BAG3 mediates chaperone-based aggresome-targeting and selective autophagy of misfolded proteins
Behl and colleagues report a novel cellular pathway utilizing the specificity of Hsp70 towards misfolded proteins as a basis for aggresome-targeting and autophagic degradation and find that this pathway is regulated by the co-chaperone BAG3.
Keywords: ALS, BAG3, dynein, p62, ubiquitin
Abstract
Increasing evidence indicates the existence of selective autophagy pathways, but the manner in which substrates are recognized and targeted to the autophagy system is poorly understood. One strategy is transport of a particular substrate to the aggresome, a perinuclear compartment with high autophagic activity. In this paper, we identify a new cellular pathway that uses the specificity of heat-shock protein 70 (Hsp70) to misfolded proteins as the basis for aggresome-targeting and autophagic degradation. This pathway is regulated by the stress-induced co-chaperone Bcl-2-associated athanogene 3 (BAG3), which interacts with the microtubule-motor dynein and selectively directs Hsp70 substrates to the motor and thereby to the aggresome. Notably, aggresome-targeting by BAG3 is distinct from previously described mechanisms, as it does not depend on substrate ubiquitination.
Introduction
Macroautophagy (referred to as autophagy) is a catabolic process by which cytoplasmic material becomes sequestered in a vesicle—the autophagosome—and shuttled to lysosomes for degradation (Klionsky et al, 2010). Selective autophagy of damaged proteins requires the concentration and isolation of substrates from other cytoplasmic components. Such sequestration occurs in a perinuclear compartment called the aggresome, by retrograde transport of substrates along microtubules through the cytoplasmic dynein motor complex (Kopito, 2000; Johnston et al, 2002). Cargo loading on to the motor necessitates a high specificity to selectively remove misfolded proteins by the aggresome-autophagy pathway. Specific aggresome-targeting is usually achieved by ubiquitination of substrates, which are then recognized by ubiquitin adaptor proteins that bind to dynein (Kawaguchi et al, 2003). However, many misfolded proteins found in aggresomes are not ubiquitinated (García-Mata et al, 1999), suggesting that selective cargo loading onto dynein also occurs independently of ubiquitin signalling. Here, we describe the identification of a new chaperone-based aggresome-targeting mechanism controlled by the heat-shock protein 70 (Hsp70) co-chaperone Bcl-2-associated athanogene 3 (BAG3), which was recently implicated in selective autophagy (Gamerdinger et al, 2009; Arndt et al, 2010).
Results And Discussion
We examined the BAG3-mediated selective autophagy pathway after proteasome inhibition, a well-known trigger for autophagy activation. Proteasomal impairment induced the upregulation of both BAG3 and its binding partner Hsp70 (Fig 1A). Other BAG family members were not induced (Fig 1A; supplementary Fig S1A online), suggesting a specific role for BAG3 in this stress response. Interestingly, BAG3 localized to juxtanuclear inclusions that were also enriched in ubiquitinated proteins and vimentin (Fig 1C; supplementary Fig S1E,F online). These structures resemble aggresomes, which sequester misfolded proteins by retrograde transport of protein aggregates along microtubules (Kopito, 2000). Accordingly, the microtubule-depolymerizing drug vinblastine completely suppressed the assembly of the BAG3-positive perinuclear structure (Fig 1B,C), although BAG3 was still upregulated by the proteasome inhibitor (Fig 1A). Instead, several BAG3- and ubiquitin-positive microaggregates were dispersed throughout the cytoplasm (Fig 1B,C; supplementary Fig S1F online). These findings indicate that BAG3 is actively transported with misfolded proteins on microtubules to the aggresome.
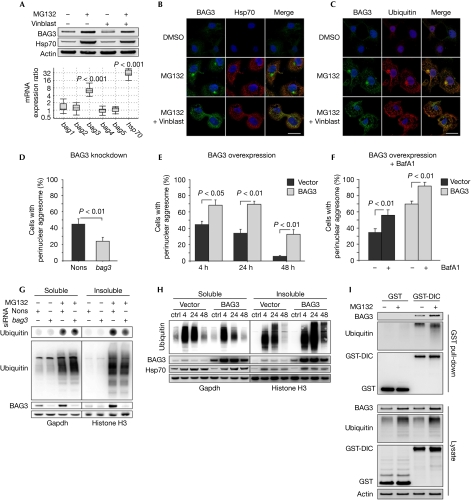
Figure 1.
BAG3 facilitates aggresome formation. (A) Upper panel: immunoblot analysis of COS7 cells treated for 12 h with MG132 (25 μM) and vinblastine (25 μM). Lower panel: real-time PCR analysis showing gene expression changes in COS7 cells following MG132 exposure (25 μM, 12 h). (B,C) Immunofluorescence staining of indicated proteins in COS7 cells treated as in (A). Scale bar, 20 μm. (D) COS7 cells were transfected for 24 h with nonsense or bag3 siRNA and treated with MG132 (25 μM, 12 h). Aggresomes were visualized by ubiquitin immunostaining (supplementary Fig S1D online). Diagram shows the percentage of aggresome-positive cells. (E) Aggresome quantification as in supplementary Fig S1D online from cells analysed in (H). Images are shown in supplementary Fig S1G online. Diagram shows the percentage of aggresome-positive cells at 4, 24 and 48 h after MG132 wash out. (F) Cells were treated as in (H); however, 4 h after MG132 wash out, BafA1 (5 μM) was added for 20 h to inhibit aggresome clearance. Diagrams show the percentage of aggresome-positive cells and the mean size of aggresomes (supplementary Fig S1H online). Images are shown in supplementary Fig S1G online. (G) Dot-blot (upper panel, quantification in supplementary Fig S1B online) and western blot (lower panel) analyses of 0.5% NP40-soluble and -insoluble proteins from cells shown in Fig 1D; supplementary Fig S1D online. (H) COS7 cells transfected with BAG3 or empty vector were treated with either MG132 (25 μM, 18 h) or DMSO as a control. After MG132 wash out, cells were allowed to recover for 4, 24 and 48 h and 0.5% NP40-soluble and -insoluble proteins were analysed by immunoblotting. (I) COS7 cells were transfected for 24 h with GST or GST fused to DIC (GST-DIC) and treated with MG132 (25 μM, 12 h). Indicated proteins in whole-cell lysates and in precipitates obtained by GST pull-down with glutathione beads were analysed by immunoblotting. BafA1, bafilomycin A1; ctrl, control; DIC, dynein intermediate chain; DMSO, dimethyl sulphoxide; GST, glutathione-S-transferase; Hsp70, heat-shock protein 70; nons, nonsense; siRNA, short-interfering RNA; vinblast, vinblastine.
To investigate the function of BAG3 in the aggresome pathway, BAG3 levels were modulated and aggresome formation was analysed microscopically after proteasomal inhibition. Formation of ubiquitin-positive aggresomes was decreased in BAG3 knockdown cells (Fig 1D; supplementary Fig S1D online) and increased when BAG3 was overexpressed (Fig 1E; supplementary Fig S1G online). In addition, we measured the cellular aggresome content on the basis of the solubility of ubiquitinated proteins, which become partly insoluble in mild detergent solvents if sequestered in aggresomes. Consistent with a reduced aggresome assembly in BAG3 knockdown cells, fewer ubiquitinated proteins became insoluble after proteasomal inhibition (Fig 1G; supplementary Fig S1B,C online). Conversely, BAG3 overexpression decreased the solubility of ubiquitinated proteins during the recovery from proteasome stress; this became particularly apparent at later time points, when endogenous BAG3 returned to basal levels in control cells (Fig 1H).
The higher aggresome content in BAG3-overexpressing cells could have resulted from either an increased assembly or a decreased clearance. To address this issue, autophagy was inhibited with bafilomycin A1 (BafA1) during the recovery period to suppress aggresome clearance. Autophagy inhibition raised the percentage of cells containing aggresomes in BAG3-overexpressing cells in a manner comparable with that of controls (Fig 1F), and contributed to a significant increase in the mean aggresomal size (supplementary Fig S1H online). These findings exclude a clearance defect and show that overexpression of BAG3 promotes the sequestration of ubiquitinated proteins in aggresomes.
Aggresome-targeting of substrates involves their coupling to the dynein motor for transport along microtubules (Johnston et al, 2002). To test whether BAG3 associates with dynein, we used a pull-down assay based on the glutathione-S-transferase (GST)-tagged dynein intermediate chain (GST-DIC), which efficiently incorporates into the cytoplasmic dynein complex (supplementary Fig S2A online; Ström et al, 2008). These analyses showed that endogenous BAG3 associates with dynein, along with ubiquitinated proteins (Fig 1I). Moreover, endogenous DIC was detected in immunoprecipitated BAG3 complexes (supplementary Fig S2B online). These results are consistent with the conclusion that BAG3 is transported with ubiquitinated substrates to the aggresome by dynein.
BAG proteins bind with high affinity to the ATPase domain of Hsp70 through the BAG domain, thereby inducing nucleotide exchange on Hsp70 and the release of the chaperone-bound substrate (Sondermann et al, 2001). Hence, it is possible that BAG3 couples chaperoned substrates to the dynein motor complex, thereby promoting aggresome assembly. To investigate this, we constructed FLAG-tagged BAG3 deletion mutants with different binding abilities towards dynein and Hsp70. BAG3 contains two WW domains, followed by a proline-rich repeat (PxxP) and the BAG domain (Fig 2A). As expected, deletion of the BAG domain completely abrogated Hsp70 binding (supplementary Fig S2C online), but only had a small effect on the interaction of BAG3 with dynein (Fig 2A). The BAG domain alone failed to interact with dynein (Fig 2A), indicating that other regions of BAG3 are crucial for motor-complex binding. Further binding studies showed that the PxxP region, located upstream from the BAG domain, is an important determinant of BAG3–dynein interaction. This is supported by the fact that the PxxP region restored dynein interaction of the BAG domain in PxxP–BAG domain fusion peptides (Fig 2A).
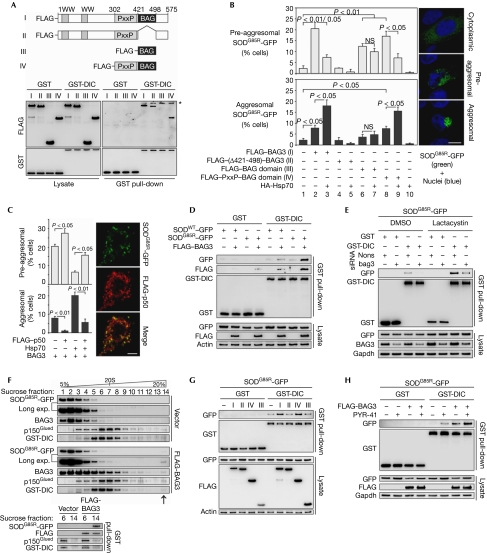
Figure 2.
BAG3 specifically directs Hsp70 substrates to the aggresome by loading misfolded proteins on to dynein. (A) GST pull-down analysis as in Fig 1I, but performed in cells transfected for 36 h with full-length FLAG-BAG3 (I) or the BAG3 deletion mutants FLAG–(Δ421–498)–BAG3 (II), FLAG–BAG-domain (III), and FLAG–PxxP–BAG-domain (IV). Asterisk, nonspecific band. (B) HEK cells were transfected for 18 h with SODG85R-GFP, along with the indicated FLAG-tagged BAG3 constructs (shown in A) and HA-tagged Hsp70 (HA-Hsp70). SODG85R-GFP showed three distinct localization patterns: cytoplasmic, pre-aggresomal and aggresomal (see representative pictures on the right and supplementary Fig S2F online). The percentages of cells bearing pre-aggresomal (upper panel) and aggresomal (lower panel) SODG85R-GFP are shown. Scale bar, 10 μm. (C) HEK cells transfected with SODG85R-GFP, BAG3 and Hsp70 were additionally transfected with FLAG-p50, as indicated. Diagrams show the percentages of cells with pre-aggresomal and aggresomal SODG85R-GFP. Right panel: representative image showing pre-aggresomal localization of FLAG-p50 and SODG85R-GFP. Scale bar, 5 μm. (D) GST pull-down analysis as in (A), but in HEK cells transfected for 18 h with SODWT-GFP or mutant SODG85R-GFP, along with FLAG-BAG3. (E) HEK cells were transfected with the indicated plasmids with nonsense or bag3 siRNA. At 6 h after transfection, lactacystin (10 μM) or DMSO as vehicle control was added for 12 h and GST pull-down analysis was performed as in (D). (F) Upper panel: sucrose-density gradient analysis of extracts from HEK cells transfected with SODG85R-GFP and GST-DIC, along with FLAG-BAG3 or vector control. Note the immunoreactive bands in high-density fraction 14 on BAG3 overexpression (arrow). Migration of a 20S marker protein is indicated. Lower panel: GST-DIC pull-down analyses of dynein complexes present in sucrose fraction 6 and 14. (G) GST pull-down analysis of SODG85R-GFP as in (D) but in cells transfected with FLAG-tagged BAG3 constructs (I–IV; indicated as in A). (H) GST pull-down analysis of SODG85R-GFP as in (D) but in cells treated with PYR41 (10 μM; 12 h), as indicated. DIC, dynein intermediate chain; DMSO, dimethyl sulphoxide; GST, glutathione-S-transferase; HA, haemagglutinin; Hsp70, heat-shock protein 70; NS, not significant; nons, nonsense; PxxP, proline-rich repeat; siRNA, short-interfering RNA; SOD, superoxide dismutase; SODWT, wild-type SOD.
To analyse whether dynein- and Hsp70-binding functions have a role in BAG3-mediated aggresome-targeting, we used amyotrophic-lateral-sclerosis-linked mutant superoxide dismutase (SOD)1G85R (mtSOD) fused to GFP, a well-known Hsp70 substrate (Wang et al, 2009), as a model protein. MtSOD expressed in human embryonic kidney (HEK) cells showed cytoplasmic, pre-aggresomal and aggresomal localization patterns (Fig 2B). BAG3 overexpression increased several cells showing pre-aggresomal and aggresomal mtSOD localization (Fig 2B). Interestingly, aggresome-targeting of mtSOD increased and pre-aggresomal structures decreased when BAG3 was expressed along with Hsp70. Hsp70 overexpression alone did not induce, but rather suppressed, aggresome formation (Fig 2B; supplementary Fig S2F online). BAG3/Hsp70 had no effect on wild-type SOD1 (SODWT), which exclusively showed cytoplasmic localization (supplementary Fig S2D online). These data indicate that BAG3 specifically directs Hsp70 substrates to the aggresome. Consistent with this conclusion, deletion of the BAG domain completely removed the aggresome-targeting activity of BAG3. Overexpression of the BAG domain alone failed to sequester mtSOD in aggresomes, but an increase in pre-aggresomal structures was observed (Fig 2B; supplementary Fig S2F online). These data indicate that the BAG domain alone contributed to local mtSOD aggregation by inhibiting Hsp70-folding reactions, but failed to stimulate the transport of protein aggregates to the aggresome. Thus, the BAG domain in BAG3 is necessary but not sufficient for the aggresome-targeting of Hsp70 substrates. By contrast, the BAG3 peptide composed of the BAG domain and the PxxP motif was sufficient to target mtSOD to aggresomes, although in a less effective manner than full-length BAG3 (Fig 2B; supplementary Fig S2F online).
Our data indicate that mtSOD is specifically directed to the aggresome pathway through the cooperative function of BAG3 and Hsp70. To test whether this involves active transport of mtSOD by dynein, we overexpressed p50, a competitive dynein inhibitor that stalls cargo from the motor (García-Mata et al, 1999). Overexpression of p50 strongly suppressed the aggresome-targeting of mtSOD by BAG3/Hsp70 (Fig 2C). Instead, mtSOD localized with p50 in pre-aggresomal structures (Fig 2C). These data show that mtSOD targeted to the aggresome is indeed transported by the dynein motor. To examine this further, we directly analysed the interaction of mtSOD with dynein. Strikingly, BAG3 overexpression strongly promoted the association of mtSOD, but not of SODWT with dynein (Fig 2D), suggesting that BAG3 specifically targets Hsp70 substrates to the motor complex. The interaction of BAG3 with dynein was enhanced in the presence of mutant substrates (Fig 2D), indicating that BAG3 is captured during the loading of misfolded proteins on to the motor and therefore found concentrated along with mtSOD in aggresomes (supplementary Fig S2F online). These results indicate that mtSOD binds to dynein in a BAG3-dependent manner. Accordingly, the mtSOD–dynein interaction was impaired in BAG3 knockdown cells (Fig 2E). Analysis of protein complexes separated by sucrose density gradient centrifugation showed that BAG3 overexpression induces the formation of high-molecular dynein complexes that are loaded with mtSOD as cargo. BAG3 was found to associate with both the cargo-loaded (fraction 14) and unloaded (fraction 6) dynein complexes (Fig 2F). These data are consistent with the model that BAG3 loads misfolded proteins onto dynein by coupling the release of Hsp70-bound substrates to the motor complex. Accordingly, the Hsp70-binding deficient BAG3 mutant—which shows residual dynein-binding capacity (Fig 2A) but no aggresome-targeting activity (Fig 2B; supplementary Fig S2F online)—was inefficient in loading mtSOD on to dynein (Fig 2G). Similarly, the BAG domain alone—which is inefficient in dynein binding (Fig 2A) and induces only pre-aggresomal structures (Fig 2B; supplementary Fig S2F online)—failed to load significant amounts of cargo onto the motor (Fig 2G). However, cargo loading was restored when the PxxP region was added to the BAG domain (Fig 2G), which is consistent with the finding that the PxxP motif in BAG3 is crucial for dynein binding (Fig 2A) and thus for the aggresome-targeting of mtSOD (Fig 2B; supplementary Fig S2F online). Together, these data indicate that BAG3 specifically directs misfolded proteins to the aggresome pathway by loading Hsp70 substrates on to the dynein motor complex. Remarkably, the absence of a characteristic molecular weight shift for ubiquitinated proteins in western blotting analyses (Fig 2D,E,G) suggested that mtSOD coupled to dynein by BAG3 was not ubiquitinated. Moreover, BAG3-mediated targeting of mtSOD to dynein was not reduced under ubiquitination-deficient conditions triggered by the E1 inhibitor PYR41 (Fig 2H). A dose of PYR41 that was sufficient to block the ubiquitination and degradation of a GFP-based proteasome reporter (supplementary Fig S2E online) strengthened the mtSOD–dynein interaction (Fig 2H), as the steady-state levels of mtSOD increased, probably because of impaired proteasomal degradation. These data indicate that BAG3-controlled aggresome targeting is distinct from the ubiquitin-dependent mechanisms described previously (Kawaguchi et al, 2003).
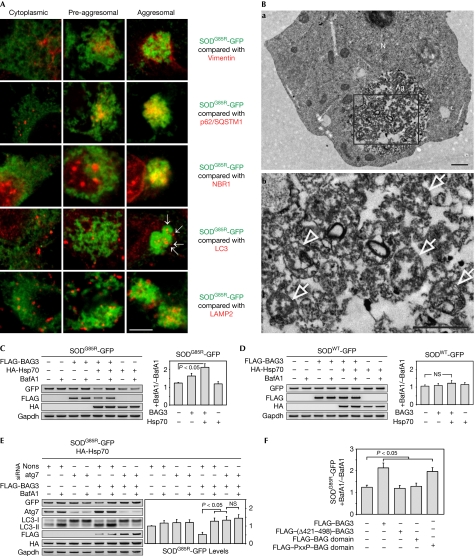
MtSOD in aggresomes was completely surrounded by a filament network composed of vimentin (Fig 3A; supplementary Fig S3A online). This isolation could be the basis for a selective degradation of mtSOD by autophagy. Consistent with the high autophagic activity of aggresomes (Kopito, 2000), we found that, specifically within this compartment, mtSOD localized with the autophagy receptors p62/SQSTM1 and NBR1 (Fig 3A; supplementary Fig S3B,C online), which are known to recruit the autophagic machinery (Kirkin et al, 2009). Accordingly, aggresomal mtSOD was extensively decorated by autophagosomal and lysosomal markers (Fig 3A; supplementary Fig S3D,E online). Transmission electron microscopic analysis of aggresomes confirmed the presence of several autophagic vesicles that enclose the surrounding granular material (Fig 3B), indicating the autophagic degradation of mtSOD in aggresomes. To analyse this further, we treated cells with BafA1 to block lysosomal degradation. This treatment led to increased levels of mtSOD, but not of SODWT in BAG3-transfected cells, particularly when BAG3 was overexpressed with Hsp70 (Fig 3C,D), correlating with the aggresome-targeting activity (Fig 2B). Moreover, genetic inhibition of autophagy by Atg7 knockdown also increased the steady-state levels of mtSOD in BAG3/Hsp70-transfected cells (Fig 3E). BafA1 treatment in autophagy-deficient cells no longer promoted the accumulation of mtSOD (Fig 3E), supporting the idea that BafA1 is an autophagy inhibitor in the analysed mtSOD degradation pathway. BAG3 deletion mutants deficient in aggresome-targeting were also inefficient in promoting autophagic degradation of mtSOD, in contrast to the dynein-binding and aggresome-targeting active PxxP–BAG domain peptide (Fig 3F; supplementary Fig S3F online). These data indicate that misfolded proteins targeted by BAG3/Hsp70 to the aggresome are subject to autophagic degradation.
Figure 3.
Aggresome-targeted proteins are degraded by autophagy. (A) Analysis of SODG85R-GFP (green) localization with aggresome marker vimentin, the autophagic receptors p62 and NBR1, the autophagosome marker LC3 and the lysosomal marker LAMP2 (all in red) in BAG3/Hsp70-transfected HEK cells. Representative merged images of cells with cytoplasmic, pre-aggresomal and aggresomal SOD distribution are shown. Arrows indicate LC3-positive autophagosomes. Scale bar, 5 μm. (B) Transmission electron microscope analysis of SODG85R-GFP aggresome in cells transfected as in (A). Marked area in (a) is shown magnified in (b). Arrows, autophagosomes. Arrowheads, autolysosomes. Scale bar, 1 μm. (C) HEK cells transfected for 8 h with SODG85R-GFP along with FLAG-BAG3 and HA-Hsp70, as indicated, were treated for 24 h with BafA1 (0.1 μM). Indicated proteins were analysed by immunoblotting. Diagram shows the ratio of SODG85R-GFP levels in BafA1-treated cells to those in untreated cells. (D) Same analysis as in (C) but with SODWT-GFP. (E) Similar analysis as in (C) but in cells transfected 24 h before with nonsense or atg7 siRNA. (F) HEK cells were transfected with SODG85R-GFP and indicated FLAG-tagged BAG3 constructs together with HA-Hsp70, and lysosomal degradation of SODG85R-GFP was examined as in (C). Corresponding immunoblots are shown in supplementary Fig S3F online. Ag, aggresome; BafA1, bafilomycin A1; GFP, green fluorescent protein; HA, haemagglutinin; Hsp70, heat-shock protein 70; N, nucleus; NS, not significant; nons, nonsense; PxxP, proline-rich repeat; siRNA, short interfering RNA; SOD, superoxide dismutase; SODWT, wild-type SOD.
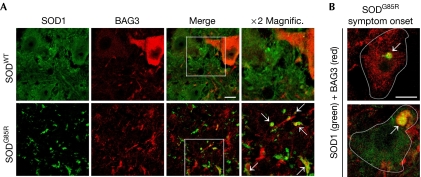
Our data reveal a new chaperone-based mechanism in cultured cells that specifically targets misfolded proteins to the aggresome pathway. To examine whether this pathway is relevant in vivo in mouse models of protein misfolding diseases, we analysed mice transgenic for SODWT and SODG85R. Mice overexpressing SODG85R develop an amyotrophic lateral-sclerosis-like neuropathology that is associated with inclusion body formation in motor neurons of the spinal cord (Johnston et al, 2000). In the spinal cord of SODWT mice, motor neurons generally showed a weak BAG3 expression; however, in some motor neurons, BAG3 expression was elevated (Fig 4A). Although the basis for the heterogeneous BAG3 expression in SODWT mice remains to be explored, it shows that BAG3 can be upregulated in motor neurons in certain conditions. In contrast to SODWT mice, SODG85R mice at disease end stage showed several SOD1-positive aggregates in the spinal cord neuropil that were also positive for BAG3 (Fig 4A). To analyse whether BAG3 localizes with mtSOD within motor neurons, we analysed SODG85R mice at symptom onset. In these mice, surviving spinal cord motor neurons were detected showing SOD1- and BAG3-positive perinuclear inclusions of different sizes (Fig 4B). As previously reported in similar mouse models (Johnston et al, 2000), these structures probably resemble aggresomes, suggesting that the BAG3-mediated aggresome pathway is induced. Accordingly, BAG3 expression was higher in the spinal cords of two mtSOD transgenic mouse models at end stage (SODG93A and SODG85R, supplementary Fig S3G online). Moreover, when spinal cord homogenates were fractionated by differential centrifugation, BAG3 was found, together with mtSOD and ubiquitinated proteins, in the insoluble fraction (supplementary Fig S3G online). Interestingly, the autophagy receptor p62/SQSTM1 and the lysosomal marker LAMP1 were also increased in this fraction (supplementary Fig S3G online), indicating a possible role for autophagy in the removal of these aggregates.
Figure 4.
BAG3 associates with aggresomes in vivo. (A) Spinal cord sections of mice overexpressing SODWT and SODG85R (disease end-stage) were immunostained for BAG3 and SOD1. Arrows indicate SOD1- and BAG3-positive aggregates in the neuropil. Scale bar, 20 μm. (B) Localization analysis of BAG3 and SODG85R in surviving spinal cord motor neurons from SODG85R mice at the time of disease symptom onset. Arrows indicate perinuclear aggresomes containing BAG3 and SODG85R. Scale bar, 10 μm. Magnific., magnification; SOD, superoxide dismutase; SODWT, wild-type SOD.
Conclusion
This study identifies a new aggresome-targeting pathway in cellular and in vivo models that uses the specificity of Hsp70 chaperones to misfolded proteins as the basis for selectivity. The main factor of this pathway is the stress-induced co-chaperone BAG3 that couples the release of Hsp70-bound substrates to the dynein motor complex, thereby mediating aggresome-targeting and autophagic degradation of chaperoned substrates. Importantly, chaperone-based aggresome-targeting by BAG3 is distinct from previously described mechanisms, as here, substrate ubiquitination does not seem to be necessary for the selective transfer of misfolded proteins to the aggresome.
Methods
Standard methods. Cell transfection, PCR, transmission electron microscopy, immunocytochemistry, immunoprecipitation and immunoblotting were carried out as described previously (Gamerdinger et al, 2007, 2009). Laser-scanning microscopy was performed using an LSM 710 microscope (Carl Zeiss). Plasmids and antibodies are described in supplementary information online.
Quantification of aggresomes. COS7 cells were immunostained and recorded as described (Gamerdinger et al, 2009). Aggresome-positive cells were counted in five random view fields containing 50–150 cells in three independent experiments. The mean aggresomal size of aggresomes was determined with Image J software using the particle analyser plug-in. In Fig 1E,F,H and supplementary Fig S1H online, cells were growth arrested by aphidicolin (5 μM) to exclude potential differential dilution of aggresomes. SODG85R-GFP-positive aggresomes and pre-aggresomal structures were directly quantified in living fluorescent HEK cells using the Axiovert 200 microscope (Carl Zeiss). Ten random view fields containing 50–100 cells were counted in three independent experiments.
Dynein GST pull-down analysis. GST-DIC pull-down was carried out as described previously (Ström et al, 2008), with some modifications (see supplementary information online).
Sucrose-density gradient analysis. Cell extracts were separated on a 2 ml linear 5–20% sucrose-density gradient by ultracentrifugation (55,000 r.p.m., 2 h, 4°C) in a TLS55 rotor (BeckmannCoulter). Fourteen fractions (150 μl) were collected and volumes of 10 μl and 50 μl of each fraction were subjected directly to immunoblot and GST pull-down analysis, respectively. Detection of the 20S proteasome complex acted as the 20S marker.
In vivo analyses. Details of animals used and the tissue extraction procedure were reported previously (Witan et al, 2009). Immunohistochemistry of spinal cord sections is described in the supplementary information online.
Statistics. Statistical significance was determined by Student's t-test. Every experiment was performed 3–5 times and the results are expressed as mean±s.e.m.
Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Supplementary Material
Acknowledgments
We are grateful to Heidrun Witan for cloning SOD1 plasmids. We thank Elisabeth Sehn for her excellent technical assistance in the transmission electron microscopy studies. A.M.K. is a member of the Neuroscience Graduate School of Mainz (DFG GRK1044). This work was supported by the Interdisziplinärer Forschungsschwerpunkt Neurowissenschaften (IFSN) of the University of Mainz (A.M.C.) and by grants from the Fritz-and-Hildegard-Berg-Foundation of the Stifterverband and the Bragard-Foundation to C.B.
Footnotes
The authors declare that they have no conflict of interest.
References
- Arndt V et al. (2010) Chaperone-assisted selective autophagy is essential for muscle maintenance. Curr Biol 20: 143–148 [DOI] [PubMed] [Google Scholar]
- Gamerdinger M, Clement AB, Behl C (2007) Cholesterol-like effects of selective cyclooxygenase inhibitors and fibrates on cellular membranes and amyloid-beta production. Mol Pharmacol 72: 141–151 [DOI] [PubMed] [Google Scholar]
- Gamerdinger M, Hajieva P, Kaya AM, Wolfrum U, Hartl FU, Behl C (2009) Protein quality control during aging involves recruitment of the macroautophagy pathway by BAG3. EMBO J 28: 889–901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Mata R, Bebök Z, Sorscher EJ, Sztul ES (1999) Characterization and dynamics of aggresome formation by a cytosolic GFP-chimera. J Cell Biol 146: 1239–1254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston JA, Dalton MJ, Gurney ME, Kopito RR (2000) Formation of high molecular weight complexes of mutant Cu, Zn-superoxide dismutase in a mouse model for familial amyotrophic lateral sclerosis. Proc Natl Acad Sci USA 97: 12571–12576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston JA, Illing ME, Kopito RR (2002) Cytoplasmic dynein/dynactin mediates the assembly of aggresomes. Cell Motil Cytoskeleton 53: 26–38 [DOI] [PubMed] [Google Scholar]
- Kawaguchi Y, Kovacs JJ, McLaurin A, Vance JM, Ito A, Yao TP (2003) The deacetylase HDAC6 regulates aggresome formation and cell viability in response to misfolded protein stress. Cell 115: 727–738 [DOI] [PubMed] [Google Scholar]
- Kirkin V, McEwan DG, Novak I, Dikic I (2009) A role for ubiquitin in selective autophagy. Mol Cell 34: 259–269 [DOI] [PubMed] [Google Scholar]
- Klionsky DJ et al. (2010) A comprehensive glossary of autophagy-related molecules and processes. Autophagy 6: 438–448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopito RR (2000) Aggresomes, inclusion bodies and protein aggregation. Trends Cell Biol 10: 524–530 [DOI] [PubMed] [Google Scholar]
- Sondermann H, Scheufler C, Schneider C, Hohfeld J, Hartl FU, Moarefi I (2001) Structure of a Bag/Hsc70 complex: convergent functional evolution of Hsp70 nucleotide exchange factors. Science 291: 1553–1557 [DOI] [PubMed] [Google Scholar]
- Ström A, Shi P, Zhang F, Gal J, Kilty R, Hayward LJ, Zhu H (2008) Interaction of amyotrophic lateral sclerosis (ALS)-related mutant copper-zinc superoxide dismutase with the dynein–dynactin complex contributes to inclusion formation. J Biol Chem 283: 22795–22805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J et al. (2009) Progressive aggregation despite chaperone associations of a mutant SOD1-YFP in transgenic mice that develop ALS. Proc Natl Acad Sci USA 106: 1392–1397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witan H, Gorlovoy P, Kaya AM, Koziollek-Drechsler I, Neumann H, Behl C, Clement AM (2009) Wild-type Cu/Zn superoxide dismutase (SOD1) does not facilitate, but impedes the formation of protein aggregates of amyotrophic lateral sclerosis causing mutant SOD1. Neurobiol Dis 36: 331–342 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.