Abstract
Transmission of HIV-1 drug resistance has important clinical and epidemiological consequences including earlier treatment failure and forward transmission of resistance strains in high-risk groups. To evaluate the prevalence and molecular epidemiology of transmitted drug resistance in Rhode Island, we collected genotypic, demographic, clinical, and laboratory data from treatment-naive individuals presenting to the largest outpatient HIV clinic in the state from January 2007 to November 2007. Sequences from 35 treatment-naive individuals were available, 83% of whom were men who had sex with men (MSM). All sequences were HIV-1 subtype B. Drug resistance mutations were identified in 7/35 [20%; 95% confidence interval (CI), 0.08–0.37] patients, six of whom had K103N. Two phylogenetic transmission clusters were found, involving 17% (6/35) of individuals, three in each cluster. We did not find an association between belonging to a cluster and age, gender, AIDS-defining illness, CD4 cell count, or viral load. Drug resistance mutations were more commonly observed in transmission clusters (p = 0.08). Individuals in one cluster all had K103N and were MSM who had attended local bathhouses. Individuals forming clusters were significantly more likely to have visited a bathhouse compared to nonclusters (p = 0.02). The prevalence of transmitted drug resistance in Rhode Island is high, further justifying genotypic testing on presentation to care and prior to treatment initiation. Molecular epidemiological analysis and association of resistance with phylogenetic networks using data obtained for clinical purposes may serve as useful tools for the prevention of drug resistance transmission and for contact tracing.
Primary drug resistance in human immunodeficiency virus group 1 (HIV-1), defined as resistance to one or more antiretroviral drugs in treatment-naive individuals, is an important public health concern.1,2 In areas in which highly active antiretroviral therapy (HAART) is widely available such as the United States, primary drug resistance is a result of viral transmission from treatment-experienced individuals and is, therefore, also referred to as transmitted drug resistance. The prevalence of transmitted drug resistance in the United States ranges from 8 to 26%.3–7 Mutations associated with nucleoside reverse transcriptase inhibitors (NRTIs) and non-nucleoside reverse transcriptase inhibitors (NNRTIs) are most commonly transmitted, notably K103N (lysine mutated to asparagine at RT position 103), which confers resistance against the entire class of NNRTIs.8
Drug resistance in HIV-infected individuals leads to fewer treatment options, decreased efficacy of treatment, and higher rates of morbidity and mortality.7,9 Furthermore, forward transmission of resistant virus by treatment-naive individuals may lead to transmission “clusters,” defined as groups of individuals that are infected with viruses that are phylogenetically close to each other and may therefore be epidemiologically linked. Transmission of HIV-1 often takes place early in the course of infection secondary to increased virus levels and contributes to a significant number of new cases.10–13 Transmission within clusters may be secondary to unknown or untreated infection, and is associated with high-risk behavior.10,14 Early identification of index cases can lead to reductions in forward transmission of HIV infection by interventions such as contact tracing, counseling, and education.
Phylogenetic analysis is a useful tool for studying the transmission dynamics of HIV-1. Phylogenetic clusters of viral sequences with similar ancestral lineages can be identified and often reflect shared virus transmission between epidemiologically linked individuals such as mother-to-child transmission, men who have sex with men (MSM),11,12 or intravenous drug users (IVDUs).15,16 HIV drug resistance testing by genotyping of the pol gene is now standard practice at initiation of HIV care in resource-rich settings to determine the presence of drug resistance mutations.17 Despite some disagreement,18,19 phylogenetic analyses of pol gene sequences have been shown to hold enough information to accurately determine epidemiological linkage20,21 and transmission networks.18,22 In addition to sexually transmitted diseases,23 transmission of HIV harboring drug resistance mutations has been reported to be significantly higher in transmission clusters, with mutations at NNRTI resistance-associated positions more commonly observed.13,14 NRTI and protease inhibitor (PI) mutations, associated with decreased viral fitness,14,24 are less frequently transmitted. Implementation of phylogenetic analyses in HIV clinical care is not standard practice and is rarely performed.
The Samuel and Esther Chester Immunology Center at The Miriam Hospital located in Providence, Rhode Island was founded in 1986 and is the largest HIV outpatient clinic in the state. It serves over 1400 patients from diverse backgrounds, including a significant number of MSM. Rhode Island has an active MSM community, as well as two bathhouses that serve as private clubs and meeting points for MSM throughout New England.25 Such bathhouses have been identified as venues in which high-risk sexual behavior occurs among MSM.26,27
During 2007, several newly diagnosed, HIV-infected, treatment-naive MSM who presented to the Immunology Center were found to have the K103N drug resistance mutation. These individuals were also noted to have attended bathhouses in the area. This prompted us to analyze available pol gene sequences from treatment-naive patients presenting to our clinic in 2007. We sought to examine transmission patterns and determine prevalence and associated factors of transmitted drug resistance.
The study population consisted of all individuals presenting for the first time to The Miriam Hospital Immunology Center in Providence, Rhode Island during 2007. Patients were referred from outpatient clinics, hospitals, the state prison system, and community-based programs that offer routine HIV screening. Patients were included in the study if they were older than 18 years of age, were treatment-naive based on self-report, and had HIV genotyping during their initial clinic visit. Drug resistance testing was performed using the Siemens TruGene HIV-1 assay (ViroMed Laboratories, Minnetonka, MN). Data were collected from paper and electronic charts and included age, gender, sexual preference, substance abuse, visitation habits to bathhouses, sexually transmitted diseases, number of sexual partners in the last year, CD4+ cell counts, plasma viral loads (PVL), and treatment histories. The study was approved by The Miriam Hospital Institutional Review Board.
Pol gene sequences were aligned using Clustal W.28 Sequences were manually edited to remove gaps and trimmed to identical sequence lengths using Bioedit.29 Phylogenetic analysis was performed using Phylip version 3.68.30 Phylogenetic trees were estimated with the neighbor-joining methodology using the F84 model of evolution with a transversion to transition ratio of 2:1. Trees were rooted with the Los Alamos Database (www.hiv.lanl.gov) HIV-1 subtype C reference sequence (accession number U46016). Robustness of the trees was determined using bootstrap analysis with 1000 replicates. Clustering was defined as high bootstrap values (>99%) and small genetic distances (<0.05). Subtyping was performed using the REGA HIV-1 Subtyping Tool Version 2.0.31 Drug resistance interpretation was performed using the Stanford HIV Sequence Database (http://hivdb.stanford.edu) and the International AIDS Society (IAS-USA) criteria.32 Statistical analyses included the Fisher's exact test for categorical variables and the Mann–Whitney nonparametric test for continuous variables. Significance was defined as p-values less than 0.05.
Thirty-five treatment-naive patients with available pol sequences presented to The Miriam Hospital Immunology outpatient HIV clinic for the first time between January 2007 and November 2007 (Table 1). All sequences were subtype B. Of the 35 patients, 86% (30/35) were male. Seventy-four percent (26/35) were MSM, which was the most prevalent risk factor for HIV acquisition. Two MSM had a one-time history of IVDU, but this was thought to be an unlikely route of HIV transmission. All five (14%) females had heterosexual sex as their only risk factor. The average person was 38 years old (range: 19–60). The average CD4+ cell count at HIV diagnosis was 335 cells/ml (range: 3–812). Thirteen (37%) individuals presented with a CD4+ cell count less than 200 cells/ml: seven (20%) between 200 and 350 cells/ml, seven (20%) between 350 and 500 cells/ml, and eight (23%) greater than 500 cells/ml. The average PVL at presentation was 159,930 copies/ml (range: 1914 to >500,000). Two (6%) individuals had less than 5000 copies/ml, one (3%) between 5000 and 10,000 copies/ml, 16 (46%) between 10,000 and 100,000 copies/ml, and 16 (46%) greater than 100,000 copies/ml. Fourteen out of 35 patients (40%) were recent (within the last year) seroconverters based on documentation of either a recent negative HIV test or clinical viral syndrome.
Table 1.
| Patient | Age (years) | Sex | Risk factor | Other history | Initial CD4 (cells/μl) | Initial PVL (copies/ml) | DRM |
|---|---|---|---|---|---|---|---|
| ID1 | 35 | M | MSM | From El Salvador | 3 | 125,855 | |
| ID2c | 23 | M | MSM | >250 partners, prostitution, group sex | 706 | 10,375 | |
| ID3 | 43 | M | MSM | Likely contracted in Las Vegas | 421 | 14,396 | D67N, K103N, K238T |
| ID4 | 53 | M | MSM | Anonymous partners, bathhouse | 363 | 87,321 | |
| ID5 | 37 | M | MSM | Non-IVDU | 16 | 140,847 | |
| ID6 | 55 | F | FSM | Unprotected sex | 48 | 163,944 | |
| ID7 | 36 | F | FSM | Likely sex with a bisexual | 333 | 36,923 | |
| ID8 | 44 | M | MSM | One incidence IVDU | 41 | >100,000 | |
| ID9 | 32 | F | FSM | From Guatemala | 93 | 384,255 | |
| ID10 | 51 | M | MSM | Male partner has HIV | 629 | 130,092 | |
| ID11 | 55 | F | FSM | Non-IVDU | 144 | 72,189 | K103N |
| ID12c | 23 | M | MSM | Bathhouse | 438 | 12,340 | |
| ID13c | 34 | M | MSM | 216 | 392,594 | ||
| ID14 | 22 | F | FSM | Non-IVDU | 572 | 148,718 | |
| ID15 | 20 | M | MSM | Multiple partners | 491 | 11,119 | |
| ID16c | 19 | M | MSM | Non-IVDU | 89 | 340,526 | |
| ID17c | 51 | M | MSM | Bathhouse | 252 | 80,935 | K103N |
| ID18 | 60 | M | MSM | Bisexual | 190 | >500,000 | |
| ID19 | 26 | M | MSM | Multiple partners | 317 | 62,746 | |
| ID20c | 34 | M | MSF | From Cape Verde | 480 | 51,098 | |
| ID21 | 23 | M | MSM | >500 partners, from Washington, DC | 283 | 1,914 | |
| ID22c | 22 | M | MSM | Born Dominican Republic, from NYC | 515 | 4,634 | |
| ID23c | 32 | M | MSM | ACI | 687 | >500,000 | |
| ID24c | 43 | M | MSM | HIV+partner | 344 | 479,729 | |
| ID25c | 24 | M | MSM | Diagnosed in WA; lived in FL, NY | 468 | 5,300 | |
| ID26 | 31 | M | MSM | >100 partners,bathhouse | 264 | 456,471 | |
| ID27 | 55 | M | MSM | Bathhouse | 142 | 105,198 | K103N |
| ID28c | 33 | M | MSM | From Miami, one incidence IVDU | 464 | 143,162 | |
| ID29c | 53 | M | MSM | Bisexual, from CT, bathhouse | 846 | 17,607 | |
| ID30 | 42 | M | MSM | >50 partners | 14 | 92,047 | |
| ID31c | 41 | M | MSF | Non-IVDU, ACI | 812 | 17,250 | M46I |
| ID32 | 38 | M | MSM | Bathhouse | 677 | 311,524 | K103N |
| ID33 | 45 | M | MSF | From Jamaica | 16 | 38,420 | K103N |
| ID34 | 48 | M | MSF | Portuguese; says only had sex with wife for 25 years, negative HIV 3–4 years prior | 19 | >500,000 | |
| ID35c | 53 | M | MSM | Bathhouse | 344 | 58,034 |
ACI, state prison correctional institute; ARS, acute retroviral syndrome; CVA, cerebral vascular accident; DRM, drug resistance mutation; F, female; FSM, female who has sex with males; IVDU, intravenous drug use; M, male; MSF, male who has sex with females; MSM, male who has sex with males; PCP, Pneumocystis jiroveci pneumonia; PVL, plasma viral load.
Individuals forming transmission clusters are in italics; cluster A from Fig. 1 is in bold.
Individuals who were recent (<12 months) seroconverters.
Seven individuals (20%; 95% CI, 0.08–0.37) had major mutations conferring resistance to antiretrovirals (Tables 1 and 2). The K103N mutation was the most common and was present in six subjects (17% of the patient sample, 86% of patients with resistance). Four of the six individuals (67%) with the K103N mutation were initially started on efavirenz-containing regimens. Of these, two were later switched to a PI-based regimen, one was lost to follow-up, and one was continued on efavirenz. Interestingly, the patient who continued on efavirenz did well clinically with an undetectable PVL and a high CD4 cell count for 2 years, despite harboring the K103N mutation.
Table 2.
Characteristics and Follow-up of Patients with Transmitted Drug Resistancea
| |
|
CD4 cell count (cells/ml) |
PVL (copies/ml) |
|
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Patient | Mutation(s) | Initial | 6-month | 1-year | 2-year | Initial | 6-month | 1-year | 2-year | Treatment history |
| ID3 | D67N, K103N, K238T | 421 | 361 | 608 | 707 | 14396 | 22453 | <75 | <75 | Started on EFV/TDF/FTC at 6 months, continues on this medication |
| ID11 | K103N | 144 | 253 | 240 | 464 | 72189 | <75 | <75 | <75 | Started on EFV/TDF/FTC initially, changed to FOS/ TDF/FTC 1 month later |
| ID17 | K103N | 252 | 440 | 434 | 448 | 80935 | <75 | <75 | <75 | Started on LPV/RTV/TDF/FTC because partners had K103N |
| ID27 | K103N | 142 | 246 | 180 | 336 | 105198 | <75 | <75 | <50 | Started on EFV/TDF/FTC initially, changed to LPV/RTV/TDF/FTC 1 week later |
| ID31 | M46I | 812 | Unavailable | 980 | 443 | 17250 | Unavailable | 546 | 14505 | Started on LPV/RTV/TDF/FTC through ACTG trial, stopped after 9 months |
| ID32 | K103N | 677 | Unavailable | 362 | Unavailable | 311524 | Unavailable | 119740 | Unavailable | No medications initially, lost to follow-up |
| ID33 | K103N | 16 | Unavailable | Unavailable | Unavailable | 38420 | Unavailable | Unavailable | Unavailable | Started on EFV/TDF/FTC initially, lost to follow-up |
ACTG, AIDS Clinical Trial Group; EFV, efavirenz; TDF, tenofovir; FTC, emtricitabine; FOS, fosamprenavir; LPV/RTV, lopinavir and ritonavir.
Other mutations included M46I (N = 1), D67N (N = 1), and K238T (N = 1), the latter two in the same patient. Of the seven patients with drug resistance, six (17%) had one-class resistance, five (14%) to NNRTIs and one (3%) to PIs. One individual (3%) had two-class resistance (D67N, K103N, and K238T), which was most likely acquired in Las Vegas. Twenty-six percent (9/35) of individuals likely acquired HIV-1 infection outside of Rhode Island, including at least five (14%) patients who were from a country with a higher incidence of HIV than the United States. Only the individual from Jamaica had a drug resistance mutation, K103N.
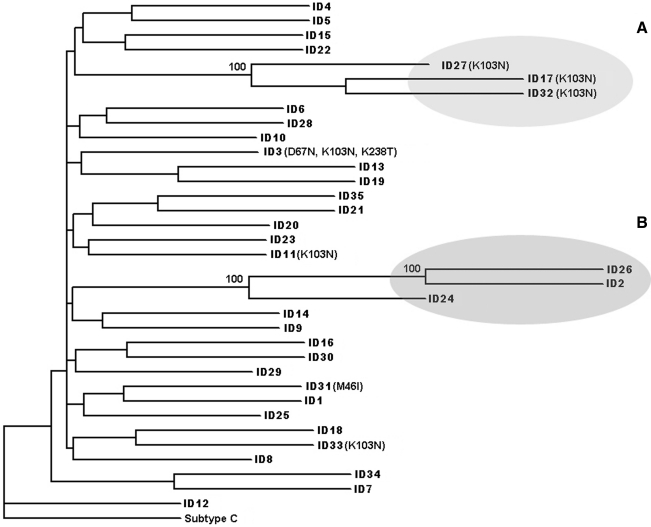
Phylogenetic analysis revealed two distinct clusters with small genetic distances and high bootstrap values. The clusters included a total of six individuals with three MSM in each (Fig. 1, Clusters A and B). Phylogenetic cluster topology remained significant after removal of positions associated with known drug resistance mutations. All sequences in cluster A, the original MSM cluster that prompted this study, had the K103N mutation. On further epidemiological investigation of this cluster, all individuals had known sexual contact with each other and reported bathhouse attendance, although the index case was unknown (personal communication with clinicians). Cluster B contained three MSM who were not known to be epidemiologically linked, but two of whom reported over 100 sexual partners and one who reported bathhouse attendance. Patients belonging to cluster B had no resistance mutations. We did not find an association between being in a cluster and age, gender, MSM status, AIDS-defining illness at initial presentation to the clinic (data not shown), acute infection, initial CD4+ cell count, or initial PVL. There was a trend toward harboring a drug resistance mutation and being in a cluster (p = 0.08). Drug resistance mutations were not more common in MSM compared to other individuals. None of the individuals who were thought to have acquired HIV outside of Rhode Island was included in transmission clusters.
FIG. 1.
Phylogenetic reconstruction of pol sequences from 35 treatment-naive individuals based on neighbor-joining methodology. Branch numbers represent bootstrapping analysis of 1000 replicates. Clusters A and B are shaded in gray and were defined by bootstrap values >99% and branch lengths <0.05.
A total of 23% (8/35) of patients reported visitation to a local bathhouse as a risk factor for HIV acquisition. Individuals forming clusters were significantly more likely to have attended a bathhouse (4/6, 67%) compared to nonclusters (4/25, 16%; p = 0.02). All three MSM who were part of cluster A containing the K103N mutation had attended a bathhouse.
Transmission of drug-resistant HIV among treatment-naive patients is common in areas in which HAART has been widely available.33 We describe a high rate (20%) of transmitted drug resistance in 2007 at the largest HIV clinic in Rhode Island, comparable to other high-prevalence areas of the United States3–7 and higher than the average rates observed in Europe and resource-poor countries.33 This high prevalence of transmitted drug resistance supports the current rationale of resistance testing at the initiation of HIV care and/or before HAART is initiated.
High rates of transmitted drug resistance are concerning and limit the number of drug classes available to treat HIV. Seventeen percent of our patient sample had one-class and 3% had two-class drug resistance. The most prevalent drug resistance-associated mutation was K103N, which confers resistance to the entire class of NNRTIs. Previous reports have also identified this mutation as being more easily transmitted than mutations conferring resistance to NRTIs or PIs, possibly due to the decreased viral fitness of the latter mutations.14,24 Other reasons could include earlier introduction or more widespread use of NNRTIs compared to PIs. Studies examining recent transmission of drug resistance are needed to investigate possible changing patterns of resistance according to current treatment guidelines and use of initial antiretroviral regimens.
Individuals with transmitted drug resistance did not appear to have worse clinical outcomes in our study, despite previous reports suggesting otherwise.7,9 Our group of individuals with transmitted drug resistance was small, but the majority were appropriately treated, either initially or based on genotypic testing. One patient harbored the NNRTI-associated mutations K103N and K238T as well as the NRTI-associated mutation D67N. This individual was adherent to a regimen containing efavirenz, tenofovir, and lamivudine, and demonstrated increasing trends of CD4+ cells as well as an undetectable PVL during the 2-year follow-up. The significance and etiology of complete and prolonged viral suppression on an efavirenz-based regimen, in the presence of K103N, remain to be determined.
Individuals visiting local bathhouses in Providence, Rhode Island were more likely to belong to a transmission cluster, as well as harbor virus with drug resistance, suggesting that resistant HIV may be transmitted by MSM through specific sexual networks. These observations remained the same after removal of positions associated with known drug resistance mutations, suggesting true epidemiological linkage rather than mutation-related clustering. There are two main bathhouses in Providence that are frequented by MSM for sexual encounters.25 The newer bathhouse is the largest in New England and caters to many out-of-state MSM. The relationship between HIV transmission and visitation to bathhouses would be difficult to prove and may represent local geographic transmission or otherwise be a marker for high-risk behavior. Previous reports have linked transmission of HIV between IVDUs to local geographic clinics,16 but not among the MSM population. To our knowledge, this is the first study to identify a specific site that may be related to transmission of drug-resistant HIV among MSM. Larger studies need to be done to substantiate this finding and study its implications and opportunities for HIV prevention.
We identified two transmission clusters by phylogenetic analysis of the pol gene, one of which was confirmed by epidemiological data. Previous studies have demonstrated the reliability of the pol gene in determining accurate phylogenetic linkages20,21 even though there is some disagreement.18,19 Our study, linking phylogenetic analysis with epidemiological data, provides further evidence that phylogenetic analysis of the pol gene can identify transmission networks of HIV infection.
The six individuals who were part of the two transmission clusters were all MSM and members of one cluster had virus containing the NNRTI-associated resistance mutation K103N. The transmission of drug resistance in this setting may reflect high-risk behavior among the MSM population. Current screening measures may not identify some of these individuals early enough to prevent further transmission of drug-resistant HIV. Despite widespread efforts of HIV education, counseling, and outreach programs, many patients reported extremely high-risk behavior, including unprotected sexual acts, anonymous encounters, and sexual partners that numbered in the hundreds. Improved public health strategies to prevent HIV transmission should target these specific populations as well as places where high-risk behavior occurs, specifically bathhouses or other sex club venues. Effective voluntary counseling and testing programs have already been implemented in bathhouse settings, including our own.25,34,35
During 2007, risk factors for HIV acquisition of patients actively enrolled at our clinic included MSM (N = 382/1144, 34%), IVDUs (N = 255/1144, 22%), and heterosexual behavior (N = 491/1144, 43%).36 In this study, a much higher proportion (74%) included MSM, which was the main risk factor for HIV acquisition, an apparent increasing trend that has also been reported among other HIV-infected cohorts.37,38 In Rhode Island, an aggressive campaign to make clean needles available to IVDUs has likely led to a decrease in both HIV and hepatitis C transmission via that route.39 Alternatively, increasing HIV rates among MSM may reflect changing attitudes and behaviors regarding HIV transmission and prevention.40
Limitations of this study include the small sample size, the availability of limited pol gene sequences, and the retrospective nature of the data collection. A high rate of potential acute seroconverters was found; however, it was difficult to accurately assess this measure due to recall bias of both clinical symptoms and previous HIV testing.
In summary, evaluation of genotypic sequence data revealed a high prevalence of transmitted drug resistance in our clinic, with epidemiologically confirmed, phylogenetic transmission clusters linked to drug resistance and bathhouse attendance in MSM. Such analytic tools, usually not implemented in clinical care, may provide important information to clinicians about HIV and drug resistance transmission patterns in the community. From a public health standpoint, such findings can and should lead to implementation of improved screening, education, and counseling strategies, specifically directed at engaging the high-risk MSM population in Rhode Island. Such targeted intervention programs may include refocusing efforts on condom use and be particularly beneficial at local bathhouses or other sex club venues.
Acknowledgments
Rami Kantor is funded by an NIH RO1 Grant AI66922. We thank Timothy Flanigan, MD, Kenneth Mayer, MD, and Joseph Hogan, PhD for their critical review of this manuscript.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Clavel F. Hance AJ. HIV drug resistance. N Engl J Med. 2004;350:1023–1035. doi: 10.1056/NEJMra025195. [DOI] [PubMed] [Google Scholar]
- 2.Deeks SG. Treatment of antiretroviral-drug-resistant HIV-1 infection. Lancet. 2003;362:2002–2011. doi: 10.1016/S0140-6736(03)15022-2. [DOI] [PubMed] [Google Scholar]
- 3.Boyd AC. Herzberg EM. Marshall MM, et al. Antiretroviral drug resistance among treatment-naive HIV-1-infected persons in Washington, D.C. AIDS Patient Care STDS. 2008;22:445–448. doi: 10.1089/apc.2007.0203. [DOI] [PubMed] [Google Scholar]
- 4.Eshleman SH. Husnik M. Hudelson S, et al. Antiretroviral drug resistance, HIV-1 tropism, and HIV-1 subtype among men who have sex with men with recent HIV-1 infection. AIDS. 2007;21:1165–1174. doi: 10.1097/QAD.0b013e32810fd72e. [DOI] [PubMed] [Google Scholar]
- 5.Grant RM. Hecht FM. Warmerdam M, et al. Time trends in primary HIV-1 drug resistance among recently infected persons. JAMA. 2002;288:181–188. doi: 10.1001/jama.288.2.181. [DOI] [PubMed] [Google Scholar]
- 6.Kim D. Wheeler W. Ziebell R, et al. Prevalence of transmitted antiretroviral drug resistance among newly-diagnosed HIV-1-infected persons, US, 2007; 17th Conference on Retroviruses and Opportunistic Infections; San Francisco. 2010. [Google Scholar]
- 7.Little SJ. Holte S. Routy JP, et al. Antiretroviral-drug resistance among patients recently infected with HIV. N Engl J Med. 2002;347:385–394. doi: 10.1056/NEJMoa013552. [DOI] [PubMed] [Google Scholar]
- 8.Soriano V. de Mendoza C. Genetic mechanisms of resistance to NRTI and NNRTI. HIV Clin Trials. 2002;3:237–248. doi: 10.1310/hct.2002.3.3.008. [DOI] [PubMed] [Google Scholar]
- 9.Deeks SG. Gange SJ. Kitahata MM, et al. Trends in multidrug treatment failure and subsequent mortality among antiretroviral therapy-experienced patients with HIV infection in North America. Clin Infect Dis. 2009;49:1582–1590. doi: 10.1086/644768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brenner BG. Roger M. Routy JP, et al. High rates of forward transmission events after acute/early HIV-1 infection. J Infect Dis. 2007;195:951–959. doi: 10.1086/512088. [DOI] [PubMed] [Google Scholar]
- 11.Lewis F. Hughes GJ. Rambaut A. Pozniak A. Leigh Brown AJ. Episodic sexual transmission of HIV revealed by molecular phylodynamics. PLoS Med. 2008;5:e50. doi: 10.1371/journal.pmed.0050050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pilcher CD. Tien HC. Eron JJ, Jr, et al. Brief but efficient: Acute HIV infection and the sexual transmission of HIV. J Infect Dis. 2004;189:1785–1792. doi: 10.1086/386333. [DOI] [PubMed] [Google Scholar]
- 13.Yerly S. Junier T. Gayet-Ageron A, et al. The impact of transmission clusters on primary drug resistance in newly diagnosed HIV-1 infection. AIDS. 2009;23:1415–1423. doi: 10.1097/QAD.0b013e32832d40ad. [DOI] [PubMed] [Google Scholar]
- 14.Brenner BG. Roger M. Moisi DD, et al. Transmission networks of drug resistance acquired in primary/early stage HIV infection. AIDS. 2008;22:2509–2515. doi: 10.1097/QAD.0b013e3283121c90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen JH. Wong KH. Li P, et al. Molecular epidemiological study of HIV-1 CRF01_AE transmission in Hong Kong. J Acquir Immune Defic Syndr. 2009;51:530–535. doi: 10.1097/QAI.0b013e3181aac516. [DOI] [PubMed] [Google Scholar]
- 16.Nguyen L. Hu DJ. Choopanya K, et al. Genetic analysis of incident HIV-1 strains among injection drug users in Bangkok: Evidence for multiple transmission clusters during a period of high incidence. J Acquir Immune Defic Syndr. 2002;30:248–256. doi: 10.1097/00042560-200206010-00014. [DOI] [PubMed] [Google Scholar]
- 17.Hirsch MS. Gunthard HF. Schapiro JM, et al. Antiretroviral drug resistance testing in adult HIV-1 infection: 2008 recommendations of an International AIDS Society–USA panel. Clin Infect Dis. 2008;47:266–285. doi: 10.1086/589297. [DOI] [PubMed] [Google Scholar]
- 18.Leitner T. Escanilla D. Franzen C. Uhlen M. Albert J. Accurate reconstruction of a known HIV-1 transmission history by phylogenetic tree analysis. Proc Natl Acad Sci USA. 1996;93:10864–10869. doi: 10.1073/pnas.93.20.10864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sturmer M. Preiser W. Gute P. Nisius G. Doerr HW. Phylogenetic analysis of HIV-1 transmission: pol gene sequences are insufficient to clarify true relationships between patient isolates. AIDS. 2004;18:2109–2113. doi: 10.1097/00002030-200411050-00002. [DOI] [PubMed] [Google Scholar]
- 20.Hue S. Clewley JP. Cane PA. Pillay D. HIV-1 pol gene variation is sufficient for reconstruction of transmissions in the era of antiretroviral therapy. AIDS. 2004;18:719–728. doi: 10.1097/00002030-200403260-00002. [DOI] [PubMed] [Google Scholar]
- 21.Kaye M. Chibo D. Birch C. Phylogenetic investigation of transmission pathways of drug-resistant HIV-1 utilizing pol sequences derived from resistance genotyping. J Acquir Immune Defic Syndr. 2008;49:9–16. doi: 10.1097/QAI.0b013e318180c8af. [DOI] [PubMed] [Google Scholar]
- 22.Bao L. Vidal N. Fang H, et al. Molecular tracing of sexual HIV type 1 transmission in the southwest border of China. AIDS Res Hum Retroviruses. 2008;24:733–742. doi: 10.1089/aid.2007.0269. [DOI] [PubMed] [Google Scholar]
- 23.Pao D. Fisher M. Hue S, et al. Transmission of HIV-1 during primary infection: Relationship to sexual risk and sexually transmitted infections. AIDS. 2005;19:85–90. doi: 10.1097/00002030-200501030-00010. [DOI] [PubMed] [Google Scholar]
- 24.Turner D. Brenner BG. Routy JP. Petrella M. Wainberg MA. Rationale for maintenance of the M184v resistance mutation in human immunodeficiency virus type 1 reverse transcriptase in treatment experienced patients. New Microbiol. 2004;27:31–39. [PubMed] [Google Scholar]
- 25.Mayer KH. Abbott D. Cavanaugh T. Case P. Sexual health services in a New England gay bathhouse: Opportunities for HIV/STD treatment, prevention; National HIV Prevention Conference; Atlanta, GA. 2009. [Google Scholar]
- 26.Binson D. Pollack LM. Blair J. Woods WJ. HIV transmission risk at a gay bathhouse. J Sex Res. 2009:1–9. doi: 10.1080/00224490903216755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Woods WJ. Binson D. Blair J. Han L. Spielberg F. Pollack LM. Probability sample estimates of bathhouse sexual risk behavior. J Acquir Immune Defic Syndr. 2007;45:231–238. doi: 10.1097/QAI.0b013e318055601e. [DOI] [PubMed] [Google Scholar]
- 28.Thompson JD. Gibson TJ. Plewniak F. Jeanmougin F. Higgins DG. The CLUSTAL_X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hall TA. Bioedit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- 30.Feldstein J. PHYLIP (Phylogeny Inference Package) version 3.69. Department of Genetics, University of Washington; Seattle: 2009. [Google Scholar]
- 31.de Oliveira T. Deforche K. Cassol S, et al. An automated genotyping system for analysis of HIV-1 and other microbial sequences. Bioinformatics. 2005;21:3797–3800. doi: 10.1093/bioinformatics/bti607. [DOI] [PubMed] [Google Scholar]
- 32.Johnson VA. Brun-Vezinet F. Clotet B, et al. Update of the drug resistance mutations in HIV-1: December 2009. Top HIV Med. 2009;17:138–145. [PubMed] [Google Scholar]
- 33.Chan PA. Kantor R. Transmitted drug resistance in nonsubtype B HIV-1 infection. HIV Ther. 2009;3:447–465. doi: 10.2217/hiv.09.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Daskalakis D. Silvera R. Bernstein K, et al. Implementation of HIV testing at 2 New York City bathhouses: From pilot to clinical service. Clin Infect Dis. 2009;48:1609–1616. doi: 10.1086/598979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huebner DM. Binson D. Woods WJ. Dilworth SE. Neilands TB. Grinstead O. Bathhouse-based voluntary counseling and testing is feasible and shows preliminary evidence of effectiveness. J Acquir Immune Defic Syndr. 2006;43:239–246. doi: 10.1097/01.qai.0000242464.50947.16. [DOI] [PubMed] [Google Scholar]
- 36.Gillani FS. Zaller ND. Zeller K, et al. Changes in demographics and risk factors among persons living with HIV in an academic medical center from 2003–2007. Med Health R I. 2009;92:237–240. [PMC free article] [PubMed] [Google Scholar]
- 37.Hall HI. Song R. Rhodes P, et al. Estimation of HIV incidence in the United States. JAMA. 2008;300:520–529. doi: 10.1001/jama.300.5.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jaffe HW. Valdiserri RO. De Cock KM. The reemerging HIV/AIDS epidemic in men who have sex with men. JAMA. 2007;298:2412–2414. doi: 10.1001/jama.298.20.2412. [DOI] [PubMed] [Google Scholar]
- 39.Rich JD. Hogan JW. Wolf F, et al. Lower syringe sharing and re-use after syringe legalization in Rhode Island. Drug Alcohol Depend. 2007;89:292–297. doi: 10.1016/j.drugalcdep.2007.02.016. [DOI] [PubMed] [Google Scholar]
- 40.Crepaz N. Marks G. Liau A, et al. Prevalence of unprotected anal intercourse among HIV-diagnosed MSM in the United States: A meta-analysis. AIDS. 2009;23:1617–1629. doi: 10.1097/QAD.0b013e32832effae. [DOI] [PubMed] [Google Scholar]