Abstract
Weather is one of the most basic factors impacting animal populations, but the typical strength of such impacts on population dynamics is unknown. We incorporate weather and climate index data into analysis of 492 time series of mammals, birds and insects from the global population dynamics database. A conundrum is that a multitude of weather data may a priori be considered potentially important and hence present a risk of statistical over-fitting. We find that model selection or averaging alone could spuriously indicate that weather provides strong improvements to short-term population prediction accuracy. However, a block randomization test reveals that most improvements result from over-fitting. Weather and climate variables do, in general, improve predictions, but improvements were barely detectable despite the large number of datasets considered. Climate indices such as North Atlantic Oscillation are not better predictors of population change than local weather variables. Insect time series are typically less predictable than bird or mammal time series, although all taxonomic classes display low predictability. Our results are in line with the view that population dynamics is often too complex to allow resolving mechanisms from time series, but we argue that time series analysis can still be useful for estimating net environmental effects.
Keywords: weather, climate, NAO, population dynamics, time series, prediction
1. Introduction
How and to what extent weather and climate affect the dynamics of populations is a question that has interested ecologists since the early days of population ecology. In fact, one of the most well-known and long-lived debates in ecology concerned whether the dynamics of wild animal populations are mainly regulated by exogenous environmental factors [1] or by endogenous density-dependent processes [2]. Although modern thinking usually views endogenous and exogenous factors in a more unified way, operating together on any population [3], we lack a macro-ecological view of the typical importance of weather compared with other factors across many populations and species. Beyond these fundamental ecological questions, the role of the environment in shaping the dynamics of populations is of interest in applied contexts. This is exemplified by the current concern about effects of climate change, which has sparked an ever-increasing interest in estimating how environmental factors affect populations.
While the plain fact that weather can and does impact populations is beyond dispute, the problem of how to integrate weather into understanding of the dynamics of specific populations has generated its own debates. Certain well-studied species have been shown to react to fluctuations in climate and weather through an intricate interplay between demographic traits and combinations of environmental factors. In soay sheep (Ovis aries) on St Kilda, UK, weather affects survival differently among different age classes, while the age distribution fluctuates independently of total population size [4]. Predicting the response of this population to weather change therefore requires detailed data on population structure or individual-based data. This has generated the idea that ‘the devil is in the details’ for understanding population change [5], implying that extensive data and background knowledge may be necessary to resolve the mechanisms by which changes in weather and climate lead to changes in population size.
Population data, however, often come in the form of time series resulting from censuses or surveys, and there is an extensive number of studies where weather variables have been included in analyses of such series (e.g. [6–10]). Apart from the potential problem of complexity, weather is often just an indirect cause of population change. For instance, herbivore species depend on vegetation for foraging, which in turn may be driven by rainfall. Inferring how rainfall affects population dynamics from time series may then require prior knowledge about the mechanisms leading from rainfall through vegetation growth to herbivores, including potential feedback from herbivores to vegetation [11]. Thus, the extent to which time series analyses are useful for inferring mechanisms in population dynamics is under debate [12]. While there certainly are limitations to time series analyses, at least part of the debate seems to stem from an over-expectation of what kind of answers they can provide. Along these lines it has been argued that time-series analysis may often be the best tool, given the limited information typically available on a population, and can serve as a starting point for examining hypotheses prior to further exploration with detailed data [13]. Time-series analysis can also be used as a tool aimed at improving predictions of abundance [14], in line with the classical role of time-series analysis in statistics.
In the absence of detailed prior knowledge of a system, there are potential risks and rewards to incorporating weather variables in time-series analysis. Environmental variance influencing population dynamics is routinely referred to in time-series analyses without any explicit inclusion of environmental factors. For instance, process error variance in discrete time population models is often equated with environmental variance [3], while in practice the importance of exogenous factors in relation to endogenous factors is nearly impossible to determine unless the exogenous factors can be identified and measured [15]. Including weather variables offers the potential to clarify other factors affecting populations by explaining part of the variation and may, for instance, improve estimates of density dependence [16]. On the other hand, because of the many potential weather variables that could be considered, explicit incorporation of weather in time-series analyses runs the risk of over-fitting in the sense that seemingly statistically significant effects may be spurious [12]. A partial remedy may be to use climate indices such as North Atlantic Oscillation (NAO), which have been found to often correlate more strongly with demographic traits or population abundance than local weather variables [17]; the hypothesis is that NAO may capture the overall fluctuations of a mix of relevant weather variables [18].
We use 492 population time series from the global population dynamics database (GPDD), large-scale weather data [19], and climate indices representing the NAO and the Southern Oscillation (SO) to address questions about the role of weather in population dynamics. The GPDD has been suggested as an important tool for exploring questions in population ecology [20]. Previous studies using the GPDD have focused on population viability [21], and regulation and density dependence [22–25], but none have included weather or climate variables. We address three main questions: first, how much do weather variables appear to improve predictions of population dynamics and how much of the apparent improvement can be attributed to over-fitting? Second, are climate indices such as NAO and SO better predictors than local weather variables? Third, to what extent are populations predictable, given knowledge of weather? We focus on short-term predictability (through residual variance) because it relates to the practical and direct impact of weather rather than its relative impact. The results show that the data are related to weather covariates but that the effects are weak and that the data are often highly unpredictable at an annual scale.
2. Material and methods
(a). Population data
The GPDD [26] consists of over 5000 time series on population abundance estimates across a wide range of taxa, geographical locations and sample periods. The database has been used to analyse density dependence [22,23,25], population cycles [27], extinction risk [28] and population variability [20]. We applied selection criteria to choose suitable datasets from the database. We removed datasets that were short (taking less than 10 unique values), sparse (more than 30% missing data), sampled at non-annual intervals, based on harvest or non-index data, collected from a location for which weather data were not available for the entirety of the study or in other ways did not meet our selection criteria (see electronic supplementary material for further details). Additional species data (such as approximate lifespan) that are not in the GPDD were obtained from [28].
(b). Weather and climate data
Weather data were obtained from a global database of gridded weather variables at 0.5 × 0.5° resolution [19]. At each grid, monthly weather data have been spatially interpolated from weather station records. For each population time series, we extracted weather data from the grid in which the population data were collected. We further obtained large-scale climate data in the form of winter NAO (http://www.cgd.ucar.edu/cas/jhurrell/indices.html) and annual SO (http://www.bom.gov.au/climate/current/soihtm1.shtml) indices. Winter NAO is indicative of winter climate in the North Atlantic region, while SO is related to El Niño and large-scale weather events mostly affecting the Pacific region [29].
From these data, we derived two collections of weather covariates (one small and one large). The small collection was chosen to reduce the risk of over-fitting while at the same time increasing the risk that an important variable would be missed. This includes summer and winter temperature and precipitation as well as winter NAO and annual SO. Weather indices were obtained by averaging the daily mean of maximum and minimum temperature, and total precipitation for summer (June–August) and winter (December–February). For the change in a population index from year t − 1 to year t, the temperature and precipitation in the summer of year t − 1 and in the winter between year t − 1 and year t were used as covariates. The NAO for the same winter and SO for year t − 1 were also used as covariates. The collection thus contains six covariates for each population dataset.
The large collection included more seasons, additional variables and more possible lags in the effect of weather. Specifically it included winter (December–February), spring (March–May), summer (June–August) and autumn (September–November) averages of daily minimum and maximum temperature, and of precipitation and frost day frequency. For the population change from year t − 1 to t, seasonal weather indices from the winter between year t − 2 and t − 1 to the autumn in year t, climate NAO indices from the winter between year t − 3 and t − 2 to the winter between year t − 1 and t, and SO indices from the years t − 3, t − 2 and t − 1 were used as covariates. In summary, the large collection contains 38 weather and climate covariates for each population time series.
All covariates were standardized to mean 0 and variance 1 to simplify the analyses.
(c). Models
We modelled log-transformed population data using linear autoregressive (AR) models having either one or two AR terms and with either no covariate or one weather or climate covariate at the time. Combinations of multiple weather or climate variables were not considered owing to the combinatorial explosion of the number of models that would need to be examined. In total, each time series was fitted to 14 different models for the small collection of weather variables and to 78 different models for the large collection. The most general model was of the form
| 2.1 |
where xt is log-transformed population index in year t, ɛt is normally distributed with standard deviation σ and wt − 1 is the weather covariate corresponding to the population transition from year t − 1 to year t. For each dataset, two models without covariates (with either one or two AR terms) and two models for each of the covariates (with either one or two AR terms) were fitted. Our main parameter of interest is σ2, which is usually referred to as the process error variance, or even the environmental variance. Here, we will refer to σ2 as the prediction error variance, to minimize ecological implications of the nomenclature. The prediction error is the variance of one-step-ahead predictions of the data on the log scale and our main aim is to see to what extent weather variables can improve predictions. Prediction error may arise because of, among other things, environmental variation, model misspecification and measurement error.
We used maximum likelihood methods to obtain the parameter estimates. For time series with no missing data, maximum likelihood estimates of AR models are identical to linear regression estimates and for these datasets we used the lm function in R [30] to fit the models. For datasets where some data were missing, we maximized the likelihood numerically using the optim routine in R. The estimate of σ2 was bias-corrected by multiplying the maximum likelihood estimate by the number of data points divided by the degrees of freedom. This bias correction is exact for linear regression but corresponds to a first-order correction for time series.
We limited our analysis to simple AR models (equation (2.1)), rather than including either state-space models to incorporate measurement error [31] or autoregressive moving average (ARMA) models [25]. Many of the GPDD time series are short, which could cause high imprecision in parameter estimates of more complex models owing to shallow likelihoods (see [32] for state-space models). Also, our results illustrate the hazards of over-fitting from model selection or model averaging, and adding several-fold model structures to the analysis could exacerbate over-fitting problems owing to the increase in the number of models that would be applied to each dataset.
(d). Model selection and model averaging
Because of the many potential weather and climate covariates possible for any of the datasets, only one covariate at a time was included in the model (2.1), and model selection was used to differentiate between covariates, as well as between one or two density dependence lags. A small-sample correction to AIC, the AICc, defined as
where k is the number of model parameters and T is the number of data points (T = n − 2 if there are no missing data), has been shown to be superior to AIC for AR time series [33]. Inference via model selection can be achieved by inference from the best model, in the sense that the model with smallest AICc value is selected and used for obtaining estimates of quantities of interest. Inference from the best model has the drawback that no account is taken for the uncertainty of the model selection process, and the performances of models other than the best one are not considered. An alternative approach to inference is model averaging. This may be done by computing model weights based on the information criterion via
where AICci is the AICc value for models i and j run over all models under consideration [34]. The model weights can then be used to estimate a parameter of interest, say θ, by
where 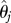 is the estimate under model j. Although model averaging is philosophically problematic in a frequentistic framework and the choice of model weights is somewhat arbitrary, AIC-based model weighting performs well by smoothing model-averaged estimators relative to best-model estimators [35]. Our results will alternately be presented in terms of model-averaged estimates and best-model estimates.
is the estimate under model j. Although model averaging is philosophically problematic in a frequentistic framework and the choice of model weights is somewhat arbitrary, AIC-based model weighting performs well by smoothing model-averaged estimators relative to best-model estimators [35]. Our results will alternately be presented in terms of model-averaged estimates and best-model estimates.
(e). Over-fitting and model selection bias
When too many models are included in a model selection process there is a risk of over-fitting in the sense that a covariate included in a selected model may only be spuriously related to the data [36]. In other words, even if no covariate had any effect on any of the datasets, the AICc best model would still be expected to include a covariate for some of the datasets and the model-averaged estimates would be expected to be derived via model weights that are non-zero for some of the models with a covariate. This is related to the multiple comparisons problem in hypothesis testing and is a more severe hazard with increasing size of the collection of datasets used for model selection. We therefore evaluated whether the reductions in σ2 owing to model selection were greater than expected by chance by resampling the covariates. For each population time series, we resampled the covariates 100 times, refitted all models to the original population data (but using the resampled covariates), and recorded the resulting model selection estimates of σ2. For each dataset, we then calculated a p-value estimate as the proportion of times the estimate of σ2 corresponding to the resampled covariates were smaller than the true estimate of σ2. The lower the p-value, the more the original covariates decrease the prediction error relative to the resampled covariates. In order to preserve some of the potential autocorrelation of the covariates, the resampling was done by repeatedly sampling blocks of 3 consecutive years with replacement [37] and retrieving the weather covariates corresponding to those years. We resampled years rather than individual environmental variables in order to maintain any correlations between the covariates.
(f). Goodness of fit
Goodness of fit was checked by analysing the residuals of the AICc best model. We tested for autocorrelation of the residuals using the Ljung–Box test and for normality using the Shapiro test.
3. Results
After removing datasets that did not meet our selection criteria (see electronic supplementary material), 66 datasets on mammals, 225 on birds and 201 on insects remained and were used in further analyses. All these datasets were from either Europe or North America, with 47 per cent of the mammal datasets, 69 per cent of the bird datasets and 96 per cent of the insect datasets originating from Europe.
Of all the datasets analysed, the AICc best model failed to pass either the residual normality or the residual autocorrelation test at the 95 per cent level for 24 per cent of the datasets for the small collection of covariates and for 20 per cent for the large. Considering the tests individually, 14 per cent for the small collection and 11 per cent for the large collection failed the normality test, and 10 per cent failed the autocorrelation test for the both the small and the large collection. Autocorrelated residuals may be a sign of observation error [31], an autocorrelated environment [15], a stage-structured population or nonlinear dynamics [25]. Rather than including more complex models in our analysis at the risk of over-fitting, we tested the sensitivity of our results to goodness of fit by comparing results using all 492 datasets to results from the collection of datasets that passed both of the goodness-of-fit tests. Restricting the analyses to these data had only minor quantitative and no qualitative effects on the results.
(a). Effects of weather and climate
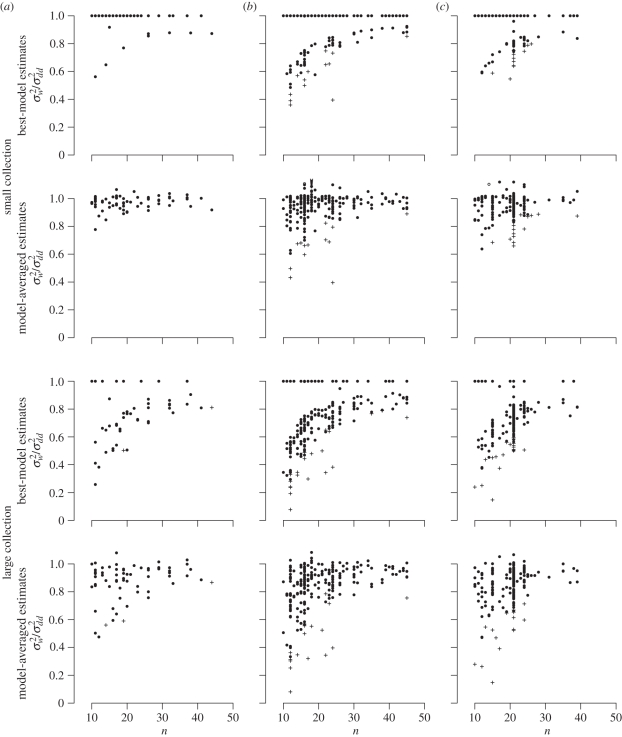
Of the total 492 datasets, the AICc best model included a covariate in 124 cases (25%) for the small collection and in 371 cases (75%) for the large collection. Within the taxonomic classes the best model included a covariate for 9 (14%) and 41 (62%) of the datasets for mammals, 64 (28%) and 170 (76%) of the datasets for birds, and 51 (25%) and 160 (80%) of the datasets for insects, for the small and large collections, respectively. Ignoring the risk of over-fitting, the best-model estimate of σ2 was reduced by up to 64 per cent for the small collection relative to the best model without covariates and up to 92 per cent for the large collection (figure 1). However, for only 19 of the datasets for the small collection and for 35 of the datasets for the large collection did the best model significantly reduce the prediction error variance relative to the resampled covariates at the 95 per cent level. The reduction in the model-averaged estimate of the prediction error variance was significant at the 95 per cent level in 22 cases for the small collection and in 31 cases for the large collection (figure 1). In general, model-averaged estimates, as expected, appeared smoother than the best-model estimates.
Figure 1.
The quotient between the variance estimates after model selection, 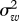 , and the variance estimate of the one of the two models without covariates with the lowest AICc value,
, and the variance estimate of the one of the two models without covariates with the lowest AICc value, 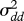 , plotted against time-series length, n. For the first and third rows, the
, plotted against time-series length, n. For the first and third rows, the 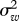 are best-model estimates, and for the second and fourth rows they are model-averaged estimates. The two upper rows show estimates under the small collection of covariates and the two lower rows show estimates under the large collection. (a) Mammal datasets, (b) birds and (c) insects. Datasets for which the reduction in variance was significant at the 95 per cent level according to the resample test are marked with a cross.
are best-model estimates, and for the second and fourth rows they are model-averaged estimates. The two upper rows show estimates under the small collection of covariates and the two lower rows show estimates under the large collection. (a) Mammal datasets, (b) birds and (c) insects. Datasets for which the reduction in variance was significant at the 95 per cent level according to the resample test are marked with a cross.
It is clear from the above that effects of the covariates were not strong enough to provide a high power of detecting their influence on any particular dataset. However, the simultaneous analyses of a large collection of datasets provided an opportunity to detect more subtle patterns. To explore this, we looked at the distribution of the resampled p-values across all datasets. Under a null hypothesis of no effect of the covariates the distribution is expected to be approximately uniform. A histogram showed that in fact the distribution of p-values corresponding to model-averaged estimates of σ2 appeared shifted to the left relative to the uniform (see electronic supplementary material, figure S1). This was confirmed by a Monte Carlo test where the distribution of p-values was compared with the distribution of p-values under the resampled covariates. As a test statistic, we used the integral of the difference between the empirical cumulative distribution function of the p-values and the cumulative distribution function of the uniform distribution (the identity function between 0 and 1). A positive value of the test statistic then indicates that the distribution of p-values is left-shifted relative to the uniform. The test came out significant for both the small and the large collection of covariates (p < 0.01). This means that the prediction error variances corresponding to the true covariates in general tended to be smaller than the variances for the resampled covariates, indicating that the weather and climate indices in fact do have an effect in reducing the prediction error for at least some datasets (although we cannot tell for which). Figure S1 in the electronic supplementary material also shows the distribution of q-values computed using the method of [38]. For a given p-value threshold to determine significance, the q-value is the estimated proportion of significant p-values that are falsely significant (type I errors)—that is, the proportion of false discoveries. For example, for a p-value threshold of 0.05, approximately 61 per cent of the 22 putatively significant weather effects are spurious for the small collection, and 17 per cent of the 31 effects for the large collection. Moreover, for the large collection of covariates the mean of the model-averaged prediction error variances across all datasets (the mean of σ2 was 0.40) was significantly smaller than the mean over each of the 100 batches of resampled covariates. The latter ranged from 0.40 to 0.42. The mean across all datasets was not significantly smaller than means for the resample batches for the small collection of covariates.
(b). Weather data or climate indices?
To investigate how NAO and SO perform relative to the weather data in reducing prediction error for the GPDD, we checked whether the climate indices were overrepresented in the AICc best model. To this end, we restricted attention to the datasets for which the best model included a covariate. We used a χ2-test to see if the proportion of datasets for which the best model had a weather, NAO or SO covariate differed from what would be expected if the covariate in the best model was uniformly distributed over all possible covariates (see electronic supplementary material, figure S2). The distribution differed significantly from the uniform only for the small collection of weather covariates (p < 0.01). This, however, was owing to an under-representation of the number of datasets for which the best model included an SO index. Such an under-representation might be expected because the SO mostly influences the Pacific region and only a few datasets were from western North America (36 of the total 492 datasets were sampled west of 100°W). Restricting these analyses only to datasets for which the best model had a weather or NAO covariate yielded no significant results. Hence there were no indications that NAO or SO was a better predictor than the weather covariates for the population data analysed here.
(c). How predictable are the population data?
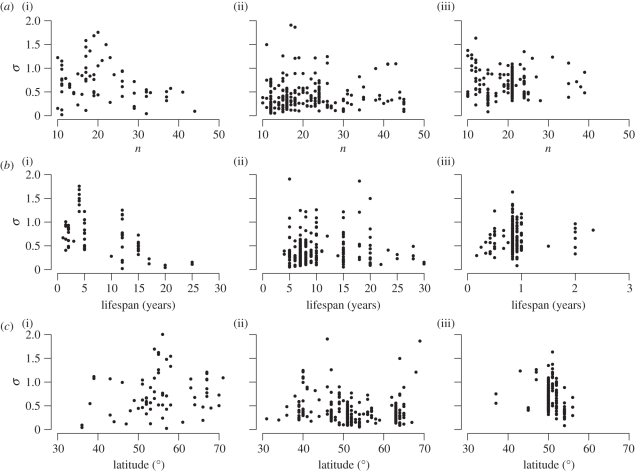
A fundamental question in population ecology and management is to what extent the dynamics of populations are predictable. It is therefore of interest to see how predictable the data in the GPDD are on an annual time scale using linear models, and whether predictability differs between mammals, birds and insects. Since the prediction errors here are modelled at the log scale, they are multiplicative at the population scale. For a prediction error standard deviation of 1 at the log scale, there is a 95 per cent probability that the population size one time step ahead will stay between μ exp (−1.96) ≈0.14 μ and μ exp (1.96) ≈7.10 μ, where μ is the median predicted population size. A prediction error standard deviation of 1 therefore means that the population size one time step ahead is highly unpredictable. For a prediction error standard deviation of 0.5 a 95 per cent one-step-ahead predictive interval is given by (0.38 μ, 2.66 μ) and for a standard deviation of 0.25 by (0.61 μ, 1.63 μ). Figure 2 shows the model-averaged estimates of σ for the large collection of covariates against time-series length, species lifespan (maximal age attained by individuals [28]) and latitude. A large portion of the datasets were only weakly predictable by the linear model at an annual scale (say σ > 0.5). While there were datasets that were highly unpredictable for all three taxonomic classes, there were fewer insect than mammal or bird time series with a small prediction error variance; for 1 per cent of the insect datasets, 14 per cent of the mammal datasets and 34 per cent of the bird datasets the model-averaged estimate of σ for the large collection was smaller than 0.25. Relations between predictability and time-series length, lifespan and latitude were generally weak, but prediction error variance decreased with length of lifespan for mammal data (figure 2).
Figure 2.
Model-averaged estimates of the prediction error standard deviation at the log scale for the large collection of covariates shown against (a) time-series length, (b) species lifespan and (c) latitude. (i) Mammal datasets, (ii) birds and (iii) insects.
4. Discussion
Our results show that the statistical signatures of weather on population dynamics, not to be confused with direct effects of weather on life-cycle components, are weak or absent in most datasets. Nonetheless, taken as a group, there is statistical evidence that weather plays a role in explaining the dynamics of at least some species. The broad picture obtained of the statistical signatures of weather effects could provide a reference against which to compare claims of relatively strong or weak effects (figure 1). Additionally, the hazards of over-fitting that have been raised in principle [12] have been broadly demonstrated in practice. For short time series, a drastic reduction in prediction error is required to separate a real effect from over-fitting and there is a relatively high probability, increasing with the number of models considered, that a covariate unrelated to the data will greatly reduce the prediction error (figure 1). At the same time, increasing the number of covariates increases the chance of including relevant variables. For any particular population, an effect of weather on population dynamics would however be difficult to detect and separate from over-fitting unless strong.
Climate indices have been shown to be strong predictors of the dynamics for a range of populations [17,39] and can outperform local weather variables by acting as weather packages [18]. For instance, winter NAO may capture different forms of severe winter weather potentially affecting winter mortality in many populations [40]. Since climate indices are to some extent predictable, there is hope that strong influence of these on populations may help ecologists to predict population dynamics [29]. Given the difficulty of detecting effects of weather even when relatively few covariates are considered, as demonstrated here, using climate indices as weather packages to replace a plethora of weather variables seems even more appealing. Unfortunately, although it is clear from previous studies that climate indices can have strong effects on populations, our results indicate that climate indices do not provide a general solution.
The biological effects of weather are likely to be stronger than the statistical signatures of weather in our analyses for several reasons. First, observation or sampling errors in the population indices or in weather covariates may have reduced the power of detecting real effects and diluted the magnitude of effects. If that is the sole cause, it is likely to be a common problem with ecological time series since many of the time series in the GPDD are taken from the scientific literature. Second, even our large set of covariates lacks nuances of weather impacts that could be important for individual populations. In a broad analysis of several hundred populations belonging to different species and taxonomic classes some compromises are unavoidable. For instance, for simplicity we chose to use the same types of covariates for all population time series (rather than selecting covariates based on species biology) in order to make results more easily interpretable and to avoid subjectivity in the selection. In an analysis of any particular population, using fewer and more carefully selected covariates will probably improve the power of detecting real effects of environmental covariates. Third, population dynamics is complex and detecting responses to environmental change may require detailed data on population structure as well as an understanding of the mechanisms leading from environmental to population change [5]. Our results are thus consistent with the view that the effects of variation in climate and weather on population dynamics are typically too complex to be readily picked up from time series on population abundance.
The variances of one-year-ahead predictions of the data are strikingly large for many of the populations we analysed. Relative to mammals and birds there are few insect datasets with a small residual variance (figure 2). Within the mammal datasets the prediction error variance decreases with lifespan. A decrease in prediction error variance with lifespan at an annual time scale may be because of an increase in overlap of generations. However, even for some long-lived birds and mammals the prediction error variance was large enough to make predictions very uncertain. This raises the question of whether population dynamics in general can be significantly better predicted when detailed demographic data and knowledge are available.
5. Conclusions
Due to their aggregate nature, time series on population abundance do not contain sufficient information to ultimately resolve mechanisms leading from environmental variation to population fluctuations [11,12]. However, time series analyses can be used to compare hypotheses and estimate the net effects of important environmental variables on populations. Our results show that, while off-the-shelf weather covariates do in general tend to have an effect on population time series, they can be difficult to pick up from any single dataset. For studies of specific populations, a priori hypotheses about which weather factors are important may reduce the number of covariates that need to be tested, thereby reducing the risk of over-fitting and increasing the power of the analysis. Hence, for detecting the impact of weather we recommend limiting analyses to just a few covariates for which there are well-founded reasons to believe they might have a strong effect on the dynamics. This applies to local weather variables as well as to climate indices.
Acknowledgements
We are indebted to Chongyang Wang for his help with extracting data for analysis and thank Chris Wilmers, Andreas Lindén and Jörgen Ripa for comments and suggestions that helped improve the manuscript. The research was supported by the Hellman Family Faculty Fund at the University of California, Berkeley.
References
- 1.Andrewartha H. G., Birch L. C. 1954. The distribution and abundance of animals. Chicago, IL: University of Chicago Press [Google Scholar]
- 2.Nicholson A. J. 1933. Supplement: the balance of animal populations. J. Anim. Ecol. 2, 131–178 [Google Scholar]
- 3.Bjørnstad O. N., Grenfell B. T. 2001. Noisy clockwork: time series analysis of population fluctuations in animals. Science 293, 638–643 10.1126/science.1062226 (doi:10.1126/science.1062226) [DOI] [PubMed] [Google Scholar]
- 4.Coulson T., Catchpole E. A., Albon S. D., Morgan B. J. T., Pemberton J. M., Clutton-Brock T. H., Crawley M. J., Grenfell B. T. 2001. Age, sex, density, winter weather, and population crashes in Soay sheep. Science 292, 1528–1531 10.1126/science.292.5521.1528 (doi:10.1126/science.292.5521.1528) [DOI] [PubMed] [Google Scholar]
- 5.Benton T. G., Plaistow S. J., Coulson T. N. 2006. Complex population dynamics and complex causation: devils, details and demography. Proc. R. Soc. B 273, 1173–1181 10.1098/rspb.2006.3495 (doi:10.1098/rspb.2006.3495) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lewellen R. H., Vessey S. H. 1998. The effect of density dependence and weather on population size of a polyvoltine species. Ecol. Monogr. 68, 571–594 10.1890/0012-9615(1998)068[0571:TEODDA]2.0.CO;2 (doi:10.1890/0012-9615(1998)068[0571:TEODDA]2.0.CO;2) [DOI] [Google Scholar]
- 7.Dennis B., Otten M. R. M. 2000. Joint effects of density dependence and rainfall on abundance of San Joaquin kit fox. J. Wildl. Manage. 64, 388–400 [Google Scholar]
- 8.Roy D. B., Rothery P., Moss D., Pollard E., Thomas J. A. 2001. Butterfly numbers and weather: predicting historical trends in abundance and the future effects of climate change. J. Anim. Ecol. 70, 201–217 10.1046/j.1365-2656.2001.00480.x (doi:10.1046/j.1365-2656.2001.00480.x) [DOI] [Google Scholar]
- 9.Stige L. C., Chan K., Zhang Z., Frank D., Stenseth N. C. 2007. Thousand-year-long Chinese time series reveals climatic forcing of decadal locust dynamics. Proc. Natl Acad. Sci. USA 104, 16 188–16 193 10.1073/pnas.0706813104 (doi:10.1073/pnas.0706813104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Saether B., Lillegård M., Grøtan V., Drever M. C., Engen S., Nudds T. D., Podruzny K. M. 2008. Geographical gradients in the population dynamics of North American prairie ducks. J. Anim. Ecol. 77, 869–882 10.1111/j.1365-2656.2008.01424.x (doi:10.1111/j.1365-2656.2008.01424.x) [DOI] [PubMed] [Google Scholar]
- 11.Månsson L., Lundberg P. 2006. An analysis of the analysis of herbivore population dynamics. Oikos 113, 217–225 10.1111/j.2006.0030-1299.14362.x (doi:10.1111/j.2006.0030-1299.14362.x) [DOI] [Google Scholar]
- 12.Krebs C. J., Berteaux D. 2006. Problems and pitfalls in relating climate variability to population dynamics. Clim. Res. 32, 143–149 10.3354/cr032143 (doi:10.3354/cr032143) [DOI] [Google Scholar]
- 13.Berryman A., Turchin P. 2001. Identifying the density-dependent structure underlying ecological time series. Oikos 92, 265–270 10.1034/j.1600-0706.2001.920208.x (doi:10.1034/j.1600-0706.2001.920208.x) [DOI] [Google Scholar]
- 14.Saether B., Grotan V., Engen S., Noble D. G., Freckleton R. P. 2009. Critical parameters for predicting population fluctuations of some British passerines. J. Anim. Ecol. 78, 1063–1075 10.1111/j.1365-2656.2009.01565.x (doi:10.1111/j.1365-2656.2009.01565.x) [DOI] [PubMed] [Google Scholar]
- 15.Jonzén N., Lundberg P., Ranta E., Kaitala V. 2002. The irreducible uncertainty of the demography–environment interaction in ecology. Proc. R. Soc. Lond. B 269, 221–225 10.1098/rspb.2001.1888 (doi:10.1098/rspb.2001.1888) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ripa J., Ives A. R. 2007. Interaction assessments in correlated and autocorrelated environments. In The impact of environmental variability on ecological systems (eds Vasseur D. A., McCann K. S.), pp. 111–113 Dordrecht, The Netherlands: Springer [Google Scholar]
- 17.Post E., Stenseth N. C. 1999. Climatic variability, plant phenology, and northern ungulates. Ecology 80, 1322–1339 10.1890/0012-9658(1999)080[1322:CVPPAN]2.0.CO;2 (doi:10.1890/0012-9658(1999)080[1322:CVPPAN]2.0.CO;2) [DOI] [Google Scholar]
- 18.Hallett T. B., Coulson T., Pilkington J. G., Clutton-Brock T. H., Pemberton J. M., Grenfell B. T. 2004. Why large-scale climate indices seem to predict ecological processes better than local weather. Nature 430, 71–75 10.1038/nature02708 (doi:10.1038/nature02708) [DOI] [PubMed] [Google Scholar]
- 19.Mitchell T. D., Jones P. D. 2005. An improved method of constructing a database of monthly climate observations and associated high-resolution grids. Int. J. Climatol. 25, 693–712 10.1002/joc.1181 (doi:10.1002/joc.1181) [DOI] [Google Scholar]
- 20.Inchausti P., Halley J. 2001. Investigating long-term ecological variability using the global population dynamics database. Science 293, 655–657 10.1126/science.293.5530.655 (doi:10.1126/science.293.5530.655) [DOI] [PubMed] [Google Scholar]
- 21.Brook B. W., O'Grady J. J., Chapman A. P., Burgman M. A., Akçakaya H. R., Frankham R. 2000. Predictive accuracy of population viability analysis in conservation biology. Nature 404, 385–387 10.1038/35006050 (doi:10.1038/35006050) [DOI] [PubMed] [Google Scholar]
- 22.Brook B. W., Bradshaw C. J. A. 2006. Strength of evidence for density dependence in abundance time series of 1198 species. Ecology 87, 1445–1451 10.1890/0012-9658(2006)87[1445:SOEFDD]2.0.CO;2 (doi:10.1890/0012-9658(2006)87[1445:SOEFDD]2.0.CO;2) [DOI] [PubMed] [Google Scholar]
- 23.Sibly R. M., Barker D., Denham M. C., Hone J., Pagel M. 2005. On the regulation of populations of mammals, birds, fish, and insects. Science 309, 607–610 10.1126/science.1110760 (doi:10.1126/science.1110760) [DOI] [PubMed] [Google Scholar]
- 24.Sibly R. M., Barker D., Hone J., Pagel M. 2007. On the stability of populations of mammals, birds, fish and insects. Ecol. Lett. 10, 970–976 10.1111/j.1461-0248.2007.01092.x (doi:10.1111/j.1461-0248.2007.01092.x) [DOI] [PubMed] [Google Scholar]
- 25.Ziebarth N. L., Abbott K. C., Ives A. R. 2010. Weak population regulation in ecological time series. Ecol. Lett. 13, 21–31 10.1111/j.1461-0248.2009.01393.x (doi:10.1111/j.1461-0248.2009.01393.x) [DOI] [PubMed] [Google Scholar]
- 26.NERC Centre for Population Biology, Imperial College 1999. The global population dynamics database. See http://www.sw.ic.ac.uk/cpb/cpb/gpdd.html. [Google Scholar]
- 27.Kendall B. E., Prendergast J., Bjørnstad O. N. 1998. The macroecology of population dynamics: taxonomic and biogeographic patterns in population cycles. Ecol. Lett. 1, 160–164 10.1046/j.1461-0248.1998.00037.x (doi:10.1046/j.1461-0248.1998.00037.x) [DOI] [Google Scholar]
- 28.Brook B. W., Traill L. W., Bradshaw C. J. A. 2006. Minimum viable population sizes and global extinction risk are unrelated. Ecol. Lett. 9, 375–382 10.1111/j.1461-0248.2006.00883.x (doi:10.1111/j.1461-0248.2006.00883.x) [DOI] [PubMed] [Google Scholar]
- 29.Stenseth N. C., Ottersen G., Hurrell J. W., Mysterud A., Lima M., Chan K., Yoccoz N. G., Adlandsvik B. 2003. Review paper. Studying climate effects on ecology through the use of climate indices: the North Atlantic Oscillation, El Niño Southern Oscillation and beyond. Proc. R. Soc. Lond. B 270, 2087–2096 10.1098/rspb.2003.2415 (doi:10.1098/rspb.2003.2415) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.R Development Core Team 2009. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing [Google Scholar]
- 31.Dennis B., Ponciano J. M., Lele S. R., Taper M. L., Staples D. F. 2006. Estimating density dependence, process noise, and observation error. Ecol. Monogr. 76, 323–341 10.1890/0012-9615(2006)76[323:EDDPNA]2.0.CO;2 (doi:10.1890/0012-9615(2006)76[323:EDDPNA]2.0.CO;2) [DOI] [Google Scholar]
- 32.Knape J. 2008. Estimability of density dependence in models of time series data. Ecology 89, 2994–3000 10.1890/08-0071.1 (doi:10.1890/08-0071.1) [DOI] [PubMed] [Google Scholar]
- 33.Hurvich C. M., Tsai C. 1989. Regression and time series model selection in small samples. Biometrika 76, 297–307 10.1093/biomet/76.2.297 (doi:10.1093/biomet/76.2.297) [DOI] [Google Scholar]
- 34.Buckland S. T., Burnham K. P., Augustin N. H. 1997. Model selection: an integral part of inference. Biometrics 53, 603–618 10.2307/2533961 (doi:10.2307/2533961) [DOI] [Google Scholar]
- 35.Hjort N. L., Claeskens G. 2003. Frequentist model average estimators. J. Am. Stat. Assoc. 98, 879–899 10.1198/016214503000000828 (doi:10.1198/016214503000000828) [DOI] [Google Scholar]
- 36.Grosbois V., Gimenez O., Gaillard J.-M., Pradel R., Barbraud C., Clobert J., Møller A. P., Weimerskirch H. 2008. Assessing the impact of climate variation on survival in vertebrate populations. Biol. Rev. 83, 357–399 10.1111/j.1469-185X.2008.00047.x (doi:10.1111/j.1469-185X.2008.00047.x) [DOI] [PubMed] [Google Scholar]
- 37.Efron B., Tibshirani R. J. 1993. An introduction to the bootstrap. New York, NY: Chapman & Hall [Google Scholar]
- 38.Storey J. D., Tibshirani R. 2003. Statistical significance for genomewide studies. Proc. Natl Acad. Sci. USA 100, 9440–9445 10.1073/pnas.1530509100 (doi:10.1073/pnas.1530509100) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Saether B., Sutherland W. J., Engen S. 2004. Climate influences on avian population dynamics. In Birds and climate change (eds Moller A. P., Fiedler W., Berthold P.), pp. 185–209 Amsterdam, The Netherlands: Academic Press [Google Scholar]
- 40.Stenseth N. C., Mysterud A. 2005. Weather packages: finding the right scale and composition of climate in ecology. J. Anim. Ecol. 74, 1195–1198 10.1111/j.1365-2656.2005.01005.x (doi:10.1111/j.1365-2656.2005.01005.x) [DOI] [Google Scholar]