Abstract
Interleukin-1 (IL-1) is a proinflammatory cytokine that signals through the Type I IL-1 receptor (IL-1RI). Novel IL-1–like cytokines were recently identified. Their functions in lung disease remain unclear. Interleukin-1 family member–9 (IL-1F9) is one such IL-1–like cytokine, expressed in the lungs of humans and mice. IL-1F9 signals through IL-1 receptor–related protein 2 (IL-1Rrp2/IL-1RL2), which is distinct from IL-1RI. We sought to determine if IL-1F9 acts as a proinflammatory cytokine in lung disease. IL-1F9 protein was increased in lung homogenates of house dust mite–challenged A/J mice compared with controls, and expression was seen in airway epithelial cells. The intratracheal administration of recombinant mouse IL-1F9 increased airway hyperresponsiveness and induced neutrophil influx and mucus production, but not eosinophilic infiltration in the lungs of mice. In addition, IL-1α protein levels in bronchoalveolar lavage fluid, chemokines, and chemokine-receptor mRNA expression in the lungs were increased after the instillation of intratracheal IL-1F9. Consistent with these changes, NF-κB transcription factor activity was increased in the lungs of mice challenged with IL-1F9 and in a macrophage cell line treated with IL-1F9. These data suggest that IL-1F9 is upregulated during inflammation, and acts as a proinflammatory cytokine in the lungs.
Keywords: IL-1F9, chemokines, lung diseases, inflammation, neutrophil
CLINICAL RELEVANCE.
This study identifies IL-1F9 as a novel IL-1–like cytokine involved in pulmonary inflammation. Importantly IL-1F9 enhances neutrophil influx and chemokine production in the lungs. Further studies are required to determine if IL-1F9 can be therapeutically targeted to attenuate inflammatory lung diseases.
Interleukin 1 (IL-1) is a proinflammatory cytokine that may have a pathogenic role in chronic inflammatory lung diseases such as asthma and chronic obstructive pulmonary disease (1). The IL-1 family of cytokines consists of a functional receptor (IL-1RI), a nonfunctional decoy receptor (IL-1RII), a receptor accessory protein (IL-1RAcP), receptor agonists (IL-1α and IL-1β), and a receptor antagonist (IL-1Ra) (2). IL-1RI agonists bind the IL-1 receptor complex, consisting of IL-1RI and IL-1RAcP, and elicit proinflammatory signals by activating transcription factors such as NF-κB (3). IL-1Ra binds the IL-1 receptor complex but fails to elicit a signaling response, thereby acting as an endogenous competitive inhibitor to IL-1RI agonists, decreasing inflammation. An IL-1 cytokine–mediated increase or decrease in inflammation ensues after imbalances between the IL-1RI agonist and antagonist cytokines (4). The proinflammatory role of IL-1 and the anti-inflammatory role of IL-1Ra in chronic inflammatory lung diseases such as asthma are widely documented (5–7).
Recently, a group of novel genes structurally similar to IL-1 was discovered and mapped to human and mouse chromosome 2 (8). The novel IL-1 family cytokines IL-1F6, IL-1F8, and IL-1F9 exert IL-1α–like and IL-1β–like activities, whereas IL-1F5 exerts IL-1Ra–like activity, suggesting a new inter-regulated cytokine network similar to that of the classic IL-1RI agonists and antagonists (1). The novel IL-1 cytokines do not bind the classic IL-1 receptor IL-1RI. Instead, they bind a distinct receptor complex consisting of the novel receptors IL-1RL2 (also called IL-1Rrp2) and IL-1RAcP (9). This constitutes an uncharacterized pathway that may exert a significant impact in inflammatory diseases.
The functions of the novel IL-1 family cytokines have not been investigated in inflammatory lung diseases. We previously reported increased IL-1F9 gene expression in ovalbumin-challenged mice in a model of experimental allergic asthma (10). In mice, the Il1f9 gene is located in Abhr1, a quantitative trait locus for allergen-induced bronchial hyperresponsiveness (11). IL-1F9 is expressed in murine and human lungs and tracheas, with its predominant expression in epithelial tissues (9). Furthermore, IL-1F9 mRNA is upregulated in human bronchial epithelial cells after infection with Pseudomonas aeruginosa (12). These data suggest that IL-1F9 is a potentially important cytokine in lung diseases. This study sought to test the hypothesis that IL-1F9 acts as a proinflammatory cytokine in the lung by increasing chemokine production and inflammatory cell recruitment.
MATERIALS AND METHODS
Mice and Allergen Challenge
Five-week-old to 6-week-old male A/J mice with a genetic predisposition toward allergen-induced airway hyperresponsiveness were purchased from Jackson Laboratory (Bar Harbor, ME) and allowed to acclimate for 1 week. For some control experiments, 5–6-week-old C3H/HeJ mice that are genetically nonresponsive to endotoxin were also purchased from Jackson laboratory and allowed to acclimate for 1 week. Animals were housed and studied under protocols approved by our Institutional Animal Care and Use Committee in the specific pathogen–free animal facility of the University of Florida. Mice were anesthetized with a mixture of xylazine (8 mg/kg) and ketamine (45 mg/kg). Mice were sensitized and challenged with an intratracheal instillation of 100 μg of house dust mite extract (HDM, D. pteronyssinus, Greer Laboratories, Lenoir, NC) in 50 μl of PBS on Days 0, 7, and 14. Control mice received 50 μl of PBS. HDM challenge doses were calculated based on dry weight. Endotoxin contamination in the extracts was < 0.01 endotoxin units (EU)/μg HDM extract, as reported in the supplier's certificate of analysis. Tissues were collected at various time points after the last challenge.
IL-1F9 Polyclonal Antibody Generation
Anti-mouse IL-1F9 antibodies are currently unavailable. Hence, custom rabbit anti-mouse polyclonal antiserum to IL-1F9 was raised against a mouse IL-1F9–specific synthetic peptide (KILDLYHHPEPMKP). The IL-1F9 antibody was affinity-purified (Invitrogen, Carlsbad, CA) from the serum, and used for the molecular investigation of murine IL-1F9. The specificity of the IL-1F9 antibody was confirmed by its ability to detect recombinant mouse IL-1F9 (rmIL-1F9) protein via Western blotting.
Histology
Mice were killed with an overdose of intraperitoneally injected pentobarbital and exsanguinated, and their lungs were inflation-fixed with 4% paraformaldehyde to 20 cm H2O and removed en bloc. Inflated lungs were stored at 4°C overnight in 4% paraformaldehyde, rinsed with PBS, sectioned, sequentially dehydrated in 30%, 50%, and 70% ethanol, and paraffin-embedded. Five-micrometer sections of paraffin-embedded tissues were deparaffinized and stained with hematoxylin and eosin or periodic acid–Schiff reagents (Richard Allen Scientific, Kalamazoo, MI).
RNA and Protein Isolation from Lung Tissues
Lung tissue was homogenized in TriZol (Invitrogen) and stored at −80°C for subsequent RNA isolation. Total RNA was isolated from lung homogenates in TriZol, according to the manufacturer's protocol, and stored at −80°C. For protein isolation, lung tissue was homogenized in a buffer containing PBS and 0.1% Tween-20. Homogenized tissue was immediately centrifuged at 10,000 rpm for 10 minutes in a tabletop centrifuge at 4°C. After centrifugation, supernatants containing the protein were removed and stored at −80°C until further analyses. Protein concentrations from lung homogenates were quantified by the Bradford assay (Bio-Rad, Hercules, CA).
Western Blotting
One hundred micrograms of total protein from lung homogenates were separated on 15% Tris-Hcl SDS-PAGE gels (Bio-Rad), transferred to polyvinylidene fluoride membranes, and probed with rabbit anti-mouse polyclonal antibody to IL-1F9 (2 μg/mL) overnight at 4°C. Horseradish peroxidase–conjugated secondary antibodies and an ECL Chemiluminescence Kit (Amersham, Piscataway, NJ) were used to detect IL-1F9. A specific band was detected around 22 kD, the predicted size of mouse IL-1F9 protein. Membranes were stripped in Restore-Plus stripping buffer (Pierce, Rockford, IL) and reprobed for β-actin to ensure equal protein loading.
Bronchoalveolar Lavage and Differential Cell Counts
Mice were killed with an overdose of pentobarbital, and their tracheas were opened and catheterized. Lungs were lavaged three times with 1 ml of ice-cold PBS. Bronchoalveolar lavage fluid (BALF) was centrifuged at 2,000 rpm for 15 minutes, and the supernatants were removed and stored at −80°C until analysis. Cell pellets were treated with 0.3 ml NH4Cl lysis buffer to remove erythrocytes, neutralized with 10 × volume of sterile PBS, and centrifuged at 2,000 rpm for 15 minutes. The supernatants were removed, the cell pellet was resuspended in PBS, and total cell counts were performed using a hemocytometer and light microscopy. Cells were cytospun onto positively charged slides, dried overnight, and stained with a Hema-3 White Cell Differential Staining Kit (Fisher, Pittsburgh, PA). Stained slides were examined using light microscopy. One hundred cells were enumerated in each slide to obtain differential cell counts.
Immunofluorescence and Confocal Microscopy
Formalin-fixed, paraffin-embedded lung sections were immunostained with a rabbit anti-mouse IL-1F9 antibody. Lung sections were heat-retrieved in 10 mM citrate buffer, pH 6.0, before blocking with normal serum and staining overnight with primary antibodies at 4°C. Positive signals were detected with a goat anti-rabbit secondary antibody conjugated to AlexaFluor488. Controls consisting of isotype and concentration-matched immunoglobulin were included for each section. Immunofluorescence images were acquired in a Leica DM2500 microscope (Leica Microsystems, Bannockburn, IL) at the Cell and Tissue Analysis Core of the University of Florida. Images were analyzed using ImageJ software (http://rsbweb.nih.gov/ij/).
Real-Time PCR and SYBR Green Real-Time PCR Miniarray
Total RNA isolated from the lungs was quantified using Nanodrop (Thermo Scientific, Waltham, MA). Total RNA was treated with amplification-grade DNaseI (Invitrogen) to remove potential genomic DNA contamination. DNaseI-treated total RNA was quantified using Nanodrop. For mouse lung tissues, 400 ng of DNaseI-treated total RNA were reverse-transcribed in a reaction volume of 20 μl. For in vitro studies, 200 ng of total RNA were reverse-transcribed in a reaction volume of 20 μl. Reverse-transcription reactions were performed with random hexamers, using Taqman Reverse transcription reagents (Applied Biosystems, Foster City, CA). Complementary DNA (cDNA) obtained from reverse transcription was used as the template for SYBR Green–based quantitative real-time PCR. Primers for the real-time PCR analysis of target genes were designed using the mouse qPrimerdepot database (http://mouseprimerdepot.nci.nih.gov/). Transcript expression patterns of 47 chemokine and chemokine receptor genes and an endogenous control (18S ribosomal RNA) were simultaneously assessed using a miniarray format. The PCR reaction consisted of 12.5 μl of Power SYBR Green PCR Mastermix (Applied Biosystems), 1 μl (∼ 4 ng) of cDNA, 1 μl each of forward and reverse primers (10 μM), and 9.5 μl of distilled, deionized, DNase-free and RNase-free water in a reaction volume of 25 μl. Gene-specific primers were subsequently added to the PCR mix. Dissociation curve analyses were performed after each run, to rule out nonspecific amplification. Quantitative real-time PCR was performed on an ABI7300 thermocycler (Applied Biosystems). Samples or primer pairs showing multiple dissociation peaks were excluded from data analyses. Four to five biological replicates were analyzed using this method for in vivo studies. Cycle threshold values obtained from real-time PCR assays were analyzed using the Global Pattern Recognition (GPR) algorithm (13). Genes that significantly (P < 0.05) differed in mRNA expression between treatment and control groups were identified according to the GPR algorithm (13).
ELISA Assays
ELISA Max standard sets for IL-1α and IL-1β were purchased from BioLegend (San Diego, CA). One milliliter of BALF from a total collection of 3 ml was concentrated to one third of the initial volume under vacuum. One hundred microliters of the concentrated sample were used in ELISA assays. Assays were performed according to the manufacturer's protocols.
Generation of Recombinant Murine IL-1F9
Total RNA was isolated from the lungs of A/J mice using TriZol (Invitrogen), and treated with amplification-grade DNaseI (Invitrogen) to remove genomic DNA contamination. Full-length mouse Il1f9 cDNA (GenBank NM_153511.1) was amplified from DNaseI-treated total RNA, using a forward primer engineered with a BglII restriction site (bold and underlined) 5′-GGAAGATCTTTTTCTAAACACCCATTTTCTACACACATCTCAGG-3′, and a reverse primer engineered with a KpnI restriction site (bold and underlined) 5′- GGGGTACCCCTCCACTTCCATGCTGAGT-3′, using the Superscript One-Step RTPCR Kit for long templates (Invitrogen). Purified insert was ligated to pBADHis_b bacterial expression vector (Invitrogen), digested with BglII and KpnI, and treated with calf intestinal phosphatase. Ligation reactions were transformed into one-shot Top10 Escherichia coli. Ampicillin-resistant colonies were selected, tested for the presence of the insert using PCR, and bidirectionally sequenced to confirm insert identity. Positive colonies were grown on LB-ampicillin broth, and the expression of IL-1F9 was induced by the addition of L-arabinose. IL-1F9 was purified from bacterial lysate by Ni2+ column chromatography (B-PER 6xHis Fusion Protein Purification Kit, Pierce), and dialyzed against PBS. Purified IL-1F9 was treated with enterokinase (EK-Max, Invitrogen) to remove purification tags and additional amino acids from recombinant IL-1F9 fusion protein. Enterokinase was removed from the tag-removed IL-1F9 preparation by treating with EK-Away (Invitrogen), according to the manufacturer's protocols. The purified, tag-removed, enterokinase-removed recombinant IL-1F9 protein preparation was treated with polymyxin-B agarose beads (Sigma, St. Louis, MO) to remove LPS contamination. LPS concentration was < 0.01 EU/μg of polymyxin-treated IL-1F9, as measured by the limulus amebocyte lysate (LAL) assay. Purification tag–removed, polymyxin-treated rmIL-1F9 was used for in vitro and in vivo studies.
Coomassie Staining
To test the purity of the rmIL-1F9 preparation, increasing concentrations of IL-1F9 were separated on an SDS-polyacrylamide gel under reducing and denaturing conditions. The gel was soaked in Fairbank's Coomassie solution A (25% isopropanol, 10% acetic acid, and 0.05% Coomassie R) and microwaved, and shaken for 10 minutes at room temperature to stain the gel. The stain was drained, and the gel was soaked in distilled water and microwaved for 10 minutes. The solution was drained and replaced with fresh distilled water, and the gel was shaken at room temperature for 10 minutes to destain the gel.
Intratracheal IL-1F9 Challenge
A single dose of 10 μg rmIL-1F9 was intratracheally instilled into mouse lungs in a volume of 50 μl PBS. In separate experiments, mice were challenged with 10 μg rmIL-1F9 twice daily for 2 days. Control mice received 50 μl of PBS. Tissues were collected for analysis 6 hours and 24 hours after the last IL-1F9 challenge. For some experiments, mice sensitized with PBS or HDM on Days 0 and 7 were challenged with PBS, HDM, or 10 μg IL-1F9 on Day 14, and BALF was collected 24 hours later.
Measurement of Lung Function in IL-1F9–Challenged Mice
Six-week-old A/J mice were purchased from Jackson Laboratories and maintained at the specific pathogen–free facility at Massachusetts General Hospital (Charlestown, MA). All procedures were performed according to protocols approved by the Subcommittee on Research Animal Care at Massachusetts General Hospital. Mice were intratracheally administered 10 μg rmIL-1F9, and lung function measurements were performed 24 hours later, using the Flexivent system (Scireq, Montreal, Quebec, Canada). Control mice received 50 μl of PBS. In brief, IL-1F9–challenged and control mice were anesthetized with an intraperitoneal injection of xylazine (12 mg/kg) and pentobarbital (70 mg/kg). Anesthetized mice were tracheostomized, cannulated, and ventilated with 6 ml/kg tidal volumes at 150 breaths per minute. To suppress spontaneous breathing during measurements of lung function, anesthetized mice were intraperitoneally injected with pancuronium bromide (2 mg/kg). Incremental doses of nebulized methacholine were used to determine total lung resistance according to the Snapshot-150 perturbation provided by the Flexivent equipment. Thirteen data points were collected for each methacholine dose, and only data with a coefficient of determination greater than 0.95 were included in analyses. The survival of mice during the procedure was simultaneously monitored using electrocardiograms.
NF-κB Activation Assays in Mouse Macrophage Cell Lines
The mouse macrophage cell line RAW264.7, stably transfected with the NF-κB–responsive E-selectin promoter driving the expression of enhanced green fluorescent protein (eGFP), was used to investigate if IL-1F9 induced NF-κB activity in alveolar macrophages. Cells were cultured in RPMI-1640 with 10% fetal bovine serum and 1% penicillin and streptomycin. Cells were plated at a density of 0.5 × 106 cells/well, 1 day before the experiments. Media were removed on the day of the experiment, and recombinant mouse IL-1F9 mixed with media was added to the cells at appropriate concentrations and incubated at 37°C for 6 hours. After incubation, media were removed and cells were washed twice with ice-cold PBS. Cells were collected, and eGFP fluorescence was measured using an Accuri C6 flow cytometer (Accuri Cytometers, Ann Arbor, MI).
Murine Alveolar Macrophage Cell Culture and IL-1F9 Stimulation of MH-S Cells
The murine alveolar macrophage cell line (MH-S) was cultured in medium containing RPMI-1640 with 10% fetal bovine serum, 0.05 mM β-mercaptoethanol, and 1% penicillin and streptomycin. MH-S cells were plated at a density of 0.5 × 106 cells/well on tissue culture plates 1 day before the experiments. Media were removed on the day of the experiment, and recombinant mouse IL-1F9 was added to the cells at appropriate concentrations and incubated at 37°C for 1–6 hours. After incubation, culture supernatants were removed and stored at −80°C until further use. Cells in tissue culture plates were lysed in TriZol and stored at −80°C for RNA extraction.
Transcription Factor Multiplex Assays
Mice were intratracheally challenged with 10 μg of rmIL-1F9. Control mice received 50 μl of PBS. Lungs were collected from mice euthanized 2 and 6 hours after the challenge. Nuclear protein was extracted from lungs, using a Panomics Nuclear extraction kit (Panomics, Fremont, CA), and quantified by a DC Protein Assay (Bio-Rad) according to the manufacturer's protocol. Nuclear extracts were prepared into aliquots and stored at −80°C until further use. Two micrograms of nuclear protein from each lung sample were used to assess transcription factor activity in the multiplexed transcription factor activity assays. Positive and negative nuclear and whole-cell lysate protein controls supplied by the manufacturer were simultaneously assayed to verify assay validity. Assay background was calculated from a blank sample in which protein was not included. A Procarta Transcription Factor Assay Kit (40-plex Panel 1; Panomics) was used to quantify transcription factor activity in PBS-treated and IL-1F9–treated mouse lung nuclear protein extracts, according to the manufacturer's protocol. Briefly, nuclear extract was incubated with a mixture of transcription factor (TF)–specific, biotinylated, double-stranded DNA probes to form protein/DNA complexes. Protein/DNA complexes were bound to a semiporous filter, and unbound probes were washed away by centrifugation. Protein-bound DNA was eluted, heat-denatured, and hybridized with TF-specific antisense conjugated beads. Streptavidin–phycoerythrin was added to the probe-bound beads, and samples were analyzed using a Luminex 100-IS instrument (Luminex, Austin, TX). Data were acquired and analyzed using Luminex 100 IS software, version 2.3. Transcription factor targets with bead counts of less than 50 were discarded from analysis. Transcription factor targets in each lung sample were included for analysis only if their median fluorescence intensity (MFI) values were at least twofold greater than the assay background MFI. Results were reported as the mean ± SEM of MFIs for each sample calculated, after subtraction of the assay background MFI.
RESULTS
IL-1F9 Is Constitutively Expressed and Inducibly Upregulated in Mouse Lungs, with Predominant Expression in Epithelial Cells
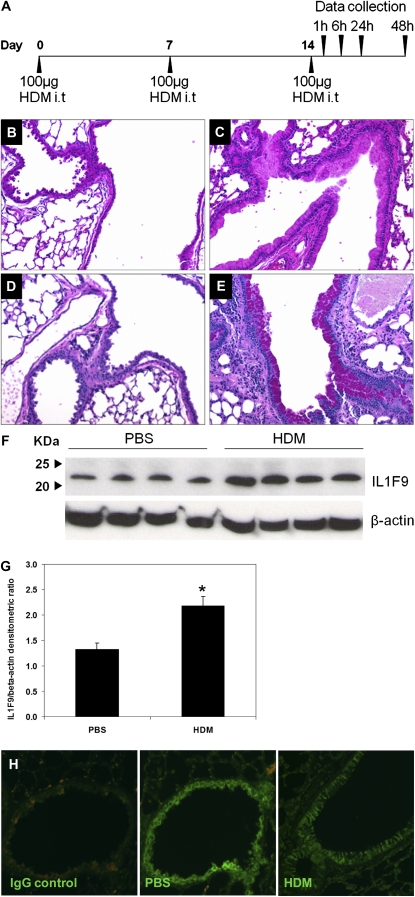
We previously demonstrated that IL-1F9 mRNA was increased in the lungs of mice that develop allergic inflammation. Here, we tested if IL-1F9 protein is increased in the lungs of mice sensitized and challenged with HDM. The challenge with HDM elicited allergic lung inflammation in the lungs of mice, including increased cellular infiltration and mucus production 24 hours after the last challenge (Figures 1B–1E). IL-1F9 protein was significantly increased in the lungs of HDM-treated mice 24 hours after the challenge with HDM, compared with PBS-treated control mice (Figures 1F and 1G). IL-1F9 protein was also detected in the lungs of both HDM-treated and PBS-treated mice collected 1, 6, and 48 hours after the challenge with HDM, but without significant differences between HDM-treated and PBS-treated groups (data not shown). These results suggest that IL-1F9 is constitutively expressed in the lung and upregulated during the development of lung inflammation. To determine cell types in lungs that express IL-1F9, paraffin-embedded lung tissue from PBS-treated and HDM-treated mice were analyzed by immunofluorescence microscopy. IL-1F9 was predominantly expressed by airway epithelial cells in PBS-treated and HDM-treated lungs (Figure 1H). No significant differences in IL-1F9 expression were evident in airway epithelial cells between PBS-treated and HDM-treated mice, suggesting that other cell types may contribute to the differences in IL-1F9 expression observed in total lung homogenates of PBS-treated and HDM-treated mice.
Figure 1.
Increased interleukin-1 family member–9 (IL-1F9) protein production in lungs of house dust mite (HDM)–treated mice, and constitutive IL-1F9 expression in airway epithelial cells. (A) Allergen sensitization and challenge protocol. Five-week-old to 6-week-old male A/J mice were intratracheally (i.t) sensitized and challenged with 100 μg of HDM extract diluted in 50 μl PBS on Days 0, 7, and 14. Control mice received 50 μl PBS. Data were collected at specific time points after the last challenge on day 14. (B and C) Hematoxylin and eosin–stained lung sections of mice were treated with PBS or HDM, respectively. Increased cellular infiltration and epithelial cell hypertrophy were observed in lungs of mice treated with HDM. (D and E) Periodic acid–Schiff–stained lung sections of mice were treated with PBS or HDM, respectively. Goblet-cell hypertrophy and increased mucus production were evident in airways of HDM-treated mice. (B–E) Representative sections from one mouse for each group (n = 5 mice/group). (F) Increased IL-1F9 protein in the mouse lung homogenates 24 hours after the last HDM challenge. IL-1F9 was detected by Western blot as an ∼ 22-kD band by a rabbit anti-mouse polyclonal antibody. Each lane represents a single mouse. (G) Densitometry of IL-1F9/β-actin ratios in samples from F, represented as mean ± SEM. *Significant differences (P < 0.05) from PBS-treated group. (H) IL-1F9 protein expression was detected in airway epithelial cells of PBS-treated, paraffin-embedded mouse lung sections by immunofluorescence microscopy, using a rabbit anti-mouse polyclonal antibody against IL-1F9 and an AlexaFluor488-conjugated secondary antibody (IL-1F9). Representative section (×20 magnification) from one mouse (n = 5 mice/group) demonstrates IL-1F9 protein expression in airway epithelial cells of both PBS-treated and HDM-treated mice.
Intratracheal IL-1F9 Challenge Increased Lung Cell Infiltrates and Mucus Production in Mouse Airways
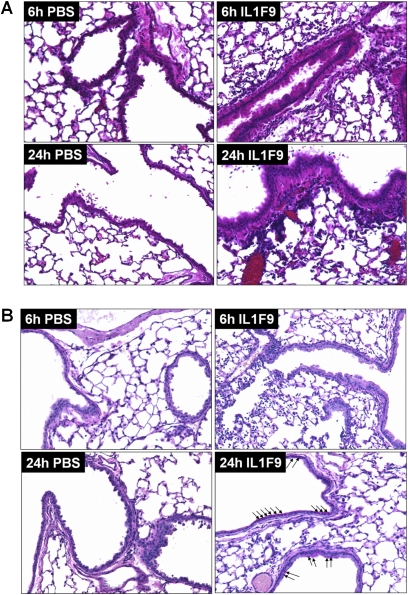
We administered rmIL-1F9 intratracheally into the lungs of A/J mice to simulate transient IL-1F9 overexpression, and to determine if the intratracheal administration of IL-1F9 alone is sufficient to elicit an inflammatory response in the lung. Mice were intratracheally challenged with 10 μg IL-1F9 twice a day for 2 days. The histopathologic examination of lung sections from mice collected 6 or 24 hours after the last dose of IL-1F9 revealed epithelial cell hypertrophy and increased cellular infiltrates (Figure 2A). Cellular infiltrates were predominantly observed in the alveolar spaces and around the airways. Mucus production was detected in mouse airways 24 hours after the last challenge with intratracheal IL-1F9 (Figure 2B). However, the mucus production observed after challenges with IL-1F9 was less than that observed after challenges with HDM (Figure 1E).
Figure 2.
Intratracheal challenge with IL-1F9 leads to cellular infiltrates and mucus production in airways of mice previously unexposed to allergens. (A) Hematoxylin and eosin staining of paraffin-embedded mouse lung sections collected 6 hours and 24 hours after the last of four intratracheal challenges with PBS or IL-1F9. (B) Periodic acid–Schiff staining of paraffin-embedded mouse lung sections collected 6 hours and 24 hours after the last of four intratracheal challenges with PBS or IL-1F9. Arrows indicate PAS positive, mucus-producing cells. Slides are representative of n = 4–5 mice/group/time point.
Intratracheal Challenge with IL-1F9 Increased Airway Hyperresponsiveness and Induced Neutrophil Influx in the Lungs
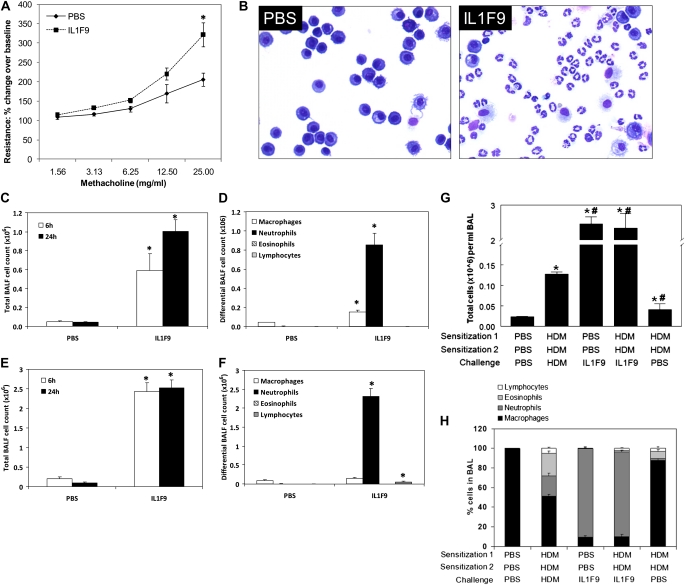
To determine if IL-1F9 challenges increased airway hyperresponsiveness, lung resistance was determined in mice intratracheally challenged with a single dose of IL-1F9 or PBS. Lung resistance increased 24 hours after a single intratracheal challenge with IL-1F9, compared with PBS-challenged mice (Figure 3A). Increased numbers of neutrophils were observed in the BALF recovered from mice 24 hours after a single intratracheal challenge with IL-1F9 (Figure 3B). To determine the infiltrating cell types, differential leukocyte counts were performed on BALF cells collected from mice after single or multiple intratracheal challenges with IL-1F9. The total number of cells in BALF increased, with a predominance of neutrophils 6 and 24 hours after the administration of a single dose of IL-1F9 to the lungs (Figures 3C and 3D). To simulate a model of IL-1F9 overexpression and determine if multiple IL-1F9 challenges change the composition of infiltrating cells, A/J mice were intratracheally challenged with IL-1F9 twice a day for 2 days. The total cell number in BALF was significantly increased after multiple challenges, compared with a single challenge of IL-1F9. However, the cellular composition remained similar, with a predominance of neutrophils and a small number of lymphocytes (Figures 3E and 3F). Eosinophils were not evident in BALF collected from mice instilled with either single or multiple doses of IL-1F9. Sensitization with HDM followed by challenge with IL-1F9 also increased total BALF cell numbers and resulted in marked neutrophil but not eosinophil or lymphocyte infiltration into mouse lungs (Figures 3G and 3H). These results demonstrate that neutrophil influx is the predominant response to instillation of IL-1F9 into the lung.
Figure 3.
Increased airway hyperresponsiveness and lung inflammation in mice after intratracheal challenge with IL-1F9. Intratracheal IL-1F9 challenge induced neutrophil influx but not eosinophils. (A) Single intratracheal challenge with 10 μg IL-1F9 significantly increased total lung resistance in mice. Data represent mean ± SEM of percent change over baseline (PBS) measurements. *Significant differences (P < 0.05) from PBS-treated group using two-factor ANOVA, n = 5 mice/group. (B) Increased number of neutrophils in bronchoalveolar lavage fluid (BALF) from mice recovered 24 hours after a single intratracheal challenge with IL-1F9. Representative images are shown of 4–5 mice/group. (C) Total leukocyte counts were increased in BAL 6 hours and 24 hours after a single intratracheal challenge with 10 μg IL-1F9. (D) Differential cell counts of BAL cells collected from mice 24 hours after a single IL-1F9 challenge. (E) Total cell counts were increased in BALF 6 hours and 24 hours after the last of four intratracheal challenges with 10 μg IL-1F9, administered twice a day for 2 days. (F) Differential cell counts of BAL cells collected 6 hours and 24 hours after the last of four intratracheal challenges with 10 μg IL-1F9, administered twice a day for 2 days. Data represent mean ± SEM. *Significant differences (P < 0.05) from PBS-treated group, n = 4–5 mice/group. (G) Total cell counts of BAL cells collected 24 hours after the last challenge. #Significant differences (P < 0.05) from HDM-sensitized and -challenged group. (H) Differential counts of BAL cells collected 24 hours after the last challenge. Data represent mean ± SEM. n = 4–5 mice/group.
Neutrophil Accumulation in IL-1F9–Challenged Mouse Lung Is Not Attributable to Endotoxin Contamination
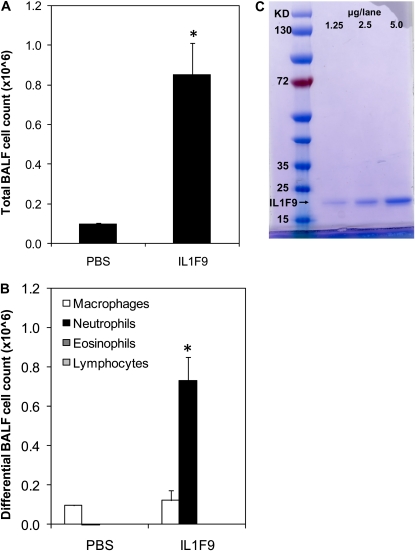
The endotoxin concentration of polymyxin-B–treated rmIL-1F9 preparations used in these studies was less than 0.01 EU/μg of rmIL-1F9 protein, as assessed by the LAL assay. This concentration was similar to endotoxin concentrations reported in commercially available HDM extracts used in the present study (< 0.01 EU/μg). To determine if this low concentration of endotoxin in the rmIL-1F9 preparation contributed to the inflammation and neutrophil influx observed in the lungs of A/J mice after challenge with rmIL-1F9, 10 μg of rmIL-1F9 were intratracheally administered to a C3H/HeJ strain of mice. C3H/HeJ mice are nonresponsive to endotoxin because of a loss-of-function mutation in the Toll-like receptor–4 gene. Control mice received PBS. Increases in total cell number (Figure 4A) and neutrophil accumulation (Figure 4B) were evident in the BALF recovered from C3H/HeJ mice 24 hours after treatment with rmIL-1F9. The increase in BALF cell number and BALF cell composition of C3H/HeJ mice was similar to that in A/J mice after a single intratracheal challenge with rmIL-1F9. This finding demonstrates that the neutrophil infiltration observed in the BALF of mice after intratracheal challenge with IL-1F9 is not attributable to endotoxin contamination. Moreover, the recombinant IL-1F9 preparation migrated as a single discrete band in an SDS–polyacrylamide gel under reducing and denaturing conditions, demonstrating the purity of the preparation (Figure 4C).
Figure 4.
Neutrophil influx observed in IL-1F9 challenged mice is not attributable to endotoxin contamination of recombinant mouse IL-1F9 preparation. Endotoxin-resistant C3H/HeJ mice were intratracheally challenged with 10 μg IL-1F9 or PBS. BAL fluid recovered from mice 24 hours after the challenge was used to assess total and differential cell counts. (A) Total cells were increased in BAL 24 hours after a single intratracheal challenge with IL-1F9. (B) Differential cell counts of BAL cells collected 24 hours after a single intratracheal challenge with IL-1F9. Data represent mean ± SEM. *Significant differences (P < 0.05) from PBS-treated group, n = 4–5 mice/group. (C) Purity of IL-1F9 protein preparation assessed by Fairbanks Coomassie staining. Recombinant IL-1F9 protein is observed as a single band between 15–25 kD.
Increased Chemokine mRNA Expression and IL-1α Protein Production after Intratracheal Challenge with IL-1F9
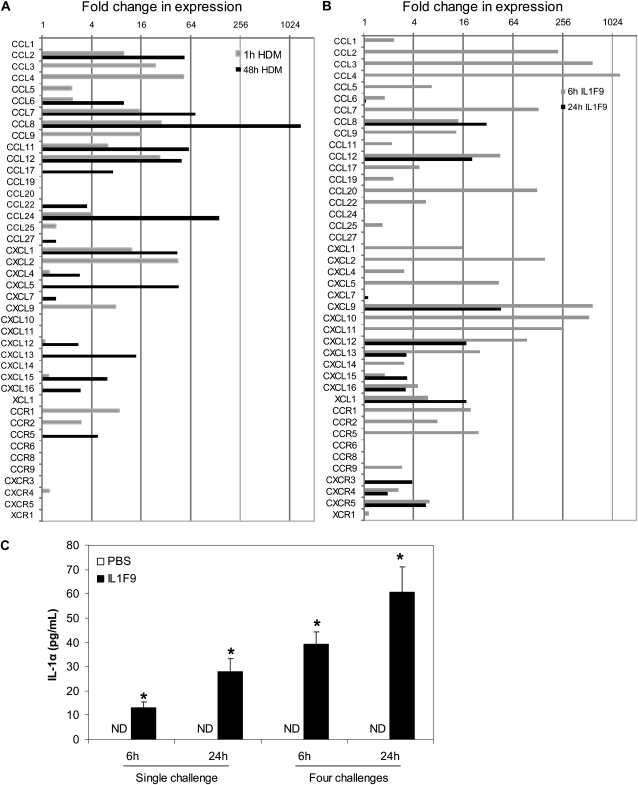
Chemokines drive leukocyte recruitment to the site of inflammation and exert a major role in the accentuation of lung inflammation after an inflammatory stimulus. As expected, the transcript expression of chemokines specific to allergic responses, such as CCL17, CCL11, and CCL24, increased after challenges with HDM (Figure 5A). Moreover, challenges with HDM also increased the mRNA expression of neutrophil chemoattractants such as CXCL1 and CXCL2 (Figure 5A). Challenges with IL-1F9 increased the expression of several chemokines, including the neutrophil chemoattractants CXCL1 and CXCL2. However, the eosinophil-specific chemokines CCL11 and CCL24 did not increase (Figure 5B). A comparison of chemokine/chemokine receptor expression profiles between HDM-challenged or IL-1F9–challenged mice displayed a strong overlap in the expression of several CCL and CXCL chemokines and chemokine receptors (as shown in Table E2 in the online supplement). IL-1α and IL-1β protein were measured in BALF to determine if the novel cytokine IL-1F9 induced the production of classic IL-1 agonists. Interestingly, an intratracheal instillation of IL-1F9 induced a time-dependent and dose-dependent increase in IL-1α protein levels in BALF, but IL-1β was not detectable in BALF at any time points tested (Figure 5C). Neither IL-1α nor IL-1β protein was detected in the BALF of control mice intratracheally challenged with PBS.
Figure 5.
HDM challenge and IL-1F9 challenge induce production of CC and CXC chemokines in the lungs of mice, whereas challenges with IL-1F9 increased IL-1α protein in BALF. RNA isolated from lung tissues 1 hour and 48 hours after challenge with HDM (A) or 6 and 24 hours after last of four IL-1F9 challenges (B) was quantified for chemokine and chemokine receptor mRNA expression, using a SYBR-Green based quantitative PCR (qPCR) miniarray. Data were analyzed using the Global Pattern Recognition (GPR) algorithm. Only the genes with significant change in expression (P < 0.05) after HDM or IL-1F9 treatment compared with control samples are presented as fold change in mRNA expression (n = 4–5 mice/group/time point). (C) BALF recovered from mice 6 and 24 hours after single or four intratracheal challenges with IL-1F9 was evaluated for IL-1α and IL-1β protein levels by ELISA. IL-1β was not detectable in BALF. Data represent mean ± SEM. *Significant differences (P < 0.05) from PBS-treated group, n = 4–5 mice/group. ND, not detected.
Increased NF-κB Activation in Mouse Lungs after Intratracheal Administration of IL-1F9
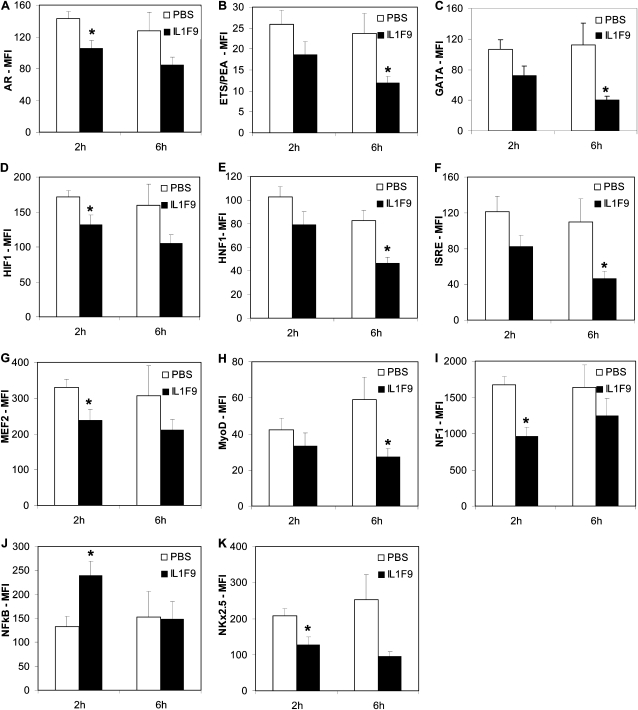
To identify transcription factors that are important in IL-1F9–mediated inflammation and chemokine production, transcription factor activity in IL-1F9–challenged lungs was investigated using multiplex transcription factor assay kits. NF-κB binding activity was significantly increased in nuclear extracts from mouse lungs collected 2 hours after the administration of IL-1F9 (Figure 6J). The NF-κB binding activity was no different between nuclear extracts from mouse lungs collected 6 hours after challenge with IL-1F9 or PBS (Figure 6J). Interestingly, the DNA probe–binding activities of several nuclear factors had decreased in lungs 2 hours after the administration of IL-1F9, including the androgen receptor (Figure 6A), hypoxia inducible factor (Figure 9E), myocyte enhancer factor–2 (Figure 6G), nuclear factor–1 (Figure 6I), and NK2 transcription factor–related locus–5 (Figure 6K). Reduced activities of ETS-binding factor PEA (Figure 6B), GATA binding (Figure 6C), interferon-stimulated response element (Figure 6F) binding, and myogenic differentiation factor (Figure 6H) were evident in nuclear extracts from lungs collected 6 hours after challenge with IL-1F9.
Figure 6.
NF-κB activity was increased, and activities of several other transcription factors were decreased, in mouse lungs after challenge with IL-1F9. Mice were intratracheally challenged with 10 μg IL-1F9, and lungs were collected 2 hours and 6 hours later. Nuclear extracts prepared from lung tissue were used to assess nuclear factor activity, using multiplex assays in a Luminex 100 IS instrument. Data are reported as mean ± SEM of median fluorescence intensity (MFI) values, corrected for blank MFI values. (J) NF-κB activity was increased in lungs 2 hours after challenge with IL-1F9. No change was evident in NF-κB activity 6 hours after challenge with IL-1F9. (A–I, K) Transcription factors with decreased consensus DNA sequence binding activities, at either 2 hours or 6 hours after challenge with IL-1F9. Data represent mean ± SEM. *Significant differences from PBS-treated group (P < 0.05), n = 5 mice/group. AR, androgen receptor; HIF, hypoxia inducible factor; MEF2, myocyte enhancer factor–2; NF1, nuclear factor–1; NkX2-5, NK2 transcription factor–related locus–5; ETS/PEA, E-twenty six binding factor polyoma enhancer activator; ISRE, interferon-stimulated response element; MyoD, binding and myogenic differentiation factor.
IL-1F9 Increased NF-κB Activity and Increased Neutrophil-Specific Chemokine mRNA Expression in Mouse Macrophages
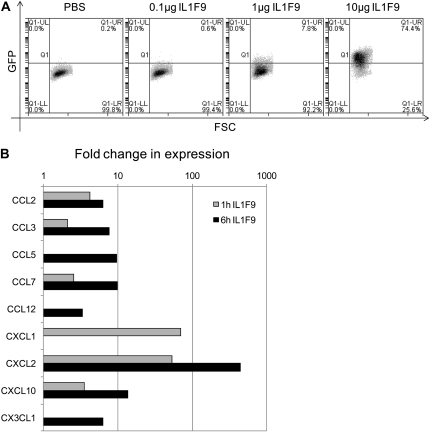
During inflammation, macrophages produce and respond to classic IL-1 agonists such as IL-1α and IL-1β. The RAW264.7 mouse macrophage cell line with NF-κB activation–dependent GFP expression was used to determine if IL-1F9 induces inflammatory responses by inducing NF-κB activity in macrophages. IL-1F9 induced a dose-dependent increase in GFP expression, indicating the activation of NF-κB (Figure 7A). Transcript expression of the neutrophil chemoattractants CXCL1 and CXCL2 was significantly increased after stimulation of the mouse alveolar macrophage cell line MH-S with IL-1F9 (Figure 7B). These data suggest that the IL-1F9–mediated induction of NF-κB activity and neutrophil-specific chemokine gene expression in macrophages may constitute an important pathway in lung inflammation.
Figure 7.
IL-1F9 stimulated NF-κB activity and induced mRNA expression of neutrophil chemoattractants in mouse macrophage cell lines. (A) RAW264.7 cells stably transfected with NF-κB–responsive E-selectin promoter driving enhanced green fluorescent protein (GFP) expression were stimulated with increasing doses of IL-1F9 for 6 hours. NF-κB activity was measured by the percentage of cells that expressed enhanced GFP after stimulation. A representative depiction is based on two independent experiments with similar results. (B) Murine alveolar macrophage (MH-S) mouse alveolar macrophage cell lines were stimulated with 10 μg IL-1F9 for 1 hour and 6 hours. RNA isolated from cells was used to measure chemokine and chemokine receptor mRNA expression, using a quantitative PCR miniarray. Data were analyzed using the GPR algorithm. Only genes with significant changes in expression (P < 0.05) after IL-1F9 treatment compared with control samples are presented here, as fold change in mRNA expression.
DISCUSSION
We demonstrate that IL-1F9 is constitutively expressed in mouse lungs, and is upregulated after challenge with HDM. The expression of IL-1F9 was observed predominantly in the airway epithelium. Challenges of naive mice with intratracheally administered IL-1F9 led to increased NF-κB activity, increased expression of neutrophil-active chemokines, neutrophil recruitment into the lungs, and increased airway hyperresponsiveness. In vitro experiments suggest that macrophages are a primary target of IL-1F9 in the lung.
IL-1α and IL-1β are proinflammatory cytokines that were shown to play a pathogenic role in lung diseases such as asthma (14). IL-1α and IL-1β increase and IL-1ra decreases airway hyperresponsiveness, T-cell proliferation, Th2 cytokine production, and airway inflammation in murine models of experimental allergic asthma (6). In addition, IL-1 induces pulmonary inflammation and fibrosis in mice (15). IL-1 cytokines induce lung inflammation by upregulating chemokine expression. IL-1–induced chemokine expression in airway epithelial cells in vitro and chemokines are increased in the lungs of IL-1β–overexpressing mice (16–19). IL-1β increased eotaxin and CCL2 expression from human airway smooth muscle cells (20). The overexpression of IL-1β in mouse lung epithelial cells increased the expression of CXCL1, CXCL2, CCL2, and CCL7 chemokines, along with increased accumulations of neutrophils and macrophages in the lungs (21).
The gene encoding IL-1F9 cytokine is located on mouse chromosome 2 in a quantitative trait locus, with linkage to allergen-induced bronchial hyperresponsiveness (11). We previously reported that IL-1F9 mRNA is upregulated in the lungs of mice in response to allergen challenge (10). To the best of our knowledge, no reports exist concerning IL-1F9 protein levels in human inflammatory lung diseases. However, several lines of recent evidence demonstrate increased mRNA expression in human lung diseases or in vitro cultures of human cell lines. Increased IL-1F9 mRNA expression was observed in human bronchial epithelial cells infected with Pseudomonas aeruginosa (12), and IL-1F9 mRNA expression was significantly increased in biopsies of recurrent respiratory papillomas (RRPs) from human patients (22). Interestingly, biopsies from patients with severe RRP had significantly higher IL-1F9 mRNA expression, compared with patients with moderate RRP disease severity (22), suggesting that the expression of IL-1F9 may be correlated with disease severity. Infection with rhinovirus also upregulated IL-1F9 mRNA expression in patients with asthma (23). Cigarette smoke condensate, tumor necrosis factor, IL-1β, IL-17, and double-stranded RNA are some of the other stimuli reported to increase mRNA expression in normal human bronchial epithelial cells (24, 25).
IL-1F9 signals through a receptor complex consisting of IL-1RL2 and IL-1RAcP, which is distinct from the receptor complex (IL-1RI and IL-1RAcP) required for IL-1α and IL-1β signaling (9). Hence, the increased IL-1F9 protein in mouse lungs after the challenge with HDM observed in this study suggests a potentially novel, uncharacterized pathway contributing to lung inflammation. The balance between agonists and antagonists in the IL-1RI–receptor–mediated or IL-1RL2–receptor–mediated signaling cascades determines if proinflammatory or anti-inflammatory responses are initiated by classic (4) and novel (26) IL-1 family members. Of the novel IL-1 family of cytokines, IL-1F6, IL-1F8, and IL-1F9 are agonists for IL-1RL2 signaling, whereas IL-1F5 is an antagonist to IL-1RL2 (9). Hence, the constitutive expression of IL-1F9 may not be proinflammatory. However, an inflammatory response may ensue when the expression of IL-1F9 is increased after a stimulus.
IL-1F9 protein was constitutively expressed in mouse airway epithelial cells. IL-1α and IL-1β are secreted by monocytes, macrophages, and dendritic cells (1). LPS stimulated the production and release of IL-1 from monocytes. Similarly, IL-1F6, IL-1F8, and IL-1F9 mRNA expression increased in monocytes after stimulation with LPS (9). The novel IL-1–like cytokines are peptides without the leader sequences necessary for protein secretion. Hence their presence in the extracellular space and their mechanisms of secretion have not been adequately characterized. A recent report demonstrated that IL-1F6 externalizes from monocytes after stimulation with LPS and ATP-dependent purinergic receptor activation (27). IL-1F9–expressing cells may externalize IL-1F9 by a similar mechanism. Analyses of IL-1F9 expression in the present study were not exhaustive. Hence IL-1F9 may also be expressed in other lung cell types, apart from airway epithelial cells.
Intratracheal instillations of IL-1F9 increased airway hyperresponsiveness and induced neutrophil influx in the lungs. Previous reports demonstrated that an intratracheal administration of IL-1 increased neutrophil accumulation in the lungs of rats (28, 29). Overlapping and distinct chemokine gene expression patterns were observed in the lungs of mice after challenges with HDM or IL-1F9. In this study, the gene expression of CCL2, CCL3, CCL4, CCL5, CCL7, CCL8, CCL9, CCL11, and CCL12 increased soon after challenge with HDM or IL-1F9. The transcript expression of the neutrophil chemoattractants CXCL1 and CXCL2 was upregulated after challenges with HDM or IL-1F9. Increased CXCL1 mRNA expression was previously reported in a murine model of ovalbumin-induced asthma, where neutrophils and eosinophils were present in BALF after challenge with allergen (30). A recent report demonstrated that IL-1F9 stimulated the induction of CXCL8 and CCL20 in primary human fibroblasts by activating mitogen-activated protein kinase (MAPK) and NF-κB (25). IL-1α, but not IL-1β, was increased in the BALF of mice after challenge with IL-1F9, suggesting that IL-1F9 may also be involved in a feedback loop leading to the upregulation of classic IL-1 agonists. Together with previous reports, data from the present study suggest that IL-1F9 may act as a proinflammatory cytokine in the lung by inducing chemokine production, neutrophil influx, and airway hyperresponsiveness.
Challenges with IL-1F9 increased the activation of NF-κB in mouse lungs. NF-κB is proinflammatory in lung disease. Persistent activation of NF-κB was reported in airway epithelial cells from humans with severe asthma (31). Blocking NF-κB activation reduced airway inflammation, mucus-cell metaplasia, circulating IgE, proinflammatory cytokine, and chemokine production (32–34). The increased NF-κB activity in the lungs of IL-1F9–challenged mice in the present study is consistent with previous in vitro studies demonstrating NF-κB activation upon stimulation with IL-1F9 (9). The activities of several transcription factors relevant to lung homeostasis and disease were reduced in mouse lungs after challenge with IL-1F9. Although nuclear extracts from whole lung demonstrate overall differences in transcription factor activity, transcription factor activities may vary in individual lung cell types. In the present study, IL-1F9 induced NF-κB activity in macrophages and increased the mRNA expression of CXCL1 and CXCL2, two major chemokines implicated in neutrophil chemotaxis. The gene expression of chemokines such as CCL2, CCL3, CCL5, CCL7, CCL11, CXCL1, CXCL2, and CXCL5 are regulated by NF-κB (17, 35–42). The transcript expression of these chemokines is also upregulated in the lungs of mice after the intratracheal instillation of IL-1F9. Taken together, these data suggest that the IL-1F9–mediated induction of NF-κB activity and neutrophil-specific chemokine gene expression in macrophages may constitute an important pathway leading to lung inflammation.
The present study demonstrates that IL-1F9 is proinflammatory in the lung by increasing airway hyperresponsiveness, and enhancing NF-κB activity, neutrophil influx, and chemokine production. Lung epithelial cells produce IL-1F9, and macrophages respond to IL-1F9. Neutrophil accumulation in the airways contributes to the pathogenesis of several acute and chronic lung diseases such as asthma, chronic obstructive pulmonary disease, acute respiratory disease syndrome, and cystic fibrosis. IL-1F9 may contribute, in part, to these diseases by recruiting neutrophils to the lung. The further characterization of IL-1F9 and other novel IL-1 family members in human lung disease will be important in understanding the pathogenesis of these inflammatory lung diseases, and in determining the therapeutic potential of novel IL-1 family member targets.
Supplementary Material
Acknowledgments
For their excellent technical support, the authors thank Marda Jorgensen (immunofluorescence microscopy), Marcus Moore and Clive Wasserfall (Luminex assays), and Malgorzata Gil (LAL assays).
This work was supported by National Institute of Health grants RO1-HL-71522 (A.M.L.) and 5T32HL007874-14 (B.D.M).
The present address of Ravisankar A. Ramadas is the Pulmonary and Critical Care Unit, Center for Immunology and Inflammatory Diseases, Massachusetts General Hospital and Harvard Medical School, Charlestown, Massachusetts.
This article has an online supplement, which is accessible from this issue's table of contents at www.atsjournals.org
Originally Published in Press as DOI: 10.1165/rcmb.2009-0315OC on March 18, 2010
Author Disclosure: A.M.L. received a sponsored grant from the National Institutes of Health for more than $100,001. B.D.M. received expert witness fees from Bengston and Jestings, LLP, for $5,001–$10,000, and a sponsored grant from the National Institutes of Health for more than $100,001. None of the other authors have a financial relationship with a commercial entity that has an interest in the subject of this manuscript.
References
- 1.Dinarello CA. Immunological and inflammatory functions of the interleukin-1 family. Annu Rev Immunol 2009;27:519–550. [DOI] [PubMed] [Google Scholar]
- 2.Arend WP, Guthridge CJ. Biological role of interleukin 1 receptor antagonist isoforms. Ann Rheum Dis 2000;59:i60–i64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dinarello CA. Biologic basis for interleukin-1 in disease. Blood 1996;87:2095–2147. [PubMed] [Google Scholar]
- 4.Arend WP. The balance between IL-1 and IL-1Ra in disease. Cytokine Growth Factor Rev 2002;13:323–340. [DOI] [PubMed] [Google Scholar]
- 5.Johnson VJ, Yucesoy B, Luster MI. Prevention of IL-1 signaling attenuates airway hyperresponsiveness and inflammation in a murine model of toluene diisocyanate–induced asthma. J Allergy Clin Immunol 2005;116:851–858. [DOI] [PubMed] [Google Scholar]
- 6.Nakae S, Komiyama Y, Yokoyama H, Nambu A, Umeda M, Iwase M, Homma I, Sudo K, Horai R, Asano M, et al. IL-1 is required for allergen-specific Th2 cell activation and the development of airway hypersensitivity response. Int Immunol 2003;15:483–490. [DOI] [PubMed] [Google Scholar]
- 7.Wang CC, Fu CL, Yang YH, Lo YC, Wang LC, Chuang YH, Chang DM, Chiang BL. Adenovirus expressing interleukin-1 receptor antagonist alleviates allergic airway inflammation in a murine model of asthma. Gene Ther 2006;13:1414–1421. [DOI] [PubMed] [Google Scholar]
- 8.Barksby HE, Lea SR, Preshaw PM, Taylor JJ. The expanding family of interleukin-1 cytokines and their role in destructive inflammatory disorders. Clin Exp Immunol 2007;149:217–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Towne JE, Garka KE, Renshaw BR, Virca GD, Sims JE. Interleukin (IL)-1F6, IL-1F8, and IL-1F9 signal through IL-1Rrp2 and IL-1RAcP to activate the pathway leading to NF-kappaB and MAPKs. J Biol Chem 2004;279:13677–13688. [DOI] [PubMed] [Google Scholar]
- 10.Ramadas RA, Li X, Shubitowski DM, Samineni S, Wills-Karp M, Ewart SL. IL-1 receptor antagonist as a positional candidate gene in a murine model of allergic asthma. Immunogenetics 2006;58:851–855. [DOI] [PubMed] [Google Scholar]
- 11.Ewart SL, Kuperman D, Schadt E, Tankersley C, Grupe A, Shubitowski DM, Peltz G, Wills-Karp M. Quantitative trait loci controlling allergen-induced airway hyperresponsiveness in inbred mice. Am J Respir Cell Mol Biol 2000;23:537–545. [DOI] [PubMed] [Google Scholar]
- 12.Vos JB, van Sterkenburg MA, Rabe KF, Schalkwijk J, Hiemstra PS, Datson NA. Transcriptional response of bronchial epithelial cells to Pseudomonas aeruginosa: identification of early mediators of host defense. Physiol Genomics 2005;21:324–336. [DOI] [PubMed] [Google Scholar]
- 13.Akilesh S, Shaffer DJ, Roopenian D. Customized molecular phenotyping by quantitative gene expression and pattern recognition analysis. Genome Res 2003;13:1719–1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Arend WP, Palmer G, Gabay C. IL-1, IL-18, and IL-33 families of cytokines. Immunol Rev 2008;223:20–38. [DOI] [PubMed] [Google Scholar]
- 15.Gasse P, Mary C, Guenon I, Noulin N, Charron S, Schnyder-Candrian S, Schnyder B, Akira S, Quesniaux VF, Lagente V, et al. IL-1R1/MyD88 signaling and the inflammasome are essential in pulmonary inflammation and fibrosis in mice. J Clin Invest 2007;117:3786–3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heiman AS, Abonyo BO, Darling-Reed SF, Alexander MS. Cytokine-stimulated human lung alveolar epithelial cells release eotaxin-2 (CCL24) and eotaxin-3 (CCL26). J Interferon Cytokine Res 2005;25:82–91. [DOI] [PubMed] [Google Scholar]
- 17.Jedrzkiewicz S, Nakamura H, Silverman ES, Luster AD, Mansharamani N, In KH, Tamura G, Lilly CM. IL-1beta induces eotaxin gene transcription in A549 airway epithelial cells through NF-kappaB. Am J Physiol Lung Cell Mol Physiol 2000;279:L1058–L1065. [DOI] [PubMed] [Google Scholar]
- 18.Manzer R, Wang J, Nishina K, McConville G, Mason RJ. Alveolar epithelial cells secrete chemokines in response to IL-1beta and lipopolysaccharide but not to ozone. Am J Respir Cell Mol Biol 2006;34:158–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lappalainen U, Whitsett JA, Wert SE, Tichelaar JW, Bry K. Interleukin-1beta causes pulmonary inflammation, emphysema, and airway remodeling in the adult murine lung. Am J Respir Cell Mol Biol 2005;32:311–318. [DOI] [PubMed] [Google Scholar]
- 20.Wuyts WA, Vanaudenaerde BM, Dupont LJ, Demedts MG, Verleden GM. Modulation by cAMP of IL-1beta–induced eotaxin and MCP-1 expression and release in human airway smooth muscle cells. Eur Respir J 2003;22:220–226. [DOI] [PubMed] [Google Scholar]
- 21.Bry K, Whitsett JA, Lappalainen U. IL-1beta disrupts postnatal lung morphogenesis in the mouse. Am J Respir Cell Mol Biol 2007;36:32–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.DeVoti JA, Rosenthal DW, Wu R, Abramson AL, Steinberg BM, Bonagura VR. Immune dysregulation and tumor-associated gene changes in recurrent respiratory papillomatosis: a paired microarray analysis. Mol Med 2008;14:608–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bochkov YA, Hanson KM, Keles S, Brockman-Schneider RA, Jarjour NN, Gern JE. Rhinovirus-induced modulation of gene expression in bronchial epithelial cells from subjects with asthma. Mucosal Immunol 3:69–80. [DOI] [PMC free article] [PubMed]
- 24.Parsanejad R, Fields WR, Steichen TJ, Bombick BR, Doolittle DJ. Distinct regulatory profiles of interleukins and chemokines in response to cigarette smoke condensate in normal human bronchial epithelial (NHBE) cells. J Interferon Cytokine Res 2008;28:703–712. [DOI] [PubMed] [Google Scholar]
- 25.Kato, A., R.A. Chustz, R.P. Schleimer. Regulation and function of newly recognized IL-1 family cytokines in human bronchial epithelial cells. J Immunol 2009;182:98.18 [abstract].
- 26.Blumberg H, Dinh H, Trueblood ES, Pretorius J, Kugler D, Weng N, Kanaly ST, Towne JE, Willis CR, Kuechle MK, et al. Opposing activities of two novel members of the IL-1 ligand family regulate skin inflammation. J Exp Med 2007;204:2603–2614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Martin U, Scholler J, Gurgel J, Renshaw B, Sims JE, Gabel CA. Externalization of the leaderless cytokine IL-1F6 occurs in response to lipopolysaccharide/ATP activation of transduced bone marrow macrophages. J Immunol 2009;183:4021–4030. [DOI] [PubMed] [Google Scholar]
- 28.Hybertson BM, Jepson EK, Cho OJ, Clarke JH, Lee YM, Repine JE. TNF mediates lung leak, but not neutrophil accumulation, in lungs of rats given IL-1 intratracheally. Am J Respir Crit Care Med 1997;155:1972–1976. [DOI] [PubMed] [Google Scholar]
- 29.Leff JA, Baer JW, Bodman ME, Kirkman JM, Shanley PF, Patton LM, Beehler CJ, McCord JM, Repine JE. Interleukin-1–induced lung neutrophil accumulation and oxygen metabolite–mediated lung leak in rats. Am J Physiol 1994;266:L2–L8. [DOI] [PubMed] [Google Scholar]
- 30.Fulkerson PC, Zimmermann N, Hassman LM, Finkelman FD, Rothenberg ME. Pulmonary chemokine expression is coordinately regulated by STAT1, STAT6, and IFN-gamma. J Immunol 2004;173:7565–7574. [DOI] [PubMed] [Google Scholar]
- 31.Gagliardo R, Chanez P, Mathieu M, Bruno A, Costanzo G, Gougat C, Vachier I, Bousquet J, Bonsignore G, Vignola AM. Persistent activation of nuclear factor–kappaB signaling pathway in severe uncontrolled asthma. Am J Respir Crit Care Med 2003;168:1190–1198. [DOI] [PubMed] [Google Scholar]
- 32.Choi IW, Kim DK, Ko HM, Lee HK. Administration of antisense phosphorothioate oligonucleotide to the p65 subunit of NF-kappaB inhibits established asthmatic reaction in mice. Int Immunopharmacol 2004;4:1817–1828. [DOI] [PubMed] [Google Scholar]
- 33.Desmet C, Gosset P, Pajak B, Cataldo D, Bentires-Alj M, Lekeux P, Bureau F. Selective blockade of NF-kappa B activity in airway immune cells inhibits the effector phase of experimental asthma. J Immunol 2004;173:5766–5775. [DOI] [PubMed] [Google Scholar]
- 34.Poynter ME, Cloots R, van Woerkom T, Butnor KJ, Vacek P, Taatjes DJ, Irvin CG, Janssen-Heininger YM. NF-kappa B activation in airways modulates allergic inflammation but not hyperresponsiveness. J Immunol 2004;173:7003–7009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Martin T, Cardarelli PM, Parry GC, Felts KA, Cobb RR. Cytokine induction of monocyte chemoattractant protein–1 gene expression in human endothelial cells depends on the cooperative action of NF-kappa B and AP-1. Eur J Immunol 1997;27:1091–1097. [DOI] [PubMed] [Google Scholar]
- 36.Ueda A, Okuda K, Ohno S, Shirai A, Igarashi T, Matsunaga K, Fukushima J, Kawamoto S, Ishigatsubo Y, Okubo T. NF-kappa B and Sp1 regulate transcription of the human monocyte chemoattractant protein–1 gene. J Immunol 1994;153:2052–2063. [PubMed] [Google Scholar]
- 37.Moriuchi H, Moriuchi M, Fauci AS. Nuclear factor–kappa B potently up-regulates the promoter activity of RANTES, a chemokine that blocks HIV infection. J Immunol 1997;158:3483–3491. [PubMed] [Google Scholar]
- 38.Keates AC, Keates S, Kwon JH, Arseneau KO, Law DJ, Bai L, Merchant JL, Wang TC, Kelly CP. ZBP-89, Sp1, and nuclear factor–kappa B regulate epithelial neutrophil–activating peptide-78 gene expression in Caco-2 human colonic epithelial cells. J Biol Chem 2001;276:43713–43722. [DOI] [PubMed] [Google Scholar]
- 39.Rezzonico R, Imbert V, Chicheportiche R, Dayer JM. Ligation of CD11b and CD11c beta(2) integrins by antibodies or soluble CD23 induces macrophage inflammatory protein 1alpha (MIP-1alpha) and MIP-1beta production in primary human monocytes through a pathway dependent on nuclear factor-kappaB. Blood 2001;97:2932–2940. [DOI] [PubMed] [Google Scholar]
- 40.Li X, Massa PE, Hanidu A, Peet GW, Aro P, Savitt A, Mische S, Li J, Marcu KB. IKKalpha, IKKbeta, and NEMO/IKKgamma are each required for the NF-kappa B–mediated inflammatory response program. J Biol Chem 2002;277:45129–45140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Feng G, Ohmori Y, Chang PL. Production of chemokine CXCL1/KC by okadaic acid through the nuclear factor–kappaB pathway. Carcinogenesis 2006;27:43–52. [DOI] [PubMed] [Google Scholar]
- 42.De Plaen IG, Han XB, Liu X, Hsueh W, Ghosh S, May MJ. Lipopolysaccharide induces CXCL2/macrophage inflammatory protein–2 gene expression in enterocytes via NF-kappaB activation: independence from endogenous TNF-alpha and platelet-activating factor. Immunology 2006;118:153–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.