This study examines how histone modification confers salt stress in Arabidopsis. The floral initiator SKB1 is found to mediate the plant’s response to salt stress by altering the methylation status of histone H4R3 and of the small nuclear ribonucleoprotein Sm-like4 (LSM4), thereby affecting the expression of stress-responsive genes.
Abstract
Plants adapt their growth and development in response to perceived salt stress. Although DELLA-dependent growth restraint is thought to be an integration of the plant’s response to salt stress, little is known about how histone modification confers salt stress and, in turn, affects development. Here, we report that floral initiator Shk1 kinase binding protein1 (SKB1) and histone4 arginine3 (H4R3) symmetric dimethylation (H4R3sme2) integrate responses to plant developmental progress and salt stress. Mutation of SKB1 results in salt hypersensitivity, late flowering, and growth retardation. SKB1 associates with chromatin and thereby increases the H4R3sme2 level to suppress the transcription of FLOWERING LOCUS C (FLC) and a number of stress-responsive genes. During salt stress, the H4R3sme2 level is reduced, as a consequence of SKB1 disassociating from chromatin to induce the expression of FLC and the stress-responsive genes but increasing the methylation of small nuclear ribonucleoprotein Sm-like4 (LSM4). Splicing defects are observed in the skb1 and lsm4 mutants, which are sensitive to salt. We propose that SKB1 mediates plant development and the salt response by altering the methylation status of H4R3sme2 and LSM4 and linking transcription to pre-mRNA splicing.
INTRODUCTION
The growth and productivity of sessile plants are greatly affected by environmental factors such as drought, low temperature, and soil salinity. To withstand environmental stresses, plants have evolved highly integrated sensing and response signaling pathways that regulate developmental processes, such as growth and flowering. In general, a stress signaling pathway of plants comprises a sensor, signal transduction, and a response. The sensor perceives adverse environmental conditions. Although few stress sensors have been identified, the subsequent signal transduction and response reactions have been studied intensively (Zhu, 2001, 2002; Xiong et al., 2002a; Chinnusamy et al., 2004; Yamaguchi-Shinozaki and Shinozaki, 2006; Munns and Tester, 2008; Cutler et al., 2010). In Arabidopsis thaliana, several perception and signaling pathways have been identified for the response to high salinity in soil, including the salt overly sensitive pathway and the phytohormone abscisic acid (ABA)-dependent and -independent transcriptional regulation pathways (Serrano and Rodriguez-Navarro, 2001; Zhu, 2002, 2003; Quan et al., 2007; Mahajan et al., 2008).
High salinity induces the accumulation of the phytohormone ABA by increasing the expression of ABA biosynthetic genes, such as ABA1, ABA3, and NCED3 (Seo et al., 2000; Xiong et al., 2001a, 2002a, 2002b; Ruggiero et al., 2004; Matsui et al., 2008). Under high salt and other osmotic stress, ABA accumulation activates ABA-dependent signaling by increasing the expression of transcription factors DEHYDRATION RESPONSIVE ELEMENT (DRE) BINDING FACTOR 2A (DREB2A), DREB2B, RESPONSIVE TO DESSICATION 22 BINDING PROTEIN (RD22BP), and ABA-RESPONSIVE ELEMENT BINDING BASIC LEUCINE ZIPPER PROTEINS (ABFs/AREBs) as well as stress-responsive genes, such as RD29A, RD29B, RD22, COLD-REGULATED47 (COR47), and COLD-INDUCIBLE1 (Zhu, 2002; Yamaguchi-Shinozaki and Shinozaki, 2006). The expression of stress-responsive genes is greatly reduced in the aba1 and aba3 mutants, as well as in the double mutant of G-PROTEIN COUPLED RECEPTOR-TYPE G PROTEINS (gtg1 gtg2) and in the quadruple mutant of PYRABACTIN RESISTANCE1 and PYRABACTIN LIKE (pyr1 pyl1 pyl2 pyl4), two types of ABA receptors with unknown functions in salt stress (Xiong et al., 2001a, 2002b; Pandey et al., 2009; Park et al., 2009). ABA-dependent signaling is negatively regulated by type 2C protein phosphatases, namely, ABI1 and ABI2, and the downstream signaling ceases in the dominant-positive ABA-insensitive mutants abi1-1 and abi2-1 (Leung et al., 1994, 1997; Meyer et al., 1994; Allen et al., 1999; Gosti et al., 1999; Murata et al., 2001; Becker et al., 2003; Saez et al., 2004). During osmotic stress, the expression of stress-responsive genes is also regulated in an ABA-independent pathway. In this pathway, high salinity stress induces the expression of transcription factors such as DREB2A and DREB2B, which activate stress response genes containing DRE cis-elements. Although ABA-dependent and -independent pathways exist in osmotic stress signaling, the expression of some response genes, such as RD29A, which contains both DRE and ABA-responsive element, is interdependent (Zhu, 2002; Yamaguchi-Shinozaki and Shinozaki, 2006).
In adapting to adverse environments, the developmental processes of flowering time and plant growth are also regulated by stress sensor and signal transduction pathways (Apse et al., 1999; Zhu, 2002). Recent studies have identified several genes that function in developmental pathways as key regulators that connect environmental sensing and plant growth. A flowering autonomous pathway gene, FVE, which regulates flowering time by affecting chromatin histone acetylation of the floral integrator FLOWERING LOCUS C (FLC), is a sensor of cold temperature. FVE perceives cold temperature and integrates cold response and flowering time by modulating the expression of FLC and cold response genes (Blázquez et al., 2003; Kim et al., 2004). Apart from FVE, another autonomous pathway gene, FCA, which encodes an RNA binding protein that functions as a thermosensor that regulates flowering time, might integrate the cold response and flowering time via small interfering RNA-mediated chromatin silencing (Blázquez et al., 2003; Bäurle et al., 2007). Nucleosomes containing H2A.Z, a histone H2A variant, perceive ambient temperature and influence flowering time independently of transcription (Kumar and Wigge, 2010). In addition to ambient temperature, high salinity in soil slows the growth of plants and delays flowering time (Achard et al., 2006). DELLA protein accumulation, which restrains growth, is critical for integrating the salt response and flowering time, and it does so by inhibiting the expression of LEAFY (LFY), which encodes a floral initiator protein. FLC transcript levels are increased in plants grown on salt but are not regulated by DELLAs (Achard et al., 2006). Compelling evidence shows that the expression of genes such as FLC and salt response genes is regulated by histone modification (He and Amasino, 2005; Sridhar et al., 2007; Zhu et al., 2008; Bond et al., 2009; Dalal et al., 2009; He, 2009). However, the key histone marker that integrates salt response and plant development remains unclear. It is possible that a chromatin remodeling factor may contribute to the expression of genes involved in both flowering time and salt response. We report that SKB1 and histone H4R3 symmetric dimethylation (H4R3sme2) are involved in the integration of the salt response and flowering time in Arabidopsis.
SKB1, also named protein arginine methyltransferase 5 (PRMT5), is a type II Arg methyltransferase that catalyzes Arg symmetric dimethylation (Bedford and Richard, 2005; Pahlich et al., 2006; Bedford, 2007; Bedford and Clarke, 2009). In mammalian cells, PRMT5 methylates a wide spectrum of substrates, including histone and nonhistone proteins, to regulate gene transcription, RNA elongation, pre-mRNA splicing, protein interaction, and protein stability (Kwak et al., 2003; Pal et al., 2003; Liu et al., 2007; Chari et al., 2008; Guo and Bao, 2010; Ren et al., 2010; Zhou et al., 2010). We and others previously reported that Arabidopsis SKB1/PRMT5 promotes flowering by suppressing the expression of FLC through H4R3sme2 in the promoter region (Wang et al., 2007; Schmitz et al., 2008). The skb1-1 and skb1-2 mutants show a reduced level of H4R3sme2 and have a late-flowering phenotype, as well as severe developmental retardation (Pei et al., 2007; Wang et al., 2007). We report that mutation of SKB1 results in hypersensitivity to salt stress and ABA. Salt stress reduces the H4R3sme2 level, which is essential for suppressing the expression of FLC and stress-responsive genes, and this finding suggests a mechanism by which transcription can be regulated by salt. We also show that SKB1 methylates U6 small nuclear ribonucleoprotein (snRNP)–specific Sm-like protein LSM4 and that mutations in SKB1 lead to splicing defects in hundreds of genes that are involved in many biological processes, including the abiotic stress response. In addition, the lsm4 mutant, similarly to skb1, is found to be hypersensitive to salt and results in the same splicing defects in some genes. Two independent groups recently described the induction of a broad range of RNA splicing defects in the skb1 mutant (Deng et al., 2010; Sanchez et al., 2010).
Taken together, our work and that of Deng et al. (2010) and Sanchez et al. (2010) provide a comprehensive picture of how SKB1-mediated protein Arg methylations master transcription and pre-mRNA splicing to regulate plant development and the plant’s response to environment cues.
RESULTS
The skb1-1 Mutant Is Hypersensitive to Salt Stress
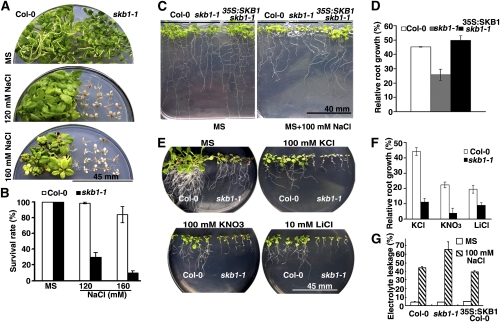
Previous studies by us and others have shown that SKB1 is a member of an autonomous pathway that regulates flowering time in Arabidopsis (Pei et al., 2007; Wang et al., 2007). The skb1-1 flc-3 double mutant suppresses the skb1-1 late-flowering phenotype. However, compared with wild-type and flc-3 mutant plants, the skb1-1 flc-3 mutant was much smaller when grown in soil (see Supplemental Figure 1 online). Linked to our previous study, the SKB1 homolog in fission yeast is involved in the KCl-induced hyperosmotic stress response (Bao et al., 2001). We investigated whether SKB1 is also required in the salt stress response in Arabidopsis. Four-day-old skb1 mutants and wild-type seedlings germinated on Murashige and Skoog (MS) medium were transferred to MS medium containing NaCl. The growth of both skb1-1 and skb1-2 mutant plants was completely inhibited by 120 or 160 mM NaCl, and the plants died. By contrast, wild-type plants under the same conditions showed slightly inhibited growth and remained alive (Figures 1A and 1B; see Supplemental Figure 2 online). The hypersensitivity of skb1-1 mutant plants to NaCl was reversed in transgenic plants (35S:SKB1 skb1-1) that constitutively expressed SKB1 in the skb1-1 mutant background (Figure 1C).
Figure 1.
Phenotypic Analysis of skb1-1 Mutant Plants Exposed to Salt Stress.
(A) and (B) Comparison of the survival rate of the wild type (Col-0) and the skb1-1 mutant grown on MS medium containing the indicated concentration of NaCl. Photographs were taken and the survival rate was determined 30 d after seedling transfer to the treatment medium. Data represent means ± (se of three independent experiments (n > 50).
(C) and (D) Comparison of root growth of wild type (Col-0), skb1-1, and 35S:SKB1 skb1-1 plants grown on MS medium with 100 mM NaCl. Four-day-old seedlings were transferred from MS medium to MS medium with 0 or 100 mM NaCl. Root growth was measured relative to controls 10 d after seedling transfer onto the treatment medium. More than 24 roots were measured for each data point. Data represent means ± se of three independent experiments.
(E) and (F) Comparison of the root growth of the wild type (Col-0) and skb1-1 mutant grown on MS medium with 100 mM KCl, 100 mM KNO3, or 10 mM LiCl. Primary root length was measured and root growth relative to controls was analyzed 10 d after seedling transfer to the treatment medium. More than 24 roots were measured for each data point. Data represent means ± se of three independent experiments.
(G) Relative electrolyte leakage of leaves from wild-type (Col-0), skb1-1, and 35S:SKB1 Col-0 plants after exposure to 100 mM NaCl. Data represent means ± se of three independent experiments.
We examined the growth of roots, a primary indicator of salt concentration. The root length of wild-type plants grown on MS medium containing 100 mM NaCl was ~40 to 50% that of plants grown on MS medium alone. By contrast, the root length of skb1 mutants was reduced to only 20% that of plants grown on MS medium, and this root growth defect of skb1 mutants was rescued in 35S:SKB1 skb1 transgenic plants (Figures 1C and 1D). In addition, compared with wild-type plants, transgenic plants overexpressing SKB1 (35S:SKB1 Columbia [Col]) showed slightly increased tolerance to NaCl-induced osmotic stress (see Supplemental Figure 3 online).
To determine whether the skb1-1 mutant is specifically hypersensitive to NaCl, wild-type and skb1-1 mutant seedlings were transferred to MS medium containing KCl, KNO3, or LiCl for 10 d. Primary root growth of the skb1-1 mutants was inhibited more severely than that of the wild type with each treatment (Figures 1E and 1F).
In addition, we determined the relative electrolyte leakage (REL) of skb1-1 seedlings treated with 100 mM NaCl and found that the REL of the skb1-1 mutant was significantly higher than that of the wild type but that the REL of plants overexpressing SKB1 was lower (Figure 1G). Thus, SKB1 is involved in this adaptive response to salt stress in Arabidopsis.
SKB1 Regulates the Salt Stress Response Independently of FLC
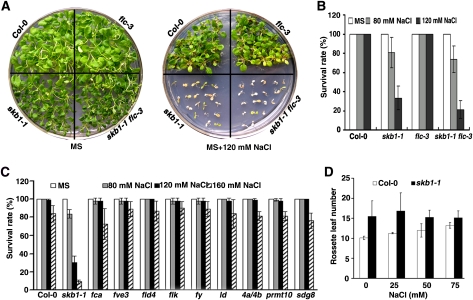
SKB1 regulates flowering time in an FLC-dependent manner (Pei et al., 2007; Wang et al., 2007; Schmitz et al., 2008). We asked whether SKB1 functions in salt stress tolerance through the expression of FLC. We examined the sensitivity to NaCl-induced salt stress of flc-3, a mutant with loss of FLC function, and the skb1-1 flc-3 double mutant. Compared with the wild type, the skb1-1 mutant was hypersensitive to NaCl, whereas the flc-3 mutant was not (Figures 2A and 2B). However, the skb1-1 flc-3 double mutant showed a sensitivity to salt stress similar to that of the skb1-1 mutant, which suggests that SKB1 functions independently of FLC in salt stress tolerance.
Figure 2.
The Sensitivity of Mutants of Genes That Regulate Flowering Time to Salt Stress.
(A) and (B) Similar to skb1-1, the skb1-1 flc-3 double mutant is hypersensitive to salt stress, and the sensitivity of flc-3 to salt stress is similar to that of the wild type.
(C) The survival rate of the other mutants of genes that regulate flowering time, including fca, fve, fld4, flk, fy, ld, prmt4a prmt4b, prmt10, and sdg8. Four-day-old seedlings were transferred from MS medium to MS medium with 0, 80, 120, or 160 mM NaCl. Survival rate was determined 30 d after seedling transfer to the treatment medium. Data represent means ± se of three independent experiments (n > 40).
(D) Flowering times of the wild type (Col-0) and the skb1-1 mutant treated on MS medium with 0, 25, 50, or 75 mM NaCl for 5 d before transfer to soil.
[See online article for color version of this figure.]
We then examined other autonomous pathway genes, FVE, FCA, FLD, FLK, FY, LD, PRMT4A, PRMT4B, and PRMT10, and the PAF1 complex component, SDG8, and found that none of the mutant plants was sensitive or more tolerant to NaCl than was the wild type (Figure 2C; see Supplemental Figure 4 online). We further examined the effects of salt stress treatment on the flowering time of wild-type and skb1-1 plants. Five-day-old seedlings germinated on MS medium were transferred to MS medium containing 0, 25, 50, or 75 mM NaCl and cultured for 5 d before transfer to soil. Whereas flowering time delay of the wild type was found to be NaCl concentration dependent, salt stress treatment did not obviously affect the flowering time of the skb1-1 mutant (Figure 2D). These results suggest that SKB1 might be a key regulator connecting flowering time and salt tolerance.
skb1-1 Is Hypersensitive to ABA
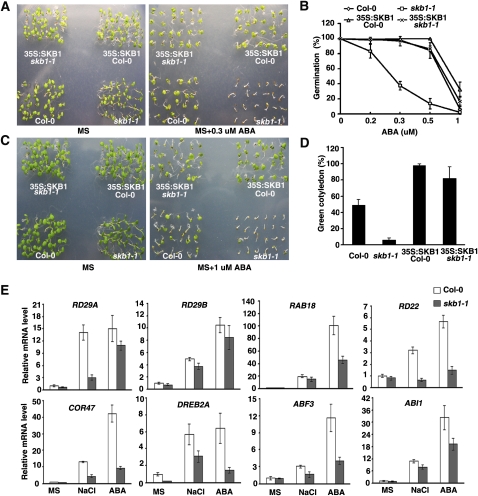
High concentrations of salt induce ABA accumulation and activate ABA signaling pathways (Nakashima et al., 2000; Xiong et al., 2002a; Zhu, 2002; Christmann et al., 2006). To examine whether SKB1 is involved in ABA signaling, wild-type and skb1-1 seeds were planted in MS medium containing various concentrations of ABA. In the absence of ABA, skb1-1 and wild-type plants showed similar seed germination rates (Figures 3A and 3B). In the presence of 0.3 μM ABA, only 40% of skb1-1 seeds germinated, and these did not show any cotyledon greening. By contrast, 100% of wild-type seeds germinated and showed cotyledon greening (Figures 3A and 3B). The 35S:SKB1 skb1-1 transgenic seeds rescued the ABA hypersensitivity of the skb1-1 mutant (Figures 3A to 3D). In the presence of ABA, 35S:SKB1 Col-0 plants exhibited more green cotyledons than did wild-type plants (Figures 3C and 3D), and the plants showed more root growth (see Supplemental Figure 5 online).
Figure 3.
Loss of SKB1 Leads to Hypersensitivity to ABA.
(A) and (B) Seed germination of the wild type (Col-0), skb1-1, 35S:SKB1 skb1-1, and 35S:SKB1 Col-0. Seeds harvested on the same day were planted on MS medium containing 0, 0.2, 0.3, 0.5, or 1 μM ABA. Plates were transferred to a growth chamber after stratification at 4°C for 3 d. Photographs were taken, and germination rate was determined 3 d after the transfer. Data represent means ± se of three independent experiments (n = 96).
(C) and (D) Cotyledon greening of the wild type (Col-0), skb1-1, 35S:SKB1 skb1-1, and 35S:SKB1/Col-0. Photographs were taken and cotyledon greening rate was determined 6 d after transfer to the growth chamber. Data represent means ± se of three independent experiments (n = 96).
(E) Real-time quantitative RT-PCR analysis of the expression of salt stress and ABA-responsive genes in the wild type (Col-0) and skb1-1 and normalized with 18S rRNA expression. Error bars indicate the relative sd of three independent experiments. Template RNA isolated from 11-d-old seedlings growing on MS agar medium was treated with 200 mM NaCl for 6 h or 100 μM ABA for 3 h.
To investigate the molecular mechanism underlying the hypersensitivity of skb1 plants to salt stress and ABA, we examined the transcript levels of genes involved in the salt stress and ABA response and signaling pathways. The expression of the stress-responsive genes RD29A, RD29B, RD22, COR47, and Response to ABA 18 (RAB18) and transcription factors DREB2A and ABF3, as well as a regulator of ABA signaling, ABI1, was strongly induced by both NaCl and ABA in wild-type plants; however, the induced expression of these genes was lower in skb1-1 seedlings (Figure 3E). Thus, loss of SKB1 function results in hypersensitivity to increased salt and ABA, and SKB1 could be involved in the ABA signaling pathway.
A High Concentration of Salt Reduces the Level of H4R3sme2 by Disrupting the Association of SKB1 with Chromatin
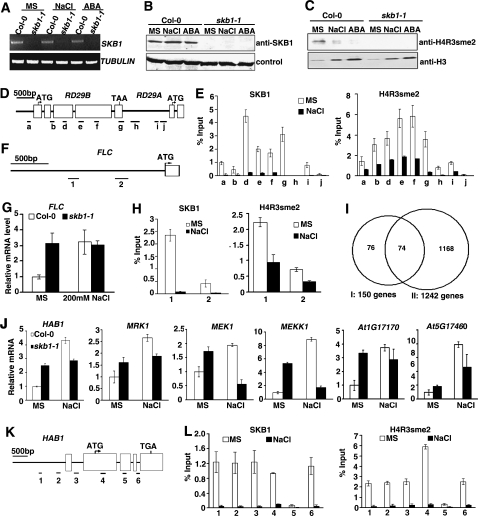
To elucidate the molecular mechanism of the involvement of SKB1 in the plant’s response to salt stress, we examined SKB1 mRNA expression. Wild-type 11-d-old (Col-0) plants were subjected to 200 mM NaCl or 100 μM ABA. Compared with the untreated control, neither mRNA (Figure 4A) nor protein (Figure 4B) levels of SKB1 differed in response to either treatment. Therefore, the expression of SKB1 was not induced by high salinity or ABA. SKB1 has been linked to the histone mark H4R3sme2 (Fabbrizio et al., 2002; Ancelin et al., 2006; Wang et al., 2007). Using an H4R3sme2-specific antibody, we examined H4R3sme2 levels under high salinity or ABA by immunoblot analysis and found that the global level of H4R3sme2 was reduced by both treatments in the wild type (Figure 4C).
Figure 4.
SKB1 Expression Profile and H4R3sme2 Level in Global and Specific Sites in Response to Salt Stress and ABA Treatments.
(A) RT-PCR analysis of the expression of SKB1 in the wild type (Col-0) in response to salt stress and ABA treatment. Template RNA isolated from 11-d-old seedlings grown on MS medium was treated with 200 mM NaCl for 6 h or 100 μM ABA for 3 h. TUBULIN was the loading control. Three biological replicates were performed with similar results.
(B) Immunoblot analysis of SKB1 expression in response to salt stress and ABA treatment. Wild-type (Col-0) and skb1-1 plants were treated as in (A).
(C) Immunoblot analysis of Arg symmetric dimethylation modification of the H4R3sme2 level in response to salt stress and ABA treatment, using histone-enriched whole protein extract and an H4R3sme2-specific antibody. Wild-type (Col-0) and skb1-1 seedlings were treated as in (A).
(D), (F), and (K) Diagrams of the RD29A and RD29B (D), FLC (F), and HAB1 (K) genes structure, with bars representing the regions examined by ChIP shown in (E), (H), and (L), respectively. White boxes represent exons or 5′ or 3′ untranslated regions, and black lines represent introns.
(E), (H), and (L) ChIP assay for RD29B and RD29A (E), FLC (H), and HAB1 (L) performed with anti-SKB1 and -H4R3sme2 antibodies. Chromatin extracted from 11-d-old seedlings grown on MS medium was treated with 200 mM NaCl for 6 h or 100 μM ABA for 3 h, untreated as a control. Data represent triplicate multiquantitative PCR measurements of immunoprecipitated DNA, and the input represents chromatin before immunoprecipitation. Error bars represent relative sd of ChIP data.
(G) Quantitative RT-PCR analysis of the expression of FLC in response to salt stress. Template RNA isolated from 11-d-old seedlings grown on MS medium was treated with 200 mM NaCl for 6 h. The experiment was performed independently three biological replicates and normalized by TUBULIN expression. Error bars indicate the relative sd of three independent experiments.
(I) Venn diagram of genes upregulated in the skb1-1 mutant in standard conditions (a) and in the wild type after treatment with 200 mM NaCl for 6 h (b), as determined by microarray analysis.
(J) Real-time quantitative RT-PCR analysis of the expression of salt stress–induced genes in the wild type and skb1-1 mutant in standard and salt stress conditions. Template RNA, experiments, and data analysis are indicated in (G).
To determine whether the decreased level of H4R3sme2 results from a change of SKB1 associative chromatin, we performed a chromatin immunoprecipitation (ChIP) assay using anti-SKB1 and -H4R3sme2 antibodies to analyze the gene locus of RD29A and RD29B, whose expression is regulated by histone modification and DNA methylation in response to salt and ABA stress (Gong et al., 2002; Sridhar et al., 2007). As shown in Figures 4D and 4E, both anti-SKB1 and -H4R3sme2 antibodies pulled down chromatin DNA at the d, e, f, g, and i regions in the wild type but not in the skb1-1 mutant (see Supplemental Figure 6 online). Compared with untreated controls, treatment with salt substantially decreased immunoprecipitated DNAs at these chromatin regions (Figure 4E).
The transcription of the flowering time suppressor FLC is suppressed by H4R3sme2 (Wang et al., 2007; Schmitz et al., 2008) and is increased in plants grown on salt (Figure 4G) (Achard et al., 2006). We then examined the H4R3sme2 and SKB1 level in the FLC promoter and found that salt treatment resulted in significantly decreased levels and even loss of both SKB1 and H4R3sme2 in the FLC promoter chromatin (Figure 4H). This indicates that salt stress increases the expression of FLC by reducing H4R3sme2 levels and decreasing the association of SKB1 with chromatin.
To further elucidate how the stress response is regulated by SKB1, we performed a transcriptome analysis using a microarray assay (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE26398). Relative to the wild type, the expression of 150 genes was upregulated in the skb1-1 mutant in unstressed conditions. The expression of 1242 genes was induced in the wild type by treatment with 200 mM NaCl for 6 h. Among these 1242 salt stress–induced genes, 74 genes, including the ABA signaling negative regulator, Homology to ABI1 (HAB1), and the protein kinase genes, MAP Kinase Kinase or Erk Kinase1 (MEK1), MAP Kinase Kinase Kinase 1 (MEKK3), and Arabidopsis protein kinase related to mammal mixed-lineage kinases and Raf protein kinases 1 (MRK1), were upregulated in unstressed conditions in the skb1-1 mutant plants (Figure 4I). The results of quantitative RT-PCR confirmed that the expression of HAB1, MEK1, MEKK1, and MRK1, and some other stress-responsive genes, such as At1G17170 and At5G17460, was increased in the skb1-1 mutant grown under normal conditions (Figure 4J). Given the critical role of HAB1 in ABA and salt stress (Saez et al., 2004, 2006), we asked whether SKB1 regulates its transcription directly. ChIP analysis showed that SKB1 associated with HAB1 chromatin, and anti-H4R3sme2 antibody pulled down HAB1 chromatin DNAs (Figures 4K and 4L). Consistently, either SKB1 association with HAB1 chromatin or the level of H4R3sme2 in HAB1 chromatin decreased under salt stress conditions (Figures 4K and 4L). These results suggest that SKB1 suppresses the expression of HAB1 by increasing the levels of H4R3sme2.
SKB1 Methylates the U6 snRNP LSM4 in Response to Salt Stress and ABA
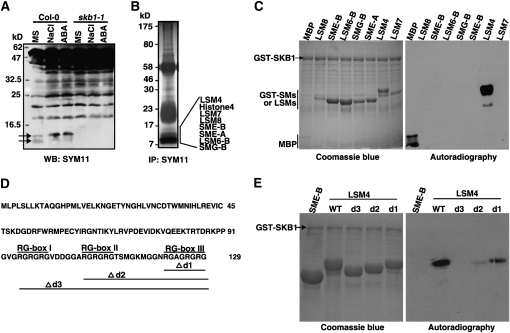
The histone H4R3sme2 is a silencing mark of transcription and its reduced levels activate transcription. The H4R3sme2 levels of RD29A and RD29B chromatin were decreased in skb1 mutant plants (see Supplemental Figure 6 online); however, the expected higher mRNA levels of RD29A and RD29B were not observed in the skb1 mutant plants (Figure 3E). This prompted us to test whether other pathways are affected in the skb1 mutant. We used an antibody, SYM11, which specifically recognizes symmetric dimethylated arginine (sDMA)–containing peptides (Boisvert et al., 2003), to test the change in sDMA at the global level in response to high salinity or ABA treatment. The methylation signal of some protein(s) of ~14 kD was increased significantly and that of other proteins of ~10 kD were decreased in the wild type after high salinity or ABA treatment; however, the corresponding bands were lost or became much weaker in the skb1-1 mutants (Figure 5A).
Figure 5.
SKB1 Methylates the U snRNP Core Component LSM4.
(A) Immunoblot analysis of Arg symmetric dimethylation modification with the SYM11 antibody in response to salt stress and ABA treatments. Total protein was extracted from 11-d-old wild-type (Col-0) or skb1-1 seedlings treated with 200 mM for 6 h or 100 μM ABA for 3 h. The experiment was performed independently three times with similar results.
(B) Total protein from wild-type seedlings treated with ABA for 5 h was immunoprecipitated with SYM11. Protein bands (7 to 17 kD) were identified by mass spectrometry and are indicated at the right.
(C) and (E) In vitro methylation assay. GST-SM or GST-LSM proteins and GST-SKB1 were purified from E. coli and incubated in the presence of [3H]-SAM. Methylated proteins were monitored by autoradiography.
(D) Full-length protein sequence of Arabidopsis LSM4, the location of three RG boxes, and the truncations in LSM4 (d1), LSM4 (d2), and LSM4 (d3).
To identify these sDMA-containing proteins, we performed immunoprecipitation with SYM11. The SYM11 immunoprecipitates isolated from the protein extracts of the wild-type plants treated with 100 μM ABA for 5 h were subjected to SDS-PAGE, and proteins of between 7 and 17 kD that could not be methylated in the skb1 mutant were identified by liquid chromatography–tandem mass spectrometry (Figure 5B). These proteins include histone H4, SM proteins (SME, SMF, and SMG-B), and SM-like proteins (LSM 4, 7, 8, and 6-B). Apart from H4, all of these proteins are core components of snRNPs (Figure 5B) (He and Parker, 2000; Pannone and Wolin, 2000; Zamski et al., 2001; Zhao et al., 2009).
To test whether SKB1 could methylate the SM and LSM proteins, the glutathione S-transferase (GST)–tagged fusion proteins were purified from Escherichia coli. SKB1 methylated only LSM4 (Figure 5C). The other SM and LSM proteins immunoprecipitated by SYM11 but were unable to be methylated by SKB1. Therefore, these unmethylated SM or LSM proteins might have formed a complex with methylated LSM4 and were coimmunoprecipitated with methylated LSM4 (Figure 5B). Protein amino acid sequence alignment showed that Arabidopsis LSM4, a 14-kD protein, is an evolutionarily highly conserved protein, with homologs in yeast, mouse, and humans (see Supplemental Figure 7 online); it contains three Arg-Gly repeats (Mazzoni et al., 2005) at the C-terminal region (Figure 5D). To define which Arg residues could be methylated, we generated three truncated mutants that deleted one (d1), two (d2), or three (d3) RG repeat(s) (Figure 5D). Compared with the methylation signal for wild-type LSM4, methylation decreased gradually in the d1 and d2 mutants and was completely absent in the d3 mutant (Figure 5E).
Loss of SKB1 Influences Pre-mRNA Splicing
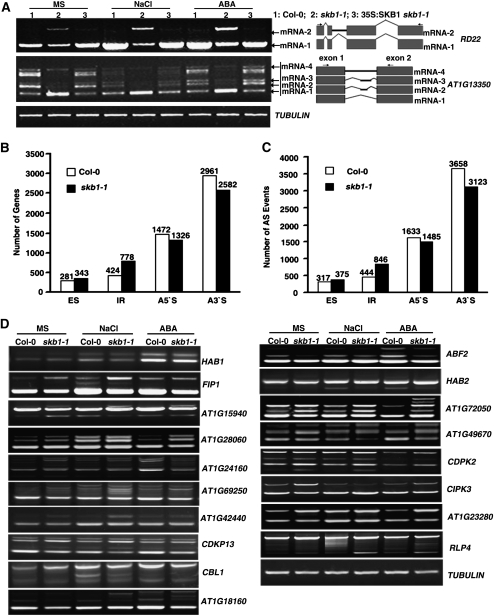
U6 snRNA-associated LSM4 is involved in pre-mRNA splicing in humans, yeast, and Xenopus laevis (Cooper et al., 1995; Tharun et al., 2000; Pannone et al., 2001; Tharun and Parker, 2001; Tomasevic and Peculis, 2002). To determine whether loss of SKB1 function influences pre-mRNA splicing, we examined the transcriptional products of over 20 genes that are or might be involved in salt stress, ABA response, or in encoding Ser/Arg-rich proteins that are important regulators of pre-mRNA splicing (Reddy, 2004; Palusa et al., 2007). These transcripts include exons interrupted by introns (Figure 6A; see Supplemental Figure 8 online; data not shown). Among these genes, the RT-PCR product of RD22 in the skb1-1 mutant differed from that in the wild type (Figure 6A). To determine whether this shifted PCR product is an alternative splicing variant in the skb1 mutant, we cloned and sequenced the PCR products of RD22 and demonstrated that the shifted PCR product in the skb1-1 mutant was RD22 with the second intron retained (Figure 6A). Furthermore, expression of SKB1 in the skb1-1 mutant rescued this pre-mRNA splicing defect of RD22 (Figure 6A), which suggests that SKB1 is required for RD22 pre-mRNA splicing. In addition to RD22, a pre-mRNA splicing defect was also found in a putative protein kinase gene, At1G13350, and the pre-mRNA splicing defect was ameliorated in 35S:SKB1 skb1-1 transgenic plants (Figure 6A). Different from RD22 with the second intron retention, alternative splicing occurred at At1G13350 in the skb1-1 mutant.
Figure 6.
RNA Splicing Analysis.
(A) RT-PCR analysis of the pre-mRNA splicing pattern of RD22 and AT1G13350 in wild-type (1), skb1-1 (2), and 35S:SKB1 skb1-1 (3) plants. Eleven-day-old seedlings grown on MS medium were treated with 200 mM NaCl for 6 h or 100 μM ABA for 3 h. Three biological replicates were performed with similar results. The splicing pattern of each alternative splicing isoform is shown at the right. Gray square, exon; black line, unspliced intron; bent line, spliced intron. Arrowheads at the right show primer locations.
(B) and (C) Statistical chart of alternative splicing (AS) genes and events. ES, exon skipping; IR, intron retention; A5′S, alternative 5′ splicing; A3′S, alternative 3′ splicing.
(D) The splicing pattern of genes in the wild type and skb1-1 mutant. Candidate genes were selected that were identified as having different splicing patterns in the wild type and skb1 mutant by transcriptome RNA sequencing.
To reveal the role of SKB1 in pre-mRNA splicing, we performed a transcriptome RNA sequencing analysis and found that many pre-mRNA splicing defects occurred in the skb1-1 mutant, including exon skipping, intron retention, alternative 5′ splicing, and alternative 3′ splicing. Intron retention events were more common in the skb1-1 mutant than in the wild type (Figures 6B and 6C). Based on the results of deep sequencing, we verified the pre-mRNA splicing of some stress-related genes by RT-PCR. We found that the pre-mRNA splicing of the genes of HAB1, FIP1 (RNA binding protein), AT1G15940 (similar to Tudor contains domain ARM repeat), AT1G28060 (snRNP family protein), AT1G24160 (unknown protein), AT1G69250 (nuclear transport factor 2 family protein), and AT1G42440 (unknown protein) in skb1 differed from that in the wild type (Figure 6D). Moreover, the splicing-specific variants of these genes in the skb1 mutant under unstressed conditions were similar to those of the wild type under salt stress and ABA treatment (Figure 6D). In addition, the splicing of other genes that are involved in the response to salt stress and ABA, such as Calcineurin B-like Protein1 (CBL1), ABF2, and HAB2, was also affected by SKB1 mutation (Figure 6D). Interestingly, two groups recently reported the pre-mRNA splicing defects in the skb1-1 or prmt5-5 mutants (Deng et al., 2010; Sanchez et al., 2010). Consistently, their studies found that genes with splicing defects in the prmt5 mutants were enriched in various biological processes, such as circadian rhythms and flowering, and largely in response to abiotic stimulus. A comparison between our deep sequence data and these previously reported results obtained by tilling arrays revealed that the same or similar splicing defects were found in 28 genes out of a total of 44 that were validated by RT-PCR. By contrast, alternative splicing of the circadian clock gene PSEUDO RESPONSE REGULATOR9 (PRR9) (Sanchez et al., 2010) was not found in our deep sequencing analysis (see Supplemental Table 2 online). This difference is most likely due to the plants being treated in a different light-dark cycle, which influences PRR9 expression levels, or to different methods of analysis being used. Thus, SKB1 is required for pre-mRNA splicing and likely mediates the methylation of LSM4.
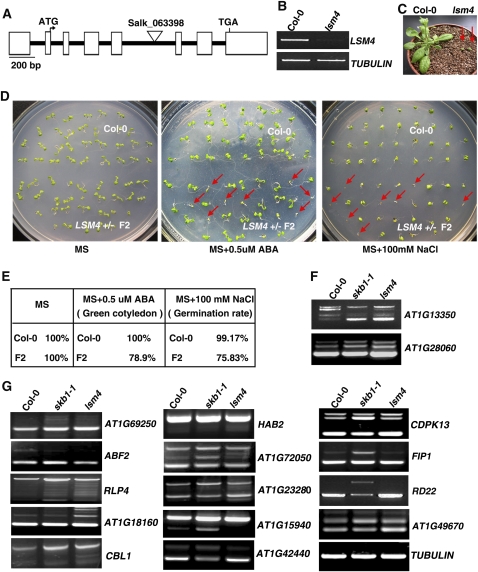
Loss of LSM4 Results in Growth Retardation and Hypersensitivity to Salt
We next investigated the function of LSM4. A T-DNA insertional mutant (SALK_063398) was identified at the AT5G27720 region of Arabidopsis in a Col genetic background from the Salk T-DNA collection (Figure 7A). Full-length LSM4 mRNA was undetectable in the lsm4 mutant (Figure 7B). The lsm4 mutant plants displayed severe developmental retardation compared with wild-type plants (Figure 7C), and lsm4 mutant plants died when grown in MS medium or in soil. Because the developmental retardation phenotype was observed only in homozygous plants, we concluded that the lsm4 mutation is recessive. Since lsm4 homozygous plants die during later development, heterozygous LSM4+/− (F1) seeds were harvested and used to examine whether LSM4 is involved in salt and ABA stress. About 25 or 21.5% of seeds (F2) exhibited germination defects in response to NaCl or ABA treatment, respectively, whereas no defects were observed in the absence of stress treatment (Figures 7D and 7E). The ratio of germination defects in the F2 seeds was close to Mendelian expectations (1:2:1), so seeds were lsm4 homozygous mutants sensitive to salt and ABA; a similar phenotype was observed in skb1 mutants (Figures 1 and 3). In addition, the splicing defect of At1G13350, At1G28060, At1G69250, ABF2, and RLP4 in the lsm4 mutant grown under unstressed conditions was similar to that in the skb1-1 mutant (Figures 7F and 7G). More surprisingly, the splicing of At1G23280, At1G18160, CBL1, HAB2, and At1G72050 in the lsm4 mutant grown under unstressed conditions was similar to that in the wild type grown under salt stress (Figures 6 and 7G). Note that the splicing of At1G15940 and At1G42440 in the lsm4 mutant was different from that in both the wild type and the skb1-1 mutant (Figure 7G), suggesting that loss of LSM4 function might disturb splicing more severely than does compromised methylation. Thus, SKB1 and LSM4 may have (at least in part) overlapping functions in the salt tolerance of Arabidopsis. Nevertheless, we could not explain why the splicing defect of RD22 was only detectable in the skb1-1 mutant (Figure 7G).
Figure 7.
LSM4 Structure and Pleiotropic Phenotypes of the lsm4 Mutant.
(A) LSM4 structure and the T-DNA insertion site of the lsm4 mutant (Salk_063398). White boxes represent exons or 5′ or 3′ untranslated regins, and black lines represent introns.
(B) RT-PCR analysis of LSM4 expression in 7-d-old wild-type (Col-0) and lsm4 seedlings. TUBULIN was used as a loading control.
(C) The phenotypes of 4-week-old wild-type (Col-0) and lsm4 mutant seedlings.
(D) The sensitivity of LSM4+/− F2 seedlings to ABA and salt stress. Photographs were taken after 8 d of culture at 23°C. Red arrows show nongreening seedlings or ungerminated seeds.
(E) Quantitative analysis of wild-type (Col-0) and LSM4+/− F2 greening cotyledon rate in MS medium containing 0.5 μM ABA and of germination rate in MS medium containing 100 mM NaCl, as shown in (D), n = 120.
(F) and (G) The pre-mRNA splicing pattern of genes, whose pre-mRNA splicing was affected by SKB1 mutation, in the lsm4 mutant.
DISCUSSION
In this study, we revealed how the Arabidopsis floral initiator SKB1, a protein Arg methyltransferase, perceives salt stress by changing the level of its catalytic substrates, H4R3 and LSM4. Whereas the level of H4R3sme2, a symmetric dimethylated H4R3 modified by SKB1, decreased with salt treatment, coincident with upregulation of mRNA transcription, the sDMA levels of LSM4, a U6 snRNP-associated protein involved in posttranscriptional regulation, increased with salt treatment. Consistently, when H4R3sme2 levels and the sDMA levels of LSM4 were inhibited in the skb1 mutant, plants displayed a constitutive salt response related to development as well as changes in transcription and posttranscription, which indicates that SKB1 provides information on the salt status of the cell.
SKB1 as a Salt-Responsive Regulator Functions in Transcription and Posttranscription
Gene expression is determined by both transcription and posttranscription. Transcription is largely regulated by histone modifications. The histone mark H4R3sme2, catalyzed by SKB1 or its homolog PRMT5, suppresses gene transcription, which is supported by the observation that decreasing H4R3sme2 levels increase gene transcription by disrupting SKB1 (Pal et al., 2004; Wang et al., 2007; Schmitz et al., 2008; Zhao et al., 2009; Majumder et al., 2010). When plants are exposed to high concentrations of salt and ABA, SKB1 disassociates from chromatin to reduce H4R3sme2 levels and the resulting high expression of genes in ABA signaling and salt response (Figures 3 and 4). Our results suggest that SKB1-mediated H4R3sme2 is also a response to salt and ABA. This result suggests a direct mechanism by which salt and ABA affect gene transcription. Posttranscriptional regulation of gene expression includes pre-mRNA splicing, RNA transport, and decay. We and others found that SKB1 methylates spliceosome component protein(s) for pre-mRNA splicing (Meister and Fischer, 2002; Bedford and Richard, 2005; Gonsalvez et al., 2006; Anne et al., 2007; Jansson et al., 2008; Bedford and Clarke, 2009; Bruns et al., 2009; Deng et al., 2010; Sanchez et al., 2010). SKB1 methylates LSM4, and its methylation is increased with salt and ABA treatment but is decreased in skb1 mutant plants (Figure 4), which suggests that LSM4 methylation levels respond to salt and ABA stress. Furthermore, skb1 and lsm4 mutant plants show defects in pre-mRNA splicing (Figures 7F and 7G), which suggests that SKB1 is linked to the posttranscriptional regulation of gene expression in response to salt. This finding also implies that the phenomena of low expression of genes in the salt response of the skb1 mutant whereby these mis-spliced RNAs such as intron retention might be degraded in the posttranscriptional process.
Cross-Link between Salt Response and Developmental Processes
High concentrations of salt delay flowering time, either by increasing the expression of FLC or decreasing LFY expression in Arabidopsis. The accumulation of DELLA proteins in the gibberellin pathway, which induces growth restrain and inhibits the expression of LFY, promotes the survival of plants under high salt conditions (Achard et al., 2006). In contrast with DELLA accumulation, which is resistant to high concentrations of salt, skb1 mutant plants, which exhibit retarded growth and late flowering, are sensitive to salt, which suggests that another mechanism exists that connects salt tolerance and developmental processes. Previous studies by us and others have shown that the expression of FLC was upregulated in the skb1-1 mutant, which downregulates LFY expression (Pei et al., 2007; Wang et al., 2007). Here, we found that SKB1 dissociates from the FLC promoter after high salt and ABA treatment and that H4R3sme2 levels at the FLC promoter decrease correspondingly (Figure 4G). This increases FLC expression (Figure 4G) and results in late flowering in the wild type. In addition, salt stress–induced late flowering does not occur in skb1 mutant plants (Figure 2D). This finding suggests that the salt response pathway activates FLC expression in response to salt stress, independently of DELLAs. However, other autonomous pathway mutants, such as fld, fve, fca, flk, fy, ld, prmt4a/4b, prmt10, and the PAF1 complex mutant sdg8, which up- or downregulate FLC expression, as well as the flc mutant, are neither more sensitive nor more tolerant to salt (Figures 2C and 2D). Consistently, the skb1 flc double mutant suppresses the skb1 late-flowering phenotype but does not rescue growth retardation and salt sensitivity (Figures 2A and 2B). This suggests that FLC expression only confers flowering time in the presence or absence of salt but does not permit salt tolerance or growth retardation. Interestingly, another salt response pathway, pre-mRNA splicing, also affects flowering time and growth in plants (Quesada et al., 2005; Terzi and Simpson, 2008; Lorković, 2009). Pre-mRNA splicing is also perturbed in skb1 mutant plants (Deng et al., 2010; Sanchez et al., 2010; this work) by reducing Arg symmetric dimethylation of LSM4 (Figure 5), a U6 snRNP protein in our study (Figure 7). Moreover, SAD1, a homolog of human LSM5 and another U6 snRNA-associated LSM protein, also confers salt and ABA tolerance and retards plant growth (Xiong et al., 2001b). These results suggest that SKB1 influences plant growth and salt tolerance by regulating pre-mRNA splicing through LSM methylation.
Regulation of SKB1 Dissociation from Chromatin in Response to Salt Stress
SKB1, or PRMT5, methylates Arg residues in multiple histone and nonhistone proteins, including H4R3 (Pei et al., 2007; Wang et al., 2007; Schmitz et al., 2008) and LSM4 in plants and H4R3, H3R8 (Pal et al., 2004), p53 (Jansson et al., 2008), p300 (Yang et al., 2009), 130-kD cis-Golgi matrix protein (GM130) (Zhou et al., 2010), slit-robo Rho GTPase activating protein 2 (srGAP2) (Guo and Bao, 2010), Ribosomal protein S10 (Rps10) (Ren et al., 2010) and SM (Friesen et al., 2001; Meister and Fischer, 2002) in animals. SKB1 localizes to both the cytoplasm and the nucleus. In the nucleus, SKB1-mediated H4R3sme2, as well as H3R8sme2 in mammalian cells, is closely associated with transcriptional repression. In the cytoplasm, SKB1 mainly exists in the spliceosomal complex (Meister and Fischer, 2002) and Golgi apparatus (Zhou et al., 2010). The subcellular distribution of SKB1 in the cytoplasm and the nucleus, which link to substrate selection, is attributed to the status of cells. Regulation of SKB1 function remains poorly understood. In mammalian cells, the association of SKB1 with Blimp1 is thought to be critical for its translocation from the nucleus to the cytoplasm (Ancelin et al., 2006). By contrast, coexpression of its binding partners, AJUBA and SNAIl, increases the redistribution of SKB1 in the nucleus (Hou et al., 2008). In addition, the association of COPR5 with SKB1 changes the balance of affinity for substrates from H3R8 toward H4R3 (Lacroix et al., 2008). In plants, the association of SKB1 with chromatin is reduced in response to salt stress. However, it is unclear if SKB1 is released from chromatin to regulate splicing by changing association partners.
Modification of SKB1 by phosphorylation, acetylation, or ubiquitylation has not been reported, although possible modification sites of human PRMT5 are reported in the human protein reference database (http://www.hprd.org). It would be interesting to determine whether plant SKB1 is indeed modified and whether this modification shifts the balance of activity from H4R3 toward LSM4 in response to salt stress.
Methylation of LSM4 Influences Its Function in Pre-mRNA Splicing
A key form of posttranscriptional regulation is RNA splicing, which occurs in the spliceosome. The Sm and Lsm proteins are core proteins of spliceosomal U snRNPs. SmB/B', SmD1, SmD3, and Lsm4 are symmetrically dimethylated by PRMT5, and these methylations increase but are not required for the binding to the SMN complex to promote the assembly of the spliceosome (Chari et al., 2008). Therefore, PRMT5-dependent Sm or Lsm methylation might modulate the kinetics or efficiency of UsnRNPs biogenesis and pre-mRNA splicing. Although direct evidence linking LSM4 symmetric dimethylation with UsnRNPs assembly is lacking in Arabidopsis, given that splicing machinery and SKB1 are evolutionarily conserved and that LSM4 symmetric dimethylation by SKB1 was found in this work and other (Deng et al., 2010), we speculate that methylation of LSM4 contributes to the efficiency of UsnRNP assembly. In addition, our results and the recent studies of Sanchez et al. (2010) and Deng et al. (2010) revealed that splicing defects in the skb1 mutants occur in several specific biological processes, indicating that LSM4 symmetric dimethylation might regulate splicing site selection. Interestingly, in addition to LSM4, other Arabidopsis proteins involved in the RNA splicing process, including SM, LSM, and RNA binding proteins, contain Gly-Arg–rich domains that are methylated by SKB1 or other methyltransferases. Determining whether and how the methylation of these Arg residues regulates splicing site recognition during different biological events will enhance our understanding of the molecular mechanism underlying pre-mRNA splicing.
We propose that SKB1 affects gene transcription by methylating H4R3 and functions posttranscriptionally by modifying LSM4. Under normal growth conditions, SKB1 associates mainly with chromatin to maintain H4R3sme2 at high levels and thereby suppresses the transcription of genes such as FLC, which regulates flowering time, and RD29A, RD29B, and HAB1, which function in stress signaling. Meanwhile, the methylation levels of LSM4 are kept at relatively low levels to regulate pre-mRNA splicing of genes such as At1G18160 for intron removal and At1G13350 for alternative splicing. When plants are exposed to salt stress, a new balance between transcription and posttranscription regulation is established by changing the levels of SKB1-mediated H4R3sme2 and LSM4. In response to salt, H4R3sme2 levels decline, which increases the transcription of FLC and stress response genes, such as RD29A, RD29B, and HAB1. Coincident with transcription, SKB1 methylates LSM4, which improves the efficiency of pre-mRNA splicing of At1G18160 or alters the splicing variants of genes such as At1g13350. Thus, SKB1 is likely an integrator of plant development and the salt response.
METHODS
Plant Materials and Growth Conditions
The Arabidopsis thaliana ecotype Col-0 was used as wild type in this study. skb1-1, skb1-2, 35S:SKB1 skb1-1, 35S:SKB1 Col-0, skb1-1 flc-3, flc-3, fld-4, and fve-3 were identified or described previously (He et al., 2003; Ausín et al., 2004; Bezerra et al., 2004; Wang et al., 2007). prmt4a prmt4b, prmt10, and fy were previously described (Niu et al., 2007, 2008) and were gifts from Xiaofeng Cao. fca (SALK_057540), flk (SALK_007750), ld (SALK_150861), sdg8 (SALK_065480), and lsm4 (SALK_063398) were isolated from the SALK collection, and the gene-specific primers used to verify these mutants and the T-DNA left border primer LBb1 are listed in Supplemental Table 1 online. Seeds of each genotype were surface sterilized with 70% ethanol for 1 min and 15% sodium hypochlorite for 15 min and washed four times with sterile water. Sterilized seeds were plated on MS medium. For experimental treatments, the MS agar medium was supplemented with NaCl, KNO3, KCl, LiCl, or ABA at concentrations listed in Results. Plants were stratified at 4°C in darkness for 3 d and then transferred to a culture room at 23°C with a 16-h-light/8-h-dark photoperiod (light intensity ~120 μE s−1m−2).
Salt Stress Tolerance and Root Growth Response to Salts
Seeds were germinated on MS agar medium. For the NaCl tolerance assay, 4-d-old seedlings were transferred from germination medium to MS agar medium containing different concentrations of NaCl. For the root growth assay, 4-d-old seedlings were transferred from germination medium to MS agar medium containing different concentrations of NaCl, and then the seedlings were grown vertically for 10 d.
ABA Sensitivity Assay
To measure ABA sensitivity at germination, seeds were plated on MS agar medium containing various concentrations of ABA. To score seed germination, the percentage of seeds showing cotyledon emergence was determined 3 d after transfer to 23°C. To measure ABA sensitivity of postgermination growth, the seedlings with green cotyledons were counted to determine the percentage of cotyledon greening, as ABA is an inhibitor of cotyledon greening.
Electrolyte Leakage Test
Relative electrolyte leakage was determined according to a previous description (Cao et al., 2007).
RNA Extraction and Real-Time Quantitative PCR Analysis
Total RNA was isolated from 11-d-old seedlings treated with MS liquid medium or MS liquid medium containing 200 mM NaCl for 6 h or 100 μM ABA for 3 h with TRIzol reagent (Invitrogen) as recommended by the manufacturer and digested with DNase I (TaKaRa). cDNAs were synthesized from 4 μg total RNA using Superscript III reverse transcriptase (Invitrogen). The cDNA was quantified using a SYBR PCR master mix (Applied Biosystems) with gene-specific primers (see Supplemental Table 1 online) in an Applied Biosystems real-time system.
Microarray Analysis
Total RNA was isolated with Trizol reagent (Invitrogen) from 11-d-old seedlings of the wild type and skb1-1 mutant without or with 200 mM NaCl for 6 h. The 29k Arabidopsis Genome Array hybridization was performed by CapitalBio Corporation. Array scanning and data analysis were performed as previously described (Zheng and Wang, 2008; Li et al., 2009). Two independent biological replicates were performed, and only genes whose expression alteration was consistent in two microarray assays were selected as differentially expressed genes. These microarray data are available at http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE26398.
Transcriptome Sequencing
Total RNA was isolated with Trizol reagent (Invitrogen) from 11-d-old seedlings of the wild type and skb1-1 mutant grown in standard conditions. Experiments were performed and data were analyzed by the Beijing Genomics Institute as previously described (Wang et al., 2009).
Immunoblot Analysis
Total protein was extracted from 11-d-old seedlings treated with MS liquid medium or MS liquid medium with 200 mM NaCl for 6 h or 100 μM ABA for 3 h. Immunoblot analysis was performed with anti-SKB1 polyclonal antibody and SYM11. Histone-enriched protein was isolated as described (Lildballe et al., 2008), and immunoblot analysis was performed with antibodies against H4 symmetric dimethyl Arg 3 (Abcam; catalog number ab5823).
ChIP
Twenty-day-old seedlings grown under long-day conditions and treated with MS medium with 0 or 200 mM NaCl for 8 h were used in a ChIP assay as previously described (Bowler et al., 2004). Primers for quantitative PCR detection of RD29A, RD29B, FLC, or HAB1 chromatin regions are listed in Supplemental Table 1 online.
Constructs, Protein Purification, and Methylation Analysis
Full-length coding sequences of Arabidopsis SMs or LSMs were amplified by PCR and cloned in frame into pGEX4T-1. Specific primers are presented in Supplemental Table 1 online. GST-SM or GST-LSM expression, purification, and methylation assays were as previously described (Bao et al., 2001).
SYM11 Immunoprecipitation
Immunoprecipitation was performed as previously described (Shalitin et al., 2002) with some modification. Briefly, seedlings were treated with MS medium with 100 μM ABA for 5 h and homogenized in ice-cold immunoprecipitation buffer (20 mM Tris-HCl, pH 8.0, 150 mM NaCl, 1 mM EDTA, 10% glycerol, 0.2% Triton X-100, 1 mM PMSF, and 1× Complete protease inhibitor cocktail [Roche]). Extracts were passed through 0.22-μm filters. The filtrate was then precleared by the addition of 20 μL protein G-sepharose beads (Merck) at 4°C for 1 h. SYM11 was added (1:150) to the filtrate, and samples were incubated at 4°C for 4 h. Then, protein G-sepharose was added (1:150), and samples were incubated at 4°C for 2 h. The immunoprecipitation complex was washed three times with ice-cold washing buffer (50 mM Tris-HCl, pH 8.0, 140 mM NaCl, 1 mM EDTA, and 0.1% Triton X-100), mixed with SDS-PAGE sample buffer, and boiled for 5 min. SDS-PAGE was performed, and the gels were stained with silver stain. Proteins were identified by mass spectrometry as previously described (Boisvert et al., 2003).
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL data libraries under the following accession numbers: SKB1, At4G31120; RD29A, At5G52310; RD29B, At5G52300; RD22, At5G25610; RAB18, At5G66400; COR47, At1G20440; DREB2A, At5G05410; DREB2B, At3G11020; ABF2, At1G45249; ABF3, At4G34000; ABI1, At4G26080; HAB1, At1G72770; HAB2, At1G17550; MRK1: At3G63260; MEK1, At4G26070; MEKK1, At4G08500; CDPK2, At1G35670; CIPK3, At2G26980; CDPK13, At3G51850; CBL1, At4G17615; RLP4, At1G28340; TUBULIN, At5G62690; SME-A, At4G30330; SME-B, At2G18740; SMG-B, At3G11500; LSM4, At5G27720; LSM6-B, At2G43810; LSM7, At2G03870; and LSM8, At1G65 700. The microarray data were submitted to the National Center for Biotechnology Information Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE26398).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. The Phenotypes of flc-3, Wild Type (Col-0), skb1-1 flc-3, and skb1-1 Grown in Soil for 35 d under Long Days.
Supplemental Figure 2. The skb1 Mutants Are Hypersensitive to Salt Stress.
Supplemental Figure 3. Overexpression of SKB1 Slightly Increases the Tolerance to Salt Stress.
Supplemental Figure 4. The Phenotypes of Flowering Time-Controlling Gene Mutants Grown on Medium with Salt.
Supplemental Figure 5. Growth of Plants on MS Medium Containing Different Concentrations of ABA.
Supplemental Figure 6. Anti-SKB1 and -H4R3sme2 Antibodies Could Not Pull Down Chromatin DNA in the skb1-1 Mutant.
Supplemental Figure 7. Sequence Alignment of LSM4 Homologs in Arabidopsis, Human, Mouse, and Yeast.
Supplemental Figure 8. RT-PCR Analysis of the Splice Variants of Ser/Arg-Rich Proteins Encoding Genes.
Supplemental Table 1. Primers for Real-Time Quantitative RT-PCR, Gene Cloning, and Verification of SALK T-DNA Insertion Mutants.
Supplemental Table 2. Alternative Splicing Changes in the Wild Type and skb1-1 Mutant.
Supplementary Material
Acknowledgments
We thank Yan Guo for critical discussion of this work. We also thank Ruiling Mu, Wei Zhang, Shanshan Zhu, Huajie Fan, and Huilan Wu for technical assistance. This work was supported by the National Basic Research Program of China Grants 2007CB948202 and 2009CB825506 (to S.B.) and the Chinese National Natural and Sciences Foundation Grants 31071120, 91019018 (to S.B.), 30800626 (to Y.Z.), and 31000129 (to D.L.).
References
- Achard P., Cheng H., De Grauwe L., Decat J., Schoutteten H., Moritz T., Van Der Straeten D., Peng J., Harberd N.P. (2006). Integration of plant responses to environmentally activated phytohormonal signals. Science 311: 91–94 [DOI] [PubMed] [Google Scholar]
- Allen G.J., Kuchitsu K., Chu S.P., Murata Y., Schroeder J.I. (1999). Arabidopsis abi1-1 and abi2-1 phosphatase mutations reduce abscisic acid-induced cytoplasmic calcium rises in guard cells. Plant Cell 11: 1785–1798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ancelin K., Lange U.C., Hajkova P., Schneider R., Bannister A.J., Kouzarides T., Surani M.A. (2006). Blimp1 associates with Prmt5 and directs histone arginine methylation in mouse germ cells. Nat. Cell Biol. 8: 623–630 [DOI] [PubMed] [Google Scholar]
- Anne J., Ollo R., Ephrussi A., Mechler B.M. (2007). Arginine methyltransferase Capsuleen is essential for methylation of spliceosomal Sm proteins and germ cell formation in Drosophila. Development 134: 137–146 [DOI] [PubMed] [Google Scholar]
- Apse M.P., Aharon G.S., Snedden W.A., Blumwald E. (1999). Salt tolerance conferred by overexpression of a vacuolar Na+/H+ antiport in Arabidopsis. Science 285: 1256–1258 [DOI] [PubMed] [Google Scholar]
- Ausín I., Alonso-Blanco C., Jarillo J.A., Ruiz-García L., Martínez-Zapater J.M. (2004). Regulation of flowering time by FVE, a retinoblastoma-associated protein. Nat. Genet. 36: 162–166 [DOI] [PubMed] [Google Scholar]
- Bao S., Qyang Y., Yang P., Kim H., Du H., Bartholomeusz G., Henkel J., Pimental R., Verde F., Marcus S. (2001). The highly conserved protein methyltransferase, Skb1, is a mediator of hyperosmotic stress response in the fission yeast Schizosaccharomyces pombe. J. Biol. Chem. 276: 14549–14552 [DOI] [PubMed] [Google Scholar]
- Bäurle I., Smith L., Baulcombe D.C., Dean C. (2007). Widespread role for the flowering-time regulators FCA and FPA in RNA-mediated chromatin silencing. Science 318: 109–112 [DOI] [PubMed] [Google Scholar]
- Becker D., Hoth S., Ache P., Wenkel S., Roelfsema M.R., Meyerhoff O., Hartung W., Hedrich R. (2003). Regulation of the ABA-sensitive Arabidopsis potassium channel gene GORK in response to water stress. FEBS Lett. 554: 119–126 [DOI] [PubMed] [Google Scholar]
- Bedford M.T. (2007). Arginine methylation at a glance. J. Cell Sci. 120: 4243–4246 [DOI] [PubMed] [Google Scholar]
- Bedford M.T., Clarke S.G. (2009). Protein arginine methylation in mammals: Who, what, and why. Mol. Cell 33: 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedford M.T., Richard S. (2005). Arginine methylation an emerging regulator of protein function. Mol. Cell 18: 263–272 [DOI] [PubMed] [Google Scholar]
- Bezerra I.C., Michaels S.D., Schomburg F.M., Amasino R.M. (2004). Lesions in the mRNA cap-binding gene ABA HYPERSENSITIVE 1 suppress FRIGIDA-mediated delayed flowering in Arabidopsis. Plant J. 40: 112–119 [DOI] [PubMed] [Google Scholar]
- Blázquez M.A., Ahn J.H., Weigel D. (2003). A thermosensory pathway controlling flowering time in Arabidopsis thaliana. Nat. Genet. 33: 168–171 [DOI] [PubMed] [Google Scholar]
- Boisvert F.M., Côté J., Boulanger M.C., Richard S. (2003). A proteomic analysis of arginine-methylated protein complexes. Mol. Cell. Proteomics 2: 1319–1330 [DOI] [PubMed] [Google Scholar]
- Bond D.M., Wilson I.W., Dennis E.S., Pogson B.J., Jean Finnegan E. (2009). VERNALIZATION INSENSITIVE 3 (VIN3) is required for the response of Arabidopsis thaliana seedlings exposed to low oxygen conditions. Plant J. 59: 576–587 [DOI] [PubMed] [Google Scholar]
- Bowler C., Benvenuto G., Laflamme P., Molino D., Probst A.V., Tariq M., Paszkowski J. (2004). Chromatin techniques for plant cells. Plant J. 39: 776–789 [DOI] [PubMed] [Google Scholar]
- Bruns A.F., Grothe C., Claus P. (2009). Fibroblast growth factor 2 (FGF-2) is a novel substrate for arginine methylation by PRMT5. Biol. Chem. 390: 59–65 [DOI] [PubMed] [Google Scholar]
- Cao W.H., Liu J., He X.J., Mu R.L., Zhou H.L., Chen S.Y., Zhang J.S. (2007). Modulation of ethylene responses affects plant salt-stress responses. Plant Physiol. 143: 707–719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chari A., Golas M.M., Klingenhäger M., Neuenkirchen N., Sander B., Englbrecht C., Sickmann A., Stark H., Fischer U. (2008). An assembly chaperone collaborates with the SMN complex to generate spliceosomal SnRNPs. Cell 135: 497–509 [DOI] [PubMed] [Google Scholar]
- Chinnusamy V., Schumaker K., Zhu J.K. (2004). Molecular genetic perspectives on cross-talk and specificity in abiotic stress signalling in plants. J. Exp. Bot. 55: 225–236 [DOI] [PubMed] [Google Scholar]
- Christmann A., Moes D., Himmelbach A., Yang Y., Tang Y., Grill E. (2006). Integration of abscisic acid signalling into plant responses. Plant Biol. (Stuttg.) 8: 314–325 [DOI] [PubMed] [Google Scholar]
- Cooper M., Johnston L.H., Beggs J.D. (1995). Identification and characterization of Uss1p (Sdb23p): A novel U6 snRNA-associated protein with significant similarity to core proteins of small nuclear ribonucleoproteins. EMBO J. 14: 2066–2075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutler S.R., Rodriguez P.L., Finkelstein R.R., Abrams S.R. (2010). Abscisic acid: Emergence of a core signaling network. Annu. Rev. Plant Biol. 61: 651–679 [DOI] [PubMed] [Google Scholar]
- Dalal M., Tayal D., Chinnusamy V., Bansal K.C. (2009). Abiotic stress and ABA-inducible Group 4 LEA from Brassica napus plays a key role in salt and drought tolerance. J. Biotechnol. 139: 137–145 [DOI] [PubMed] [Google Scholar]
- Deng X., Gu L., Liu C., Lu T., Lu F., Lu Z., Cui P., Pei Y., Wang B., Hu S., Cao X. (2010). Arginine methylation mediated by the Arabidopsis homolog of PRMT5 is essential for proper pre-mRNA splicing. Proc. Natl. Acad. Sci. USA 107: 19114–19119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabbrizio E., El Messaoudi S., Polanowska J., Paul C., Cook J.R., Lee J.H., Negre V., Rousset M., Pestka S., Le Cam A., Sardet C. (2002). Negative regulation of transcription by the type II arginine methyltransferase PRMT5. EMBO Rep. 3: 641–645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friesen W.J., Paushkin S., Wyce A., Massenet S., Pesiridis G.S., Van Duyne G., Rappsilber J., Mann M., Dreyfuss G. (2001). The methylosome, a 20S complex containing JBP1 and pICln, produces dimethylarginine-modified Sm proteins. Mol. Cell. Biol. 21: 8289–8300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong Z., Morales-Ruiz T., Ariza R.R., Roldán-Arjona T., David L., Zhu J.K. (2002). ROS1, a repressor of transcriptional gene silencing in Arabidopsis, encodes a DNA glycosylase/lyase. Cell 111: 803–814 [DOI] [PubMed] [Google Scholar]
- Gonsalvez G.B., Rajendra T.K., Tian L., Matera A.G. (2006). The Sm-protein methyltransferase, dart5, is essential for germ-cell specification and maintenance. Curr. Biol. 16: 1077–1089 [DOI] [PubMed] [Google Scholar]
- Gosti F., Beaudoin N., Serizet C., Webb A.A., Vartanian N., Giraudat J. (1999). ABI1 protein phosphatase 2C is a negative regulator of abscisic acid signaling. Plant Cell 11: 1897–1910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo S., Bao S. (2010). srGAP2 arginine methylation regulates cell migration and cell spreading through promoting dimerization. J. Biol. Chem. 285: 35133–35141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He W., Parker R. (2000). Functions of Lsm proteins in mRNA degradation and splicing. Curr. Opin. Cell Biol. 12: 346–350 [DOI] [PubMed] [Google Scholar]
- He Y. (2009). Control of the transition to flowering by chromatin modifications. Mol. Plant 2: 554–564 [DOI] [PubMed] [Google Scholar]
- He Y., Amasino R.M. (2005). Role of chromatin modification in flowering-time control. Trends Plant Sci. 10: 30–35 [DOI] [PubMed] [Google Scholar]
- He Y., Michaels S.D., Amasino R.M. (2003). Regulation of flowering time by histone acetylation in Arabidopsis. Science 302: 1751–1754 [DOI] [PubMed] [Google Scholar]
- Hou Z., Peng H., Ayyanathan K., Yan K.P., Langer E.M., Longmore G.D., Rauscher F.J., III (2008). The LIM protein AJUBA recruits protein arginine methyltransferase 5 to mediate SNAIL-dependent transcriptional repression. Mol. Cell. Biol. 28: 3198–3207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansson M., Durant S.T., Cho E.C., Sheahan S., Edelmann M., Kessler B., La Thangue N.B. (2008). Arginine methylation regulates the p53 response. Nat. Cell Biol. 10: 1431–1439 [DOI] [PubMed] [Google Scholar]
- Kim H.J., Hyun Y., Park J.Y., Park M.J., Park M.K., Kim M.D., Kim H.J., Lee M.H., Moon J., Lee I., Kim J. (2004). A genetic link between cold responses and flowering time through FVE in Arabidopsis thaliana. Nat. Genet. 36: 167–171 [DOI] [PubMed] [Google Scholar]
- Kumar S.V., Wigge P.A. (2010). H2A.Z-containing nucleosomes mediate the thermosensory response in Arabidopsis. Cell 140: 136–147 [DOI] [PubMed] [Google Scholar]
- Kwak Y.T., Guo J., Prajapati S., Park K.J., Surabhi R.M., Miller B., Gehrig P., Gaynor R.B. (2003). Methylation of SPT5 regulates its interaction with RNA polymerase II and transcriptional elongation properties. Mol. Cell 11: 1055–1066 [DOI] [PubMed] [Google Scholar]
- Lacroix M., El Messaoudi S., Rodier G., Le Cam A., Sardet C., Fabbrizio E. (2008). The histone-binding protein COPR5 is required for nuclear functions of the protein arginine methyltransferase PRMT5. EMBO Rep. 9: 452–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung J., Bouvier-Durand M., Morris P.C., Guerrier D., Chefdor F., Giraudat J. (1994). Arabidopsis ABA response gene ABI1: features of a calcium-modulated protein phosphatase. Science 264: 1448–1452 [DOI] [PubMed] [Google Scholar]
- Leung J., Merlot S., Giraudat J. (1997). The Arabidopsis ABSCISIC ACID-INSENSITIVE2 (ABI2) and ABI1 genes encode homologous protein phosphatases 2C involved in abscisic acid signal transduction. Plant Cell 9: 759–771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., et al. (2009). Gene expression-based classification and regulatory networks of pediatric acute lymphoblastic leukemia. Blood 114: 4486–4493 [DOI] [PubMed] [Google Scholar]
- Lildballe D.L., Pedersen D.S., Kalamajka R., Emmersen J., Houben A., Grasser K.D. (2008). The expression level of the chromatin-associated HMGB1 protein influences growth, stress tolerance, and transcriptome in Arabidopsis. J. Mol. Biol. 384: 9–21 [DOI] [PubMed] [Google Scholar]
- Liu Z., Zhou Z., Chen G., Bao S. (2007). A putative transcriptional elongation factor hIws1 is essential for mammalian cell proliferation. Biochem. Biophys. Res. Commun. 353: 47–53 [DOI] [PubMed] [Google Scholar]
- Lorković Z.J. (2009). Role of plant RNA-binding proteins in development, stress response and genome organization. Trends Plant Sci. 14: 229–236 [DOI] [PubMed] [Google Scholar]
- Mahajan S., Pandey G.K., Tuteja N. (2008). Calcium- and salt-stress signaling in plants: Shedding light on SOS pathway. Arch. Biochem. Biophys. 471: 146–158 [DOI] [PubMed] [Google Scholar]
- Majumder S., Alinari L., Roy S., Miller T., Datta J., Sif S., Baiocchi R., Jacob S.T. (2010). Methylation of histone H3 and H4 by PRMT5 regulates ribosomal RNA gene transcription. J. Cell. Biochem. 109: 553–563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui A., et al. (2008). Arabidopsis transcriptome analysis under drought, cold, high-salinity and ABA treatment conditions using a tiling array. Plant Cell Physiol. 49: 1135–1149 [DOI] [PubMed] [Google Scholar]
- Mazzoni C., Herker E., Palermo V., Jungwirth H., Eisenberg T., Madeo F., Falcone C. (2005). Yeast caspase 1 links messenger RNA stability to apoptosis in yeast. EMBO Rep. 6: 1076–1081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meister G., Fischer U. (2002). Assisted RNP assembly: SMN and PRMT5 complexes cooperate in the formation of spliceosomal UsnRNPs. EMBO J. 21: 5853–5863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer K., Leube M.P., Grill E. (1994). A protein phosphatase 2C involved in ABA signal transduction in Arabidopsis thaliana. Science 264: 1452–1455 [DOI] [PubMed] [Google Scholar]
- Munns R., Tester M. (2008). Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 59: 651–681 [DOI] [PubMed] [Google Scholar]
- Murata Y., Pei Z.M., Mori I.C., Schroeder J. (2001). Abscisic acid activation of plasma membrane Ca(2+) channels in guard cells requires cytosolic NAD(P)H and is differentially disrupted upstream and downstream of reactive oxygen species production in abi1-1 and abi2-1 protein phosphatase 2C mutants. Plant Cell 13: 2513–2523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashima K., Shinwari Z.K., Sakuma Y., Seki M., Miura S., Shinozaki K., Yamaguchi-Shinozaki K. (2000). Organization and expression of two Arabidopsis DREB2 genes encoding DRE-binding proteins involved in dehydration- and high-salinity-responsive gene expression. Plant Mol. Biol. 42: 657–665 [DOI] [PubMed] [Google Scholar]
- Niu L., Lu F., Pei Y., Liu C., Cao X. (2007). Regulation of flowering time by the protein arginine methyltransferase AtPRMT10. EMBO Rep. 8: 1190–1195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu L., Zhang Y., Pei Y., Liu C., Cao X. (2008). Redundant requirement for a pair of PROTEIN ARGININE METHYLTRANSFERASE4 homologs for the proper regulation of Arabidopsis flowering time. Plant Physiol. 148: 490–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pahlich S., Zakaryan R.P., Gehring H. (2006). Protein arginine methylation: Cellular functions and methods of analysis. Biochim. Biophys. Acta 1764: 1890–1903 [DOI] [PubMed] [Google Scholar]
- Pal S., Vishwanath S.N., Erdjument-Bromage H., Tempst P., Sif S. (2004). Human SWI/SNF-associated PRMT5 methylates histone H3 arginine 8 and negatively regulates expression of ST7 and NM23 tumor suppressor genes. Mol. Cell. Biol. 24: 9630–9645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal S., Yun R., Datta A., Lacomis L., Erdjument-Bromage H., Kumar J., Tempst P., Sif S. (2003). mSin3A/histone deacetylase 2- and PRMT5-containing Brg1 complex is involved in transcriptional repression of the Myc target gene cad. Mol. Cell. Biol. 23: 7475–7487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palusa S.G., Ali G.S., Reddy A.S. (2007). Alternative splicing of pre-mRNAs of Arabidopsis serine/arginine-rich proteins: regulation by hormones and stresses. Plant J. 49: 1091–1107 [DOI] [PubMed] [Google Scholar]
- Pandey S., Nelson D.C., Assmann S.M. (2009). Two novel GPCR-type G proteins are abscisic acid receptors in Arabidopsis. Cell 136: 136–148 [DOI] [PubMed] [Google Scholar]
- Pannone B.K., Kim S.D., Noe D.A., Wolin S.L. (2001). Multiple functional interactions between components of the Lsm2-Lsm8 complex, U6 snRNA, and the yeast La protein. Genetics 158: 187–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pannone B.K., Wolin S.L. (2000). Sm-like proteins wRING the neck of mRNA. Curr. Biol. 10: R478–R481 [DOI] [PubMed] [Google Scholar]
- Park S.Y., et al. (2009). Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science 324: 1068–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei Y., Niu L., Lu F., Liu C., Zhai J., Kong X., Cao X. (2007). Mutations in the Type II protein arginine methyltransferase AtPRMT5 result in pleiotropic developmental defects in Arabidopsis. Plant Physiol. 144: 1913–1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quan R., Lin H., Mendoza I., Zhang Y., Cao W., Yang Y., Shang M., Chen S., Pardo J.M., Guo Y. (2007). SCABP8/CBL10, a putative calcium sensor, interacts with the protein kinase SOS2 to protect Arabidopsis shoots from salt stress. Plant Cell 19: 1415–1431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quesada V., Dean C., Simpson G.G. (2005). Regulated RNA processing in the control of Arabidopsis flowering. Int. J. Dev. Biol. 49: 773–780 [DOI] [PubMed] [Google Scholar]
- Reddy A.S. (2004). Plant serine/arginine-rich proteins and their role in pre-mRNA splicing. Trends Plant Sci. 9: 541–547 [DOI] [PubMed] [Google Scholar]
- Ren J., Wang Y., Liang Y., Zhang Y., Bao S., Xu Z. (2010). Methylation of ribosomal protein S10 by protein-arginine methyltransferase 5 regulates ribosome biogenesis. J. Biol. Chem. 285: 12695–12705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruggiero B., Koiwa H., Manabe Y., Quist T.M., Inan G., Saccardo F., Joly R.J., Hasegawa P.M., Bressan R.A., Maggio A. (2004). Uncoupling the effects of abscisic acid on plant growth and water relations. Analysis of sto1/nced3, an abscisic acid-deficient but salt stress-tolerant mutant in Arabidopsis. Plant Physiol. 136: 3134–3147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saez A., Apostolova N., Gonzalez-Guzman M., Gonzalez-Garcia M.P., Nicolas C., Lorenzo O., Rodriguez P.L. (2004). Gain-of-function and loss-of-function phenotypes of the protein phosphatase 2C HAB1 reveal its role as a negative regulator of abscisic acid signalling. Plant J. 37: 354–369 [DOI] [PubMed] [Google Scholar]
- Saez A., Robert N., Maktabi M.H., Schroeder J.I., Serrano R., Rodriguez P.L. (2006). Enhancement of abscisic acid sensitivity and reduction of water consumption in Arabidopsis by combined inactivation of the protein phosphatases type 2C ABI1 and HAB1. Plant Physiol. 141: 1389–1399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez S.E., et al. (2010). A methyl transferase links the circadian clock to the regulation of alternative splicing. Nature 468: 112–116 [DOI] [PubMed] [Google Scholar]
- Schmitz R.J., Sung S., Amasino R.M. (2008). Histone arginine methylation is required for vernalization-induced epigenetic silencing of FLC in winter-annual Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 105: 411–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo M., Peeters A.J., Koiwai H., Oritani T., Marion-Poll A., Zeevaart J.A., Koornneef M., Kamiya Y., Koshiba T. (2000). The Arabidopsis aldehyde oxidase 3 (AAO3) gene product catalyzes the final step in abscisic acid biosynthesis in leaves. Proc. Natl. Acad. Sci. USA 97: 12908–12913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano R., Rodriguez-Navarro A. (2001). Ion homeostasis during salt stress in plants. Curr. Opin. Cell Biol. 13: 399–404 [DOI] [PubMed] [Google Scholar]
- Shalitin D., Yang H., Mockler T.C., Maymon M., Guo H., Whitelam G.C., Lin C. (2002). Regulation of Arabidopsis cryptochrome 2 by blue-light-dependent phosphorylation. Nature 417: 763–767 [DOI] [PubMed] [Google Scholar]
- Sridhar V.V., Kapoor A., Zhang K., Zhu J., Zhou T., Hasegawa P.M., Bressan R.A., Zhu J.K. (2007). Control of DNA methylation and heterochromatic silencing by histone H2B deubiquitination. Nature 447: 735–738 [DOI] [PubMed] [Google Scholar]
- Terzi L.C., Simpson G.G. (2008). Regulation of flowering time by RNA processing. Curr. Top. Microbiol. Immunol. 326: 201–218 [DOI] [PubMed] [Google Scholar]
- Tharun S., He W., Mayes A.E., Lennertz P., Beggs J.D., Parker R. (2000). Yeast Sm-like proteins function in mRNA decapping and decay. Nature 404: 515–518 [DOI] [PubMed] [Google Scholar]
- Tharun S., Parker R. (2001). Targeting an mRNA for decapping: displacement of translation factors and association of the Lsm1p-7p complex on deadenylated yeast mRNAs. Mol. Cell 8: 1075–1083 [DOI] [PubMed] [Google Scholar]
- Tomasevic N., Peculis B.A. (2002). Xenopus LSm proteins bind U8 snoRNA via an internal evolutionarily conserved octamer sequence. Mol. Cell. Biol. 22: 4101–4112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Zhang Y., Ma Q., Zhang Z., Xue Y., Bao S., Chong K. (2007). SKB1-mediated symmetric dimethylation of histone H4R3 controls flowering time in Arabidopsis. EMBO J. 26: 1934–1941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Gerstein M., Snyder M. (2009). RNA-Seq: A revolutionary tool for transcriptomics. Nat. Rev. Genet. 10: 57–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong L., Gong Z., Rock C.D., Subramanian S., Guo Y., Xu W., Galbraith D., Zhu J.K. (2001b). Modulation of abscisic acid signal transduction and biosynthesis by an Sm-like protein in Arabidopsis. Dev. Cell 1: 771–781 [DOI] [PubMed] [Google Scholar]
- Xiong L., Ishitani M., Lee H., Zhu J.K. (2001a). The Arabidopsis LOS5/ABA3 locus encodes a molybdenum cofactor sulfurase and modulates cold stress- and osmotic stress-responsive gene expression. Plant Cell 13: 2063–2083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong L., Lee H., Ishitani M., Zhu J.K. (2002b). Regulation of osmotic stress-responsive gene expression by the LOS6/ABA1 locus in Arabidopsis. J. Biol. Chem. 277: 8588–8596 [DOI] [PubMed] [Google Scholar]
- Xiong L., Schumaker K.S., Zhu J.K. (2002a). Cell signaling during cold, drought, and salt stress. Plant Cell 14(suppl.): S165–S183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K., Shinozaki K. (2006). Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annu. Rev. Plant Biol. 57: 781–803 [DOI] [PubMed] [Google Scholar]
- Yang M., Sun J., Sun X., Shen Q., Gao Z., Yang C. (2009). Caenorhabditis elegans protein arginine methyltransferase PRMT-5 negatively regulates DNA damage-induced apoptosis. PLoS Genet. 5: e1000514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamski E., Guo W.W., Yamamoto Y.T., Pharr D.M., Williamson J.D. (2001). Analysis of celery (Apium graveolens) mannitol dehydrogenase (Mtd) promoter regulation in Arabidopsis suggests roles for MTD in key environmental and metabolic responses. Plant Mol. Biol. 47: 621–631 [DOI] [PubMed] [Google Scholar]
- Zhao Q., Rank G., Tan Y.T., Li H., Moritz R.L., Simpson R.J., Cerruti L., Curtis D.J., Patel D.J., Allis C.D., Cunningham J.M., Jane S.M. (2009). PRMT5-mediated methylation of histone H4R3 recruits DNMT3A, coupling histone and DNA methylation in gene silencing. Nat. Struct. Mol. Biol. 16: 304–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Q., Wang X.J. (2008). GOEAST: A web-based software toolkit for Gene Ontology enrichment analysis. Nucleic Acids Res. 36(Web Server issue): W358––W363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z., et al. (2010). PRMT5 regulates Golgi apparatus structure through methylation of the golgin GM130. Cell Res. 20: 1023–1033 [DOI] [PubMed] [Google Scholar]
- Zhu J., Jeong J.C., Zhu Y., Sokolchik I., Miyazaki S., Zhu J.K., Hasegawa P.M., Bohnert H.J., Shi H., Yun D.J., Bressan R.A. (2008). Involvement of Arabidopsis HOS15 in histone deacetylation and cold tolerance. Proc. Natl. Acad. Sci. USA 105: 4945–4950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J.K. (2001). Cell signaling under salt, water and cold stresses. Curr. Opin. Plant Biol. 4: 401–406 [DOI] [PubMed] [Google Scholar]
- Zhu J.K. (2002). Salt and drought stress signal transduction in plants. Annu. Rev. Plant Biol. 53: 247–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J.K. (2003). Regulation of ion homeostasis under salt stress. Curr. Opin. Plant Biol. 6: 441–445 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.