Abstract
Kaposi sarcoma–associated herpesvirus (KSHV) is a B-lymphotropic virus whose primary site of replication is the oropharynx. KSHV can infect both T and B cells from primary tonsillar explant cultures. However, T cells do not support lytic replication, while B cells spontaneously produce substantial amounts of infectious virus. Here, we provide evidence for a mechanism by which activated T cells may promote or stabilize latency of KSHV infection in B cells. When mixed cultures of B cells and activated T cells were exposed to KSHV, little spontaneous virus production was observed. Removing T cells from the mix or treating the mixed culture with immune suppressants enhanced virus production. Adding back activated T cells to purified infected B cells efficiently suppressed KSHV production, primarily due to CD4+ T cells. This suppressive activity required T cell activation and direct cell-cell contact, but not prior exposure to KSHV antigen. Suppression was not MHC restricted and did not result in killing of the target cell. We therefore propose that oropharyngeal T cells activated by a variety of stimuli can recognize ligands on infected target B cells, leading to signaling events that prevent spontaneous lytic activation and promote latent infection in this compartment.
Introduction
Kaposi sarcoma–associated herpesvirus (KSHV, also called human herpesvirus 8 [HHV-8]) is the most recently identified human herpesvirus. Herpesviruses are large, enveloped DNA viruses that can engage 1 of 2 transcriptional programs upon infection — latency and lytic replication. In latency, most viral genes are silenced and the genome persists in a cryptic state as a nuclear episome; latency is considered the default pathway for KSHV infection in most cell types (1, 2). A hallmark of latency is reversibility: when the correct signals are provided to a latently infected cell, the lytic program can be triggered. During the lytic cycle, virtually all KSHV genes are expressed in a temporally regulated program that results in the release of progeny viruses and killing of the host cell (reviewed in ref. 3).
KSHV was initially identified as the causative agent of Kaposi sarcoma (KS) (4), a neoplasm of endothelial cells (5, 6). Phylogenetically, however, KSHV (like its distant relative EBV) is a member of the gammaherpesvirus subfamily (7), which is characterized by tropism for lymphoid cells. Indeed, KSHV DNA is found principally in circulating B cells in infected subjects (8, 9), and viral infection is strongly linked to 2 B cell lymphoproliferative disorders: primary effusion lymphoma (PEL) and multicentric Castleman diseases (MCD) (10–12). Paradoxically, despite this clear evidence of lymphotropism in vivo, all established B cell lines tested to date are resistant to KSHV infection in culture (2, 13, 14) — a fact that has greatly retarded studies of the biology of lymphoid infection by KSHV.
Recently, 2 groups have developed systems for studying KSHV infection in primary B cells. Rappocciolo et al. showed (15) that B cells isolated from PBMCs can be infected in vitro, but only if the cells are first activated by IL-4 and CD40 ligand. Our group has taken a different approach, based on the fact that, in infected humans, the principal site of virus replication is the oropharynx, with shedding of virus in the saliva (16) (salivary virus is critical to KSHV epidemiology, as it is the principal source of infectivity in person-to-person transmission of KSHV). The presumed source of KSHV in saliva is replication in tonsillar and other oropharyngeal lymph node tissue and perhaps in oral epithelial cells as well (17). For this reason, we examined KSHV infection in explants from human tonsils removed at surgery, using a genetically marked recombinant KSHV that bears a constitutively active GFP gene to mark cells in which entry and latency have been achieved (18). Our initial studies (19) showed that, surprisingly, both T and B cells in the tonsil can support viral entry and GFP expression, though neither population becomes immortalized. The T cell infection, however, is abortive — that is, it cannot lead to production of infectious viral progeny. B cells, in contrast, produced abundant infectivity, even in the absence of inducing stimuli (19).
Here, we have exploited this culture system to examine lytic virus replication in tonsillar B cells. As before, we find that de novo infection of primary B cells is associated with high rates of spontaneous virus production, indicative of active lytic replication. Importantly, KSHV replication in infected B cells is strikingly suppressed by activated CD4+ T cells, by a noncytolytic mechanism, which we believe to be novel, that requires T cell activation and cell-cell contact — but is not antigen primed or MHC restricted. This mechanism is very different from other forms of T cell control that have been described in EBV or other herpesviral infections. The effect of this suppression is to prevent spontaneous lytic reactivation in B cells and to promote latent infection in this compartment.
Results
rKSHV.219 infection of primary lymphoid cells: induction of lytic replication.
rKSHV.219 is a recombinant KSHV bearing a GFP gene expressed from a constitutively active promoter (18). Therefore, GFP expression after infection demonstrates successful viral entry and gene expression. In standard cultured cell lines, GFP-positive cells generally have all the hallmarks of latency — nuclear covalently closed circular DNA (cccDNA) episomes, restricted viral gene expression, and the absence of cytopathic effect (CPE). Treatment of such cells with chemical inducers such as the histone deacetylase complex (HDAC) inhibitors butyrate and valproate can trigger lytic reactivation, with full viral gene expression and production of infectious progeny virus; the latter can be quantitated by its ability to transduce naive cells to GFP positivity (18).
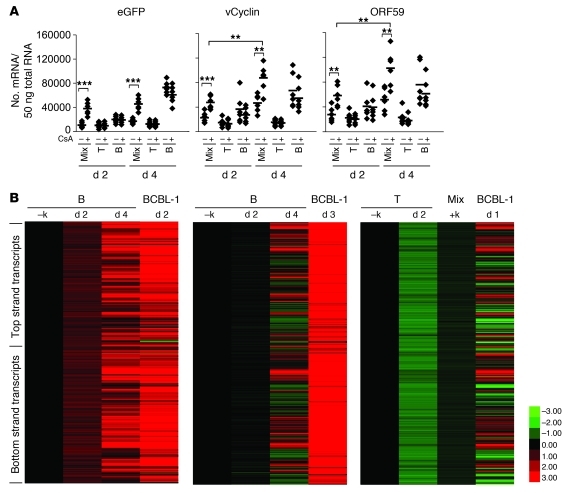
In our earlier work (19), we focused primarily on de novo infection of T and B cells in tonsillar cultures. (For convenience, representative studies of that type are synopsized in Supplemental Figure 1 [supplemental material available online with this article; doi: 10.1172/JCI43755DS1], which shows that only about 4%–5% of B cells but 20%–40% of T cells are competent to support viral entry, as judged by GFP expression.) Here, we focus on the ability of such cultures to support lytic KSHV replication. Accordingly, we carried out the experiment shown in Figure 1, in which infected primary tonsillar cells were examined for the release of infectious KSHV by testing culture supernatants for their ability to transduce GFP expression into 293 cells. Figure 1 shows the results for a mixed (T+B cell) culture in which T cells have been activated with phytohemagglutinin (PHA); these cells produce very little GFP-transducing viable virus at baseline, but show modest induction of virus release when treated with the known inducer, valproic acid (Val). This affirms our prior observation that primary tonsillar cultures are capable of supporting the complete replicative cycle of KSHV. More importantly, however, when similar mixed cultures are treated with the drug cyclosporin A (CsA), an inhibitor of T cell activation, induction is substantially more robust — so much so that it cannot be further augmented by valproate. Next, we asked whether similar results would be obtained if separated populations of T and B cells were similarly infected — and observed a dramatically different result. As previously noted (19), isolated PHA-activated T cells, although readily infectable with KSHV (Supplemental Figure 1), produce little or no infectious virus, either spontaneously or following treatment with valproate or CsA (Figure 1). In contrast, isolated infected B cells spontaneously release high levels of KSHV infectivity, even in the absence of known inducers; these levels of virus production are especially remarkable when one considers that in this setting, typically only approximately 5% of B cells become infected in the first place (see Supplemental Figure 1). Moreover, this virus production cannot be further augmented by valproate or CsA (Figure 1). The last result is a major surprise, since in most established cell lines the default pathway for KSHV infection is latency (1, 2). This result suggests that either lytic infection is the default pathway in primary B cells or that latently infected cells have a high rate of spontaneous activation of the lytic cycle. The fact that quantitative RT-PCR for latent KSHV transcripts (e.g., v-cyclin) revealed expression of such transcripts in infected B cells, especially by day 4 after infection (Figure 2A), may favor the latter explanation. As expected, RNAs for the lytic marker ORF59 were well expressed in infected B cells, but not in T cells (Figure 2A).
Figure 1. Isolated B cells support spontaneous lytic replication of KSHV.
PHA-stimulated unfractionated tonsillar cells and fractionated T or B cells were infected by rKSHV.219 at MOI 3 for 6 to 12 hours (the length of viral infection did not affect the outcomes). Excess unbound viruses were extensively washed. Infected cultures were left uninduced or induced by valproate (600 μM) and/or CsA (1 μg/ml) for 5 days. Culture supernatants were titrated on 293 cells, and IUs (see Methods) were plotted with background subtracted. Each dot represents IUs in the culture supernatant of each individual tonsillar cell. The mean of data from 6 different tonsils is indicated by the horizontal bar. *P < 0.05; ***P < 0.001 by Student’s t test.
Figure 2. Viral gene expression in infected tonsillar cells.
(A) Copy numbers of eGFP, vCyclin, and ORF59 mRNA were determined by real-time quantitative PCR after gene-specific reverse transcriptase reactions on 50 ng of DNase-treated total RNA from fractionated T or B cells or unfractionated tonsillar cells (Mix) at the indicated time points; the cells were infected as described in Figure 1. Cells were treated with CsA for the indicated duration. Dots represent data from each individual tonsil. The mean of data from 6 different tonsils is indicated by the horizontal bar. **P < 0.01; ***P < 0.001 by Student’s t test. (B) Mixed cells or purified CD19+ B and CD3+ T cells were infected with rKSHV.219 as described in Figure 1 and Methods. At day 2 and day 4 after infection, total RNA was extracted and subjected to tiling microarray with linear amplification (for details, see Methods). –k, no viral infection. Total RNA from BCBL-1 cells, which were induced to lytic replication by Val (600 μM) for 1–3 days, was included as a positive control. KSHV tiling microarray data (ordered by genome position) are displayed for 13,444 unique probes. The color bar indicates the fold change relative to the level for the uninfected B cells. 2 independent experiments for B cells are shown. Array data for T and mixed cells are 1 representative of 2 independent experiments.
To get a more complete picture of viral gene expression in infected B cells, we prepared RNA from 2 independent B cell infections and examined it by hybridization to a custom KSHV genomic tiling array. In this array, overlapping 60-mer probes, offset by 20 nt, are deployed to represent each strand of the entire KSHV genome (save for the noncoding terminal repeats). As shown in Figure 2B, both preps displayed extensive transcription of both stands of KSHV and closely resembled the pattern seen in lytically reactivated BCBL-1 cells. The kinetics of viral transcription differed in the 2 preparations, with the first prep showing more rapid induction of viral transcription, while the second infection showed after 4 days a pattern resembling BCBL-1 cells 1 day after chemical induction (Figure 2B). It is not unexpected that primary cells from different donors might display such differences, though the mechanisms underlying them are not known with certainty. Nonetheless, the expression profiling data strongly reinforce the conclusion, drawn from the infectivity analysis of Figure 1, that B cells display prominent lytic infection, be it from de novo entry into the lytic cycle or high-level spontaneous reactivation from latency.
Finally, we wish to note that this PHA-induced inhibition of viral replication in infected unfractionated cultures, while very reproducible in individual samples, was not invariably observed with all samples: it occurred in 52 out of 83 (63%) tonsils used in this study. In 32% of tonsils, we observed virus replication in unfractionated cultures in spite of PHA stimulation; in the remaining 5% of specimens, no infectious virus could be detected in the culture supernatant with or without CsA treatment. This variability among samples may reflect the differing states of disease in the tonsils taken from ill patients as well as variability in the genetic backgrounds of the donors. In the following sections, we explore the mechanisms by which viral replication was suppressed in those PHA-activated cultures in which it was observed.
Activated T cells suppress KSHV production by B cells.
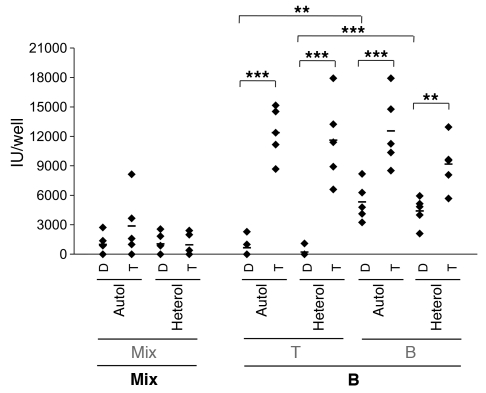
The most striking finding of the data shown in Figure 1 is that although B cells actively produce virus when separated from other lymphoid cells, virus production is effectively shut off in mixed cultures harboring PHA-activated T cells in addition to B cells. Moreover, an inhibitor of T cell activation (CsA) reverses this shutoff in the mixed culture. These facts led us to determine whether T cells were able to suppress the spontaneous production of KSHV by B cells. Isolated T or B cells were first infected with rKSHV.219. Then, a suspension of either uninfected T cells or uninfected B cells was added to each infected culture; 5 days later, culture supernatants were harvested for infectivity analysis. Because the CsA effect pointed to a role for T cell activation (see below), in all cases, the uninfected cells were first activated by exposure to PHA. As shown in Figure 3, infected T cells produced little virus, and therefore no suppression was detectable following coculture with PHA-treated B or T cells. However, when infected B cells were cocultured with uninfected PHA-activated T cells, strong suppression of virus production ensued. This suppression was evident when T cells were preactivated by PHA before coculture and was irrespective of whether the T cells were autologous (and therefore MHC matched) or heterologous (in most cases of which, cells would be mismatched at MHC loci; this mismatch was confirmed by demonstration of IL-2 production in the culture supernatant of a mixed lymphocyte reaction [MLR] in separate experiments; data not shown). As expected, no suppression was observed from cocultivating infected B cells with uninfected B cells. To determine the molecular level at which this suppression operates, we examined mixed (T+B cell) primary cultures cultivated in the presence of PHA for accumulation of viral transcripts, using our custom KSHV tiling array. Figure 2 shows that this suppression was associated with a profound block to viral transcript accumulation.
Figure 3. Activated T cells inhibit spontaneous KSHV lytic replication from B cells in an MHC-unrestricted manner.
rKSHV.219 was infected into fractionated (T or B cells) or unfractionated (Mix) cells at MOI 3 for 6 to 12 hours. After extensive wash, uninfected cells (in gray font), from the same (Autol) or different (Heterol) donors were added into infected cells (in bold font). All the cells used here were prestimulated by PHA. Relevant controls without addition of uninfected cells (left panels) were included for comparison. IUs in the culture supernatants were evaluated on 293 cells after 5 days of culture. Dots represent data from individual tonsils with background subtracted. The mean of data from 7 different tonsils is indicated by the horizontal bar. **P < 0.01; ***P < 0.001 by Student’s t test.
In hepatitis B virus infection, cytokine production by activated T cells is known to suppress virus replication, and this suppression is due to soluble mediators like IFN-γ and TNF-α (refs. 20–22, and reviewed in ref. 23). To ask whether the suppression of KSHV replication by T cells is similarly dependent upon soluble factors, we performed the Transwell experiment shown in Figure 4. Infected B cells were plated in the bottom of a Transwell dish; then uninfected, PHA-stimulated T cells were added to either the same compartment (in which case direct cell-cell contact would occur) or to the upper chamber of the Transwell; in the latter case, the infected B cells can contact soluble factors released from T cells, but not the T cells themselves. As shown in Figure 4, direct contact of T cells with infected B cells strongly suppressed virus release, while no such inhibition was observed when the T cells were placed in trans. Again, suppression was achieved when direct contact was allowed with T cells of either homologous or heterologous origin. In this experiment, weak suppression was observed when uninfected B cells were admixed with infected B cells even though the suppression was not statistically significant; we do not know the origin of this activity, but note that it was not observed in the similar experiment in Figure 3.
Figure 4. PHA-activated T cells inhibit KSHV replication in B cells via a direct cell-cell contact.
Tonsillar cells were fractionated and infected as in Figure 3. When mixed, uninfected cells, of autologous or heterologous origin, were added in the same compartment with infected cells (direct contact, D) or separated in the upper chamber of a Transwell plate (T) and cultured for 5 days. IUs were determined as above. Note that infected cells in a given culture are in bold font, while uninfected cells are in gray font. Dots represent data from individual tonsils with background subtracted. The mean of data from 5 different tonsils is indicated by the horizontal bar. **P < 0.01; ***P < 0.001 by Student’s t test.
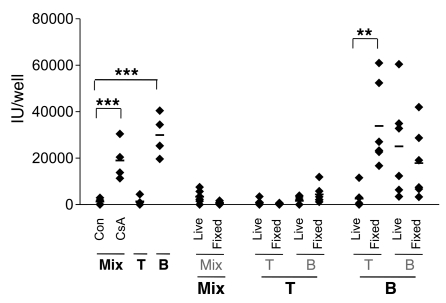
Because KSHV suppression requires cell-cell contact, we presume that one or more ligands on the surface of effector CD4+ T cells must recognize infected B cells. Such recognition could trigger an inhibitory signaling cascade in recipient B cells or could trigger an exocytosis event in the effector T cell to direct products to the infected B cell. The latter model would require that the T cell be viable, while the former model might not. Accordingly, we asked whether suppression could be reproduced by T cells that were gently fixed at room temperature (RT) with 0.05% paraformaldehyde for 1 minute, conditions which are known to block exocytosis and cytokine release from antigen-presenting cells but still preserve the native structure of MHC-I on antigen-presenting cells, as judged by their ability to stimulate CTL (24, 25). As shown in Figure 5, suppression was abolished in fixed cells, while unfixed cells that were harvested in parallel were fully active in KSHV suppression. We also attempted to induce inhibitory activity by coincubating infected B cells with crude membrane preparations derived from PHA-activated T cells (cf. Methods); however, these membrane fractions failed to demonstrate suppressive effect (data not shown). These data suggest that viable T cells are required to evoke suppression of viral replication
Figure 5. Fixed T cells are incapable of conferring inhibition on viral replication in B cells.
PHA-prestimulated fractionated (T or B cells) and unfractionated tonsillar cells (Mix) were left untreated (live) or fixed by 0.05% paraformaldehyde (fixed) for 1 minute at RT. Cells were washed thoroughly and added into infected cells (MOI 3). Infected cells are in bold font, while uninfected cells are in gray font. IUs in the culture supernatants were determined 5 days after culture. Cell fractionation was performed as described in Methods. Con denotes infected mix cells without induction. Dots represent data from individual tonsils with background subtracted. The mean of data from 6 different tonsils is indicated by the horizontal bar. **P < 0.01; ***P < 0.001 by Student’s t test.
Suppression is mediated by CD4+ T cells and requires T cell activation.
To further assess the role of PHA activation in this phenotype, we conducted the experiment shown in Figure 6. Isolated infected B cells were used as the source of KSHV production; to these producer cells was added an equal number of uninfected T or B cells, with or without prior PHA stimulation. The uninfected suppressor cells were either of autologous origin or heterologous origin. In the absence of prior PHA treatment, T cells could not suppress KSHV replication in B cells; PHA treatment of such T cells restored the ability to suppress KSHV when added back to infected B cell cultures. As before, PHA treatment did not induce suppression by uninfected B cells. Clearly, T cell activation is necessary for suppression.
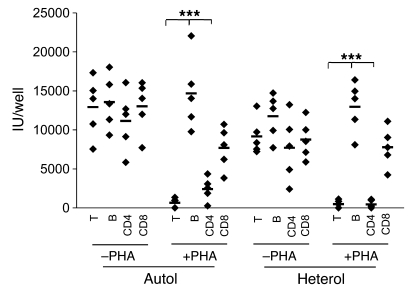
Figure 6. Activated CD4+ T cells are responsible for the inhibition by T cells on viral replication in B cells.
CD3+ T and CD19+ B cells were purified by negative selection as described before. CD4+ T and CD8+ T cells were isolated by positive selection from unfractionated tonsillar cells using beads as in Methods. Purified B cells were infected with rKSHV.219 at MOI 3 for 6 to 12 hours. Excess uninfected viruses were extensively washed. Purified total T, CD4+ or CD8+ T cells, or B cells were activated by PHA or left untreated and added into infected B cells from the same (Autol) or different (Heterol) donors. The determination of IUs was performed as described above. Dots represent data from individual tonsils with background subtracted. The mean of data from 5 different tonsils is indicated by the horizontal bar. ***P < 0.001 by Student’s t test.
Since we expected an MLR reaction to activate T cells when heterologous T cells were added to infected B cells, we were initially surprised by the failure of PHA-naive heterologous T cells to suppress lytic replication in this experiment. However, as will be outlined below, the lesser magnitude and especially the slower kinetics of T cell activation in the MLR compared with PHA treatment likely explains this result (see Figure 7B and discussion below).
Figure 7. TCR stimulation confers inhibitory activity on T cells.
(A) Unfractionated tonsillar cells were prestimulated (left panel) or left untreated (right panel) by PHA (10 μg/ml) for 6 hours and infected with rKSHV.219 at MOI 3 for 6 or 12 hours. After extensive wash, infected cultures were treated with stimulatory antibodies (10 μg/ml) to different cell surface molecules for 3 successive days as indicated. Different isotype antibody controls and medium alone resulted in similar results, and medium alone is shown. IUs in the culture supernatants, with background subtracted, were determined as before. The mean of data from 5 different tonsils is indicated by the horizontal bar. (B) MLR was induced by mixing tonsillar cells from 2 different individuals (see Methods) for the indicated days. T cells were purified from the mixed cultures and added into infected B cells. Relevant control cells from the same donor were included (left panel) for comparison. IUs were determined as before and normalized to those of the medium alone control and plotted as percentage of control (y axis). Dots represent data from individual tonsils. The mean of data from 7 different tonsils is indicated by the horizontal bar. *P < 0.05; ***P < 0.001 by Student’s t test.
In the experiment shown in Figure 6, we also addressed which major subset of tonsillar T cells (CD4+ or CD8+) was responsible for suppression upon cocultivation with infected B cells. Uninfected T cells were fractionated into CD4+ or CD8+ subpopulations as described in Methods. The resulting populations were then examined for their ability to suppress KSHV production following cocultivation with infected B cells. As shown in Figure 6, CD4+ T cells — but not CD8+ T cells — were competent to completely suppress KSHV production. As expected from examination of bulk T cell populations, this suppression also required prior activation of the cells by PHA treatment.
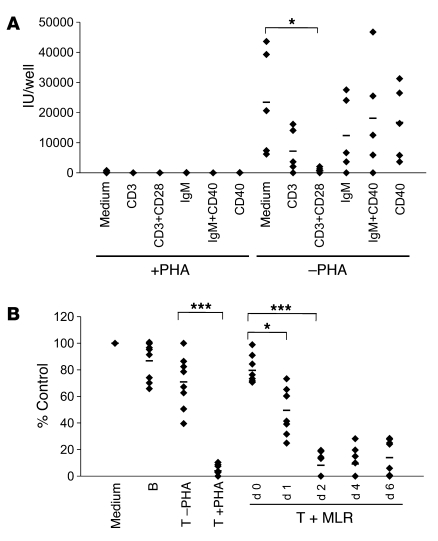
PHA is a plant lectin that crosslinks human T cell surface proteins, including the TCR, thereby inducing polyclonal T cell activation (26, 27). To further examine the requirement for activation with more specific reagents, we examined the ability of other activating ligands to evoke suppression activity. PHA-stimulated or -unstimulated mixed (T+B) tonsillar cultures were infected with rKSHV.219 at MOI 3 for 6–12 hours. Unbound excess viruses were washed off extensively before cells received the following antibody treatments: (a) anti-CD3 antibody; (b) anti-CD3 plus anti-CD28; (c) anti-IgM (anti-BCR); (d) anti-IgM plus anti-CD40 activating antibodies; or (e) activating anti-CD40 alone. Figure 7A shows that, as expected, when no PHA treatment was given to activate T cells in the mixture, spontaneous virus production was observed, presumably from infected B cells. Partial suppressive activity could be evoked by anti-CD3 antibody treatment, but full suppressive activity required crosslinking of both CD3 and the costimulatory ligand CD28. As expected, none of the B cell stimulatory ligands (anti-IgM, anti-CD40) resulted in suppressive activity; conversely, when T cells were first activated with PHA, full suppression was induced, and no further effects could be observed from subsequent treatment with any of the above ligands. These results, together with the ablation of suppression by CsA, strongly indicate that the suppressive factor or factors produced by CD4+ T cells are under the control of the TCR signaling pathway.
Consistent with this, T cells activated by an MLR are also capable of generating the suppressive activity, albeit with slower kinetics than PHA treatment. T cells were purified by negative selection from mixed tonsillar cultures of 2 unrelated donors for varying periods, then added to freshly infected B cells without PHA stimulation (Figure 7B). By 2 days of cocultivation, MLR-stimulated T cells demonstrated an inhibitory activity on KSHV replication in B cells that was only slightly less potent than that of PHA-activated T cells. However, this activity is achieved by PHA treatment in as little as 6–12 hours, while 48 hours of MLR is required to generate comparable levels of inhibition.
Since direct contact with viable T cells is required for suppression, a simple model would be that the activated T cells trigger apoptotic cell death in the target infected B cells, e.g., via release of perforin and granzymes. However, the experiment shown in Figure 8 argues strongly against such a mechanism. Infected B cells were admixed with either (a) no cells; (b) unactivated T cells; or (c) PHA-activated T cells, then examined for annexin V staining by flow cytometry (gating on CD19+ cells). As shown in Figure 8, in the absence of any T cells, there is demonstrable annexin V staining on the B cells, in accord with the known propensity of cultured primary B cells to undergo cell death in vitro. However, this apoptosis was not accelerated by addition of activated T cells — indeed, if anything B cell survival was better in the presence of such cells than in their absence. Note that addition of staurosporine further accentuated annexin staining, indicating that the spontaneous rate of apoptosis was not already so high that a further increase would be undetectable. These data strongly suggest that the suppressive activity of activated T cells is noncytotoxic.
Figure 8. Activated T cells do not induce apoptotic death in KSHV-infected B cells.
Purified CD19+ B cells were infected with rKSHV.219 at MOI 3 for 6 or 12 hours. After extensive wash, infected B cells were mixed with uninfected T cells, prestimulated or unstimulated by PHA. Infected B cells, receiving medium alone or treated with staurosporine for 4 hours, were included as controls. At 2 days of culture, cells were stained and infected (GFP+) B cells were gated for annexin V staining intensity. Shaded histogram represents medium-alone control and thick histogram staurosporine, thin gray histogram unstimulated T cells, and thin histogram PHA-stimulated T cells, as indicated.
Further evidence in support of this notion is shown in Figure 9. The idea behind this experiment is that if activated T cells are killing infected B cells, then the number of B cells capable of supporting viral replication should decline faster in the presence of activated T cells than in their absence. Accordingly, we set up mixed (T+B) cultures and stimulated them in the presence or absence of PHA for 6–12 hours prior to rKSHV.219 infection; following KSHV infection, cells were extensively washed, then cultured. On days 0, 1, or 2 after infection, the ability of the B cells in the culture to support virus production was evaluated by assaying the supernatant of each culture for infectious KSHV following induction by CsA or valproate. Virus production from cultures maintained in the absence of PHA provides an index of the number of productively infected B cells and their natural decline with time in culture (from intrinsic apoptosis). If PHA-activated T cells kill infected B cells, then the production of virus from these cultures should decline even more steeply than in cultures maintained in the absence of PHA. However, as shown in Figure 9, this is not the case: virus production from B cells cultivated with activated T cells remains inducible by CsA to levels identical to those released from B cells grown for the same duration in the absence of activated T cells. This experiment strongly supports the noncytolytic nature of the T cell–mediated suppression of KSHV growth — and rules out the possibility that B cells are being killed by a nonapoptotic mechanism.
Figure 9. Infected B cells are not killed by activated T cells in the coculture.
Purified B cells were infected as for Figure 7. Infected B cells received uninfected T cells, prestimulated or unstimulated by PHA, or were treated with CsA or Val as in Figure 1 at the indicated days of coculture. Cells were further cultured for 5 days, and IUs in the culture supernatants were determined and plotted as before. The dotted line indicates detection limit. Dots represent data from each individual tonsil. The mean of data from 7 different tonsils is indicated by the horizontal bar. ***P < 0.001 by Student’s t test.
The fact that B cells whose virus output has been suppressed by activated T cells can nonetheless be subsequently induced to produce virus with valproate also implies another remarkable feature: that this suppression most likely acts by promoting viral latency, that is, in the presence of T cell inhibitory factors, infected B cells return to a state in which no virus is released, but the cell survives with a persistent, cryptic infection from which lytic replication can be evoked by HDAc inhibitors — the experimental definition of herpesviral latency. The implications of this will be discussed below.
Discussion
Difficulties in infecting established lymphoid cells in vitro have severely retarded our understanding of the biology of KSHV in its natural target cell, the B lymphocyte. However, our development of a system for KSHV infection in primary tonsillar explants has allowed us to begin to ask basic questions about this aspect of KSHV biology. The present work establishes several findings that advance our understanding of KSHV infection in the lymphoid compartment. First, a surprising feature of KSHV infection of primary lymphoid cells was the production of infectious KSHV virions from isolated infected B cells that had not been treated with chemical inducers (Figure 1). These B cells released significant quantities of infectious virus and did not produce additional virus when treated with known inducers such as butyrate or valproate (aside: virus produced either (a) constitutively from primary B cells or (b) by mixed primary cultures in the presence of chemical inducers displayed comparable particle/infectivity ratios; Supplemental Figure 2). Such constitutive production of KSHV has not been noted in conventional adherent cell lines and suggests that either (a) virus can directly enter the lytic cycle in these cells or (b) spontaneous reactivation from latency is extremely frequent when infected B cells are isolated in pure culture. It is difficult to know with certainty whether this finding reflects B cell infection in vivo or is a feature of primary B cell culture, but it certainly impels us to rethink the adage — itself derived from even more artificial forms of cell culture — that the default pathway of KSHV infection is always latency (see discussion below).
This finding, coupled to the poor inducibility of PHA-treated mixed cultures infected with KSHV, led us to our study’s second finding: that uninfected but activated T cells in many (but not all) tonsillar explants are capable of suppressing virus production by infected B cells. This suppression is mediated primarily by CD4+ T cells and requires (a) that the cells be activated via stimulation of the TCR and (b) that the effector CD4+ T cells make contact with their infected B cell targets. This suppression of KSHV replication does not proceed via killing of the infected target and is not subject to MHC restriction. As such, it does not need to be mediated by classical, antigen-specific TCR recognition of KSHV peptides presented on the B cell surface (though we have little doubt that this form of TCR activation would also suppress replication). In accord with this, we note that the effector cells in our study came from the tonsils of American children, most of whom were less than 6 years old — epidemiologic studies indicate that over 96% of such children are seronegative for KSHV (28) and therefore would be expected to have no KSHV-specific T cells as well. In accord with this, we have directly examined cultures from 22 of these children by nested PCR for KSHV DNA; this revealed no evidence of KSHV infection in any sample, while EBV DNA was found, as expected, in 45% of the cases (Supplemental Figure 3).
Some aspects of this regulation are superficially reminiscent of certain classical observations made in EBV infection. In the EBV field, it has long been known that, in seropositive individuals, the outgrowth of EBV-immortalized lymphoblastoid cell lines from primary lymphoid cultures is facilitated by either T cell depletion or treatment with CsA (29–32). However, mechanistically that process is completely different: it is a result of an adaptive immune response that (a) requires specific anti-viral CD8+ T cells, (b) is mediated by killing of the EBV-infected target cell, (c) is MHC class I restricted, and (d) is primarily directed against latently infected cells (not lytically infected cells) (33–37). In contrast, the process we describe here is not dependent upon virus-specific adaptive immunity. The vast majority of the cultures studied here are derived from KSHV-naive hosts (as noted above), and the process is not MHC restricted.
The suppression of KSHV replication by T cells recalls several other T cell–mediated antiviral processes that have previously been described, but is likely also distinct from them. In HIV infection, CD8+ T cells have been described to produce a soluble antiviral factor (CD8 antiviral factor [CAF]) with potent inhibitory effects on HIV replication, though the molecular identity of CAF remains unknown (38–41). Since the KSHV suppression is mediated principally by CD4+ T cells, it is clearly distinct from this process; also, CAF elaboration and action do not require cell-cell contact. A more interesting analogy exists with hepatitis B virus infection, in which it has been shown that activated T cells (either CD4– or CD8+, though primarily the latter) can lead to suppression of viral replication and gene expression in infected hepatocytes. Interestingly, this includes T cells activated by heterologous infectious agents (e.g., LCMV, CMV, or adenovirus) as well as those responding to HBV-specific antigens (42, 43). This process has largely been characterized by adoptive transfer experiments in HBV transgenic mice; as such, it has been harder to assess the requirements for cell-cell contact, making direct comparison with our observations difficult. But it is tantalizing that (a) both mechanisms are noncytolytic and act via suppression of virus replication; and (b) in both cases, T cells activated by heterologous stimuli can confer the suppression. However, in the case of HBV, these mechanisms are thought to lead to viral clearance (44, 45). In contrast, the experiment shown in Figure 9 suggests that in KSHV, these mechanisms, by preventing lytic replication, instead appear to promote viral latency, that is, viral genomes are not eradicated, but their expression is suppressed; replication can be evoked from these persistent (latent) genomes by classical inducers like valproate.
These findings, in turn, raise new questions about KSHV latency in the lymphoid compartment. Current thinking about the molecular biology of latency in both EBV and KSHV, which is largely derived from cell culture models, is that latency is the default pathway of infection. Lytic replication requires that the latently infected cell receive an instructive signal (typically mimicked by phorbol esters or HDAc inhibitors) that turns on lytic genes. Our results suggest that in primary B cells, it is latency that may require an instructive signal, here delivered by contact with activated T cells. All that is necessary to trigger lytic infection is release from that signal.
It is, of course, impossible to know at this stage whether B cell infection in the intact host in vivo behaves as it does in primary culture. One possible interpretation of our in vitro results is that, because B cell death is so prominent in lymphoid cultures, this proapoptotic program is itself a trigger of lytic reactivation; in this view, if the activated T cells simply deliver a survival signal this may “reset” the B cells back to their physiologic state, in which latency is the default pathway. We cannot entirely exclude this idea, but note that, as shown in Figure 8, unactivated T cells delivered just as strong a survival signal to B cells as did PHA-activated cells (as judged by annexin V staining), yet only the latter are capable of viral suppression — arguing against such an interpretation. Additionally, as shown in Figure 7A, activating anti-IgM antibodies and soluble CD40 ligand, which in concert provide proliferation and survival signals to B cells, could not suppress viral replication in treated infected B cells. These data imply that simple prevention of cell death may not satisfactorily explain the suppression of viral replication.
The phenomena described here also allow for new or revised interpretations of earlier findings in KSHV biology. First, early experiments of Decker et al. (46) indicated that in infected subjects, viral DNA in PBMCs was predominantly linear — the genomic form associated with lytic replication and virion production. Since B cells are the prime reservoir of KSHV DNA in the blood, this suggests that lytic infection of B cells occurs prominently in vivo, in accord with our in vitro observations. This inference was later confirmed by an independent group (47). The fact that lytic replication is also prominent in B cells in lymph nodes from MCD is also consistent with our findings in vitro (48–50). In contrast, KSHV latency is routinely observed in PEL tumors in the absence of accompanying T cells; if latency isn’t the default pathway in vivo in B cells, it would suggest that such cells must have undergone epigenetic or genetic changes that autonomously promote latency. We propose that the rarity of PEL tumors may, in part, be explained by the rarity with which such secondary events occur in nature.
Our findings may also allow reinterpretation of certain clinical observations in KSHV infection. Prior clinical studies demonstrate the correlation between the use of CsA and the risk of KS development in organ transplant recipients (51–53). Although this finding is often interpreted in terms of loss of immune surveillance, our findings that CsA may release lytic KSHV replication from control by T cells could also help to explain those observations, as it is well established that KS development depends on continued viral replication (54–57). Similarly, the progression of KSHV viral load that accompanies HIV-related CD4 depletion may be due not only to the loss of adaptive immunity to KSHV, but also to the loss of the KSHV-suppressive activity in the lymphoid compartment.
How might the antiviral suppression we observe in tonsillar cultures work? We propose that TCR stimulation (from any source) leads to upregulation of one or more molecules on CD4+ cell surface that allows recognition of infected B cells. (Whether the ligand for such recognition is of viral or host origin is, of course, unknown at present.) Following such recognition, the activated T cell could (a) secrete soluble mediators, and/or (b) directly trigger signaling events in the infected B cell; the effect of either pathway would be to block the progression of the lytic cycle and stabilize latency. (Of course, these 2 possibilities are not mutually exclusive.) We note with interest that tonsillar nodes are frequently activated by exposure to a plethora of mucosal bacterial and viral pathogens. If oropharyngeal lymphoid tissue is indeed the main reservoir of KSHV infection, then heterologous T cell activation in this site may be an important determinant of viral persistence.
Methods
Cells and human lymphoid aggregate culture.
QBI293A cells (Q-Biogene) were used to titrate rKSHV.219 stocks prepared from stable Vero cells harboring the recombinant virus, which was described before (18). Tonsillar tissues were obtained from the Cooperative Human Tissue Network (CHTN). All experiments with human tonsils were performed in compliance with the UCSF Committee on Human Research. Human lymphoid aggregate culture (HLAC) was prepared as described before (58, 59). Briefly, tonsils were minced into small blocks and passed through a 40-μm cell strainer in tonsil medium (RPMI medium supplemented with 15% FBS and 1% glutamate and nonessential amino acid). HLAC was cultured and maintained in a U-bottomed 96-well plate (1–2 × 106/well in 200 μl tonsil medium) and used within a week of culture.
Reagents and antibodies.
Antibodies used were purchased from various vendors. Conjugated antibodies for FACS staining (anti-CD3, anti-CD8a, and anti-CD19) were from Biolegend; anti-CD4 antibody was from eBioscience; purified antibodies for lymphocyte receptor stimulation (anti-CD3, anti-CD28, and anti-CD40) were from eBioscience, and anti-human IgM antibody (μ chain–specific mouse F[ab′]2) was from Southernbiotech. APC-conjugated annexin V was purchased from Invitrogen. Staurosporine solution from Streptomyces sp (1 mM) was purchased from Sigma-Aldrich. Lyophilized PHA and CsA were purchased from Sigma-Aldrich, and 1 mg/ml stocks of each chemical were prepared and stored at –20°C in aliquots to avoid repeated freeze-thaw cycles. ELISA MAX Set Deluxe for Human IL-2 was purchased from Biolegend to assess IL-2 expression in the MLR culture supernatants.
Virus stock preparation and titration.
rKSHV.219 stocks were prepared from stable Vero cells (18) with minor modifications, which are described in detail elsewhere (19). In Brief, rKSHV.219-harboring Vero cells were induced by valproate (1.2 mM) and adenovirus-expressing RTA (MOI 100), a lytic switch protein of KSHV. At 24 hours after induction, culture medium was changed with complete DMEM containing 10% FBS in the absence of viral inducers and puromycin, which was to maintain stable Vero cells. When 50% cells were floating, a sign of clear CPE by KSHV replication and release, culture supernatants were collected and cleared of cells and debris by low-speed centrifugation (950 g for 10 minutes) and filtration through a 0.45-μm filter. Virus particles were pelleted by ultracentrifugation (27,000 g for 2 hours). Virus was resuspended in tonsil medium, and aliquots were stored at –80°C until use. Infectious units (IU) of recombinant virus stocks and various culture supernatants were determined on QBI293A cells by spinoculation, as described before (18, 60). Briefly, serially diluted virus solution was infected on QBI293A cells by low-speed centrifugation (950 g for 90 minutes). Fresh DMEM containing 2% FBS was added and further cultured for 2 days before being analyzed by FACS. IUs were calculated by multiplying the percentage of GFP+ QBI293A cells by the total number of QBI293Q cells at the time of analysis.
Culture of tonsillar cells and virus infection.
1 × 106 tonsillar cells were stimulated with PHA (Sigma-Aldrich) for 6–12 hours and washed extensively before use. PHA-stimulated cells were infected at a total of 200 μl/well at MOI 1 (for infection studies) or 3 (for inhibition-of-virus-replication studies) in a 96-well U-bottomed plate (MOI is defined by number of IUs per cell). For infection studies, cells were cultured for 2 days before being stained for flow-cytometric analysis. For studies of virus replication in infected lymphocytes, cells were extensively washed at 6–12 hours after infection at least 3 times in warm tonsil medium and cultured further for 5 days before virus titer in the culture supernatants was assessed on QBI293A cells. When necessary, T cells stimulated either by PHA or MLR (see below), were fixed by 0.05% paraformaldehyde at RT for 1 minute to investigate the ability of fixed cells to inhibit virus replication in infected B cells. Membrane fractions were prepared by rupturing cells by repeated freeze-thaw cycles in PBS or by hypotonic cell lysis buffer (5 mM Tris, pH 7.5, 5 mM MgCl2, 1 mM EGTA, and 1 mM dithiothreitol). Membrane fractions were collected by centrifugation (16000 g, 30 minutes). In some experiments, isolated T or B cells (for cell isolation, see below) were added to infected B cells to evaluate suppressive activity of those cells. When indicated, purified T or B cells were added in the same compartment with infected B cells, allowing direct cell-cell contact, or to the upper chamber of a Transwell plate, separated by a 0.22-μm membrane (Nunc). Tonsillar cultures were treated with antibodies to various cell-surface receptors, e.g., CD3, CD28, IgM, CD40, etc., at day 0, day 1, and day 2 after infection at 10 μg/ml to study their efficacy in suppressing KSHV replication.
MLR.
To investigate whether heterologous T cells, either activated by PHA or untreated, inhibit KSHV replication in infected B cells, purified T cells, by magnetic beads (see below), were added to infected B cells from the same donor (autologous) or the different donor (heterologous) on the day of coculture. To study whether T cells, preactivated by MLR, suppress KSHV replication in infected B cells, stimulated T cells were isolated from mixed tonsillar cultures from 2 different individuals, which were cocultured in equal numbers in a U-bottom 96-well plate (2 × 106 cells /well) for various periods of time. Purified MLR-stimulated T cells were added to infected tonsillar B cells to investigate their ability to inhibit virus replication in infected tonsillar B cells.
Cell purification using magnetic beads.
Untouched B or T cells were isolated by negative selection utilizing B Cell Isolation Kit II and Pan T cell Isolation Kit II (Miltenyi Biotec). Antibody labeling and column purification were performed according to the manufacturer’s instructions. Briefly, 108 cells were labeled with 100 μl of biotin-antibody cocktail followed by 200 μl of anti-biotin MicroBeads. Unwanted cells were removed through a column and eluents were washed extensively in tonsil medium before use. CD4+ or CD8+ T cells were positively selected by employing CD4+ and CD8+ T MicroBeads (Miltenyi), respectively. All reagents used for cell purification were purchased from Miltenyi Biotec.
Extraction and enumeration of viral DNA in the culture supernatants.
Viral DNA was extracted from culture supernatants as described previously (61). In brief, culture supernatants were treated with DNase I (20 U/ml) to remove free viral DNA in the medium at 37°C for 1 hour. Virions were pelleted down at 27,000 g for 2 hours at 4°C and resuspended and incubated in lysis buffer (20 mM Tris-HCl at pH 8, 10 mM EDTA, 100 mM NaCl, 0.5% SDS) at RT for 10 minutes. 0.7 mg/ml proteinase K and linearized plasmid encoding an unrelated malaria gene (cysteine protease gene of Plasmodium falciparum) were added and incubated at 37°C overnight with subsequent extraction with phenol/chloroform/isoamyl alcohol. Viral DNA was precipitated and resuspended in water, which was used in the subsequent TaqMan and SYBR Green assays for PAN promoter and spiked-in malaria gene as a normalizing factor as described previously (61) using BAC36 as a standard (62). TaqMan (for Pan promoter) and SYBR Green (for malaria DNA) reactions were performed in triplicate using TaqMan Universal Master Mix (Applied Biosystems) and SYBR Green ER Master Mix (Invitrogen), respectively, according to the manufacturer’s instructions.
RNA extraction and quantitative real-time PCR.
Total RNA was extracted by RNA-Bee (Tel-Test Inc.) according to the manufacturer’s instructions. DNase I–treated 50 ng of total RNA was used to synthesize cDNA using gene-specific primers for eGFP, ORF59, and vCyclin. Primers and probes were as follows: eGFP, forward: 5′-CTGCTGCCCGACAACCA-3′, reverse: 5′-TGTGATCGCGCTTCTCGTT-3′, probe: 5′-FAM-CCCAGTCCGCCCTGAGCAAAGAC-TAMRA-3′; ORF59, forward: 5′-GTTTACCCCCGGGCTGAT-3′, reverse: 5′-GGGCACACCTTCCACTTCTAAT-3′, probe: 5′-FAM-TGGCACTCCAACGAA-MGB-3′; vCyclin, forward: 5′-CGCGGCATAGCAAAGTGAA-3′, reverse: 5′-GCCTGTTAGTGGCCAGTAAGCT-3′, probe: 5′-FAM-TGGTAGAAATAGGCGTGAGGCTTCT-MGB-3′. Reverse primers were used to generate gene-specific cDNA in reverse transcriptase reactions using SuperScript III Reverse Transcriptase (Invitrogen). Resulting cDNA was used for quantitative real-time PCR by employing TaqMan Universal PCR Master Mix (Applied Biosystems). Plasmids encoding each gene were used as a standard.
Flow cytometry.
Tonsillar cells, infected or left uninfected, were stained with antibodies at 4°C for 30–60 minutes. Stained cells were washed twice in 2 ml incomplete PBS supplemented with 4% FBS and 0.09% sodium azide before being fixed in 0.5% paraformaldehyde, followed by wash. Cells were then analyzed on LSR II (BD). Data analysis was performed utilizing FlowJo software.
Assessment of apoptosis by annexin V staining.
Apoptotic death of infected B cells in various cultures was determined by their affinity to annexin V proteins according to the manufacturer’s instructions (Invitrogen). Briefly, cells were washed in annexin V binding buffer, containing 10 mM HEPES, pH 7.4, 140 mM NaCl, 25 mM CaCl2. Cells were resuspended in 100 μl annexin-binding buffer, and 5 μl of APC-conjugated annexin V was added and incubated at RT for 15 minutes. 400 μl of binding buffer was added into the staining reaction, and annexin V staining was immediately analyzed on LSR II. Apoptotic death in B cells was induced with staurosporine as a positive control at 1 μM for 4 hours before FACS analysis. Vehicle-only control (0.1% DMSO) was also included.
Viral gene expression profiling by tiling microarray.
Total RNA from tonsillar cells was extracted using RNA-Bee (Tel-Test Inc.) and further purified using the RNeasy Mini Kit (QIAGEN) according to the manufacturer’s protocols. The reference RNA corresponds to a mixture of total purified RNA from infected and uninfected endothelial, B, and T cells. The quality and concentration of the RNA was analyzed prior to labeling using the 2100 Bioanalyzer (Agilent) and the ND1000 spectophotometer (Nanodrop) All RNA samples displayed an RNA integrity number (RIN) higher than 9. Labeled cRNA was generated from 250 ng of total RNA using the 2-color Quick Amp Labeling (Agilent) according to the manufacturer’s protocol. Experimental samples (labeled with Cyanine 5-CTP) and reference samples (labeled with Cyanine 3-CTP) were competitively hybridized to custom viral microarrays (Agilent) according to the manufacturer’s instructions. The design of the viral microarrays has been previously described (63) and can be obtained from Agilent (Amadid 029238). Washed and processed arrays were scanned using the GenePix 4000B Scanner (Axon Instruments) and the feature intensities extracted using GenePix Pro 6.0 software. TIFF images of scanned slides were analyzed using the Feature Extraction Software, Ver 9.5.3 (Agilent), and a graphical presentation of processed data (heat map) was generated with Java Treeview.
Detection of KSHV DNA in tonsillar cells by nested PCR.
Total DNA was extracted from 22 tonsils using DNeasy Blood and Tissue Kit (QIAGEN) according to the manufacturer’s instructions. DNA was eluted in final 200 μl elution buffer. 100 ng of extracted total DNA was used for the first PCR reactions, and 2 μl of the first PCR reaction was used as template for the second PCR reactions. To analyze the amount and the quality of extracted DNA used, human GAPDH gene was amplified by Phusion Hot Start II High Fidelity DNA polymerase (Finnzymes) at the following conditions: denaturation at 98°C for 10 minutes, annealing at 58°C for 10 minutes, and extension at 72°C for 10 minutes for 25 cycles using the forward primer ACCCGGGTTCATAACTGTCTGCTT and the reverse primer AGGCATTGCTGCAAAGAAAGAGGG. First and second PCR of Nested PCR to detect EBV or KSHV genome was performed using Phusion Hot Start II High Fidelity DNA polymerase at the following conditions: denaturation at 98°C for 10 minutes, annealing at 58°C for 10 minutes, and extension at 72°C for 10 minutes for 35 cycles/PCR using the following sets of primers. To detect KSHV DNA, 3 different sets of primers were employed targeting different regions of the viral genome. LANA forward primer for the first PCR: AGCTCAGAAGCCTCACGCCTATTT, LANA reverse primer for the first PCR: TAGCTGTCCAGAATGCGCAGATCA, LANA forward primer for the second PCR: AGAACTCCTTGAGAAGTTGGCGTG, LANA reverse primer for the second PCR: AATGACGTTGGCAGGAACCAACAG; 91287 forward primer for the first PCR: TAAATCTCTGGGATGCAGGCCCTT, 91287 reverse primer for the first PCR: TTTCCTAGGCGGTTGGGATTCACT, 91287 forward primer for the second PCR: TTGACCTGTCAGAATGGTCTGCCT, 91287 reverse primer for the second PCR: ATACCCTGTTTCAAAGCTGGTCCG.
To detect EBV DNA in tonsillar samples, the following primers were used: LMP-1 forward primer for the first PCR: TTCTCTGTCCACTTGGAGCCCTTT, LMP-1 reverse primer for the first PCR: GGCCAGAATCATCGGTAGCTTGT, LMP-1 forward primer for the second PCR: TTGGAGATTCTCTGGCGACTTGGT, LMP-1 reverse primer for the second PCR: TGGTGGTGTTCATCACTGTGTCGT.
Statistics.
Data represented in this study are shown as the mean ± SD of at least 3 independent experiments. The significance of differences in the mean values was evaluated by 2-tailed Student’s t test. P < 0.05 was considered statistically significant.
Supplementary Material
Acknowledgments
This work was supported by a grant from the NIH (P01DE019085). We thank Lewis Lanier and Richard Locksley for critical reading of the manuscript.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J Clin Invest. 2011;121(3):1130–1140. doi:10.1172/JCI43755.
See the related Commentary beginning on page 838.
References
- 1. Vieira J, O’Hearn P, Kimball L, Chandran B, Corey L. Activation of Kaposi’s sarcoma-associated herpesvirus (human herpesvirus 8) lytic replication by human cytomegalovirus. J Virol. 2001;75(3):1378–1386. doi: 10.1128/JVI.75.3.1378-1386.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bechtel JT, Liang Y, Hvidding J, Ganem D. Host range of Kaposi’s sarcoma-associated herpesvirus in cultured cells. J Virol. 2003;77(11):6474–6481. doi: 10.1128/JVI.77.11.6474-6481.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ganem D. KSHV infection and the pathogenesis of Kaposi’s sarcoma. Annu Rev Pathol. 2006;1:273–296. doi: 10.1146/annurev.pathol.1.110304.100133. [DOI] [PubMed] [Google Scholar]
- 4. Chang Y, et al. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi’s sarcoma. Science. 1994;266(5192):1865–1869. doi: 10.1126/science.7997879. [DOI] [PubMed] [Google Scholar]
- 5. Dupin N, et al. Herpesvirus-like DNA sequences in patients with Mediterranean Kaposi’s sarcoma. Lancet. 1995;345(8952):761–762. doi: 10.1016/S0140-6736(95)90642-8. [DOI] [PubMed] [Google Scholar]
- 6. Moore PS, Chang Y. Detection of herpesvirus-like DNA sequences in Kaposi’s sarcoma in patients with and without HIV infection. N Engl J Med. 1995;332(18):1181–1185. doi: 10.1056/NEJM199505043321801. [DOI] [PubMed] [Google Scholar]
- 7. Russo JJ, et al. Nucleotide sequence of the Kaposi sarcoma-associated herpesvirus (HHV8). Proc Natl Acad Sci U S A. 1996;93(25):14862–14867. doi: 10.1073/pnas.93.25.14862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ambroziak JA, et al. Herpes-like sequences in HIV-infected and uninfected Kaposi’s sarcoma patients. Science. 1995;268(5210):582–583. doi: 10.1126/science.7725108. [DOI] [PubMed] [Google Scholar]
- 9. Mesri EA, et al. Human herpesvirus-8/Kaposi’s sarcoma-associated herpesvirus is a new transmissible virus that infects B cells. J Exp Med. 1996;183(5):2385–2390. doi: 10.1084/jem.183.5.2385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cesarman E, Chang Y, Moore PS, Said JW, Knowles DM. Kaposi’s sarcoma-associated herpesvirus-like DNA sequences in AIDS-related body-cavity-based lymphomas. N Engl J Med. 1995;332(18):1186–1191. doi: 10.1056/NEJM199505043321802. [DOI] [PubMed] [Google Scholar]
- 11. Cesarman E, Moore PS, Rao PH, Inghirami G, Knowles DM, Chang Y. In vitro establishment and characterization of two acquired immunodeficiency syndrome-related lymphoma cell lines (BC-1 and BC-2) containing Kaposi’s sarcoma-associated herpesvirus-like (KSHV) DNA sequences. Blood. 1995;86(7):2708–2714. [PubMed] [Google Scholar]
- 12. Soulier J, et al. Kaposi’s sarcoma-associated herpesvirus-like DNA sequences in multicentric Castleman’s disease. Blood. 1995;86(4):1276–1280. [PubMed] [Google Scholar]
- 13. Renne R, Blackbourn D, Whitby D, Levy J, Ganem D. Limited transmission of Kaposi’s sarcoma-associated herpesvirus in cultured cells. J Virol. 1998;72(6):5182–5188. doi: 10.1128/jvi.72.6.5182-5188.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Blackbourn DJ, et al. The restricted cellular host range of human herpesvirus 8. AIDS. 2000;14(9):1123–1133. doi: 10.1097/00002030-200006160-00009. [DOI] [PubMed] [Google Scholar]
- 15. Rappocciolo G, et al. Human herpesvirus 8 infects and replicates in primary cultures of activated B lymphocytes through DC-SIGN. J Virol. 2008;82(10):4793–4806. doi: 10.1128/JVI.01587-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Pauk J, et al. Mucosal shedding of human herpesvirus 8 in men. N Engl J Med. 2000;343(19):1369–1377. doi: 10.1056/NEJM200011093431904. [DOI] [PubMed] [Google Scholar]
- 17. Webster-Cyriaque J, Duus K, Cooper C, Duncan M. Oral EBV and KSHV infection in HIV. Adv Dent Res. 2006;19(1):91–95. doi: 10.1177/154407370601900118. [DOI] [PubMed] [Google Scholar]
- 18. Vieira J, O’Hearn PM. Use of the red fluorescent protein as a marker of Kaposi’s sarcoma-associated herpesvirus lytic gene expression. Virology. 2004;325(2):225–240. doi: 10.1016/j.virol.2004.03.049. [DOI] [PubMed] [Google Scholar]
- 19. Myoung J, Ganem D. Infection of primary human tonsillar lymphoid cells by KSHV reveals frequent but abortive infection of T cells. Virology. doi: 10.1016/j.virol.2010.12.036. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kakimi K, et al. Blocking chemokine responsive to gamma-2/interferon (IFN)-gamma inducible protein and monokine induced by IFN-gamma activity in vivo reduces the pathogenetic but not the antiviral potential of hepatitis B virus-specific cytotoxic T lymphocytes. J Exp Med. 2001;194(12):1755–1766. doi: 10.1084/jem.194.12.1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Guidotti LG, Chisari FV. Noncytolytic control of viral infections by the innate and adaptive immune response. Annu Rev Immunol. 2001;19:65–91. doi: 10.1146/annurev.immunol.19.1.65. [DOI] [PubMed] [Google Scholar]
- 22. Cavanaugh VJ, Guidotti LG, Chisari FV. Interleukin-12 inhibits hepatitis B virus replication in transgenic mice. J Virol. 1997;71(4):3236–3243. doi: 10.1128/jvi.71.4.3236-3243.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ganem D, Prince AM. Hepatitis B virus infection--natural history and clinical consequences. N Engl J Med. 2004;350(11):1118–1129. doi: 10.1056/NEJMra031087. [DOI] [PubMed] [Google Scholar]
- 24. Scala G, Oppenheim JJ. Antigen presentation by human monocytes: evidence for stimulant processing and requirement for interleukin 1. J Immunol. 1983;131(3):1160–1166. [PubMed] [Google Scholar]
- 25. Moreno J, Lipsky PE. Differential ability of fixed antigen-presenting cells to stimulate nominal antigen-reactive and alloreactive T4 lymphocytes. J Immunol. 1986;136(10):3579–3587. [PubMed] [Google Scholar]
- 26. Chilson OP, Boylston AW, Crumpton MJ. Phaseolus vulgaris phytohaemagglutinin (PHA) binds to the human T lymphocyte antigen receptor. EMBO J. 1984;3(13):3239–3245. doi: 10.1002/j.1460-2075.1984.tb02285.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Chilson OP, Kelly-Chilson AE. Mitogenic lectins bind to the antigen receptor on human lymphocytes. Eur J Immunol. 1989;19(2):389–396. doi: 10.1002/eji.1830190225. [DOI] [PubMed] [Google Scholar]
- 28. Martro E, et al. Comparison of human herpesvirus 8 and Epstein-Barr virus seropositivity among children in areas endemic and non-endemic for Kaposi’s sarcoma. J Med Virol. 2004;72(1):126–131. doi: 10.1002/jmv.10548. [DOI] [PubMed] [Google Scholar]
- 29. Bejarano MT, Masucci MG, Ernberg I, Klein E, Klein G. Effect of cyclosporin-A (CsA) on the ability of T lymphocyte subsets to inhibit the proliferation of autologous EBV-transformed B cells. Int J Cancer. 1985;35(3):327–333. doi: 10.1002/ijc.2910350308. [DOI] [PubMed] [Google Scholar]
- 30. Bird AG, McLachlan SM, Britton S. Cyclosporin A promotes spontaneous outgrowth in vitro of Epstein-Barr virus-induced B-cell lines. Nature. 1981;289(5795):300–301. doi: 10.1038/289300a0. [DOI] [PubMed] [Google Scholar]
- 31. Shapiro RS, et al. Epstein-Barr virus associated B cell lymphoproliferative disorders following bone marrow transplantation. Blood. 1988;71(5):1234–1243. [PubMed] [Google Scholar]
- 32. Zutter MM, et al. Epstein-Barr virus lymphoproliferation after bone marrow transplantation. Blood. 1988;72(2):520–529. [PubMed] [Google Scholar]
- 33. Rickinson AB, Moss DJ, Pope JH. Long-term C-cell-mediated immunity to Epstein-Barr virus in man. II. Components necessary for regression in virus-infected leukocyte cultures. Int J Cancer. 1979;23(5):610–617. doi: 10.1002/ijc.2910230505. [DOI] [PubMed] [Google Scholar]
- 34. Rickinson AB, Moss DJ, Pope JH, Ahlberg N. Long-term T-cell-mediated immunity to Epstein-Barr virus in man. IV. Development of T-cell memory in convalescent infectious mononucleosis patients. Int J Cancer. 1980;25(1):59–65. doi: 10.1002/ijc.2910250108. [DOI] [PubMed] [Google Scholar]
- 35. Moss DJ, Rickinson AB, Pope JH. Long-term T-cell-mediated immunity to Epstein-Barr virus in man. III. Activation of cytotoxic T cells in virus-infected leukocyte cultures. Int J Cancer. 1979;23(5):618–625. doi: 10.1002/ijc.2910230506. [DOI] [PubMed] [Google Scholar]
- 36. Callan MF, et al. Large clonal expansions of CD8+ T cells in acute infectious mononucleosis. Nat Med. 1996;2(8):906–911. doi: 10.1038/nm0896-906. [DOI] [PubMed] [Google Scholar]
- 37. Gudgeon NH, Taylor GS, Long HM, Haigh TA, Rickinson AB. Regression of Epstein-Barr virus-induced B-cell transformation in vitro involves virus-specific CD8+ T cells as the principal effectors and a novel CD4+ T-cell reactivity. J Virol. 2005;79(9):5477–5488. doi: 10.1128/JVI.79.9.5477-5488.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Levy JA, Mackewicz CE, Barker E. Controlling HIV pathogenesis: the role of the noncytotoxic anti-HIV response of CD8+ T cells. Immunol Today. 1996;17(5):217–224. doi: 10.1016/0167-5699(96)10011-6. [DOI] [PubMed] [Google Scholar]
- 39. Mackewicz CE, Blackbourn DJ, Levy JA. CD8+ T cells suppress human immunodeficiency virus replication by inhibiting viral transcription. Proc Natl Acad Sci U S A. 1995;92(6):2308–2312. doi: 10.1073/pnas.92.6.2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Mackewicz CE, Ortega H, Levy JA. Effect of cytokines on HIV replication in CD4+ lymphocytes: lack of identity with the CD8+ cell antiviral factor. Cell Immunol. 1994;153(2):329–343. doi: 10.1006/cimm.1994.1032. [DOI] [PubMed] [Google Scholar]
- 41. Levy JA. The search for the CD8+ cell anti-HIV factor (CAF). Trends Immunol. 2003;24(12):628–632. doi: 10.1016/j.it.2003.10.005. [DOI] [PubMed] [Google Scholar]
- 42. Guidotti LG, et al. Viral cross talk: intracellular inactivation of the hepatitis B virus during an unrelated viral infection of the liver. Proc Natl Acad Sci U S A. 1996;93(10):4589–4594. doi: 10.1073/pnas.93.10.4589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Cavanaugh VJ, Guidotti LG, Chisari FV. Inhibition of hepatitis B virus replication during adenovirus and cytomegalovirus infections in transgenic mice. J Virol. 1998;72(4):2630–2637. doi: 10.1128/jvi.72.4.2630-2637.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Heise T, Guidotti LG, Cavanaugh VJ, Chisari FV. Hepatitis B virus RNA-binding proteins associated with cytokine-induced clearance of viral RNA from the liver of transgenic mice. J Virol. 1999;73(1):474–481. doi: 10.1128/jvi.73.1.474-481.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Heise T, Guidotti LG, Chisari FV. Characterization of nuclear RNases that cleave hepatitis B virus RNA near the La protein binding site. J Virol. 2001;75(15):6874–6883. doi: 10.1128/JVI.75.15.6874-6883.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Decker LL, et al. The Kaposi sarcoma-associated herpesvirus (KSHV) is present as an intact latent genome in KS tissue but replicates in the peripheral blood mononuclear cells of KS patients. J Exp Med. 1996;184(1):283–288. doi: 10.1084/jem.184.1.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Monini P, et al. Reactivation and persistence of human herpesvirus-8 infection in B cells and monocytes by Th-1 cytokines increased in Kaposi’s sarcoma. Blood. 1999;93(12):4044–4058. [PubMed] [Google Scholar]
- 48. Dupin N, et al. Distribution of human herpesvirus-8 latently infected cells in Kaposi’s sarcoma, multicentric Castleman’s disease, and primary effusion lymphoma. Proc Natl Acad Sci U S A. 1999;96(8):4546–4551. doi: 10.1073/pnas.96.8.4546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Staskus KA, et al. Cellular tropism and viral interleukin-6 expression distinguish human herpesvirus 8 involvement in Kaposi’s sarcoma, primary effusion lymphoma, and multicentric Castleman’s disease. J Virol. 1999;73(5):4181–4187. doi: 10.1128/jvi.73.5.4181-4187.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Parravicini C, et al. Differential viral protein expression in Kaposi’s sarcoma-associated herpesvirus-infected diseases: Kaposi’s sarcoma, primary effusion lymphoma, and multicentric Castleman’s disease. Am J Pathol. 2000;156(3):743–749. doi: 10.1016/S0002-9440(10)64940-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Penn I. Sarcomas in organ allograft recipients. Transplantation. 1995;60(12):1485–1491. doi: 10.1097/00007890-199560120-00020. [DOI] [PubMed] [Google Scholar]
- 52. Farge D, et al. Human herpes virus-8 and other risk factors for Kaposi’s sarcoma in kidney transplant recipients. Groupe Cooperatif de Transplantation d’ Ile de France (GCIF). Transplantation. 1999;67(9):1236–1242. doi: 10.1097/00007890-199905150-00007. [DOI] [PubMed] [Google Scholar]
- 53. Cattaneo D, Gotti E, Perico N, Bertolini G, Kainer G, Remuzzi G. Cyclosporine formulation and Kaposi’s sarcoma after renal transplantation. Transplantation. 2005;80(6):743–748. doi: 10.1097/01.tp.0000173803.97398.31. [DOI] [PubMed] [Google Scholar]
- 54. Martin DF, Kuppermann BD, Wolitz RA, Palestine AG, Li H, Robinson CA. Oral ganciclovir for patients with cytomegalovirus retinitis treated with a ganciclovir implant. Roche Ganciclovir Study Group. N Engl J Med. 1999;340(14):1063–1070. doi: 10.1056/NEJM199904083401402. [DOI] [PubMed] [Google Scholar]
- 55. Engels EA, et al. Detection and quantification of Kaposi’s sarcoma-associated herpesvirus to predict AIDS-associated Kaposi’s sarcoma. AIDS. 2003;17(12):1847–1851. doi: 10.1097/00002030-200308150-00015. [DOI] [PubMed] [Google Scholar]
- 56. Quinlivan EB, Zhang C, Stewart PW, Komoltri C, Davis MG, Wehbie RS. Elevated virus loads of Kaposi’s sarcoma-associated human herpesvirus 8 predict Kaposi’s sarcoma disease progression, but elevated levels of human immunodeficiency virus type 1 do not. J Infect Dis. 2002;185(12):1736–1744. doi: 10.1086/340652. [DOI] [PubMed] [Google Scholar]
- 57. Cannon MJ, et al. Blood-borne and sexual transmission of human herpesvirus 8 in women with or at risk for human immunodeficiency virus infection. N Engl J Med. 2001;344(9):637–643. doi: 10.1056/NEJM200103013440904. [DOI] [PubMed] [Google Scholar]
- 58. Eckstein DA, et al. HIV-1 actively replicates in naive CD4(+) T cells residing within human lymphoid tissues. Immunity. 2001;15(4):671–682. doi: 10.1016/S1074-7613(01)00217-5. [DOI] [PubMed] [Google Scholar]
- 59. Kreisberg JF, Yonemoto W, Greene WC. Endogenous factors enhance HIV infection of tissue naive CD4 T cells by stimulating high molecular mass APOBEC3G complex formation. J Exp Med. 2006;203(4):865–870. doi: 10.1084/jem.20051856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Yoo SM, et al. Centrifugal enhancement of Kaposi’s sarcoma-associated virus infection of human endothelial cells in vitro. J Virol Methods. 2008;154(1–2):160–166. doi: 10.1016/j.jviromet.2008.07.026. [DOI] [PubMed] [Google Scholar]
- 61. Grossmann C, Ganem D. Effects of NFkappaB activation on KSHV latency and lytic reactivation are complex and context-dependent. Virology. 2008;375(1):94–102. doi: 10.1016/j.virol.2007.12.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Zhou FC, et al. Efficient infection by a recombinant Kaposi’s sarcoma-associated herpesvirus cloned in a bacterial artificial chromosome: application for genetic analysis. J Virol. 2002;76(12):6185–6196. doi: 10.1128/JVI.76.12.6185-6196.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Chandriani S, Ganem D. Array-based transcript profiling and limiting-dilution reverse transcription-PCR analysis identify additional latent genes in Kaposi’s sarcoma-associated herpesvirus. J Virol. 2010;84(11):5565–5573. doi: 10.1128/JVI.02723-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.