Abstract
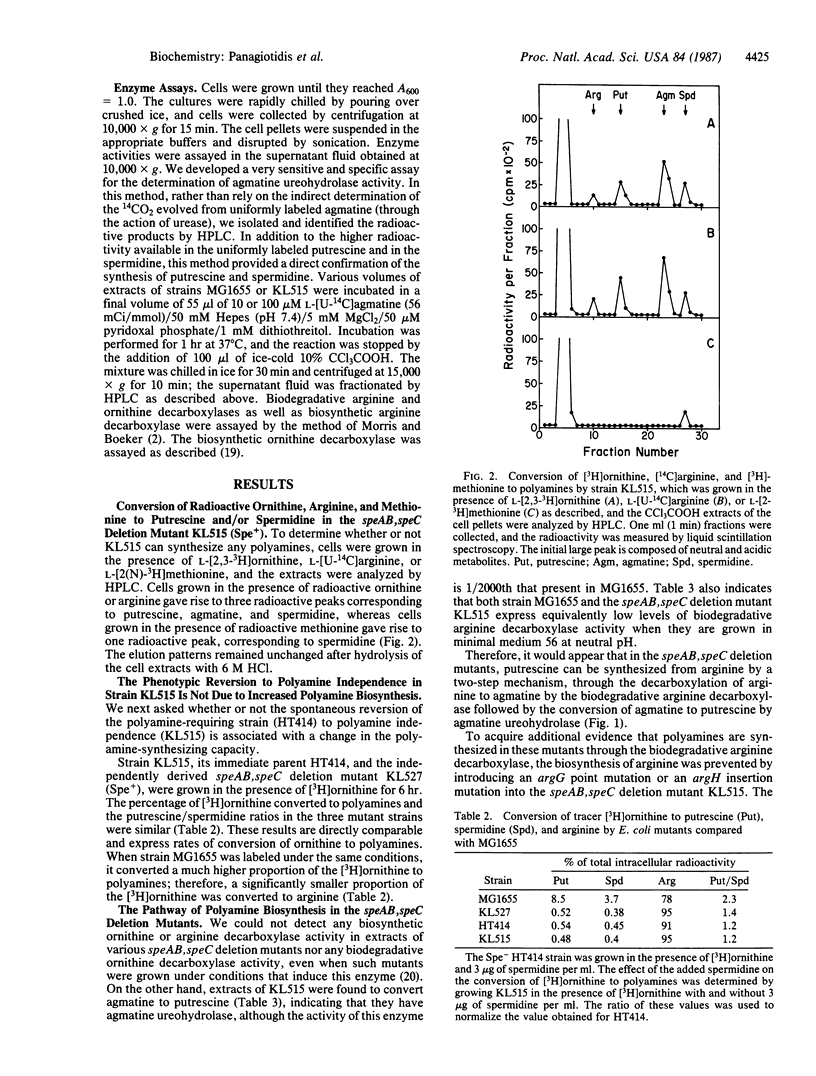
Escherichia coli K-12 mutants that carry deletions in their genes for ornithine decarboxylase (L-ornithine carboxy-lyase, EC 4.1.1.17) (speC), arginine decarboxylase (L-arginine carboxy-lyase, EC 4.1.1.19) (speA), and agmatine ureohydrolase (agmatinase or agmatine amidinohydrolase, EC 3.5.3.11) (speB) can still synthesize very small amounts of putrescine and spermidine. The putrescine concentration in these mutants was found to be 1/2500th that in spe+ cells. The pathway of putrescine synthesis appears to be through the biodegradative arginine decarboxylase, which converts arginine to agmatine, in combination with a low agmatine ureohydrolase activity--1/2000th that in spe+ strains. These results suggest that even such low levels of polyamines permit a low level of protein synthesis. Evidence is presented that the polyamine requirement for the growth of the polyamine-dependent speAB, speC deletion mutants, which are also streptomycin resistant, is not due to a decreased ability to synthesize polyamines.
Full text
PDF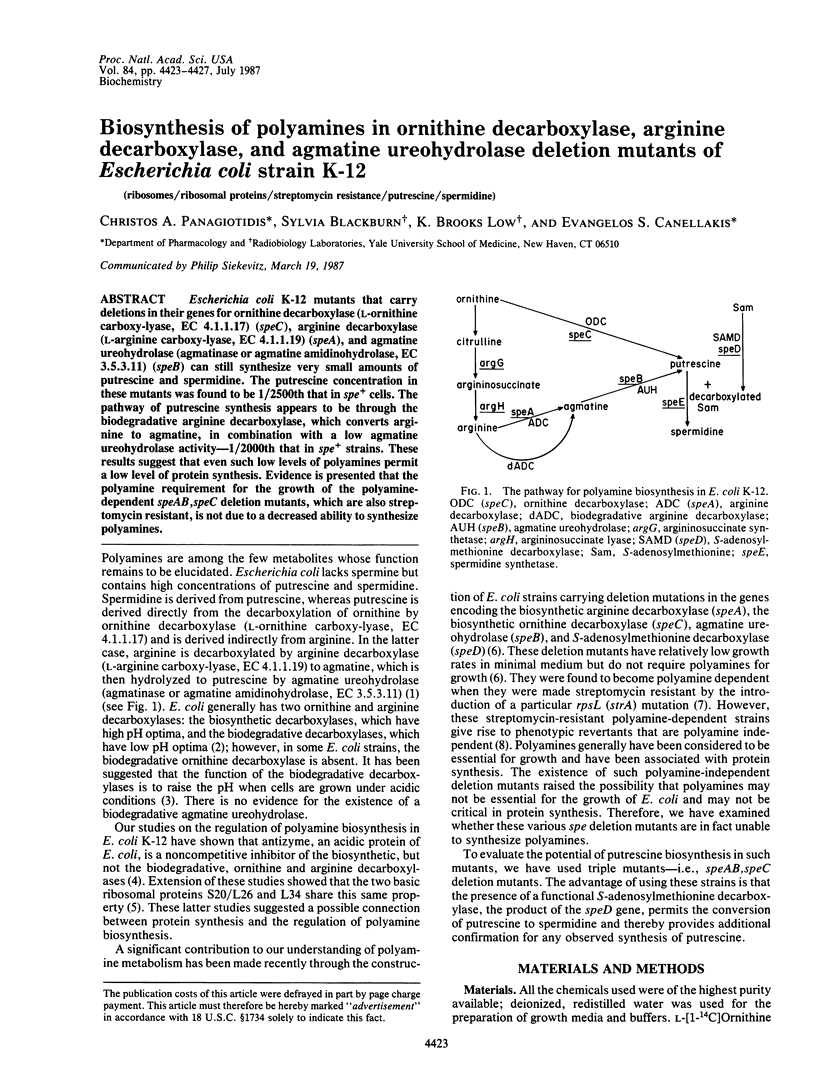



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Applebaum D., Sabo D. L., Fischer E. H., Morris D. R. Biodegradative ornithine decarboxylase of Escherichia coli. Purification, properties, and pyridoxal 5'-phosphate binding site. Biochemistry. 1975 Aug 12;14(16):3675–3681. doi: 10.1021/bi00687a025. [DOI] [PubMed] [Google Scholar]
- Bachmann B. J. Linkage map of Escherichia coli K-12, edition 7. Microbiol Rev. 1983 Jun;47(2):180–230. doi: 10.1128/mr.47.2.180-230.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canellakis E. S., Kyriakidis D. A., Rinehart C. A., Jr, Huang S. C., Panagiotidis C., Fong W. F. Regulation of polyamine biosynthesis by antizyme and some recent developments relating the induction of polyamine biosynthesis to cell growth. Review. Biosci Rep. 1985 Mar;5(3):189–204. doi: 10.1007/BF01119588. [DOI] [PubMed] [Google Scholar]
- Cunningham-Rundles S., Maas W. K. Isolation, characterization, and mapping of Escherichia coli mutants blocked in the synthesis of ornithine decarboxylase. J Bacteriol. 1975 Nov;124(2):791–799. doi: 10.1128/jb.124.2.791-799.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guyer M. S., Reed R. R., Steitz J. A., Low K. B. Identification of a sex-factor-affinity site in E. coli as gamma delta. Cold Spring Harb Symp Quant Biol. 1981;45(Pt 1):135–140. doi: 10.1101/sqb.1981.045.01.022. [DOI] [PubMed] [Google Scholar]
- Hafner E. W., Tabor C. W., Tabor H. Mutants of Escherichia coli that do not contain 1,4-diaminobutane (putrescine) or spermidine. J Biol Chem. 1979 Dec 25;254(24):12419–12426. [PubMed] [Google Scholar]
- Heller J. S., Rostomily R., Kyriakidis D. A., Canellakis E. S. Regulation of polyamine biosynthesis in Escherichia coli by basic proteins. Proc Natl Acad Sci U S A. 1983 Sep;80(17):5181–5184. doi: 10.1073/pnas.80.17.5181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyriakidis D. A., Heller J. S., Canellakis E. S. Modulation of ornithine decarboxylase activity in Escherichia coli by positive and negative effectors. Proc Natl Acad Sci U S A. 1978 Oct;75(10):4699–4703. doi: 10.1073/pnas.75.10.4699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Low B. Rapid mapping of conditional and auxotrophic mutations in Escherichia coli K-12. J Bacteriol. 1973 Feb;113(2):798–812. doi: 10.1128/jb.113.2.798-812.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MONOD J., COHEN-BAZIRE G., COHN M. Sur la biosynthèse de la beta-galactosidase (lactase) chez Escherichia coli; la spécificité de l'induction. Biochim Biophys Acta. 1951 Nov;7(4):585–599. doi: 10.1016/0006-3002(51)90072-8. [DOI] [PubMed] [Google Scholar]
- Morris D. R., Boeker E. A. Biosynthetic and biodegradative ornithine and arginine decarboxylases from Escherichia coli. Methods Enzymol. 1983;94:125–134. doi: 10.1016/s0076-6879(83)94020-x. [DOI] [PubMed] [Google Scholar]
- Morris D. R., Fillingame R. H. Regulation of amino acid decarboxylation. Annu Rev Biochem. 1974;43(0):303–325. doi: 10.1146/annurev.bi.43.070174.001511. [DOI] [PubMed] [Google Scholar]
- Roth M., Hampaï A. Column chromatography of amino acids with fluorescence detection. J Chromatogr. 1973 Aug 29;83:353–356. doi: 10.1016/s0021-9673(00)97051-1. [DOI] [PubMed] [Google Scholar]
- Seiler N., Knödgen B. High-performance liquid chromatographic procedure for the simultaneous determination of the natural polyamines and their monoacetyl derivatives. J Chromatogr. 1980 Dec 12;221(2):227–235. doi: 10.1016/s0378-4347(00)84307-8. [DOI] [PubMed] [Google Scholar]
- Tabor C. W., Tabor H. Polyamines in microorganisms. Microbiol Rev. 1985 Mar;49(1):81–99. doi: 10.1128/mr.49.1.81-99.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabor H., Tabor C. W., Cohn M. S., Hafner E. W. Streptomycin resistance (rpsL) produces an absolute requirement for polyamines for growth of an Escherichia coli strain unable to synthesize putrescine and spermidine [delta(speA-speB) delta specC]. J Bacteriol. 1981 Aug;147(2):702–704. doi: 10.1128/jb.147.2.702-704.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu W. H., Morris D. R. Biosynthetic arginine decarboxylase from Escherichia coli. Purification and properties. J Biol Chem. 1973 Mar 10;248(5):1687–1695. [PubMed] [Google Scholar]
- Zengel J. M., Mueckl D., Lindahl L. Protein L4 of the E. coli ribosome regulates an eleven gene r protein operon. Cell. 1980 Sep;21(2):523–535. doi: 10.1016/0092-8674(80)90490-0. [DOI] [PubMed] [Google Scholar]


