Abstract
Background
Ionizing radiation is a well-known mutagen and a risk factor for thyroid cancer. MicroRNAs (miRNAs) play an important role in the regulation of gene expression on post-transcriptional level and are dysregulated in thyroid cancer. The goal of this study was to investigate the effects of acute exposure to 1 and 10 Gy of γ-irradiation on miRNA expression in normal human thyroid cells.
Methods
Expression of 319 miRNAs was studied in primary cultures of normal human thyroid cells 4 and 24 hours postirradiation using a miRNA expression array with further confirmation of miRNAs expression by reverse transcription-polymerase chain reaction.
Results
We identified 30 miRNAs that were unregulated or downregulated more than twofold after irradiation as compared to nonirradiated thyroid cells, with no significant difference found between 1 and 10 Gy of radiation. Four distinct patterns of miRNA expression change were observed: miRNAs downregulated at 4 hours and returned to normal levels at 24 hours, miRNAs upregulated at 4 hours and returned to normal levels at 24 hours, and miRNAs either upregulated or downregulated at both time points. No dysregulation of miRNAs known to occur in thyroid cancer was observed.
Conclusions
Acute exposure of thyroid cells to γ-radiation results in several specific patterns of miRNA response. It appears that alteration in miRNA expression seen 4–24 hours after irradiation has no direct association with carcinogenesis. However, it is likely to affect other cell functions, such as DNA repair.
Introduction
Ionizing radiation is a well-established risk factor for various types of cancer, including thyroid cancer. Numerous reports have documented an increased incidence of thyroid tumors in populations exposed to ionizing radiation, including survivors of atomic bomb explosions in Japan in 1945 (1,2), residents of the Marshall Islands exposed to radioactive fallout in 1954 (3,4), patients exposed to therapeutic irradiation (5,6), and children exposed to radiation after the Chernobyl nuclear power accident (7,8). In all of these populations, thyroid papillary carcinoma has been identified as a type of thyroid cancer associated with radiation exposure.
Ionizing radiation induces damage to various cell components, whereas damage to cellular DNA is primarily responsible for mutagenesis and carcinogenesis, and double-strand breaks are likely to be the initial events in the generation of specific carcinogenic mutations (9,10). However, it remains unclear whether these mutations are formed immediately after radiation exposure or later. Thyroid papillary carcinoma developed after radiation exposure at Chernobyl and in atomic bomb survivors in Japan is characterized by a very high frequency of RET/PTC chromosomal rearrangement, which was found to directly correlate with radiation dose in some studies (11–13).
To repair radiation damage, a complex cellular response is initiated, including alteration in gene expression, especially in genes involved in stress response, cell cycle control, and DNA synthesis/repair (14–16). It has been recently discovered that microRNAs (miRNAs) play an important role in the regulation of gene expression on post-transcriptional level (17,18). However, the role of miRNAs in cell response to radiation damage in human cells remains largely unknown.
miRNAs are small endogenous noncoding RNAs that act as negative regulators of the protein-coding gene expression through the complementary binding to 3′ untranslated region (UTR) of target mRNA, which lead to translational repression and inhibition of protein synthesis. miRNA expression is deregulated in many types of human cancers, including thyroid cancer. Several studies have demonstrated that normal thyroid cells have a unique profile of miRNA expression and many miRNAs are dysregulated in thyroid cancer cells. A subset of these miRNAs, including miR-221, miR-222, miR-146b, miR-155, and miR-187, have been consistently found to be upregulated in thyroid papillary carcinoma (19–21). Moreover, strong correlation was found between miRNA expression and somatic mutations found in this tumor type. Specifically, upregulation of miR-187, miR-146b, and miR-155 was found to be significantly more pronounced in papillary carcinomas carrying RET/PTC rearrangements (20), a genetic event characteristically found in radiation-induced thyroid tumors.
Several studies have reported the association between miRNAs and radiation exposure. He et al. found upregulation of miR-34 miRNA family (miR-34a, −34b, and −34c) in a variety of mouse tissues after exposure to ionizing radiation (22). Interestingly, they showed that these miRNAs were transcriptionally activated by TP53 in response to DNA damage, and their upregulation, in turn, led to downregulation of their target genes (i.e., CDK4 and MET) and to cell cycle arrest. Weidhaas et al. showed the dysregulation of let-7 family miRNAs in response to radiation in lung cancer cells and demonstrated that overexpression of some of these miRNAs altered radiosensitivity of these cells (23). In another study, dysregulation of miR-521 and miR-34c was observed in prostate cancer cells subjected to external beam radiation (24). However, according to our knowledge, no studies of miRNA expression in normal human thyroid cells exposure to ionizing radiation have been reported to date. In this observation, we report the results of our analysis of miRNA expression in human normal thyroid cells subjected to different doses of γ-irradiation and followed for the first 24 hours after irradiation.
Materials and Methods
Cell culture and irradiation
Normal thyroid tissue from surgically removed thyroid samples was collected at the Department of Pathology, University of Cincinnati, after the University of Cincinnati Institutional Review Board (IRB) approval. Primary cell cultures were established from the two freshly collected normal thyroid tissue samples as previously described (25). In both cases, the tissue was obtained from an opposite lobe of total thyroidectomy specimens containing a discrete nodule diagnosed pathologically as either an encapsulated papillary carcinoma or a hyperplastic nodule. Cells were cultured for 3–4 days at 37°C in 95% humidity and 5% CO2 in RPMI-1640 medium containing 10% of fetal bovine serum. The presence of epithelial thyroid cells was confirmed by reverse transcription-polymerase chain reaction (RT-PCR) detection of thyroglobulin expression performed as previously described (26). Confluent cell cultures were exposed to a single dose of γ-irradiation from a cesium-137 source at a dose rate of 1.7 Gy/min. Cells were exposed to 1 or 10 Gy γ-irradiation in triplicate for each dose and collected at 4 and 24 hours after exposure.
RNA isolation
Total RNA was isolated using Trizol reagent (Invitrogen, Carlsbad, CA) as previously described (27). RNA yield was determined using the NanoDrop 1000 spectrophotometer (ThermoScientific, Wilmington, DE). RNA integrity was assessed with the Agilent 6000 NANO kit for the 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA) according to manufacture's protocol.
miRNA expression array
Quantitation of expression of 319 mature miRNA was performed using FlexmiR™ human microRNA pool, version 8 (Exiqon, Vedbaek, Denmark), and analyzed on Luminex 200 (Luminex Corporation, Austin, TX) according to the manufacturer's instructions. Specifically, 2.5 μg of total RNA was labeled with biotin using the FlexmiR™ MicroRNA Labeling Kit (Luminex) followed by hybridization to beads coated with locked nucleic acids probes complementary to mature miRNA sequences. All samples were assayed in duplicate. The system calibration was performed using the xMAP™ calibration control reagents (Luminex). A blank control set of beads was used to normalize the background value for every individual miRNA. Five synthetic controls and four small nucleolar RNA (snoRNA) normalization controls were used to adjust mean fluorescence intensities between samples and between runs using the average correction factor for these controls as recommended by the manufacturer. Finally, the miRNA expression in the irradiated cells was quantitated relatively to nonirradiated cells using Luminex IS™ software v.2.3 (Luminex).
miRNA real-time RT-PCR
Expression of individual miRNAs was detected by real-time RT-PCR using miRNA sequence-specific primers (Applied Biosystems, Inc., Foster City, CA). Briefly, 10 ng of total RNA was reverse transcribed using High-Capacity cDNA Archive kit (Applied Biosystems, Inc.) followed by amplification on ABI 7500 Real-Time PCR System (Applied Biosystems, Inc.). All RT-PCRs were performed in triplicate. Small nucleolar RNA RNU44 was used as endogenous control for the normalization of RNA input. miRNA expression levels were calculated by relative quantitation using the ABI 7500 Real-Time PCR SDS 1.2 software (Applied Biosystems, Inc.) and the fold change of expression was determined by 2−ΔΔCT method (28). No template reaction was used as a negative control.
Statistical analysis
One-way analysis of variance was initially used to identify a subset of miRNAs significantly altered after 1 and 10 Gy of γ-irradiation. Student's t-test was used to determine statistical significance of differentially expressed miRNAs between irradiated and nonirradiated cells and for comparison of miRNA expression between different doses of irradiation. Agglomerative hierarchical clustering analysis was performed using Cluster software and TreeView software (http://genome-www5.stanford.edu/resources/restech.shtml) (29).
Search for target genes
Putative miRNA target genes were identified using miRBase (http://microrna.sanger.ac.uk), TargetScan (http://genes.mit.edu/targetscan) (30), and PicTar (http://pictar.bio.nyu.edu) (31) target prediction programs.
Results
First, primary cultures of normal human thyroid cells exposed to 1 and 10 Gy of γ-irradiation were collected 4 and 24 hours postirradiation and studied for expression of 319 human mature miRNAs by miRNA expression array. The analysis revealed a significant number of miRNAs that were dysregulated after radiation exposure as compared to nonirradiated thyroid cells. Of those, 30 miRNAs were upregulated or downregulated more than twofold either after 1 or 10 Gy of irradiation at each time point. These 30 miRNAs were further studied by real-time RT-PCR to confirm the results and determine the expression levels with higher degree of precision. Some of these miRNAs were up to 24-fold upregulated and other were up to 50-fold downregulated at different time intervals after cell irradiation (Table 1).
Table 1.
Differentially Expressed MicroRNAs in Normal Human Thyroid Cells After 1 and 10 Gy of γ-Radiation
| |
Irradiated cells, 1 Gy |
Irradiated cells, 10 Gy |
||
|---|---|---|---|---|
| miRNA | 4 hours | 24 hours | 4 hours | 24 hours |
| hsa-miR-409-5p | 17.3 | 0.3 | 6.8 | 0.5 |
| has-miR-520a | 13.3 | 11.5 | 6.3 | 8.6 |
| has-let-7d | 12.4 | 13.5 | 24.4 | 16.1 |
| has-miR-489 | 10.1 | 13.7 | 11.3 | 9.1 |
| has-miR-193a | 6.8 | 4.0 | 6.2 | 3.9 |
| has-let-7g | 5.7 | 0.5 | 9.4 | 0.8 |
| has-miR-188 | 5.5 | 7.5 | 1.7 | 7.2 |
| has-miR-122a | 4.3 | 2.9 | 2.2 | 2.6 |
| has-let-7c | 2.2 | 2.1 | 2.3 | 1.2# |
| has-miR-146a | 2.0 | 0.5 | 2.9# | 0.2 |
| has-miR-96 | 2.0 | 0.9# | 1.5 | 0.8# |
| has-miR-203 | 2.0 | 2.1 | 2.3 | 2.3 |
| has-miR-365 | 1.7# | 0.9 | 1.6# | 0.9# |
| has-miR-34b | 1.6# | 3.2 | 1.8 | 3.3 |
| has-miR-34a | 1.4# | 0.3 | 1.8 | 0.7# |
| has-miR-452* | 0.7# | 4.5 | 0.4 | 4.4 |
| has-miR-527 | 0.6# | 1.6# | 0.6# | 1.0# |
| has-miR-377 | 0.5 | 1.1# | 0.2 | 1.2# |
| has-miR-526b | 0.5 | 1.7 | 0.1 | 1.3 |
| has-miR-522 | 0.4 | 0.3 | 0.4 | 0.5 |
| has-miR-181c | 0.3 | 0.1 | 0.2 | 0.2 |
| has-miR-326 | 0.3 | 0.3 | 0.4 | 0.2 |
| has-miR-186 | 0.2 | 0.5 | 0.1 | 0.1 |
| has-miR-384 | 0.2 | 0.4 | 0.3 | 0.4 |
| has-miR-453 | 0.1 | 11.1 | 0.5 | 9.8 |
| has-miR-520e | 0.1 | 5.2 | 0.1 | 1.7# |
| has-miR-1 | 0.1 | 0.2 | 0.02 | 0.1 |
| has-miR-93 | 0.1 | 0.3 | 0.1 | 0.2 |
| hsa-miR-338 | 0.1 | 0.2 | 0.2 | 0.1 |
| hsa-let-7f | 0.1 | 0.2 | 0.1 | 0.3 |
Values represent fold change relative to nonirradiated cells as detected by reverse transcription-polymerase chain reaction; all values have a significant difference as compared to nonirradiated cells (p < 0.05) except those labeled with #.
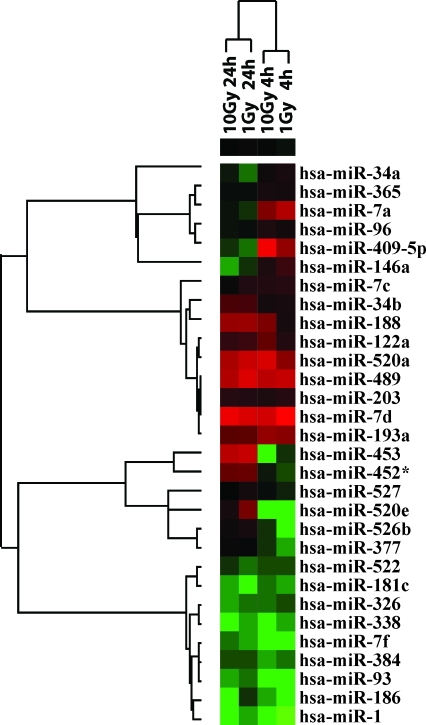
Next, the unsupervised hierarchical clustering analysis of miRNA expression was performed. The analysis grouped samples by time of collection after radiation (4 hours vs. 24 hours), but not by radiation dose (Fig. 1). It revealed four distinct groups of miRNAs: (i) miRNAs downregulated at 4 hours and returned to normal or to upregulated level at 24 hours; (ii) miRNAs upregulated at 4 hours and returned to normal or downregulated level at 24 hours; (iii) miRNAs upregulated at both time points; and (iv) miRNAs downregulated at both time points (Fig. 1).
FIG. 1.
Hierarchical cluster analysis of microRNA (miRNA) expression detected 4 and 24 hours after 1 and 10 Gy of γ-radiation. The upregulated miRNAs are depicted in shades of red, and downregulated miRNAs are depicted in shades of green. Color images available online at www.liebertonline.com/thy.
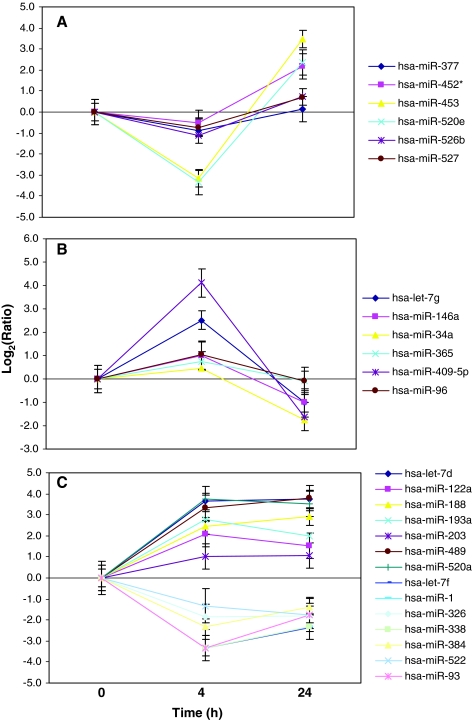
Six miRNAs constituted the first group: mir-377, miR-452*, miR-453, miR-520e, miR-526b, and miR-527. They showed more than twofold downregulation at 4 hours postexposure as compared to nonirradiated cells. At 24 hours, their expression was either at normal levels or became elevated (Fig. 2A). The patterns of expression were similar between 1 and 10 Gy doses of radiation. The second group also contained six miRNAs (miR-409-5p, let-7g, miR-146a, miR-96, miR-365, and miR-34a), which were more than twofold upregulated early after irradiation and returned to their normal levels of expression or became slightly downregulated at 24 hours postirradiation with 1 and 10 Gy (Fig. 2B). Two other groups showed consistent upregulation after 4 and 24 hours postirradiation with 1 or 10 Gy (miR-520a, let-7d, miR-489, miR-193a, miR-188, miR-122a, let-7c, miR-203, and miR-34b) or downregulation (miR-1, miR-93, let-7f, miR-338, miR-186, miR-384, miR-181c, miR-326, and miR-522) as compared to nonirradiated cells (Fig. 2C).
FIG. 2.
Temporal patterns of miRNA expression after 1 Gy of γ-radiation. (A) miRNAs downregulated at 4 hours and returned to normal or to upregulated level at 24 hours; (B) miRNAs upregulated at 4 hours and returned to normal or downregulated level at 24 hours; (C) miRNAs either upregulated or downregulated at both time points. Bars, mean ± standard error. Color images available online at www.liebertonline.com/thy.
Among miRNAs found to be significantly dysregulated after the acute exposure to radiation, there was no miRNAs known to be strongly upregulated in thyroid papillary carcinomas, such as miR-221, miR-222, miR-146b, miR-155, and miR-187. The latter three of those are particularly frequently dysregulated in tumors carrying a radiation-related RET/PTC rearrangement.
Discussion
In this study, we investigated the expression levels of miRNAs after acute exposure of human normal thyroid cells to 1 and 10 Gy of γ-radiation. These doses of radiation were in general comparable to those received by individuals who subsequently developed thyroid cancer in human populations. For example, thyroid doses in those affected by the Chernobyl accident were in the range of 0.05–4 Gy and thyroid cancer patients previously exposed to therapeutic or environmental radiation received 0.09–12.5 Gy (5). We found a significant overall change in the levels of miRNA expression at both 4 and 24 hours time intervals after exposure. Similar to our findings, early changes in miRNA expression after irradiation were found in human T-cell and B-cell lymphoblast cell lines (32), human prostate cell lines (24), and lung cancer cells (23), which showed miRNAs dysregulation at 0.5 to 4 hours postexposure.
We observed several specific patterns of miRNA response to γ-radiation, including some miRNAs that were consistently upregulated or downregulated at both time intervals. However, other miRNAs showed a time-dependant response, with specific miRNAs being downregulated or upregulated at 4 hours, the time of most active DNA repair, with the levels of expression returning to their baseline levels by 24 hours. Similar patterns in miRNA dysregulation were observed in lung epithelial cells after exposure to 2.5 Gy of radiation (23) and in human fibroblasts after low-linear energy transfer radiation (33), suggesting that these time-dependent patterns represent a common paradigm of miRNA response of various types of normal human cells to radiation exposure. The difference in miRNA expression patterns points to the complexity of cellular response to radiation. However, the exact function of miRNAs in the cell response to radiation remains unclear. If miRNA plays an active role in this process, the downregulated miRNAs may act to increase expression of genes responsible for DNA repair, whereas the upregulated miRNAs may aim to decrease the expression levels of pro-apoptotic genes to prevent cell death. On the other hand, it is possible that after radiation exposure miRNAs react on the changed levels of the coding gene expression aiming to return those to the baseline levels. In this case, they would be expected to have a limited biological role, but still can be exploited as biomarkers of radiation exposure and DNA damage repair.
Several miRNAs found to be dysregulated in thyroid cells in this study, such as let-7 family, miR-34, and miR-520, have been previously found to be dysregulated after radiation exposure in other cell types, suggesting that they participate in radiation response across the different cell types (23,34). Other miRNAs found to be dysregulated in this study (such as miR-409-5b, miR-452, miR-489, and miR-203) have not been previously reported in association with radiation exposure and may be unique for thyroid cells. It is important to note that this study was based on the analysis of 319 mature miRNAs and more recently a significant number of additional miRNAs has been discovered (35). Therefore, additional miRNAs whose expression is altered in thyroid cells after exposure to ionizing radiation may be discovered in the future.
Many upregulated and downregulated miRNAs found in this study are predicted to target genes involved in DNA repair. In fact, using three prediction programs, miRBase, TargetScan, and PicTar, the dysregulated miRNAs may target several key components of this process such as ATM, ATR, RAD51, and HRCC4 genes. Interestingly, miR-203 is predicted to target several genes, RAD51, HRCC4, and ATM, following the basic principle of miRNA action where no perfect complementarity between the miRNA sequence and 3′-UTR of a gene is required for inducing the translational repression, and, therefore, a single miRNA molecule can regulate expression of multiple protein coding genes. On the other hand, many individual coding genes have at their 3′-UTRs the predicted target sites for multiple miRNAs, indicating the likelihood of a combinatory action of several miRNAs on gene activity. In our study, 6 dysregulated miRNAs, miR-520a, miR-520e, miR-526b, miR-203, miR-186, and miR-96, are predicted to target the RAD51 gene.
This study also explored the possibility that miRNAs may directly participate in radiation-induced carcinogenesis by influencing the generation of cancer-specific chromosomal rearrangements such as RET/PTC. This genetic abnormality is a common feature of radiation-induced thyroid cancer, and evidence exists suggesting that it may be formed as a direct consequence of radiation-induced DNA breaks (36). Importantly, the presence of this chromosomal rearrangement in thyroid cancer cells correlates with strong upregulation of miR-187, miR-146b, and miR-155 (20). If upregulation of those miRNA was found in thyroid cells after radiation exposure, it would suggest that miRNAs play a direct role in the generation of carcinogenic chromosomal rearrangements in human cells. However, none of those miRNAs were found to be upregulated either after 4 or 24 hours after exposure. This indicates that miRNAs are unlikely to participate directly in the generation of carcinogenic chromosomal rearrangements after radiation exposure, but more likely to play a role in the cellular response to radiation by modulating the expression of DNA repair genes.
Acknowledgment
This work was supported by the National Institute of Health grant R01 CA88041.
Disclosure Statement
The authors declare no conflict of interest related to this study.
References
- 1.Parker LN. Belsky JL. Yamamoto T. Kawamoto S. Keehn RJ. Thyroid carcinoma after exposure to atomic radiation. A continuing survey of a fixed population, Hiroshima and Nagasaki, 1958–1971. Ann Intern Med. 1974;80:600–604. doi: 10.7326/0003-4819-80-5-600. [DOI] [PubMed] [Google Scholar]
- 2.Nagataki S. Shibata Y. Inoue S. Yokoyama N. Izumi M. Shimaoka K. Thyroid diseases among atomic bomb survivors in Nagasaki. JAMA. 1994;272:364–370. [PubMed] [Google Scholar]
- 3.Hamilton TE. van Belle G. LoGerfo JP. Thyroid neoplasia in Marshall Islanders exposed to nuclear fallout. JAMA. 1987;258:629–635. [PubMed] [Google Scholar]
- 4.Cronkite EP. Bond VP. Conard RA. Medical effects of exposure of human beings to fallout radiation from a thermonuclear explosion. Stem Cells. 1995;13(Suppl 1):49–57. [PubMed] [Google Scholar]
- 5.Ron E. Lubin JH. Shore RE. Mabuchi K. Modan B. Pottern LM. Schneider AB. Tucker MA. Boice JD., Jr. Thyroid cancer after exposure to external radiation: a pooled analysis of seven studies. Radiat Res. 1995;141:259–277. [PubMed] [Google Scholar]
- 6.Winship T. Rosvoll RV. Cancer of the thyroid in children. Proc Natl Cancer Conf. 1970;6:677–681. [PubMed] [Google Scholar]
- 7.Kazakov VS. Demidchik EP. Astakhova LN. Thyroid cancer after Chernobyl. Nature. 1992;359:21. doi: 10.1038/359021a0. [DOI] [PubMed] [Google Scholar]
- 8.Cardis E. Howe G. Ron E. Bebeshko V. Bogdanova T. Bouville A. Carr Z. Chumak V. Davis S. Demidchik Y. Drozdovitch V. Gentner N. Gudzenko N. Hatch M. Ivanov V. Jacob P. Kapitonova E. Kenigsberg Y. Kesminiene A. Kopecky KJ. Kryuchkov V. Loos A. Pinchera A. Reiners C. Repacholi M. Shibata Y. Shore RE. Thomas G. Tirmarche M. Yamashita S. Zvonova I. Cancer consequences of the Chernobyl accident: 20 years on. J Radiol Prot. 2006;26:127–140. doi: 10.1088/0952-4746/26/2/001. [DOI] [PubMed] [Google Scholar]
- 9.Goodhead DT. Initial events in the cellular effects of ionizing radiations: clustered damage in DNA. Int J Radiat Biol. 1994;65:7–17. doi: 10.1080/09553009414550021. [DOI] [PubMed] [Google Scholar]
- 10.Ward JF. Radiation mutagenesis: the initial DNA lesions responsible. Radiat Res. 1995;142:362–368. [PubMed] [Google Scholar]
- 11.Rabes HM. Demidchik EP. Sidorow JD. Lengfelder E. Beimfohr C. Hoelzel D. Klugbauer S. Pattern of radiation-induced RET and NTRK1 rearrangements in 191 post-chernobyl papillary thyroid carcinomas: biological, phenotypic, and clinical implications. Clin Cancer Res. 2000;6:1093–1103. [PubMed] [Google Scholar]
- 12.Nikiforov YE. Rowland JM. Bove KE. Monforte-Munoz H. Fagin JA. Distinct pattern of ret oncogene rearrangements in morphological variants of radiation-induced and sporadic thyroid papillary carcinomas in children. Cancer Res. 1997;57:1690–1694. [PubMed] [Google Scholar]
- 13.Hamatani K. Eguchi H. Ito R. Mukai M. Takahashi K. Taga M. Imai K. Cologne J. Soda M. Arihiro K. Fujihara M. Abe K. Hayashi T. Nakashima M. Sekine I. Yasui W. Hayashi Y. Nakachi K. RET/PTC rearrangements preferentially occurred in papillary thyroid cancer among atomic bomb survivors exposed to high radiation dose. Cancer Res. 2008;68:7176–7182. doi: 10.1158/0008-5472.CAN-08-0293. [DOI] [PubMed] [Google Scholar]
- 14.Chaudhry MA. Chodosh LA. McKenna WG. Muschel RJ. Gene expression profile of human cells irradiated in G1 and G2 phases of cell cycle. Cancer Lett. 2003;195:221–233. doi: 10.1016/s0304-3835(03)00154-x. [DOI] [PubMed] [Google Scholar]
- 15.Fachin AL. Mello SS. Sandrin-Garcia P. Junta CM. Ghilardi-Netto T. Donadi EA. Passos GA. Sakamoto-Hojo ET. Gene expression profiles in radiation workers occupationally exposed to ionizing radiation. J Radiat Res. 2009;50:61–71. doi: 10.1269/jrr.08034. [DOI] [PubMed] [Google Scholar]
- 16.Yin E. Nelson DO. Coleman MA. Peterson LE. Wyrobek AJ. Gene expression changes in mouse brain after exposure to low-dose ionizing radiation. Int J Radiat Biol. 2003;79:759–775. doi: 10.1080/09553000310001610961. [DOI] [PubMed] [Google Scholar]
- 17.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 18.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 19.He H. Jazdzewski K. Li W. Liyanarachchi S. Nagy R. Volinia S. Calin GA. Liu CG. Franssila K. Suster S. Kloos RT. Croce CM. de la Chapelle A. The role of microRNA genes in papillary thyroid carcinoma. Proc Natl Acad Sci U S A. 2005;102:19075–19080. doi: 10.1073/pnas.0509603102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nikiforova MN. Tseng GC. Steward D. Diorio D. Nikiforov YE. MicroRNA expression profiling of thyroid tumors: biological significance and diagnostic utility. J Clin Endocrinol Metab. 2008;93:1600–1608. doi: 10.1210/jc.2007-2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pallante P. Visone R. Ferracin M. Ferraro A. Berlingieri MT. Troncone G. Chiappetta G. Liu CG. Santoro M. Negrini M. Croce CM. Fusco A. MicroRNA deregulation in human thyroid papillary carcinomas. Endocr Ret Cancer. 2006;13:497–508. doi: 10.1677/erc.1.01209. [DOI] [PubMed] [Google Scholar]
- 22.He L. He X. Lim LP. de Stanchina E. Xuan Z. Liang Y. Xue W. Zender L. Magnus J. Ridzon D. Jackson AL. Linsley PS. Chen C. Lowe SW. Cleary MA. Hannon GJ. A microRNA component of the p53 tumour suppressor network. Nature. 2007;447:1130–1134. doi: 10.1038/nature05939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Weidhaas JB. Babar I. Nallur SM. Trang P. Roush S. Boehm M. Gillespie E. Slack FJ. MicroRNAs as potential agents to alter resistance to cytotoxic anticancer therapy. Cancer Res. 2007;67:11111–11116. doi: 10.1158/0008-5472.CAN-07-2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Josson S. Sung SY. Lao K. Chung LW. Johnstone PA. Radiation modulation of microRNA in prostate cancer cell lines. Prostate. 2008;68:1599–1606. doi: 10.1002/pros.20827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nikiforova MN. Stringer JR. Blough R. Medvedovic M. Fagin JA. Nikiforov YE. Proximity of chromosomal loci that participate in radiation-induced rearrangements in human cells. Science. 2000;290:138–141. doi: 10.1126/science.290.5489.138. [DOI] [PubMed] [Google Scholar]
- 26.Caudill CM. Zhu Z. Ciampi R. Stringer JR. Nikiforov YE. Dose-dependent generation of RET/PTC in human thyroid cells after in vitro exposure to gamma-radiation: a model of carcinogenic chromosomal rearrangement induced by ionizing radiation. J Clin Endocrinol Metab. 2005;90:2364–2369. doi: 10.1210/jc.2004-1811. [DOI] [PubMed] [Google Scholar]
- 27.Nikiforova MN. Caudill CM. Biddinger P. Nikiforov YE. Prevalence of RET/PTC rearrangements in Hashimoto's thyroiditis and papillary thyroid carcinomas. Int J Surg Pathol. 2002;10:15–22. doi: 10.1177/106689690201000104. [DOI] [PubMed] [Google Scholar]
- 28.Livak KJ. Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 29.Eisen MB. Spellman PT. Brown PO. Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci U S A. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lewis BP. Shih IH. Jones-Rhoades MW. Bartel DP. Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 31.Krek A. Grun D. Poy MN. Wolf R. Rosenberg L. Epstein EJ. MacMenamin P. da Piedade I. Gunsalus KC. Stoffel M. Rajewsky N. Combinatorial microRNA target predictions. Nat Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- 32.Chaudhry MA. Real-time PCR analysis of micro-RNA expression in ionizing radiation-treated cells. Cancer Biother Radiopharm. 2009;24:49–56. doi: 10.1089/cbr.2008.0513. [DOI] [PubMed] [Google Scholar]
- 33.Maes OC. An J. Sarojini H. Wu H. Wang E. Changes in MicroRNA expression patterns in human fibroblasts after low-LET radiation. J Cell Biol. 2008;105:824–834. doi: 10.1002/jcb.21878. [DOI] [PubMed] [Google Scholar]
- 34.Johnson SM. Grosshans H. Shingara J. Byrom M. Jarvis R. Cheng A. Labourier E. Reinert KL. Brown D. Slack FJ. RAS is regulated by the let-7 microRNA family. Cell. 2005;120:635–647. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 35.Ryan BM. Robles AI. Harris CC. Genetic variation in microRNA networks: the implications for cancer research. Nat Rev Cancer. 2010;10:389–402. doi: 10.1038/nrc2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gandhi M. Evdokimova V. Nikiforov YE. Mechanisms of chromosomal rearrangements in solid tumors: the model of papillary thyroid carcinoma. Mol Cell Endocrinol. 2010;321:36–43. doi: 10.1016/j.mce.2009.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]