Abstract
The fusion domain of the influenza coat protein hemagglutinin HA2, bound to dodecyl phosphocholine (DPC) micelles, was recently shown to adopt a structure consisting of two anti-parallel α-helices, packed together in an exceptionally tight hairpin configuration. Four interhelical Hα to C=O aliphatic H-bonds were identified as factors stabilizing this fold. Here, we report evidence for an additional stabilizing force: a strong charge dipole interaction between the N-terminal Gly1 amino group and the dipole moment of helix 2. pH titration of the amino-terminal 15N resonance, using a methylene-TROSY based 3D NMR experiment, and observation of Gly1 13C′ show a strongly elevated pK value of 8.8, considerably higher than expected for an N-terminal amino group in a lipophilic environment. Chemical shifts of three C-terminal carbonyl carbons of helix 2 titrate with the protonation state of Gly1-N, indicative of a close proximity between the N-terminal amino group and the axis of helix 2, thereby providing an optimal charge-dipole stabilization of the antiparallel hairpin fold. pK values of the side chain carboxylate groups of Glu11 and Asp19 are higher by about one and 0.5 unit, respectively, than commonly seen for solvent-exposed side chains in water-soluble proteins, indicative of dielectric constants of ε = ~30 (Glu11) and ε = ~60 (Asp19), which places these groups in the headgroup region of the phospholipid micelle.
The driving force for the formation of the three-dimensional fold of water soluble proteins often is dominated by burial of the hydrophobic surfaces of its secondary structure elements, α-helices and β-sheets, which in turn are stabilized by internal H-bonding.1 In the lipophilic environment of membranes, burial of hydrophobic surfaces does not provide a driving force for stabilizing the tertiary structure of membrane proteins. Instead, electrostatic interactions between elements of secondary structure and aliphatic H-bonds have been proposed as stabilizing factors,2 although the magnitude of the energetic contribution of Cα-H···O H-bonds remains a matter of debate.3 Multiple Cα-H···O H-bonds were recently identified in the tight helical hairpin fold of the fusion domain of the HA2 domain of the influenza virus surface glycoprotein hemagglutinin.4 Here, we provide evidence for an additional important stabilizing force resulting from the charge dipole interaction between the N-terminal NH3+ group of Gly1 and the dipole moment of helix 2, as evidenced by an elevated pK of the Gly1 amino group and the impact of its deprotonation on the 13C′ chemical shifts of C-terminal residues of helix 2. Stabilizing interactions between a helix dipole moment and a positively charged titratable group have long been recognized to increase the pK value of the titratable group,5 and chemical shift changes remote in sequence from the titratable group can provide unambiguous evidence for electrostatic interactions.6
Hemagglutinin is solely responsible for mediating the fusion of the viral and the host-cell endosomal membranes during infection.7 A ‘spring-loaded’ conformational change of hemagglutinin is triggered by the low pH of the endosome, thereby exposing the N-terminal segment of HA2.8 Following this conformational change, the first 23 residues of HA2, referred to as HAfp23, become embedded in the target membrane.9 HAfp23 is both quite hydrophobic and glycine-rich, and represents the most conserved region of the hemagglutinin protein.10
Pioneering work by Lear and DeGrado demonstrated that synthetic peptides composed of the first 20 residues of HA2 (HAfp20) were sufficient to induce fusion of lipid vesicles.11 However, HAfp20, which has been the topic of numerous biophysical and biochemical studies over the past 15 years, lacks the three C-terminal residues, Trp21-Tyr22-Gly23, which not only are hydrophobic and completely conserved across all serotypes but also interact with the membrane.9,12 Moreover, it was recently found that these additional residues significantly impact the tertiary structure of the peptide.4 In zwitterionic dodecyl phosphatidylcholine (DPC) micelles, HAfp23 adopts a structure consisting of two α-helices packed together in a very tight helical-hairpin arrangement. Intermolecular NOEs between detergent and HAfp23 indicate that its hydrophobic side faces the core of the detergent micelle, while the more polar side is exposed to solvent,4 a conclusion reinforced by paramagnetic relaxation enhancement observed when spin-labeled lipids are added to the detergent (SI Fig. S1). Two “Gly-ridges”, part of a GXXGXXXG and its inverse GXXXGXXG motif, are located at the interhelical interface and enable their very tight packing. We previously identified four interhelical aliphatic CαHα ···O=C hydrogen bonds, between residue pairs Gly1/Trp21, Ala5/Met17 and Phe9/Gly13, which are known to stabilize helical packing in membrane proteins.13
The HAfp23 structure suggested that the N-terminal amino group of Gly1 may also form a number of important hydrogen bonds that position its positive charge at the C-terminal end of the peptide’s second α-helix, implying a charge-dipole interaction. Together, these interactions may be responsible for the strict conservation of Gly1 in all serotypes.10,14,15 In much of the literature, the putative protonation state of Gly1 has been marked as NH2, however, and few studies have looked at this topic more closely.16 Ambiguity regarding the protonation state of Gly1 has also permeated the molecular modeling literature, where simulations were conducted either for the protonated state,17 for multiple protonation states,18 or for an unspecified or deprotonated state.19
Zhou et al. found a pK of 8.69 for Gly1 of the truncated fusion peptide,16 which is elevated compared to values for peptides in solution (pK: 7.5–8.0).20 An elevated pK for this N-terminal amino group is surprising, considering that even in aqueous solution the pK value of the N-terminal amino group of an α-helix is depressed by about 0.5 units by the positive potential imposed by the helix dipole moment.21 Embedding of the peptide in the hydrophobic environment of neutral membranes would be expected to decrease its pK value even further,22,23 but a potential α-NH3+ interaction with one of the carboxyl groups of the peptide or with the phosphate of the lipid headgroup was proposed as a possible reason for the elevated pK of Gly1.16 This truncated fusion peptide was subsequently reported to adopt an open ‘boomerang’ structure,24 while embedding its α-NH3+ in the lipid bilayer, without revealing any interactions that could stabilize its protonated state. We recently found that the open boomerang structure of truncated HAfp20, which also contains a C-terminal “solubilization tag”, is dynamic and transiently adopts the helical-hairpin structure of HAfp23.4 In an effort to evaluate whether stabilizing interactions in this hairpin can account for the elevated pK value, we set out to measure the pK values of titratable groups in full-length HAfp23. Furthermore, we measure pK values of the only two negatively charged side chains in HAfp23 of serotype H1: Glu11 and Asp19, with Glu11 found to change protonation states at the pH where fusion is activated.
Solution NMR of membrane-binding proteins, usually carried out on detergent-solubilized systems,25 is a powerful approach for obtaining detailed structural information. The protonation state of an amine group is probed most easily by its 15N chemical shift, which undergoes a substantial ~12 ppm upfield change upon deprotonation.26 Hydrogen exchange rates with solvent of the amino hydrogens of Gly1 are too high to permit observation of their resonance with a standard 15N HSQC experiment. In order to avoid ambiguity between the Gly1 amino group and the side chains of multiple Lys residues in the C-terminal solubilization tag of HAfp23, we probed the Gly1 15N chemical shift indirectly, using a new three-dimensional CH2-TROSY HACAN experiment, which is particularly well suited for Gly residues (SI, Fig. S2). This experiment correlates the 15N chemical shift of residues i and i+1 to the 1Hα and 13Cα chemical shifts of residue i, assigned previously.4 Gly methylene groups in D2O solution constitute well isolated 1Hα2/1Hα3/13Cα three-spin systems, which can be detected in a resolution- and sensitivity-optimized manner by the CH2-TROSY scheme.27
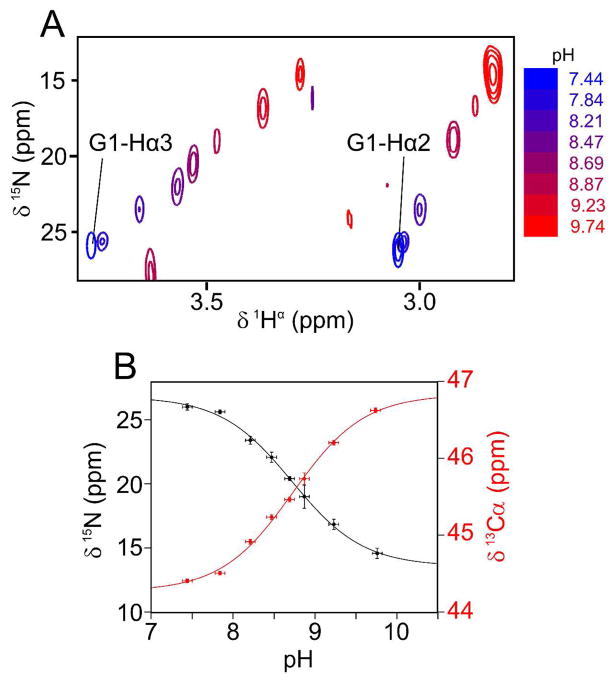
The pH dependence of the amino terminal 15N resonance is readily observed from cross sections through the 3D CH2-TROSY HACAN spectra (Figure 1A), and fits well to the standard Henderson-Hasselbalch equation (Figure 1B). The 15N chemical shift moves from 27 ppm (NH3+ form) to 13.6 ppm (NH2 form) when increasing the pH, while the Gly1 13Cα chemical shift moves downfield by 2.5 ppm from 44.3 ppm to 46.8 ppm. The best-fit pK from these measurements and those of 13C′ shifts on a separate sample (Fig. 2) is 8.80±0.04, a value slightly above that of the truncated peptide in DPC (pK = 8.63; SI Fig. S3).
Figure 1.
pH titration of the N-terminal amino 15N resonance of Gly1 with the HACAN CH2-TROSY experiment. (A) Superimposed small regions of 1H/15N cross sections taken through the 3D CH2-TROSY HACAN spectrum of HAfp23, showing the correlation between the Gly1 1Hα2/1Hα3 and amino 15N chemical shifts. (B) The Gly1 backbone amino 15N (black) and 13Cα (red) chemical shift dependence on pH is shown. A non-linear least-squares regression was used to fit the Henderson-Hasselbalch equation to the titration curves. The fitted values are: pK = 8.73±0.07, 15N and 13Cα chemical shifts of 27.0±0.6. ppm and 44.3±0.1 ppm, respectively, for the protonated form (NH3+), and 13.6±0.4 ppm and 46.8±0.1 ppm, respectively, for the deprotonated form (NH2). The fitted pK value when measuring the Gly1 13C′ shift on a separate sample (Fig. 2) is 8.84±0.05.
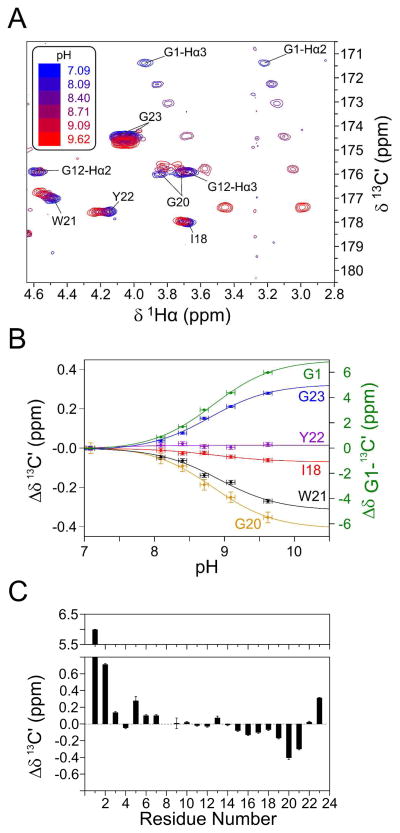
Figure 2.
Changes in the 13C′ chemical shifts on titrating the amino group of Gly1. (A) Superimposed small regions of cross-sections taken through the 3D HCACO spectra of HAfp23, recorded at pH values ranging from pH 7.09 to 9.62, with 13C′ chemical shift changes summarized in (C). The Gly8 13C′ chemical shifts could not be identified uniquely due to spectral congestion and are not reported. (B) pH dependence of Gly1 13C′ (green, right axis) and 13C′ chemical shifts of various residues near the end of helix 2. Reported chemical shift changes are relative to values measured at pH 7.09. Traces shown represent nonlinear regression fits to the Henderson-Hasselbalch equation, yielding: Δδ = 7.1±0.2 ppm and pK = 8.84±0.05 for Gly1; Δδ = −0.08±0.02 ppm and pK = 8.9±0.2 for Ile18; Δδ = −0.46±0.02 ppm and pK = 8.84±0.09 for Gly20; Δδ = −0.35±0.06 ppm and pK = 8.9±0.2 for Trp21; Δδ = +0.37±0.04 ppm and pK = 8.87 ±0.10 for Gly23.
Two factors are known to significantly impact the pK values of titratable moieties that interact with micelles: the surface potential of the micelle and the difference in partitioning between aqueous and apolar membrane environments for the neutral and charged forms of the titratable group.28 The first factor is particularly pertinent for charged micelles and membranes, as illustrated by the increased pK of the α-NH3 group of the M13 coat protein in anionic micelles.29 The second factor accounts for the high energy associated with burial of charged groups and the stabilization of neutralized species in membranes. So, compared to being immersed in water, population of the charged form of a titratable group will be disfavored in a medium of moderate dielectric constant, ε, found at the lipid-water interface or the even lower ε in the aliphatic core of the micelle.28
Literature values for the N-terminal amino group pK values of peptides and proteins in free aqueous solution mostly fall in the 7.5–8.0 range.20 In a neutral micelle environment, the pK for the amino group of Gly1 would be expected to shift towards lower values because the charged NH3+ species is destabilized relative to the neutral NH2 species in the membrane. For example, for a somatostatin analogue peptide, an amino-terminal pK shift of −0.55 units was observed between bulk solution and binding to a neutral bilayer surface.23 Accordingly, based solely on the lipid-binding induced pK shift caused by the low-dielectric micelle environment, a pK value lower than ca 7.5 is expected for Gly1. The observed Gly1 pK of 8.73 is higher by at least 0.7 pH units than values expected in aqueous solution; the difference is even greater when the expected destabilization of NH3+ in the decreased dielectric environment of the micelle is taken into account, or the additional increase expected for its location at the N-terminus of an α-helix.21 An increase in the pK by one pH unit is indicative of an interaction that stabilizes the NH3+ form by 1.4 kcal/mol — the increase by at least 1.5 pH units therefore corresponds to a stabilization by ca 2 kcal/mol.
In the absence of an observable 1H NMR signal for the N-terminal amino group, its potential interaction partners previously could only be inferred from the approximate position of the NH3+ group, calculated without its stabilizing restraints.4 However, pH titration allows for straightforward evaluation of potential interaction partners, which are expected to be significantly impacted by a change in protonation state of the amino group. As can be seen (Figure 2), besides upfield changes in the chemical shifts of Gly1, Leu2 and Ala5 13C′ located proximate to the N-terminal amino group, substantial changes are also observed for Gly20, Trp21, and Gly23 carbonyl resonances, pointing to potential H-bonds to the Gly1 amino protons. The pH values corresponding to the midpoints of the chemical shift change observed for these C-terminal carbonyl resonances (Figure 2B) match those of Gly1, which further substantiates a connection between these groups. Indeed, such interactions are geometrically allowed, and would stabilize the observed tight anti-parallel packing of the two helices. Moreover, H-bonding of the three amino protons to Gly20, Trp21, and Gly23 carbonyl oxygens positions the positively charged amino group close to the center of helix 2, resulting in a favorable interaction with the dipole moment of this helix, rather than the unfavorable charge dipole interaction that would occur if the NH3+ were positioned on the helix 1 axis. A small impact (0.1–0.2 ppm 13C′ upfield shift) of the deprotonation of Gly1 is also seen for the three carbonyls (Gly16, Met17 and Asp19) that make α-helical H-bonds to these carboxy-terminal residues. Even though a weakening of an H-bond is known to be associated with an upfield 13C′ shift, it should be noted that quantitative interpretation of the observed 13C′ shift changes is difficult, because the loss of an intramolecular H-bond to a carbonyl oxygen almost certainly will be replaced by an H-bond to a water molecule or to a choline headgroup of DPC, with an interaction strength and impact on the 13C′ shift that is difficult to calculate quantitatively.
Further evidence for destabilization of the helical hairpin structure at high pH can be found in the 13Cα chemical shift changes that occur upon deprotonation of Gly1-NH3+. These changes correlate fairly well with the 13Cα chemical shift difference seen between the tightly folded HAfp23 sequence at pH 7.4, and 13Cα chemical shifts observed for a G8A mutant, designed to disrupt the tight antiparallel packing of the two helices (Figure S4). Interestingly, the spectral data observed for G8A also correlate well with those of the truncated HAfp20 fusion peptide (data not shown), which presumably is destabilized because it lacks the three helical C-terminal residues found to interact with the Gly1 amino group in HAfp23.
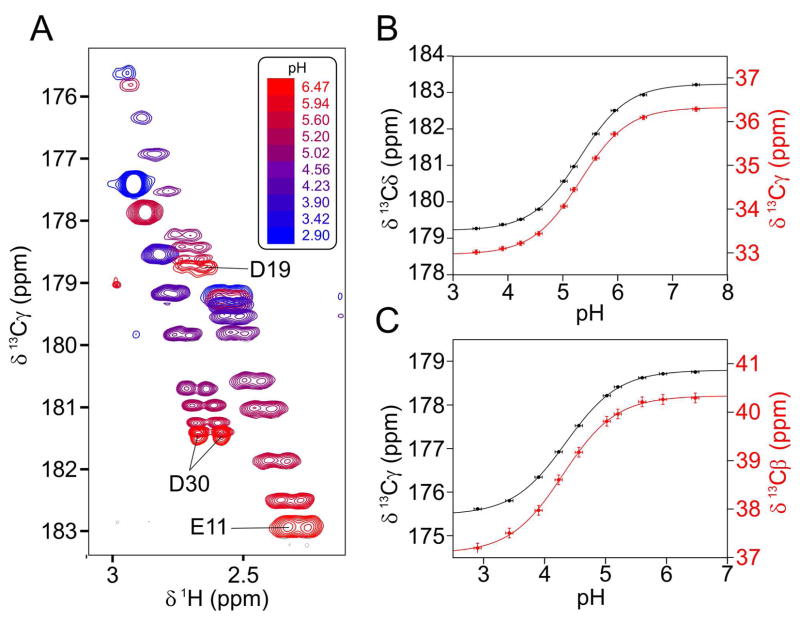
Titration plots for Glu11 and Asp19 show that 13C chemical shifts move upfield as the carboxylates are protonated: by 3.3 ppm for Glu11, and by 3.2 ppm for Asp19 (Figure 3). A non-linear fit of the titrations to the Henderson-Hasselbalch equation yields pK values of 5.31 for Glu11 and 4.35 for Asp19. A satisfactory fit was achieved without recourse to a Hill coefficient. Compared to model pentapeptides, which have side chain pK values of 4.25 ± 0.05 (Glu) and 3.90 ± 0.02 (Asp),21 the pK values in HAfp23 are higher by 1.1 (Glu11) and 0.5 (Asp19) units.
Figure 3.
Chemical shift titrations of the carboxylate groups of Glu11 and Asp19. (A) Superimposed small regions of cross sections taken through the 3D HCCO spectra taken over the range from pH 6.5 to 2.9, showing the correlation between the carboxylic acid 13C and its vicinal methylene 1H chemical shifts. Chemical shift versus pH for (B) the Glu11 side chain 13Cδ (black) and 13Cγ (red), and (C) the Asp19 side chain 13Cγ (black) and 13Cβ (red) are fit to the Henderson-Hasselbalch equation using non-linear least-squares regression. Fitted values for (B) are: pK = 5.31±0.01, with 13Cδ and 13Cγ chemical shift of 179.2 ppm and 33.0 ppm, respectively, for the protonated form (COOH), and a 13Cδ and 13Cγ chemical shift of 183.2 ppm and 36.3 ppm for the deprotonated form (COO−). Fitted values for Asp19 are: pK = 4.35±0.02, with 13Cγ and 13Cβ chemical shift of 175.5 ppm and 37.1 ppm, respectively, for the protonated form, and 13Cγ and 13Cβ chemical shifts of 178.8 ppm and 40.3 ppm for the deprotonated form (COO−).
In the absence of stabilizing interactions, a shift in pK can be attributed to the decreased local dielectric constant near the titratable group, resulting from its location with respect to the water lipid interface. For small indicator molecules, a one unit change in pK between water and a neutral micelle solution has been translated into a shift from ε =80 to ε =32.28 pK values we find for the side chain carboxylate groups of Glu11 and Asp19 are higher by about one and 0.5 unit, respectively, than commonly seen for short reference peptides in water, and if these changes are attributed to the change in dielectric constant for the neutral DPC micelle, one similarly obtains ε = ~30 for Glu11, and ε = ~60 for Asp19, which places these side chain carboxylates in the headgroup region.
A previous study of the 25-residue H3 subtype fusion peptide in SDS micelles reported pK values for Glu11, Glu15 and Asp19 of 5.91, 5.25, and 5.19, respectively.30 These pK values in negative micelles are 0.6–0.8 units higher than those found by us in neutral micelles, an effect that may be attributed to the negative charge of the SDS sulfate head-groups, giving rise to a strong negative surface potential for the micelle, which further destabilizes the negatively charged carboxylate state. The increase in pK between charged and neutral micelles can be quite significant: a pK increase as high as +2.3 has been observed between species bound to neutral and negatively charged micelles.28
The functional roles of Glu11 and Asp19 in the fusion peptide are not entirely clear. Serotypes with Ala and Asn substitutions for Asp19 also occur,10 while Glu11 is strictly conserved. However, hemagglutinin protein with an E11V mutation is fusogenic and shows no change in the optimal pH of 5.5 for fusion.15 Interestingly, however, the pK of Glu11 coincides with this optimal pH of fusion, and even though the monomeric structure of HAfp23 does not change significantly between pH 4.0 and 7.4,4 the protonated state of Glu11 may be required for switching to the postulated oligomeric structure required for actual fusion.
Supplementary Material
Acknowledgments
This work was supported by the Intramural Research Program of the NIDDK, NIH, and by the Intramural Antiviral Target Program of the Office of the Director, NIH. We thank Dennis A. Torchia for useful discussions.
Footnotes
Supporting Information Available: Experimental details regarding sample preparation and NMR measurements; one figure showing PRE in the presence of 5-doxylstearic acid; a pulse diagram of the HACAN CH2-TROSY experiment; a pH titration of Gly1 13Cα and 15N chemical shifts in HAfp20; a plot with 13C′ chemical shift changes between pH 9.5 and 7.6 in HAfp20; a plot comparing the 13Cα chemical shift differences between the wild type HAfp23 and its G8A mutant with the difference between HAfp23 at pH 9.6 and pH 7.1; a jack-knife analysis of the data shown in Fig. 2B. This information is available free of charge via the Internet at http://pubs.acs.org/.
References
- 1.Creighton TE. Proteins: structures and molecular properties. Macmillan; New York: 1993. [Google Scholar]
- 2.Senes A, Ubarretxena-Belandia I, Engelman DM. Proc Natl Acad Sci U S A. 2001;98:9056–9061. doi: 10.1073/pnas.161280798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yohannan S, Faham S, Yang D, Grosfeld D, Chamberlain AK, Bowie JU. J Am Chem Soc. 2004;126:2284–2285. doi: 10.1021/ja0317574. [DOI] [PubMed] [Google Scholar]
- 4.Lorieau JL, Louis JM, Bax A. Proc Natl Acad Sci U S A. 2010;107:11341–11346. doi: 10.1073/pnas.1006142107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Perutz MF, Gronenborn AM, Clore GM, Fogg JH, Shih DTB. J Mol Biol. 1985;183:491–498. doi: 10.1016/0022-2836(85)90016-6. [DOI] [PubMed] [Google Scholar]; Sali D, Bycroft M, Fersht AR. Nature. 1988;335:740–743. doi: 10.1038/335740a0. [DOI] [PubMed] [Google Scholar]
- 6.Tomlinson JH, Ullah S, Hansen PE, Williamson MP. J Am Chem Soc. 2009;131:4674–4684. doi: 10.1021/ja808223p. [DOI] [PubMed] [Google Scholar]
- 7.Wiley DC, Skehel JJ. Annu Rev Biochem. 1987;56:365–394. doi: 10.1146/annurev.bi.56.070187.002053. [DOI] [PubMed] [Google Scholar]
- 8.Carr CM, Chaudhry C, Kim PS. Proc Natl Acad Sci U S A. 1997;94:14306–14313. doi: 10.1073/pnas.94.26.14306. [DOI] [PMC free article] [PubMed] [Google Scholar]; Cross KJ, Langley WA, Russell RJ, Skehel JJ, Steinhauer DA. Protein Pept Lett. 2009;16:766–778. doi: 10.2174/092986609788681715. [DOI] [PubMed] [Google Scholar]
- 9.Durrer P, Galli C, Hoenke S, Corti C, Gluck R, Vorherr T, Brunner J. J Biol Chem. 1996;271:13417–13421. doi: 10.1074/jbc.271.23.13417. [DOI] [PubMed] [Google Scholar]
- 10.Nobusawa E, Aoyama T, Kato H, Suzuki Y, Tateno Y, Nakajima K. Virology. 1991;182:475–485. doi: 10.1016/0042-6822(91)90588-3. [DOI] [PubMed] [Google Scholar]
- 11.Lear JD, Degrado WF. J Biol Chem. 1987;262:6500–6505. [PubMed] [Google Scholar]
- 12.Gray C, Tatulian SA, Wharton SA, Tamm LK. Biophys J. 1996;70:2275–2286. doi: 10.1016/S0006-3495(96)79793-X. [DOI] [PMC free article] [PubMed] [Google Scholar]; Dubovskii PV, Zhmak MN, Maksaev GI, Arseniev AS. Russ J Bioorg Chem. 2004;30:196–198. [Google Scholar]
- 13.MacKenzie KR, Prestegard JH, Engelman DM. Science. 1997;276:131–133. doi: 10.1126/science.276.5309.131. [DOI] [PubMed] [Google Scholar]; Russ WP, Engelman DM. J Mol Biol. 2000;296:911–919. doi: 10.1006/jmbi.1999.3489. [DOI] [PubMed] [Google Scholar]; Kleiger G, Grothe R, Mallick P, Eisenberg D. Biochemistry. 2002;41:5990–5997. doi: 10.1021/bi0200763. [DOI] [PubMed] [Google Scholar]
- 14.Gething MJ, Doms RW, York D, White J. J Cell Biol. 1986;102:11–23. doi: 10.1083/jcb.102.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]; Wu CW, Cheng SF, Huang WN, Trivedi VD, Veeramuthu B, Kantchev AB, Wu WG, Chang DK. Biochimica Et Biophysica Acta-Biomembranes. 2003;1612:41–51. doi: 10.1016/s0005-2736(03)00084-1. [DOI] [PubMed] [Google Scholar]
- 15.Steinhauer DA, Wharton SA, Skehel JJ, Wiley DC. J Virol. 1995;69:6643–6651. doi: 10.1128/jvi.69.11.6643-6651.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou Z, Macosko JC, Hughes DW, Sayer BG, Hawes J, Epand RM. Biophys J. 2000;78:2418–2425. doi: 10.1016/S0006-3495(00)76785-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Spassov VZ, Yan L, Szalma S. J Phys Chem B. 2002;106:8726–8738. [Google Scholar]
- 18.Huang Q, Chen CL, Herrmann A. Biophys J. 2004;87:14–22. doi: 10.1529/biophysj.103.024562. [DOI] [PMC free article] [PubMed] [Google Scholar]; Sammalkorpi M, Lazaridis T. Biochimica Et Biophysica Acta-Biomembranes. 2007;1768:30–38. doi: 10.1016/j.bbamem.2006.08.008. [DOI] [PubMed] [Google Scholar]; Panahi A, Feig MJ. Phys Chem B. 2010;114:1407–1416. doi: 10.1021/jp907366g. [DOI] [PubMed] [Google Scholar]
- 19.Sammalkorpi M, Lazaridis T. Biophys J. 2007;92:10–22. doi: 10.1529/biophysj.106.092809. [DOI] [PMC free article] [PubMed] [Google Scholar]; Lague P, Roux B, Pastor RW. J Mol Biol. 2005;354:1129–1141. doi: 10.1016/j.jmb.2005.10.038. [DOI] [PubMed] [Google Scholar]; Vaccaro L, Cross KJ, Kleinjung J, Straus SK, Thomas DJ, Wharton SA, Skehel JJ, Fraternali F. Biophys J. 2005;88:25–36. doi: 10.1529/biophysj.104.044537. [DOI] [PMC free article] [PubMed] [Google Scholar]; Jang H, Michaud-Agrawal N, Johnston JM, Woolf TB. Proteins-Structure Function and Bioinformatics. 2008;72:299–312. doi: 10.1002/prot.21925. [DOI] [PubMed] [Google Scholar]; Li JY, Das P, Zhou RH. J Phys Chem B. 2010;114:8799–8806. doi: 10.1021/jp1029163. [DOI] [PubMed] [Google Scholar]; Kasson PM, Lindahl E, Pande VS. PLoS Comput Biol. 2010;6:1137–1147. doi: 10.1371/journal.pcbi.1000829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nozaki Y, Tanford C. Meth Enzymol. 1967;11:715–734. [Google Scholar]; Thurlkill RL, Grimsley GR, Scholtz JM, Pace CN. Protein Sci. 2006;15:1214–1218. doi: 10.1110/ps.051840806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lockhart DJ, Kim PS. Science. 1993;260:198–202. doi: 10.1126/science.8469972. [DOI] [PubMed] [Google Scholar]; Sitkoff D, Lockhart DJ, Sharp KA, Honig B. Biophys J. 1994;67:2251–2260. doi: 10.1016/S0006-3495(94)80709-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Woolley G, Deber C. Biopolymers. 1987;26:S109–S121. doi: 10.1002/bip.360260012. [DOI] [PubMed] [Google Scholar]
- 23.Beschiaschvili G, Seelig J. Biochemistry. 1992;31:10044–10053. doi: 10.1021/bi00156a026. [DOI] [PubMed] [Google Scholar]
- 24.Han X, Bushweller JH, Cafiso DS, Tamm LK. Nature Structural Biology. 2001;8:715–720. doi: 10.1038/90434. [DOI] [PubMed] [Google Scholar]
- 25.Opella SJ, Marassi FM. Chem Rev (Washington, DC, U S) 2004;104:3587–3606. doi: 10.1021/cr0304121. [DOI] [PMC free article] [PubMed] [Google Scholar]; Krueger-Koplin RD, Sorgen PL, Krueger-Koplin ST, Rivera-Torres AO, Cahill SM, Hicks DB, Grinius L, Krulwich TA, Girvin ME. J Biomol NMR. 2004;28:43–57. doi: 10.1023/B:JNMR.0000012875.80898.8f. [DOI] [PubMed] [Google Scholar]; Van Horn WD, Kim HJ, Ellis CD, Hadziselimovic A, Sulistijo ES, Karra MD, Tian CL, Sonnichsen FD, Sanders CR. Science. 2009;324:1726–1729. doi: 10.1126/science.1171716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhu LY, Kemple MD, Yuan P, Prendergast FG. Biochemistry. 1995;34:13196–13202. doi: 10.1021/bi00040a035. [DOI] [PubMed] [Google Scholar]
- 27.Miclet E, Williams DC, Clore GM, Bryce DL, Boisbouvier J, Bax A. J Am Chem Soc. 2004;126:10560–10570. doi: 10.1021/ja047904v. [DOI] [PubMed] [Google Scholar]
- 28.Fernandez MS, Fromherz P. J Phys Chem. 1977;81:1755–1761. [Google Scholar]
- 29.Henry GD, Weiner JH, Sykes BD. Biochemistry. 1986;25:590–598. doi: 10.1021/bi00351a012. [DOI] [PubMed] [Google Scholar]
- 30.Chang DK, Cheng SF, Lin CH, Kantchev EAB, Wu CW. Biochimica Et Biophysica Acta-Biomembranes. 2005;1712:37–51. doi: 10.1016/j.bbamem.2005.04.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.