Abstract
Here we analyze the essential process of X-chromosome dosage compensation (DC) to elucidate mechanisms that control the assembly, genome-wide binding, and function of gene regulatory complexes that act over large chromosomal territories. We demonstrate that a subunit of Caenorhabditis elegans MLL/COMPASS, a gene activation complex, acts within the DC complex (DCC), a condensin complex, to target the DCC to both X chromosomes of hermaphrodites for chromosome-wide reduction of gene expression. The DCC binds to two categories of sites on X: rex (recruitment element on X) sites that recruit the DCC in an autonomous, sequence-dependent manner, and dox (dependent on X) sites that reside primarily in promoters of expressed genes and bind the DCC robustly only when attached to X. We find that DC mutations that abolish rex site binding greatly reduce dox site binding but do not eliminate it. Instead, binding is diminished to the low level observed at autosomal sites in wild-type animals. Changes in DCC binding to these non-rex sites occur throughout development and correlate directly with transcriptional activity of adjacent genes. Moreover, autosomal DCC binding is enhanced by rex site binding in cis in X-autosome fusion chromosomes. Thus, dox and autosomal sites have similar binding potential but are distinguished by linkage to rex sites. We propose a model for DCC binding in which low-level DCC binding at dox sites is dictated by intrinsic properties correlated with high transcriptional activity. Sex-specific DCC recruitment to rex sites then enhances the magnitude of DCC binding to dox sites in cis, which lack high affinity for the DCC on their own. We also show that the DCC balances X-chromosome gene expression between sexes by controlling transcription.
Keywords: dosage compensation, MLL/COMPASS, condensin, epigenetics, gene expression, C. elegans
Regulation of gene expression in multicellular organisms requires diverse strategies that operate over dramatically different distances along chromosomes. Local control of gene expression involves recruitment of specific transcription factors (TFs) or chromatin-modifying enzymes to the particular gene targets they regulate. Long-range gene control mechanisms involve targeting of regulatory proteins to sites throughout the genome, and coordinating the action of those proteins to regulate gene expression within large chromosomal domains or across entire chromosomes. Such long-range regulatory strategies often require changes in chromosome structure or nuclear location to reposition genes and thereby mediate expression at a distance.
In the present study, we analyzed the essential process of dosage compensation (DC) to understand the complex mechanisms that regulate gene expression across large chromosomal territories. DC has evolved to fine-tune gene expression across an entire chromosome, in part by co-opting molecules and mechanisms employed by other regulatory processes that act either locally or over long range (Kind et al. 2008; Payer and Lee 2008; Meyer 2010). The need for DC arises in organisms that use chromosome-based mechanisms to determine sex (e.g., XX female and XY or XO male) (Charlesworth 1996). The potential imbalance in X-chromosome gene expression created by the disparity in X dose between sexes must be offset by a chromosome-wide mechanism of gene regulation to prevent sex-specific lethality. Strategies to achieve this balance in expression differ. Fruit flies double expression of the male's single X (Gelbart and Kuroda 2009), mammals inactivate one of the female's two X chromosomes (Payer and Lee 2008), and nematodes halve expression of both hermaphrodite X chromosomes to equal that of the male's single X (Meyer 2010). Regardless of mechanism, these different DC strategies all rely on specific targeting of regulatory complexes to discrete regions along the X chromosome of one sex to alter gene expression chromosome-wide.
The Caenorhabditis elegans DC complex (DCC) is composed not just of subunits unique to the DCC, some of which confer both X and sex specificity to DC, but also of subunits shared with other molecular machines. Four C. elegans DC proteins function in both the DCC and condensin complexes, highly conserved complexes shown to mediate chromosome compaction, resolution, and segregation in mitosis and meiosis and control crossover recombination between homologous meiotic chromosomes (Csankovszki et al. 2009; Mets and Meyer 2009; Meyer 2010). A fifth DCC subunit, DPY-27, is homologous to condensin SMC proteins, yet is specific to the DCC (Chuang et al. 1994). The direct participation of DCC subunits in condensin complexes that restructure chromosomes suggests that the mechanism of DC involves changes in chromosome structure, perhaps to reposition DCC-binding sites near genes controlled by the DC process. Beyond C. elegans DC, condensin subunits participate in other forms of gene regulation: transcriptional silencing of fly homeotic genes and yeast mating type loci, position-effect variegation in flies, and T-cell development in mice (for review, see Wood et al. 2010). Thus, lessons learned from C. elegans DC will enhance our general understanding of gene regulation and chromosome structure.
Recent experiments revealed part of the mechanism by which the DCC is targeted to X chromosomes and the means by which the DCC influences gene expression once bound. The DCC binds to two distinct classes of cis-acting regulatory elements on X: rex (recruitment element on X) sites recruit the DCC in an autonomous manner, even when detached from X (McDonel et al. 2006; Jans et al. 2009), whereas dox (dependent on X) sites bind the DCC robustly only when attached to X (Jans et al. 2009). Analysis of rex sites revealed a 12-base-pair (bp) DNA consensus motif that is enriched on X (MEX [motif enriched on X]) and is essential for binding at most, but not all, rex sites (Jans et al. 2009). The MEX confers much of X specificity to DCC binding. Unlike rex sites, dox sites lack a specific DNA motif and are instead enriched in the promoters of highly expressed genes (Jans et al. 2009). dox sites are fivefold to 10-fold more prevalent than rex sites (100–200 sites). Although the DCC is greatly enriched on X chromosomes, it also binds to sites throughout the genome that, like dox sites, occur in promoters of expressed genes (Jans et al. 2009). These autosomal sites occur at a lower density than dox sites and have less DCC binding. Prior studies have shown that binding of the DCC to a gene on X does not determine whether that gene is dosage-compensated, as genes can be dosage-compensated without the DCC bound nearby, and DCC-bound genes may not be compensated (Jans et al. 2009). Thus, the DCC must act over long distances to modulate expression of the genes that lack DCC binding, and it may act locally to regulate expression of the genes it binds.
In this study, we addressed fundamental questions about the composition, binding, and functioning of the DCC. We first asked whether the DCC shares components with other dedicated gene regulatory complexes and, if so, whether an overlap exists in the DNA-binding sites of regulatory complexes that share proteins. Pivotal for the DC process is the protein DPY-30, the C. elegans ortholog of a subunit of the conserved MLL/COMPASS gene regulatory complexes, which catalyze trimethylation of histone H3 at Lys 4 (H3K4me3) at 5′ ends of genes and contribute to local gene activation (Hsu et al. 1995; Miller et al. 2001; Nagy et al. 2002). We examined the capacity in which DPY-30 functions in DC, including whether DPY-30 is a physical component of the DCC or whether it acts separately in a regulatory fashion to control the expression of genes that implement DC. We then explored the mechanism by which the DCC is assembled onto X. Although two classes of cis-acting X regulatory elements had been identified, the molecular requirements for DCC recruitment to rex sites and DCC binding to dox sites had not been determined. Models had been proposed in which DCC occupancy of dox sites is dependent on DCC recruitment to rex sites (Jans et al. 2009), but the relationship between DCC binding at the two classes of sites had not been tested directly. Lastly, we assessed the specific aspect of gene regulation controlled by the DC process. While C. elegans DC had been shown to reduce the overall level of transcripts from X-linked genes in XX animals (Meyer and Casson 1986), it was not known whether this reduction occurred through an effect on transcription, or instead an effect on mRNA stability or mRNA processing.
Here we show DPY-30 to be a subunit of two distinct regulatory complexes: the MLL/COMPASS gene-activating complex and the DCC gene-repressing complex. Remarkably, the DCC and MLL/COMPASS complexes bind to the same sites genome-wide, but have opposite effects on gene regulation. DPY-30 is critical for recruitment of the DCC to X. We also describe fundamental differences in DCC binding to rex, dox, and autosomal sites. DCC binding to rex sites requires not just DPY-30, but also SDC-2 and SDC-3, essential components of the C. elegans DC regulatory hierarchy that triggers DCC binding specifically to X in XX embryos. dox sites also lack robust DCC-binding ability without these recruitment proteins, but, unlike rex sites, many retain intrinsic, low-level DCC-binding ability that is similar in level to DCC binding at autosomal sites in wild-type animals. Changes in DCC binding at both dox and autosomal sites occur throughout development and are positively correlated with changes in gene expression. Our data support a model in which the location of both dox and autosomal sites is dictated by intrinsic DCC-binding properties correlated with transcriptional activity. Proximity to rex sites in cis then strongly influences the magnitude of binding. Indeed, even DCC binding to autosomes can be enhanced when rex sites are attached in cis by fusion of an X chromosome to an autosome (see also Ercan et al. 2009). Finally, we show that C. elegans DC reduces X-chromosome transcript levels by regulating transcription.
Results
DPY-30 functions in two distinct gene regulatory complexes: the MLL/COMPASS gene-activating complex and the DCC gene-repressing complex
Like other genes essential for DC, dpy-30 was identified in screens for suppressors of the XO-specific lethality caused by mutation of xol-1, the gene that blocks the DCC from functioning in males (Hsu and Meyer 1994). Mutations in dpy-30 cause XX-specific lethality (Hsu and Meyer 1994; Hsu et al. 1995) and prevent all DCC components, except SDC-2, from assembling onto hermaphrodite X chromosomes, as shown by immunofluorescence studies (Chuang et al. 1996; Lieb et al. 1996, 1998; Davis and Meyer 1997; Dawes et al. 1999; Yonker and Meyer 2003). Unlike most DC genes, dpy-30 functions in other developmental processes—including vulval development, male mating behavior, and aging—suggesting that dpy-30 might also function in a second gene regulatory capacity (Hsu and Meyer 1994; Greer et al. 2010). Indeed, studies in yeast and mammals identified DPY-30 homologs as subunits of MLL/COMPASS complexes, which are recruited to 5′ ends of actively transcribed genes to catalyze H3K4me3, a signature of active chromatin (Miller et al. 2001; Roguev et al. 2001; Shilatifard 2008). The pleiotropic effects of dpy-30 mutations in XO and XX animals, along with the diffuse nuclear localization of DPY-30 instead of the punctate, X-specific localization of the DCC, suggested that DPY-30 might function indirectly in DC (Hsu et al. 1995).
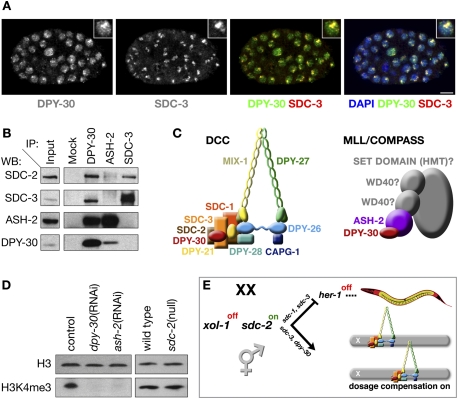
Production of two DPY-30-specific antibodies (Supplemental Fig. S1A) permitted us to re-evaluate the roles of DPY-30 and show that DPY-30 is a bona fide member of the DCC and the MLL/COMPASS complex. Immunofluorescence studies using both antibodies confirmed that DPY-30 exhibits a more diffuse nuclear localization pattern than other DCC components. However, in contrast to prior results, our results show that DPY-30 is enriched on X chromosomes of XX embryos and colocalizes with the DCC (Fig. 1A). Moreover, DPY-30 antibodies coimmunoprecipitate DCC subunits SDC-2 and SDC-3 (Fig. 1B), consistent with DPY-30 being a DCC component.
Figure 1.
DPY-30 participates in two gene regulatory complexes: the DCC and MLL/COMPASS. (A) DPY-30 exhibits both diffuse nuclear localization and enrichment on X colocalized with DCC subunits. Confocal images of an XX embryo costained with 4,6-diamidino-2-phenylindole (DAPI, blue), and antibodies against DPY-30 (green in merge) and SDC-3 (red in merge). DPY-30 and SDC-3 colocalize (yellow). The diffuse nuclear localization of DPY-30 supports a function beyond DC. Inset is an enlarged nucleus. Bar, 5 μm. (B) DPY-30 interacts with subunits of the DCC and the MLL/COMPASS complex. Immunoprecipitation (IP) and Western blot (WB) analysis using embryo extracts confirm association of DPY-30 with the DCC subunits SDC-2 and SDC-3 as well as the MLL/COMPASS subunit ASH-2. Immunoprecipitation of ASH-2 fails to recover SDC-3 and very weakly recovers SDC-2, indicating that DPY-30 likely functions in two distinct complexes. (C) Schematics of the C. elegans DCC and C. elegans MLL/COMPASS complexes with known subunits identified (in color). DPY-30 (red) is shared between both complexes. (D) C. elegans DPY-30 and ASH-2 are required for H3K4me3, consistent with their participation in MLL/COMPASS. Shown are Western blots of either RNAi empty vector control (L4440), dpy-30(RNAi), ash-2(RNAi), wild-type, or sdc-2 (null) mutant embryos blotted with antibodies to histone H3 or H3K4me3. H3K4me3 is undetectable in embryos depleted of DPY-30 or ASH-2, while H3 levels are unaffected. H3K4me3 levels are not reduced in sdc-2 mutant embryos, confirming that participation of DPY-30 in MLL/COMPASS, and not the DCC, is responsible for H3K4me3, and that disruption of DC does not affect H3K4me3. (E) Assembly of the DCC onto X is controlled by a genetic pathway that regulates both sex determination and DC in which repression of xol-1 activity in XX animals permits activation of the XX-specific gene sdc-2. sdc-2 turns on the hermaphrodite pathway of sexual differentiation in collaboration with sdc-1 and sdc-3 by repressing the male sex determination gene her-1, and sdc-2 triggers loading of the DCC onto X chromosomes in concert with sdc-3 and dpy-30 (Meyer 2010).
DPY-30 also associates physically with the C. elegans homolog of ASH-2, an MLL/COMPASS subunit, consistent with its diffuse nuclear localization (Fig. 1B; Supplemental Fig. S1B for ASH-2 antibody specificity). DPY-30 and C. elegans ASH-2 coimmunoprecipitate in a reciprocal manner from embryonic extracts. In contrast, ASH-2 antibodies fail to coimmunoprecipitate SDC-3 and only very weakly coimmunoprecipitate SDC-2, suggesting that ASH-2 interacts with DPY-30 in a complex that is distinct from the DCC. Furthermore, reduction of dpy-30 or ash-2 activities through RNAi greatly reduces the level of H3K4me3, whereas complete loss of sdc-2 activity does not alter H3K4me3 levels (Fig. 1D; Simonet et al. 2007; Greer et al. 2010). Thus, DPY-30 promotes histone modification through its participation in a C. elegans MLL/COMPASS complex rather than in the DCC, and disruption of DC does not affect H3K4me3 levels. DPY-30 acts in both a chromosome-wide gene repression complex and a genome-wide gene activation complex (Fig. 1C).
DPY-30 binds to all DCC-binding sites throughout the genome, yet the level of DPY-30 binding at autosomal sites is higher than that for other DCC subunits
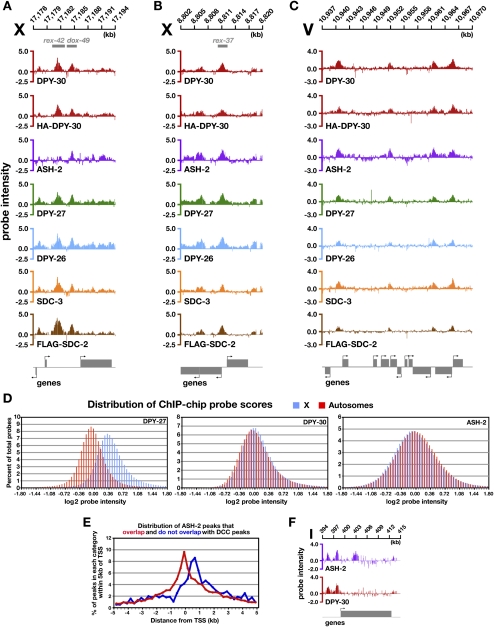
DPY-30 has a binding profile on X expected for a DCC component. The profile was determined by chromatin immunoprecipitation (ChIP) reactions followed by hybridization to genome-wide tiling arrays (ChIP–chip) using two combinations of protein extracts and antibodies: extracts from wild-type embryos and antibodies to endogenous DPY-30, or extracts from a dpy-30-null mutant strain expressing a rescuing Hemagglutinin (HA)-tagged DPY-30 protein and HA antibodies. DPY-30-binding profiles from both approaches closely resemble each other and those of other DCC subunits (Fig. 2A–C; Supplemental Fig. S2A,B). More than 98.5% of DPY-30 peaks on X called in either experiment correspond to peaks called in DPY-27 or SDC-3 ChIP–chip experiments. DPY-30 binds to nearly all rex (97%) and dox (99%) sites, and most DPY-30 sites are in promoters of expressed genes, as are DCC sites (Supplemental Fig. S3B,D). Thus, not only is DPY-30 essential for DC, it functions as part of the DCC and binds to virtually all DCC sites on X.
Figure 2.
The DPY-30-binding profile on X and autosomes is coincident with the DCC profile, but the level of DPY-30 binding on autosomal sites is higher than other DCC subunits. (A–C) Representative ChIP–chip profiles of five DCC subunits (DPY-30, DPY-27, DPY-26, SDC-3, and SDC-2) and one MLL/COMPASS subunit (ASH-2) on X and an autosome. Gene locations are indicated below the ChIP profiles, and the direction of transcription is indicated by an arrow. Both endogenous DPY-30 and a rescuing HA-tagged DPY-30 bind to all previously identified DCC-binding sites on X and autosomes. The ASH-2-binding profile is similar to that of the DCC, but differs in an important way at rex sites. ASH-2 binds to most rex sites within promoters of genes but rarely at intergenic rex sites. (A) DPY-30, but not ASH-2, binds to the intergenic rex-42 site. ASH-2 binds the nearby dox site. (B) DPY-30 and ASH-2 bind to rex-37, a site overlapping divergent promoters. (C) On autosomes, DPY-30-binding sites are indistinguishable from those of other DCC subunits. ASH-2 binding is coincident with DPY-30 and DCC subunits, except for a small number of ASH-2-specific binding sites (see E,F). (D) Histograms depicting the distribution of DPY-27, DPY-30, or ASH-2 ChIP–chip probe scores across the X chromosome and autosomes in wild-type XX embryos. Probe scores were grouped into bins with a step size of 0.06 (log2 scale), and the percentage of total probes in each bin was graphed on the Y-axis. DPY-27 binds more strongly to X than to autosomes, as shown by the higher average probe intensity. Consistent with DPY-30 functioning in both DCC and MLL/COMPASS complexes, DPY-30 binding is more evenly distributed across X and autosomes than DPY-27 binding, with a slight enrichment on X. ASH-2 binding is also more equally distributed between X and autosomes, with a slight enrichment on autosomes, consistent with its more singular role in the MLL/COMPASS complex. (E) ASH-2 binds to a small set of sites independently from DPY-30 and other DCC subunits, consistent with its participation in a separate complex. Unique ASH-2 peaks were defined as those not overlapping with DPY-30-, DPY-27-, DPY-26-, SDC-3-, or SDC-2-binding sites. The distance from the center of the unique or nonunique peaks to the nearest transcriptional start site (TSS) was determined. These distances were grouped into 250-bp bins, and the percentage of peaks within each bin was calculated for each category (negative X-axis values correspond to peaks that lie upstream of the TSS). The unique ASH-2 peaks occur 3′ of the TSS, within the coding region, while the peaks that overlap with DCC-binding sites are enriched within promoters. (F) ChIP–chip binding profile of a unique ASH-2-binding site. ASH-2 binds within the coding region of the gene, unlike DPY-30 or other DCC subunits.
The DCC also binds to autosomes, primarily in promoters of expressed genes, but at a lower density (one-fifth) of sites than on X and with less occupancy per site (Jans et al. 2009). DPY-30-binding sites on autosomes overlap robustly with DCC-binding sites (Fig. 2C; Supplemental Figs. S2B, S3D). However, the level of DPY-30 binding at X and autosomal sites is equivalent, unlike that of the DCC-specific subunit DPY-27, which exhibits greater binding to sites on X than autosomes. Both the distribution of ChIP–chip probe scores across X and autosomes (Fig. 2D) and the ratio of average peak scores for autosomes versus X support this conclusion. The autosome to X ratio is 1.1 for DPY-30, but 0.51 for DPY-26, 0.6 for SDC-2, and 0.69 for DPY-27. The higher level of DPY-30 binding to autosomes might reflect an association with the MLL/COMPASS complex.
Genome-wide binding profile of MLL/COMPASS subunit ASH-2
To assess whether DPY-30’s participation in MLL/COMPASS accounts for the quantitative differences in autosomal binding between DPY-30 and other DCC subunits, we determined the genome-wide binding profile of ASH-2 (Fig. 2A–C; Supplemental Fig. S2A,B). Like DPY-30, ASH-2 binds preferentially to the promoters of expressed genes (Supplemental Fig. S3A), and the distribution of ASH-2 ChIP–chip probe scores is very similar for X and autosomes. Furthermore, the ratio of average peak scores for autosomes versus X is greater for ASH-2 (0.97) than for DCC subunits (∼0.60), supporting the view that DPY-30’s participation in both MLL/COMPASS and the DCC contributes to the level of DPY-30 autosomal binding.
The binding profiles of ASH-2 and DPY-30 do differ in significant ways, however. On X, ASH-2 binds to fewer rex sites than DPY-30 (53% vs. 97%, respectively), and the rex sites not bound by ASH-2 are typically in intergenic regions rather than in promoters (Fig. 2A,B; Supplemental Fig. S2A). On autosomes, binding of both ASH-2 and DPY-30 is strongly biased toward the promoters of expressed genes, but ASH-2 binds to ∼10% more sites per autosome than DPY-30 (Fig. 2E,F). The unique ASH-2 sites occur mostly in coding regions, suggesting that ASH-2 may participate in distinct complexes with and without DPY-30 (Fig. 2E,F; Supplemental Fig. S3C). The disparity between ASH-2- and DCC-binding profiles suggests that binding of DPY-30 and DCC subunits would have overlapping yet distinct responses to recruitment factors like SDC-2 that direct DCC loading onto X.
Targeting the DCC to X: different principles govern DCC binding to rex and dox sites
Precise knowledge of DCC-binding sites allowed us to determine the contributions of individual genes in the DC hierarchy toward targeting the DCC to X and assess the relationship between DCC binding at rex and dox sites. Until the present study, the DC hierarchy had been inferred solely from genetic and immunofluorescence data (Fig. 1E). SDC-2, a 350-kDa XX-specific protein with a large coiled-coil domain, confers both sex and X specificity to DCC binding (Nusbaum and Meyer 1989; Dawes et al. 1999). Although SDC-2 can bind to X independently of other DCC components, it collaborates with the zinc finger protein SDC-3 and DPY-30 to achieve full DCC loading onto X (for review, see Meyer 2010).
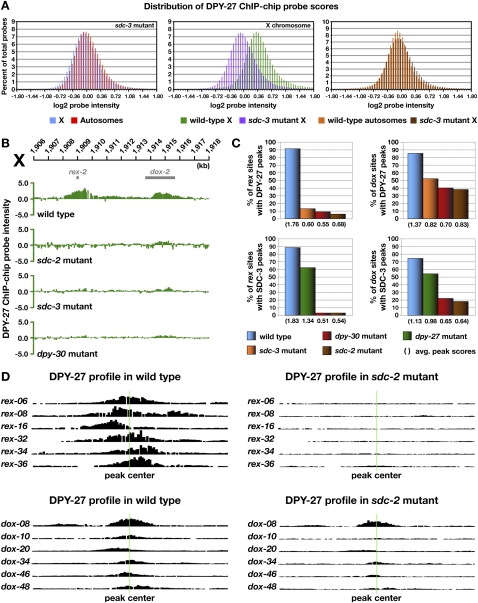
Genome-wide binding profiles of DCC subunits in DC-defective mutant embryos showed that the overall binding of DPY-27 and SDC-3 on X chromosomes in either sdc-2- or sdc-3-null mutant embryos was dramatically decreased, as measured by the distribution of ChIP–chip probe scores (Fig. 3A; Supplemental Fig. S4A,B). Binding at rex sites was nearly eliminated in both null mutants: Limited DPY-27 binding was retained on only 6% of rex sites in sdc-2 mutants and 13% of rex sites in sdc-3 mutants (Fig. 3B–D; Supplemental Fig. S5A–C). Similarly, SDC-3 binding was retained on 3% of rex sites in sdc-2 mutants. DCC binding at dox sites was more complex. DPY-27 binding was greatly depleted at 62% of dox sites in sdc-2 mutants and 48% of dox sites in sdc-3 mutants (Fig. 3B–D; Supplemental Fig. S5A–C). Of the dox sites that retained DPY-27 binding, the average peak scores were substantially reduced (Fig. 3C). The level of DPY-27 protein was not changed in sdc-2 mutants (Supplemental Fig. S6A), indicating that reduced binding was not due to reduced protein levels.
Figure 3.
Differential requirements for DCC binding to rex and dox sites. (A) Histograms show distribution of DPY-27 ChIP–chip probe scores across X and autosomes in wild-type versus sdc-3 mutant embryos. Probe scores were analyzed as in Figure 2D. The probe score distribution for X in sdc-3 mutant embryos is greatly reduced compared with that in wild-type embryos and closely resembles that of wild-type autosomes (see Fig. 2D). Additional histograms of probe intensities from DPY-27 ChIP–chip experiments for other DCC mutants are in Supplemental Figure S4. (B) ChIP–chip profile of DPY-27 binding in wild-type XX embryos compared with embryos mutant for dpy-30, sdc-3, or sdc-2. rex site binding is nearly eliminated in these mutants, while dox site binding is greatly reduced but present. (C) Histograms showing quantification of DPY-27 and SDC-3 binding at rex and dox sites in wild-type versus DC mutant embryos. The height of the bar corresponds to the percentage of bound rex and dox sites with a peak score of 0.75 or better in DC mutants. The average peak scores shown in parentheses below the histogram were calculated from the scores of all called peaks at rex and dox sites. DCC binding to rex sites is dependent on SDC-2, SDC-3, and DPY-30, whereas a low-level dox site binding is independent of these proteins. (D) Graphical representations of DPY-27 ChIP–chip probe intensities along 5-kb regions centered on representative rex and dox sites (green line) in wild-type versus sdc-2 mutant embryos further show that binding to rex sites is eliminated and binding to dox sites is severely reduced. Additional graphical representations of probe intensities from SDC-3 and DPY-27 ChIP–chip experiments for other DCC mutants are in Supplemental Figure S5.
Similar results were obtained for SDC-3: 82% of dox sites had little or no SDC-3 binding in sdc-2 mutants, and the average peak score of dox sites that retained SDC-3 was reduced (Fig. 3C). SDC-3 stability depends, in part, on SDC-2 (Davis and Meyer 1997), and the level of SDC-3 protein was reduced somewhat in sdc-2 mutants, but the reduced level is unlikely to cause a preferential reduction in binding to higher-affinity sites (rex) than lower-affinity sites (dox) (Supplemental Fig. S6A).
We conclude that rex site binding has a nearly absolute requirement for SDC-2 and SDC-3. In contrast, maximal binding at all dox sites requires SDC-2 and SDC-3, but binding at many dox sites can occur at reduced levels in an SDC-independent manner. That is, dox sites have an inherent ability to bind some DCC subunits independently of the genetic hierarchy that governs sex-specific DCC loading onto X. The inability to eliminate dox site binding while retaining rex site binding prevents us from assessing the specific role of dox sites in DC.
DCC binding on autosomes is similar to SDC-independent binding at dox sites on X
DPY-27 binding on autosomes changed very little compared with that on X in sdc-2 and sdc-3 mutant embryos, as assessed by ChIP–chip profiles (Supplemental Fig. S7), the distribution of ChIP–chip probe scores (Supplemental Fig. S4A,B), or the average peak scores. The average DPY-27 autosomal peak score in wild-type embryos was 0.58 compared with 0.60 in sdc-2 mutants and 0.55 in sdc-3 mutants. Furthermore, in sdc-3 (Fig. 3A) and sdc-2 (Supplemental Fig. S4B) mutants, the density of DPY-27 binding on X and the level of occupancy at bound sites mimicked that on autosomes, suggesting that SDC-independent binding at dox sites on X is governed by similar principles as DCC binding to autosomes.
The similarity in DCC binding at autosomal sites in wild-type embryos and dox sites in sdc-2 mutant embryos suggested that this intrinsic, low-level DCC binding might reflect the general binding properties of mitotic condensin in interphase chromosomes. Indeed, the mitotic condensin-specific subunit SMC-4, a paralog of the DC-specific protein DPY-27, has a profile of X and autosomal binding in wild-type embryos that closely resembles the pattern for DPY-27 and SDC-3 in sdc-2 mutant embryos (Supplemental Fig 7A,B). We inferred that the SMC-4 binding is on interphase chromosomes because the embryo population was skewed to post-mitotic animals. We found that, of dox sites that retain some DPY-27 binding in sdc-2-null mutants, 91% also have SMC-4 binding in wild-type embryos. Similarly, of dox sites that have SMC-4 binding in wild-type embryos, 89% retain DPY-27 binding in sdc-2-null mutants. Moreover, in sdc-2 mutants, 78% of all DPY-27 autosomal peaks and 93% of the strongest DPY-27 autosomal peaks (score >0.75) correspond to sites of SMC-4 binding in wild-type embryos.
In summary, the DCC has low, intrinsic binding capability at promoters of expressed genes throughout the genome that is SDC-independent, but the robust binding of the DCC to X at both rex and dox sites requires SDC-2 and SDC-3. The low-level, intrinsic DCC promoter binding may reflect the inherent binding ability of condensin complexes in general. Whether this low-level binding affects gene expression is not known.
The shared DCC and MLL/COMPASS subunit DPY-30 is essential for DCC binding to rex sites
Immunofluorescence studies showed that DPY-30, like SDC-2 and SDC-3, is essential for the assembly of other DCC components onto X. However, the dual roles of DPY-30 in DC and gene activation through an MLL/COMPASS complex led to speculation that DPY-30, unlike SDC-2 and SDC-3, might specifically control dispersal of the DCC from rex sites to dox sites (Ercan and Lieb 2009). We assessed this hypothesis directly by determining the genome-wide binding profile of SDC-3 and DPY-27 in dpy-30 mutant embryos (Fig. 3B,C; Supplemental Figs. S4C, S5A–C). We found DCC binding in dpy-30 mutants to be similar to that in sdc-2 or sdc-3 mutants. DPY-27 and SDC-3 binding at all rex sites was nearly abolished in dpy-30 mutants. At dox sites, binding was nearly eliminated at 60% of sites for DPY-27 and 78% of sites for SDC-3. dox sites that retained binding showed reduced binding for both proteins (Fig. 3C). The loss of rex site binding argues against a selective role for DPY-30 in directing DCC binding to dox sites, and instead establishes that DPY-30, like SDC-2 and SDC-3, is essential for binding to rex sites and optimal, but not all, binding at dox sites.
DPY-30 binding to rex sites is facilitated by both DCC and MLL/COMPASS complexes
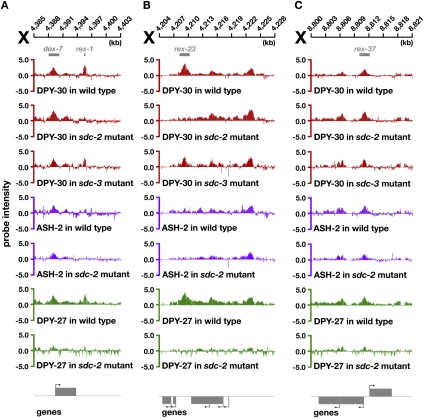
To clarify the contributions of MLL/COMPASS and DC complexes in DPY-30 binding to rex sites, we analyzed genome-wide DPY-30 and ASH-2 binding in sdc-2 mutants. Unlike other DCC subunits, DPY-30 was retained on a subset of rex sites and almost all dox and autosomal sites in sdc-2 mutants (Fig. 4A–C; Supplemental Fig. S8A–D). DPY-30 was retained on 13% (two of 15) of rex sites in intergenic regions and 47% (nine of 19) of rex sites in promoters or coding regions in sdc-2 mutants. Unlike binding of other DCC subunits, DPY-30 binding to rex and dox sites was not perturbed by an sdc-3 mutation (Fig. 4A–C; Supplemental Fig. S8A). The partial SDC-2 dependence of DPY-30 binding at some rex sites is consistent with DPY-30’s participation in the DCC. In contrast, the SDC-2-independent DPY-30 binding at rex sites likely reflects the association of DPY-30 with MLL/COMPASS. The ability of DPY-30 to bind rex sites without SDC-3 suggests that SDC-2 acts alone rather than in concert with SDC-3 to mediate DPY-30 binding to rex sites.
Figure 4.
DPY-30 binds to rex sites through participation in two distinct complexes. ChIP–chip profiles of DPY-30, ASH-2, or DPY-27 in wild-type or DC mutant embryos, with genes depicted below and arrows showing direction of transcription. DPY-30 binding is independent of sdc-3 at all rex sites, but is dependent on sdc-2 at some rex sites. (A,B) At most intergenic rex sites, such as rex-1 and rex-23, binding of DPY-30 and DPY-27 is eliminated in an sdc-2 mutant, and ASH-2 is not bound in any genotype, consistent with DPY-30 binding to these sites as a member of the DCC. (C) At most rex sites within promoters, such as rex-37, DPY-30 binding is retained in an sdc-2 mutant even though DPY-27 binding is greatly reduced. At these rex sites, ASH-2 binds, suggesting that DPY-30 binding reflects its participation in the MLL/COMPASS complex.
A specific prediction of these results is that the MLL/COMPASS subunit ASH-2 should bind in wild-type embryos to rex sites that retain DPY-30 binding in sdc-2 mutants and not to rex sites that lose DPY-30 binding in sdc-2 mutants. Moreover, ASH-2 binding to all sites throughout the genome should not be perturbed by mutation of sdc-2. These predictions were met (Fig. 4A–C; Supplemental Figs. S8A–D, S9). Nearly 85% (11 of 13) of intergenic rex sites with reduced DPY-30 binding in sdc-2 mutants lack ASH-2 binding in wild-type embryos. All genic (nine of nine) and intergenic (two of two) rex sites with robust DPY-30 binding in sdc-2 mutants have robust ASH-2 binding in wild-type embryos. Lastly, ASH-2 binding is retained in sdc-2 mutants at all rex sites strongly bound by ASH-2 in wild-type embryos. We conclude that DPY-30 binds to rex sites as part of the DCC but can also bind to a subset of rex sites as part of MLL/COMPASS. It is currently unknown whether MLL/COMPASS and the DCC bind simultaneously to this subset of rex sites.
Condensin subunits are required for DCC stability and binding but not for X-chromosome recognition
Prior immunofluorescence studies suggested that loss of the DCC condensin subunit DPY-27 caused other DCC condensin subunits and SDC-3 to be unstable and be degraded (Chuang et al. 1996; Davis and Meyer 1997). Nonetheless, SDC-2 binds to X in dpy-27 mutants, implying that DPY-27 may not be essential for the DCC to recognize X (Dawes et al. 1999). To assess the role of DPY-27 in DCC binding to X more directly, we measured SDC-3 binding in dpy-27-null mutant embryos. Without DPY-27, SDC-3 binding was reduced equally at both rex and dox sites, yet was not eliminated at either. Only 30% of rex sites and 27% of dox sites exhibited severe reduction of SDC-3 binding in dpy-27 mutants (Fig. 3C; Supplemental Fig. S5C). Of rex and dox sites bound by SDC-3, the average intensity of binding was changed only modestly in dpy-27 mutant versus wild-type embryos (Fig. 3C). Thus, DPY-27, and perhaps other DCC condensin subunits, is required for DCC stability and full SDC binding at both rex and dox sites, but DPY-27 is unlikely to participate in sequence-specific recognition of rex sites.
Prediction of rex sites
A subset of rex sites was identified previously from DCC-binding sites on X by their ability to bind the DCC in vivo when detached from X and incorporated into an extrachromosomal array (Jans et al. 2009). The abundance of DCC-binding sites on X, and the low ratio of rex to dox sites, meant that a less labor-intensive strategy was needed to identify rex sites from among the 90% of DCC sites not yet surveyed. We used two criteria to predict additional rex sites. (1) Sites had to exhibit greatly diminished DCC occupancy in sdc-2 mutants. (2) Sites could not be enriched for the modified histone H3K4me3 in wild-type embryos (data from Gerstein et al. 2010; Liu et al. 2010). This mark occurs in promoters of highly expressed genes, the locations for most dox sites and few rex sites. Indeed, 37% of known rex sites and only 2% of known dox sites fulfill these stringent criteria.
As proof of principle, four of 37 peaks that fulfilled these criteria were tested for recruitment in our extrachromosomal array assay. All four recruited the DCC (rex-39 to rex-42), verifying the utility of this method for rex site prediction (Supplemental Fig. S10A–C). As a control, we assayed a fifth site that had severely reduced DCC binding in sdc-2 mutants but also had the H3K4me3 mark. That site (dox-50) failed to recruit, as predicted (Supplemental Fig. S10C). The addition of 37 new rex sites brings the number of rex sites (tested and predicted) to 75.
Of the previously identified rex sites, 74% have a 12-bp consensus motif (MEX) that is enriched on the X chromosomes (P-value of ≤e−10) (Jans et al. 2009). Of the newly predicted rex sites, 16 of 24 most highly bound sites (67%) contain MEXs, which is a further indication that these are bona fide rex sites.
Proximity to rex sites enhances DCC binding to autosomal sites
The discovery of autonomous (rex) and dependent (dox) DCC-binding sites on X suggested a model by which the DCC loads onto X: The DCC binds to rex sites, which facilitate DCC binding to dox sites distributed along X (Jans et al. 2009). Such a model makes two predictions. First, binding to dox sites should be dependent, at least in part, on the proteins responsible for recruiting the DCC to rex sites. Second, attachment of rex sites to autosomes should enhance DCC binding to autosomal sites.
The first prediction was met by our finding that DCC binding to nearly all rex sites and the majority of dox sites was essentially eliminated by mutations in sdc-2, sdc-3, or dpy-30, and binding to the remaining dox sites was substantially reduced (Fig. 3B–D). The residual DCC binding at dox sites on X in mutant embryos resembled DCC binding on autosomes in wild-type embryos, in both the density of bound sites and the level of occupancy at the sites (Fig. 3A; Supplemental Fig. S4A–C).
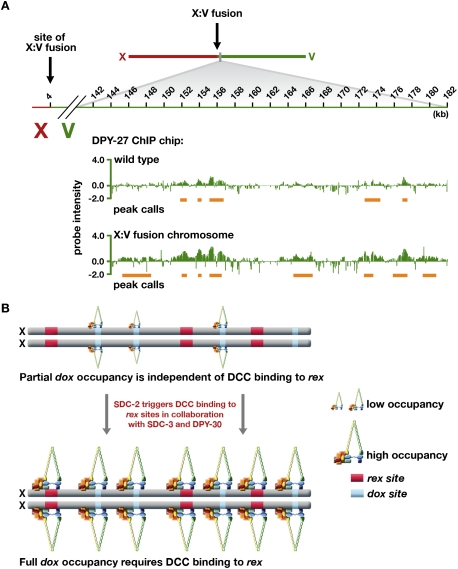
The second prediction was met by genome-wide analysis of DPY-27 binding in a strain carrying a fusion of chromosomes X and V (ypT28) (Lowden et al. 2008). Fusion of X to V enhanced DCC binding on the autosomal portion of the fusion over a 2-Mb region from the fusion breakpoint. Binding decreased progressively as distance from the breakpoint increased (Fig. 5A; Supplemental Fig. S11A,B). Two types of DCC binding were found on the V portion of the X:V fusion chromosome: enhanced binding at sites also bound by the DCC on the wild-type chromosome V, and new sites of binding. The new sites occurred preferentially in the promoters of active genes, as is typical of both dox and autosomal sites, and most showed a low level of binding below peak detection on wild-type V. Of the 224 peaks called in the first 2 Mb of V in the X:V fusion chromosome, 96 (43%) were also called on wild-type chromosome V, and the average peak score rose by 0.26 on the fusion chromosome. Furthermore, 78% (49 of 63) of the largest peaks (score >0.75) on V in the fusion chromosome were represented by smaller peaks on wild-type V. In contrast, DCC binding along X in the X:V fusion chromosome appeared unchanged from that on wild-type X (Supplemental Fig. S11C,D). Increased DCC binding to sites on V in the X:V fusion was confirmed in a separate assay: ChIP followed by quantitative real-time PCR (Supplemental Table S1).
Figure 5.
DCC binding to autosomes is increased when rex sites are attached in cis in an X:autosome fusion chromosome. (A) ChIP–chip profile of DPY-27 binding to chromosome V in wild-type embryos compared with those carrying an X:V chromosomal fusion (strain ypT28). Peak calls are underlined in orange. Attaching X in cis to an autosome enhances the level of DCC binding at sites bound on wild-type autosomes and also creates new sites of binding. (B) Model for loading the DCC onto X based on DC mutant and fusion chromosome data. dox sites possess a low intrinsic binding capability that is comparable with autosomal binding and is positively correlated with transcriptional activity of nearby genes. The DCC loads onto rex sites in a sequence-specific manner dependent on SDC-2, SDC-3, and DPY-30. The intrinsic low-level dox site binding, specified at least in part by transcriptional activity, is enhanced by proximity in cis to DCC-bound rex sites.
Thus, DCC binding at autosomal sites can be enhanced by the proximity of rex sites attached in cis, consistent with the model that DCC binding to rex sites confers X-chromosome specificity to DC and facilitates DCC binding along X at dox sites (Fig. 5B). Fulfillment of both predictions indicates that the proximity of rex sites in cis to dox sites accounts for the preference in DCC binding to dox versus autosomal sites.
The increase in DCC binding to autosomal territories located on X-to-autosome fusion chromosomes was also seen by Ercan et al. (2009) with ChIP–chip experiments. However, in their study, DCC binding to wild-type autosomes seemed negligible, and DCC binding to autosomal territories adjacent to X was interpreted as the establishment of new DCC-binding sites. Comparisons of DCC binding to autosomes in their and our data sets, which are presented in Supplemental Figure S12A–D and analyzed in the Discussion, support our results that DCC binding occurs even on wild-type autosomes.
Dynamic binding of the DCC: Changes in DCC binding at dox and autosomal sites throughout development correlate directly with changes in transcriptional activity of adjacent genes
DCC binding at dox and autosomal sites is positively correlated with expression of nearby genes, and the higher the level of gene expression, the greater the probability of a DCC-binding site (Jans et al. 2009). If the transcriptional state of a gene influences DCC binding, then the pattern of DCC binding should change dynamically throughout development, as genes are turned on and off.
We profiled DCC binding to rex, dox, and autosomal sites in response to changes in gene activity by correlating the level of DCC binding with the level of gene expression in embryos and synchronous fed L1 larvae. DCC binding to rex sites remained constant across the two developmental stages (Fig. 6B; Supplemental Fig. S13). In contrast, DCC binding at X-linked promoters and defined dox sites was positively correlated with changes in gene expression. Genes expressed more highly in embryos than in fed L1 larvae had greater DCC occupancy in promoters of embryos, and vice versa. The positive correlation was evident from ChIP–chip profiles (Fig. 6A), scatter plots (Supplemental Fig. S14D), and plots of moving averages (Fig. 6C; Supplemental Fig. S14A–C). The statistical significance of the correlation was shown by both Pearson's and Spearman's statistical tests (Supplemental Fig. S14F).
Figure 6.
DCC binding at both X and autosomal promoters changes as gene expression changes. (A,B) DPY-27 ChIP–chip profiles from embryos (green) or fed L1 larvae (magenta), with corresponding gene expression data from microarrays. Genes (below profiles) with higher expression in embryos are shown in orange, and those with higher expression in fed L1s are shown in blue. Genes with less than a fourfold difference in expression are shown in gray. The direction of transcription is indicated by an arrow. (A) DPY-27 binding changes as gene expression changes on both X and autosomes. Genes that are more highly expressed in embryos tend to have more DCC binding in embryos, and genes more highly expressed in fed L1s have more DCC binding in fed L1s. (B) Binding of DPY-27 to rex sites is constant between embryos and fed L1 larvae, as exemplified by the profile of DPY-27 at rex-24 in both embryos and fed L1s. Additional examples are in Supplemental Figure S13. (C) Graph shows the positive correlation for both X and autosomes between changes in gene expression and changes in DPY-27 binding at promoters. For each gene that is expressed in embryos or fed L1s, the third highest ChIP–chip probe score within the promoter was used for the comparative analysis of DCC binding. The fed L1 ChIP–chip and gene expression values were subtracted from those of embryos and sorted by the difference in DPY-27 binding. A moving average of 100 genes with a step size of 1 was determined for the difference in DPY-27 binding and gene expression. The data for both X and chromosome II are compared. The positive quadrants of the axes correspond to higher binding or expression in embryos compared with fed L1s.
A previous study reached a similar conclusion about the dynamic properties of DCC binding on X through comparisons of RNA polymerase II (RNA Pol II) binding and DCC binding defined by ChIP–chip analysis in embryos and L3 larvae (Ercan et al. 2009). However, the study also concluded that the rare DCC binding to autosomal sites did not respond to changes in gene expression during development. In contrast, our study shows the opposite: DCC binding to promoters of autosomal genes, like DCC binding to promoters on X, is positively correlated with changes in gene expression throughout development. The positive correlation on autosomes was evident using the same criteria as for that for X (Fig. 6A; Supplemental Fig. S14A–C,E,F). As an example, of 517 autosomal genes that were expressed at least fourfold higher in embryos than in fed L1 larvae and also had a DCC peak in one or both developmental stages, 76% had a DCC peak solely in embryos, and only 7% had a DCC peak solely in fed L1 larvae.
Several lines of evidence presented in this study show that DCC binding to dox and autosomal sites is governed by similar principles: Levels of occupancy are correlated with levels of gene expression, and occupancy is enhanced by proximity to rex sites in cis.
DC acts at the level of transcription
We showed previously that DC in C. elegans equalizes X-chromosome gene expression between the sexes by a mechanism that reduces transcript levels from both hermaphrodite X chromosomes (Meyer and Casson 1986). Not known was whether the reduction in transcripts occurs through the regulation of transcription or is instead imposed by a post-transcriptional mechanism. We addressed this question by measuring the genome-wide binding of RNA Pol II in wild-type and sdc-2 mutant XX embryos to determine whether the elevated X transcript levels caused by disrupting DC correlate with an increase in RNA Pol II occupancy and hence a change in transcription.
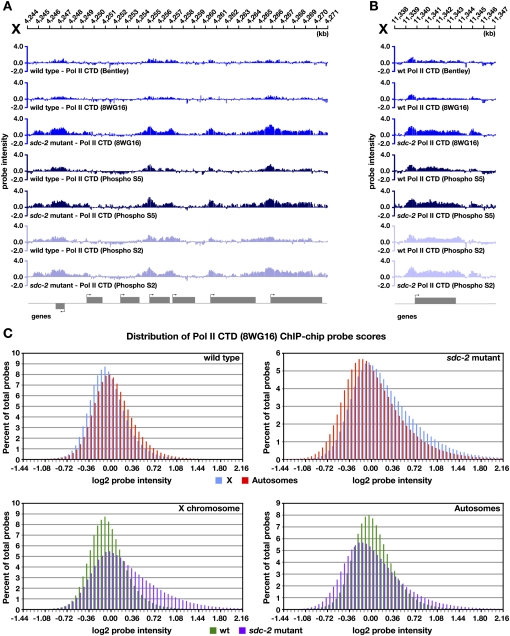
RNA Pol II occupancy was first measured through ChIP–chip experiments using an antibody (8WG16) raised against the unphosphorylated form of the C-terminal heptapeptide repeat domain (CTD) in the largest RNA Pol II subunit. This antibody primarily recognizes RNA Pol II bound at promoters in preinitiation complexes but also cross-reacts with phosphorylated forms of polymerase engaged in transcription initiation or elongation. In wild-type embryos, we found the level of RNA Pol II binding at a gene to be directly correlated with the expression level of the gene (Supplemental Fig. S15A,B). The expected, positive correlation validates our ChIP–chip experiments. On autosomal genes, we found the binding of RNA Pol II to be changed only modestly in sdc-2 mutant embryos compared with wild-type embryos, as demonstrated by ChIP–chip profiles (Supplemental Fig. S16A) and the distribution of probe scores (Fig. 7C). In contrast, RNA Pol II binding increased across the X chromosome in both gene promoters and coding regions of sdc-2 mutants (Fig. 7A–C), consistent with the elevation in X-chromosome transcript levels. These results provide the first indication that DC in C. elegans acts at the level of transcription.
Figure 7.
DC acts at the level of transcription. (A,B) ChIP–chip binding profiles on X using antibodies raised to different forms of RNA Pol II in wild-type and sdc-2 mutant embryos: RNA Pol II unphosphorylated CTD form (8WG16 and Bentley), initiating RNA Pol II (phospho S5), or elongating/terminating RNA Pol II (phospho S2). Binding of all three forms of RNA Pol II is increased at genes across X in sdc-2 mutant embryos, indicating that DC must function at least at the level of transcription. (C) Histograms showing distribution of RNA Pol II (8WG16) ChIP–chip probe scores across X and autosomes in wild-type XX embryos compared with sdc-2 mutants. Probe scores were analyzed as in Figure 2D. In wild-type embryos, the probe score distribution on X is slightly lower than on autosomes. Conversely, in sdc-2 mutant embryos, probe scores on X are generally higher than on autosomes, indicative of increased RNA Pol II binding across X.
Transcriptional regulation occurs at different steps. The DC process could impede RNA Pol II binding, transcription initiation, the transition from initiation to elongation, or the elongation rate. To assess the step of transcription controlled by the DC process, we performed ChIP–chip analysis of RNA Pol II in wild-type and sdc-2 mutant embryos using antibodies that recognize the CTD phosphorylated on either Ser 5 (phospho S5) or Ser 2 (phospho S2). Phospho S5 is a hallmark of initiating RNA Pol II, but it also persists on the CTD during the early steps of elongation. Phospho S2 marks elongating RNA Pol II committed to making full-length mRNAs (Buratowski 2009).
ChIP–chip binding profiles of all three forms of RNA Pol II were surprisingly similar to each other in wild-type embryos (Fig. 7A), although some expected enhancement was seen of phospho S5 in the promoter and of phospho S2 in the coding region (Fig. 7B). The probe scores at genes for all the experiments correlated directly with the expression levels of the genes (Supplemental Fig. S15A,B). In sdc-2 mutants, profiles and probe scores for phospho S5 and phospho S2 showed a significant increase on X-linked genes, but not autosomal genes, when compared with those in wild-type embryos (Fig. 7A,B; Supplemental Fig. S16A–C). We conclude that DC regulates transcription, and that more RNA Pol II is recruited to the promoter and coding regions of genes on X in DC mutants. The coordinate increase in phospho S5 and phospho S2 in sdc-2 mutants would be expected from a general increase in RNA Pol II recruitment, and prevents us from determining whether DC mutations only increase RNA Pol II recruitment or also permit more RNA Pol II to be released from the promoter and thereby increase either the proportion or rate of elongating polymerases.
Discussion
Our work has revealed mechanisms by which the C. elegans DCC assembles, binds to discrete sites throughout the genome, and regulates gene expression across the X chromosome. We showed that the DCC co-opted a subunit from MLL/COMPASS, a gene-activating complex that modifies chromatin, to target the DCC to X and thereby reduce X-chromosome-wide gene expression. We also defined distinct targeting principles that govern DCC binding to two classes of sites on X. This knowledge enabled us to predict additional DCC recruitment sites on X and to refine a model for genome-wide DCC binding. In this model, the location of dox and autosomal sites is dictated by intrinsic DCC-binding properties correlated with regions of high transcriptional activity. Sex-specific, sequence-dependent DCC recruitment to rex sites then increases the magnitude of DCC binding to sites in cis that lack robust DCC-binding ability by themselves. Three lines of evidence support this model. First, in DC mutants that exhibit complete loss of rex site binding, dox site binding is reduced, but only to the low level comparable with autosomal binding in wild-type animals. Second, attachment of rex sites in cis to an autosome increases autosomal DCC binding near the site of fusion. Third, changes in DCC binding at dox and autosomal sites during development are positively correlated with changes in expression of nearby genes. Thus, dox and autosomal sites have similar intrinsic DCC binding ability, and the higher preference for binding to dox sites is dictated primarily by their linkage to rex sites in cis. Our studies also showed that the DCC reduces gene expression on X by regulating transcription.
A role for DPY-30 in two distinct complexes with opposing effects on transcription
Having found that DPY-30 participates not only in the DCC to establish DCC binding on X, but also in MLL/COMPASS to activate genes, we dissected DPY-30’s role in regulating DCC binding to rex and dox sites. Others had speculated that DPY-30 might specifically control DCC spreading from rex sites to dox sites, but not binding to rex sites (Ercan and Lieb 2009). Our finding that dpy-30, like sdc-2 and sdc-3, is essential for DCC binding to both rex and dox sites argues against a selective role for dpy-30 in directing DCC binding only to dox sites.
Our binding data also suggest how DPY-30 might function in DCC recruitment and MLL/COMPASS binding. DPY-30 requires SDC-2, but not SDC-3, for binding to rex sites, while SDC-3 requires both DPY-30 and SDC-2 for rex site binding, and DPY-27 requires all three. In contrast, SDC-2 binds to rex sites without other DCC subunits. These data implicate SDC-2 as the initial DCC protein to recognize and bind X chromosomes. DPY-30 then likely assists SDC-3 to interact stably with SDC-2, and thereby recruits DPY-27 and other DCC components to X. Consistent with this model, overexpression of sdc-3 can partially rescue the DC defects of dpy-30 mutants (Davis and Meyer 1997), perhaps by enhancing the interaction between SDC-3 and SDC-2 in the absence of DPY-30.
If DPY-30 stabilizes protein–protein interactions within the DCC, it may serve a similar role for MLL/COMPASS. In both yeast and worms, DPY-30 is essential for the production of H3K4me3, even though it does not catalyze the trimethylation reaction itself (Schneider et al. 2005). Mass spectrometric analysis suggested that each yeast COMPASS complex contains at least three DPY-30 molecules (Schneider et al. 2005). Perhaps DPY-30 facilitates interactions among MLL/COMPASS subunits, and thereby promotes complex assembly and catalytic activity.
The binding sites for DCC and MLL/COMPASS are remarkably similar along X chromosomes and autosomes. However, differences exist. All DPY-30 sites overlap with DCC sites, but not all sites for ASH-2, an MLL/COMPASS subunit, coincide with DPY-30 or DCC sites. ASH-2 only binds to a subset of rex sites, primarily those located in the promoters of genes. Some ASH-2 sites lack DPY-30 binding and are strongly enriched in the coding regions of genes, unlike DPY-30 sites, which are in promoters. The DPY-30-independent ASH-2-binding sites suggest that ASH-2 may function in other complexes. Precedent exists in other organisms: ASH-2 homologs interact with the H3K27 demethylase Jmjd3 (De Santa et al. 2007).
Targeting the DCC to X: the connection between DCC binding at rex and dox sites
The existence of two classes of DCC-binding sites on X led to a model in which the DCC is recruited to X via rex sites and disperses to dox sites in cis (Jans et al. 2009). We tested this model directly. First, by assessing genome-wide binding of DCC subunits in mutants defective in the DC regulatory hierarchy, we found that rex sites absolutely require the recruitment proteins SDC-2, SDC-3, and DPY-30 for DCC binding. Most dox sites lack robust DCC binding without these recruitment proteins, but many retain an intrinsic, low-level binding comparable with that observed on wild-type autosomes. Thus dox sites have low-level DCC-binding capability, but full dox-site occupancy requires the DCC recruitment proteins and, presumably, their binding to rex sites. Since rex and dox sites can be separated by distances up to 90 kb (Jans et al. 2009), long-range communication between rex and dox sites appears to be important for full DCC binding to dox sites. Second, we tested whether rex sites can influence DCC binding over long distance by asking whether the low-level DCC binding at sites on autosomes could be enhanced by proximity in cis to rex sites using a strain carrying a fusion of X to chromosome V. We found that DCC binding on V in the X:V fusion chromosome was increased across a 2-Mb region near the point of fusion. In this region, 43% of binding sites were also present on wild-type chromosomes and showed enhanced DCC binding on the fusion chromosome. Of the new binding sites that arose, most showed binding just below the level of peak calling on wild-type autosomes. Thus, rex sites can strongly influence DCC binding over long range.
Increased autosomal DCC binding on X-to-autosome fusion chromosomes has also been reported by Ercan et al. (2009). However, in this study, the majority of DCC binding on autosomal portions of X-to-autosome fusion chromosomes was interpreted as new sites of binding, since they found very few DCC-binding sites on wild-type autosomes. Since we detected significant DCC binding on wild-type autosomes, we compared our DPY-27 ChIP–chip data sets from wild-type autosomes with the DPY-27 ChIP–chip data sets from X:II fusion chromosomes in Ercan et al. (2009) and from wild-type autosomes in Ercan et al. (2007) following the same normalization and peak-calling methods used in their study (Supplemental Fig. S12A–D). None of the 65 DPY-27 sites called by Ercan et al. (2009) in the first 2.5 Mb on II near the X:II fusion site were called as peaks in the wild-type chromosome II data set from Ercan et al. (2007). However, we found that 13 of these 65 DPY-27 peaks were called as peaks on our wild-type chromosome II. Of those not called as peaks, 71% showed appreciable binding by visual inspection in our wild-type chromosome II profiles. The significant overlap in DCC-binding locations on II between our wild-type chromosome II data set and the X:II fusion data set from Ercan et al. (2009) suggests that many of the binding sites called by Ercan et al. (2009) as new sites of DCC binding instead represent increased binding at sites already bound at a lower level on wild-type autosomes, yet were below the threshold of detection in their assay. Although the interpretations of both studies support the view that proximity in cis to rex sites strengthens dox site binding, they differ in important mechanistic interpretations. Our data support a model in which the location of dox and autosomal sites is dictated by intrinsic binding properties that are correlated with transcriptional activity (see below), and linkage to rex sites strongly influences the magnitude of binding. The alternative model by Ercan et al. (2009), which is less supported by our data, is that rex sites create adjacent DCC-binding sites de novo.
DCC binding at dox and autosomal sites is dynamic and positively correlated with changes in gene expression
Both dox and autosomal sites occur preferentially in the promoters of expressed genes in embryos, but the extent to which transcriptional activity of genes assists this intrinsic binding was unknown. Our comparison of DCC binding across developmental stages showed that a promoter could switch from little or no DCC binding when the gene was inactive to significant DCC binding when the gene became active. Also, binding of the DCC to promoters could be transient: When a gene was reduced in expression, DCC occupancy at its promoter was also reduced. dox and autosomal sites responded similarly to changes in transcription. These results further validate our autosomal DCC-binding sites, ones that were infrequently detected in two other studies (Ercan et al. 2007, 2009). The results also suggest that the low-level intrinsic binding at dox sites in DCC mutants and at autosomal sites in wild-type animals is dependent on the transcriptional activity of adjacent genes. Defining the salient features of active genes that promote binding is an important next step.
DCC binding to promoters on X and autosomes
ChIP analyses have identified 5′ ends of active genes as common sites for TF binding, raising the concern that features of promoters, such as open chromatin, cause spurious results. We think this premise is unlikely to account for the prevalence of DCC sites in promoters, because the level of DCC binding is not static: It changes in response to linkage of rex sites in cis and the level of transcription. Furthermore, >1000 5′ autosomal sites with strong DCC binding do not correspond to HOT (high-occupancy target) regions bound by numerous C. elegans TFs (Gerstein et al. 2010), and 33% of autosomal HOT regions lack DCC peaks.
We do not ascribe DC function to the low-level DCC binding at dox and autosomal sites that occurs without DCC-bound rex sites in cis. The lack of gene DC in sdc-2 mutants and the concomitant elimination of rex site binding suggests that the residual low-level DCC binding at dox sites has little, if any, impact on gene expression. We interpret the residual binding as an indication of specific, but lower-affinity binding. No known condition eliminates dox site binding while retaining rex site binding; thus, we cannot assess the specific contribution to DC of fully bound dox sites. However, mutant analyses establish that rex sites are pivotal.
Although no evidence supports a direct role for autosomal-bound DCC on gene expression in cis, DC disruption does reduce expression of 23% of autosomal genes, the opposite effect as on X. Whether this decrease is an indirect consequence of the DC mechanism itself or of changes in X gene expression (see Jans et al. 2009), or is instead a reflection of direct DCC action on wild-type autosomes, is not yet known.
Evolution of DCC binding on X
The presence of genome-wide DCC-binding sites that respond similarly to changes in gene expression and to linkage to rex sites, coupled with the fact that four of five DCC subunits function in mitotic condensin, has implications for the evolutionary origins of the DCC. Low-level, non-sex-specific binding of mitotic condensin at gene promoters might have preceded the evolution of sex chromosomes and DC. Indeed, the mitotic/meiotic-specific condensin subunit SMC-4, a paralog of the DC-specific protein DPY-27, has a profile of binding on all interphase chromosomes that closely resembles the pattern for DPY-27 binding in an sdc-2 mutant. Through the creation of a DCC-specific condensin subunit (DPY-27) and a protein like SDC-2 that recruits the DCC to X in a sex-specific manner, mitotic condensin subunits could have been co-opted by the DC process for sex-specific, sequence-dependent binding on X to regulate gene expression over long distances by altering chromosome structure. This theory could help explain why the DCC is bound to genes that escape DC and, similarly, why dosage-compensated genes often lack DCC binding nearby.
The XX-specific DCC recruitment protein SDC-2 binds to both rex and dox sites
Understanding the principles that underlie DCC binding to rex and dox sites requires knowledge of the binding sites for each DCC subunit and an assessment of whether DC complexes bound at rex and dox sites are heterogeneous in composition. Using ChIP–chip analysis, we showed previously that SDC-2 binds to both rex and dox sites at levels similar to SDC-3. As all SDC-2 antibodies generated do not immunoprecipitate SDC-2 very well, we used anti-Flag antibodies and extracts from a strain expressing Flag-tagged SDC-2 from an extrachromosomal array that rescues sdc-2 mutants. In contrast, the study by Ercan et al. (2009) reported SDC-2 ChIP–chip profiles using different SDC-2 antibodies and found that SDC-2, unlike SDC-3, bound primarily to rex sites with little accumulation at known dox sites or other gene promoters on X. They concluded that SDC-2 and SDC-3 differ in their role for promoting DCC binding at dox sites, and proposed that the disparity in their and our SDC-2 results is due to overexpression of the tagged-SDC-2 protein. Our biochemical and genetic data show this speculation to be highly unlikely. Western blots of SDC-2 in wild-type embryos and embryos expressing Flag-tagged SDC-2 showed the SDC-2 levels to be indistinguishable (Supplemental Fig. 6B). Furthermore, the tagged strain showed no male lethality, which is a hallmark of SDC-2 overexpression. Finally, our DPY-30 ChIP–chip profiles from an HA-tagged DPY-30 strain using HA antibodies and from wild-type embryos using antibodies to the endogenous protein were indistinguishable, showing that expression of tagged proteins from extrachromosomal arrays need not cause an erroneous profile. These results argue strongly against Flag-SDC-2 binding being artificially elevated due to SDC-2 overexpression, and instead suggest that the lower levels of SDC-2 binding seen by Ercan et al. (2009) across X, and particularly at dox sites, were due to inefficient ChIP by their SDC-2 antibodies.
DC regulates transcription
While it has long been known that C. elegans DC regulates the abundance of X-linked transcripts in XX animals, it was not known whether this regulation occurred at the level of transcription or was imposed post-transcriptionally. Here we report that binding of unphosphorylated and phosphorylated forms of RNA Pol II—representing the binding, initiating, and elongating forms of RNA Pol II—is dramatically increased throughout the promoter and coding regions of genes on X chromosomes in DC mutants. Therefore, we conclude that DC regulates gene expression transcriptionally, at least in part by regulating recruitment of RNA Pol II to genes across X. Given the coordinate increase in initiating and elongating forms of RNA Pol II, we could not determine whether subsequent steps such as the release from transcription initiation or the rate of transcription elongation are also specifically regulated.
Comparison of DCC binding in nematodes and flies
Flies and nematodes have evolved opposite strategies for regulating X-linked gene expression. While nematodes reduce gene expression of the hermaphrodite's two X chromosomes, flies increase gene expression of the male's single X (Gelbart and Kuroda 2009). Despite this fundamental difference, similarities have emerged in the principles that govern binding of the DC complexes in these two organisms. Like the recruitment of the worm DCC to X via 100–200 rex sites, the fly MSL (male-specific lethal) complex is recruited to X via ∼150–300 chromatin entry sites in a sequence-dependent manner using a motif enriched on X (Alekseyenko et al. 2008; Straub et al. 2008; Jans et al. 2009). A second sequence-independent mode of binding also exists for both organisms (Kind and Akhtar 2007; Larschan et al. 2007; Kind et al. 2008). The latter class of binding is strongly correlated with high levels of gene expression, suggesting that transcription-related features may facilitate this additional binding on X. However, this sequence-independent MSL binding on X is enriched at the 3′ end of fly genes (Alekseyenko et al. 2006; Gilfillan et al. 2006; Legube et al. 2006), while sequence-independent DCC binding on both X and autosomes is enriched at the 5′ end of worm genes (Ercan et al. 2007; Jans et al. 2009). Full binding to transcriptionally active sites in both organisms requires proximity in cis to the sequence-dependent recruitment/entry sites (Gorchakov et al. 2009; this study). Finally, both fly and worm DC complexes have co-opted subunits from other regulatory complexes. The worm DCC recruited condensin subunits and a subunit of the gene-activating MLL/COMPASS complex. The fly MSL complex recruits the H4K16 acetyltransferase protein MOF, which functions in a more global transcription activation complex present on X and autosomes in females and on autosomes in males (Akhtar and Becker 2000; Smith et al. 2000; Kind et al. 2008; Prestel et al. 2010). The global gene activation potential of MOF appears to be constrained by its association with the male-specific MSL complex to permit only the twofold increase in transcription on X that is necessary for DC. Thus, an emerging theme in the evolution of DC strategies is the adaptation of subunits from other regulatory complexes already involved in genome-wide control mechanisms for the selective purpose of X-chromosome gene regulation by the addition of one or more sex-specific proteins.
Materials and methods
Nematode strains
The nematode strains used were as follows: TY125, wild-type XX (N2). sdc-2 partial loss-of-function: TY0810, sdc-2(y93,RNAi) X XX. sdc-2-null: TY2222, her-1(hv1y101) V; xol-1(y9) sdc-2(y74) unc-9(e101) X XO. TY2205 her-1(e1520) sdc-3(y126) V; xol-1(y9) X XO. TY2470 unc-32(e189) dpy-27(y167) III; flu-2(e1003) xol-1(y9) X XO. TY1119 dpy-30(y130ts) V XX. TY2095 dpy-30(y228ts) V; yEx90[rol-6 + HA-dpy-30] XX. TY4573 sdc-2(y74) X; yEx992[Flag-sdc-2 + myo2::gfp] XX. YA0936 VL:XR fusion chromosome (ypT28) XX.
ChIP extract preparation
Embryo extracts were prepared as described (Jans et al. 2009), except that HA-tagged DPY-30 (TY2095) ChIP extracts were precleared with protein G sepharose. L1 larvae that had been hatched into medium without food and then fed for 3 h were cross-linked in 2% formaldehyde in M9 for 30 min at 20°C and quenched with 100 mM Tris-HCl (pH 7.5). Samples were then washed twice with M9 and once with homogenization buffer (50 mM HEPES-KOH at pH 7.6, 0.5% NP-40, 140 mM KCl, 1 mM EDTA, 5 mM DTT, 1 mM PMSF, PI inhibitor cocktail [EMD Biosciences]) and snap-frozen. Samples were ground in liquid nitrogen by mortar and pestle, resuspended in 1 vol (w/v) of homogenization buffer, and snap-frozen. Samples were sonicated to a range of 200 bp to 1 kb using a Heat System XL2020 Sonicator with a Misonix 419 tip twice for 15 sec at 10%, then eight times for 30 sec at 8%. Extracts were centrifuged at 20,000g for 15 min at 4°C, and the supernatant was precleared with protein A sepharose for 30 min.
ChIP reactions
Embryo ChIP experiments were performed as described (Jans et al. 2009) with the following changes. ChIPs using 5 μg of SDIX DPY-30 antibody were incubated for 4 h. HA-tagged DPY-30 ChIP used 5 mg of total nucleic acid and 15 μg of HA antibody. Five micrograms of antibody was used for all polymerase and DPY-26 ChIPs, while 10 μg of antibody was used for all other ChIPs. ChIPs from fed L1s were also performed as described in Jans et al. (2009) with the following changes. The equivalent of 2 mg of nucleic acid of extract was incubated with DPY-27 antibody for 4 h.
ChIP–chip platform and data analysis
ChIP–chip experiments were hybridized to 2.1 million feature high-density (HD2), genome-wide (WormBase release WS180), isothermal (tm = 76°C) tiling arrays from Nimblegen (Design ID #6737). Median probe spacing was 40 bp, and probe length varied from 50 to 75 bp, with repeat masking. ChIPs were hybridized at the Fred Hutchinson Cancer Research Center Genomics Facility, which supplied the ratio GFF and annotation files. Data were analyzed and peaks were called using NimbleScan software,and were viewed using SignalMap software as described (Jans et al. 2009). All ChIP–chip experiments were performed in duplicate except RNA Pol II CTD phospho S5 and RNA Pol II CTD phospho S2 in wild-type and sdc-2 embryos and HA-DPY-30 in wild type. Gene locations and transcriptional start sites (Start position) are per release WS180.
Accession number
ChIP–chip and gene expression data in this study can be accessed in the NCBI Gene Expression Omnibus public repository with the accession number GSE25834.
Acknowledgments
We thank A. Blardone for help with computational analysis; K. Brejc for Western blots; J. Gunther for figure design; C. Hassig for DPY-30 and ASH-2 antibodies and initial work on MLL/COMPASS; D. Bentley for RNA Pol II antibodies; C. Schartner for identification of rex-39; J. Wilhelmy for expression microarray processing; and T. Cline, T. Lo, A. Severson, B. Wheeler, and A. Wood for critical comments on the manuscript. R.R.P. and W.S.K. were supported by NIH training grant T32GM07127. R.R.P. was also supported by a Genentech Foundation Predoctoral Fellowship. Research was supported by NIH grant RO1-GM30702 to B.J.M., an Investigator of the Howard Hughes Medical Institute.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.2016011.
Supplemental material is available for this article.
References
- Akhtar A, Becker PB 2000. Activation of transcription through histone H4 acetylation by MOF, an acetyltransferase essential for dosage compensation in Drosophila. Mol Cell 5: 367–375 [DOI] [PubMed] [Google Scholar]
- Alekseyenko AA, Larschan E, Lai WR, Park PJ, Kuroda MI 2006. High-resolution ChIP-chip analysis reveals that the Drosophila MSL complex selectively identifies active genes on the male X chromosome. Genes Dev 20: 848–857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alekseyenko AA, Peng S, Larschan E, Gorchakov AA, Lee OK, Kharchenko P, McGrath SD, Wang CI, Mardis ER, Park PJ, et al. 2008. A sequence motif within chromatin entry sites directs MSL establishment on the Drosophila X chromosome. Cell 134: 599–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buratowski S 2009. Progression through the RNA polymerase II CTD cycle. Mol Cell 36: 541–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth B 1996. The evolution of chromosomal sex determination and dosage compensation. Curr Biol 6: 149–162 [DOI] [PubMed] [Google Scholar]
- Chuang PT, Albertson DG, Meyer BJ 1994. DPY-27: a chromosome condensation protein homolog that regulates C. elegans dosage compensation through association with the X chromosome. Cell 79: 459–474 [DOI] [PubMed] [Google Scholar]
- Chuang PT, Lieb JD, Meyer BJ 1996. Sex-specific assembly of a dosage compensation complex on the nematode X chromosome. Science 274: 1736–1739 [DOI] [PubMed] [Google Scholar]
- Csankovszki G, Collette K, Spahl K, Carey J, Snyder M, Petty E, Patel U, Tabuchi T, Liu H, McLeod I, et al. 2009. Three distinct condensin complexes control C. elegans chromosome dynamics. Curr Biol 19: 9–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis TL, Meyer BJ 1997. SDC-3 coordinates the assembly of a dosage compensation complex on the nematode X chromosome. Development 124: 1019–1031 [DOI] [PubMed] [Google Scholar]
- Dawes HE, Berlin DS, Lapidus DM, Nusbaum C, Davis TL, Meyer BJ 1999. Dosage compensation proteins targeted to X chromosomes by a determinant of hermaphrodite fate. Science 284: 1800–1804 [DOI] [PubMed] [Google Scholar]
- De Santa F, Totaro MG, Prosperini E, Notarbartolo S, Testa G, Natoli G 2007. The histone H3 lysine-27 demethylase Jmjd3 links inflammation to inhibition of Polycomb-mediated gene silencing. Cell 130: 1083–1094 [DOI] [PubMed] [Google Scholar]
- Ercan S, Lieb JD 2009. C. elegans dosage compensation: a window into mechanisms of domain-scale gene regulation. Chromosome Res 17: 215–227 [DOI] [PubMed] [Google Scholar]
- Ercan S, Giresi PG, Whittle CM, Zhang X, Green RD, Lieb JD 2007. X chromosome repression by localization of the C. elegans dosage compensation machinery to sites of transcription initiation. Nat Genet 39: 403–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ercan S, Dick LL, Lieb JD 2009. The C. elegans dosage compensation complex propagates dynamically and independently of X chromosome sequence. Curr Biol 19: 1777–1787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelbart ME, Kuroda MI 2009. Drosophila dosage compensation: a complex voyage to the X chromosome. Development 136: 1399–1410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerstein MB, Lu ZJ, Van Nostrand EL, Cheng C, Arshinoff BI, Liu T, Yip KY, Robilotto R, Rechtsteiner A, Ikegami K, et al. 2010. Integrative analysis of the Caenorhabditis elegans genome by the modENCODE Project. Science 330: 1775–1787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilfillan GD, Straub T, de Wit E, Greil F, Lamm R, van Steensel B, Becker PB 2006. Chromosome-wide gene-specific targeting of the Drosophila dosage compensation complex. Genes Dev 20: 858–870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorchakov AA, Alekseyenko AA, Kharchenko P, Park PJ, Kuroda MI 2009. Long-range spreading of dosage compensation in Drosophila captures transcribed autosomal genes inserted on X. Genes Dev 23: 2266–2271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greer EL, Maures TJ, Hauswirth AG, Green EM, Leeman DS, Maro GS, Han S, Banko MR, Gozani O, Brunet A 2010. Members of the H3K4 trimethylation complex regulate lifespan in a germline-dependent manner in C. elegans. Nature 466: 383–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu DR, Meyer BJ 1994. The dpy-30 gene encodes an essential component of the Caenorhabditis elegans dosage compensation machinery. Genetics 137: 999–1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu DR, Chuang PT, Meyer BJ 1995. DPY-30, a nuclear protein essential early in embryogenesis for Caenorhabditis elegans dosage compensation. Development 121: 3323–3334 [DOI] [PubMed] [Google Scholar]
- Jans J, Gladden JM, Ralston EJ, Pickle CS, Michel AH, Pferdehirt RR, Eisen MB, Meyer BJ 2009. A condensin-like dosage compensation complex acts at a distance to control expression throughout the genome. Genes Dev 23: 602–618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kind J, Akhtar A 2007. Cotranscriptional recruitment of the dosage compensation complex to X-linked target genes. Genes Dev 21: 2030–2040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kind J, Vaquerizas JM, Gebhardt P, Gentzel M, Luscombe NM, Bertone P, Akhtar A 2008. Genome-wide analysis reveals MOF as a key regulator of dosage compensation and gene expression in Drosophila. Cell 133: 813–828 [DOI] [PubMed] [Google Scholar]
- Larschan E, Alekseyenko AA, Gortchakov AA, Peng S, Li B, Yang P, Workman JL, Park PJ, Kuroda MI 2007. MSL complex is attracted to genes marked by H3K36 trimethylation using a sequence-independent mechanism. Mol Cell 28: 121–133 [DOI] [PubMed] [Google Scholar]
- Legube G, McWeeney SK, Lercher MJ, Akhtar A 2006. X-chromosome-wide profiling of MSL-1 distribution and dosage compensation in Drosophila. Genes Dev 20: 871–883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieb JD, Capowski EE, Meneely P, Meyer BJ 1996. DPY-26, a link between dosage compensation and meiotic chromosome segregation in the nematode. Science 274: 1732–1736 [DOI] [PubMed] [Google Scholar]
- Lieb JD, Albrecht MR, Chuang PT, Meyer BJ 1998. MIX-1: an essential component of the C. elegans mitotic machinery executes X chromosome dosage compensation. Cell 92: 265–277 [DOI] [PubMed] [Google Scholar]
- Liu T, Rechtsteiner A, Egelhofer TA, Vielle A, Latorre I, Cheung MS, Ercan S, Ikegami K, Jensen M, Kolasinska-Zwierz P, et al. 2010. Broad chromosomal domains of histone modification patterns in C. elegans. Genome Res. doi: 10.1101/gr.115519.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowden MR, Meier B, Lee TW, Hall J, Ahmed S 2008. End joining at Caenorhabditis elegans telomeres. Genetics 180: 741–754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonel P, Jans J, Peterson BK, Meyer BJ 2006. Clustered DNA motifs mark X chromosomes for repression by a dosage compensation complex. Nature 444: 614–618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mets DG, Meyer BJ 2009. Condensins regulate meiotic DNA break distribution, thus crossover frequency, by controlling chromosome structure. Cell 139: 73–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer BJ 2010. Targeting X chromosomes for repression. Curr Opin Genet Dev 20: 179–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer BJ, Casson LP 1986. Caenorhabditis elegans compensates for the difference in X chromosome dosage between the sexes by regulating transcript levels. Cell 47: 871–881 [DOI] [PubMed] [Google Scholar]
- Miller T, Krogan NJ, Dover J, Erdjument-Bromage H, Tempst P, Johnston M, Greenblatt JF, Shilatifard A 2001. COMPASS: a complex of proteins associated with a trithorax-related SET domain protein. Proc Natl Acad Sci 98: 12902–12907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy PL, Griesenbeck J, Kornberg RD, Cleary ML 2002. A trithorax-group complex purified from Saccharomyces cerevisiae is required for methylation of histone H3. Proc Natl Acad Sci 99: 90–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nusbaum C, Meyer BJ 1989. The Caenorhabditis elegans gene sdc-2 controls sex determination and dosage compensation in XX animals. Genetics 122: 579–593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payer B, Lee JT 2008. X chromosome dosage compensation: how mammals keep the balance. Annu Rev Genet 42: 733–772 [DOI] [PubMed] [Google Scholar]
- Prestel M, Feller C, Straub T, Mitlohner H, Becker PB 2010. The activation potential of MOF is constrained for dosage compensation. Mol Cell 38: 815–826 [DOI] [PubMed] [Google Scholar]
- Roguev A, Schaft D, Shevchenko A, Pijnappel WW, Wilm M, Aasland R, Stewart AF 2001. The Saccharomyces cerevisiae Set1 complex includes an Ash2 homologue and methylates histone 3 lysine 4. EMBO J 20: 7137–7148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider J, Wood A, Lee JS, Schuster R, Dueker J, Maguire C, Swanson SK, Florens L, Washburn MP, Shilatifard A 2005. Molecular regulation of histone H3 trimethylation by COMPASS and the regulation of gene expression. Mol Cell 19: 849–856 [DOI] [PubMed] [Google Scholar]
- Shilatifard A 2008. Molecular implementation and physiological roles for histone H3 lysine 4 (H3K4) methylation. Curr Opin Cell Biol 20: 341–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonet T, Dulermo R, Schott S, Palladino F 2007. Antagonistic functions of SET-2/SET1 and HPL/HP1 proteins in C. elegans development. Dev Biol 312: 367–383 [DOI] [PubMed] [Google Scholar]
- Smith ER, Pannuti A, Gu W, Steurnagel A, Cook RG, Allis CD, Lucchesi JC 2000. The Drosophila MSL complex acetylates histone H4 at lysine 16, a chromatin modification linked to dosage compensation. Mol Cell Biol 20: 312–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straub T, Grimaud C, Gilfillan GD, Mitterweger A, Becker PB 2008. The chromosomal high-affinity binding sites for the Drosophila dosage compensation complex. PLoS Genet 4: e1000302 doi: 10.1371/journal.pgen.1000302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood AJ, Severson AF, Meyer BJ 2010. Condensin and cohesin complexity: the expanding repertoire of functions. Nat Rev Genet 11: 391–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonker SA, Meyer BJ 2003. Recruitment of C. elegans dosage compensation proteins for gene-specific versus chromosome-wide repression. Development 130: 6519–6532 [DOI] [PubMed] [Google Scholar]