Abstract
Metal enhanced fluorescence (MEF) increased total photon emission of Cy3- and Cy5-labeled ribosomal initiation complexes near 50 nm silver particles 4- and 5.5-fold respectively. Fluorescence intensity fluctuations above shot noise, at 0.1 – 5 Hz, were greater on silver particles. Overall signal to noise ratio was similar or slightly improved near the particles. Proximity to silver particles did not compromise ribosome function, as measured by codon-dependent binding of fluorescent tRNA, dynamics of fluorescence resonance energy transfer between adjacent tRNAs in the ribosome, and tRNA translocation induced by elongation factor G.
Keywords: Single molecule fluorescence, Metal enhanced fluorescence, Silver colloids, Ribosome, Protein translation
Introduction
Fluorescence technology is widely applicable in biological research and medical/clinical diagnostics.1,2 In the last decade, single molecule fluorescence (SMF) detection has been developed to selectively address individual molecules in order to detect processes that are hidden in ensemble measurements and to avoid averaging effects.3-8 SMF has been used to study biologically important processes such as the motions of molecular motors,9-13 transcription,14,15 and translation.16-18 In single molecule fluorescence studies of localization or orientation, the target macromolecule is labeled with an organic fluorophore, such as rhodamine, cyanine dye, or a semiconductor quantum dot (QD), or ligated to a variant of green fluorescent protein.19-21 For distance measurements and dynamics, these probes are often used in pairs (e.g., Cy3 and Cy5) that exhibit distance-dependent Fluorescence Resonance Energy Transfer (FRET).
However, low emission intensity, fluctuations and photobleaching of organic fluorophores and fluorescent proteins often limit the signal-to-noise ratios achievable in SMF measurements.22-23 The local environment of the probe on the macromolecule, the imaging buffer and its oxygen content, and the illumination intensity all strongly influence photostability. Stronger illumination increases fluorescence intensity but typically with a proportional worsening of the photobleaching rate. Other characteristics also need to be taken into consideration. For instance, quantum dots are very bright and resistant to photobleaching, but they are physically larger than organic fluorophores (2-10 nm diameter) and they blink extensively.24
Many approaches have been used to improve signal-to-noise ratios in SMF measurements, including advanced confocal microscopy, total internal reflection fluorescence (TIRF) and zero-mode waveguides (ZMWs) recently used for real-time DNA sequencing.25 A common characteristic of these approaches is that they limit the volume from which fluorescence signals are acquired. An attractive alternative is addressed in this paper, an enhancement of fluorescence due to the presence of a metal nanoparticle in the immediate vicinity of the fluorophore. The method relies on using surface plasmons of noble metals to modify the spectral properties of fluorophores so as to enhance their fluorescence.26 Coupling between the fluorescence resonance of the fluorophore and the plasmon resonance of nearby metal particles forms an oscillating dipole that increases the radiative rate of fluorescence.27-29 This phenomenon is termed metal-enhanced fluorescence (MEF). Increasing the rate of fluorescence emission increases brightness, while also improving photostability and reducing blinking.30,31 In addition, as the fluorescence is enhanced only in a small region surrounding the metallic particle, MEF can effectively limit the volume from which fluorescence is collected, or, equivalently, greatly limit the contribution of background fluorescence originating from freely diffusing molecules in the sample. Importantly, the application of MEF does not exclude using many of the above mentioned techniques that limit the volume from which the fluorescence is acquired, greatly enhancing the choice of techniques available to improve signal-to-noise in SMF.
We recently reported on MEF for a series of the individual molecular components of protein synthesis.32 Although excellent applications have been found for MEF in various other biomedical assays, such as DNA hybridization,1,2,26 and cellular imaging,33 MEF has not yet been used extensively for determining biological mechanisms involving supramolecular complexes. This may partly be due to alterations of biological activity of macromolecules within the enhancement distance of these noble metals. For instance, chemical energy production in a photosynthetic reaction center is strongly altered in presence of metal nanoparticles.34
Here we tested aspects of metal enhanced fluorescence of organic fluorophores bound to components of a supramolecular complex of the ribosome and tRNAs, parts of the protein synthesis machinery. A 4–7-fold MEF enhancement in fluorescence intensity is accompanied by only minor alterations in photobleaching rate, leading to increased photon collection. The enhancement in intensity is achieved with maintenance of ribosome functionality. These results demonstrate that MEF is a promising technology for improving single molecule fluorescence and FRET monitoring in functional studies of biomacromolecular complexes such as the ribosome.
Results
Characterization of Colloidal Silver Nanoparticles
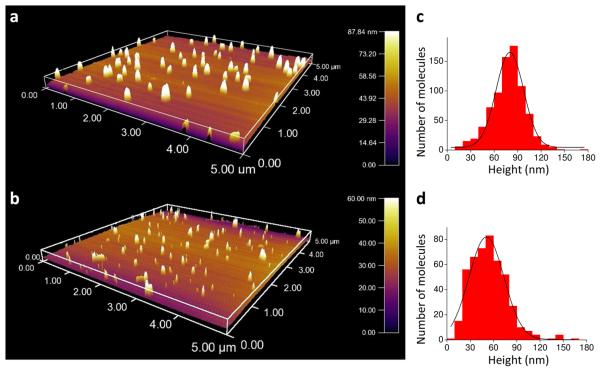
Colloidal silver particles, prepared by reducing silver nitrate with sodium citrate, were attached to glass microscope slides as described in Methods. Two sizes of particles were made by varying the rate of reduction. The size of the silver particles was measured using AFM giving average diameters of 50 ± 16 and 85 ± 18 nm (mean ± s.d., n = 463 and 789) from the heights of the small and large particles respectively (Fig. 1). Some of the particles aggregated into larger spherical or rod shaped structures which are evident in the AFM images. Surfaces coated with colloidal particles and illuminated by visible (e.g. green 532 nm) laser light appeared as discrete, focused spots in images recorded at longer wavelengths (e.g. yellow-orange, 560-620 nm, and red, 660-720 nm, Table S1).
FIGURE 1.
AFM 3-D rendering of large (a) and small (b) colloidal silver particles and their corresponding size distributions (c) and (d).
As such background emission might interfere with detection of single fluorophores on a biological sample, we further characterized its relationship to the size of the silver particles and its spectral characteristics (Supporting Information text and Figs. S1 and S2). The emission was colocalized with the silver particles detected either by AFM or by backscattered visible light and the intensity from the larger particles was greater and extended to longer wavelengths than that from the smaller particles. Fluorescence of silver clusters has been described before,35-37 but commonly after oxidation or higher intensity illumination than used here. Other possible causes of this background may be surface-enhanced Raman scattering of adsorbed material or fluorescence from other optical components, although the sharp focus of the red-shifted luminescence (Fig. S1 in Supporting Information) renders the latter explanation unlikely.
The background from silver particles either does not photobleach or else bleaches slowly and gradually, whereas single organic fluorophores used for biological labeling photobleach in a single step. As a result, stepwise photobleaching to a steady background enables fluorophores to be identified at the site of a colloidal particle or on plain glass.
Cy3 and Cy5 Fluorescence Enhancement near Colloidal Silver Nanoparticles
The smaller, 50 nm silver particles produced less background, so this material was used to characterize the enhancement of fluorescence from Cy3 and Cy5 probes. Fluorescently labeled ribosomes were attached to the microscope slides through a short biotinylated test mRNA linked to a layer of polyethylene glycol (PEG) covering sparsely distributed colloidal particles (Methods). Ribosomal initiation complexes (ICs) were labeled specifically with Cy3 or Cy5 on the large subunit protein L11,18 or pre-translocation complexes contained Cy5-labeled arginine tRNA bound to the A-site. The labeled ribosomes attached to the surface via a biotinylated mRNA, in positions that were random with respect to the silver particles. The Cy3- or Cy5-labeled complexes were excited in TIRF mode by green (532 nm) or red (640 nm) lasers, respectively. Single molecules were selected on the basis of single step photobleaching to avoid interference from colloidal background, as described above, and intensities were calculated by fitting 2D Gaussian profiles to the intensity distributions as described in Methods.
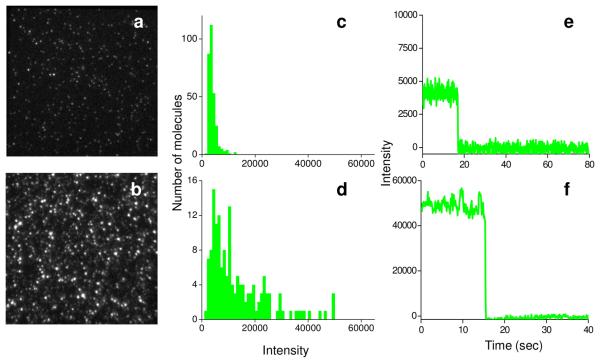
Comparison of fluorescence micrographs of Cy3-labeled initiation complexes bound to PEG-coated glass slides without and with silver particles (Fig. 2a and 2b, respectively) shows marked enhancement of fluorescence intensity by the colloidal particles. The median intensity on plain glass was 3,900 camera intensity units, with very few spots having intensities higher than 10,000 units (Fig. 2c). In contrast, a large proportion of the single molecule complexes had intensities of >10,000 units on slides containing silver particles (Fig. 2d). As determined from the quantum yield of the camera and its gain (measured as described in Methods), 10,000 intensity units correspond to ~160 photons striking a camera pixel during each 100 ms recording period. The higher intensity spots in the presence of the colloidal particles are not aggregates of more than one labeled ribosome, because the fluorescence bleaches to the background level in a single step (intensity traces, Fig. 2e and f). As expected, quite similar results were found for Cy5-labeled initiation complexes (Fig. S3 in Supporting Information).
FIGURE 2.
Comparison of Cy3 labeled initiation complexes (ICs) on the plain glass surface and small silver particle coated glass surface. Fluorescence images (a, b), intensity histograms (c, d), and single molecule traces (e, f), on plain glass (a, c, e) and on silver particle coated glass (b, d, f).
The higher fluorescence on the colloidal particles often displayed 0.1 – 5 Hz fluctuations above photon counting shot noise, as in Fig. 2f and S3. Noise in the traces on plain glass was slightly higher than expected from photon counting statistics, presumably due to excess noise of the camera gain multiplier and fluorophore's photochemical processes. For ICs colocalized with 50 nm silver particles, the noise above the expected shot noise was higher, 2 – 3-fold above shot noise (Table S2 in Supporting Information). After photobleaching of both Cy3 and Cy5 high intensity spots, the background intensity was slightly higher (~500 intensity units) than on plain glass, presumably due to background emission of the nearby colloid particle.
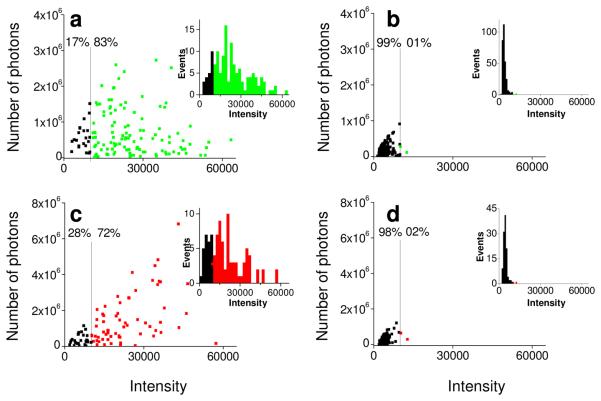
To confirm that enhancement of the fluorescence was due to the proximity of the ribosomes to the silver particles, we captured images of backscattered light from the silver particles and fluorescence from labeled ribosomes in the same area, registered the images to each other using Matlab and ImageJ scripts, and sorted the spots according to colocalization with a silver particle, within one pixel. For Cy3-labeled ICs, this analysis showed that 83% of the molecules that were colocalized with a silver particle had high (>10,000) intensity compared to 1% of the spots on plain glass (Fig. 3a and 3b, respectively). The corresponding values for Cy5-labeled ICs were 72% and 2% (Fig. 3c and 3d respectively). Median intensities for the fluorophores colocalized with colloids (Table S2 in Supporting Information) were enhanced, 6.7-and 4.7-fold for Cy3 and Cy5, respectively, relative to values on plain glass.
FIGURE 3.
Photostability-intensity plots of Cy3 and Cy5 labeled ICs on plain glass and small silver particle coated glass. The number of photons emitted from each single spot before photobleaching is plotted vs. intensity of Cy3 labeled ICs colocalized (within 1 pixel) with silver particles (a) and on plain glass (b), Cy5 labeled ICs colocalized silver particles (c) and on plain glass (d). Intensity distributions for respective Cy3 and Cy5 labeled ICs are in the insets.
Assuming that binding of ribosome ICs to the PEG coated surface was random, irrespective of whether a colloidal particle was nearby, the density of colloidal particles and the proportion of fluorophores with enhanced fluorescence allow an estimate of the area around a particle that leads to enhancement. The density of colloidal particles on the surface, detected by AFM was 2.3 ± 0.3 per μm2. 52% of Cy5 labeled complexes on the silver-treated slides exhibited high intensity (>10,000 camera units). These values lead to an area with apparent radius of 270 nm surrounding each particle that gave fluorescent enhancement. This value is very approximate because the density of colloids was measured by AFM on separate slides from those used for fluorescence measurements. In addition, fewer particles, presumably only the larger ones, were identified by optical backscattering (Fig. S1 in Supporting Information).
For the fluorescence enhancement to be of practical benefit in a dynamic biophysical experiment, the total number of photons collected from the fluorophore before it photobleaches must be enhanced. Recording times before photobleaching of the fluorescence enhanced Cy3- and Cy5-labeled ICs close to silver particles were compared to ones bound to plain glass. The product of the recording time (determined by the rate of photobleaching) and the intensity gives a relative measure of the total number of photons captured. Under the present recording conditions, the average recording times for Cy3 and Cy5 labeled initiation complexes were unchanged or slightly increased by the presence of silver particles (31 s for Cy3 near particles vs. 30 s on plain glass and 43 s for Cy5 near particles vs. 33 s on glass (Table S2, Fig. S4 in Supporting Information). The total number of photons emitted by Cy3- and Cy5-labeled initiation complexes (the products of intensity and recording time) were 4.2- and 5.5-fold higher on average, respectively, near silver particles compared to plain glass (Table S2 in Supporting Information, Fig. 3).
Due to higher scattering, large silver colloidal particles have been reported to produce greater enhancement of fluorescence than small ones.1 Median intensity was increased more (6.7-fold, Fig. S5, Table S3 in Supporting Information) on 85 nm particles than on 50 nm ones (4.7-fold, Fig. 3, Table S2 in Supporting Information). It is worth noting, however, that the average recording time before photobleaching was shorter near the 85 nm particles than the 50 nm particles: 30s (Fig. S6 in Supporting Information) and 43s (Fig. S4 in Supporting Information), respectively. The enhancement of total number of photons collected from Cy5 near colloidal particles, above those on plain glass, was very similar on large particles (4.8-fold, Table S3 in Supporting Information) to that on small ones (5.5-fold, Table S2 in Supporting Information).
Activity Assays
The ribosome contains three tRNA binding locations: the aminoacyl (A) site, the peptidyl (P) site, and the exit (E) site. During polypeptide synthesis, the state after formation of a peptide bond, but before translocation of the messenger RNA and two transfer RNAs (tRNAs) along the ribosome is termed the pretranslocation complex. This state (containing a deacylated tRNA in the P-site and peptidyl tRNA in the A-site) has two conformational states, denoted as the classic (A/A, P/P) and hybrid (A/P, P/E) tRNA/ribosome pretranslocation states.38,39 The classic and hybrid states have been found to interconvert dynamically in single molecule FRET (smFRET) studies.40-42 In separate single molecule FRET (smFRET) studies conducted on plain glass and reported elsewhere42, we found both complexes in the 2nd elongation cycle, and containing tRNAArg(Cy5) and fMetArgPhe-tRNAPhe(Cy3) bound to adjacent codons in the ribosome (termed PRE2-tt)43, and complexes in the first elongation cycle, containing Cy3-labeled L11 and fMet-Arg-tRNAArg(Cy5) in the A-site (termed PRE1-Lt), exhibited fluctuations between two conformations consistent with the classic and hybrid states. We also showed that addition of EF-G·GTP to PRE1-Lt complexes leads to a large decrease in FRET efficiency, consistent with the expected increase in L11-tRNA distance that accompanies translocation of A-site tRNA to the P-site.
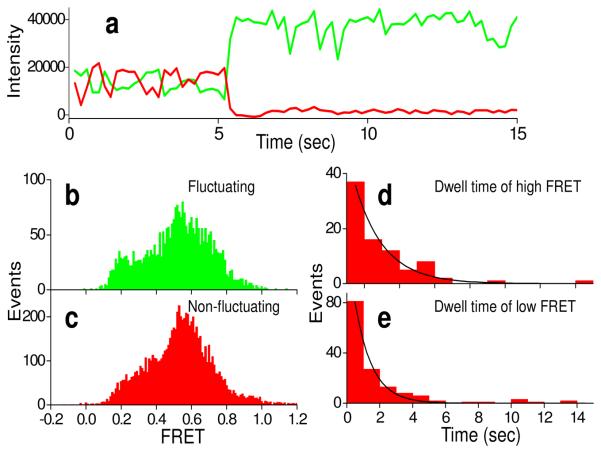
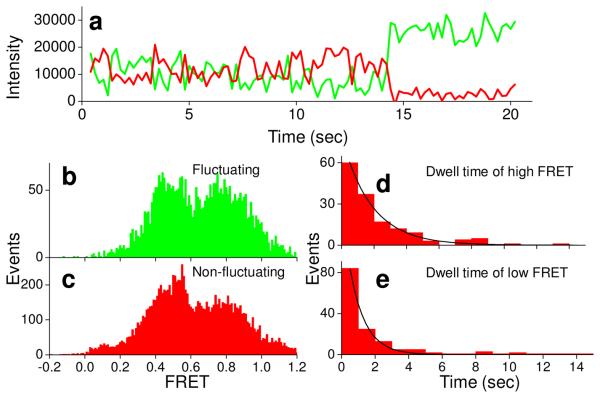
Quite similar results were found here for ribosome complexes bound close to silver particles, providing strong evidence that such complexes retain functional activity. Of ~900 ribosomes prepared as PRE2-tt complexes and containing colocalized Cy3 and Cy5 fluorophores (collected from at least 6 different regions from two different experiments, Methods), 65% gave high Cy5 intensity (>10,000 counts for Cy5 upon direct excitation, assumed to be close to the silver particles) and 35% gave lower Cy5 intensity (≤10,000 counts, assumed to be away from silver particles). Of the high intensity colocalized spots, 21% showed FRET efficiency >0.35 between the two adjacent tRNAs, compared to 30% for lower intensity ones, suggesting that slightly fewer of the ribosomes are active in this assay near the silver particles than on PEG-coated plain glass. Both the high and low intensity traces showed two FRET components (corresponding to the classic and hybrid states) (high intensity, efficiencies 0.65 and 0.36, Fig. 4; low intensity, efficiencies 0.66 and 0.43, Fig. S7 in Supporting Information) those were very similar to each other and to the FRET components found on plain glass (ref. 42, 0.69 and 0.38). In many of the ribosomes, the Cy5-tRNAArg and Cy3-tRNAPhe fluctuate between these two states. Time courses of anti-correlated Cy3 donor and Cy5 acceptor fluorescence intensities (Figs. 4, Fig. S7 in Supporting Information), show similar characteristics except for the higher total intensity on silver particles. Average dwell times at high and low FRET were 2.2 ± 0.2 s and 1.5 ± 0.2 s, respectively, for high intensity traces (Fig. 4d,e), 2.8 ± 0.4 s and 1.8 ± 0.2 s, respectively, for low intensity traces (Fig. S7 in Supporting Information) similar to the values of 2.5 ± 0.3 and 2.1 ± 0.2 s on plain glass.42 Thus the FRET efficiency ratios and dynamics are not affected markedly by proximity to colloidal particles.
FIGURE 4.
Time courses of fluorescence intensity of pre-complex (PRE-tt, having tRNAArg(Cy3) in P site and fMet-Arg-Phe-tRNAPhe(Cy5) in the A site) close to particles on a small silver particle coated surface (a). Cy3 (green) and Cy5 (red) fluorescence intensity traces under 532 nm laser illumination show fluctuation between high and low FRET and photobleaching of the Cy5 at ~5 s. FRET efficiency distributions from fluctuating and non-fluctuating complexes are shown in (b) and (c), respectively. (d) and (e) are dwell time distributions for high and low FRET states, respectively.
Results obtained for ribosomes prepared as PRE-Lt complexes also parallel those seen for the study using plain glass.42 Many ribosomes oscillated back and forth between high and low L11-tRNA FRET efficiency values (assigned to classic and hybrid states), and some of them remained at stable high or low FRET values. The L11-tRNA FRET efficiencies for pretranslocation complexes near silver particles averaged 0.82 and 0.48 with average dwell times of 2.2 ± 0.3 s and 2.0 ± 0.2 s in the high and low FRET states, respectively (Fig. 5b-e). These values were similar to high and low FRET values of 0.80 and 0.44 on plain glass and dwell times of 2.1 ± 0.1 s and 1.9 ± 0.2 s, respectively.42 Further, infusion of EF-G·GTP into surfaces containing the PRE1-Lt complex led to a large decrease (92%) in the number of molecules near silver colloidal particles containing colocalized Cy3 and Cy5 with FRET efficiencies greater than 0.2. As on the plain glass,42 this is consistent with translocation of fMet-Arg-tRNAArg from the A-site to the P-site.
FIGURE 5.
Time courses of fluorescence intensity of Cy3-L11 labeled pre-complex (PRE-Lt, having tRNAfMet in P site and fMet-Arg-tRNAArg(Cy5) in the A site) close to a particle on a small silver particle coated surface. Cy3 (green) and Cy5 (red) fluorescence intensity traces of PRE-Lt complex close to silver particles (a) under 532 nm laser illumination show photobleaching of Cy5 at ~25 s. FRET efficiency distributions from fluctuating and non-fluctuating complexes are shown in b and c, respectively. (d) and (e) are dwell time distributions for high and low FRET states, respectively.
Discussion
We present here an application of metal enhanced fluorescence (MEF) for single molecule studies of protein synthesis. Although MEF has previously been shown for many fluorophores, including Cy3 and Cy5, prior to the present work it was unknown whether proximity to colloidal silver particles affects the recording time before photobleaching. Our results clearly show a 4 - 7-fold enhancement of fluorescence intensity of labeled ribosomal initiation complexes (ICs) near silver colloidal particles compared to that seen with PEG over plain glass. Changes in photobleaching rate are minor. The enhancement of fluorescence intensity leads to a 4- and 5-fold increase in total number of photons collected for Cy3 and Cy5 labeled (ICs), respectively. Larger colloids enhance the fluorescence signal more than smaller ones, as expected from earlier reports,1 but in the size range of 50-85 nm tested here, enhancement of total numbers of photons from Cy5 fluorescence does not depend on the particle size. Despite this large enhancement in fluorescence intensity, a concomitant increase in 0.1 – 5 Hz fluctuations in fluorescence intensity from surfaces coated with colloidal silver particles led to an overall signal to noise ratio that was either similar to or slightly enhanced relative to fluorescent labeled ICs on plain glass.
The recording time before photobleaching under laser illumination is inversely proportional to laser intensity.44 Therefore, we expect that the enhancement can be used effectively to extend the recording time before photobleaching. Our work represents the first application of MEF to the study of a supramolecular complex, in this case the ribosome. The similarities in the results obtained for pretranslocation and posttranslocation complexes bound near silver colloidal particles compared to those bound to plain glass strongly suggest that MEF did not significantly compromise ribosome function.
Ribosomes bound randomly to the surface, either near or far from the colloids. In principle, virtually all of the labeled ribosomes that co-localize with the silver particles should give MEF, while all those that do not co-localize should not. In practice we found that 70 - 80% of the co-localized labeled ribosomes had high intensity, as compared with 20-30% of the noncolocalized ribosomes. The higher than expected intensities for some of the non-localized ribosomes (non-colocalized spots, Fig. S6 in Supporting Information) are most likely attributable to incomplete identification of the colloidal particles by light scattering due to the noise and limited sensitivity of the camera. This explanation is supported by the very small number of high intensity spots found on plain glass and the 3 - 4-fold higher density of particles that were detected by AFM than by light scattering. The low intensity observed for some of the co-localized ribosomes most likely reflects the limited resolution of co-localization (~250 nm), as well as the expected quenching of fluorescence when fluorophores are within 2 nm of colloids.45 The latter effect should be limited however, since the 5,000 Da PEG coating the surface was approximately 10 nm thick. In addition the mRNA strand linking the ribosome to the PEG was ~5 nm long.
The estimate of the radius surrounding the colloidal particles that provided fluorescence enhancement, according to the density of particles and the proportion of enhanced fluorophores, ~270 nm, would also need to be adjusted if a central region is quenched, rather than enhanced. An earlier study45 reported smaller distances for enhancement, peak enhancement at 5-9 nm between the particle and the fluorescent probe and extending to ~50 nm. Differences between that study and ours are that the silver particles were (~10-fold) smaller, more densely applied to the surface to form a film, and the metal particle-fluorophore distance was determined by an intervening protein layer rather than non-specific attachment.
Another major issue in the use of MEF is background intensity from bare colloids that overlaps the emission spectra of the fluorophores, thereby potentially interfering with single molecule measurements. For the smFRET results reported in this study, we selected molecules that showed only single step photobleaching to a stable background. This criterion distinguishes fluorescence emission of Cy3 and Cy5 from colloidal background intensity, since the luminescence signal from colloidal aggregates either does not bleach or bleaches slowly and gradually, and never in a single step.
In conclusion, this study shows that metal enhanced fluorescence with 50-85 nm silver colloidal particles could be successfully applied in studies of biological processes involving supramolecular complexes. We found that ribosomal complexes are fully active near these particles. We observed long wavelength luminescence background and excess noise associated with many of the silver particles. The background could be eliminated as a problem by using single step photobleaching to select organic fluorophores. These results engender confidence that metal enhanced fluorescence will be useful in further studies of ribosomes, and quite possibly, of many other types of supramolecular complexes.
Methods
Small and Large Colloids
Small and large colloids were prepared according to ref. 46 with slight modifications. Trisodium citrate (2 ml at 34 mM) was added dropwise to a stirred solution of AgNO3 (100 ml at 1 mM) at 90 °C. The reaction mixture was heated to 90-95 °C, and stirring was continued for 15 min or until the reaction mixture turned pale yellow. For large colloids, the trisodium citrate was added in 4 aliquots of 0.5 ml each every 15 min. The resulting mixture was then incubated in an ice bath for 15-20 min. The small and large colloids were then purified by centrifugation three times at 8000 and 3500 rpm respectively for 8 min each time; the precipitate was then suspended in 1 ml of 1 mM trisodium citrate.
Buffers
All single molecule experiments were carried out in TAM15 buffer (20 mM Tris-HCl (pH 7.5), 15 mM Mg(OAc)2, 30 mM NH4Cl, 70 mM KCl, 0.75 mM EDTA, 1 mM DTT, and 0.2% (w/v) Tween 20). A deoxygenation enzyme system of 3 mg/mL glucose, 100 μg/mL glucose oxidase (Sigma-Aldrich), 40 μg/ml catalase (Roche), and 1.5 mM 6-hydroxy-2,5,7,8-tetramethylchromane-2-carboxylic acid (Trolox, Sigma-Aldrich), initially dissolved at 150 mM in DMSO, was added during single-molecule measurements to decrease photobleaching and photoblinking.
Proteins, Ribosomes and tRNAs
Proteins were prepared as described in ref. 42. 70SCy3 ribosomes were prepared by incubating a 2-fold molar excess of C38S/S87C-L11Cy3 with 2 μM AM77 ribosomes lacking L11 as described,42 yielding a ribosome pellet with Cy3/ribosome ratio of 0.8. fMet-tRNAfMet(Cy5), Arg-tRNAArg(Cy3), Arg-tRNAArg(Cy5), and Phe-tRNAPhe(Cy5) were prepared using the reduction, charging, and labeling protocol as described.42,43
mRNA
The mRNA, was purchased from Dharmacon, Inc. 5′-biotin labeled- GGG AAU UCA AAA AUU UAA AAG UUA AUA AGG AUA CAU ACU AUG CGU UUC UUC CGU UUC UAU CGU UUC. The underlined sequence is a strong Shine-Dalgarno region and italized sequence codes for MRFFRFYRF (single letter amino acid code).
Complex Formation
70S initiation complex was formed by incubating 1 μM 70S ribosome, 4 μM 5′-biotinylated mRNA, 1.5 μM each of IF1, IF2, IF3 and fMet-tRNAfMet, and 1 mM GTP in TAM15 buffer for 25 min at 37 °C. Ternary complex was formed by incubating 4 μM EF-Tu, 2 μM dye-labeled and charged tRNA, 3 mM GTP, 1.3 mM phosphoenolpyruvate, and 5 mg/L pyruvate kinase in TAM15 buffer for 15 min at 37 °C.
Immobilization Method
Pre-cleaned glass coverslips (Fisher Scientific) were aminosilanized by 3-aminopropyltriethoxysilane (United Chemical Technologies, Inc.). Colloids were then reacted with the aminosilane surface for 3 hr, followed by incubation with polyethylene glycol (PEG, Laysan Bio, mixture of 5000 MW PEG-succinimidyl valerate and biotin-PEG-succinimidyl valerate at a molar ratio of 100:1 unbiotinylated:biotinylated PEG) for an additional 3 hr. Inititation complexes were bound to the surface by applying streptavidin solution (0.5 mg/mL), and biotinylated initiation complex solution (1-10 nM) for 3 min each followed by a wash with. To prepare immobilized pre-translocation complexes EF-Tu/tRNA ternary complex (10 nM) was applied for 3 min followed by a TAM15 buffer wash. The translocation reaction was started by injecting 2 μM EF-G and 3 mM GTP into immobilized pre-translocation complexes. All single molecule studies were carried out at 21 °C.
Fluorescence intensity, FRET, and function of ribosomes were compared on slides containing colloidal silver particles as above with corresponding measurements on identically prepared slides except the colloids were not applied. In the text, the slides without colloids are termed “plain glass” but all of the layers (e.g. silane and PEG) except the colloids were the same.
TIRF measurements on immobilized ribosomes
A custom-built objective-type total internal reflection fluorescence (TIRF) microscope was based on a commercial inverted microscope (Eclipse Ti, Nikon) with a 1.49 N.A. 100x oil immersion objective (Apo TIRF; Nikon, Tokyo, Japan). Direct excitation (for fluorescence enhancement experiments) or Alternating-Laser EXcitation (ALEX)47 (for FRET in activity assays) was used with a 532 nm laser (CrystaLaser, Inc.) and a 640 nm laser (Coherent, Inc.) to obtain Cy3 and Cy5 fluorescence intensities, and the FRET signal between Cy3 and Cy5. Fluorescence emission from Cy3 and Cy5 were separated by a Quad View splitter (Photometrics, Tucson, AZ) and recorded with an electron multiplying charge-coupled device (EM-CCD) camera (Cascade II, Photometrics) at 100 ms integration time as described.42
For recording, several areas of 50 × 50 μm each were scanned from different slide chambers. Single molecules were then selected on the basis of single step photobleaching to avoid colloidal luminescence and intensities were calculated by fitting the intensity distribution to 2D Gaussian profiles within a 9 × 9 pixel area
| Equation 1 |
where A is the amplitude, x0, y0 is the center and σx, σy are the x and y standard deviations of the intensity distribution. The total number of camera digitizer units in the image (the intensity) was calculated as I = 2π A σx, σy. σx = σy = 1.15 and 1.2 for Cy3 and Cy5, respectively. For fluorescence enhancement studies, intensities were then converted into number of photons using camera gain measured for the EMCCD camera by the procedure described in the next section. For FRET measurements, intensity traces were directly used and the FRET efficiency was calculated using equation 2
| Equation 2 |
where ID is fluorescence intensity of donor, IA is fluorescence intensity of acceptor, χ is crosstalk of donor into acceptor channel, and γ accounts for the differences in quantum yield and detection efficiency between the donor and the acceptor. γ is calculated as the ratio of change in the acceptor intensity, ΔIA to change in the donor intensity, ΔID upon acceptor photobleaching or change of FRET efficiency (γ =ΔIA/ΔID).48,49
Camera gain measurement
The gain of the EM-CCD camera was measured in order to calculate the conversion factor relating the spot intensities to the number of collected photons. Measured pixel intensity A is related to the number of photons, N, by A = Q N C / B, where Q = quantum yield of the camera detector, C = gain (output current/input current) of the camera electron multiplier, and B = number of electrons (after the cascade amplifier) converted to each digital intensity unit (ADUs). B is often termed “Gain” in EM-CCD camera literature. The variance (V) of a pixel intensity is given by V = 2 Q N C2 / B2, where the factor of 2 is an approximation of the excess noise introduced by the cascade amplifier.50 Values of variance and intensities were obtained by measuring pixel intensities over time with a stable light source, or from successive difference images. Plots of V vs. A were linear with slope (V / A = 2 C / B) corresponding to C / B = 70 at the experimental cascade amplifier setting. Taking Q as 0.9 from the camera specifications, the ratio of pixel output signal to number of collected photons is thus 63.
HaMMy
The Hidden Markov Model based software HaMMy51 was used to analyze the FRET traces. The software was set model two FRET states. Traces with only one FRET state were considered to be stable traces, whereas the traces with two FRET states were considered to be fluctuating traces. Dwell times of high and low FRET of fluctuating traces were calculated by HaMMy.
AFM
An atomic force microscope (MFP 3D-BIO Model, Asylum Research, Inc.) was used to characterize the size of colloidal silver particles. Silver particles were prepared and deposited as described above and dry sample imaging was performed in tapping mode. An AC 240TS cantilever with resonant frequency ~60 kHz was used. The AFM system was integrated with an inverted objective-type TIRF microscope similar to the instrument described earlier, based on a Nikon Eclipse Ti platform and a 1.49 N.A 100x (Apo TIRF; Nikon, Inc.) oil immersion objective. For data analysis of the AFM images, we used Igor-based MFP 3D and ARgyle Light software supplied by Asylum Research, Inc.
Supplementary Material
Acknowledgements
The work was supported by National Institutes of Health grants GM080376 (to BSC and YEG) and HG004364 (to WM) and NIST grant 70NANB7H7011 (to Anima Cell metrology). We thank Dr. Yujie Sun and Taniya Das for help with the AFM and John F. Beausang and Shawn Pfeil for helpful discussions.
Footnotes
Author contributions
W.M., I.G., Z.G., B.S.C. and Y.E.G designed the research. S.B., C.C., B.S., and J.K., conducted the research. Z.S. supplied specialized software. S.B., C.C., B.S., B.S.C. and Y.E.G wrote the paper. All authors discussed and commented on the manuscript.
Supporting Information Available
Characterization of wavelength-shifted background emission of silver particles, intensity distribution and noise of Cy3 and Cy5 labeled ICs on plain glass, small and large silver colloids and Materials & Methods. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Lakowicz JR. Principles of Fluorescence Spectroscopy. Kluwer Academic/Plenum Publishers; New York: 2006. [Google Scholar]
- 2.Mason W. Fluorescent and Luminescent Probes for Biological Activity: A Practical Guide to Technology for Quantitative Real-Time Analysis. Academic Press; New York: 1999. [Google Scholar]
- 3.Kapanidis AN, Strick T. Biology, One Molecule at a Time. Trends Biochem Sci. 2009;34:234–243. doi: 10.1016/j.tibs.2009.01.008. [DOI] [PubMed] [Google Scholar]
- 4.Aleman EA, Lamichhane R, Rueda D. Exploring RNA Folding One Molecule at a Time. Curr Opin Chem Biol. 2008;12:647–654. doi: 10.1016/j.cbpa.2008.09.010. [DOI] [PubMed] [Google Scholar]
- 5.Deniz AA, Mukhopadhyay S, Lemke EA. Single-Molecule Biophysics: at the Interface of Biology, Physics and Chemistry. J R Soc Interface. 2008;5:15–45. doi: 10.1098/rsif.2007.1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Peters R. Single-Molecule Fluorescence Analysis of Cellular Nanomachinery Components. Annu Rev Biophys Biomol Struct. 2007;36:371–394. doi: 10.1146/annurev.biophys.36.040306.132715. [DOI] [PubMed] [Google Scholar]
- 7.Bustamante C, Bryant Z, Smith SB. Ten Years of Tension: Single-Molecule DNA Mechanics. Nature. 2003;421:423–427. doi: 10.1038/nature01405. [DOI] [PubMed] [Google Scholar]
- 8.Beausang JF, Sun Y, Quinlan ME, Forkey JN, Goldman YE. Orientation and Rotational Motions of Single Molecules by Polarized Total Internal Reflection Fluorescence Microscopy. In: Selvin PR, Ha T, editors. Single-Molecule Techniques: A Laboratory Manual. Cold Spring Harbor Press; New York: 2008. pp. 121–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Peterman EJG, Sosa H, Moerner WE. Single-Molecule Fluorescence Spectroscopy and Microscopy of Biomolecular Motors. Annu Rev Phys Chem. 2004;55:79–96. doi: 10.1146/annurev.physchem.55.091602.094340. [DOI] [PubMed] [Google Scholar]
- 10.Goldman YE. Imaging and Molecular Motors. In: Yanagida T, Ishii Y, editors. Single Molecule Dynamics in Life Sciences. Wiley-VCH; Weinheim: 2009. pp. 41–86. [Google Scholar]
- 11.Ross JL, Wallace K, Shuman H, Goldman YE, Holzbaur EL. Processive Bidirectional Motion of Dynein-Dynactin Complexes in vitro. Nat Cell Biol. 2006;8:562–570. doi: 10.1038/ncb1421. [DOI] [PubMed] [Google Scholar]
- 12.Sun Y, Schroeder HW, 3rd., Beausang JF, Homma K, Ikebe M, Goldman YE. Myosin VI Walks “Wiggly” on Actin with Large and Variable Tilting. Mol Cell. 2007;28:954–964. doi: 10.1016/j.molcel.2007.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dixit R, Ross JL, Goldman YE, Holzbaur EL. Differential Regulation of Dynein and Kinesin Motor Proteins by Tau. Science. 2008;22:1086–1089. doi: 10.1126/science.1152993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Galburt EA, Grill SW, Bustamante C. Single Molecule Transcription Elongation. Methods. 2009;48:323–332. doi: 10.1016/j.ymeth.2009.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Davenport RJ, Wuite GJ, Landick R, Bustamante C. Single-Molecule Study of Transcriptional Pausing and Arrest by E. coli RNA Polymerase. Science. 2000;287:2497–2500. doi: 10.1126/science.287.5462.2497. [DOI] [PubMed] [Google Scholar]
- 16.Blanchard SC. Single-Molecule Observations of Ribosome Function. Curr Opin Struct Biol. 2009;19:103–109. doi: 10.1016/j.sbi.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vanzi F, Takagi Y, Shuman H, Cooperman BS, Goldman YE. Mechanical Studies of Single Ribosome/mRNA Complexes. Biophys J. 2005;89:1909–1919. doi: 10.1529/biophysj.104.056283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang Y, Qin H, Kudaravalli RD, Kirillov SV, Dempsey GT, Pan D, Cooperman BS, Goldman YE. Single-Molecule Structural Dynamics of EF-G-Ribosome Interaction during Translocation. Biochemistry. 2007;46:10767–10775. doi: 10.1021/bi700657d. [DOI] [PubMed] [Google Scholar]
- 19.Resch-Genger U, Grabolle M, Cavaliere-Jaricot S, Nitschke R, Nann T. Quantum Dots Versus Organic Dyes as Fluorescent Labels. Nature methods. 2008;5:763–775. doi: 10.1038/nmeth.1248. [DOI] [PubMed] [Google Scholar]
- 20.Shaner NC, Steinbach PA, Tsien RY. A Guide to Choosing Fluorescent Proteins. Nature Methods. 2005;2:905–909. doi: 10.1038/nmeth819. [DOI] [PubMed] [Google Scholar]
- 21.Miyawaki A, Sawano A, Kogure T. Lighting up Cells: Labeling Proteins with Fluorophores. Nat Cell Biol. 2003;(Suppl):S1–7. [PubMed] [Google Scholar]
- 22.Weiss S. Fluorescence Spectroscopy of Single Biomolecules. Science. 1999;283:1676–1683. doi: 10.1126/science.283.5408.1676. [DOI] [PubMed] [Google Scholar]
- 23.Gell C, Brockwell D, Smith A. Handbook of Single Molecule Fluorescence Spectroscopy. Oxford Univ. Press; Oxford: 2006. Preparation of Samples for Single Molecule Fluorescence Spectroscopy. [Google Scholar]
- 24.Lee SF, Osborne MA. Brightening, Blinking, Bluing and Bleaching in the Life of a Quantum Dot: Friend or Foe? Chemphyschem. 2009;10:2174–2191. doi: 10.1002/cphc.200900200. [DOI] [PubMed] [Google Scholar]
- 25.Eid J, Fehr A, Gray J, Luong K, Lyle J, et al. Real-time DNA Sequencing from Single Polymerase Molecules. Science. 2009;323:133–138. doi: 10.1126/science.1162986. [DOI] [PubMed] [Google Scholar]
- 26.Aslan K, Gryczynski I, Malicka J, Matveeva E, Lakowicz JR, Geddes CD. Metal-Enhanced Fluorescence: An Emerging Tool in Biotechnology. Curr Opin Biotechnol. 2005;16:55–62. doi: 10.1016/j.copbio.2005.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lakowicz JR. Radiative Decay Engineering 5: Metal-Enhanced Fluorescence and Plasmon Emission. Anal Biochem. 2005;337:171–194. doi: 10.1016/j.ab.2004.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lakowicz JR. Radiative Decay Engineering: Biophysical and Biomedical Applications. Anal Biochem. 2001;298:1–24. doi: 10.1006/abio.2001.5377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ghosh SK, Pal T. Interparticle Coupling Effect on the Surface Plasmon Resonance of Gold Nanoparticles: from Theory to Applications. Chem Rev. 2007;107:4797–4862. doi: 10.1021/cr0680282. [DOI] [PubMed] [Google Scholar]
- 30.Malicka J, Gryczynski I, Fang J, Lakowicz JR. Fluorescence Spectral Properties of Cyanine Dye-Labeled DNA Oligomers on Surfaces Coated with Silver Particles. Anal Biochem. 2003;317:136–146. doi: 10.1016/S0003-2697(03)00005-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fu Y, Lakowicz JR. Single-Molecule Studies of Enhanced Fluorescence on Silver Island Films. Plasmonics. 2007;2:1–4. doi: 10.1007/s11468-007-9023-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mandecki W, Bharill S, Borejdo J, Cabral D, Cooperman BS, Farrell I, Fetter L, Goldman E, Gryczynski Z, Jakubowski H, Liu H, Luchowski R, Matveeva E, Pan D, Qin H, Tennant D, Gryczynski I. Fluorescence Enhancement on Silver Nanostructures: Studies of Components of Ribosomal Translation in vitro. Proc Soc Photo Opt Instrum Eng. 2008;6862:68620T. [Google Scholar]
- 33.Zhang J, Fu Y, Liang D, Zhao RY, Lakowicz JR. Fluorescent Avidin-Bound Silver Particle: A Strategy for Single Target Molecule Detection on a Cell Membrane. Anal Chem. 2009;81:883–889. doi: 10.1021/ac801932m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Govorov AO, Carmeli I. Hybrid Structures Composed of Photosynthetic System and Metal Nanoparticles: Plasmon Enhancement Effect. Nano lett. 2007;7:620–625. doi: 10.1021/nl062528t. [DOI] [PubMed] [Google Scholar]
- 35.Kreibig U. Optical Properties of Metal Clusters. Springer; New York: 1994. [Google Scholar]
- 36.Peyser LA, Vinson AE, Bartko AP, Dickson RM. Photoactivated Fluorescence from Individual Silver Nanoclusters. Science. 2001;291:103–106. doi: 10.1126/science.291.5501.103. [DOI] [PubMed] [Google Scholar]
- 37.Geddes CD, Parfenov A, Gryczynski I, Lakowicz JR. Luminescent Blinking from Silver Nanostructures. J Phys Chem B. 2003;18:9989–9993. doi: 10.1021/jp030290g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moazed D, Noller HF. Intermediate States in the Movement of Transfer RNA in the Ribosome. Nature. 1989;342:142–148. doi: 10.1038/342142a0. [DOI] [PubMed] [Google Scholar]
- 39.Agirrezabala X, Lei J, Brunelle JL, Ortiz-Meoz RF, Green R, Frank J. Visualization of the Hybrid State of tRNA Binding Promoted by Spontaneous Ratcheting of the Ribosome. Mol Cell. 2008;32:190–197. doi: 10.1016/j.molcel.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Munro JB, Altman RB, O'Connor N, Blanchard SC. Identification of Two Distinct Hybrid State Intermediates on the Ribosome. Mol Cell. 2007;25:505–517. doi: 10.1016/j.molcel.2007.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Blanchard SC, Kim HD, Gonzalez RL, Jr., Puglisi JD, Chu S. tRNA Dynamics on the Ribosome During Translation. Proc Natl Acad Sci USA. 2004;101:12893–12898. doi: 10.1073/pnas.0403884101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cooperman BS, Chen C, Stevens B, Liu H, Cabral D, Zhang H, Kaur J, Wang Y, Pan D, Smilansky Z, Goldman YE. Single Molecule Structural Dynamics before and during Translocation. FASEB J. 2010;24:838.3. [Google Scholar]
- 43.Pan D, Qin H, Cooperman BS. Synthesis and Functional Activity of tRNAs Labeled with Fluorescent Hydrazides in the D-loop. RNA. 2009;15:346–354. doi: 10.1261/rna.1257509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Quinlan ME, Forkey JN, Goldman YE. Orientation of the Myosin Light Chain Region by Single Molecule Total Internal Reflection Fluorescence Polarization Microscopy. Biophys J. 2005;89:1132–1142. doi: 10.1529/biophysj.104.053496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Malicka J, Gryczynski I, Gryczynski Z, Lakowicz JR. Effects of Fluorophore-to-Silver Distance on the Emission of Cyanine–Dye-Labeled Oligonucleotides. Anal Biochem. 2003;315:57–66. doi: 10.1016/S0003-2697(02)00702-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lukomska J, Gryczynski I, Malicka J, Makowiec S, Lakowicz JR, Gryczynski Z. Oneand Two-Photon Induced Fluorescence of Pacific Blue-Labeled Human Serum Albumin Deposited on Different Core Size Silver Colloids. Biopolymers. 2005;81:249–255. doi: 10.1002/bip.20407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kapanidis AN, Laurence TA, Lee NK, Margeat E, Kong X, Weiss S. Alternating-Laser Excitation of Single Molecules. Acc Chem Res. 2005;38:523–533. doi: 10.1021/ar0401348. [DOI] [PubMed] [Google Scholar]
- 48.Sabanayagam CR, Eid JS, Meller A. High-Throughput Scanning Confocal Microscope for Single Molecule Analysis. Appl Phys Lett. 2004;84:1216–1218. [Google Scholar]
- 49.Ha TJ, Ting AY, Liang J, Caldwell WB, Deniz AA, Chemla DS, Schultz PG, Weiss S. Single-Molecule Fluorescence Spectroscopy of Enzyme Conformational Dynamics and Cleavage Mechanism. Proc Natl Acad Sci USA. 1999;96:893–898. doi: 10.1073/pnas.96.3.893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Robbins MS, Hadwen BJ. The Noise Performance of Electron Multiplying Charge Coupled Devices. IEEE Transactions on Electron Devices. 2003;50:1227–1232. [Google Scholar]
- 51.McKinney SA, Joo C, Ha T. Analysis of Single-Molecule FRET Trajectories Using Hidden Markov Modeling. Biophys J. 2006;91:1941–1951. doi: 10.1529/biophysj.106.082487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bike SG. Measuring Colloidal Forces Using Evanescent Wave Scattering. Curr Opin Colloid Interface Sci. 2000;5:144–150. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.