Abstract
Amide hydrogen/deuterium exchange (HDX) rate constants of bovine ubiquitin in an ammonium acetate solution containing 1% of the electrospray ionization (ESI) “supercharging” reagent m-nitrobenzyl alcohol (m-NBA) were obtained using top-down, electron transfer dissociation (ETD) tandem mass spectrometry (MS). The supercharging reagent replaces the acid and temperature “quench” step in the conventional MS approach to HDX experiments by causing rapid protein denaturation to occur in the ESI droplet. The higher charge state ions that are produced with m-NBA are more unfolded, as measured by ion mobility, and result in higher fragmentation efficiency and higher sequence coverage with ETD. Single amino acid resolution was obtained for 44 of 72 exchangeable amide sites, and summed kinetic data were obtained for regions of the protein where adjacent fragment ions were not observed, resulting in an overall spatial resolution of 1.3 residues. Comparison of these results with previous values from NMR indicates that the supercharging reagent does not cause significant structural changes to the protein in the initial ESI solution, and that scrambling or back-exchange is minimal. This new method for top-down HDX-MS enables real-time kinetic data measurements under physiological conditions, similar to those obtained using NMR, with comparable spatial resolution and significantly better sensitivity.
INTRODUCTION
The rates of backbone amide hydrogen deuterium exchange (HDX) in solution can provide a wealth of information about the structures and conformational dynamics of proteins and protein complexes. There is a substantial reduction of the exchange rates of different amide sites as a result of either intramolecular hydrogen bonding or from sequestration of the amide hydrogen atoms from bulk solvent, making these exchange rates a useful probe of higher-order structure and thermodynamics of local or global unfolding events.1 Since the early studies of Linderstrom-Lang,2 measurements of HDX have been made using a variety of different analytical techniques, including NMR,3 IR,4 and Raman5 spectroscopies, and mass spectrometry (MS).6–9 NMR has the advantage that exchange rate information about individual amides is obtained, which is the ultimate spatial resolution possible with HDX. Mass spectrometry has the advantage of high sensitivity, and it can be applied to much larger proteins. However, the spatial resolution with which information about HDX is obtained with mass spectrometry is often limited to relatively large regions of the protein defined by peptides formed by proteolysis.
HDX coupled to MS is frequently performed with the “bottom-up” approach,6,10,11 although the more recently developed “top-down” approach appears promising.12 In the former, fully protonated or deuterated protein is rapidly diluted into either D2O or H2O, respectively, allowed to exchange for a fixed period of time, and then “quenched” by lowering the pH (and temperature) into a range where the intrinsic exchange rate is a minimum (pH ~3.0).1 The protein is then proteolyzed with an enzyme that is active in acidic solution, such as pepsin, and the resulting peptides are separated by liquid chromatography (LC) and either mass analyzed directly or separated further in the gas phase and fragmented by tandem MS methods. During the proteolysis and LC steps, amides that were protected during the exchange period become exposed to the bulk solvent, which can cause some back-exchange and concomitant loss of HDX information. This effect can be minimized, though not eliminated completely, with online pepsin columns, short chromatography methods, and/or back-exchange correction factors.9,13,14 A significant advantage of the bottom-up approach is the theoretically unlimited mass range for the analyte, including large multimeric assemblies,15–17 although there is an irreversible loss of HDX information for two amides for each proteolytic cleavage.18
In the top-down approach, the exchange and quench steps are the same, but rather than proteolysis and LC-MS or LC-MS/MS, the intact protein is fragmented in the gas phase. A significant advantage of the top-down approach is the potential for single amino acid spatial resolution compared to the traditional bottom-up approach where the spatial resolution is limited by the length and overlap of the peptides from proteolysis. Slow heating methods that are commonly used to fragment gas-phase peptides and proteins, such as collisional activation, may cause scrambling, where intramolecular proton or hydrogen atom transfer can lead to loss of information about where exchange originally occurred in solution.19–21 In contrast, recent work by a number of researchers has shown that scrambling is largely reduced when intact proteins or large peptides are fragmented with either electron capture dissociation18,22–24 (ECD) or electron transfer dissociation (ETD).25–27 The minimal scrambling that occurs with either ECD or ETD may be due to either nonergodic dissociation, where cleavage occurs before the recombination energy has time to dissipate in the molecule,28,29 or the reduced precursors may dissociate before scrambling occurs because of low activation barriers for dissociation.30,31 In either case, there is a growing consensus that HDX coupled to ECD or ETD mass spectrometry can be a viable approach to obtain accurate amide exchange rate information18,22–27 with close to single amino acid spatial resolution possible.
With both the bottom-up and top-down approach, the quench step affects the results of the experiment in multiple ways. As described above, the primary effect is to reduce the pH to near the intrinsic exchange rate minimum, although this rate is still fast enough at ~6 × 10−3 min−1 at 0 °C (attenuated by the adjacent sidechains and local environment)1 to cause some back-exchange during any delay between the quench step and transfer to the gas phase. Decreasing the pH can have the additional advantage of unfolding the protein and increasing the charge states of ions formed by electrospray ionization (ESI), which can dramatically improve both the sequence coverage and capture efficiency in ECD32,33 or ETD34 experiments.
Increased analyte charging in ESI can also be obtained by adding small amounts of some “supercharging” reagents to the initial solution from which these ions are formed.35–37 For example, ESI of a water/methanol/acetic acid solution containing 1% m-nitrobenzyl alcohol (m-NBA) and cytochrome c resulted in an increase in the average charge state from 17.3+ to 20.8+ and the maximum observed charge state from 21+ to 24+ compared to the same solution without m-NBA.36 Originally used with “denaturing” solutions consisting of water, methanol and acetic acid,35–37 m-NBA and other supercharging reagents were shown by Loo and coworkers to increase the charge states of proteins and protein complexes formed from aqueous solutions.38,39 Although many factors, including proton transfer reactivity,40 instrumental conditions,41 surface tension,37 etc., can affect charging in ESI, the charge enhancement observed for proteins and protein complexes in aqueous solutions containing a supercharging reagent appears to be predominantly a result of chemical and/or thermal denaturation that occurs in the electrospray droplet prior to ion formation.42,43 Sulfolane, a supercharging reagent found to be effective by Loo et al.,38 destabilizes the native form of myoglobin by 1.5 kcal/mol/M, but has little effect on the native structure in the initial solution owing to the typically low concentrations used.43 Because of the high boiling points of these supercharging reagents compared to water (b.p. of m-NBA and sulfolane are 177 °C at 3 Torr44 and 287 °C at 760 Torr,44 respectively), the concentration of these supercharging reagents in the ESI droplets increases as water preferentially evaporates from the droplet, and can cause significant unfolding of the proteins in the droplet due to chemical and/or thermal denaturation.42,43 It is well known that the structure of the protein in solution can significantly affect the charging observed in ESI, where more unfolded structures become more highly charged.45–47
Here, we demonstrate that adding 1% m-NBA to aqueous solutions containing the protein ubiquitin can significantly increase charging. Traveling wave ion mobility spectrometry (TWIMS) results show that the higher charge state ions that are formed with m-NBA are more unfolded than the lower charge state ions that are formed without this supercharging reagent, and ETD of these more highly charged and unfolded ions results in significantly improved fragment ion abundance and sequence coverage. When ESI supercharging is combined with top-down HDX-MS, the quench step and the concomitant loss of information due to back-exchange can be eliminated, and the experiments can be performed continuously at physiological pH and temperature, as is done with NMR, but has the advantage of higher sensitivity. This new method could make high spatial resolution, top-down HDX-MS a much simpler and faster experiment to perform, without HDX information loss due to back-exchange or scrambling.
EXPERIMENTAL
All solutions of bovine ubiquitin were prepared from lyophilized solid (Sigma, St. Louis, MO, USA) without additional purification. Mass spectra for all HDX-MS experiments were acquired using an LTQ-orbitrap hybrid mass spectrometer (Thermo Fischer Scientific, Waltham, MA, USA). ETD experiments were performed by reacting mass-selected precursor ions with fluoranthene anions for 30 ms. Fully deuterated bovine ubiquitin was prepared (~99% deuterium incorporation measured using mass spectrometry). Exchange for hydrogen was initiated by diluting a 1 µL aliquot of the deuterated ubiquitin solution into 99 µL of 200 mM ammonium acetate, pH = 6.2, containing 1.0% m-nitrobenzyl alcohol (Sigma, St. Louis, MO, USA). The sample was mixed for a few seconds before a ~7 µL aliquot was loaded into a nanospray capillary, ESI was established, and the first ETD spectrum was acquired within approximately two minutes. The same capillary was used to obtain data up to 93 minutes, and new capillaries were used for the 185 and 385 minute timepoints. Fragment ions were identified using a m/z uncertainty constraint of ± 0.005. ESI mass spectra and TWIMS arrival time distributions of individual charge states of ubiquitin were acquired using a hybrid quadrupole/ion mobility/time-of-flight instrument (Synapt™ High Definition Mass Spectrometer; Waters, Milford, MA, USA) equipped with a Z-spray ion source. Experimental procedures are described in detail in Supporting Information.
RESULTS AND DISCUSSION
Supercharged ESI-TWIMS-MS
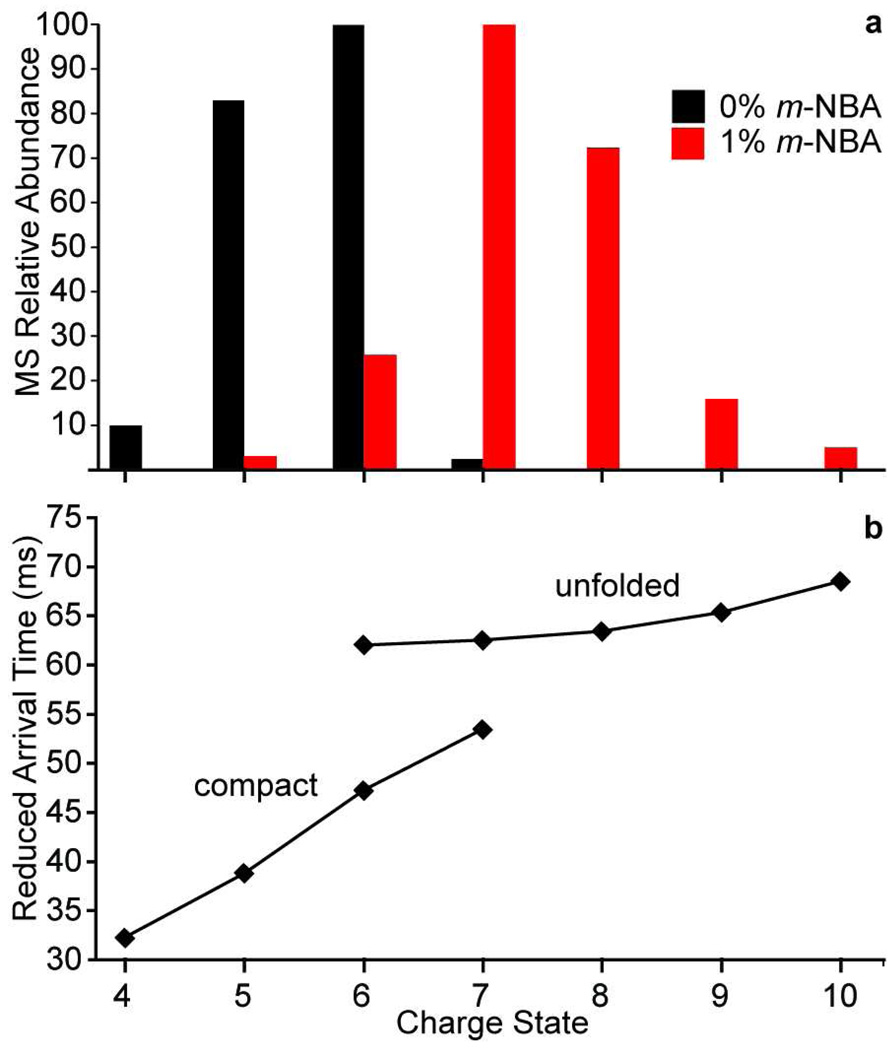
Relative abundances of charge states of ubiquitin formed by ESI (20 mM aqueous ammonium acetate, pH 7.0) from solutions that contain either 0% or 1.0% m-NBA are shown in Figure 1a. Without m-NBA, charge states ranging from 4+ to 7+ are formed. With 1% m-NBA, the maximum charge state increases from 7+ to 10+, and the average charge increases from 5.5+ to 7.4+. The corresponding arrival time distributions obtained by TWIMS for each of these charge states are used to calculate the reduced arrival times by multiplying the centroid of a given TWIMS peak by the ion charge.48 These reduced arrival times as a function of charge are shown in Figure 1b. The data are consistent with two generally separate families of conformers with the more compact conformations observed at lower charge states and the more unfolded or elongated conformers observed at higher charge states, with two distinct conformers, or families of conformations, observed for the 6+ and 7+ charge states. These data also indicate a trend of additional unfolding with increasing charge state within these conformer families. Koeniger and Clemmer measured the absolute collision cross sections of ubiquitin ions formed by ESI from a 49:49:2 water:acetonitrile:acetic acid solution and found that the 5+ – 7+ charge states had nearly identical collision cross sections, consistent with a compact conformation or family of conformers.49 A transition to multiple partially folded and elongated conformations was observed for the 8+ and 9+ ions, and multiple elongated conformations were observed for 10+ – 13+ ions. Here, the transition to a more unfolded structure(s) occurs at a lower charge state (between 6+ and 7+). This may be due to ion heating in the traveling wave apparatus that can cause unfolding,50 and/or due to the different solution compositions or ESI source conditions used. Nonetheless, these TWIMS results indicate that the higher charge state ions obtained by supercharging with m-NBA are significantly more unfolded than the low charge state ions that are formed with or without this supercharging reagent. Similar results were obtained for myoglobin ions formed by electrospray from a purely aqueous solution containing either m-NBA or sulfolane,43 suggesting that the supercharging effect is not protein-specific and should be broadly applicable for inducing rapid protein unfolding (and/or analyte heating) in an ESI droplet. The ESI mass spectra and TWIMS arrival time distributions from which the results in Figure 1 are derived are given in the Supporting Information.
Figure 1.
Plot of the (a) ESI-MS relative charge state abundances of 10 µM ubiquitin (20 mM aqueous ammonium acetate, pH 7.0) formed with and without 1% m-NBA, and (b) TWIMS reduced arrival times as a function of charge state, which shows two distinct families of conformers.
Supercharged ESI-MS/MS
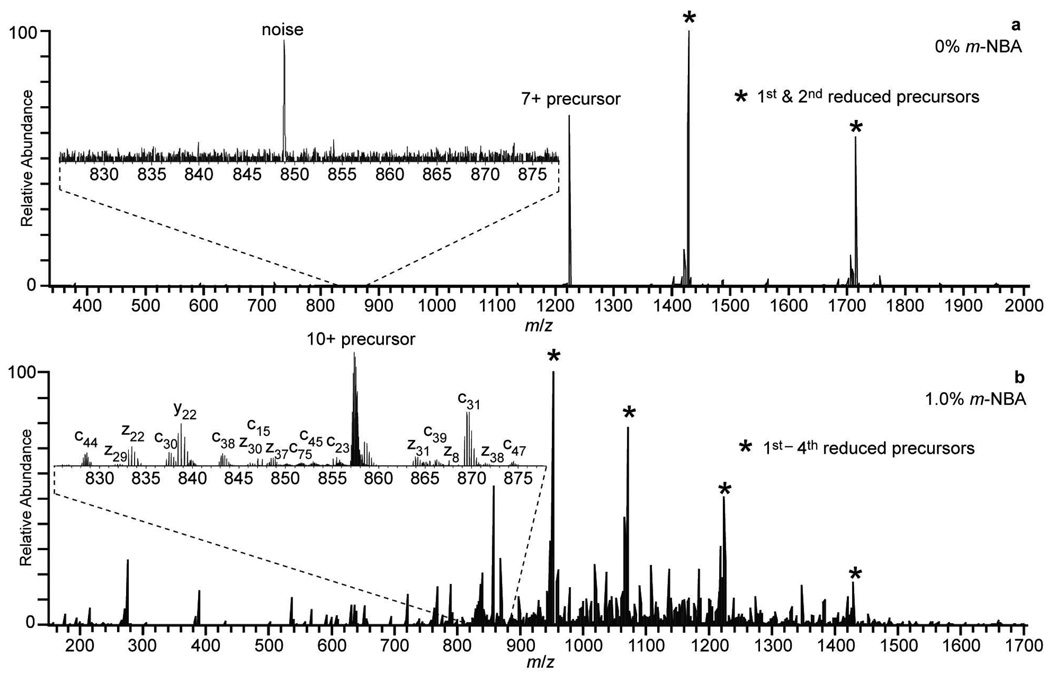
The effects of charge state on ETD sequence coverage with supercharging reagent were investigated. ETD of the isolated 7+, the maximum charge state observed without m-NBA, results primarily in formation of the 6+ and 5+ charge-reduced precursors (Figure 2a). Some c and z• ions are observed, albeit in very low abundance. Charge-reduced precursors are also formed upon ETD of the isolated 10+, the highest charge state observed with 1.0% m-NBA, but in striking contrast to the results without m-NBA, significantly more fragmentation is observed (Figure 2b). These results clearly show the advantage of combining supercharging reagents, such as m-NBA, with ETD experiments even when these ions are formed from buffered aqueous solutions.
Figure 2.
Electron transfer dissociation mass spectra of the isolated (a) 7+ and (b) 10+ ions obtained from a 3.75 µM ubiquitin, 200 mM ammonium acetate, pH 6.2, solution containing 0% and 1% m-NBA, respectively. In both cases, these were the highest charge states that could be reliably isolated with sufficient S/N to perform ETD. The reduced precursors are indicated with a *. Insets are expansions of the m/z 827 – 877 regions to illustrate differences in fragmentation efficiencies.
The sequence coverage results obtained from the c and z• ions formed by ETD of these two charge states are compared in Figure 3. For the 7+ formed without m-NBA, there are six c and 17 z• ions resulting in 25% sequence coverage. There are no cleavages between residues 35 and 74, so only limited sequence information is obtained from the N-terminal end of the protein. For the 10+ charge state formed with 1.0% m-NBA, there are 41 c and 47 z• ions, with cleavage at 62 of 72 unique sites resulting in 86% sequence coverage. For the 18 common fragment ions that are observed in both the 10+ and 7+ ETD spectra, the abundances, normalized for charge, are ~12× greater in the 10+ ETD spectrum indicating a substantially higher fragmentation efficiency for the higher charge state ion. Five of the 23 fragment ions produced by ETD of the 7+ are not observed in the ETD spectrum of the 10+, but the sequence coverage obtained with the 10+ is significantly higher and is more uniformly distributed. The unique fragmentation observed for the 7+ is likely due to conformer dependent dissociation as has been observed previously with ECD of different ubiquitin conformers separated by ion mobility.51 The sequence coverage values obtained here are somewhat higher than the 17% and 59% sequence coverage for 7+ and 10+ ions, respectively, obtained by McLafferty and coworkers using ECD.32
Figure 3.
Sequence coverage of ubiquitin obtained from ETD of the 7+ and 10+ charge states of 3.75 µM ubiqutin formed by electrospray from a 200 mM ammonium acetate, pH 6.2, solution containing 0% and 1% m-NBA, respectively. 25% and 86% sequence coverage was obtained for the 7+ and 10+, respectively.
Even higher charge states can be obtained from solutions containing organic solvents and acid. For example, the 13+ of ubiquitin is formed with the “quench” solution conditions used by Konermann and coworkers in their top-down HDX experiment (1:1 water:acetonitrile with 0.2% formic acid).24 To determine if this higher charge state results in greater sequence coverage, ETD of the 13+ and 10+ were compared, adjusting the fluoranthene anion abundances so that the precursors were depleted to a similar extent. The sequence coverage for the 13+ formed from the water/acetonitrile/formic acid solution was 81% compared to the 86% obtained from the 10+ formed by supercharging from aqueous solution, consistent with previous results that show protein shape can play a greater role in fragmentation efficiency than charge state.51 The similar sequence coverage for these two ions formed from solutions where the protein conformations differ indicates that the extent of unfolding caused by the supercharging reagent during the electrospray process is adequate for high sequence coverage with ETD.
Amide HDX Kinetics at Individual Residues
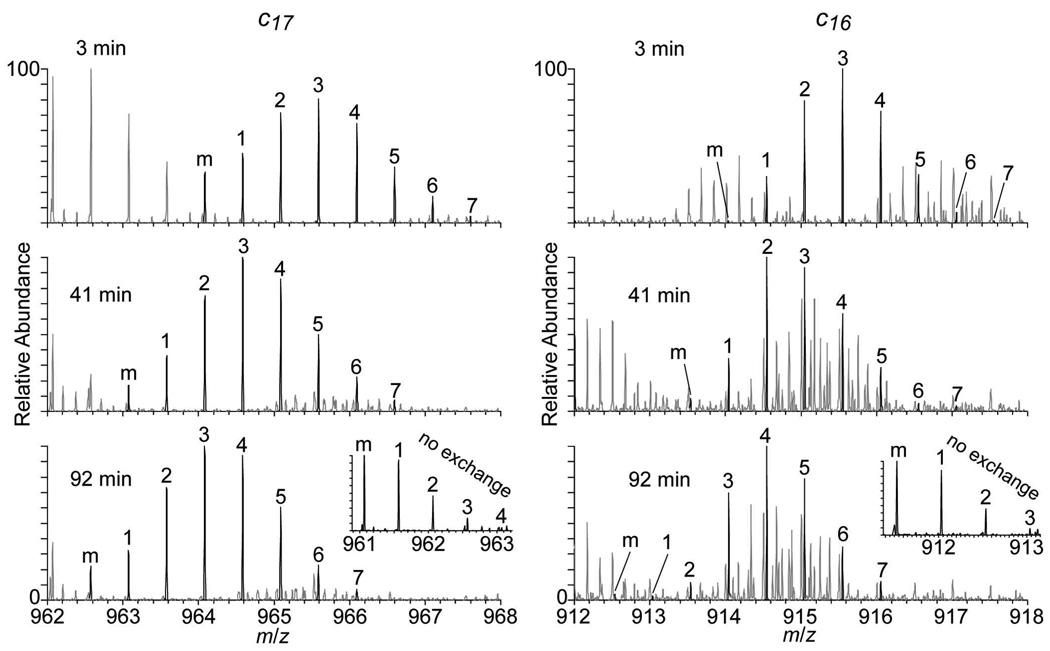
ETD spectra of fully deuterated 10+ ubiquitin (formed with 1.0% m-NBA) exchanging with hydrogen as a function of time were obtained for times up to 385 minutes using a single nanospray capillary (~7 µL) to acquire data from 2 to 94 minutes and two additional capillaries to obtain data at 185 and 385 minutes. Amide exchange kinetics for individual residues were obtained from these kinetic data. An example of partial ETD mass spectra showing the c17 and c16 fragments measured at 3, 41, and 92 minutes are shown in Figure 4. The isotopic distributions of both ions shift to lower m/z with longer times as deuterium is exchanged for hydrogen. The isotopic distributions of the fragment ions in these exchange experiments are significantly broader than the natural isotopic distribution without exchange (Figure 4, inset) owing to the statistical nature of the HDX process. The average deuterium content for each fragment ion is obtained from the difference in the average molecular weights of the exchanged and unexchanged fragments calculated using the intensity weighted average of the observed isotopic peaks. The difference in the average deuterium content in two sequential fragment ions provides information about the extent of exchange at the individual amide site.
Figure 4.
Partial ETD mass spectra of fully deuterated 10+ ubiquitin showing the c17 (left) and c16 (right) fragment ions obtained at 3, 41, and 92 minutes during exchange for hydrogen. The monoisotopic peak (m) and each isotope peak in the distribution are labeled. Partial ETD spectra for the fully protonated c17 and c16 fragment ions obtained without HDX are inset.
For example, the difference in the average deuterium content of the c17 and c16 fragments as a function of time provides information about the amide exchange kinetics at residue 18 (Glu18). The average deuterium content for the Glu18 amide obtained from these data as a function of time is shown in Figure 5a (open squares). It is apparent from these data that this site is essentially completely exchanged by ~100 minutes. The scatter in these data, as well as the slightly negative values at the two longer times (185 and 385 min.) can be attributed to subtracting two relatively large numbers corresponding to all the exchange that occurs in the c17 and c16 fragments, where a small relative error in either or both average exchange values lead to a large relative error for the difference in these values. These data can be reasonably fit with a single exponential function from which an HDX rate constant of 4.5 × 10−2 min−1 is obtained for the amide of Glu18.
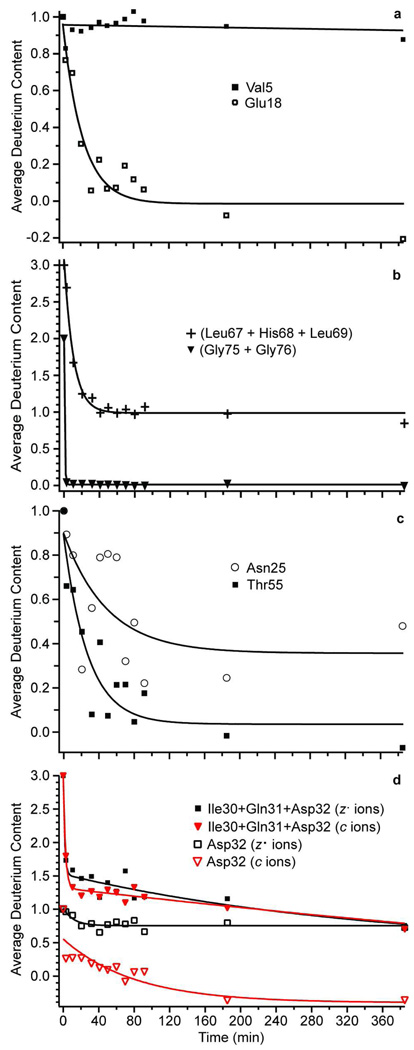
Figure 5.
Example hydrogen/deuterium exchange kinetics, where single residue resolution was obtained for (a) Val5 and Glu18 and (b) a three-residue sum and a two-residue sum were obtained for Leu67, His68, and Leu69, and Gly75 and Gly76, respectively. Single residue resolution results for (c) Asn25 and Thr55 illustrate decay to different average deuterium content endpoints. Exchange kinetics for (d) Asp 32 plotted with c ion data (red) and z• ion data (black), and as part of a three-residue in silico digestion (Ile30 + Gln31 + Asp32). Data were fit to single or double exponential functions where appropriate.
Exchange rate constants with single residue resolution were obtained for 44 of the 72 backbone amides (76 residues – the N-terminus and three prolines) residues. This resolution is slightly higher than that obtained by Konermann and coworkers (39 of 72) using ECD of all 5+ through 12+ charge states.24 The exchange rate constants for these residues varied between >1.4 and <3.5 × 10−3 min−1, which represents a three orders of magnitude kinetic window for this experiment as implemented here (Table S-1; Supporting Information). For example, single residue resolution was obtained for Val5, but in contrast to Glu18, very little exchange for hydrogen was observed over the entire 385 minute experiment (filled squares, Figure 5a) resulting in a lower limit to the exchange rate constant of 3.5 × 10−3 min−1. Significant protection of this amide from the bulk solvent can be inferred from this very slow exchange constant and is consistent with its protected position within a beta strand.52 Adjacent to Val5 on both the N- and C-terminal sides, very fast exchange occurs at residues 1–2 and 7–12 where essentially complete exchange occurs in less than three minutes. The vastly different rates of exchange at these neighboring amide exchange sites clearly indicate that neither H/D scrambling nor back-exchange occurs to any significant extent at these sites.
Amide HDX Kinetics at Contiguous Residues
For residues where sequential fragment ions are not observed, the next nearest neighbor is used to calculate the summed HDX kinetics for the residues between where cleavages occur. Examples of summed kinetic data are shown in Figure 5b for a two-amide sum, and a three-amide sum. The summed HDX kinetics of Leu67, His68, and Leu69 (+ symbol) are best fit to a single exponential with a half-life of ~7 minutes and with a constant offset corresponding to ~1 deuterium. The data for these three residues are consistent with one well-protected amide and two amides with similar rates of exchange. Although three exchange rate constants can be obtained for these three residues, no information about which of the three residues is the slow exchanger is obtained. For Gly75 and Gly76 (filled triangles), nearly complete exchange occurred by the first measurement, indicating both amides exchange on a timescale near the upper limit of exchange for the experiment (>1.4 min−1).
There are 12 regions of the protein where summed kinetics were obtained, with the longest section corresponding to five residues (Lys48 – Asp53). The average spatial resolution observed with this supercharged ESI HDX-MS approach is calculated as the average of all the segment lengths (including single residues) and is equal to 1.3 residues. This is significantly higher spatial resolution than was previously obtained for HDX-MS analysis of ubiquitin using either pepsin proteolysis and direct mass analysis of the proteolytic fragments53 or using collisionally activated dissociation of the intact protein,54 and somewhat higher resolution than was reported for a single time-point analysis of ubiquitin exchange kinetics using an acid-quenched, top-down approach.24 To the best of our knowledge, this is the first example of using top-down mass spectrometry to report exchange rate constants for H/D exchange of a protein, whereas reporting rate constants (or protection factors) is common with NMR experiments.55–58
Redundant Information to Reduce Uncertainty
Some of the HDX kinetic data can lead to ambiguities in interpretation, but there is redundant information in these data that can eliminate some uncertainty in individual rate constants. For example, Asn25 and Thr55 have similar initial exchange rate constants, but Asn25 appears to be biexponential with ~40% fully exchanged by ~200 minutes (open circles; Figure 5c), with little exchange occurring after that, whereas Thr55 can be fit to a single exponential function. The apparent biexponential behavior observed for Asn25 could either be due to scatter in the data and error induced by subtracting two relatively large numbers, where a small relative uncertainty in one or both values leads to a large relative uncertainty in the subtracted value. Or it could indicate two or more different conformations in solution and/or dynamical events that reside in the EX2 regime54 on a much longer timescale than the duration of this experiment. If these data were truly biexponential, adjacent residues should have similar offsets. The HDX kinetics for the residues adjacent to Asn25 do not show a similar ~0.4 Da offset, suggesting it is more likely due to a bias in one (or both) sets of fragment ion data.
Redundant data can also occur in regions of the protein where both c and z• ions overlap. This occurred for four residues, and statistically indistinguishable rate constants were obtained from both the c and z• ions data for Thr12, Lys29, and Leu56 (Table S-1; Supporting Information), whereas different kinetic data was obtained for Asp32 (Figure 5d). An offset value of –0.4 provided a best fit for the c ion data (open triangles), whereas the z• ion data (open square) is biexponential and indicates a significant fraction of this population does not exchange on the time frame of this experiment. The discrepancy between the results obtained for these c and z• ions at this one site may be due to different conformations of the protein that are probed individually by the respective c and z• ions,51 or it could be due to the previously discussed error induced by the subtraction.
One approach to determine if the information obtained from the both c and z• ions for Asp32 is different is to use summed kinetics to investigate the region of the primary sequence immediately adjacent to this residue to see how they are affected. The summed kinetic data for Ile30 + Gln31 + Asp32 are shown in Figure 5d. The resulting summed data is analogous to results obtained from the bottom-up method where proteolytic digestion defines the length of the fragments. This in silico digestion makes it possible to choose the residues at which to “digest” the protein to provide comparable data. Results from in silico digestion at residues before and after residues 30 and 32, respectively, are shown in Figure 5d and show that the summed kinetics using c and z• ions are essentially the same. This result suggests that the different rate constants obtained from the c and z• ion data for Asp32 do not reflect different solution-phase conformers. Based on this result, the value we report for Asp32 in Table S-1 is the average of the two single residue experimental values for this residue.
Comparison to NMR Data
Amide HDX rate constants obtained in these MS experiments and values reported from previous NMR experiments56,57 are given in Table S-1 for comparison. On average, the rate constants obtained from our MS measurements are higher than those determined by NMR. However, a direct comparison of the rate constants is complicated by differences in experimental conditions used to obtain all three sets of data. Slightly different solution conditions were used in each of the three experiments, which may affect the absolute exchange rate constants due to local structural differences influenced by pH, buffer ions, or ionic strength. Different solution temperatures were also used, which can affect protein structure and dynamics and the resulting HDX kinetics. For example, the generally faster exchange kinetics observed here could be due to faster “breathing” dynamics that occur at 38 °C compared to 30 °C56 or 25 °C57 for the two NMR experiments.
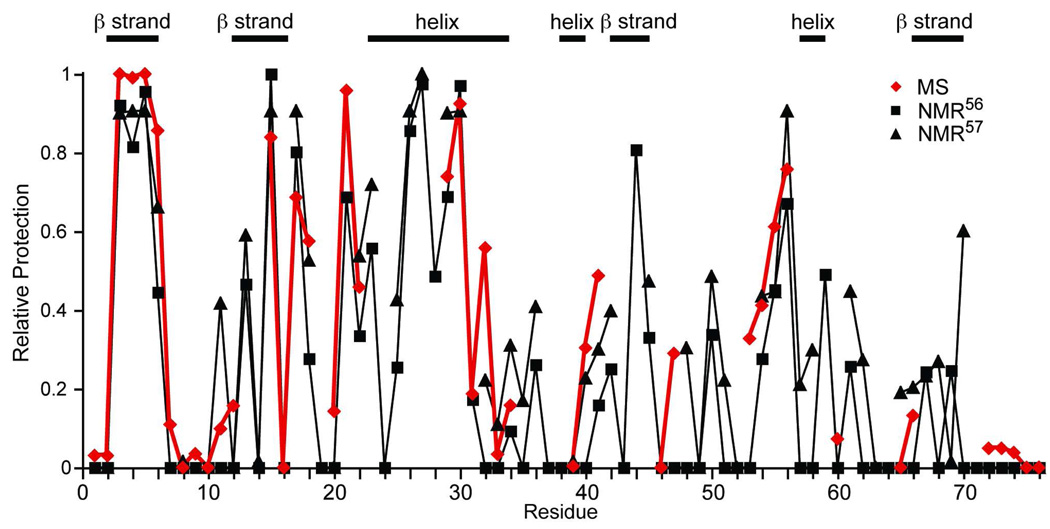
A more meaningful comparison is to compare the relative rates of exchange at each site in the protein in order to observe the regions of the protein that are either more or less protected from exchange. To do this, we normalized the (negative) logarithms of the rate constants for each of the three data sets and plotted these “relative protection” values against the amino acid sequence, where a value of 1 indicates the slowest exchange rate observed and a value of 0 indicates the fastest exchange rate observed within a set of experimental data (Figure 6). The absence of a point for the MS data (red diamond) or the NMR data (black square56 or black triangle57) indicates single residue resolution was not obtained for that amide. Overall, the MS results agree very well with the NMR results as indicated by comparable “relative protection” in regions of high protection (e.g. residues 3–6), in regions of intermediate protection (e.g. residues 54–56), and regions of low protection (e.g. both termini). Individual discrepant data points are observed randomly across the protein, but this is also the case when just the two NMR data sets are compared. For example, results from one NMR experiment57 show 0.2 relative protection for residue 65, whereas MS and the other NMR experiment56 show zero relative protection at this site. To more thoroughly evaluate these comparisons, the relative protection factors obtained by MS were plotted versus those from the individual NMR experiments, and the two NMR data sets with each other (Figure S-2, in Supporting Information). The slopes of the two MS versus NMR plots ((slope = 0.90; R2 = 0.84)56 or (slope = 0.90; R2 = 0.84)57) are somewhat better than the two NMR data sets against each other (slope = 0.85; R2 = 0.83) and the correlation coefficients are essentially equal for all three comparisons. The generally good agreement between the MS protection factors reported here and those obtained by NMR indicates that this supercharging MS method provides meaningful HDX values with nearly single amino acid spatial resolution as is typically achieved with NMR.
Figure 6.
Comparison of “Relative Protection” from HDX for individual residues obtained by MS (red diamonds), NMR56 (black squares), and NMR57 (black triangles) across the protein sequence. “Relative Protection” is calculated as the (negative) logarithm of the exchange rate constants normalized within a data set.
CONCLUSIONS
By combining ESI supercharging with top-down HDX-MS, real-time amide exchange rate constants for individual residues can be obtained with significantly higher sensitivity than NMR. The generally good agreement between values obtained with this MS method and previous NMR values indicates that H/D scrambling does not occur to any significant extent, and these results provide additional evidence that the ESI supercharging reagent does not cause significant changes to the protein structure in the initial solution, but causes proteins to denature in the electrospray droplet. Because the droplet lifetime is short (ms), this method significantly reduces the potential for back-exchange to occur when HDX is performed near physiological pH because even the fastest intrinsic exchange rates are significantly slower. The supercharging reagents have the additional advantage of increasing the charge states formed by ESI, which increases the fragmentation efficiency and the resulting sequence coverage obtained using ECD or ETD. Although not done here, including results from other precursor charge states would likely improve the spatial resolution of this method, and this may become important for larger proteins where ETD fragmentation may become more limited.
The fastest reported rate constants are ~1.4 min−1, and were limited by the long dead time in these initial experiments. Data at significantly shorter times could be obtained using a mixing apparatus, such as those used by Konermann and coworkers,18 to measure significantly faster rate constants. By using a supercharging reagent in the diluant, only a single mixing “T” is required to initiate H/D exchange and back-exchange would be essentially eliminated. An advantage of using a mixing system is the ability to continuously acquire data for a single time point so that scan averaging can be used to improve the signal-to-noise ratios of these measurements. This could result in more precise rate constants than were obtained in this experiment, where two minutes of real-time data were averaged for a given experimental time point. This type of continuous-flow approach requires much more protein than one based on a few nanoelectrospray capillaries as demonstrated here, but the significantly better sensitivity of mass spectrometry compared to NMR could make this an attractive method when a large quantity of the protein of interest is not readily available.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Michael P. Daly, Don Harris, and the Waters Corporation for generously making available a Synapt™ High Definition Mass Spectrometer demonstration instrument for the TWIMS studies, thank the National Institutes of Health RO1GM064712 and T32GM08295 (training grant H.J.S) for financial support, and thank James S. Prell for helpful discussion.
REFERENCES
- 1.Bai YW, Milne JS, Mayne L, Englander SW. Prot. Struct. Funct. Gen. 1993;17:75–86. doi: 10.1002/prot.340170110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berger A, Linderstrom-Lang K. Arch Biochem and Biophys. 1957;69:106–118. doi: 10.1016/0003-9861(57)90478-2. [DOI] [PubMed] [Google Scholar]
- 3.Dempsey CE. Prog. Nucl Mag. Res. Sp. 2001;39:135–170. [Google Scholar]
- 4.Barth A. Biochim Biophys Acta. 2007;1767:1073–1101. doi: 10.1016/j.bbabio.2007.06.004. [DOI] [PubMed] [Google Scholar]
- 5.Shashilov VA, Lednev IK. J. Raman Spectrosc. 2009;40:1749–1758. [Google Scholar]
- 6.Englander SW. J. Am. Soc. Mass Spectrom. 2006;17:1481–1489. doi: 10.1016/j.jasms.2006.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Konermann L, Tong X, Pan Y. J. Mass Spectrom. 2008;43:1021–1036. doi: 10.1002/jms.1435. [DOI] [PubMed] [Google Scholar]
- 8.Wales TE, Engen JR. Mass Spectrom. Rev. 2006;25:158–170. doi: 10.1002/mas.20064. [DOI] [PubMed] [Google Scholar]
- 9.Wang LT, Smith DL. Anal. Biochem. 2003;314:46–53. doi: 10.1016/s0003-2697(02)00620-6. [DOI] [PubMed] [Google Scholar]
- 10.Engen JR, Smith DL. Anal. Chem. 2001;73:256A–265A. doi: 10.1021/ac012452f. [DOI] [PubMed] [Google Scholar]
- 11.Smith DL, Deng YZ, Zhang ZQ. J. Mass Spectrom. 1997;32:135–146. doi: 10.1002/(SICI)1096-9888(199702)32:2<135::AID-JMS486>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 12.Kaltashov IA, Bobst CE, Abzalimov RR. Anal. Chem. 2009;81:7892–7899. doi: 10.1021/ac901366n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang HM, Bou-Assaf GM, Emmett MR, Marshall AG. J. Am. Soc. Mass Spectrom. 2009;20:520–524. doi: 10.1016/j.jasms.2008.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Feng LM, Orlando R, Prestegard JH. Anal. Chem. 2006;78:6885–6892. doi: 10.1021/ac060948x. [DOI] [PubMed] [Google Scholar]
- 15.Zencheck WD, Xiao H, Nolen BJ, Angeletti RH, Pollard TD, Almo SC. J. Mol. Biol. 2009;390:414–427. doi: 10.1016/j.jmb.2009.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cheng GL, Basha E, Wysocki VH, Vierling E. J. Biol. Chem. 2008;283:26634–26642. doi: 10.1074/jbc.M802946200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kang SB, Prevelige PE. J. Mol. Biol. 2005;347:935–948. doi: 10.1016/j.jmb.2005.02.021. [DOI] [PubMed] [Google Scholar]
- 18.Pan JX, Han J, Borchers CH, Konermann L. J. Am. Chem. Soc. 2009;131:12801–12808. doi: 10.1021/ja904379w. [DOI] [PubMed] [Google Scholar]
- 19.Hoerner JK, Xiao H, Dobo A, Kaltashov IA. J. Am. Chem. Soc. 2004;126:7709–7717. doi: 10.1021/ja049513m. [DOI] [PubMed] [Google Scholar]
- 20.Jørgensen TJD, Bache N, Roepstorff P, Gardsvoll H, Ploug M. Mol. Cell. Proteomics. 2005;4:1910–1919. doi: 10.1074/mcp.M500163-MCP200. [DOI] [PubMed] [Google Scholar]
- 21.Ferguson PL, Konermann L. Anal. Chem. 2008;80:4078–4086. doi: 10.1021/ac8001963. [DOI] [PubMed] [Google Scholar]
- 22.Charlebois JP, Patrie SM, Kelleher NL. Anal. Chem. 2003;75:3263–3266. doi: 10.1021/ac020690k. [DOI] [PubMed] [Google Scholar]
- 23.Rand KD, Adams CM, Zubarev RA, Jørgensen TJD. J. Am. Chem. Soc. 2008;130:1341–1349. doi: 10.1021/ja076448i. [DOI] [PubMed] [Google Scholar]
- 24.Pan J, Han J, Borchers CH, Konermann L. J. Am. Chem. Soc. 2008;130:11574–11575. doi: 10.1021/ja802871c. [DOI] [PubMed] [Google Scholar]
- 25.Zehl M, Rand KD, Jensen ON, Jørgensen TJD. J. Am. Chem. Soc. 2008;130:17453–17459. doi: 10.1021/ja805573h. [DOI] [PubMed] [Google Scholar]
- 26.Rand KD, Zehl M, Jensen ON, Jørgensen TJD. Anal. Chem. 2009;81:5577–5584. doi: 10.1021/ac9008447. [DOI] [PubMed] [Google Scholar]
- 27.Abzalimov RR, Kaplan DA, Easterling ML, Kaltashov IA. J. Am. Soc. Mass Spectrom. 2009;20:1514–1517. doi: 10.1016/j.jasms.2009.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Breuker K, Oh HB, Lin C. Proc. Natl. Acad. Sci. U.S.A. 2004;101:14011–14016. doi: 10.1073/pnas.0406095101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zubarev RA, Kelleher NL, McLafferty FW. J. Am. Chem. Soc. 1998;120:3265–3266. [Google Scholar]
- 30.Syrstad EA, Turecek F. J. Am. Soc. Mass Spectrom. 2005;16:208–224. doi: 10.1016/j.jasms.2004.11.001. [DOI] [PubMed] [Google Scholar]
- 31.Prell JS, O'Brien JT, Holm AIS, Leib RD, Donald WA, Williams ER. J. Am. Chem. Soc. 2008;130:12680–12689. doi: 10.1021/ja8022434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Breuker K, Oh HB, Horn DM, Cerda BA, McLafferty FW. J. Am. Chem. Soc. 2002;124:6407–6420. doi: 10.1021/ja012267j. [DOI] [PubMed] [Google Scholar]
- 33.Iavarone AT, Paech K, Williams ER. Anal. Chem. 2004;76:2231–2238. doi: 10.1021/ac035431p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Syka JEP, Coon JJ, Schroeder MJ, Shabanowitz J, Hunt DF. Proc. Natl Acad. Sci. U.S.A. 2004;101:9528–9533. doi: 10.1073/pnas.0402700101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Iavarone AT, Jurchen JC, Williams ER. Anal. Chem. 2001;73:1455–1460. doi: 10.1021/ac001251t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Iavarone AT, Williams ER. Int. J. Mass Spectrom. 2002;219:63–72. [Google Scholar]
- 37.Iavarone AT, Williams ER. J. Am. Chem. Soc. 2003;125:2319–2327. doi: 10.1021/ja021202t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lomeli SH, Yin S, Ogorzalek Loo RR, Loo JA. J. Am. Soc. Mass Spectrom. 2009;20:593–596. doi: 10.1016/j.jasms.2008.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lomeli SH, Peng IX, Yin S, Loo RRO, Loo JA. J. Am. Soc. Mass Spectrom. 2010;21:127–131. doi: 10.1016/j.jasms.2009.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Iavarone AT, Jurchen JC, Williams ER. J. Am. Soc. Mass Spectrom. 2000;11:976–985. doi: 10.1016/S1044-0305(00)00169-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Page JS, Kelly RT, Tang K, Smith RD. J. Am. Soc. Mass Spectrom. 2007;18:1582–1590. doi: 10.1016/j.jasms.2007.05.018. [DOI] [PubMed] [Google Scholar]
- 42.Sterling HJ, Williams ER. J. Am. Soc. Mass Spectrom. 2009;20:1933–1943. doi: 10.1016/j.jasms.2009.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sterling HJ, Daly MP, Feld GK, Thoren KL, Kintzer AF, Krantz BA, Williams ER. J. Am. Soc. Mass Spectrom. 2010 doi: 10.1016/j.jasms.2010.06.012. 10.1016/j.jasms.2010.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lide DR, editor. CRC Handbook of Chemistry and Physics; In [Online] 89th ed. 2008–2009. [Google Scholar]
- 45.Chowdhury SK, Katta V, Chait BT. J. Am. Chem. Soc. 1990;112:9012–9013. [Google Scholar]
- 46.Loo JA, Loo RRO, Udseth HR, Edmonds CG, Smith RD. Rapid Commun. Mass Spectrom. 1991;5:101–105. doi: 10.1002/rcm.1290050303. [DOI] [PubMed] [Google Scholar]
- 47.Konermann L, Douglas DJ. Rapid Commun. Mass Spectrom. 1998;12:435–442. doi: 10.1002/(SICI)1097-0231(19980430)12:8<435::AID-RCM181>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 48.Shelimov KB, Clemmer DE, Hudgins RR, Jarrold MF. J. Am. Chem. Soc. 1997;119:2240–2248. [Google Scholar]
- 49.Koeniger SL, Clemmer DE. J. Am. Soc. Mass Spectrom. 2007;18:322–331. doi: 10.1016/j.jasms.2006.09.025. [DOI] [PubMed] [Google Scholar]
- 50.Shvartsburg AA, Smith RD. Anal. Chem. 2008;80:9689–9699. doi: 10.1021/ac8016295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Robinson EW, Leib RD, Williams ER. J. Am. Soc. Mass Spectrom. 2006;17:1469–1479. doi: 10.1016/j.jasms.2006.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Apweiler R, Martin MJ, et al. Nucleic Acids Res. 38:D142–D148. doi: 10.1093/nar/gkp846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liuni P, Rob T, Wilson DJ. Rapid Commun. Mass Spectrom. 2010;24:315–320. doi: 10.1002/rcm.4391. [DOI] [PubMed] [Google Scholar]
- 54.Hoerner JK, Xiao H, Kaltashov IA. Biochemistry. 2005;44:11286–11294. doi: 10.1021/bi0509548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pan YQ, Briggs MS. Biochemistry. 1992;31:11405–11412. doi: 10.1021/bi00161a019. [DOI] [PubMed] [Google Scholar]
- 56.Johnson EC, Lazar GA, Desjarlais JR, Handel TM. Struct. Fold. Des. 1999;7:967–976. doi: 10.1016/s0969-2126(99)80123-3. [DOI] [PubMed] [Google Scholar]
- 57.Bougault C, Feng LM, Glushka J, Kupce E, Prestegard JH. J. Biomol. NMR. 2004;28:385–390. doi: 10.1023/B:JNMR.0000015406.66725.30. [DOI] [PubMed] [Google Scholar]
- 58.Schanda P, Forge V, Brutscher B. Proc. Natl. Acad. Sci. U.S.A. 2007;104:11257–11262. doi: 10.1073/pnas.0702069104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.