Abstract
Plant antibacterial peptides have been isolated from a wide variety of species. They consist of several protein groups with different features, such as the overall charge of the molecule, the content of disulphide bonds, and structural stability under environmental stress. Although the three-dimensional structures of several classes of plant peptides are well determined, the mechanism of action of some of these molecules is still not well defined. However, further studies may provide new evidences for their function on bacterial cell wall. Therefore, this paper focuses on plant peptides that show activity against plant-pathogenic and human-pathogenic bacteria. Furthermore, we describe the folding of several peptides and similarities among their three-dimensional structures. Some hypotheses for their mechanisms of action and attack on the bacterial membrane surface are also proposed.
1. Introduction
The first antibacterial peptide isolated from a plant species was a purothionin from wheat flour (Triticum aestivum), which has the ability to inhibit the growth of some phytopathogens such as Pseudomonas solanacearum, Xanthomonas campestris and Corynebacterium michiganense [1]. Almost 40 years later, several additional peptides with antibacterial activity have been characterized, represented not only by thionins, now named defensins, but also by other groups of proteins such as cyclotides, glycine-rich proteins, snakins, 2S albumins, and hevein-type proteins [2–5]. Peptides have been isolated from roots, seeds, flowers, stems, and leaves and have demonstrated activities towards phytopathogens, as well as against bacteria pathogenic to humans [2, 3, 6]. Over the years, antibacterial peptides have become an interesting tool for the development of new techniques in the control of crop losses and in the production of novel antibiotics for the treatment of diverse human infections [7, 8].
However, there is still little information about how these peptides affect the pathogen to cause cell death or growth inhibition. The fact that only a few peptide structures have been studied makes it more difficult to clarify the mechanism of action used to cause damage in bacterial cells [9–12]. Furthermore, it is not clear whether plant antibacterial peptides from different protein families present similar sequences, structures, and modes of action, or whether each group behaves in a different manner. Accordingly, this paper intends to explain some of these features of antibacterial peptides from plant sources. Herein, biochemical and structural properties of several peptides from different protein groups demonstrating antibacterial activity are described. Three-dimensional structures already obtained for plant antibacterial peptides are evaluated, and some possible mechanisms of action including cell membrane disruption, growth inhibition, and death are also proposed. Finally, the similarities among many antibacterial peptides are compared, and their most conserved attributes are evaluated.
2. What Are Plant Antibacterial Peptides?
Plant antibacterial peptides are of great importance as components of barrier defence and as a constitutive defense response induced upon infection in a wide variety of plants. Based on primary sequence similarity and activity towards bacteria, these peptides are classified into different groups, as described in Table 1.
Table 1.
Plant antibacterial peptides: physical and antibacterial characteristics.
| Peptide | Source | Family | Length | Molecular Mass (Da) | Activity | References |
|---|---|---|---|---|---|---|
| Cn-AMP1 | Cocos nucifera | — | 9 | 876 | Gram+/Gram− | [13] |
| Cn-AMP2 | Cocos nucifera | — | 11 | 1266.4 | Gram+/Gram− | [13] |
| Cn-AMP3 | Cocos nucifera | — | 8 | 905 | Gram+/Gram− | [13] |
| Cy-AMP1 | Cycas revoluta | — | 44 | 4591.4 | Gram+/Gram− | [14] |
| Cy-AMP2 | Cycas revoluta | — | 44 | 4577.4 | Gram+/Gram− | [14] |
| Dendrocin | Dendrocalamus latiflora | — | 35 | 3863.5 | Gram+/Gram− | [15] |
| Ginkbilobin | Ginkgo biloba | — | 40 | 4213.8 | Gram+/Gram− | [16] |
| Lunatusin | Phaseolus lunatus | — | 20 | 2178.5 | Gram+/Gram− | [17] |
| Circulin A | Chassalia parviflora | Cyclotide | 30 | 3175.8 | Gram+/Gram− | [12] |
| Circulin B | Chassalia parviflora | Cyclotide | 31 | 3308 | Gram+/Gram− | [7] |
| Cyclopsychotride A | Psychotria longipes | Cyclotide | 31 | 3255 | Gram+/Gram− | [18] |
| Kalata B2 | Oldenlandia affinis | Cyclotide | 29 | 2979.4 | Gram+ | [19] |
| Ah-AMP1 | Aesculus hippocastanum | Defensin | 50 | Gram+ | [20] | |
| Cp-DefensinII | Vigna unguiculata | Defensin | 46 | 5242.3 | Gram+/Gram− | [21] |
| Fabatin-1 | Vicia faba | Defensin | 47 | 5229.2 | Gram+/Gram− | [22] |
| Fabatin-2 | Vicia faba | Defensin | 47 | 5206.2 | Gram+/Gram− | [22] |
| Pp-AMP1 | Phyllostachys pubescens | Defensin | 44 | 4697.4 | Gram+/Gram− | [23] |
| Pp-AMP2 | Phyllostachys pubescens | Defensin | 45 | 4919.8 | Gram+/Gram− | [23] |
| Pp-Defensin | Pyrularia pubera | Defensin | 47 | 5288.2 | Gram+/Gram− | [24] |
| Pth-St1 | Solanum tuberosum | Defensin | 19 | 2207.4 | Gram+/Gram− | [25] |
| So-D1 | Spinacia oleracea | Defensin | 21 | 2296.6 | Gram+/Gram− | [26] |
| So-D2 | Spinacia oleracea | Defensin | 52 | 5803.8 | Gram+/Gram− | [26] |
| So-D3 | Spinacia oleracea | Defensin | 25 | 2778.3 | Gram+/Gram− | [26] |
| So-D4 | Spinacia oleracea | Defensin | 23 | 2623.2 | Gram+/Gram− | [26] |
| So-D5 | Spinacia oleracea | Defensin | 24 | 2737.3 | Gram+/Gram− | [26] |
| So-D6 | Spinacia oleracea | Defensin | 24 | 2552.9 | Gram+/Gram− | [26] |
| So-D7 | Spinacia oleracea | Defensin | 38 | 4230.7 | Gram+/Gram− | [26] |
| Tu-AMP 2 | Tulipa gesneriana | Defensin | 20 | 2259.6 | Gram+/Gram− | [27] |
| Tu-AMP-1 | Tulipa gesneriana | Defensin | 46 | 4992.9 | Gram+/Gram− | [27] |
| VaD1 | Vigna angularis | Defensin | 45 | 5009 | Gram+/Gram− | [28] |
| VrD1 | Vigna radiata | Defensin | 46 | 5140.88 | Gram− | [11] |
| VrD2 | Vigna radiata | Defensin | 47 | 5503.2 | Gram+/Gram− | [29] |
| White cloud bean defensin | Phaseolus vulgaris | Defensin | 47 | 5472.2 | Gram+/Gram− | [30] |
| Brazzein | Pentadiplandra brazzeana | Defensin | 54 | 6498.4 | Gram+/Gram− | [31] |
| Sesquin | Vigna sesquipedalis | Defensin-like | 10 | 1157.3 | Gram+/Gram− | [32] |
| Coconut antifungal Peptide | Cocos nucifera | Glutamic acid-rich | 10 | 1308.3 | Gram+/Gram− | [33] |
| Pg-AMP1 | Psidium guajava | Glycine-rich | 55 | 6029.4 | Gram− | [8] |
| Ac-AMP1 | Amaranthus caudatus | Hevein-like | 29 | 3033.6 | Gram+ | [34] |
| Ac-AMP2 | Amaranthus caudatus | Hevein-like | 30 | 3189.8 | Gram+ | [34] |
| Ee-CBP | Euonymus europaeus | Hevein-like | 45 | 5019.6 | Gram+ | [35] |
| Fa-AMP1 | Fagopyrum esculentum | Hevein-like | 40 | 3887.4 | Gram+/Gram− | [36] |
| Fa-AMP2 | Fagopyrum esculentum | Hevein-like | 3972.5 | Gram+/Gram− | [36] | |
| Pn-AMP1 | Pharbitis nil | Hevein-like | 41 | 4325.9 | Gram+/Gram− | [37] |
| Pn-AMP2 | Pharbitis nil | Hevein-like | 40 | 4238.8 | Gram+/Gram− | [37] |
| WjAMP1 | Eutrema wasabi | Hevein-like | 40 | 4094.5 | Gram+/Gram− | [38] |
| Ib-AMP1 | Impatiens balsamina | Impatiens | 20 | 2558 | Gram+/Gram− | [39] |
| Ib-AMP4 | Impatiens balsamina | Impatiens | 20 | 2549 | Gram+ | [39] |
| Mc-AMP1 | Mesembryanthemum crystallinum | Knottin | 38 | 4306.59 | Gram+ | [40] |
| Mj-AMP1 | Mirabilis jalapa | Knottin | 37 | 4000.5 | Gram+ | [41] |
| Mj-AMP2 | Mirabilis jalapa | Knottin | 36 | 3893.4 | Gram+ | [41] |
| Pa-AMP1 | Phytolacca americana | Knottin | 38 | 3935.5 | Gram+ | [42] |
| Pa-AMP2 | Phytolacca americana | Knottin | 37 | 3837.5 | Gram+ | [42] |
| MBP-1 | Zea mays | MBP-1 | 33 | 4130.7 | Gram+/Gram− | [43] |
| Shepherin I | Capsella bursa-pastoris | Shepherin | 28 | 2362.3 | Gram+/Gram− | [44] |
| Shepherin II | Capsella bursa-pastoris | Shepherin | 38 | 3259.2 | Gram+/Gram− | [44] |
| Snakin-1 | Solanum tuberosum | Snakins | 63 | 6934.2 | Gram+/Gram− | [45] |
| Snakin-2 | Solanum tuberosum | Snakins | 66 | 7037.2 | Gram+/Gram− | [46] |
| Vicilin-like Antimicrobial peptide 2a | Macadamia integrifolia | Vicilin-like | 49 | 6103.6 | Gram+ | [47] |
| Vicilin-like Antimicrobial peptide 2b | Macadamia integrifolia | Vicilin-like | 41 | 5110.6 | Gram+ | [47] |
| Vicilin-like Antimicrobial peptide 2c-1 | Macadamia integrifolia | Vicilin-like | 45 | 5905.5 | Gram+ | [47] |
| Vicilin-like Antimicrobial peptide 2c-2 | Macadamia integrifolia | Vicilin-like | 47 | 6189.9 | Gram+ | [47] |
| Vicilin-like Antimicrobial peptide 2c-3 | Macadamia integrifolia | Vicilin-like | 67 | 8864.7 | Gram+ | [47] |
| Vicilin-like Antimicrobial peptide 2d | Macadamia integrifolia | Vicilin-like | 35 | 4642.1 | Gram+ | [47] |
Members of the defensin family represent the highest number of antibacterial peptides described so far. They are able to inhibit a wide variety of bacterial species, especially the phytopathogenic ones (Table 1). Plant antibacterial peptides are active against bacteria at low concentrations and have been identified in peripheral cell layers of seed and vegetative tissues, in accordance with their function as a primary defense of vulnerable tissues. Most peptides share some general characteristics such as positively charged residues and high cysteine content for the formation of disulphide bonds. However, some antibacterial peptides, such as the peptides isolated from coconut water and the glycine-rich peptide from guava seeds, respectively, have acidic properties and no disulphide bridges. Most peptides have demonstrated activity against a broad range of different bacterial species and are therefore promising candidates for control of bacterial infections.
Many antibacterial peptide families have been isolated from plants. Pp-Thionin, for example, shows activity against Rhizobium meliloti, Xanthomonas campestris, Micrococcus luteus, and C. michiganensis at an IC50 < 50 μg/mL. Moreover, Pp-AMP1 and Pp-AMP2 have potent activity against several phytopathogens, including Erwinia carotovora, Agrobacterium radiobacter, Agrobacterium rhizogenes, Clavibacter michiganensis and Curtobacterium flaccumfaciens, at a concentration varying from 13 to 25 μg/mL. In addition, Circulins A-B and Cyclopsychotride A from the Cyclotides family show antibacterial effects against human pathogens such as Staphylococcus aureus, Micrococcus luteus, Escherichia coli, Pseudomonas aeruginosa, Proteus vulgaris and Klebsiella oxytoca at micromolar concentrations [7, 12, 18]. Furthermore, hevein-like proteins Ac-AMP1 and Ac-AMP1 cause growth inhibition of Bacillus megaterium and Sarcina lutea at concentrations of 40 and 250 μg/mL, respectively [34]. The same was observed earlier for peptides from the knottin family such as Mj-AMP1 and Mj-AMP2 [41]. Furthermore, two members of the Impatiens family, Ib-AMP1 and Ib-AMP4, were able to inhibit the growth of Bacillus subtilis, Micrococcus luteus, Staphylococcus aureus, and Streptococcus faecalis at very low concentrations [39]. Finally, Lc-LTPs peptides member (Lipid-transfer) inhibits the Gram-negative bacterium Agrobacterium tumefaciens [48].
3. What Are Some Characteristic Aspects of the Structures of Antibacterial Peptides?
Several studies of the structure of individual antibacterial peptides from plant sources have been performed, but only a few reports have made a comparison of their structural similarities and differences [49]. Studies comparing the primary sequences and tertiary structures of antimicrobial peptides from plants show that 33% of them present activity against bacteria, and around 59% are formed by 30 to 50 amino acid residues [50]. Moreover, it was observed that one key characteristic of antibacterial peptides is a high content of cysteine and/or glycine residues [50, 51]. The occurrence of disulphide bridges is also important for enhancing structural stability under diverse stress conditions [49–51]. Additionally, it was observed that the percentage of cysteine residues is higher in peptides with known β-sheet structures [51]. This can be compared to an antibacterial peptide belonging to the glycine-rich family and isolated from guava seeds [8]. The structure of this peptide, inferred by molecular modelling studies, consists only of α-helices and lacks β-sheets. Analysis of the primary sequence revealed no cysteine residues, and thus the peptide is unable to form disulphide bonds [8]. Therefore, evidence suggests that the presence of cysteine residues and β-sheet structures may go together, but this does not imply that these are relevant for antibacterial activity. Similar conclusions can be made concerning the presence of glycine residues. Glycine can provide flexibility to peptide structures, but nothing has been confirmed about its possible importance for antimicrobial activity [8].
However, there are implications that charged amino acids are relevant for activity against microorganisms. Around 17% of the amino acids in plant antimicrobial peptides are positively or negatively charged. Specifically, arginines and/or lysines comprise more than 70% of all charged residues found in these peptides, while the remaining 30% consists of the negatively charged aspartic acid and glutamic acid [51]. As will be described further in this paper, charged residues seem to have an essential role in activity towards pathogenic bacteria.
Among all antibacterial peptides isolated and characterised from plant sources, only eight have been evaluated in terms of their tertiary structures. Three of these peptides belong to the cyclotide family, and four others are from the defensin group [10–12, 19, 20, 29, 31, 52]. The last peptide is a hevein-like member [34].
Defensins have a typical three-dimensional structure composed of a α-helix followed by 2-3 β-strands that are stabilised by 3-4 disulphide bridges [19, 20, 29, 53]. This structure can be observed in all members of this group, even among those with different functionality.
Cyclotides are a unique type of peptide in which the N- and C-termini interact to form a cyclic structure [54]. They can be divided into the following two groups: the bracelets, the main feature of which is a three-dimensional structure composed of α-helices and strands, and the Mobius, composed mainly of β-sheets and turns [28, 54]. Circulin A is a bracelet member, while Circulin B and Kalata B2 are Mobius members. They share conserved cysteine and glycine residues, but their primary sequence identity is not high, especially for comparisons between bracelets and Mobius peptides [54]. Nevertheless, although they present a cyclic conformation, cyclotide tertiary structure is very similar to that of peptides from the defensin family. Earlier studies suggested that cyclotides are mutant variations of defensin genes, leading to structural changes in the peptides over the years [55]. In comparison, the hevein-like member Ac-AMP1 is also composed of a α-helix and two β-sheets containing 6 cysteine residues that form 3 disulphide bonds [34, 56, 57]. Although it shows characteristics of a chitin binding protein, Ac-AMP's tertiary structure is very similar to that of the defensins, giving it the classification of a hevein-like peptide [34, 56, 57].
An alignment of their primary sequences demonstrates that they present conserved cysteine residues, especially between members of the same family. They also show conservation of glycine residues and a high content of positively charged amino acids (Figures 1(a) and 1(b)).
Figure 1.
(a) Alignment of antibacterial peptides with defined three-dimensional structure: circulin A (1BH4); circulin B (2ERI); kalata B2 (1PT4); VrD1 (1BK8); VrD2 (2GL1); Ac-AMP2 (1MMC); brazzein (1BRZ). (b) Alignment of several antibacterial peptides from plant sources with sequences available at the Protein Data Bank. Hevein-like: Ac-AMP1 (AAB22103.1); knottin peptides: Mc-AMP1 (081338.1), Pa-AMP1 (P81418.1), Mj-AMP1 (P25403.4), Mj-AMP2 (P25404.2); snakins: snakin1 (AAD01518.1), snakin2 (ABL74292.1); defensins and thionins: pp-Thionin (P07504.1), Pth-St1 (AAB31351.1), and Cp-thionin (P84920.1); cyclotide: cyclopsychotride A (P56872.2); other peptides: MBP-1 (AAB23306.1). Asterisks show conserved cysteine residues. Sequences in bold represent peptides with cyclic three-dimensional conformations, indicating that they do not have N- and C-termini. Therefore, the alignment of cyclotides was based through comparison with the N- and C-termini of the other peptide groups.
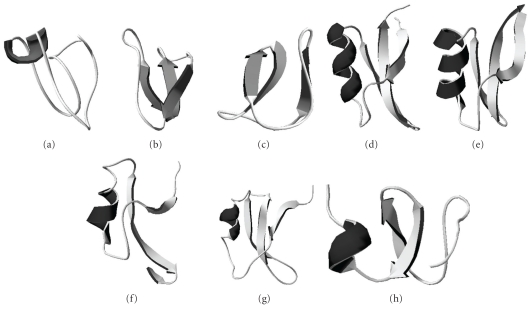
As these peptides belong mainly to 2 families with many features in common, it is possible to see that their three-dimensional conformations are also very similar (Figure 2). When the tertiary structures of all 8 antibacterial peptides for which an NMR structure has been determined are analysed, all are found to be comprised of 1 α-helix followed by 2-3 β-sheets, with the exception of Circulin B and Kalata B2. In Circulin A, β-sheets are replaced by loops (Figure 2(a)). Furthermore, defensins and hevein-like peptides show higher structural similarity to each other than to the cyclotides. Thus, it has not yet become possible to suggest which part of the structure is responsible for antibacterial activity or for conferring specificity against Gram-positive or Gram-negative bacteria.
Figure 2.
Three-dimensional structure of cyclotides (a) circulin A (PDB: 1BH4); (b) circulin B (PDB: 2ERI); (c) kalata B2 (PDB: 1PT4); defensins (d) VrD1 (PDB: 1TI5); (e) VrD2 (PDB: 2GL1); (f) Ah-AMP1 (PDB: 1BK8); (g) brazzein (1BRZ); and hevein-like (h) Ac-AMP2 (1MMC).
Among these 8 peptides, Ah-AMP1 is specific towards Gram-positive bacteria, while VrD1 only inhibits Gram-negative bacteria [19, 58]. There is a difference of one β-sheet in their structures, but this does not prove that the lack or presence of a β-sheet leads to some specific function. Nevertheless, the other 6 peptides display inhibitory activity against both Gram-positive and Gram-negative bacteria, confirming our inability to use only the tertiary conformation to predict peptide functionality. Further studies are necessary to enable the identification of a peptide's function by means of its three-dimensional structure.
4. How Do These Peptides Act against Bacteria?
Until now, there have been few reports about the mechanism of action of plant antibacterial peptides. However, these molecules have some important features responsible for antimicrobial activity, including their amphipathic structures and cationic charge at physiological pH [59–62].
The main hypothesis for their mechanism of action involves the ability of AMPs to cause membrane collapse by interacting with lipid molecules on the bacterial cell surface [59–62]. According to this hypothesis, the cationic peptides are attracted electrostatically to negatively charged molecules such as anionic phospholipids, lipopolysaccharides (LPS) (Gram-negative) and teichoic acid (Gram-positive), which are located asymmetrically in the membrane architecture. The positively charged residues can also interact with membrane lipids through specific receptors at the surface of the cell [60, 62]. Consequently, peptide binding to the membrane can activate several pathways that will cause cell death.
However, one general mechanism of action for antibacterial peptides is observed for most peptides. When they reach a threshold concentration, cationic peptides accumulate on the membrane surface in order to direct inner targets for cell lyses. Intrinsic and extrinsic parameters have been reported to influence the threshold peptide concentration. Intrinsic factors include the ability of the peptides to self-assemble and oligomerize, while extrinsic determinants include phospholipid membrane composition, membrane fluidity and head group size; these factors all influence membrane potential, which is critical for determining threshold peptide concentration [62].
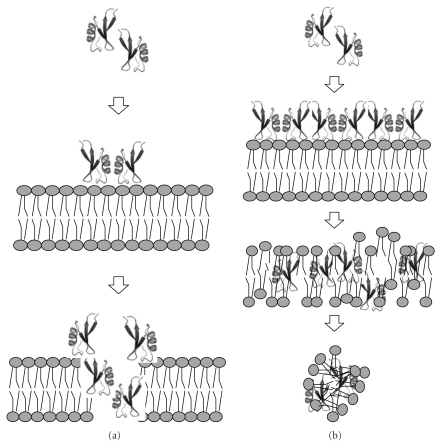
The following 3 processes of pore formation have been reported for plant antibacterial peptides: the barrel-stave mechanism, the toroid pore or wormhole mechanism and the carpet mechanism [59–62]. The barrel-stave mechanism consists of peptide aggregates forming a barrel-ring around an aqueous pore (Figure 3(a)). Peptides interact with the membrane, forcing one thin and hydrophobic portion to bind the phospholipid acyl-chains. After reaching threshold concentration, peptides from the barrel-ring open a pore in the membrane. Their hydrophilic portions comprise the core of the barrel, while the hydrophobic portion interacts with bacterial membrane phospholipids [61, 62].
Figure 3.
Scheme of the action mechanism for antibacterial peptides. (a) Barrel-stave model; (i) peptides in monomer or oligomer form come close to the membrane target; (ii) positively-charged residues from the peptides interact with the head group of the phospholipids from the membrane; (iii) at a threshold concentration of peptides, the pores are formed. In toroidal model, the major difference is the type of pore formed, where lipids and peptides are overlapped. (b) Carpet model; (i) peptides in monomer or oligomers come close to the membrane target; (ii) hydrophilic regions of peptides are exposed to solvent and hydrophobic regions to membrane; (iii) at threshold concentration of peptides, the permeability of the membrane increases, facilitating pore formation; (iv) membrane disintegration. Adapted from Shai, 2002.
The toroidal pore or wormhole hypothesis also postulates the formation of pores in a barrel-stave shape. However, in this case, these pores are composed of overlapping peptides and membrane lipids, generating one supramolecular complex. In this structure, the transmembrane pore is formed by peptide and phospholipid head groups. Therefore, the displacement of polar head groups from the peptides induces a positive curvature strain in the membrane by breaching the hydrophobic region [62].
The last mode of action suggested is the carpet mechanism [61, 62]. Initially, peptides in monomeric or oligomeric form bind to the cell surface in an electrostatic manner, covering all the membrane (Figure 3(b)) and giving an appearance of a peptide carpet on the bacterial membrane surface [61]. Consequently, the carpet causes a phospholipid displacement that alters membrane fluidity and/or reduces the barrier properties of membrane. It also leads to membrane disruption and, further, to cell death [62]. Due to the unfavourable energy observed after the membrane bilayer becomes curved, cell rupture and lysis will occur [61]. In this process, the membrane damage occurs in a dispersion-like manner without channel formation [62].
In general, peptides act by formation of membrane pores, resulting in leakage of ions and metabolites, depolarisation and interruption of the respiration process, biopolymer synthesis and cell death [62]. Plant antimicrobial peptides possibly act in the same way as other well-studied antimicrobial peptides [8]. Their folding, an overall positive charge and their amphipathic or amphiphilic nature are essential for antibacterial activity [49]. Furthermore, some studies have shown that antimicrobial peptides from plants can act on intracellular targets [63].
5. Concluding Remarks
Antibacterial peptides have been described in many different plant species. They belong to a wide range of protein families, varying from typical antimicrobial members to newly discovered ones. Some peptides show specificity towards Gram-positive or Gram-negative bacteria, but most of them are able to inhibit the activity of both. Therefore, there is at present no way to predict the specificity of any given antimicrobial peptide.
There are few reports describing the tertiary structures of such peptides. However, in silico analyses have shown that plant antibacterial peptides present similarities in their three-dimensional structures, although their primary amino acid sequences vary according to the protein family to which they belong. Knowledge of the tertiary structure could yield new insights into the mechanism of action against pathogenic bacteria. Moreover, the description of the mechanism of action for these antibacterial peptides suggests that it may involve a strong interaction with phospholipids from the pathogen's membrane. Parameters such as molecular volume, aggregation ability, and autoassembly onto the membrane surface are essential for activity against bacteria. Indeed, although the mode of action of antibacterial peptides is well-characterised, investigations of the relative importance of specific amino acid residues and their binding with the bacterial cell wall are still in progress.
References
- 1.de Caleya RF, Gonzalez-Pascual B, García-Olmedo F, Carbonero P. Susceptibility of phytopathogenic bacteria to wheat purothionins in vitro. Applied microbiology. 1972;23(5):998–1000. doi: 10.1128/am.23.5.998-1000.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Selitrennikoff CP. Antifungal Proteins. Applied and Environmental Microbiology. 2001;67(7):2883–2894. doi: 10.1128/AEM.67.7.2883-2894.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pelegrini PB, Franco OL. Plant γ-thionins: novel insights on the mechanism of action of a multi-functional class of defense proteins. International Journal of Biochemistry and Cell Biology. 2005;37(11):2239–2253. doi: 10.1016/j.biocel.2005.06.011. [DOI] [PubMed] [Google Scholar]
- 4.Witkowska D, Bartyś A, Gamian A. Defensins and cathelicidins as natural peptide antibiotics. Postępy Higieny i Medycyny Doświadczalnej. 2008;62:694–707. [PubMed] [Google Scholar]
- 5.Daly NL, Rosengren KJ, Craik DJ. Discovery, structure and biological activities of cyclotides. Advanced Drug Delivery Reviews. 2009;61(11):918–930. doi: 10.1016/j.addr.2009.05.003. [DOI] [PubMed] [Google Scholar]
- 6.Terras FR, Eggermont K, Kovaleva V, et al. Small cysteine-rich antifungal proteins from radish: their role in host defense. Plant Cell. 1995;7(5):573–588. doi: 10.1105/tpc.7.5.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tam JP, Lu Y-A, Yang J-L, Chiu K-W. An unusual structural motif of antimicrobial peptides containing end-to-end macrocycle and cystine-knot disulfides. Proceedings of the National Academy of Sciences of the United States of America. 1999;96(16):8913–8918. doi: 10.1073/pnas.96.16.8913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pelegrini PB, Murad AM, Silva LP, et al. Identification of a novel storage glycine-rich peptide from guava (Psidium guajava) seeds with activity against Gram-negative bacteria. Peptides. 2008;29(8):1271–1279. doi: 10.1016/j.peptides.2008.03.013. [DOI] [PubMed] [Google Scholar]
- 9.Saether O, Craik DJ, Campbell ID, Sletten K, Juul J, Norman DG. Elucidation of the primary and three-dimensional structure of the uterotonic polypeptide kalata B1. Biochemistry. 1995;34(13):4147–4158. doi: 10.1021/bi00013a002. [DOI] [PubMed] [Google Scholar]
- 10.Koltay A, Daly NL, Gustafson KR, Craik DJ. Structure of circulin B and implications for antimicrobial activity of the cyclotides. International Journal of Peptide Research and Therapeutics. 2005;11(1):99–106. [Google Scholar]
- 11.Liu YJ, Cheng CS, Lai SM, Hsu MP, Chen CS, Lyu PC. Solution structure of the plant defensin VrD1 from mung bean and its possible role in insecticidal activity against bruchids. Proteins. 2006;63(4):777–786. doi: 10.1002/prot.20962. [DOI] [PubMed] [Google Scholar]
- 12.Daly NL, Koltay A, Gustafson KR, Boyd MR, Casas-Finet JR, Craik DJ. Solution structure by NMR of circulin A: a macrocyclic knotted peptide having anti-HIV activity. Journal of Molecular Biology. 1999;285(1):333–345. doi: 10.1006/jmbi.1998.2276. [DOI] [PubMed] [Google Scholar]
- 13.Mandal SM, Dey S, Mandal M, Sarkar S, Maria-Neto S, Franco OL. Identification and structural insights of three novel antimicrobial peptides isolated from green coconut water. Peptides. 2009;30(4):633–637. doi: 10.1016/j.peptides.2008.12.001. [DOI] [PubMed] [Google Scholar]
- 14.Yokoyama S, Kato K, Koba A, Minami Y, Watanabe K, Yagi F. Purification, characterization, and sequencing of antimicrobial peptides, Cy-AMP1, Cy-AMP2, and Cy-AMP3, from the Cycad (Cycas revoluta) seeds. Peptides. 2008;29(12):2110–2117. doi: 10.1016/j.peptides.2008.08.007. [DOI] [PubMed] [Google Scholar]
- 15.Wang HX, Ng TB. Dendrocin, a distinctive antifungal protein from bamboo shoots. Biochemical and Biophysical Research Communications. 2003;307(3):750–755. doi: 10.1016/s0006-291x(03)01229-4. [DOI] [PubMed] [Google Scholar]
- 16.Wang H, Ng TB. Ginkbilobin, anovel antifungal protein from Ginkgo biloba seeds with sequence similarity to embryo-abundant protein. Biochemistry and Biophysics Research Communications. 2000;279:407–411. doi: 10.1006/bbrc.2000.3929. [DOI] [PubMed] [Google Scholar]
- 17.Wong JH, Ng TB. Lunatusin, a trypsin-stable antimicrobial peptide from lima beans (Phaseolus lunatus L.) Peptides. 2005;26(11):2086–2092. doi: 10.1016/j.peptides.2005.03.004. [DOI] [PubMed] [Google Scholar]
- 18.Witherup KM, Bogusky MJ, Anderson PS, et al. Cyclopsychotride A, a biologically active, 32-residue cyclic peptide isolated from Psychotria longipes. Journal of Natural Products. 1994;57(12):1619–1625. doi: 10.1021/np50114a002. [DOI] [PubMed] [Google Scholar]
- 19.Jennings CV, Rosengren KJ, Daly NL, et al. Isolation, solution structure, and insecticidal activity of kalata B2, a circular protein with a twist: do Möbius strips exist in nature? Biochemistry. 2005;44(3):851–860. doi: 10.1021/bi047837h. [DOI] [PubMed] [Google Scholar]
- 20.Fant F, Vranken WF, Borremans FAM. The three-dimensional solution structure of Aesculus hippocastanum antimicrobial protein 1 determined by 1H nuclear magnetic resonance. Proteins. 1999;37(3):388–403. doi: 10.1002/(sici)1097-0134(19991115)37:3<388::aid-prot7>3.3.co;2-6. [DOI] [PubMed] [Google Scholar]
- 21.Franco OL, Murad AM, Leite JR, Mendes PAM, Prates MV, Bloch C., Jr. Identification of a cowpea γ-thionin with bactericidal activity. FEBS Journal. 2006;273(15):3489–3497. doi: 10.1111/j.1742-4658.2006.05349.x. [DOI] [PubMed] [Google Scholar]
- 22.Zhang Y, Lewis K. Fabatins: new antimicrobial plant peptides. FEMS Microbiology Letters. 1997;149(1):59–64. doi: 10.1111/j.1574-6968.1997.tb10308.x. [DOI] [PubMed] [Google Scholar]
- 23.Fujimura M, Ideguchi M, Minami Y, Watanabe K, Tadera K. Amino acid sequence and antimicrobial activity of chitin-binding peptides, Pp-AMP 1 and Pp-AMP 2, from Japanese bamboo shoots (Phyllostachys pubescens) Bioscience, Biotechnology and Biochemistry. 2005;69(3):642–645. doi: 10.1271/bbb.69.642. [DOI] [PubMed] [Google Scholar]
- 24.Vernon LP, Evett GE, Zeikus RD, Gray WR. A toxic thionin from Pyrularia pubera: purification, properties, and amino acid sequence. Archives of Biochemistry and Biophysics. 1985;238(1):18–29. doi: 10.1016/0003-9861(85)90136-5. [DOI] [PubMed] [Google Scholar]
- 25.Moreno M, Segura A, Garcia-Olmedo F. Pseudothionin-St1, a potato peptide active against potato pathogens. European Journal of Biochemistry. 1994;223(1):135–139. doi: 10.1111/j.1432-1033.1994.tb18974.x. [DOI] [PubMed] [Google Scholar]
- 26.Segura A, Moreno M, Molina A, García-Olmedo F. Novel defensin subfamily from spinach (Spinacia oleracea) FEBS Letters. 1998;435(2-3):159–162. doi: 10.1016/s0014-5793(98)01060-6. [DOI] [PubMed] [Google Scholar]
- 27.Fujimura M, Ideguchi M, Minami Y, Watanabe K, Tadera K. Purification, characterization, and sequencing of novel antimicrobial peptides, Tu-AMP 1 and Tu-AMP 2, from bulbs of tulip (Tulipa gesneriana L.) Bioscience, Biotechnology and Biochemistry. 2004;68(3):571–577. doi: 10.1271/bbb.68.571. [DOI] [PubMed] [Google Scholar]
- 28.Chen B, Colgrave ML, Daly NL, Rosengren KJ, Gustafson KR, Craik DJ. Isolation and characterization of novel cyclotides from Viola hederaceae: solution structure and anti-HIV activity of vhl-1, a leaf-specific expressed cyclotide. Journal of Biological Chemistry. 2005;280(23):22395–22405. doi: 10.1074/jbc.M501737200. [DOI] [PubMed] [Google Scholar]
- 29.Lin KF, Lee TR, Tsai PH, Hsu MP, Chen CS, Lyu PC. Structure-based protein engineering for α-amylase inhibitory activity of plant defensin. Proteins. 2007;68(2):530–540. doi: 10.1002/prot.21378. [DOI] [PubMed] [Google Scholar]
- 30.Wong JH, Zhang XQ, Wang HX, Ng TB. A mitogenic defensin from white cloud beans (Phaseolus vulgaris) Peptides. 2006;27(9):2075–2081. doi: 10.1016/j.peptides.2006.03.020. [DOI] [PubMed] [Google Scholar]
- 31.Cornilescu CC, Tonelli M, DeRider ML, Markley JL, Assadi-Porter FM. Refined solution structure of des-pyro Glu brazzein. to appear. [Google Scholar]
- 32.Wong JH, Ng TB. Sesquin, a potent defensin-like antimicrobial peptide from ground beans with inhibitory activities toward tumor cells and HIV-1 reverse transcriptase. Peptides. 2005;26(7):1120–1126. doi: 10.1016/j.peptides.2005.01.003. [DOI] [PubMed] [Google Scholar]
- 33.Wang HX, Ng TB. An antifungal peptide from the coconut. Peptides. 2005;26(12):2392–2396. doi: 10.1016/j.peptides.2005.05.009. [DOI] [PubMed] [Google Scholar]
- 34.Martins JC, Maes D, Loris R, et al. H NMR study of the solution structure of Ac-AMP2, a sugar binding antimicrobial protein isolated from Amaranthus caudatus. Journal of Molecular Biology. 1996;258(2):322–333. doi: 10.1006/jmbi.1996.0253. [DOI] [PubMed] [Google Scholar]
- 35.Van Den Bergh KPB, Proost P, Van Damme J, Coosemans J, Van Damme EJM, Peumans WJ. Five disulfide bridges stabilize a hevein-type antimicrobial peptide from the bark of spindle tree (Euonymus europaeus L.) FEBS Letters. 2002;530(1–3):181–185. doi: 10.1016/s0014-5793(02)03474-9. [DOI] [PubMed] [Google Scholar]
- 36.Fujimura M, Minami Y, Watanabe K, Tadera K. Purification, characterization, and sequencing of a novel type of antimicrobial peptides, Fa-AMP1 and Fa-AMP2, from seeds of buckwheat (Fagopyrum esculentum Moench.) Bioscience, Biotechnology and Biochemistry. 2003;67(8):1636–1642. doi: 10.1271/bbb.67.1636. [DOI] [PubMed] [Google Scholar]
- 37.Koo JC, Lee SY, Chun HJ, et al. Two hevein homologs isolated from the seed of Pharbitis nil L. exhibit potent antifungal activity. Biochimica et Biophysica Acta. 1998;1382(1):80–90. doi: 10.1016/s0167-4838(97)00148-9. [DOI] [PubMed] [Google Scholar]
- 38.Kiba A, Saitoh H, Nishihara M, Omiya K, Yamamura S. C-terminal domain of a hevein-like protein from Wasabia japonica has potent antimicrobial activity. Plant and Cell Physiology. 2003;44(3):296–303. doi: 10.1093/pcp/pcg035. [DOI] [PubMed] [Google Scholar]
- 39.Tailor RH, Acland DP, Attenborough S, et al. A novel family of small cysteine-rich antimicrobial peptides from seed of Impatiens balsamina is derived from a single precursor protein. Journal of Biological Chemistry. 1997;272(39):24480–24487. doi: 10.1074/jbc.272.39.24480. [DOI] [PubMed] [Google Scholar]
- 40.Michalowski CB, Bohnert HJ. Antimicrobial peptide 1 from the common ice plant. The EMBL/GenBank/DDBJ Databases. In press. [Google Scholar]
- 41.Cammue BPA, De Bolle MFC, Terras FRG, et al. Isolation and characterization of a novel class of plant antimicrobial peptides from Mirabilis jalapa L. seeds. Journal of Biological Chemistry. 1992;267(4):2228–2233. [PubMed] [Google Scholar]
- 42.Liu Y, Luo J, Xu C, et al. Purification, characterization, and molecular cloning of the gene of a seed-specific antimicrobial protein from pokeweed. Plant Physiology. 2000;122(4):1015–1024. doi: 10.1104/pp.122.4.1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Duvick JP, Rood T, Rao AG, Marshak DR. Purification and characterization of a novel antimicrobial peptide from maize (Zea mays L.) kernels. Journal of Biological Chemistry. 1992;267(26):18814–18820. [PubMed] [Google Scholar]
- 44.Park CJ, Park CB, Hong SS, Lee HS, Lee SY, Kim SC. Characterization and cDNA cloning of two glycine- and histidine-rich antimicrobial peptides from the roots of shepherd’s purse, Capsella bursa-pastoris. Plant Molecular Biology. 2000;44(2):187–197. doi: 10.1023/a:1006431320677. [DOI] [PubMed] [Google Scholar]
- 45.Segura A, Moreno M, Madueño F, Molina A, García-Olmedo F. Snakin-1, a peptide from potato that is active against plant pathogens. Molecular Plant-Microbe Interactions. 1999;12(1):16–23. doi: 10.1094/MPMI.1999.12.1.16. [DOI] [PubMed] [Google Scholar]
- 46.Berrocal-Lobo M, Segura A, Moreno M, López G, García-Olmedo F, Molina A. Snakin-2, an antimicrobial peptide from potato whose gene is locally induced by wounding and responds to pathogen infection. Plant Physiology. 2002;128(3):951–961. doi: 10.1104/pp.010685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Marcus JP, Green JL, Goulter KC, Manners JM. A family of antimicrobial peptides is produced by processing of a 7S globulin protein in Macadamia integrifolia kernels. Plant Journal. 1999;19(6):699–710. doi: 10.1046/j.1365-313x.1999.00569.x. [DOI] [PubMed] [Google Scholar]
- 48.Yang X, Li J, Li X, She R, Pei Y. Isolation and characterization of a novel thermostable non-specific lipid transfer protein-like antimicrobial protein from motherwort (Leonurus japonicus Houtt) seeds. Peptides. 2006;27(12):3122–3128. doi: 10.1016/j.peptides.2006.07.019. [DOI] [PubMed] [Google Scholar]
- 49.Jenssen H, Hamill P, Hancock REW. Peptide antimicrobial agents. Clinical Microbiology Reviews. 2006;19(3):491–511. doi: 10.1128/CMR.00056-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hammami R, Ben Hamida J, Vergoten G, Fliss I. PhytAMP: a database dedicated to antimicrobial plant peptides. Nucleic Acids Research. 2009;37(1):D963–D968. doi: 10.1093/nar/gkn655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang Z, Wang G. APD: the antimicrobial peptide database. Nucleic Acids Research. 2004;32:D590–D592. doi: 10.1093/nar/gkh025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Craik DJ, Daly NL, Bond T, Waine C. Plant cyclotides: a unique family of cyclic and knotted proteins that defines the cyclic cystine knot structural motif. Journal of Molecular Biology. 1999;294(5):1327–1336. doi: 10.1006/jmbi.1999.3383. [DOI] [PubMed] [Google Scholar]
- 53.Carvalho ADO, Gomes VM. Plant defensins—prospects for the biological functions and biotechnological properties. Peptides. 2009;30(5):1007–1020. doi: 10.1016/j.peptides.2009.01.018. [DOI] [PubMed] [Google Scholar]
- 54.Pelegrini PB, Quirino BF, Franco OL. Plant cyclotides: an unusual class of defense compounds. Peptides. 2007;28(7):1475–1481. doi: 10.1016/j.peptides.2007.04.025. [DOI] [PubMed] [Google Scholar]
- 55.Conlan BF, Gillon AD, Craik DJ, Anderson MA. Circular proteins and mechanisms of cyclization. doi: 10.1002/bip.21422. Biopolymers. In press. [DOI] [PubMed] [Google Scholar]
- 56.Padovan L, Scocchi M, Tossi A. Structural aspects of plant antimicrobial peptides. Current Protein and Peptide Science. 2010;11(3):210–219. doi: 10.2174/138920310791112093. [DOI] [PubMed] [Google Scholar]
- 57.Broekaert WF, Cammue BPA, De Bolle MFC, Thevissen K, De Samblanx GW, Osborn RW. Antimicrobial Peptides from Plants. Critical Reviews in Plant Sciences. 1997;16(3):297–323. [Google Scholar]
- 58.Osborn RW, De Samblanx GW, Thevissen K, et al. Isolation and characterisation of plant defensins from seeds of Asteraceae, Fabaceae, Hippocastanaceae and Saxifragaceae. FEBS Letters. 1995;368(2):257–262. doi: 10.1016/0014-5793(95)00666-w. [DOI] [PubMed] [Google Scholar]
- 59.Epand RM, Vogel HJ. Diversity of antimicrobial peptides and their mechanisms of action. Biochimica et Biophysica Acta. 1999;1462(1-2):11–28. doi: 10.1016/s0005-2736(99)00198-4. [DOI] [PubMed] [Google Scholar]
- 60.Sitaram N, Nagaraj R. Interaction of antimicrobial peptides with biological and model membranes: structural and charge requirements for activity. Biochimica et Biophysica Acta. 1999;1462(1-2):29–54. doi: 10.1016/s0005-2736(99)00199-6. [DOI] [PubMed] [Google Scholar]
- 61.Shai Y. Mode of action of membrane active antimicrobial peptides. Biopolymers. 2002;66(4):236–248. doi: 10.1002/bip.10260. [DOI] [PubMed] [Google Scholar]
- 62.Yeamn MR, Yount NY. Mechanisms of atnimicrobial peptide action and resistance. Pharmacoloy Reviews. 2003;55:27–55. doi: 10.1124/pr.55.1.2. [DOI] [PubMed] [Google Scholar]
- 63.Aerts AM, François IEJA, Cammue BPA, Thevissen K. The mode of antifungal action of plant, insect and human defensins. Cellular and Molecular Life Sciences. 2008;65(13):2069–2079. doi: 10.1007/s00018-008-8035-0. [DOI] [PMC free article] [PubMed] [Google Scholar]