Abstract
During infection with the hepatitis A virus (HAV), most patients develop mild or asymptomatic disease. However, a small number of patients develop serious, life-threatening hepatitis. We investigated this variability in disease severity by examining 30 Argentinean patients with HAV-induced acute liver failure in a case-control, cross-sectional, observational study. We found that HAV-induced severe liver disease was associated with a 6-amino-acid insertion in TIM1/HAVCR1 (157insMTTTVP), the gene encoding the HAV receptor. This polymorphism was previously shown to be associated with protection against asthma and allergic diseases and with HIV progression. In binding assays, the TIM-1 protein containing the 157insMTTTVP insertion polymorphism bound HAV more efficiently. When expressed by human natural killer T (NKT) cells, this long form resulted in greater NKT cell cytolytic activity against HAV-infected liver cells, compared with the shorter TIM-1 protein without the polymorphism. To our knowledge, the 157insMTTTVP polymorphism in TIM1 is the first genetic susceptibility factor shown to predispose to HAV-induced acute liver failure. Furthermore, these results suggest that HAV infection has driven the natural selection of shorter forms of the TIM-1 protein, which binds HAV less efficiently, thereby protecting against severe HAV-induced disease, but which may predispose toward inflammation associated with asthma and allergy.
Introduction
Hepatitis A virus (HAV) infection, one of the most common viral infections in humans, affects approximately 10 million people annually worldwide. HAV, a single-stranded RNA picornavirus, is transmitted by the fecal-oral route in contaminated food or drinking water and is endemic in many countries, particularly those with poor sanitation. Prior to 1960, HAV infected nearly every person in the world, and the seroprevalence of antibodies against HAV approached 100% in all parts of the globe (1). However, improved hygiene and vaccination have reduced the infection rate in most Western countries to approximately 5%–10%. Nevertheless, in developing countries, HAV infection remains widespread and is generally contracted in early childhood. Most children infected with HAV develop mild or asymptomatic disease. A small number of children develop self-limited liver inflammation characterized by jaundice, liver enzyme elevation, fatigue, abdominal pain, and diarrhea. Very rarely, HAV infection can evolve into acute liver failure. In most developing countries, HAV infection remains the major cause of acute liver failure and liver transplantation in pediatric patients (2–4), although improvements in hygiene and the introduction of HAV vaccination have significantly reduced the incidence of HAV infection and HAV-associated liver failure. The reasons for the wide range in the severity of HAV infection are not understood but have been thought to be associated with viral virulence factors (5, 6), although this is controversial (6); with the age of the patient, with older individuals developing more severe disease; or with coinfection with other hepatitis viruses.
Although genetic susceptibility factors predisposing to severe forms of HAV-induced hepatitis have not been previously described, it is possible that the variability in HAV disease severity might be regulated by polymorphisms in the gene encoding the receptor for HAV, HAVCR1, also known as TIM1 (7), a member of the TIM (T cell, immunoglobulin, and mucin domain) gene family. The TIM gene family was positionally cloned from within the Tapr (T cell and airway phenotype regulator) locus and consists of a family of novel genes encoding type 1 transmembrane glycoproteins with a conserved structure (8), which includes an immunoglobulin (IgV) domain and a mucin domain, and its members (TIM-1, TIM-3, and TIM-4) serve as receptors for phosphatidylserine (PtdSer) (9–12). TIM1, the first identified member of this TIM family, is expressed in hepatocytes and lymphoid cells, and its gene product, TIM-1, has been found to be an important T cell costimulatory molecule that regulates the activation of, and the induction of tolerance in, T cells (13, 14).
Analysis of the sequence of TIM1 has demonstrated a high degree of polymorphisms within exon 4 of the human (and mouse) TIM1 gene (15, 16). These polymorphisms include nonsynonymous substitutions and insertion/deletion variants that occur much more frequently than synonymous substitutions. Furthermore, a 6-amino-acid insertion in TIM-1 encoded by polymorphisms in exon 4 has been associated in a number of studies with allergic diseases (15, 17, 18) and with progression to AIDS in HIV-infected patients (19). Since TIM1 encodes the receptor of HAV, the high degree of polymorphisms in exon 4 suggests that natural selection by infection with HAV might have driven the development of these polymorphisms (16). To examine this issue, we performed an epidemiological study with a population of children with severe HAV infection and found that the severity of HAV-induced liver disease is indeed associated with exon 4 polymorphisms in TIM1.
Results
A polymorphic variant of TIM1 is associated with increased risk for the development of severe hepatitis.
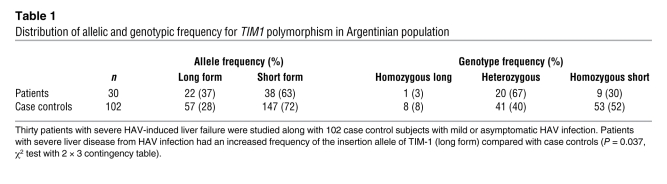
We examined 30 Argentinean patients with HAV-induced acute liver failure and 102 healthy case controls (Argentinean blood donors with a past history of mild or asymptomatic HAV infection and positive IgG, negative IgM anti-HAV serology) in a case-control, cross-sectional, observational study. The population in Argentina is genetically heterogeneous, with a predominantly European ethnic background (mainly Spanish and Italian) and with low levels of Amerindian and African admixture. Different geographic areas in the country, as well as urban/rural populations, have differences in the percentage of European, Amerindian, and African admixtures, and therefore the cases and controls in our study were matched with regard to birthplace and urban/rural distribution (20). The 30 patients in our study had severe liver insufficiency due to HAV infection and were diagnosed with positive IgM anti-HAV serology, without evidence of other causes of acute liver failure. Although the HAV was genotyped in only a few of our patients, the HAV genotype in this region has been shown to be uniformly group IA (6). These patients and the 102 healthy case controls were genotyped for TIM1 by bidirectional sequencing of the highly polymorphic exon 4 (mucin domain) of TIM1. We focused on the 157insMTTTVP insertion in exon 4 of TIM1, since it has previously been associated with allergic diseases and HIV infection (17–19) and has been hypothesized to be the target of natural selection (16). In our total population, 7% of the individuals were homozygous for the long form of TIM1 (encoding TIM-1, containing the 6-amino-acid 157insMTTTVP insertion), 46% were heterozygous, and 47% were homozygous for the short-form TIM1 (encoding TIM-1, without insertion), consistent with the frequencies observed in other studies (15, 16) and in Hardy-Weinberg equilibrium. Table 1 shows that having one or two copies of the long form of TIM1 was associated with a greater risk of developing severe HAV-induced liver failure (P = 0.037, χ2 test). On the other hand, having the short form of TIM1, which is the evolutionarily newer form of TIM1, was associated with protection against severe HAV-induced liver disease.
Table 1 .
Distribution of allelic and genotypic frequency for TIM1 polymorphism in Argentinian population
TIM1 polymorphisms affect binding of HAV.
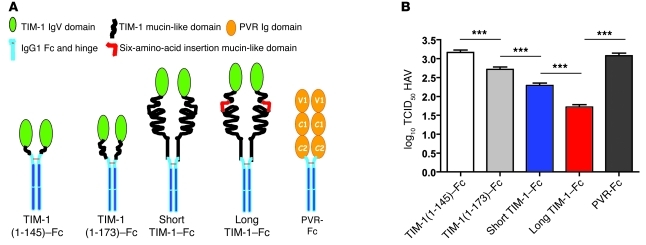
We examined possible mechanisms by which TIM1 might regulate the severity of HAV-induced hepatitis, including the binding of HAV to TIM-1. HAV binds specifically to the IgV domain of TIM-1, but the presence of the mucin domain of TIM-1 is required to uncoat the viral genome, a step that is essential for HAV infectivity (21). To determine whether the length of the TIM-1 mucin domain (containing the 157insMTTTVP insertion polymorphism) affected the ability of HAV to infect cells, we examined neutralization of HAV with soluble receptor to assess the HAV-binding function of several TIM-1–Fc fusion protein constructs containing the entire IgV domain of TIM-1 and different lengths of the mucin domain: (a) IgV domain plus the first 19 residues of the mucin domain (TIM-1[1–145]–Fc); (b) IgV domain plus the first 42 residues of the mucin domain (TIM-1[1–173]–Fc) without the 157insMTTTVP insertion; (c) IgV domain plus mucin domain (TIM-1[1–296]–Fc) without the 157insMTTTVP insertion polymorphism (short form of TIM-1); and (d) IgV domain plus mucin domain with the 157insMTTTVP polymorphism (long form of TIM-1) (Figure 1A). As a negative control, we used an Fc fusion protein containing the extracellular domain of the poliovirus receptor PVR (PVR-Fc), which does not interact with HAV (21). As shown in Figure 1B, the fusion protein containing the TIM-1 IgV domain with only 19 amino acids of the mucin domain (TIM-1[1–145]–Fc) did not neutralize HAV (same 50% tissue culture infective dose [TCID50] HAV titer as with the PVR-Fc control). In contrast, the fusion proteins with the entire mucin domain greatly enhanced HAV neutralization, with the long TIM-1–Fc having the greatest neutralization effect, a full log greater activity than that of the short TIM-1–Fc. These data indicated that the length of the TIM-1 mucin domain significantly affected the binding of HAV, and that the long allele of TIM-1 containing the 157insMTTTVP insertion polymorphism was a more effective receptor for HAV than the short allele.
Figure 1. TIM1 polymorphisms affect HAV binding and uncoating.
(A) The TIM-1–Fc fusion protein constructs used. Soluble TIM-1 fusion protein constructs containing the TIM-1 IgV domain and different lengths of the mucin domain were generated. The constructs contained the TIM-1 IgV domain and (a) the first 19 residues of the mucin domain (TIM-1[1–145]–Fc); (b) the first 42 residues of the mucin domain without the 157insMTTTVP insertion polymorphism (TIM-1[1–173]–Fc); (c) the entire mucin allele without the 157insMTTTVP insertion polymorphism (Short TIM-1–Fc); and (d) the entire mucin allele containing the 6-amino-acid insertion polymorphism (157insMTTTVP) (Long TIM-1–Fc). A Fc fusion protein containing the extracellular domain of the poliovirus receptor (PVR-Fc) with 3 Ig-like domains (V1, C1, and C2) was used as a negative control. (B) The long form of TIM-1 binds HAV more effectively than the shorter form. HAV was treated with the purified soluble fusion proteins described in A, which resulted in HAV neutralization. Residual HAV infectivity was determined by end-point ELISA titration assay in African green monkey kidney cells, and expressed as log10 of TCID50 ± SD (***P < 0.001 [ANOVA]). Lower infectivity indicates more effective binding of HAV to the designated TIM-1 form.
HAV activates natural killer T cells through TIM-1.
Since HAV infection does not itself cause major cytopathological effects in hepatic cells, the liver injury that occurs during HAV infection is presumed to result from an immune response to the virus. MHC class I–restricted CD8+ T lymphocytes that infiltrate the liver have been suggested to cause this liver injury (22). However, peripheral blood anti-HAV CD8+ CTLs peak late in infection, 2–3 weeks after onset of symptoms (23), suggesting that cells of the innate immune system play more important roles in controlling the disease. For example, natural killer T (NKT) cells, which are present in high numbers in the liver (24, 25), have been thought to play a major role in the pathophysiology of several forms of hepatitis, although the specific mechanisms by which this occurs is poorly understood (26). Therefore, we hypothesized that NKT cells might be activated by HAV infection and cause liver injury.
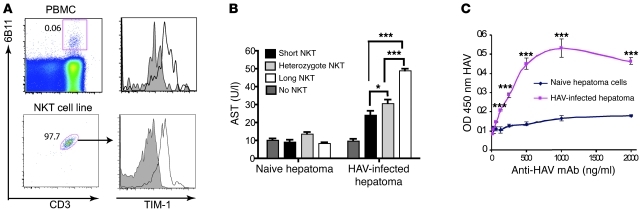
Since TIM-1 is a well-known receptor for HAV, we examined TIM-1 expression on NKT cells. NKT cells constitutively expressed TIM-1, as shown by flow cytometric analysis of peripheral blood NKT cells or primary NKT cell lines using a TIM-1–specific mAb (Figure 2A), consistent with studies of murine NKT cells (27, 28). Although human CD4+ T cells also expressed TIM-1 (data not shown), the level of expression of TIM-1 by NKT cells was considerably higher. In addition, we found that the primary NKT cell lines lysed HAV-infected but not uninfected hepatoma cells (Figure 2B), presumably by recognizing HAV on the surface of the infected cell, since infected but not uninfected cells expressed HAV particles at the cell surface, as determined by a cell surface ELISA (Figure 2C). Importantly, analysis of a panel of NKT cell lines showed that the NKT cell lines homozygous for the long form of TIM1 had greater cytotoxic activity for the HAV-infected hepatoma cells than did the NKT cell lines homozygous for the short form of TIM1 (Figure 2B). The cytotoxicity of NKT cell lines from donors heterozygous for TIM1 was less than that of homozygous long-form lines, but greater than that of homozygous short-form lines (Figure 2B). However, the mean fluorescence intensity of TIM-1 expression on the NKT cell lines from donors with long and short forms of TIM-1 did not differ (data not shown). Taken together, these results suggest that NKT cells having the long form of TIM1 can be activated by HAV more efficiently, and as a result cause severe HAV-induced liver damage.
Figure 2. HAV induces the activation of NKT cells.
(A) NKT cells constitutively express TIM-1. NKT cells were identified by flow cytometry by gating on 6B11+CD3+ NKT cells in total PBMCs (upper panels) or in an NKT cell line (lower panels). TIM-1 expression was then analyzed on gated NKT cells (right panels). The numbers in the dot plot represent the percentage of NKT cells after gating on lymphocytes. Shaded histogram represents background staining with isotype control (mIgG1). (B) NKT cells are cytotoxic for HAV-infected hepatoma cells. Naive or HAV-infected hepatoma cells were cocultured with NKT cell lines from different donors (black: homozygous short form of TIM-1 [n = 6], light gray: heterozygous form of TIM-1 [n = 10], and white: homozygous long form of TIM-1 [n = 2]). AST levels of noninfected and HAV-infected hepatoma cells cultured alone are shown. After 48 hours, culture supernatants were analyzed for AST level. Greater AST indicates greater hepatoma cell lysis. Data are given as mean ± SEM; *P < 0.05, ***P < 0.001 (ANOVA). (C) WT HAV is found at the cell surface of HAV-infected hepatoma cells. Human hepatoma Huh7-A-I cells were infected with WT HAV (HAV-infected) or mock infected (Naive) and examined with a cell surface ELISA using an anti-HAV mAb, detected with peroxidase-labeled anti-IgG. Absorbance at 450 nm was determined in an ELISA plate reader, and the mean absorbance of duplicate wells ± SD was plotted versus the anti-HAV mAb concentration (***P < 0.001 [ANOVA]). The assay was performed twice, and the data represent 1 experiment.
Activation of NKT cells by HAV is TIM-1 dependent.
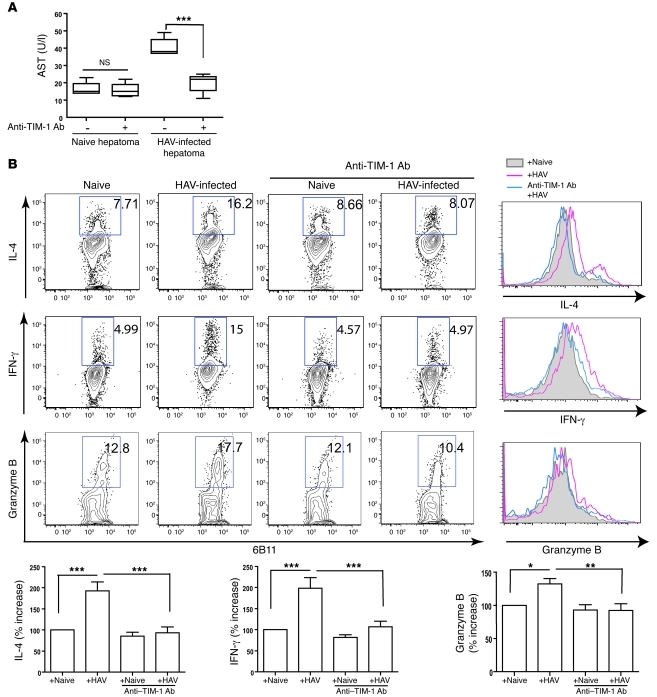
We confirmed that the activation of the NKT cells by HAV was TIM-1 dependent. First, the cytolytic activity of the NKT cells was TIM-1 specific, since only HAV-infected and not uninfected hepatoma cells were lysed (Figure 3A). Second, addition of a blocking anti–TIM-1 mAb (1D12) to cultures of NKT cells and HAV-infected hepatoma cells blocked the killing of HAV-infected hepatoma cells. Furthermore, the production of several cytokines (IL-4, IFN-γ, and granzyme B, which likely mediated the cytotoxic activity against the HAV-infected liver cells) was increased in the NKT cells cultured with HAV-infected hepatoma cells, and this production was blocked by the anti–TIM-1 mAb (Figure 3B). Note that while the NKT cells became activated by HAV, the NKT cells were not productively infected, since HAV did not grow in the NKT cell lines (data not shown). Taken together, these results strongly suggest that the recognition of HAV by the NKT cells through TIM-1 was directly responsible for NKT cell activation.
Figure 3. Activation of NKT cells by HAV is TIM-1 dependent.
(A) Anti–TIM-1 mAb blocks the cytotoxicity of NKT cells. NKT cells were incubated with anti–TIM-1 Ab (1D12), which blocks the interaction of TIM-1 with HAV for 1 hour, and then cocultured with naive or HAV-infected hepatoma cells. AST levels were measured using culture supernatant as in Figure 2B. The boundaries of boxes represent the lower and upper quartiles; the lines within the boxes represent the median; and the whiskers represent the lowest and highest value. ***P < 0.001 (ANOVA). (B) NKT cells produce cytokines in a TIM-1–dependent manner. The NKT cell lines were cocultured with HAV-infected or naive hepatoma cells, and HAV–TIM-1 interaction was blocked by anti–TIM-1 mAb (1D12). Intracellular IL-4, IFN-γ, and granzyme B from NKT cells were analyzed. The numbers shown in each dot plot represent the percentage of cytokine-producing NKT cells (within the box). The data in the dot plots are also shown as histograms, where the shaded histogram represents background cytokine production; the red histogram represents cytokine production induced with HAV-infected hepatoma cells; and the blue histogram represents cytokine production after culture with HAV-infected hepatoma cells and with anti–TIM-1 mAb. The data from 4–8 experiments are plotted in the bar graphs (mean ± SEM), as the percent increase in cytokine production compared with cytokine from NKT cells with naive hepatoma cells. *P < 0.05, **P < 0.01, ***P < 0.001 (ANOVA).
Discussion
The results of these studies together help to explain the variability in the severity of HAV-induced liver disease, which ranges from very mild, often asymptomatic disease to fulminant hepatic failure. We studied a population of children in Argentina with severe HAV-induced liver disease and found that the 157insMTTTVP polymorphism (the long form) of TIM1 was associated with severe infection. The association of TIM1, which encodes the receptor for HAV, with severe HAV-induced liver disease may be explained in part by the fact that the long form of TIM-1 binds HAV more efficiently than the short form of TIM-1 and by the observation that NKT cells expressing the long form of TIM-1 were more cytotoxic for HAV-infected liver cells. These results strongly suggest that genetic variation in TIM1 underlies at least a part of the variability in the severity of HAV-induced hepatitis.
In almost all infections in humans, there is considerable inter-individual phenotypic variability in the severity of the infection (29). There are a number of genetic explanations for this type of variability in disease severity. First, the immune response to the infection might be analogous to a complex trait, in which disease is affected by environmental factors (e.g., differences in the virus or viral load) interacting with multiple genes that are polymorphic, each contributing modestly to disease severity (one type of infection, multiple genes). In other cases, variability in disease severity may be due to variation in genes associated with immunodeficiency (e.g., IL-12/IL-23/IFN-γ family genes, including IL12B, IL12RB1, IFNGR1, IFNGR2, STAT1, NEMO, or TYK2), resulting in susceptibility to several specific types of infections, such as mycobacterial disease (one gene, few types of infections) (30). In still other cases, variation in a specific gene may confer variability in disease severity to a single commonly occurring infectious disease (one gene, one type of infection). For example, variations in the chemokine receptor CCR5 gene, which encodes a coreceptor for HIV, are associated with resistance to HIV infection (31); variations in the FUT2 gene, which encodes a receptor for noroviruses (causative agent for epidemic acute gastroenteritis), afford resistance to norovirus infection (32); and mutations in UNC-93B are associated with severe or recurrent herpes simplex virus meningoencephalitis (33). These studies of infectious diseases from a genetic point of view offer critical insight into the biological processes of infection and immunity and suggest that regulation of the severity of infection, particularly in children, is often due to genetic variation (29).
Just as mutations in many viral receptors affect the severity of the infection (31–34), we found that polymorphisms in TIM1, which encodes the receptor used by HAV to infect human cells, affects the severity of HAV-induced hepatitis. The majority of the polymorphisms in TIM1 are located in exon 4, which encodes a large part of the mucin domain of TIM-1 that is required for HAV uncoating (21), a required step leading to infectivity. Moreover, we showed that the long form of TIM-1 binds HAV more efficiently than the short form of TIM-1, and that NKT cells expressing the long form of TIM-1 had greater cytotoxicity for HAV-infected cells, suggesting that HAV might induce more vigorous immune responses and greater liver injury in individuals with the long form of TIM-1. Importantly, our observation that the long form of TIM1, the evolutionarily ancestral form, is overrepresented among children with liver failure suggests that the extensive polymorphisms in exon 4 of TIM1 gene have been driven by natural selection, as has been previously suggested (16). Thus, we believe that HAV has favored the natural selection of shorter TIM1 alleles, which may reduce the severity of HAV-induced hepatitis.
Since HAV does not by itself cause cytopathic effects in hepatic cells, liver injury in HAV infection is presumed to result from an immune response to viral antigens. Previous clinical observations that patients with greater degrees of HAV-induced liver failure have lower levels of HAV viremia (6, 35) support this idea. Furthermore, our studies now provide mechanistic evidence that an innate immune response against HAV-infected cells may also contribute to the hepatic injury. We showed that TIM-1 is constitutively expressed by NKT cells, which are present in high numbers in the liver, patrolling for pathogens in this important filtration organ (24, 25), and which are required for liver injury in several murine models of hepatitis (36). We suggest that following the initiation of HAV infection of hepatocytes, cytotoxic NKT cells recognize HAV on the surface of the hepatocyte, via TIM-1, and injure the HAV-infected hepatocyte. Furthermore, NKT cells homozygous for the long form of TIM-1 expressed greater cytotoxic activity against HAV-infected liver cells than NKT cells homozygous for the short form of TIM-1, suggesting that the degree of hepatocyte injury during HAV infection is modulated by the polymorphic variant of TIM-1 expressed by the NKT cell. It is unlikely that TIM1 polymorphisms affect the capacity of HAV to infect hepatocytes, since HAV could infect cells transfected with either form of TIM1 with equal efficiency (data not shown), and since prior to 1960 essentially all individuals worldwide, regardless of the TIM1 allele, were infected by HAV. However, additional studies with NKT cells from a greater number of individuals expressing homozygous short and long versions of TIM-1 will be required to confirm our finding.
The insertion polymorphism in exon 4 of TIM1 is associated not only with severe HAV-induced acute liver failure as shown in this study, but also with disease progression in HIV infection (19) and with protection against asthma and allergy (15). The reasons for the association between TIM1 and HIV are not clear, but are not likely to be related to direct NKT cell activation by the HIV. The relationship among HAV, allergy, and TIM1 was suggested by previous epidemiological observations showing that HAV infection protected against the development of asthma and allergy (37–39). Thus, HAV is one of the few specific infectious agents that can explain the hygiene hypothesis. The association of TIM1 both with severe HAV-induced hepatitis and with protection from atopy therefore suggests that the specific mechanisms by which TIM-1 regulates the severity of HAV-induced disease are related to those that protect against asthma and allergy and to those that connect HAV with the hygiene hypothesis. For example, NKT cells, a component of innate immunity, might mediate these relationships, in a process whereby greater activation of NKT cells by HAV through TIM-1 might trigger the hepatic injury that occurs with HAV infection and also result in greater protection against asthma and allergy. An important regulatory role for NKT cells in asthma has already been proposed, based on studies in mice, monkeys, and humans (40, 41). The mechanisms by which HAV-activated NKT cells might dampen chronic inflammation might be similar to those suggested by studies of lymphocytic choriomeningitis virus (LCMV) in a model of type 1 diabetes, in which NKT cells activated by LCMV prevented subsequent pancreatic β cell destruction (42), or to those suggested by studies of IL-4–producing NKT cells that induced Tregs in the prevention of graft-versus-host disease (43). On the other hand, HAV may directly costimulate the activation of Tregs, which express TIM-1 (44). Alternatively, since recent studies indicate that TIM-1 is a receptor for PtdSer and can mediate phagocytosis of apoptotic cells (9–12), a process that is critical for the development of tolerance (45), it is possible that HAV and TIM-1 might regulate immunity and tolerance by affecting the engulfment of apoptotic cells. In any case, we believe that further study of TIM-1 and the immune system will lead to greater insight into how HAV might protect against atopic diseases, thus explaining the hygiene hypothesis, and how infections in general regulate chronic inflammatory diseases.
In summary, our studies of the epidemiology of TIM-1 and HAV infection in humans demonstrated that in addition to their association with protection against asthma and allergy, polymorphisms in TIM1 are associated with the severity of HAV-induced hepatitis. Based on these results, we suggest that HAV has provided the selective pressure for the exceptional degree of polymorphisms found in TIM1. In addition, we propose a pathophysiological molecular mechanism by which NKT cells mediate the liver injury that develops with HAV infection, a mechanism that may be related to one that protects against asthma and allergy.
Methods
Patients.
We studied 30 Argentinean children (15 males, 15 females; mean age, 62 months; range, 20–152 months) who underwent liver transplantation due to HAV-induced acute liver failure at the Hospital Nacional de Pediatría J. P. Garrahan in Buenos Aires, a public institution that serves as a national referral center for pediatric liver transplantation. The diagnosis of HAV-induced acute liver failure was made in our children using internationally accepted criteria (46): (a) evidence of coagulation abnormality, usually an INR (international normalized ratio) of at least 1.5, and any decrease in mental status (indicating encephalopathy), without preexisting cirrhosis, with an illness of as 26 weeks duration, and (b) evidence of acute HAV infection, defined as a positive IgM anti-HAV serology. In our patients, the median aspartate aminotransferase (AST) value before transplantation was 1,430 U/l (range, 389–5,467) (29.8 times the normal value for age), and the median alanine aminotransferase (ALT) value was 1,893 U/l (range, 616–4,850) (33.8 times the normal ALT values for age), indicating severe liver disease. Other causes for acute liver failure were sought and ruled out (e.g., viral, metabolic, toxic, and autoimmune, with negative testing for HBsAg, HBeAg, IgM HBc antibodies, HBsAg antibodies, HBeAg antibodies, HCV, and negative tests for copper or ceruloplasmin, autoantibodies, drug screening [when clinically indicated], and with an otherwise negative abdominal ultrasound).
Case controls.
102 ethnically matched Argentinean healthy blood donors (74 males, 28 females; mean age, 435 months; range, 168–768 months) with a clinical history of mild or asymptomatic HAV infection (with IgG+/IgM– anti-HAV serology) were also studied. Subjects receiving HAV vaccination or receiving blood derivatives in the previous 12 months or with any kind of liver disease, primary or secondary immune deficiency, or other genetic diseases were excluded from the study. The age distribution and the male/female ratio between patients and healthy case controls were significantly different (age distribution, patients vs. healthy case controls: P < 0.0001, Student’s t test; male/female ratio, patients vs. healthy case controls: P = 0.03, χ2 test). The non-pediatric healthy case control population was selected because HAV vaccination became mandatory in Argentina almost 3 years before the study was initiated, preventing the use of a pediatric control population. All patients and case controls gave consent for this protocol, which was approved by the Ethics Committee at Hospital Nacional de Pediatría J. P. Garrahan (protocol 397/2006).
TIM1 gene sequencing.
Genomic DNA, isolated from PBMCs using standard techniques, was analyzed for TIM1 polymorphisms by amplifying the TIM1 exon 4 (108 nucleotides) and sequencing the products bidirectionally (performed at Correlagen).
NKT cell lines.
Human NKT cells from normal volunteers were separated from PBMCs using a PE-conjugated anti-human NKT cell antibody (clone 6B11, which recognizes the CDR3 loop of the conserved Vα24 TCR found on invariant NKT [iNKT] cells) (BD Biosciences — Pharmingen) and anti-PE MACS beads (Miltenyi Biotec). Cells retained on the MACS column were then eluted. DCs were generated by stimulation of CD14+ cells, obtained using anti-CD14 MACS beads, with GM-CSF (2.5 μg/ml) and rIL-4 (2 μg/ml) for 1 week. NKT cells were cocultured with DCs (ratio, 10:1) in RPMI 1640 medium containing 10% FCS, 100 U/ml rIL-2 (R&D Systems), and 1 ng/ml rIL-15 (Peprotech). NKT cell lines were 96%–98% pure.
Cytotoxicity assay.
NKT cell cytotoxicity was determined by coculture of hepatoma cells with NKT cells for 48 hours, and assay of culture supernatants for AST (a liver-derived enzyme) performed in the clinical laboratories at Children’s Hospital Boston. Human hepatoma Huh7-A-I cells, which have been shown previously to support the stable growth of WT HAV (47), were infected with a cDNA-derived WT HM-175 strain of HAV containing no cell culture–adapting mutations and were used as target cells. For detection of cytokines, NKT cells were incubated with naive hepatoma cells or HAV-infected hepatoma cells for 5 days, and then activated with CD3 (1 μg/ml) and CD28 (0.1 μg/ml) for 5 hours to visualize cytokine production. To block the HAV-NKT cell interaction, anti-human TIM-1 mAb (1D12) (5 μg/ml), which has been shown to block HAV infection (10), was added to the NKT cell cultures 1 hour before HAV stimulation. This mAb (1D12) did not activate NKT or T cells (data not shown). For intracellular staining, cells were fixed and permeabilized using Cytofix/Cytoperm kits (BD Biosciences — Pharmingen) according to the manufacturer’s instructions. Cells were incubated with APC-conjugated IL-4 (8D4-8, eBioscience), PE-conjugated granzyme B (GB11, eBioscience), Alexa Fluor 700–conjugated IFN-γ (45.B3, Biolegend), APC-conjugated anti–TIM1–, FITC- or PE-conjugated 6B11 (BD Biosciences — Pharmingen), and Alexa Fluor 750–conjugated CD3 (eBioscience). The respective isotype control mAbs were used as a control. Flow cytometry was performed on a FACSCanto instrument (BD) and analyzed with FlowJo 8.3.3 software (Tree Star Inc).
ELISA for cell surface HAV.
Monolayers of unfixed human hepatoma Huh7-A-I cells infected with cDNA-derived WT HM-175 strain of HAV or mock infected were treated in 96-well plates with dilutions of anti-HAV mAb K3-4C8 (Commonwealth Laboratories). After incubation at room temperature for 1 hour, wells were washed extensively, treated with peroxidase-labeled anti-mouse IgG antibody (KPL Inc.), and stained with SureBlue substrate (Invitrogen). Plates were read in a Synergy HT automated plate reader (BioTek). The mean absorbance at 450 nm of duplicate wells/dilution and the standard deviation were calculated and plotted versus the mAb concentration using the Excel program (Microsoft).
Neutralization of HAV with soluble TIM-1–Fc.
Soluble proteins containing different lengths of human TIM-1 fused to the hinge and Fc of human IgG1 were constructed as described previously (21). TIM-1–Fc fusion proteins containing amino acids 1–145, 1–173, 1–296, and 1–296 plus an insertion of 6 amino acids at position 157 (157insMTTTVP) of human TIM-1 joined to the hinge and Fc regions of human IgG1 were purified by chromatography on protein A–agarose columns (Supplemental Figure 1; supplemental material available online with this article; doi: 10.1172/JCI44182DS1). Supernatants containing live HAV were treated with equimolar amounts of purified Fc fusion proteins, and the residual HAV infectivity was determined by an end-point ELISA titration assay in plates containing monolayers of African green monkey kidney cells (tissue culture infectious doses 50% [TCID50]), as described previously (21).
Statistics.
Case and control TIM1 polymorphism data were compared using the χ2 test in 2 × 3 contingency tables using Prism software (GraphPad). For other studies, data are given as mean ± SEM analyzed by 1-way ANOVA, or 2-way ANOVA with Bonferroni’s post-test using Prism Software. Differences were considered significant at P < 0.05.
Supplementary Material
Acknowledgments
We thank Leslie Kalish for help with the statistical analysis; and Mirta Ciocca, Miriam Cuarterolo, Oscar Imventarza, Javier Goñi, Ana Del Pozo, Jorge Sasbón, Stefania Pittaluga, and María T. de Dávila for valuable comments and support. These studies were supported by the de Fortabat Endowment of the David Rockefeller Center for Latin American Studies at Harvard University, by a grant from the NIH (P01 AI054456), and with FDA intramural funds. The findings and conclusions in this article have not been formally disseminated by the FDA and should not be construed to represent any Agency determination or policy.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J Clin Invest. 2011;121(3):1111–1118. doi:10.1172/JCI44182.
See the related Commentary beginning on page 848.
References
- 1. Bach J. The effect of infections on susceptibility to autoimmune and allergic diseases. N Engl J Med. 2002;347(12):911–920. doi: 10.1056/NEJMra020100. [DOI] [PubMed] [Google Scholar]
- 2. Ciocca M, Ramonet M, Cuarterolo M, Lopez S, Cernadas C, Alvarez F. Prognostic factors in paediatric acute liver failure. Arch Dis Child. 2008;93(1):48–51. doi: 10.1136/adc.2006.115113. [DOI] [PubMed] [Google Scholar]
- 3. Shah U, Habib Z, Kleinman RE. Liver failure attributable to hepatitis A virus infection in a developing country. Pediatrics. 2000;105(2):436–438. doi: 10.1542/peds.105.2.436. [DOI] [PubMed] [Google Scholar]
- 4. Fujiwara K, et al. Analysis of hepatitis A virus protein 2B in sera of hepatitis A of various severities. J Gastroenterol. 2007;42(7):560–566. doi: 10.1007/s00535-007-2039-7. [DOI] [PubMed] [Google Scholar]
- 5. Fujiwara K, et al. Association between severity of type A hepatitis and nucleotide variations in the 5′ non-translated region of hepatitis A virus RNA: strains from fulminant hepatitis have fewer nucleotide substitutions. Gut. 2002;51(1):82–88. doi: 10.1136/gut.51.1.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sasbon JS, et al. Clinical implications of hepatitis A virus ribonucleic acid detection and genotyping in acute liver failure in children in Argentina. Pediatr Crit Care Med. 2010;11(3):385–389. doi: 10.1097/PCC.0b013e3181ceadca. [DOI] [PubMed] [Google Scholar]
- 7. Feigelstock D, Thompson P, Mattoo P, Zhang Y, Kaplan GG. The human homolog of HAVcr-1 codes for a hepatitis A virus cellular receptor. J Virol. 1998;72(8):6621–6628. doi: 10.1128/jvi.72.8.6621-6628.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. McIntire JJ, et al. Identification of Tapr (an airway hyperreactivity regulatory locus) and the linked Tim gene family. Nat Immunol. 2001;2(12):1109–1116. doi: 10.1038/ni739. [DOI] [PubMed] [Google Scholar]
- 9. Santiago C, et al. Structures of T cell immunoglobulin mucin protein 4 show a metal-ion-dependent ligand binding site where phosphatidylserine binds. Immunity. 2007;27(6):941–951. doi: 10.1016/j.immuni.2007.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kobayashi N, et al. TIM-1 and TIM-4 glycoproteins bind phosphatidylserine and mediate uptake of apoptotic cells. Immunity. 2007;27(6):927–940. doi: 10.1016/j.immuni.2007.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Miyanishi M, Tada K, Koike M, Uchiyama Y, Kitamura T, Nagata S. Identification of Tim4 as a phosphatidylserine receptor. Nature. 2007;450(7168):435–439. doi: 10.1038/nature06307. [DOI] [PubMed] [Google Scholar]
- 12. Ichimura T, Asseldonk EJ, Humphreys BD, Gunaratnam L, Duffield JS, Bonventre JV. Kidney injury molecule-1 is a phosphatidylserine receptor that confers a phagocytic phenotype on epithelial cells. J Clin Invest. 2008;118(5):1657–1668. doi: 10.1172/JCI34487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Umetsu S, et al. TIM-1 induces T cell activation and inhibits the development of peripheral tolerance. Nat Immunol. 2005;6(5):447–454. doi: 10.1038/ni1186. [DOI] [PubMed] [Google Scholar]
- 14. Rodriguez-Manzanet R, DeKruyff R, Kuchroo VK, Umetsu DT. The costimulatory role of TIM molecules. Immunol Rev. 2009;229(1):259–270. doi: 10.1111/j.1600-065X.2009.00772.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. McIntire JJ, et al. Immunology: hepatitis A virus link to atopic disease. Nature. 2003;425(6958):576. doi: 10.1038/425576a. [DOI] [PubMed] [Google Scholar]
- 16. Nakajima T, et al. Evidence for natural selection in the HAVCR1 gene: high degree of amino-acid variability in the mucin domain of human HAVCR1 protein. Genes Immun. 2005;6(5):398–406. doi: 10.1038/sj.gene.6364215. [DOI] [PubMed] [Google Scholar]
- 17. Graves PE, Siroux V, Guerra S, Klimecki WT, Martinez FD. Association of atopy and eczema with polymorphisms in T-cell immunoglobulin domain and mucin domain-IL-2-inducible T-cell kinase gene cluster in chromosome 5 q 33. J Allergy Clin Immunol. 2005;116(3):650–656. doi: 10.1016/j.jaci.2005.05.004. [DOI] [PubMed] [Google Scholar]
- 18. Gao PS, et al. Genetic variants of the T-cell immunoglobulin mucin 1 but not the T-cell immunoglobulin mucin 3 gene are associated with asthma in an African American population. J Allergy Clin Immunol. 2005;115(5):982–988. doi: 10.1016/j.jaci.2005.01.035. [DOI] [PubMed] [Google Scholar]
- 19. Wichukchinda N, et al. TIM1 haplotype may control the disease progression to AIDS in a HIV-1-infected female cohort in Thailand. AIDS. 2010;24(11):1625–1631. doi: 10.1097/QAD.0b013e32833a8e6d. [DOI] [PubMed] [Google Scholar]
- 20. Martinez Marignac VL, Bertoni B, Parra EJ, Bianchi NO. Characterization of admixture in an urban sample from Buenos Aires, Argentina, using uniparentally and biparentally inherited genetic markers. Hum Biol. 2004;76(4):543–557. doi: 10.1353/hub.2004.0058. [DOI] [PubMed] [Google Scholar]
- 21. Silberstein E, Xing L, van de Beek W, Lu J, Cheng H, Kaplan GG. Alteration of hepatitis A virus (HAV) particles by a soluble form of HAV cellular receptor 1 containing the immunoglobin-and mucin-like regions. J Virol. 2003;77(16):8765–8774. doi: 10.1128/JVI.77.16.8765-8774.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Vallbracht A, Maier K, Stierhof YD, Wiedmann KH, Flehmig B, Fleischer B. Liver-derived cytotoxic T cells in hepatitis A virus infection. J Infect Dis. 1989;160(2):209–217. doi: 10.1093/infdis/160.2.209. [DOI] [PubMed] [Google Scholar]
- 23. Vallbracht A, et al. Cell-mediated cytotoxicity in hepatitis A virus infection. Hepatology. 1986;6(6):1308–1314. doi: 10.1002/hep.1840060614. [DOI] [PubMed] [Google Scholar]
- 24. Geissmann F, et al. Intravascular immune surveillance by CXCR6+ NKT cells patrolling liver sinusoids. PLoS Biol. 2005;3(4):e113. doi: 10.1371/journal.pbio.0030113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Exley MA, Koziel MJ. To be or not to be NKT: natural killer T cells in the liver. Hepatology. 2004;40(5):1033–1040. doi: 10.1002/hep.20433. [DOI] [PubMed] [Google Scholar]
- 26. Crispe IN. Hepatic T cells and liver tolerance. Nat Rev Immunol. 2003;3(1):51–62. doi: 10.1038/nri981. [DOI] [PubMed] [Google Scholar]
- 27. Lee HH, et al. Apoptotic cells activate NKT cells through T cell Ig-like mucin-like-1 resulting in airway hyperreactivity. J Immunol. 2010;185(9):5225–5235. doi: 10.4049/jimmunol.1001116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kim HS, Lee CW, Chung DH. T cell Ig domain and mucin domain 1 engagement on invariant NKT cells in the presence of TCR stimulation enhances IL-4 production but inhibits IFN-gamma production. J Immunol. 2010;184(8):4095–4106. doi: 10.4049/jimmunol.0901991. [DOI] [PubMed] [Google Scholar]
- 29. Casanova JL, Abel L. Human genetics of infectious diseases: a unified theory. EMBO J. 2007;26(4):915–922. doi: 10.1038/sj.emboj.7601558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Rosenzweig SD, Holland SM. Defects in the interferon-gamma and interleukin-12 pathways. Immunol Rev. 2005;203:38–47. doi: 10.1111/j.0105-2896.2005.00227.x. [DOI] [PubMed] [Google Scholar]
- 31. Samson M, et al. Resistance to HIV-1 infection in Caucasian individuals bearing mutant alleles of the CCR-5 chemokine receptor gene. Nature. 1996;382(6593):722–725. doi: 10.1038/382722a0. [DOI] [PubMed] [Google Scholar]
- 32. Lindesmith L, et al. Human susceptibility and resistance to Norwalk virus infection. Nat Med. 2003;9(5):548–553. doi: 10.1038/nm860. [DOI] [PubMed] [Google Scholar]
- 33. Casrouge A, et al. Herpes simplex virus encephalitis in human UNC-93B deficiency. Science. 2006;314(5797):308–312. doi: 10.1126/science.1128346. [DOI] [PubMed] [Google Scholar]
- 34. Dean M, et al. Genetic restriction of HIV-1 infection and progression to AIDS by a deletion allele of the CKR5 structural gene. Hemophilia Growth and Development Study, Multicenter AIDS Cohort Study, Multicenter Hemophilia Cohort Study, San Francisco City Cohort, ALIVE Study. Science. 1996;273(5283):1856–1862. doi: 10.1126/science.273.5283.1856. [DOI] [PubMed] [Google Scholar]
- 35. Rezende G, et al. Viral and clinical factors associated with the fulminant course of hepatitis A infection. Hepatology. 2003;38(3):613–618. doi: 10.1053/jhep.2003.50366. [DOI] [PubMed] [Google Scholar]
- 36. Toyabe S, et al. Requirement of IL-4 and liver NK1+ T cells for concanavalin A-induced hepatic injury in mice. J Immunol. 1997;159(3):1537–1542. [PubMed] [Google Scholar]
- 37. Matricardi PM, Rosmini F, Panetta V, Ferrigno L, Bonini S. Hay fever and asthma in relation to markers of infection in the United States. J Allergy Clin Immunol. 2002;110(3):381–387. doi: 10.1067/mai.2002.126658. [DOI] [PubMed] [Google Scholar]
- 38. Linneberg A, et al. IgG antibodies against microorganisms and atopic disease in Danish adults: the Copenhagen Allergy Study. J Allergy Clin Immunol. 2003;111(4):847–853. doi: 10.1067/mai.2003.1335. [DOI] [PubMed] [Google Scholar]
- 39. Kocabas E, Yapicioglu H, Yildizdas D, Guneser Kendirli S, Burgut R. The prevalence of atopy in children with antibodies against hepatitis A virus and hepatitis B virus. Turk J Pediatr. 2006;48(3):189–196. [PubMed] [Google Scholar]
- 40. Akbari O, et al. Essential role of NKT cells producing IL-4 and IL-13 in the development of allergen-induced airway hyperreactivity. Nat Med. 2003;9(5):582–588. doi: 10.1038/nm851. [DOI] [PubMed] [Google Scholar]
- 41. Matangkasombut P, Pichavant M, Dekruyff RH, Umetsu DT. Natural killer T cells and the regulation of asthma. Mucosal Immunol. 2009;2(5):383–392. doi: 10.1038/mi.2009.96. [DOI] [PubMed] [Google Scholar]
- 42. Diana J, et al. NKT cell-plasmacytoid dendritic cell cooperation via OX40 controls viral infection in a tissue-specific manner. Immunity. 2009;30(2):289–299. doi: 10.1016/j.immuni.2008.12.017. [DOI] [PubMed] [Google Scholar]
- 43. Pillai AB, George TI, Dutt S, Strober S. Host natural killer T cells induce an interleukin-4-dependent expansion of donor CD4+CD25+Foxp3+ T regulatory cells that protects against graft-versus-host disease. Blood. 2009;113(18):4458–4467. doi: 10.1182/blood-2008-06-165506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ueno T, et al. The emerging role of T cell Ig mucin 1 in alloimmune responses in an experimental mouse transplant model. J Clin Invest. 2008;118(2):742–751. doi: 10.1172/JCI32451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Nagata S. Autoimmune diseases caused by defects in clearing dead cells and nuclei expelled from erythroid precursors. Immunol Rev. 2007;220:237–250. doi: 10.1111/j.1600-065X.2007.00571.x. [DOI] [PubMed] [Google Scholar]
- 46. Taylor RM, et al. Fulminant hepatitis A virus infection in the United States: Incidence, prognosis, and outcomes. Hepatology. 2006;44(6):1589–1597. doi: 10.1002/hep.21439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Konduru K, Kaplan GG. Stable growth of wild-type hepatitis A virus in cell culture. J Virol. 2006;80(3):1352–1360. doi: 10.1128/JVI.80.3.1352-1360.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.