Abstract
Diphthamide, the target of diphtheria toxin, is a unique posttranslational modification on eukaryotic and archaeal translation elongation factor 2 (EF2). Although diphthamide modification was discovered three decades ago, in vitro reconstitution of diphthamide biosynthesis using purified proteins has not been reported. The proposed biosynthesis pathway of diphthamide involves three steps. Our laboratory has recently showed that in Pyrococcus horikoshii (P. horikoshii), the first step uses an [4Fe-4S] enzyme PhDph2 to generate a 3-amino-3-carboxypropyl radical from S-adenosyl-L-methionine (SAM) to form a C-C bond. The second step is the trimethylation of an amino group to form the diphthine intermediate. This step is catalyzed by a methyltransferase called diphthine synthase or Dph5. Here we report the in vitro reconstitution of the second step using P. horikoshii Dph5 (PhDph5). Our results demonstrate that PhDph5 is sufficient to catalyze the mono-, di-, and trimethylation of P. horikoshii EF2 (PhEF2). Interestingly, the trimethylated product from PhDph5-catalyzed reaction can easily eliminate the trimethylamino group. The potential implication of this unexpected finding on the diphthamide biosynthesis pathway is discussed.
Keywords: Diphthamide, diphthine, methyltransferase, SAM, elongation factor 2
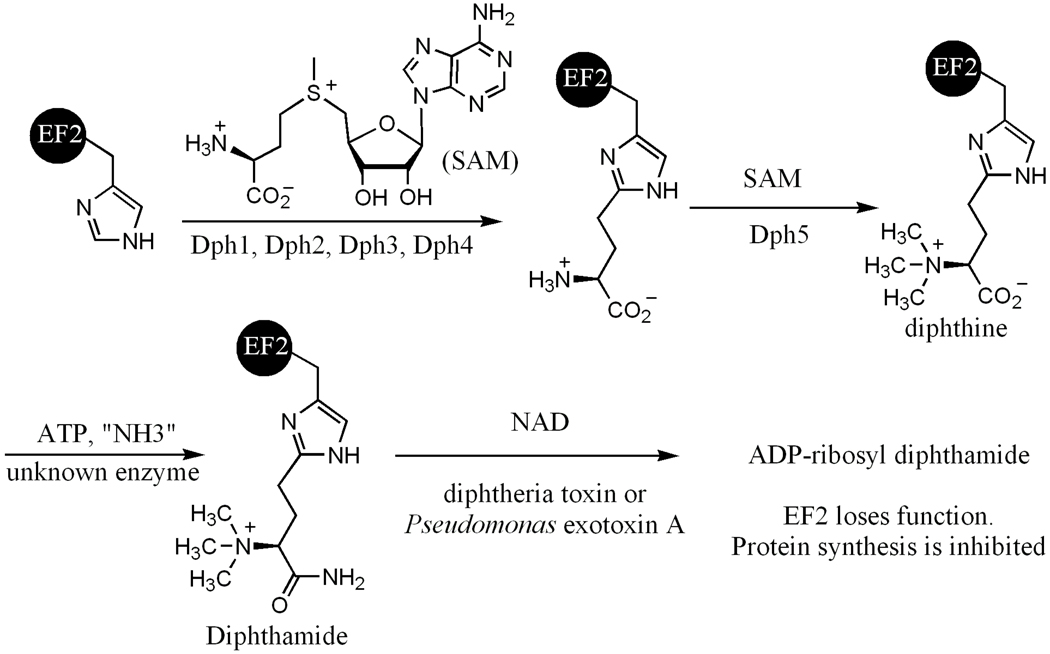
Diphthamide, found in both eukaryotes and archaea(1–3), is a unique posttranslationally modified histidine residue on translational elongation factor 2 (EF2), a GTPase required for ribosomal protein synthesis (4). The histidine residues that are modified are His699 in yeast EF2, His715 in mammalian EF2, and His600 in Pyrococcus horikoshii EF2. Diphthamide is the target of diphtheria toxin(5), which ADP-ribosylates diphthamide and inhibits protein synthesis, leading to host cell death(6). It has been indicated that diphthamide may prevent the -1 frame shift during protein synthesis(7). However, the physiological function and biosynthesis of the modification are still not completely understood(8), despite of the fact that the modification has been known for over three decades. Diphthamide modification has been proposed to involve three steps (Figure 1).(9) The first step is the transfer of the 3-amino-3-carboxypropyl group from S-adenosyl-L-methionine (SAM) to the C-2 position of the imidazole ring of the target histidine residue in EF2. The second step is the trimethylation of the amino group to form an intermediate called diphthine. The last step is the ATP-dependent amidation of the carboxyl group of diphthine. Genetic studies have identified five proteins in eukaryotes required for the biosynthesis of diphthamide, Dph1, Dph2, Dph3, Dph4, and Dph5. Dph1-4 are known to be required for the first step (6, 10–15), whereas Dph5 (also called diphthine synthase) is known to be required for the second step(16). The enzyme that catalyzes the last step has not been identified yet. Diphthamide is also found in archaea. However, among the five eukaryotic proteins required for diphthamide biosynthesis, only two orthologs can be found in archaea by BLAST search. One of them, Dph2, is homologous to eukaryotic Dph1 and Dph2 (Dph1 and Dph2 are homologous to each other), and the other one is the diphthine synthase, Dph5.
Figure 1.
Proposed diphthamide biosynthesis pathway. The diphthamide residue is the target of bacterial ADP-ribosyltransferases, such as diphtheria toxin and Pseudomonas exotoxin A. The ADP-ribosylation of diphthamide by these toxins leads to inhibition of protein synthesis in the eukaryotic host cells.
Although five genes are known to be required for the first two steps of diphthamide biosynthesis, it is not clear whether these genes are sufficient. In fact, another gene, WDR85 (17), was recently identified to be required for the first step of diphthamide biosynthesis in eukaryotes, further demonstrating the complexity of diphthamide biosynthesis. The same can be said about the second step: one can similarly ask whether Dph5 alone is sufficient to catalyze the trimethylation or additional proteins are needed. Therefore, to fully understand diphthamide biosynthesis, it is important to reconstitute the biosynthesis in vitro using purified proteins. Recently, we have successfully reconstituted the first step of diphthamide biosynthesis using the Pyrococcus horikoshii Dph2 (PhDph2) and EF2 (PhEF2)(18). We found that PhDph2 forms a homodimer and can bind a [4Fe-4S] cluster in each monomer with three conserved cysteine residues. PhDph2 is similar to the radical SAM superfamily(19) in that both contain [4Fe-4S] clusters and are SAM-dependent. However, PhDph2 does not contain the CXXCXXXC motif(20) that is found in most radical SAM enzymes. Furthermore, we showed that PhDph2 likely generates a 3-amino-3-carboxypropyl radical in the first step of diphthamide biosynthesis, instead of a 5’-adeoxyadenosyl radical. The successful reconstitution of the first step of diphthamide biosynthesis sets the stage for us to investigate the second step of diphthamide biosynthesis by providing the substrate to test whether PhDph5 is sufficient for the trimethylation step in vitro. Herein we report the reconstitution of PhDph5 activity. Our data suggest that PhDph5 is sufficient for the second step of diphthamide biosynthesis and that it catalyzes the trimethylation in a highly processive manner. Interestingly, we found that after the trimethylation step, the resulting diphthine product is not stable and can easily eliminate the trimethylamino group in a reaction similar to Hofmann elimination or Cope elimination(21).
EXPERIMENTAL PROCEDURES
Cloning, expression, and purification of PhDph2, PhDph5, and PhEF2
The cloning, expression, and purification of PhDph2 and PhEF2 were reported(22). PhDph5 was amplified by PCR from P. horikoshii genomic DNA (ATCC® 700860D-5TM) with AccuPrime Pfx DNA polymerase (Invitrogen) and the primers (AGTCAGCATATGATGGTTTTGTACTTTATTGGATTG&AGTCAGCTCGAGTTAAACATTAACCC TTAATATCTC). Amplified PhDph5 was digested by NdeI and XhoI (New England BioLabs) and then ligated into the pET-28a (+) vector by T4 DNA ligase (Invitrogen). The recombinant plasmid was transformed to TOP10 competent cells (Invitrogen), and colonies containing the plasmid were selected by colony PCR with EconoTaq® DNA polymerase (Lucigen). The amplified plasmid was purified using QIAprep® Spin Miniprep Kit (QIAGEN) and its sequence was confirmed by DNA Sequencing (performed by Cornell University Life Sciences Core Laboratories Center).
The plasmid containing PhDph5 was transformed into the E. coli expression strain BL21 (DE3) with pRARE2. The cells were grown in LB media with 100 µg/ml ampicillin at 37 °C and induced at an OD600 of 0.8 with 0.1 mM isopropyl-β-D-thiogalactopyranoside (IPTG). The induced cells were incubated in a shaker (New Brunswick Scientific Excella E25) at 37 °C and 200 rpm for 3 h. Cells were harvested by centrifugation at 6,371 ×g (Beckman Coulter Avanti J-E) and 4 °C for 10 min. Cell pellets from 2L of LB culture were re-suspended in 30 ml lysis buffer (500 mM NaCl, 10 mM MgCl2, 5 mM imidazole, and 20 mM Tris-HCl at pH 8.0). Cells were lysed using a cell disruptor (EmulsiFlex). Cell debris was removed by centrifugation at 48,400 ×g (Beckman Coulter Avanti J-E) for 30 min. The supernatant was incubated for 1 hour with 1.2 ml Ni-NTA resin (Qiagen) that was pre-washed and equilibrated with the lysis buffer. The resin was then loaded onto a polypropylene column and washed with 20 ml lysis buffer. PhDph5 was eluted from the column with 1.5 mL aliquots of buffers containing 100 mM, 150 mM and 200 mM imidazole in the lysis buffer. The protein was buffer-exchanged to 50mM Tris-HCl, pH 8.0, with 50 mM NaCl using a Bio-Rad 10–DG desalting column. The protein was further purified by heating at 65 °C for 10 min and centrifugation at 39,191 ×g to remove the precipitate. Purified PhDph5 was concentrated using an Amicon Ultra-4 centrifugal filter device (Millipore).
Reconstitution of PhDph5 activity in vitro and detection of formation of S-adenosyl-L- homocystein (SAH) by HPLC
The first step of PhEF2 modification was performed in an anaerobic chamber (5% Hydrogen, 95% Nitrogen) (Coy Laboratory Products). PhDph2 (240 µM) was incubated with 10 mM dithionite for 10 min first. PhEF2 (100 µM) and SAM (200 µM) were added and reaction was incubated at 65 °C for 40 mins. After the first step of modification, the reaction mixture was buffer exchanged to PhDph5 reaction buffer (23) (100 mM Tris-HCl, pH 8.0, 75 mM NaCl, 50 mM KCl, 1 mM EDTA, 5 mM DTT, 5 mM MgCl2, with or without 2 mM ADP, 10 mM creatine phosphate and 80 pg/ml phosphocreatine kinase) with a 10 kDa Amicon (Millipore) filter and then with a Micro Bio-Spin 6 column (BioRad) to get rid of the leftover SAM from the last step reaction. The PhDph5 activity assay was carried out with 30 µM modified PhEF2, 60 µM PhDph5, and SAM at different concentrations (0 µM, 5 µM, 10 µM, 20 µM, 30 µM and 50 µM) in a total volume of 50 µl and incubating the reaction mixtures at 37°C for 30 min. The reactions were stopped by 5% TFA. The precipitated proteins were separated from the reaction mixture by centrifugation. The supernatant was analyzed by HPLC (Shimadzu) on a C18 column (HαSprite) monitored at 260 nm absorbance, using a linear gradient from 0 to 40% buffer B in 20 min at a flow rate of 0.3 mL/min (buffer A: 50 mM ammonium acetate, pH 5.4; buffer B, 50% (v/v) methanol/water). The PhDph5 reaction buffer used this experiment was from reference 23. We found that the ADP, creatine phosphate and phosphocreatine kinase are not required for the PhDph5 reaction. They are probably required for the amidation step of diphthamide biosynthesis. Therefore, in later reactions with PhDph5, these reagents were not included.
Detection of PhEF2 modification by methyl-14C-SAM
Enzymatic reactions with methyl-14C SAM followed the similar procedure as described above. Reactions were incubated at 37°C for 30 min. The reaction mixtures were resolved by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE, 12% acrylamide gel) without heating to denature protein. The dried gel was exposed to a PhosphorImaging screen (GE Healthcare, Piscataway, NJ) which was then scanned using a STORM860 phosphorimager (GE Healthcare, Piscataway, NJ).
Characterization of PhEF2 modification with matrix-assisted laser desorption/ionization mass spectrometry (MALDI-MS) and MS/MS
PhDph5 activity assay with normal SAM were carried out following the same procedure as that used for the activity assay with 14C-labeled SAM. The mixtures were separated by SDS-PAGE (12% acrylamide gel) without heating to denature protein. The PhEF2 band from the Coomassie blue-stained gel were cut out and washed with water, 50% Ambic/acetonitrile and pure acetonitrile. Gel pieces dried in ventilated fume hood were digested by trypsin (10µg/mL) overnight in a 30°C incubator (Fisher Scientific Inc.) to cleave protein at carboxyl side of lysine or arginine into peptide fragments. Digested products were extracted and cleaned up by Ziptip C4 (Millipore). MALDI-MS was performed at the Proteomics and Mass Spectrometry Facility of Cornell University on a 4700 Proteomics Analyzer (Applied Biosystems). The instrument was operated in positive ion reflector mode (20kV). The matrix used was alpha-cyano-4-hydroxycinnamic acid at 3 mg/ml in 60% acetonitrile/0.1%TFA with 1 mM ammonium phosphate added to suppress low m/z matrix adducts. The instrument was calibrated using calibration mixture obtained from the instrument manufacturer. 1200 laser shots (80 shots/location, 15 different locations and uniform firing pattern) were used to acquire the survey spectrum from m/z 700–4000 Da. The peak of 1629.8 was further analyzed by MS/MS.
RESULTS
Initial PhDph5 activity assay with 14C-labeled SAM gave no labeling
PhEF2 (83 kDa), PhDph2 (38 kDa) and PhDph5 (30 kDa) were expressed in E. coli and purified as described in the Experimental Procedures. Their purity and sizes were checked by SDS-PAGE (Supplementary Figure 1).
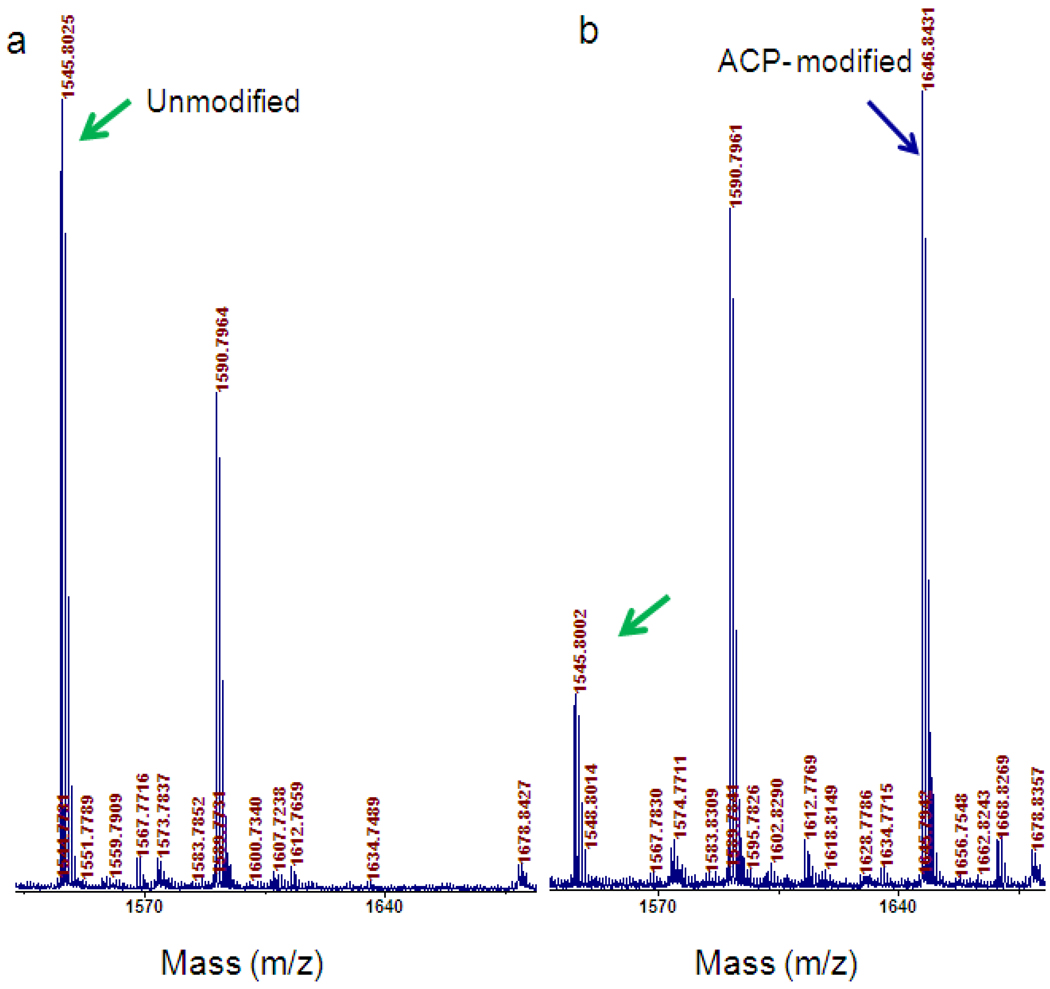
By incubating PhEF2 with PhDph2 and SAM under anaerobic conditions, we first obtained PhEF2 with the 3-amino-3-carboxypropyl (ACP) group attached to His600. The reaction was monitored by MALDI-MS (Figure 2). The major observed masses in Figure 2 are consistent with the calculated masses of predicted peptide fragments as shown in Supplementary Table 1. The peptide fragment containing the His600 residue (LLDAQVHEDNVHR) in unmodified PhEF2 has an m/z of 1545.80 (MH+, calculated 1545.78, Figure 2a). This peak was almost gone after reaction with PhDph2 and a new peak with an m/z of 1646.84 appeared (Figure 2b), which corresponds to the product of the PhDph2-catalyzed reaction with the ACP-modified histidine residue (MH+, calculated 1646.83).
Figure 2.
Monitoring PhDph2-catalyzed PhEF2 modification using MALDI-MS. a, unmodified PhEF2 peptide residue with m/z 1545.8; b, PhEF2 modified with 3-amino-3-carboxypropyl (ACP) group. Two peaks showed in the spectrum: unmodified PhEF2 peptide with m/z 1545.8; ACP-modified PhEF2 peptide with m/z 1646.8.
To test the activity of the purified PhDph5, the first step reaction mixtures were buffer exchanged to PhDph5 reaction buffer. PhDPh5 and excess methyl-14C SAM were then added to initiate the trimethylation reaction. To our surprise, no labeling on PhEF2 was found (data not shown) by autoradiography. We reasoned that additional proteins or other molecules other than PhDph5 may be needed to reconstitute the second step, since all the reported Dph5 activity assay were performed in vivo or by using crude cell lysate (16, 23). However, it is also possible that PhDph5 alone can catalyze the second step, but our reaction conditions need to be optimized to get the reaction to work.
Detection of SAH shows SAM-degrading activity catalyzed by PhDph5
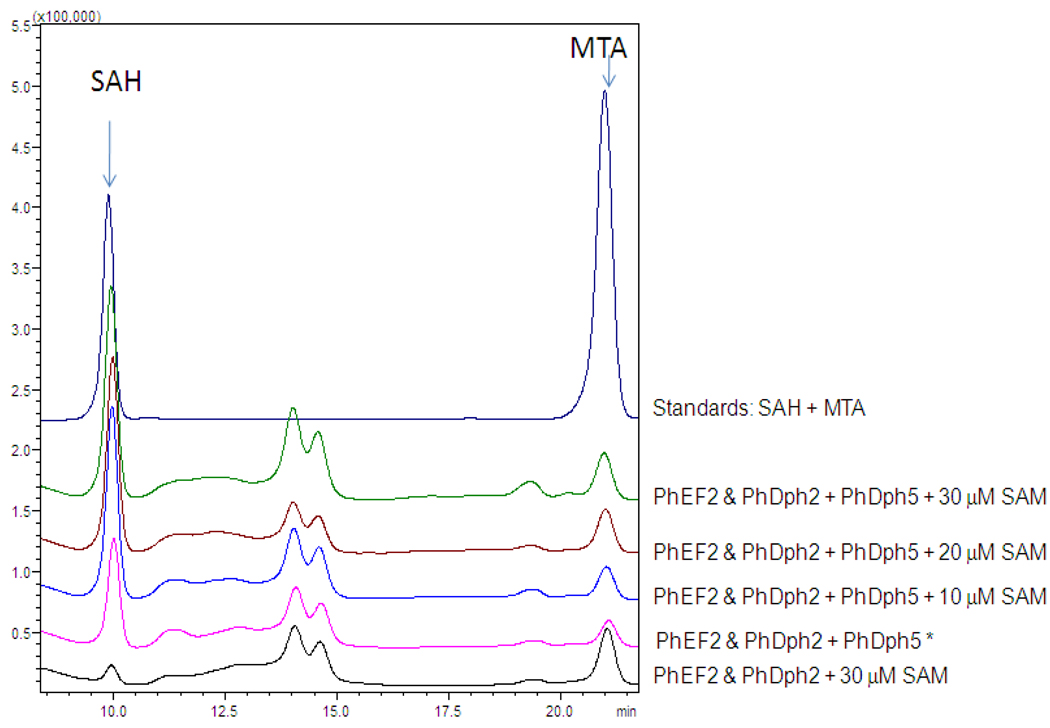
In the process to find out why no 14C-methyl group can be transferred to PhEF2, we tried other methods to detect the product of the reaction catalyzed by PhDph5. One product of methyl transfer reaction by SAM-dependent methyltransferase is S-adenosylhomocysteine (SAH), which can be used to indicate whether the methyltransfer reaction occurred or not. We monitored the SAH formation with HPLC (Figure 3). Standard SAH was eluted at 10 min, as shown in Figure 3 (dark blue line). In the reaction without PhDph5, no SAH was detected (Figure 3, black line). In contrast, when PhDph5 was present, the SAH peak increased (Figure 3, pink line) even when no additional SAM was added (some left-over SAM molecules were present from the first step PhDph2-catalyzed reaction). With increasing concentrations of SAM, the SAH peak increased first then remained unchanged (Figure 3, brown and green lines). Taken together, the data suggest that PhDph5 is active since it can remove the methyl group from SAM in the presence of the PhEF2 substrate.
Figure 3.
HPLC analysis of the reaction product showed that PhDph5 catalyzes the formation of SAH. Absorption was monitored at 260 nm. The description of each overlaid trace is provided on the right, and the identities of major peaks were indicated. In the reaction marked by *, no extra SAM was added. However, some SAM (less than 10 µM) was left from the PhDph2-catalyzed reaction to make ACP-modified PhEF2 and led to the formation of SAH when PhDph5 was added.
MALDI-MS/MS of PhEF2 revealed an elimination reaction that occurred to diphthine
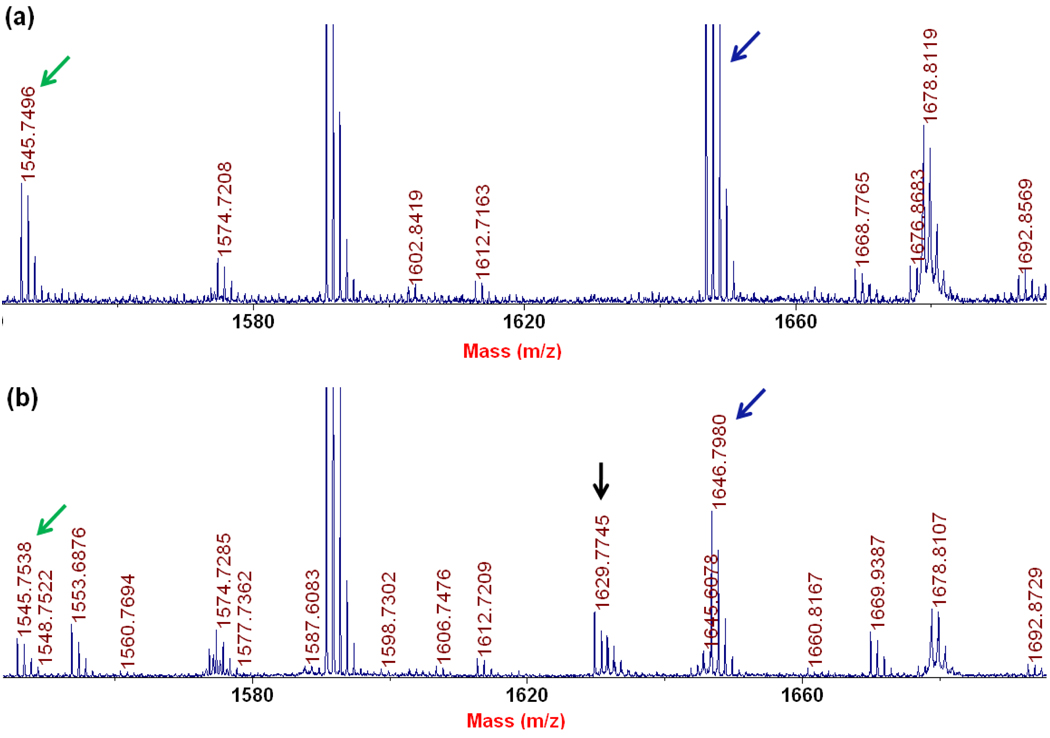
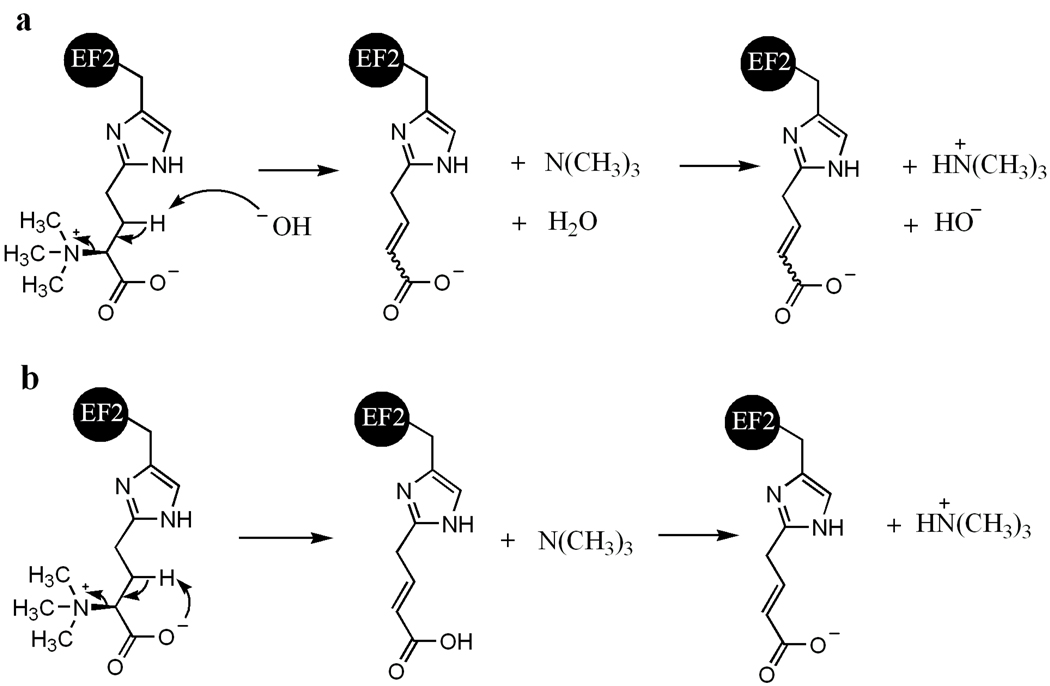
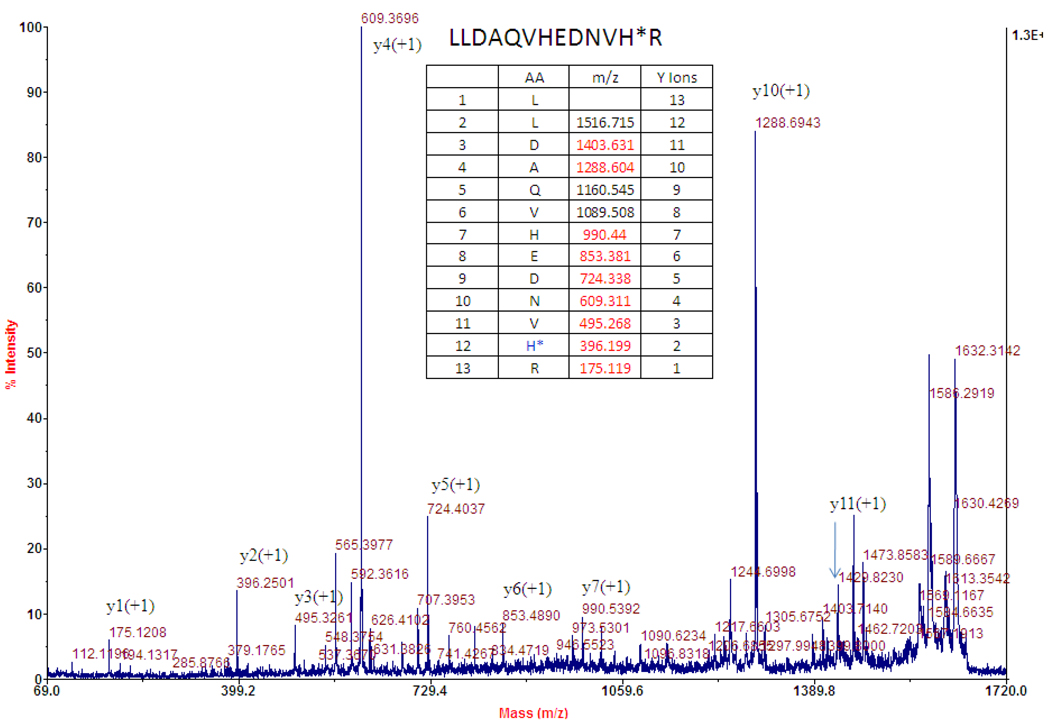
The detection of the SAM-cleavage activity suggests that PhDph5 is active under the conditions used, although it remained unclear where the methyl group of SAM ends up. To find out whether the methyl group is transferred to PhEF2, we decided to use MALD-MS to monitor the mass of PhEF2. After incubating PhEF2 with PhDph5 and SAM, the reaction mixture was resolved by SDS-PAGE and the PhEF2 band was excised, digested, and analyzed by MALDI-MS. We could not see any peaks for the mono-, di- or tri-methylation products, which is consistent with our previous results obtained with methyl-14C SAM. However, a new peak with an m/z of 1629.77 was detected (Figure 4b) which was not present in the control, the ACP-modified PhEF2 not subjected to the Dph5 reaction (Figure4a). Since the diphthine-containing peptide has a calculated m/z of 1688.87 (M+) and a trimethylamino group has an m/z of 59.1, the peak of 1629.77 corresponds to the loss of the trimethylamino group from diphthine, the trimethylated product. The peptide fragment containing the elimination product has a calculated m/z of 1629.80 (MH+). To confirm that the trimethylamino group was eliminated and a 3-carboxy-2-propenyl group is formed on the histidine residue after elimination (Figure 8), we analyzed the 1629.77 peak by tandem MS. The mass difference between Y1 and Y2 ions is 221, which is consistent with the presence of a 3-carboxy-2-propenyl on the histidine residue (Figure 5). This result suggested that PhDph5 can catalyze the trimethylation reaction but the diphthine product cannot be detected due to the elimination of the trimethylamino group.
Figure 4.
The MALDI-MS analysis of PhEF2 in PhDph5-catalyzed reaction. (a) PhEF2 from control reaction with PhDph5; (b) PhEF2 after PhDph5-catalyzed reaction. The peak with m/z 1545.8 corresponds to unmodifed PhEF2 peptide, 1646.8 corresponds to ACP-modified PhEF2 peptide, and 1629.8 corresponds to the elimination product.
Figure 8.
Proposed mechanisms for the elimination reaction of diphthine.
Figure 5.
MS/MS of precursor 1629.81, which is from the MALDI spectrum of PhEF2 modified by PhDph5. The table lists calculated Y ions. Observed Y ions in the spectrum are colored in red.
P. horikoshii grows optimally at 98°C. For the in vitro reconstitution, we normally carry out the reaction at 65°C, which we found to be optimal for the activity of PhDph2. One concern was that the high temperature may contribute to the elimination. Thus to minimize the elimination reaction, we later carried out the Dph5-catalyzed reactions at 37°C. Similarly, when analyzing the reaction by SDS-PAGE, we did not heat the sample to denature the protein. However, even under these milder conditions, the elimination always occurred based on both MS and the 14C-labeling experiments (data not shown). The other concern is that the elimination could occur during MALDI-MS from absorbing the energy of laser. This is also unlikely because the elimination also happened in the 14C-labeling experiments in which the reaction was detected by autoradiography. These observations suggest the elimination occurs readily under the reactions conditions.
Decreasing SAM concentrations allows the detection of 14C-labeling on PhEF2
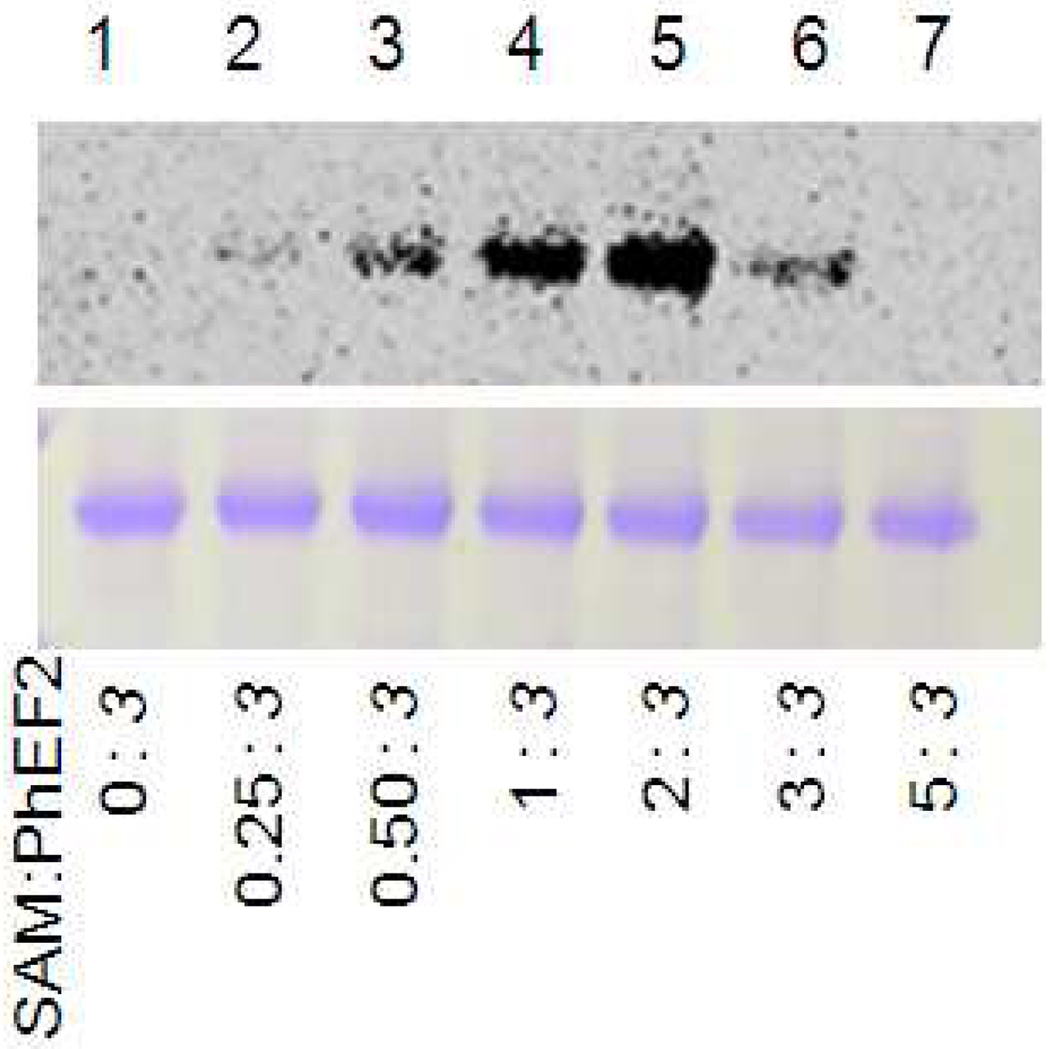
Although the above result suggested that PhDph5 can catalyze the trimethylation reaction, the actual methyltranfer to PhEF2 still need to be proven. The elimination of the trimethylamino group is similar to the Hofmann elimination reaction that can occur to quaternary ammonium hydroxide salts or the Cope elimination reaction that can happen to tertiary amine oxides(21). In Hofmann or Cope elimination, a quaternary ammonium functional group is required for the elimination to occur. Thus, we reasoned that if we can stop the PhDph5-catalyzed reaction at mono- and di-methylation stage, then no elimination reaction will occur and we may be able to detect the methyl transfer to PhEF2. Therefore we revisited the labeling experiment using different concentrations of methyl-14C SAM, hoping to accumulate the mono- and dimethylated PhEF2. After the first modification reaction, the buffer was exchanged to the methylation reaction buffer to make sure the amount of leftover SAM from the previous step was as little as possible. Different concentrations of 14C-SAM and PhDph5 were then added to allow the methylation to occur. The reaction mixtures were resolved by SDS-PAGE and the labeling was detected by autoradiography. When no methyl-14C SAM was added, no labeling was detected (Figure 6, lane 1). Labeling of PhEF2 was observed when methyl-14C-SAM was added and the labeling increased when the ratio of SAM:PhEF2 increased from 0.25:3 to 2:3 (Figure 6, lane 2 to lane 5). However, the intensity of labeling decreased when the ratio reached to 3:3 (Figure 6, lane 6). Further increasing the ratio of SAM:PhEF2 to 5:3, the labeling disappeared (Figure 6, lane 7). These results demonstrated that PhDph5 can transfer the methyl group from SAM to PhEF2. In addition, the loss of the 14C label on PhEF2 at high concentrations of methyl-14C SAM is consistent with the prediction that the trimethylated PhEF2 can undergo the elimination reaction while the mono- and dimethylated PhEF2 cannot.
Figure 6.
PhDph5-catalyzed PhEF2 methylation monitored using methyl-14C SAM. The PhDph5 activity assays were set up with 30 µM ACP-modified PhEF2, 60 µM PhDph5, and different SAM concentrations (0 µM, 2.5 µM, 5 µM, 10 µM, 20 µM, 30 µM and 50 µM). The reactions were incubated at 37°C for 30 min. Bottom panel shows the Coomassie blue-stained gel; top panel shows the autoradiography. The ratios of SAM to PhEF2 were shown at the bottom of the image.
MALDI-MS of reaction products confirmed the formation of mono- and dimethylated PhEF2
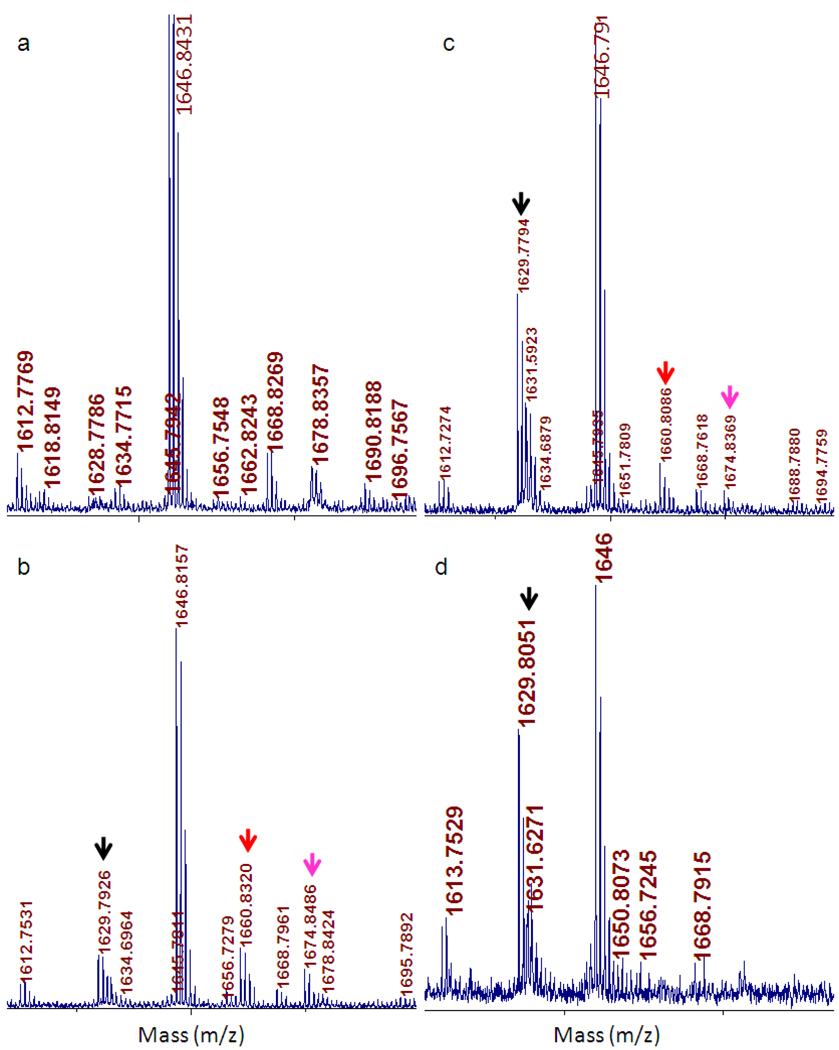
To confirm the formation of mono- and dimethylated PhEF2, we again relied on MALDI-MS. The peptide fragment containing the ACP-modified His600 residue after PhDph2-catalyzed modification has an m/z of 1646.84 (Figure 7a). After the reaction catalyzed by PhDph5 with 2:3 and 3:3 ratio of SAM:PhEF2, two new peaks appeared with m/z of 1660.83 and 1674.84 (Figure 7b and 7c), corresponding to the masses of PhEF2 with the addition of one (MH+, calculated m/z 1660.84) and two methyl groups (MH+, calculated m/z 1674.86). However, when the concentration of SAM was higher than that of PhEF2 (5:3), no methylated product was observed (Figure 7d). These results firmly established that PhDph5 can catalyze the methyltransfer to PhEF2. When one equivalent of SAM is used (Figure 7c), the major product is the eliminated product after trimethylation of the ACP group while a significant amount of unmethylated substrate is still present. This result suggests that the trimethylation reaction catalyzed by PhDph5 is highly processive.
Figure 7.
Detecting PhDph5-catalyzed PhEF2 methylation using MALDI-MS. Different ratios of PhEF2 to SAM were used to minimize the formation and elimination of the trimethylated product. a, PhEF2 modified with 3-amino-3-carboxypropyl (ACP) group. ACP-modified PhEF2 peptide has an m/z of 1646.8. b, PhEF2 modified by PhDph5 with a SAM to PhEF2 ration of 2:3. Three new peaks with m/z values of 1629.8; 1660.8; 1674.8 were observed, which correspond to diphthine with the trimethylamino group eliminated, monomethylated, and dimethylated intermediates, respectively. c, PhEF2 modified by PhDph5 with a SAM to PhEF2 ratio of 3:3. d, PhEF2 modified by PhDph5 with a SAM to PhEF2 ratio of 5:3. Only the elimination product with m/z of 1629.8 was observed.
DISCUSSION
P. horikoshii Diphthine is not stable in vitro and readily eliminates the trimethylamino group
Initially, when we used methyl-14C SAM to detect the methylation of PhEF2, no methylation was detected. HPLC analysis of the small molecule product showed that SAH was formed in the reaction, suggesting that PhDph5 is active. Using MALDI-MS, we detected a new product with m/z of 1629.77. We attributed this peak to the product resulting from the elimination of the trimethylamino group from diphthine. The structure of the elimination product was further confirmed by MS/MS. The mass difference of the Y2 and Y1 ions is 221, which is consistent with the presence of a 3-carboxy-2-propenyl group on His600, the product of diphthine after elimination of the trimethylamino group. The elimination reaction is similar to the Hofmann elimination reaction or Cope elimination reaction.(21, 24) Given that such elimination reaction will require the quaternary ammonium salt, we predicted that the mono- and dimethylated PhEF2 should not undergo the elimination product and thus should be stable and detectable. Indeed, by lowering the concentration of SAM, mono- and dimethylated PhEF2 were detected using methyl-14C-SAM (Figure 6) and MALDI-MS (Figure 7). This result further confirmed that elimination requires the trimethylamino group and that Dph5 can catalyze the trimethylation reaction. There are two possible mechanisms for the elimination reaction (Figure 8). One mechanism uses an external base to attack the proton on the β-carbon (Figure 8a) and the other mechanism uses the carboxyl group as the intra-molecular base to deprotonate the β-carbon (Figure 8b). At present, it is not known whether a similar elimination reaction also occurs to eukaryotic diphthine. If it does happen to eukaryotic diphthine, we would favor the second mechanism for the elimination reaction based on the fact that when the carboxylate is converted to the amide as in diphthamide, the trimethylamino group becomes stable given that eukaryotic diphthamide has been isolated and structurally determined(2, 25, 26).
PhDph5 is sufficient for the trimethylation step of diphthamide biosynthesis in vitro
Genetic studies have shown that PhDph5 is required for the trimethylation step of diphthamide biosynthesis. However, whether PhDph5 is sufficient for the trimethylation step was not clear(11). Our data presented above demonstrated that PhDph5 is sufficient to catalyze the trimethylation step. Mono- and dimethylated PhEF2 was detected by MALDI-MS, while the trimethylated product cannot be detected due to the facile elimination of the trimethylamino group.
The implication of the instability of diphthine on the diphthamide biosynthesis pathway
Diphthamide structure was determined using eukaryotic EF2 (2, 25, 26). Whether the final structure in archaea is the same or not is not clear. At this point, we do not know whether the elimination reaction also occurs physiologically. The elimination readily occurs in vitro even though we have taken extra care to avoid harsh conditions, such as heat denaturation, to minimize it. However, it is still possible that the elimination reaction only occurs in vitro due to the lack of the enzyme for the amidation step in the reaction. If diphthamide is the final structure in P. horikoshii, given that P. horikoshii diphthine readily eliminate, in order to form the final structure, diphthamide, the last amidation step should occur very quickly in P. horikoshii cells to avoid the elimination reaction. Alternatively, it is possible that the amidation step occurs before the trimethylation step.
It would be interesting to know whether a similar elimination reaction also occurs to eukaryotic diphthine, in vitro and in vivo, and whether the elimination reaction affects the function of EF2 in protein synthesis. The genes required for diphthamide biosynthesis, dph1-dph5, were identified in yeast and mammalian cells by screening for mutants that are resistant to diphtheria toxin, which can ADP-ribosylate diphthamide and inhibit protein synthesis (6, 10–15). The enzyme for the amidation step has not been identified yet using this genetic screening. The explanation for the inability to isolate the amidation enzyme is that diphtheria toxin is able to ADP-ribosylate both diphthamide and diphthine(11). The elimination of the trimethylamino group from diphthine may provide an alternative explanation. If EF2 loses its normal function in protein synthesis after the elimination of the trimethylamino group from diphthine, then the disruption of the gene required for the amidation step would lead to the accumulation of diphthine, which will eliminate and stop protein synthesis, giving a lethal phenotype. The lethality of disrupting the gene may explain why it has not been identified in the genetic screen. This hypothesis, if correct, may help the identification of the gene required for the amidation step in eukaryotes.
Supplementary Material
ACKNOWLEDGMENT
We thank Dr. Sheng Zhang and Robert Sherwood in the Proteomics and Mass Spectrometry of Cornell University for help with the MS analysis.
This work is partly supported by the Camille and Henry Dreyfus New Faculty Award Program and NIH (R01GM088276)
ABBREVIATIONS
- eEF2
eukaryotic translation elongation factor 2
- PhEF2
Pyrococcus horikoshii translation elongation factor 2
- PhDph2
Pyrococcus horikoshii Dph2 (a protein required for the first step of diphthamide synthesis)
- PhDph5
Pyrococcus horikoshii Dph5 or diphthine synthase
- SAM
S-adenosyl-L-methionine
- SAH
S-adenosylhomocysteineadenosylhomocysteine
Footnotes
SUPPORTING INFORMATION AVAIABLE
Supplementary Figure 1 and Table 1. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Robinson EA, Henriksen O, Maxwell ES. Elongation factor 2. amino acid sequence at the site of adenosine diphosphate ribosylation. J. Biol. Chem. 1974;249:5088–5093. [PubMed] [Google Scholar]
- 2.Van Ness BG, Howard JB, Bodley JW. ADP-ribosylation of elongation factor 2 by diphtheria toxin. NMR spectra and proposed structures of ribosyl-diphthamide and its hydrolysis products. J. Biol. Chem. 1980;255:10710–10716. [PubMed] [Google Scholar]
- 3.Van Ness BG, Howard JB, Bodley JW. ADP-ribosylation of elongation factor 2 by diphtheria toxin. Isolation and properties of the novel ribosyl-amino acid and its hydrolysis products. J. Biol. Chem. 1980;255:10717–10720. [PubMed] [Google Scholar]
- 4.Gomez-Lorenzo MG, Spahn CMT, Agrawal RK, Grassucci RA, Penczek P, Chakraburtty K, Ballesta JPG, Lavandera JL, Garcia-Bustos JF, Frank J. Three-dimensional cryo-electron microscopy localization of EF2 in the Saccharomyces cerevisiae 80S ribosome at 17.5 Å resolution. EMBO J. 2000;19:2710–2718. doi: 10.1093/emboj/19.11.2710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Collier RJ. Understanding the mode of action of diphtheria toxin: a perspective on progress during the 20th century. Toxicon. 2001;39:1793–1803. doi: 10.1016/s0041-0101(01)00165-9. [DOI] [PubMed] [Google Scholar]
- 6.Liu S, Milne GT, Kuremsky JG, Fink GR, Leppla SH. Identification of the proteins required for biosynthesis of diphthamide, the target of bacterial ADP-ribosylating toxins on translation elongation factor 2. Mol. Cell. Biol. 2004;24:9487–9497. doi: 10.1128/MCB.24.21.9487-9497.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ortiz PA, Ulloque R, Kihara GK, Zheng H, Kinzy TG. Translation elongation factor 2 anticodon mimicry domain mutants affect fidelity and diphtheria toxin resistance. J. Biol. Chem. 2006;281:32639–32648. doi: 10.1074/jbc.M607076200. [DOI] [PubMed] [Google Scholar]
- 8.Walsh CT. Posttranslational modifications of proteins: Expanding nature's inventory. Englewood, Colorado: Roberts and Company Publishers; 2006. [Google Scholar]
- 9.Dunlop PC, Bodley JW. Biosynthetic labeling of diphthamide in Saccharomyces cerevisiae. J. Biol. Chem. 1983;258:4754–4758. [PubMed] [Google Scholar]
- 10.Moehring JM, Moehring TJ, Danley DE. Posttranslational modification of elongation factor 2 in diphtheriatoxin-resistant mutants of CHO-K1 cells. Proc. Natl. Acad. Sci. USA. 1980;77:1010–1014. doi: 10.1073/pnas.77.2.1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moehring TJ, Danley DE, Moehring JM. In vitro biosynthesis of diphthamide, studied with mutant Chinese hamster ovary cells resistant to diphtheria toxin. Mol. Cell. Biol. 1984;4:642–650. doi: 10.1128/mcb.4.4.642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen JY, Bodley JW, Livingston DM. Diphtheria toxin-resistant mutants of Saccharomyces cerevisiae. Mol. Cell. Biol. 1985;5:3357–3360. doi: 10.1128/mcb.5.12.3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mattheakis LC, Sor F, Collier RJ. Diphthamide synthesis in Saccharomyces cerevisiae: structure of the DPH2 gene. Gene. 1993;132:149. doi: 10.1016/0378-1119(93)90528-b. [DOI] [PubMed] [Google Scholar]
- 14.Phillips NJ, Ziegler MR, Deaven LL. A cDNA from the ovarian cancer critical region of deletion on chromosome 17p13.3. Cancer Lett. 1996;102:85. doi: 10.1016/0304-3835(96)04169-9. [DOI] [PubMed] [Google Scholar]
- 15.Schultz DC, Balasara BR, Testa JR, Godwin AK. Cloning and localization of a human diphthamide biosynthesis-like protein-2 gene,DPH2L2. Genomics. 1998;52:186. doi: 10.1006/geno.1998.5420. [DOI] [PubMed] [Google Scholar]
- 16.Mattheakis LC, Shen WH, Collier RJ. DPH5, a methyltransferase gene required for diphthamide biosynthesis in Saccharomyces cerevisiae. Mol Cell Biol. 1992;12:4026–4037. doi: 10.1128/mcb.12.9.4026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carette JE, Guimaraes CP, Varadarajan M, Park AS, Wuethrich I, Godarova A, Kotecki M, Cochran BH, Spooner E, Ploegh HL, Brummelkamp TR. Haploid genetic screens in human cells identify host factors used by pathogens. Science. 2009;326:1231–1235. doi: 10.1126/science.1178955. [DOI] [PubMed] [Google Scholar]
- 18.Zhang Y, Zhu X, Torelli AT, Lee M, Dzikovski B, Koralewski RM, Wang E, Freed J, Krebs C, Ealick SE, Lin H. Diphthamide biosynthesis requires an organic radical generated by an iron-sulphur enzyme. Nature. 2010;465:891–896. doi: 10.1038/nature09138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Frey PA, Hegeman AD, Ruzicka FJ. The radical SAM superfamily. Crit. Rev. Biochem. Mol. Biol. 2008;43:63–88. doi: 10.1080/10409230701829169. [DOI] [PubMed] [Google Scholar]
- 20.Sofia HJ, Chen G, Hetzler BG, Reyes-Spindola JF, Miller NE. Radical SAM, a novel protein superfamily linking unresolved steps in familiar biosynthetic pathways with radical mechanisms: functional characterization using new analysis and information visualization methods. Nucl. Acids Res. 2001;29:1097–1106. doi: 10.1093/nar/29.5.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bruice PY. Organic Chemistry. Upper Saddle River, NJ: Pearson Prentice Hall; 2007. [Google Scholar]
- 22.Zhang Y, Zhu X, Torelli AT, Lee M, Dzikovski B, Koralewski RM, Wang E, Freed J, Krebs C, Ealick SE, Lin H. Diphthamide biosynthesis requires an organic radical generated by an iron-sulphur enzyme. Nature. 465:891–896. doi: 10.1038/nature09138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen JY, Bodley JW. Biosynthesis of diphthamide in Saccharomyces cerevisiae. Partial purification and characterization of a specific S-adenosylmethionine:elongation factor 2 methyltransferase. J Biol Chem. 1988;263:11692–11696. [PubMed] [Google Scholar]
- 24.Cope AC, Trumbull ER. Olefins from Amines: The Hofmann Elimination Reaction and Amine Oxide Pyrolysis. Organic Reactions. 1960;11:317–493. [Google Scholar]
- 25.Jorgensen R, Yates SP, Teal DJ, Nilsson J, Prentice GA, Merrill AR, Andersen GR. Crystal structure of ADP-ribosylated ribosomal translocase from Saccharomyces cerevisiae. J. Biol. Chem. 2004;279:45919–45925. doi: 10.1074/jbc.M406218200. [DOI] [PubMed] [Google Scholar]
- 26.Jorgensen R, Merrill AR, Yates SP, Marquez VE, Schwan AL, Boesen T, Andersen GR. Exotoxin A-eEF2 complex structure indicates ADP ribosylation by ribosome mimicry. Nature. 2005;436:979–984. doi: 10.1038/nature03871. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.