Abstract
A mandatory step in the formation of an infectious retroviral particle is the acquisition of its envelope glycoprotein (Env). This step invariably occurs by Env positioning itself in the host membrane at the location of viral budding and being incorporated along with the host membrane into the viral particle. In some ways, this step of the viral life cycle would appear to be imprecise. There is no specific sequence in Env or in the retroviral structural protein, Gag, that is inherently required for the production of an infectious Env-containing particle. Additionally, Env-defective proviruses can efficiently produce infectious particles with any of a number of foreign retroviral Env glycoproteins or even glycoproteins from unrelated viral families, a process termed pseudotyping. However, mounting evidence suggests that Env incorporation is neither passive nor random. Rather, several redundant mechanisms appear to contribute to the carefully controlled process of Env acquisition, many of which are apparently used by a wide variety of enveloped viruses. This review presents and discusses the evidence for these different mechanisms contributing to incorporation.
Introduction
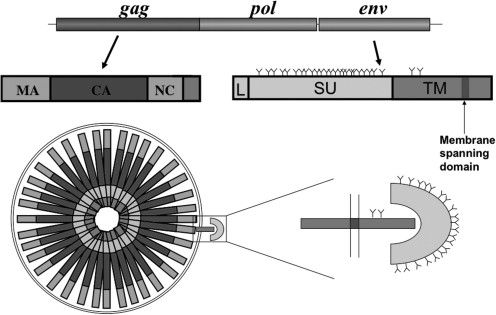
While all retroviral genomes encode the three major proteins Gag, Pol, and Env, some complex retroviruses encode additional proteins. For instance, HIV-1 also encodes the proteins, Vif, Vpr, Tat, Rev, Nef, and Vpu. For all retroviruses except the spumaretroviruses, Gag is the only viral protein required to assemble virus-like particles (VLPs) that bud through a host-derived membrane to exit the cell. Gag is expressed and assembled as a polyprotein, which is cleaved into multiple domains by a virus-derived protease during or shortly after budding (Fig. 1). The major domains of Gag are (from N- to C- terminus) matrix (MA), capsid (CA), and nucleocapsid (NC). MA is believed to be the primary membrane-targeting domain of Gag. Both N-terminal fatty acid modification of MA and patches of charged amino acids contribute to membrane binding.1 CA is the primary structural domain, which forms a curved lattice of tightly packed CA hexamers.2–4 NC is the nucleic acid binding domain of Gag and is responsible for binding and incorporating the viral RNA genome. Retroviral assembly essentially comes about through a combination of protein–protein, protein–membrane, and protein–nucleic acid interactions. Each of these interactions can be studied individually, but each can also be enhanced through cooperativity. For instance, CA hexamers can form de novo at high concentrations, but this multimerization is greatly enhanced by NC–nucleic acid interactions.5 In the cell, it is not entirely clear which interaction initiates the assembly process and in what order they proceed.
FIG. 1.
Schematic of HIV-1 genome and genes. Matrix (MA), capsid (CA), nucleocapsid (NC), leader peptide (L), surface protein (SU), transmembrane protein (TM).
Gag proteins are all produced in the cytoplasm of cells and bud through a cellular membrane to form the enveloped virus particles. For HIV-1, it is still debated where assembly is initiated, but most evidence suggests that the entire assembly process occurs on the plasma membrane.6,7 The most compelling evidence supporting this conclusion comes from imaging studies in living cells using fluorescently labeled Gag where the entire assembly process is viewed on the plasma membrane.8,9 It has often been suggested that the site of HIV-1 assembly is cell-type dependent. In macrophages, for instance, assembling VLPs have often been viewed in what appears to be internal endosomal vesicles.10,11 However, recent studies have demonstrated that these vesicles are, in fact, elaborate membrane invaginations that are continuous with the plasma membrane.12,13 In contrast to HIV-1, betaretroviruses such as Mason–Pfizer monkey virus (MPMV) assemble in the apparent absence of membrane near the microtubule organizing center before trafficking to and budding through the plasma membrane.14,15 With other retroviruses the location of viral assembly initiation has not been studied as extensively.
The question of how HIV-1 Gag specifically targets the plasma membrane, which represents a minority of all membranes in the cell, has received considerable attention in recent years. One proposed mechanism is that this targeting occurs through binding of HIV-1 MA to phosphatidylinositol-(4,5)-bisphosphate [PI(4,5)P2],16,17 a lipid found predominantly at the plasma membrane. Purified HIV-1 MA is reported to have a specific binding pocket for PI(4,5)P2.18 The functional significance of this pocket is suggested by the observation that destroying the PI(4,5)P2 in cells, by overexpression of a PI(4,5)P2-specific phosphatase, greatly reduces HIV-1 assembly and budding at the plasma membrane.16,17 Murine leukemia virus (MLV) and MPMV budding have also been reported to be altered by PI(4,5)P2 modulation.19,20 However, distinguishing primary from secondary effects of such experiments is challenging. In addition, all retroviral MA domains have clustered basic residues on one surface,1 and electrostatic attraction of MA to the acidic inner leaflet of the plasma membrane also plays a role in MA-membrane binding,21,22 as it does for numerous cellular proteins. The negative charge on the inner leaflet of the plasma membrane is contributed largely by phosphatidylserine and PI(4,5)P2. Thus, destroying the latter also affects electrostatics. For retroviral Gag-membrane targeting in general, it remains to be established what the relative contributions are of a possible specific PI(4,5)P2-binding pocket in MA (as found in HIV-1) and of simple electrostatic interactions.
The Env proteins of all retroviruses are type-1 membrane glycoproteins (Fig. 1). All have a signal peptide that is cleaved during translation in the endoplasmic reticulum (ER). Env glycoproteins are generally believed to form trimers in the ER. After exiting the ER, the glycoproteins are transported through the Golgi where they are variably glycosylated. The Env proteins are all cleaved by furin or a furin-like protease into surface (SU) and transmembrane (TM) subunits, which remain associated with each other. Most retroviral Env proteins traffic to the plasma membrane but are rapidly recycled such that at steady state they are found to be predominantly associated with intracellular membranes. To be incorporated into VLPs, the Env proteins must translocate to the membrane through which retroviral budding occurs and to the budding site. The specific mechanisms for how Env recognizes and traffics to these locations are poorly understood.
Passive Incorporation
Passive incorporation is the simplest explanation for how retroviruses acquire their surface glycoproteins. If Env is expressed at high levels on the membrane through which budding occurs, a degree of Env incorporation will occur through random diffusion to viral budding sites. The simplest evidence that supports this notion is the fact that viral pseudotyping (assembly with a foreign viral glycoprotein) occurs fairly readily.23 For example, HIV-1 can form infectious particles with Env glycoproteins from most if not all of the retroviral subfamilies including alpharetroviruses,24 betaretroviruses,25 gammaretroviruses,26 deltaretroviruses,27 spumaretroviruses,28 and other lentiviruses.29 HIV-1 can also be pseudotyped with glycoproteins from other viral families, such as rhabdoviruses,30 filoviruses,31 arenaviruses,32 togaviruses,33 coronaviruses,34 paramyxoviruses,35 flaviviruses,36 orthomyxoviruses,31 and even insect baculoviruses.37 Even more surprising, HIV-1 often produces a considerably higher titer of infectious particles with certain foreign glycoproteins such as VSV-G than it produces with its native Env glycoprotein. The fact that HIV-1 assembly can occur with a wide breadth of unrelated viral glycoproteins suggests that either all compatible viruses utilize the same mechanism to achieve glycoprotein incorporation or there is no specific mechanism required for Env incorporation.
Analyses of cellular proteins incorporated into VLPs also lend support to passive incorporation. Studies that have examined the composition of host cellular proteins incorporated into HIV-1 and MLV particles produced using an overexpression system found minimal exclusion of host proteins from VLPs.38,39 Later studies that sought to catalog the proteins incorporated into HIV-1 particles also found an abundance of host proteins incorporated into viral particles.40
Finally, passive incorporation is also supported by the finding that the MA domain of Gag is not strictly required for Env incorporation or infectivity. The MA domain of HIV-1 Gag can be replaced by nonviral membrane-binding protein domains without the virus losing its ability to form infectious particles with certain viral glycoproteins.41–43 Because MA is believed to be the targeting domain of Gag, and is the only domain likely to interact with a transmembrane glycoprotein either directly or indirectly, the observation that MA is not strictly required for glycoprotein incorporation severely restricts the notion of specific recruitment to viral budding sites.
Lessons from Exceptions
Generally, viral glycoproteins are enriched in VLPs and cellular proteins are not. Still, plenty of exceptions to this rule exist. An early noted exception is the cellular glycoprotein CD4. Although CD4 is not usually incorporated into retroviral particles, it is efficiently and selectively incorporated into Rous sarcoma virus (RSV) particles when expressed in quail cells.44 CD4, like most cellular surface proteins, interacts with networks of other host proteins in the cytoplasm. It has been hypothesized that these interactions can anchor host proteins to the cytoplasm, preventing their accumulation in VLPs.44 However, when introduced into a nonmammalian cell type, CD4 could behave more like a viral glycoprotein without binding partners that prevent incorporation. When considering mechanisms of glycoprotein incorporation, this observed exception illustrates that some mechanisms may modulate not only inclusion, but also exclusion from VLPs.
Another extensively studied exception is the exclusion of HIV-1 Env from most types of retroviruses. Unlike most retroviral Env proteins, which have short 30–50 amino acid cytoplasmic tails, lentiviral Env proteins, such as HIV-1 Env, have long cytoplasmic tails of 150 amino acids or more. The lentiviral Env proteins are generally able to form infectious particles with only their native Gag or Gag from closely related viruses. HIV-1 Env is also prevented from being incorporated into HIV-1 particles that lack their native MA domain41,42 and even into viruses with certain discrete point mutations in their MA domain.45–47 Interestingly, modest truncations from the C-terminus of HIV-1 Env generally reduce Env incorporation and viral infectivity.48–51 However, large truncations removing most or all of the HIV-1 Env cytoplasmic tail result in a protein that can form infectious particles in certain situations with wild-type HIV-1 Gag,52–54 HIV-1 Gag mutants that were incompatible with the full length HIV-1 Env,42,45,55 and even Gag from other retroviral families.56,57 Similarly, SIVagm Env and RSV Env are also rendered more compatible with foreign retroviruses if their cytoplasmic tails are truncated.24,58 It has been proposed that Env cytoplasmic tails can cause steric hindrance, which prevents incorporation into VLPs, unless the Env cytoplasmic tails and MA are matched.56,57 These examples as well as the example with human CD4 suggest that all glycoproteins on the plasma membrane can be incorporated into retroviral VLPs (perhaps passively) unless restricted through some sort of dominant interference.
Two additional examples of Gag/Env incompatibility come from the Env proteins of gammaretroviruses RD114 and Gibbon ape leukemia virus (GaLV). Simian immunodeficiency virus (SIV) and HIV-1 have been reported to be incompatible with RD114 and GaLV Env, respectively.32,59,60 These combinations stand out because most gammaretroviral Env proteins are compatible with lentiviruses. Taking advantage of the similarity between these proteins and the lentivirus-compatible Env from MLV, chimeras were engineered to pinpoint the sequences responsible for the incompatibility. In both cases, the incompatibility was mapped to the C-terminal 35 amino acid cytoplasmic tail of Env.32,59 With RD114 Env, the cytoplasmic tail appears to alter the trafficking of Env, making it inaccessible to SIV.59,61,62 With GaLV Env, the glycoprotein cytoplasmic tail is modulated by sensitivity to the HIV-1 accessory protein Vpu.63,64 Deletion of Vpu abolishes the incompatibility between these proteins. Both cases support the generalization that all viral glycoproteins are compatible with all enveloped viruses, as long as they traffic to the same cellular membrane, unless something prevents their incorporation into VLPs.
Evidence for Specificity
Despite data suggesting that glycoprotein acquisition is a passive phenomenon, there are several notable counterexamples. For example, when polarized monolayers of Madin–Darby canine kidney (MDCK) cells are infected with HIV-1, viral particles are released predominantly from the basolateral surface, and this polarized viral release is dependent on Env.65–68 In the absence of Env, Gag is released equally from apical and basolateral surfaces. Amazingly, the polarized budding of HIV-1 could also be imparted by MLV Env and human T cell leukemia virus 1 (HTLV-1) Env.65 In all cases, the basolateral targeting capability of Env is dictated by the cytoplasmic tail. These findings are based on studies performed in canine epithelial cells with extremely low levels of viral output that was measured primarily using a sensitive ELISA assay for p24. Notably, similar studies using alternative polarized models have not found the same basolateral release of infectious viral particles.69,70 It also has been demonstrated that HIV-1 Env cannot direct the budding of Gag to any membrane in the cell, as retention of HIV-1 Env in the ER does not redirect budding away from the plasma membrane.71
Two analogous examples show that the cellular distribution of Gag can be modulated by Env. The first example comes from work with rat neuronal cells where HIV-1 Env and MLV Env expression cause the relocalization of HIV-1 Gag or MLV Gag, respectively, from the axons to the somatic region of the cell.72 Unlike the example in polarized MDCK cells, this redistribution occurs only among homologous pairs; MLV Env does not redistribute HIV Gag and HIV Env does not redistribute MLV Gag. More recently, a similar phenomenon has been observed where MLV assembly is directed toward sites of cell–cell contact.73 While MLV Env is independently targeted to sites of cell–cell contact, MLV Gag targeting is Env dependent. This targeted assembly was further shown to be dependent on the cytoplasmic tail of MLV Env (Fig. 2A and B).
FIG. 2.
(A, B) Merged time lapse video imaging the localization of murine leukemia virus (MLV) Gag (red) relative to locations of cell–cell contact (green). Cells expressed (A) wild-type MLV Env or (B) MLV Env Δ16 C-terminal amino acids (image adapted from Jin et al.73). (C, E, G) Secondary electron images illustrating the topology of the cell surface. (D, F, H) Backscatter images illustrating the distribution of the MLV Env on the cell surface. (E–H) Express late domain defective HIV-1 Gag; (C–F) express wild-type MLV Env; (G, H) express MLV Env Δ33 C-terminal amino acids. (C–H) Scale bars 200 nm.
A final line of evidence for specificity of Env incorporation comes from imaging studies that visualize the distribution of glycoproteins on the surface of cells by scanning electron microscopy. These studies illustrate that viral glycoproteins appear relatively randomly distributed on the surface of cells when expressed alone, but are robustly recruited to viral budding sites of compatible structural proteins (Fig. 2C–F).63,74,75 For instance, when MLV Env is expressed along with HIV-1 Gag, the surface MLV Env is tightly clustered at HIV-1 budding sites and is essentially absent from the rest of the plasma membrane. Surprisingly, this enrichment of MLV Env to HIV-1 budding sites is not dependent on the Env cytoplasmic tail (Fig. 2G and H).75
Mechanisms of Specificity
From the experiments described above, at least in some circumstances Env acquisition by retroviruses is an active and specific process, but this information does not identify the mechanism used for incorporation. This section reviews the evidence for three types of interaction that could contribute to specificity: direct interaction among viral proteins, indirect interaction and/or convergent trafficking, and lipid microdomains as assembly platforms.
Direct interaction
At least among lentiviruses, several lines of evidence suggest a direct interaction between the cytoplasmic tail of Env and the MA domain of Gag. First, direct in vitro binding between MA and the C-terminal domain (CTD) of Env has been demonstrated for both HIV-1 and SIV.76,77 Second, a number of mutations have been characterized in both MA and Env that prevent incorporation of HIV-1 Env into HIV-1 particles (discussed in “Lessons from Exceptions”). Importantly, in one example a deletion in HIV-1 Env blocks incorporation into particles, but compensatory mutations in the MA domain of HIV-1 Gag restore incorporation.78 Third, as with many viral glycoproteins, HIV-1 Env is prevented from becoming fully fusogenic prior to viral maturation.79 With betaretroviruses and gammaretroviruses, fusogenic activity is enabled by cleavage of the C-terminal R-peptide by the viral protease.80,81 HIV-1 Env is not cleaved by protease; yet, the fusogenicity of Env is dictated by the protease processing of HIV-1 Gag,79 suggesting that a direct interaction between Gag and Env in the immature particle maintains Env in a nonfusogenic form. In summary, for retroviruses there is little doubt that at least in some instances, direct interactions occur between Gag proteins and their native Env glycoprotein. This conclusion is consistent with knowledge about other enveloped viruses, such as alphaviruses, for which direct interactions between structural and transmembrane proteins are required for assembly.82 However, it is unlikely that these interactions alone promote incorporation into retroviruses, and it is unlikely that direct interactions play any role among heterologous Gag–Env combinations.
Protein intermediates/convergent trafficking
Independent of direct interactions, both Gag and Env interact with cellular trafficking machinery that has the potential to deliver both proteins to the same microenvironment for assembly, or even to serve as a bridge between the two types of proteins. The most extensively studied interaction motif, which is found in many if not most Env proteins, is the membrane proximal YxxL motif.83 This sequence interacts with the cellular trafficking adapter protein complex 2 (AP-2).84–86 Mutation of this motif in HIV-1 Env blocks AP-2 binding,85,86 reduces the rate of Env endocytosis,87 eliminates polarized budding in MDCK cells,67 and reduces viral replication in certain situations.88 Most lentiviral Env proteins have a conserved YxxL near their transmembrane domain. Some nonlentiviruses have this conserved feature as well, but not necessarily in the same place. For instance, all gammaretroviral Env proteins also have a YxxL motif, but further from the membrane-spanning domain than in lentiviral Envs. Nonetheless, mutation of this motif in MLV Env can alter protein distribution in the cell89 and abolish polarized budding of HIV-1 Gag in MDCK cells.65 At the very C-terminus of the HIV Env CTD is a conserved dileucine motif that binds to the adapter protein complex 1 (AP-1) and may contribute to Env trafficking.90,91 Mutation of this motif eliminates AP-1 binding and alters the intracellular distribution of Env. Because HIV-1 Gag has also been reported to interact with various AP complexes,92,93 it is conceivable that this type of complex serves as a transient intermediate between Gag and Env.
A peculiar trafficking phenotype is observed with the gammaretrovirus Env RD114. As mentioned above, the cytoplasmic tail of RD114 causes Env proteins to become incompatible with the lentivirus SIV but compatible with the gammaretrovirus MLV.59 This loss of compatibility correlates with a loss of Env association with late endosomes. MLV Env partially associates with late endosomes and is associated with MLV and SIV Gags at these compartments. In contrast, RD114 Env by itself does not associate with these compartments but is relocalized to them in the presence of MLV Gag, but not SIV Gag. Both Env proteins display a mixture of furin processed and unprocessed proteins on the cell surface, but only fully processed Env is incorporated into VLPs. These experiments have been interpreted to indicate that retroviral assembly occurs, or at least commences, at these late endosome compartments, and that the incompatibility with SIV occurs because Env does not reach this location. Further dissection of the RD114 cytoplasmic tail revealed that a cluster of acidic amino acids at the C-terminus is responsible both for the loss of association with late endosomes and the loss of infectivity.62 The cellular protein PACS-1 is implicated in binding to this acidic cluster in RD114 and causing retrograde transport to the trans-Golgi network. Knockdown of PACS-1 partially relieves the RD114 Env incompatibility with SIV Gag. However, since viral particles can appear to be associated with late endosomes as a result of being endocytosed by the cell after budding,43,94 these data do not necessarily prove that assembly occurs at late endosomes. It is nonetheless intriguing that alterations in Env trafficking can affect incorporation and infectivity in a virus-specific manner.
It is almost universally agreed that furin cleavage of Env, to create the mature SU and TM proteins, is required for fusogenic activity and viral infectivity.75,95–98 At least in some cases, furin cleavage is also required for incorporation into viral particles.99,100 Still, whether lack of incorporation is due to the protein being unable to reach the cell surface is under dispute. Several studies have observed unprocessed Env expressed on the cell surface but only processed Env in viral particles.100,101 In fact, a point mutation in MLV Env that disrupts furin cleavage results in a protein that is abundant and clustered on the cell surface, but is no longer enriched at HIV-1 budding sites.102 This finding is particularly telling since the cytoplasmic tail of this protein is identical to that of wild-type MLV Env. Thus, the incorporation of Env proteins into viral particles appears to involve multiple interactions, and cannot simply be the result of random incorporation or direct interaction.
Other cellular proteins that have been reported to interact with the HIV-1 Env cytoplasmic tail include tail interacting protein 47 (TIP47),103 the prenylated Rab acceptor,104 and p115-RhoGEF.105 TIP47, in particular, has been proposed to be a physical connector between the cytoplasmic domain of HIV-1 Env and the MA domain of HIV-1 Gag. Depletion of this cellular protein by RNAi reduces HIV-1 Env incorporation into HIV-1 particles and viral infectivity but does not affect the infectivity of HIV-1 particles pseudotyped with VSV-G.103
Membrane microdomains
Many lines of evidence suggest that retroviruses bud from membrane microdomains with properties consistent with lipid rafts.6,7,106,107 Because many viral glycoproteins also associate with these domains, it has been proposed that lipid raft association is one factor that leads to selective Env incorporation and pseudotyping.108–112 However, HIV-1 Gag and Env have also been shown to associate with tetraspanin-enriched microdomains (TEMs),113–115 which are distinct from membrane rafts.116 Interestingly, the hemagglutinin (HA) protein of influenza virus, which strongly associates with membrane rafts, does not colocalize with TEMs.113,117 When HIV-1 and influenza are produced in the same cell, the two viruses appear at distinct locations on the surface, but only the HIV-1 particles are found to colocalize with TEM markers.117 An analysis of the dynamics of both TEM and raft markers relative to viral budding sites revealed that both raft and TEM markers associate with HIV-1 budding sites, and the mobility of TEM (and not raft) markers is altered by HIV-1 Gag.115 As TEM and raft markers generally do not cosegregate in cells and as the two have distinct properties, these data may suggest that retroviral assembly leads to the creation of a lipid microdomain distinct from any preexisting microdomain and is favored by many viral glycoproteins.
An interesting finding that further confuses the notion of assembly at microdomains comes from experiments in which cells express HIV-1 Gag along with two different types of viral glycoproteins, HIV-1 Env and Ebola glycoprotein. Infectious HIV-1 particles are produced with both types of glycoprotein, but the assembly is quantal with each particle containing either one type of glycoprotein or the other, but not both.118 These data are interpreted as evidence that HIV-1 assembly occurs quantally on a single lipid raft that contains a single type of viral glycoprotein. However, these data lack an explanation for how the different kinds of glycoproteins segregate themselves into distinct microdomains prior to assembly.
Conclusions
In summary, the following generalizations can be made about the factors that modulate viral glycoprotein incorporation into retroviral particles. First, incorporation requires that Env traffics to the membrane of the same subcellular compartment through which budding occurs. Viral glycoproteins are entirely dependent on cellular machinery for transportation within the cell, which means that either Env has to be delivered to the correct membrane where assembly occurs or Gag has to be recruited to assemble at the membrane where Env resides. Both scenarios probably occur under certain circumstances. Alterations in the virus or the cell that change the localization of either of these proteins have the potential to abolish incorporation. Second, on reaching the appropriate membrane, a combination of factors causes viral glycoproteins to be recruited to or excluded from the budding virus. Recruitment to retroviral particles could occur from direct interaction among viral proteins or from indirect interactions through cellular proteins, lipid microdomains, or other factors that are yet to be elucidated. Exclusion could occur through steric hindrance or through interactions that prevent incorporation into particles. The final extent of Env inclusion into a retrovirus can be thought of as a summation of all of these forces. The resulting complexity of the process confounds experiments to test the contribution of any one force. With a clearer understanding of the various forces that contribute to this elegant process, the mechanism used by each type of virus will become easier to discern.
Acknowledgments
I thank M.M. Kroll, V.M. Vogt, T.M. Lucas, S.L. Liu, D. Gregory, and S. Janaka for helpful comments. SEM studies were performed at the University of Missouri Electron Microscopy Core facility. This research was supported by U.S. Public Health Service Grant AI73098.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Murray PS. Li Z. Wang J. Tang CL. Honig B. Murray D. Retroviral matrix domains share electrostatic homology: Models for membrane binding function throughout the viral life cycle. Structure. 2005;13(10):1521–1531. doi: 10.1016/j.str.2005.07.010. [DOI] [PubMed] [Google Scholar]
- 2.Pornillos O. Ganser-Pornillos BK. Kelly BN, et al. X-ray structures of the hexameric building block of the HIV capsid. Cell. 2009;137(7):1282–1292. doi: 10.1016/j.cell.2009.04.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mortuza GB. Haire LF. Stevens A. Smerdon SJ. Stoye JP. Taylor IA. High-resolution structure of a retroviral capsid hexameric amino-terminal domain. Nature. 2004;431(7007):481–485. doi: 10.1038/nature02915. [DOI] [PubMed] [Google Scholar]
- 4.de Marco A. Davey NE. Ulbrich P, et al. Conserved and variable features of Gag structure and arrangement in immature retrovirus particles. J Virol. 84(22):11729–11736. doi: 10.1128/JVI.01423-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ganser BK. Li S. Klishko VY. Finch JT. Sundquist WI. Assembly and analysis of conical models for the HIV-1 core. Science. 1999;283(5398):80–83. doi: 10.1126/science.283.5398.80. [DOI] [PubMed] [Google Scholar]
- 6.Ono A. HIV-1 Assembly at the plasma membrane: Gag trafficking and localization. Future Virol. 2009;4(3):241–257. doi: 10.2217/fvl.09.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ono A. Relationships between plasma membrane microdomains and HIV-1 assembly. Biol Cell. 2010;102(6):335–350. doi: 10.1042/BC20090165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jouvenet N. Bieniasz PD. Simon SM. Imaging the biogenesis of individual HIV-1 virions in live cells. Nature. 2008;454(7201):236–240. doi: 10.1038/nature06998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ivanchenko S. Godinez WJ. Lampe M, et al. Dynamics of HIV-1 assembly and release. PLoS Pathog. 2009;5(11):e1000652. doi: 10.1371/journal.ppat.1000652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pelchen-Matthews A. Marsh M. Electron microscopy analysis of viral morphogenesis. Methods Cell Biol. 2007;79:515–542. doi: 10.1016/S0091-679X(06)79020-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deneka M. Pelchen-Matthews A. Byland R. Ruiz-Mateos E. Marsh M. In macrophages, HIV-1 assembles into an intracellular plasma membrane domain containing the tetraspanins CD81, CD9, and CD53. J Cell Biol. 2007;177(2):329–341. doi: 10.1083/jcb.200609050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bennett AE. Narayan K. Shi D, et al. Ion-abrasion scanning electron microscopy reveals surface-connected tubular conduits in HIV-infected macrophages. PLoS Pathog. 2009;5(9):e1000591. doi: 10.1371/journal.ppat.1000591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Welsch S. Keppler OT. Habermann A. Allespach I. Krijnse-Locker J. Krausslich HG. HIV-1 buds predominantly at the plasma membrane of primary human macrophages. PLoS Pathog. 2007;3(3):e36. doi: 10.1371/journal.ppat.0030036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sfakianos JN. LaCasse RA. Hunter E. The M-PMV cytoplasmic targeting-retention signal directs nascent Gag polypeptides to a pericentriolar region of the cell. Traffic. 2003;4(10):660–670. doi: 10.1034/j.1600-0854.2003.00125.x. [DOI] [PubMed] [Google Scholar]
- 15.Sfakianos JN. Hunter E. M-PMV capsid transport is mediated by Env/Gag interactions at the pericentriolar recycling endosome. Traffic. 2003;4(10):671–680. doi: 10.1034/j.1600-0854.2003.00126.x. [DOI] [PubMed] [Google Scholar]
- 16.Ono A. Ablan SD. Lockett SJ. Nagashima K. Freed EO. Phosphatidylinositol(4,5)bisphosphate regulates HIV-1 Gag targeting to the plasma membrane. Proc Natl Acad Sci USA. 2004;101(41):14889–14894. doi: 10.1073/pnas.0405596101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chukkapalli V. Hogue IB. Boyko V. Hu WS. Ono A. Interaction between the human immunodeficiency virus type 1 Gag matrix domain and phosphatidylinositol-(4,5)-bisphosphate is essential for efficient gag membrane binding. J Virol. 2008;82(5):2405–2417. doi: 10.1128/JVI.01614-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Saad JS. Miller J. Tai J. Kim A. Ghanam RH. Summers MF. Structural basis for targeting HIV-1 Gag proteins to the plasma membrane for virus assembly [see comment] Proc Natl Acad Sci USA. 2006;103(30):11364–11369. doi: 10.1073/pnas.0602818103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stansell E. Apkarian R. Haubova S. Diehl WE. Tytler EM. Hunter E. Basic residues in the Mason-Pfizer monkey virus gag matrix domain regulate intracellular trafficking and capsid-membrane interactions. J Virol. 2007;81(17):8977–8988. doi: 10.1128/JVI.00657-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chan R. Uchil PD. Jin J, et al. Retroviruses human immunodeficiency virus and murine leukemia virus are enriched in phosphoinositides. J Virol. 2008;82(22):11228–11238. doi: 10.1128/JVI.00981-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dalton AK. Ako-Adjei D. Murray PS. Murray D. Vogt VM. Electrostatic interactions drive membrane association of the human immunodeficiency virus type 1 Gag MA domain. J Virol. 2007;81(12):6434–6445. doi: 10.1128/JVI.02757-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dalton AK. Murray PS. Murray D. Vogt VM. Biochemical characterization of rous sarcoma virus MA protein interaction with membranes. J Virol. 2005;79(10):6227–6238. doi: 10.1128/JVI.79.10.6227-6238.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zavada J. The pseudotypic paradox. J Gen Virol. 1982;63(Pt 1):15–24. doi: 10.1099/0022-1317-63-1-15. [DOI] [PubMed] [Google Scholar]
- 24.Lewis BC. Chinnasamy N. Morgan RA. Varmus HE. Development of an avian leukosis-sarcoma virus subgroup A pseudotyped lentiviral vector. J Virol. 2001;75(19):9339–9344. doi: 10.1128/JVI.75.19.9339-9344.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu SL. Halbert CL. Miller AD. Jaagsiekte sheep retrovirus envelope efficiently pseudotypes human immunodeficiency virus type 1-based lentiviral vectors. J Virol. 2004;78(5):2642–2647. doi: 10.1128/JVI.78.5.2642-2647.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reiser J. Harmison G. Kluepfel-Stahl S. Brady RO. Karlsson S. Schubert M. Transduction of nondividing cells using pseudotyped defective high-titer HIV type 1 particles. Proc Natl Acad Sci USA. 1996;93(26):15266–15271. doi: 10.1073/pnas.93.26.15266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Landau NR. Page KA. Littman DR. Pseudotyping with human T-cell leukemia virus type I broadens the human immunodeficiency virus host range. J Virol. 1991;65(1):162–169. doi: 10.1128/jvi.65.1.162-169.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mochizuki H. Schwartz JP. Tanaka K. Brady RO. Reiser J. High-titer human immunodeficiency virus type 1-based vector systems for gene delivery into nondividing cells. J Virol. 1998;72(11):8873–8883. doi: 10.1128/jvi.72.11.8873-8883.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zeilfelder U. Bosch V. Properties of wild-type, C-terminally truncated, and chimeric maedi-visna virus glycoprotein and putative pseudotyping of retroviral vector particles. J Virol. 2001;75(1):548–555. doi: 10.1128/JVI.75.1.548-555.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Naldini L. Blomer U. Gallay P, et al. In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector [see comment] Science. 1996;272(5259):263–267. doi: 10.1126/science.272.5259.263. [DOI] [PubMed] [Google Scholar]
- 31.Kobinger GP. Weiner DJ. Yu QC. Wilson JM. Filovirus-pseudotyped lentiviral vector can efficiently and stably transduce airway epithelia in vivo. Nat Biotechnol. 2001;19(3):225–230. doi: 10.1038/85664. [DOI] [PubMed] [Google Scholar]
- 32.Christodoulopoulos I. Cannon PM. Sequences in the cytoplasmic tail of the gibbon ape leukemia virus envelope protein that prevent its incorporation into lentivirus vectors. J Virol. 2001;75(9):4129–4138. doi: 10.1128/JVI.75.9.4129-4138.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Morizono K. Bristol G. Xie YM. Kung SK. Chen IS. Antibody-directed targeting of retroviral vectors via cell surface antigens. J Virol. 2001;75(17):8016–8020. doi: 10.1128/JVI.75.17.8016-8020.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hofmann H. Hattermann K. Marzi A, et al. S protein of severe acute respiratory syndrome-associated coronavirus mediates entry into hepatoma cell lines and is targeted by neutralizing antibodies in infected patients. J Virol. 2004;78(12):6134–6142. doi: 10.1128/JVI.78.12.6134-6142.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kobinger GP. Deng S. Louboutin JP, et al. Transduction of human islets with pseudotyped lentiviral vectors. Hum Gene Ther. 2004;15(2):211–219. doi: 10.1089/104303404772680010. [DOI] [PubMed] [Google Scholar]
- 36.Bartosch B. Dubuisson J. Cosset FL. Infectious hepatitis C virus pseudo-particles containing functional E1-E2 envelope protein complexes. J Exp Med. 2003;197(5):633–642. doi: 10.1084/jem.20021756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kumar M. Bradow BP. Zimmerberg J. Large-scale production of pseudotyped lentiviral vectors using baculovirus GP64. Hum Gene Ther. 2003;14(1):67–77. doi: 10.1089/10430340360464723. [DOI] [PubMed] [Google Scholar]
- 38.Hammarstedt M. Wallengren K. Pedersen KW. Roos N. Garoff H. Minimal exclusion of plasma membrane proteins during retroviral envelope formation. Proc Natl Acad Sci USA. 2000;97(13):7527–7532. doi: 10.1073/pnas.120051597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hammarstedt M. Garoff H. Passive and active inclusion of host proteins in human immunodeficiency virus type 1 gag particles during budding at the plasma membrane. J Virol. 2004;78(11):5686–5697. doi: 10.1128/JVI.78.11.5686-5697.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chertova E. Chertov O. Coren LV, et al. Proteomic and biochemical analysis of purified human immunodeficiency virus type 1 produced from infected monocyte-derived macrophages. J Virol. 2006;80(18):9039–9052. doi: 10.1128/JVI.01013-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang CT. Zhang Y. McDermott J. Barklis E. Conditional infectivity of a human immunodeficiency virus matrix domain deletion mutant. J Virol. 1993;67(12):7067–7076. doi: 10.1128/jvi.67.12.7067-7076.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reil H. Bukovsky AA. Gelderblom HR. Gottlinger HG. Efficient HIV-1 replication can occur in the absence of the viral matrix protein. EMBO J. 1998;17(9):2699–2708. doi: 10.1093/emboj/17.9.2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jouvenet N. Neil SJD. Bess C, et al. Plasma membrane is the site of productive HIV-1 particle assembly. PLoS Biol. 2006;4(12):2296–2310. doi: 10.1371/journal.pbio.0040435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Young JA. Bates P. Willert K. Varmus HE. Efficient incorporation of human CD4 protein into avian leukosis virus particles. Science. 1990;250(4986):1421–1423. doi: 10.1126/science.2175047. [DOI] [PubMed] [Google Scholar]
- 45.Freed EO. Martin MA. Virion incorporation of envelope glycoproteins with long but not short cytoplasmic tails is blocked by specific, single amino acid substitutions in the human immunodeficiency virus type 1 matrix. J Virol. 1995;69(3):1984–1989. doi: 10.1128/jvi.69.3.1984-1989.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Freed EO. Martin MA. Domains of the human immunodeficiency virus type 1 matrix and gp41 cytoplasmic tail required for envelope incorporation into virions. J Virol. 1996;70(1):341–351. doi: 10.1128/jvi.70.1.341-351.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yu X. Yuan X. Matsuda Z. Lee TH. Essex M. The matrix protein of human immunodeficiency virus type 1 is required for incorporation of viral envelope protein into mature virions. J Virol. 1992;66(8):4966–4971. doi: 10.1128/jvi.66.8.4966-4971.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gabuzda DH. Lever A. Terwilliger E. Sodroski J. Effects of deletions in the cytoplasmic domain on biological functions of human immunodeficiency virus type 1 envelope glycoproteins. J Virol. 1992;66(6):3306–3315. doi: 10.1128/jvi.66.6.3306-3315.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yu X. Yuan X. McLane MF. Lee TH. Essex M. Mutations in the cytoplasmic domain of human immunodeficiency virus type 1 transmembrane protein impair the incorporation of Env proteins into mature virions. J Virol. 1993;67(1):213–221. doi: 10.1128/jvi.67.1.213-221.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Piller SC. Dubay JW. Derdeyn CA. Hunter E. Mutational analysis of conserved domains within the cytoplasmic tail of gp41 from human immunodeficiency virus type 1: Effects on glycoprotein incorporation and infectivity. J Virol. 2000;74(24):11717–11723. doi: 10.1128/jvi.74.24.11717-11723.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kalia V. Sarkar S. Gupta P. Montelaro RC. Rational site-directed mutations of the LLP-1 and LLP-2 lentivirus lytic peptide domains in the intracytoplasmic tail of human immunodeficiency virus type 1 gp41 indicate common functions in cell-cell fusion but distinct roles in virion envelope incorporation. J Virol. 2003;77(6):3634–3646. doi: 10.1128/JVI.77.6.3634-3646.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Murakami T. Freed EO. The long cytoplasmic tail of gp41 is required in a cell type-dependent manner for HIV-1 envelope glycoprotein incorporation into virions. Proc Natl Acad Sci USA. 2000;97(1):343–348. doi: 10.1073/pnas.97.1.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Akari H. Fukumori T. Adachi A. Cell-dependent requirement of human immunodeficiency virus type 1 gp41 cytoplasmic tail for Env incorporation into virions. J Virol. 2000;74(10):4891–4893. doi: 10.1128/jvi.74.10.4891-4893.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wilk T. Pfeiffer T. Bosch V. Retained in vitro infectivity and cytopathogenicity of HIV-1 despite truncation of the C-terminal tail of the env gene product. Virology. 1992;189(1):167–177. doi: 10.1016/0042-6822(92)90692-i. [DOI] [PubMed] [Google Scholar]
- 55.Mammano F. Kondo E. Sodroski J. Bukovsky A. Gottlinger HG. Rescue of human immunodeficiency virus type 1 matrix protein mutants by envelope glycoproteins with short cytoplasmic domains. J Virol. 1995;69(6):3824–3830. doi: 10.1128/jvi.69.6.3824-3830.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mammano F. Salvatori F. Indraccolo S. De Rossi A. Chieco-Bianchi L. Gottlinger HG. Truncation of the human immunodeficiency virus type 1 envelope glycoprotein allows efficient pseudotyping of Moloney murine leukemia virus particles and gene transfer into CD4+ cells. J Virol. 1997;71(4):3341–3345. doi: 10.1128/jvi.71.4.3341-3345.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schnierle BS. Stitz J. Bosch V, et al. Pseudotyping of murine leukemia virus with the envelope glycoproteins of HIV generates a retroviral vector with specificity of infection for CD4-expressing cells. Proc Natl Acad Sci USA. 1997;94(16):8640–8645. doi: 10.1073/pnas.94.16.8640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Stitz J. Steidl S. Merget-Millitzer H, et al. MLV-derived retroviral vectors selective for CD4-expressing cells and resistant to neutralization by sera from HIV-infected patients. Virology. 2000;267(2):229–236. doi: 10.1006/viro.1999.0121. [DOI] [PubMed] [Google Scholar]
- 59.Sandrin V. Muriaux D. Darlix JL. Cosset FL. Intracellular trafficking of Gag and Env proteins and their interactions modulate pseudotyping of retroviruses. J Virol. 2004;78(13):7153–7164. doi: 10.1128/JVI.78.13.7153-7164.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Stitz J. Buchholz CJ. Engelstadter M, et al. Lentiviral vectors pseudotyped with envelope glycoproteins derived from gibbon ape leukemia virus and murine leukemia virus 10A1. Virology. 2000;273(1):16–20. doi: 10.1006/viro.2000.0394. [DOI] [PubMed] [Google Scholar]
- 61.Sandrin V. Cosset FL. Intracellular versus cell surface assembly of retroviral pseudotypes is determined by the cellular localization of the viral glycoprotein, its capacity to interact with Gag, and the expression of the Nef protein. J Biol Chem. 2006;281(1):528–542. doi: 10.1074/jbc.M506070200. [DOI] [PubMed] [Google Scholar]
- 62.Bouard D. Sandrin V. Boson B, et al. An acidic cluster of the cytoplasmic tail of the RD114 virus glycoprotein controls assembly of retroviral envelopes. Traffic. 2007;8(7):835–847. doi: 10.1111/j.1600-0854.2007.00581.x. [DOI] [PubMed] [Google Scholar]
- 63.Lucas TM. Lyddon TD. Cannon PM. Johnson MC. Pseudotyping incompatibility between HIV-1 and gibbon ape leukemia virus Env is modulated by Vpu. J Virol. 2010;84(6):2666–2674. doi: 10.1128/JVI.01562-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Christodoulopoulos I. Droniou-Bonzom ME. Oldenburg JE. Cannon PM. Vpu-dependent block to incorporation of GaLV Env into lentiviral vectors. Retrovirology. 2010;7:4. doi: 10.1186/1742-4690-7-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lodge R. Delamarre L. Lalonde JP, et al. Two distinct oncornaviruses harbor an intracytoplasmic tyrosine-based basolateral targeting signal in their viral envelope glycoprotein. J Virol. 1997;71(7):5696–5702. doi: 10.1128/jvi.71.7.5696-5702.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lodge R. Gottlinger H. Gabuzda D. Cohen EA. Lemay G. The intracytoplasmic domain of gp41 mediates polarized budding of human immunodeficiency virus type 1 in MDCK cells. J Virol. 1994;68(8):4857–4861. doi: 10.1128/jvi.68.8.4857-4861.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lodge R. Lalonde JP. Lemay G. Cohen EA. The membrane-proximal intracytoplasmic tyrosine residue of HIV-1 envelope glycoprotein is critical for basolateral targeting of viral budding in MDCK cells. EMBO J. 1997;16(4):695–705. doi: 10.1093/emboj/16.4.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lodge R. Subbramanian RA. Forget J. Lemay G. Cohen EA. MuLV-based vectors pseudotyped with truncated HIV glycoproteins mediate specific gene transfer in CD4+ peripheral blood lymphocytes. Gene Ther. 1998;5(5):655–664. doi: 10.1038/sj.gt.3300646. [DOI] [PubMed] [Google Scholar]
- 69.Huang YT. Wright A. Gao X. Kulick L. Yan H. Lamm ME. Intraepithelial cell neutralization of HIV-1 replication by IgA. J Immunol. 2005;174(8):4828–4835. doi: 10.4049/jimmunol.174.8.4828. [DOI] [PubMed] [Google Scholar]
- 70.Wright A. Yan H. Lamm ME. Huang YT. Immunoglobulin A antibodies against internal HIV-1 proteins neutralize HIV-1 replication inside epithelial cells. Virology. 2006;356:165–170. doi: 10.1016/j.virol.2006.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Salzwedel K. West JT., Jr. Mulligan MJ. Hunter E. Retention of the human immunodeficiency virus type 1 envelope glycoprotein in the endoplasmic reticulum does not redirect virus assembly from the plasma membrane. J Virol. 1998;72(9):7523–7531. doi: 10.1128/jvi.72.9.7523-7531.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Weclewicz K. Ekstrom M. Kristensson K. Garoff H. Specific interactions between retrovirus Env and Gag proteins in rat neurons. J Virol. 1998;72(4):2832–2845. doi: 10.1128/jvi.72.4.2832-2845.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Jin J. Sherer NM. Heidecker G. Derse D. Mothes W. Assembly of the murine leukemia virus is directed towards sites of cell-cell contact. PLoS Biol. 2009;7(7):e1000163. doi: 10.1371/journal.pbio.1000163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jorgenson RL. Vogt VM. Johnson MC. Foreign glycoproteins can be actively recruited to virus assembly sites during pseudotyping. J Virol. 2009;83(9):4060–4067. doi: 10.1128/JVI.02425-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lucas TM. Lyddon TD. Grosse SA. Johnson MC. Two distinct mechanisms regulate recruitment of murine leukemia virus envelope protein to retroviral assembly sites. Virology. 2010;405(2):548–555. doi: 10.1016/j.virol.2010.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Cosson P. Direct interaction between the envelope and matrix proteins of HIV-1. EMBO J. 1996;15(21):5783–5788. [PMC free article] [PubMed] [Google Scholar]
- 77.Manrique JM. Affranchino JL. Gonzalez SA. In vitro binding of simian immunodeficiency virus matrix protein to the cytoplasmic domain of the envelope glycoprotein. Virology. 2008;374(2):273–279. doi: 10.1016/j.virol.2008.01.015. [DOI] [PubMed] [Google Scholar]
- 78.Murakami T. Freed EO. Genetic evidence for an interaction between human immunodeficiency virus type 1 matrix and alpha-helix 2 of the gp41 cytoplasmic tail. J Virol. 2000;74(8):3548–3554. doi: 10.1128/jvi.74.8.3548-3554.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wyma DJ. Jiang J. Shi J, et al. Coupling of human immunodeficiency virus type 1 fusion to virion maturation: a novel role of the gp41 cytoplasmic tail. J Virol. 2004;78(7):3429–3435. doi: 10.1128/JVI.78.7.3429-3435.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Brody BA. Hunter E. Mutations within the env gene of Mason-Pfizer monkey virus: Effects on protein transport and SU-TM association. J Virol. 1992;66(6):3466–3475. doi: 10.1128/jvi.66.6.3466-3475.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Henderson LE. Sowder R. Copeland TD. Smythers G. Oroszlan S. Quantitative separation of murine leukemia virus proteins by reversed-phase high-pressure liquid chromatography reveals newly described gag and env cleavage products. J Virol. 1984;52(2):492–500. doi: 10.1128/jvi.52.2.492-500.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Suomalainen M. Liljestrom P. Garoff H. Spike protein-nucleocapsid interactions drive the budding of alphaviruses. J Virol. 1992;66(8):4737–4747. doi: 10.1128/jvi.66.8.4737-4747.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Egan MA. Carruth LM. Rowell JF. Yu X. Siliciano RF. Human immunodeficiency virus type 1 envelope protein endocytosis mediated by a highly conserved intrinsic internalization signal in the cytoplasmic domain of gp41 is suppressed in the presence of the Pr55gag precursor protein. J Virol. 1996;70(10):6547–6556. doi: 10.1128/jvi.70.10.6547-6556.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Berlioz-Torrent C. Shacklett BL. Erdtmann L, et al. Interactions of the cytoplasmic domains of human and simian retroviral transmembrane proteins with components of the clathrin adaptor complexes modulate intracellular and cell surface expression of envelope glycoproteins. J Virol. 1999;73(2):1350–1361. doi: 10.1128/jvi.73.2.1350-1361.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Boge M. Wyss S. Bonifacino JS. Thali M. A membrane-proximal tyrosine-based signal mediates internalization of the HIV-1 envelope glycoprotein via interaction with the AP-2 clathrin adaptor. J Biol Chem. 1998;273(25):15773–15778. doi: 10.1074/jbc.273.25.15773. [DOI] [PubMed] [Google Scholar]
- 86.Ohno H. Aguilar RC. Fournier MC. Hennecke S. Cosson P. Bonifacino JS. Interaction of endocytic signals from the HIV-1 envelope glycoprotein complex with members of the adaptor medium chain family. Virology. 1997;238(2):305–315. doi: 10.1006/viro.1997.8839. [DOI] [PubMed] [Google Scholar]
- 87.Rowell JF. Stanhope PE. Siliciano RF. Endocytosis of endogenously synthesized HIV-1 envelope protein. Mechanism and role in processing for association with class II MHC. J Immunol. 1995;155(1):473–488. [PubMed] [Google Scholar]
- 88.Day JR. Van Damme N. Guatelli JC. The effect of the membrane-proximal tyrosine-based sorting signal of HIV-1 gp41 on viral infectivity depends on sequences within gp120. Virology. 2006;354(2):316–327. doi: 10.1016/j.virol.2006.06.023. [DOI] [PubMed] [Google Scholar]
- 89.Blot V. Lopez-Verges S. Breton M. Pique C. Berlioz-Torrent C. Grange MP. The conserved dileucine- and tyrosine-based motifs in MLV and MPMV envelope glycoproteins are both important to regulate a common Env intracellular trafficking. Retrovirology. 2006;3:62. doi: 10.1186/1742-4690-3-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wyss S. Berlioz-Torrent C. Boge M, et al. The highly conserved C-terminal dileucine motif in the cytosolic domain of the human immunodeficiency virus type 1 envelope glycoprotein is critical for its association with the AP-1 clathrin adaptor [correction of adapter] J Virol. 2001;75(6):2982–2992. doi: 10.1128/JVI.75.6.2982-2992.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Byland R. Vance PJ. Hoxie JA. Marsh M. A conserved dileucine motif mediates clathrin and AP-2 dependent endocytosis of HIV-1 envelope protein. Mol Biol Cell. 2007;18:414. doi: 10.1091/mbc.E06-06-0535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Batonick M. Favre M. Boge M. Spearman P. Honing S. Thali M. Interaction of HIV-1 Gag with the clathrin-associated adaptor AP-2. Virology. 2005;342(2):190–200. doi: 10.1016/j.virol.2005.08.001. [DOI] [PubMed] [Google Scholar]
- 93.Dong X. Li H. Derdowski A, et al. AP-3 directs the intracellular trafficking of HIV-1 Gag and plays a key role in particle assembly. Cell. 2005;120(5):663–674. doi: 10.1016/j.cell.2004.12.023. [DOI] [PubMed] [Google Scholar]
- 94.Harila K. Prior I. Sjoberg M. Salminen A. Hinkula J. Suomalainen M. Vpu and Tsg101 regulate intracellular targeting of the human immunodeficiency virus type 1 core protein precursor Pr55gag. J Virol. 2006;80(8):3765–3772. doi: 10.1128/JVI.80.8.3765-3772.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Freed EO. Risser R. The role of envelope glycoprotein processing in murine leukemia virus infection. J Virol. 1987;61(9):2852–2856. doi: 10.1128/jvi.61.9.2852-2856.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Machida CA. Kabat D. Role of partial proteolysis in processing murine leukemia virus membrane envelope glycoproteins to the cell surface. A viral mutant with uncleaved glycoprotein. J Biol Chem. 1982;257(23):14018–14022. [PubMed] [Google Scholar]
- 97.McCune JM. Rabin LB. Feinberg MB, et al. Endoproteolytic cleavage of gp160 is required for the activation of human immunodeficiency virus. Cell. 1988;53(1):55–67. doi: 10.1016/0092-8674(88)90487-4. [DOI] [PubMed] [Google Scholar]
- 98.Freed EO. Myers DJ. Risser R. Mutational analysis of the cleavage sequence of the human immunodeficiency virus type 1 envelope glycoprotein precursor gp160. J Virol. 1989;63(11):4670–4675. doi: 10.1128/jvi.63.11.4670-4675.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Dubay JW. Dubay SR. Shin HJ. Hunter E. Analysis of the cleavage site of the human immunodeficiency virus type 1 glycoprotein: Requirement of precursor cleavage for glycoprotein incorporation. J Virol. 1995;69(8):4675–4682. doi: 10.1128/jvi.69.8.4675-4682.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Apte S. Sanders DA. Effects of retroviral envelope-protein cleavage upon trafficking, incorporation, and membrane fusion. Virology. 2010;405(1):214–224. doi: 10.1016/j.virol.2010.06.004. [DOI] [PubMed] [Google Scholar]
- 101.Dubay JW. Roberts SJ. Hahn BH. Hunter E. Truncation of the human immunodeficiency virus type 1 transmembrane glycoprotein cytoplasmic domain blocks virus infectivity. J Virol. 1992;66(11):6616–6625. doi: 10.1128/jvi.66.11.6616-6625.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Lucas TM. Lyddon TD. Cannon PM. Johnson MC. Pseudotyping incompatibility between HIV-1 and gibbon ape leukemia virus Env is modulated by Vpu. J Virol. 2010;84(6):2666–2674. doi: 10.1128/JVI.01562-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Lopez-Verges S. Camus G. Blot G. Beauvoir R. Berlioz-Torrent C. Tail-interacting protein TIP47 is a connector between Gag and Env and is required for Env incorporation into HIV-1 virions. Proc Natl Acad Sci USA. 2006;103:14947–14952. doi: 10.1073/pnas.0602941103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Evans DT. Tillman KC. Desrosiers RC. Envelope glycoprotein cytoplasmic domains from diverse lentiviruses interact with the prenylated Rab acceptor. J Virol. 2002;76(1):327–337. doi: 10.1128/JVI.76.1.327-337.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Zhang H. Wang L. Kao S, et al. Functional interaction between the cytoplasmic leucine-zipper domain of HIV-1 gp41 and p115-RhoGEF. Curr Biol. 1999;9(21):1271–1274. doi: 10.1016/s0960-9822(99)80511-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Waheed AA. Freed EO. The role of lipids in retrovirus replication. Viruses. 2010;2(5):1146–1180. doi: 10.3390/v2051146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Waheed AA. Freed EO. Lipids and membrane microdomains in HIV-1 replication. Virus Res. 2009;143(2):162–176. doi: 10.1016/j.virusres.2009.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Briggs JA. Wilk T. Fuller SD. Do lipid rafts mediate virus assembly and pseudotyping? J Gen Virol. 2003;84(Pt 4):757–768. doi: 10.1099/vir.0.18779-0. [DOI] [PubMed] [Google Scholar]
- 109.Graham DR. Chertova E. Hilburn JM. Arthur LO. Hildreth JE. Cholesterol depletion of human immunodeficiency virus type 1 and simian immunodeficiency virus with beta-cyclodextrin inactivates and permeabilizes the virions: Evidence for virion-associated lipid rafts. J Virol. 2003;77(15):8237–8248. doi: 10.1128/JVI.77.15.8237-8248.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Nguyen DH. Hildreth JE. Evidence for budding of human immunodeficiency virus type 1 selectively from glycolipid-enriched membrane lipid rafts. J Virol. 2000;74(7):3264–3272. doi: 10.1128/jvi.74.7.3264-3272.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Pickl WF. Pimentel-Muinos FX. Seed B. Lipid rafts and pseudotyping. J Virol. 2001;75(15):7175–7183. doi: 10.1128/JVI.75.15.7175-7183.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Ono A. Freed EO. Plasma membrane rafts play a critical role in HIV-1 assembly and release. Proc Natl Acad Sci USA. 2001;98(24):13925–13930. doi: 10.1073/pnas.241320298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Nydegger S. Khurana S. Krementsov DN. Foti M. Thali M. Mapping of tetraspanin-enriched microdomains that can function as gateways for HIV-1. J Cell Biol. 2006;173(5):795–807. doi: 10.1083/jcb.200508165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Jolly C. Sattentau QJ. Human immunodeficiency virus type 1 assembly, budding, and cell-cell spread in T cells take place in tetraspanin-enriched plasma membrane domains. J Virol. 2007;81(15):7873–7884. doi: 10.1128/JVI.01845-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Krementsov DN. Rassam P. Margeat E, et al. HIV-1 assembly differentially alters dynamics and partitioning of tetraspanins and raft components. Traffic. 2010;11(11):1401–1414. doi: 10.1111/j.1600-0854.2010.01111.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Hemler ME. Tetraspanin functions and associated microdomains. Nat Rev Mol Cell Biol. 2005;6(10):801–811. doi: 10.1038/nrm1736. [DOI] [PubMed] [Google Scholar]
- 117.Khurana S. Krementsov DN. de Parseval A. Elder JH. Foti M. Thali M. Human immunodeficiency virus type 1 and influenza virus exit via different membrane microdomains. J Virol. 2007;81(22):12630–12640. doi: 10.1128/JVI.01255-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Leung K. Kim JO. Ganesh L. Kabat J. Schwartz O. Nabel GJ. HIV-1 assembly: Viral glycoproteins segregate quantally to lipid rafts that associate individually with HIV-1 capsids and virions. Cell Host Microbe. 2008;3(5):285–292. doi: 10.1016/j.chom.2008.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]