 4A). In contrast, the ROS level was markedly enhanced by PrxII knockdown, indicating that PrxII eliminates the intracellular ROS produced by etoposide treatment. In addition, the etoposide treatment did not induce the hyperoxidation of 2-Cys Prxs, a result of oxidative stress overwhelming cellular peroxidase capacity (31), although 100 μm H2O2 induced it (supplemental Fig. S2). This result suggests that the amount of ROS produced by etoposide is likely to be what PrxII can accommodate. We then utilized the BHA as a general antioxidant compound to examine whether the ROS increased by PrxII knockdown is involved in cell death. As expected, BHA prevented the increase of ROS level (Fig. 4B). However, the BHA pretreatment did not affect the etoposide-induced cell death both in the control and PrxII knockdown cells (Fig. 4C). Although ROS are produced by DNA damage, we concluded that they may not contribute to the cell death.
FIGURE 4.
Induction of cell death by etoposide is independent of ROS generation. A, measurement of ROS production in etoposide-treated HeLa cells. B, elimination of etoposide-induced ROS by BHA. Cells were pretreated with 100 or 200 μm BHA (two times) 1 h before etoposide (ETO) treatment. Data in the graph are means ± S.D. of 2′,7′-dichlorodihydrofluorescein fluorescence values calculated from about 20–30 cells (n = 3; *, p < 0.0001). Representative images are shown (upper panel). C, effect of BHA treatment on etoposide-induced cell death in HeLa cells that had been transfected with control or PrxII siRNA. Data in the graph are means ± S.D. of results from three independent death assays as done in Fig. 2 (N.S., not significant). N/T, not treated.
JNK-c-Jun Pathway Is Down-regulated by PrxII Knockdown
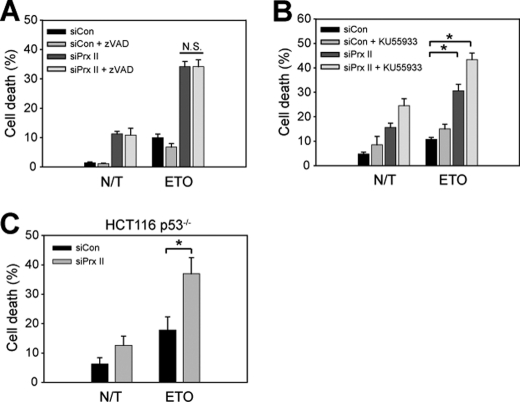
Next, we studied the signaling mechanism underlying the protective role of PrxII in DNA damage-induced cell death. First, we tested whether the etoposide-induced cell death is caspase-dependent. A pan-caspase inhibitor, benzyloxycarbonyl-VAD-fluoromethyl ketone, did not block the etoposide-induced cell death both in the control and PrxII knockdown HeLa cells (Fig. 5A), suggesting that etoposide induces cell death in a caspase-independent manner. Second, we examined whether ATM, the master kinase that induces p53-dependent apoptosis in response to DNA double strand breaks (32, 33), is involved. When the HeLa cells were pretreated with a specific ATM inhibitor KU55933, the ATM inhibition additively increased the etoposide-induced cell death in the PrxII knockdown cells (Fig. 5B). Furthermore, like parental HCT116 cells, the cell death of p53-deficient HCT116 cells induced by etoposide treatment was also augmented by PrxII knockdown compared with that of control cells (Fig. 5C). These results suggest that the PrxII-mediated protection against DNA damage-induced cell death may be independent of ATM and p53 function.
FIGURE 5.
Etoposide induces cell death in PrxII knockdown cells independently of ATM-p53 pathway. A, effect of a pan-caspase inhibitor, benzyloxycarbonyl (Z)-VAD-fluoromethyl ketone (10 μm), in etoposide (ETO)-induced cell death. B, effect of ATM inhibitor in etoposide-induced cell death. The cells were pretreated with KU55933 (2 μm) for 1 h before etoposide treatment. C, etoposide-induced cell death in parental and p53-deficient HCT116 colon cells. Data in the graph are means ± S.D. of results from three independent death assays as done in Fig. 2 (n = 3; *, p < 0.001, N.S., not significant). N/T, not treated.
Because MAPKs are known to participate in cell cycle checkpoint, apoptosis, and DNA repair under cellular stresses such as ultraviolet and DNA damage (34, 35), we explored the activation of MAPKs in etoposide-treated HeLa cancer cells. Interestingly, etoposide induced a persistent activation of all three MAPKs, among which JNK and ERK activations were apparently down-regulated by the PrxII knockdown (Fig. 6A). To dissect which kinase is involved in the cell death, we pretreated HeLa cells with ERK and JNK inhibitors. Clearly, the JNK inhibition enhanced the etoposide-induced cell death, whereas the ERK inhibition did not (Fig. 6B). Furthermore, the JNK inhibition synergistically augmented cell death in PrxII knockdown cells. Then, to ascertain how PrxII regulates JNK activation, we looked at the location of PrxII and JNK in the etoposide-treated cells by immunostaining. Interestingly, the etoposide treatment induced the nuclear accumulation of phosphorylated JNK for which an immunoreactive signal was completely blocked by a synthetic phospho-JNK antigenic peptide (Fig. 6C). As a control experiment, H2O2 treatment also induced nuclear accumulation of phosphorylated JNK. Interestingly, the nuclear level of PrxII was increased by etoposide treatment in a timely manner (Fig. 6D and supplemental Fig. S3). Moreover, the level of phosphorylated JNK in the nuclei of etoposide-treated cells was notably diminished by the PrxII knockdown (Fig. 6D), suggesting that PrxII may preserve the phosphorylated JNK in the nucleus. Collectively, these results indicate that JNK activation is essential for cell survival under DNA damage, and it is positively regulated by PrxII.
FIGURE 6.
PrxII protect cancer cells against DNA damage via activation JNK/c-Jun pathway. A, reduction of etoposide (ETO)-induced activation of MAPKs by PrxII knockdown in HeLa cells. The activation of each MAPK was assessed by phospho-specific antibodies. The phospho-specific bands were quantified and normalized by the intensities of the corresponding nonphospho bands. Data in the graphs are means ± S.D. of fold increase of the band intensities versus untreated control one (n = 3; *, p < 0.005; **, p < 0.002). One representative blot of three experiments is shown. IB, immunoblot. B, effect of ERK and JNK inhibitors on etoposide-induced cell death. Cells were pretreated with PD98059 (PD, 100 μm) or SP600125 (SP, 30 μm) for 1 h before etoposide treatment. Data in the graph are means ± S.D. of three independent death assays as done in Fig. 2 (n = 6 or 8; *, p < 0.001, N.S., not significant). C, nuclear translocation of phosphorylated JNK (P-JNK) by etoposide or H2O2 treatment. The specificity of anti-p-JNK antibody was assessed using antigenic p-JNK peptide. N/T, not treated. D, reduction of nuclear accumulation of p-JNK by PrxII knockdown. Nuclei are labeled with DAPI (blue). E, impairment of c-Jun expression and phosphorylation by PrxII knockdown. The immunoreactive bands were quantified and normalized by the intensities of the corresponding tubulin bands. Data in the graphs are means ± S.D. of fold increase of the band intensities versus an untreated control one. One representative blot of two experiments is shown. F, effect of ERK and JNK inhibitors on c-Jun expression. G, impairment of c-Jun expression by PrxII knockdown in HCT116 and U2OS cancer cells. C–G, representative image and blot of three or four experiments are shown.
Next, we examined the phosphorylation and expression of c-Jun as a direct substrate of the activated JNK. The etoposide treatment in HeLa cancer cells markedly induced c-Jun expression and phosphorylation following the time course of JNK activation, which was reduced by PrxII knockdown (Fig. 6E). As expected from the result in Fig. 5B, the inhibition of JNK, not ERK, also reduced c-Jun expression in etoposide-treated control HeLa cells and more severely reduced it in the cell with PrxII knockdown (Fig. 6F). Furthermore, the PrxII knockdown also abolished the increase of c-Jun expression in other cancer cells, such as HCT116 and U2OS, that had exhibited the enhanced cell death by the PrxII knockdown in response to etoposide (Fig. 6G). These results collectively indicate that PrxII is an essential component of the JNK-dependent survival pathway in response to DNA damage.
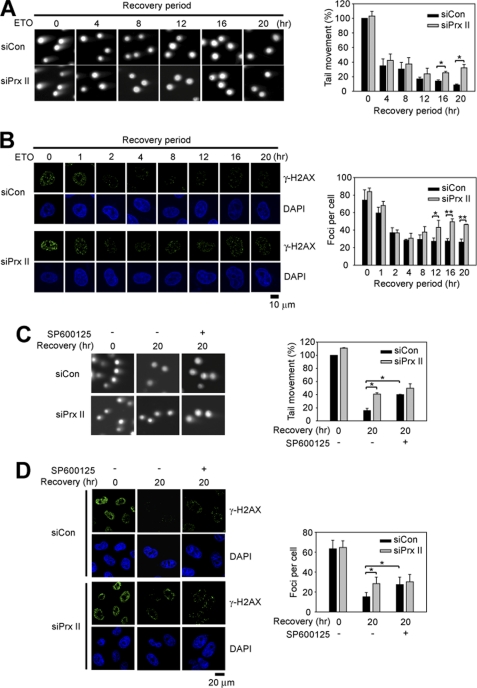
PrxII Knockdown Results in a Defect in DNA Repair
Recent reports have shown that c-Jun and ATF play a protective role in DNA damage by inducing the genes involved in the DNA repair process (36, 37). Therefore, we examined the influence of PrxII knockdown during DNA repair after etoposide treatment. The extent of DNA repair was estimated as the amount of DNA double strand breaks measured by a neutral Comet assay and γ-H2AX focus formation assay. The DNA repair process was biphasic; therefore, the extent of damaged DNA was rapidly dropped by 40% within several hours after removal of etoposide and then slowly decreased close to the zero level (Fig. 7). As seen at zero time of recovery, the extent of DNA damage induced by etoposide was not affected by the PrxII knockdown. However, the DNA repair was retarded by the PrxII knockdown, as evident in a later phase (Fig. 7A). A similar effect was observed in the γ-H2AX focus formation (Fig. 7B). These results suggest that down-regulation of the JNK-c-Jun pathway by the PrxII knockdown affects completion of the DNA repair process. To support this, we performed a comet assay either in the presence or absence of the JNK inhibitor. The JNK inhibition resulted in the retarded DNA repair as much as was done by the PrxII knockdown (Fig. 7C). Similar results were obtained by the γ-H2AX focus formation assay (Fig. 7D). Consequently, the dual blockade of JNK and PrxII was not an additive effect, indicating that they are in the same pathway.
FIGURE 7.
PrxII knockdown retards DNA repair in JNK-dependent manner. A and B, effect of PrxII knockdown on repair of DNA damage induced by etoposide (ETO). HeLa cells were treated with etoposide for 1 h, and further cultured in fresh media for the indicated times after removal of etoposide. The amount of double strand breaks was indirectly assessed by comet assay (A) and γ-H2AX focus assay (B). Data in A are means ± S.D. of the percent of averaged tail length versus zero time of recovery (n = 3; *, p < 0.01). Data in B are means ± S.D. of number of foci per cell averaged from 25 to 35 cells versus zero time of recovery (n = 3; *, p < 0.05; **, p < 0.001). C and D, effect of JNK inhibition on repair of DNA damage induced by etoposide. HeLa cells were pretreated with or without SP600125 (10 μm) for 1 h before etoposide treatment. Comet assay (C) and γ-H2AX focus assay (D) were done zero time and 20 h after removal of etoposide (n = 3; *, p < 0.01). Representative images are shown (DAPI for nuclei).
DISCUSSION
Cancer has the highest mortality rate throughout the world. Thus, its eradication is the most crucial medical and scientific issue that must be resolved. Although radio- and chemotherapies are powerful treatments based on the unlimited hyper-proliferative characteristics of cancer cells, the cytotoxic side effect to normal cells is debilitating to patients with cancer. To ensure a full recovery for a patient with cancer, alternative strategies have long been sought for combinatorial therapy. One promising example is anti-VEGF therapy that blocks the formation of angiogenic vasculature essential for tumor growth (38). However, recent studies have reported an adverse effect wherein the cancer cells that survived after short term anti-VEGF treatment became more aggressive and invasive (39, 40). Undeniably, efforts must be continued to selectively and efficiently kill the cancerous cells.
In this study, we explored the role of nuclear PrxI and PrxII in DNA damage-induced cell death and made several important discoveries. 1) Nuclear PrxII specifically confers the resistance against DNA damage-induced death to cancer cells and not to normal cells. 2) DNA-damaging agents induce cancer cell death independently of ROS. 3) Nuclear PrxII positively regulates JNK activation in response to DNA damage in opposition to its negative role in TNF-α-induced JNK activation (41). 4) PrxII-dependent regulation of JNK/c-Jun pathway is essential for completion, rather than initiation, of DNA repair. These findings implicate PrxII as a novel anti-cancer target that regulates DNA repair in cancer cells against DNA-damaging agents. The biological function of PrxI and PrxII related to tumorigenesis is somewhat complex. Knock-out mice studies have shown that the PrxII-deficient mice did not produce any phenotype related to tumorigenesis (21), whereas the deficiency of PrxI resulted in the development of malignant tumors (18) and increased DNA oxidation in various tissues of aged mice (17). Although the PrxI-deficient mice were produced by transposon or retroviral insertion, which could have been accompanied with additional mutations, the evidence suggests that PrxI can suppress the tumorigenesis. Some in vitro evidence provided mechanistic explanation for this tumor suppressor function such that PrxI inhibits c-Abl kinase and c-Myc by direct interaction (15, 42). However, it is paradoxical that the expression of both PrxI and PrxII was increased in various tumor types (10). Moreover, decreasing PrxI or PrxII expression enhances radiosensitivity of cancer cells (43–46). Collectively, this means that the function of cytosolic 2-Cys Prxs in tumorigenesis and tumor growth may differ. Our study provided new data that PrxII knockdown sensitizes cancer cells to cell death induced by chemotherapeutic agents through regulating DNA repair. Mechanistically, down-regulation of ERK and JNK activation by PrxII knockdown is a particularly salient result. Numerous studies have shown that both ERK1/2 and JNK are activated by DNA damage stresses. Etoposide-induced ERK activation was shown to mediate cell cycle arrest in an ATM-dependent but p53-independent manner (47). In the case of other stresses, such as hydroxyurea and ionizing radiation, ERK activation was required for S phase and G2/M checkpoint arrest, respectively (48, 49). Moreover, Yan et al. reported that ionizing radiation-induced ERK1/2 activation required a direct interaction with BRCA1 for G2/M cell cycle checkpoint arrest (50). Thus, the evidence suggests that ERK activation by DNA damage may preferentially be involved in cell cycle arrest before cell death. However, the JNK activation pathway has a dual function of pro- and anti-apoptotic properties (51). In other words, the sustained JNK activation is predominantly associated with induction or enhancement of apoptosis, whereas transient JNK activation is involved in the cell survival response (52). In the DNA damage response, JNK activation was shown to promote drug resistance and cell survival by enhancing DNA repair (37). Furthermore, Hayakawa et al. (36) have also shown that the activated JNK leads to phosphorylation of downstream transcription factors, ATF2 and c-Jun, which induce the expression of components of DNA repair machinery. Our data shows that inhibition of JNK, not ERK, was linked to etoposide-induced cell death. Furthermore, the down-regulation of JNK activation by PrxII knockdown led to the reduced expression and phosphorylation of the c-Jun transcription factor, which in turn results in the defective repair of etoposide-induced double strand breaks. Although we have not analyzed the DNA repair machinery in etoposide-treated cells, our data clearly support that PrxII positively regulates JNK/c-Jun-dependent DNA repair in cancer cells.
The mechanism of how PrxII positively regulates JNK activation in DNA damage response remains questionable. Few studies have shown the interaction of Prx with the MAPKs. In lung cancer cells, PrxI interacted with a GSTpi-JNK complex and suppressed ionizing radiation-induced JNK activation and apoptosis (12). On the contrary, a yeast 2-Cys Prx called thioredoxin peroxidase 1 activated a homolog of mammalian p38/JNK MAPK, Sty1, via the peroxide-induced disulfide formation (53). We have failed to determine through our experiments the direct protein-protein interaction between PrxII and JNK. Nonetheless, because the nuclearly targeted PrxII rescued the cancer cells against etoposide-induced cell death and the level of phosphorylated JNK in the nucleus was reduced by the PrxII knockdown, it is most likely that PrxII contributes to maintain phosphorylated JNK in the nucleus.
In summary, we found that absence of nuclear PrxII sensitizes cancer cells to chemotherapeutic agents by impairing the JNK-dependent DNA repair. It was previously shown that a dominant negative mutant of PrxII also sensitizes cancer cells to TNF-α-induced cell death (54). The former was ROS-independent, and the latter was ROS-dependent. Given that most cancer cells produce a large amount of ROS, the present finding leads us to propose that inhibiting PrxII can be a powerful means to kill the cancer cells by bursting death signaling in the cytoplasm and by impairing DNA repair in the nucleus.
Supplementary Material
Acknowledgments
We thank Drs. Bert Vogelstein and Deug-Yong Shin for providing p53-deficient HCT116 cells.
This work was supported in part by Mid-career Researcher Program through National Research Foundation of Korea Grant 2009-0054572 funded by the Ministry of Education, Science and Technology, by 21C Frontier Functional Proteomics Project FPR08-B1-190 funded by the Ministry of Education, Science and Technology, and by KOSEF through the Center for Cell Signaling and Drug Discovery Research Grant R15-2006-020 at Ewha Womans University.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3.
- ROS
- reactive oxygen species
- Prx
- peroxiredoxin
- H2O2
- hydrogen peroxide
- BHA
- butylated hydroxyanisole
- MEF
- mouse embryonic fibroblast
- PI
- propidium iodide
- NLS
- nuclear localization signal
- ATM
- ataxia telangiectasia mutated.
REFERENCES
- 1. Marnett L. J., Riggins J. N., West J. D. (2003) J. Clin. Invest. 111, 583–593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Cerutti P. A. (1994) Lancet 344, 862–863 [DOI] [PubMed] [Google Scholar]
- 3. Finkel T., Holbrook N. J. (2000) Nature 408, 239–247 [DOI] [PubMed] [Google Scholar]
- 4. Janssen-Heininger Y. M., Mossman B. T., Heintz N. H., Forman H. J., Kalyanaraman B., Finkel T., Stamler J. S., Rhee S. G., van der Vliet A. (2008) Free Radic. Biol. Med. 45, 1–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Rhee S. G., Bae Y. S., Lee S. R., Kwon J. (2000) Sci. STKE 2000, pe1. [DOI] [PubMed] [Google Scholar]
- 6. Halliwell B., Gutteridge J. M. C. (2007) Free Radicals in Biology and Medicine, 4th Ed., pp. 70–144, Oxford University Press, Inc., New York [Google Scholar]
- 7. Brigelius-Flohé R. (1999) Free Radic. Biol. Med. 27, 951–965 [DOI] [PubMed] [Google Scholar]
- 8. Rhee S. G., Chae H. Z., Kim K. (2005) Free Radic. Biol. Med. 38, 1543–1552 [DOI] [PubMed] [Google Scholar]
- 9. Wood Z. A., Schröder E., Robin Harris J., Poole L. B. (2003) Trends Biochem. Sci. 28, 32–40 [DOI] [PubMed] [Google Scholar]
- 10. Kang S. W., Rhee S. G., Chang T. S., Jeong W., Choi M. H. (2005) Trends Mol. Med. 11, 571–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Szatrowski T. P., Nathan C. F. (1991) Cancer Res. 51, 794–798 [PubMed] [Google Scholar]
- 12. Kim Y. J., Lee W. S., Ip C., Chae H. Z., Park E. M., Park Y. M. (2006) Cancer Res. 66, 7136–7142 [DOI] [PubMed] [Google Scholar]
- 13. Park S. H., Chung Y. M., Lee Y. S., Kim H. J., Kim J. S., Chae H. Z., Yoo Y. D. (2000) Clin. Cancer Res. 6, 4915–4920 [PubMed] [Google Scholar]
- 14. Chang T. S., Cho C. S., Park S., Yu S., Kang S. W., Rhee S. G. (2004) J. Biol. Chem. 279, 41975–41984 [DOI] [PubMed] [Google Scholar]
- 15. Mu Z. M., Yin X. Y., Prochownik E. V. (2002) J. Biol. Chem. 277, 43175–43184 [DOI] [PubMed] [Google Scholar]
- 16. Cao J., Schulte J., Knight A., Leslie N. R., Zagozdzon A., Bronson R., Manevich Y., Beeson C., Neumann C. A. (2009) EMBO J. 28, 1505–1517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Egler R. A., Fernandes E., Rothermund K., Sereika S., de Souza-Pinto N., Jaruga P., Dizdaroglu M., Prochownik E. V. (2005) Oncogene 24, 8038–8050 [DOI] [PubMed] [Google Scholar]
- 18. Neumann C. A., Krause D. S., Carman C. V., Das S., Dubey D. P., Abraham J. L., Bronson R. T., Fujiwara Y., Orkin S. H., Van Etten R. A. (2003) Nature 424, 561–565 [DOI] [PubMed] [Google Scholar]
- 19. Manta B., Hugo M., Ortiz C., Ferrer-Sueta G., Trujillo M., Denicola A. (2009) Arch. Biochem. Biophys. 484, 146–154 [DOI] [PubMed] [Google Scholar]
- 20. Low F. M., Hampton M. B., Winterbourn C. C. (2008) Antioxid. Redox. Signal. 10, 1621–1630 [DOI] [PubMed] [Google Scholar]
- 21. Lee T. H., Kim S. U., Yu S. L., Kim S. H., Park D. S., Moon H. B., Dho S. H., Kwon K. S., Kwon H. J., Han Y. H., Jeong S., Kang S. W., Shin H. S., Lee K. K., Rhee S. G., Yu D. Y. (2003) Blood 101, 5033–5038 [DOI] [PubMed] [Google Scholar]
- 22. Choi M. H., Lee I. K., Kim G. W., Kim B. U., Han Y. H., Yu D. Y., Park H. S., Kim K. Y., Lee J. S., Choi C., Bae Y. S., Lee B. I., Rhee S. G., Kang S. W. (2005) Nature 435, 347–353 [DOI] [PubMed] [Google Scholar]
- 23. Kang S. W., Chae H. Z., Seo M. S., Kim K., Baines I. C., Rhee S. G. (1998) J. Biol. Chem. 273, 6297–6302 [DOI] [PubMed] [Google Scholar]
- 24. Chae H. Z., Kim H. J., Kang S. W., Rhee S. G. (1999) Diabetes Res. Clin. Pract. 45, 101–112 [DOI] [PubMed] [Google Scholar]
- 25. Kudo N., Matsumori N., Taoka H., Fujiwara D., Schreiner E. P., Wolff B., Yoshida M., Horinouchi S. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 9112–9117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ghosh S., Karin M. (2002) Cell 109, S81–S96 [DOI] [PubMed] [Google Scholar]
- 27. Wang S., Konorev E. A., Kotamraju S., Joseph J., Kalivendi S., Kalyanaraman B. (2004) J. Biol. Chem. 279, 25535–25543 [DOI] [PubMed] [Google Scholar]
- 28. Kurosu T., Fukuda T., Miki T., Miura O. (2003) Oncogene 22, 4459–4468 [DOI] [PubMed] [Google Scholar]
- 29. Sentürker S., Tschirret-Guth R., Morrow J., Levine R., Shacter E. (2002) Arch. Biochem. Biophys. 397, 262–272 [DOI] [PubMed] [Google Scholar]
- 30. Mizutani H., Tada-Oikawa S., Hiraku Y., Oikawa S., Kojima M., Kawanishi S. (2002) J. Biol. Chem. 277, 30684–30689 [DOI] [PubMed] [Google Scholar]
- 31. Yang K. S., Kang S. W., Woo H. A., Hwang S. C., Chae H. Z., Kim K., Rhee S. G. (2002) J. Biol. Chem. 277, 38029–38036 [DOI] [PubMed] [Google Scholar]
- 32. Shiloh Y. (2003) Nat. Rev. Cancer 3, 155–168 [DOI] [PubMed] [Google Scholar]
- 33. Zhou B. B., Elledge S. J. (2000) Nature 408, 433–439 [DOI] [PubMed] [Google Scholar]
- 34. Dent P., Yacoub A., Fisher P. B., Hagan M. P., Grant S. (2003) Oncogene 22, 5885–5896 [DOI] [PubMed] [Google Scholar]
- 35. Tournier C., Hess P., Yang D. D., Xu J., Turner T. K., Nimnual A., Bar-Sagi D., Jones S. N., Flavell R. A., Davis R. J. (2000) Science 288, 870–874 [DOI] [PubMed] [Google Scholar]
- 36. Hayakawa J., Mittal S., Wang Y., Korkmaz K. S., Adamson E., English C., Ohmichi M., Omichi M., McClelland M., Mercola D. (2004) Mol. Cell 16, 521–535 [DOI] [PubMed] [Google Scholar]
- 37. Hayakawa J., Depatie C., Ohmichi M., Mercola D. (2003) J. Biol. Chem. 278, 20582–20592 [DOI] [PubMed] [Google Scholar]
- 38. Ellis L. M., Hicklin D. J. (2008) Nat. Rev. Cancer 8, 579–591 [DOI] [PubMed] [Google Scholar]
- 39. Pàez-Ribes M., Allen E., Hudock J., Takeda T., Okuyama H., Viñals F., Inoue M., Bergers G., Hanahan D., Casanovas O. (2009) Cancer Cell 15, 220–231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ebos J. M., Lee C. R., Cruz-Munoz W., Bjarnason G. A., Christensen J. G., Kerbel R. S. (2009) Cancer Cell 15, 232–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kang S. W., Chang T. S., Lee T. H., Kim E. S., Yu D. Y., Rhee S. G. (2004) J. Biol. Chem. 279, 2535–2543 [DOI] [PubMed] [Google Scholar]
- 42. Wen S. T., Van Etten R. A. (1997) Genes. Dev. 11, 2456–2467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Smith-Pearson P. S., Kooshki M., Spitz D. R., Poole L. B., Zhao W., Robbins M. E. (2008) Free Radic. Biol. Med. 45, 1178–1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Chen M. F., Keng P. C., Shau H., Wu C. T., Hu Y. C., Liao S. K., Chen W. C. (2006) Int. J. Radiat. Oncol. Biol. Phys. 64, 581–591 [DOI] [PubMed] [Google Scholar]
- 45. Zhang B., Su Y., Ai G., Wang Y., Wang T., Wang F. (2005) J. Radiat. Res. 46, 305–312 [DOI] [PubMed] [Google Scholar]
- 46. Wang T., Tamae D., LeBon T., Shively J. E., Yen Y., Li J. J. (2005) Cancer Res. 65, 10338–10346 [DOI] [PubMed] [Google Scholar]
- 47. Tang D., Wu D., Hirao A., Lahti J. M., Liu L., Mazza B., Kidd V. J., Mak T. W., Ingram A. J. (2002) J. Biol. Chem. 277, 12710–12717 [DOI] [PubMed] [Google Scholar]
- 48. Wu D., Chen B., Parihar K., He L., Fan C., Zhang J., Liu L., Gillis A., Bruce A., Kapoor A., Tang D. (2006) Oncogene 25, 1153–1164 [DOI] [PubMed] [Google Scholar]
- 49. Yan Y., Black C. P., Cowan K. H. (2007) Oncogene 26, 4689–4698 [DOI] [PubMed] [Google Scholar]
- 50. Yan Y., Black C. P., Cao P. T., Haferbier J. L., Kolb R. H., Spieker R. S., Ristow A. M., Cowan K. H. (2008) Cancer Res. 68, 5113–5121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Davis R. J. (2000) Cell 103, 239–252 [DOI] [PubMed] [Google Scholar]
- 52. Ventura J. J., Hübner A., Zhang C., Flavell R. A., Shokat K. M., Davis R. J. (2006) Mol. Cell 21, 701–710 [DOI] [PubMed] [Google Scholar]
- 53. Veal E. A., Findlay V. J., Day A. M., Bozonet S. M., Evans J. M., Quinn J., Morgan B. A. (2004) Mol. Cell 15, 129–139 [DOI] [PubMed] [Google Scholar]
- 54. Lee J. Y., Jung H. J., Song I. S., Williams M. S., Choi C., Rhee S. G., Kim J., Kang S. W. (2009) Free Radic. Biol. Med. 47, 1162–1171 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.