Abstract
Typically, pathogen-associated molecular patterns (PAMPs) are considered to be conserved throughout classes of microbes and to contribute to general microbial fitness, whereas effectors are species, race, or strain specific and contribute to pathogen virulence. Both types of molecule can trigger plant immunity, designated PAMP-triggered and effector-triggered immunity (PTI and ETI, respectively). However, not all microbial defense activators conform to the common distinction between PAMPs and effectors. For example, some effectors display wide distribution, while some PAMPs are rather narrowly conserved or contribute to pathogen virulence. As effectors may elicit defense responses and PAMPs may be required for virulence, single components cannot exclusively be referred to by one of the two terms. Therefore, we put forward that the distinction between PAMPs and effectors, between PAMP receptors and resistance proteins, and, therefore, also between PTI and ETI, cannot strictly be maintained. Rather, as illustrated by examples provided here, there is a continuum between PTI and ETI. We argue that plant resistance is determined by immune receptors that recognize appropriate ligands to activate defense, the amplitude of which is likely determined by the level required for effective immunity.
INTRODUCTION: A CONTEMPORARY VIEW OF PLANT IMMUNITY
Early in the 20th century, by performing genetic experiments, Harold Flor showed that inheritance of plant immunity to pathogens, as well as the ability of the pathogen to cause disease, is controlled by corresponding gene pairs (Flor, 1942). The plant genetic factor was referred to as the resistance (R) gene, while the pathogen genetic factor that determined the inability to cause disease was referred to as the avirulence (Avr) gene. A plant producing an R protein is resistant toward a pathogen strain that produces the corresponding Avr protein, and it was originally believed that gene-for-gene resistance was conferred by a direct interaction between the R and Avr proteins (Keen, 1990). However, experiments designed to show such direct ligand–receptor interactions often produced negative results, which led to the formulation of the guard hypothesis, stating that R proteins monitor the state of host components that are targeted by pathogen molecules (Van der Biezen and Jones, 1998). This model recognizes that pathogen molecules have intrinsic functions to promote pathogen virulence, which requires the modulation of host components that have thus become virulence targets. Rather than the presence of the pathogen molecules themselves, it is the manipulation of their host targets that is sensed by the R proteins (Chisholm et al., 2006; Jones and Dangl, 2006). Thus, the pathogen molecules that were originally referred to as avirulence factors genuinely are virulence factors. Presently, the term “effector” is commonly used for this type of molecule (Bent and Mackey, 2007; Boller and Felix, 2009).
In parallel to research on gene-for-gene resistance, the existence of inducers of plant defense responses that were not determinants of race or cultivar specificity became evident (Ebel and Cosio, 1994). These non-race-specific inducers of defense were termed elicitors and harbored a wide range of different types of molecules (Boller, 1995). However, for a long time it was debated whether elicitor-induced defense responses were physiologically relevant to plant immunity and what the relationship between gene-for-gene resistance and elicitor-induced defense would be. With the identification of the first elicitor receptor (FLS2; Gómez-Gómez and Boller, 2000), subsequent proof for its role in plant immunity (Zipfel et al., 2004), and identification of microbial effectors that suppress this type of immunity (Hauck et al., 2003), proof that both types of defense contribute to plant immunity was eventually provided.
A simple but elegant view of innate immunity in plant pathogen interactions is depicted by the so-called zigzag model introduced by Jones and Dangl (2006). This model proposes that the first line of active plant defense is formed by pattern recognition receptors (PRRs). These are cell surface receptors that recognize pathogen-associated molecular patterns (PAMPs). PAMPs were originally defined as highly conserved molecules within a class of microbes that have an essential function in microbial fitness or survival (Medzhitov and Janeway, 1997; Nürnberger and Brunner, 2002). As they may also occur in nonpathogenic microorganisms, the alternative term microbe-associated molecular pattern also is used (Boller and Felix, 2009). PRRs activate an innate immune response upon detection of PAMPs, so-called PAMP-triggered immunity (PTI). Successful pathogens are able to overcome PTI by means of secreted effectors that suppress PTI responses, resulting in effector-triggered susceptibility. Pathogenic bacteria typically inject such effectors directly into the host cytoplasm by means of their type III secretion machinery, and during evolution, plants have responded to these effectors through the development of cytoplasmic R proteins that recognize (the presence or activity of) single effectors. The majority of these R proteins are intracellular receptor proteins of the nucleotide binding–leucine-rich repeat (NB-LRR) type that activate so-called effector-triggered immunity (ETI). Typically, the propensity to trigger ETI is pathogen strain or race specific and is associated with programmed cell death, a response which is referred to as the hypersensitive response (HR), and systemic acquired resistance (SAR) in the host. As a result of selection pressure, pathogen isolates evolved that have either lost or altered the effector that is recognized or that have gained novel effectors to suppress the ETI response. In turn, new plant receptors evolved that either recognize the obvious effectors or the newly acquired effectors, resulting again in ETI. This coevolution proceeds, with continuous selection for novel pathogen isolates that overcome ETI and new plant genotypes that resurrect ETI.
In accordance with the zigzag model, plant pathologists discriminate two phases of plant immunity: PTI triggered by PAMPs and ETI triggered by effectors, with the paradigm that activated immune responses in ETI occur quicker and are more prolonged and more robust than those in PTI (Tao et al., 2003; Jones and Dangl, 2006; Tsuda and Katagiri, 2010). As a consequence, the molecules that were historically termed elicitors or avirulence factors have more recently been renamed PAMPs and effectors, respectively (Chisholm et al., 2006; Jones and Dangl, 2006; Bent and Mackey, 2007). However, accumulating evidence indicates that the separation between PAMPs and effectors, and between PRRs and R proteins, and thus also between PTI and ETI, cannot strictly be maintained. Rather, as illustrated by examples provided here, there is a continuum between PTI and ETI.
THE AMBIGUOUS PAMP-EFFECTOR DICHOTOMY
Classical examples of PAMPs are structural molecules, such as bacterial flagellin, peptidoglycan, and lipopolysaccharides, oomycete glucans, and fungal chitin (Ayers et al., 1976; Felix et al., 1993, 1999; Dow et al., 2000; Gust et al., 2007; Erbs et al., 2008). While PAMPs are widely conserved across genera, effectors are specific to single or a few related species (Chisholm et al., 2006; Jones and Dangl, 2006; Bent and Mackey, 2007). However, based on their widespread occurrence, several groups of effector proteins qualify to be designated as PAMPs.
Fungal LysM Effectors Suppress Chitin-Triggered Immunity
Chitin, a β-(1,4)-linked homopolymer of N-acetylglucosamine, is an essential component of the cell walls of all fungi that is not found in plants and vertebrates. Chitin fragments have long been known to act as elicitors of defense responses in plants (Felix et al., 1993). Taking the widespread, conserved, and intrinsic structural nature of this polysaccharide into account, chitin is an undisputed fungal PAMP. Presently, a number of plant immune receptors for chitin have been characterized. A high-affinity chitin binding protein, the Chitin Oligosaccharide Elicitor Binding Protein (CEBiP) receptor, was isolated from the plasma membranes of rice (Oryza sativa) cells (Kaku et al., 2006). Knockdown of CEBiP expression resulted in suppression of chitin-induced defense responses, showing that CEBiP plays a key role in perception of chitin oligosaccharides. CEBiP is a receptor-like protein (RLP) that contains extracellular LysM domains but lacks an obvious intracellular signaling domain, suggesting that additional components are required for subsequent initiation of defense signaling. A potential coreceptor of CEBiP is the recently identified LysM-containing Chitin Elicitor Receptor Kinase-1 (Os-CERK1) that, similar to CEBiP, is required for chitin signaling in rice cells (Shimizu et al., 2010). In addition, a similar LysM-containing plasma membrane receptor required for chitin-triggered immunity was identified in Arabidopsis thaliana (Miya et al., 2007; Wan et al., 2008), and recent evidence suggests that it directly binds chitin fragments (Petutschnig et al., 2010; Iizasa et al., 2010).
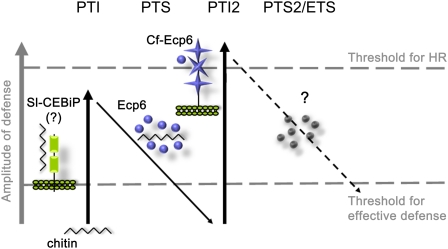
Based on the zigzag model, it must be anticipated that successful fungal pathogens have developed effectors to interfere with chitin-triggered immunity (Figure 1). From the fungal tomato pathogen Cladosporium fulvum, the abundantly secreted protein Ecp6 (for extracellular protein 6) was identified that, like the plant chitin receptors CEBiP and CERK1, also contains LysM domains (Bolton et al., 2008). Ecp6 was found to bind chitin fragments to suppress chitin-triggered immune responses and, moreover, to compete with the plant LysM-containing CEBiP receptor for the binding of chitin oligomers (de Jonge et al., 2010). Thus, the Ecp6 protein prevents activation of host immunity by sequestering chitin fragments that are released during host colonization (de Jonge et al., 2010). Interestingly, the Ecp6 gene is found in all strains of C. fulvum that have been analyzed thus far, and only little sequence variation is observed (Bolton et al., 2008). Moreover, conserved Ecp6 orthologs, termed LysM effectors, widely occur in the fungal kingdom (Bolton et al., 2008; de Jonge and Thomma, 2009), suggesting that scavenging of chitin oligosaccharides is fundamental to the lifestyle of fungal pathogens upon colonization of their hosts (de Jonge et al., 2010). Indeed, several LysM effectors from other pathogenic fungi may contribute to virulence through suppression of chitin-triggered host immunity (B.P.H.J. Thomma, unpublished data). Although these orthologs have been named LysM effectors (de Jonge and Thomma, 2009), their widespread functional conservation in the fungal kingdom is reminiscent of typical PAMPs. This aspect is further substantiated by the recent identification of tomato (Solanum lycopersicum) genotypes that specifically recognize C. fulvum Ecp6 (B.P.H.J. Thomma, unpublished data; Figure 1).
Figure 1.
A Variation on the Zigzag Model to Describe the Evolution of Chitin Signaling in the Interaction between C. fulvum and Tomato.
The C. fulvum PAMP chitin activates PTI in tomato plants, presumably upon perception by the tomato homolog of the rice cell surface receptor CEBiP. Thus far, a chitin-triggered HR has not been observed in tomato. To overcome PTI, C. fulvum employs the abundantly secreted LysM effector Ecp6 that binds chitin, thereby preventing activation of Sl-CEBiP. Since LysM effectors are widely conserved in the fungal kingdom, they qualify as PAMPs, and Ecp6-mediated PTI suppression therefore should be referred to as PAMP-triggered susceptibility (PTS). Tomato genotypes that have evolved to recognize Ecp6 develop an HR upon Ecp6 infiltration and presumably carry a cell surface receptor for this molecule, tentatively called C. fulvum resistance to Ecp6 (Cf-Ecp6), again resulting in PTI (PTI2). The question mark indicates that subsequent susceptibility can again be provoked by C. fulvum, either through mutation of the Ecp6 protein such that it still sequesters chitin fragments but is no longer recognized by Cf-Ecp6 or by producing an effector that suppresses Sl-CEBiP signaling in an alternative manner. (Adapted by permission from Macmillan Publishers Ltd.: Nature; Jones and Dangl [2006].)
The LysM effectors are not the only group of effectors that qualify to be designated as PAMPs based on their widespread occurrence. This similarly applies to, for instance, necrosis- and ethylene-inducing peptide 1 (Nep1)-like proteins that are conserved among bacteria, fungi, and oomycetes (Gijzen and Nürnberger, 2006; Kamoun, 2006), crinklers that are produced by oomycete species (Torto et al., 2003; Haas et al., 2009), and harpins that are produced by Gram-negative bacteria (Tampakaki et al., 2010).
PAMPs with Narrow Distribution
In contrast with the groups of widely conserved effectors mentioned above, several studies have shown that some PAMPs are only narrowly conserved (Brunner et al., 2002; Lee et al., 2009).
The rice Xa21 gene, which encodes a receptor-like kinase, confers immunity to most strains of the bacterium Xanthomonas oryzae pv oryzae (Xoo; Song et al., 1995). Resistance is activated by avrXa21 (avirulence protein corresponding to XA21), a protein recently renamed Ax21 (activator of Xa21-mediated immunity; Lee et al., 2009). Several bacterial genes that are required for Ax21 activity were identified that either encode components of a bacterial type I secretion system or are involved in sulfation (da Silva et al., 2004). A sulfated, 17–amino acid synthetic peptide (axYS22) derived from the N-terminal region of Ax21 was found to be sufficient for biological activity and to bind to Xa21, whereas peptides lacking sulfation were inactive (Lee et al., 2009). Interestingly, axYS22 is fully conserved in strains of Xanthomonas campestris pv campestris, Xanthomonas axonopodis pv glycinea, X. axonopodis pv vesicatoria, and X. oryzae pv oryzicola.
Another PAMP with a similarly narrow distribution spectrum is Pep-13, a surface-exposed fragment of a calcium-dependent cell wall transglutaminase (TGase) that is conserved among Phytophthora species and that activates defense in various plants (Nürnberger et al., 1994; Brunner et al., 2002). Thus, in contrast with the current paradigm that PAMPs are broadly conserved, at least some PAMPs have a rather narrow distribution. In addition to this, despite being widespread, some PAMPs are only recognized by a narrow range of plant hosts as recognition of the ubiquitous bacterial PAMPs EF-Tu and cold-shock protein is restricted to the Brassicaceae and Solanaceae, respectively, which again is an effector-like characteristic (Felix and Boller, 2003; Zipfel et al., 2006).
PAMPs and Virulence
Apart from their distribution, the intrinsic function for the pathogen has been a parameter to distinguish PAMPs from effectors. Whereas PAMPs are essential for microbial fitness and survival, effectors specifically contribute to virulence by targeting host (defense) physiology. Nevertheless, several examples indicate that the distinction between requirement for fitness and survival on the one hand and for virulence on the other hand is difficult to maintain. For example, one of the most widely studied PAMPs, flagellin, also plays a role in virulence. It has been shown that specific Pseudomonas syringae pv tabaci flagellin mutants affected in elicitor activity also display reduced virulence in planta due to reduced motility (Taguchi et al., 2006, 2010; Naito et al., 2008). The importance of flagellar motility in bacterial virulence has also been reported for other P. syringae pathovars (Panopoulos and Schroth, 1974; Haefele and Lindow, 1987; Hattermann and Ries, 1989) and other bacterial species, including Erwinia carotovora (Hossain et al., 2005) and Ralstonia solanacearum (Tans-Kersten et al., 2004). Interfering with biological activity through changes in composition of the lipopolysaccharide (LPS) or peptidoglycan envelope may similarly affect bacterial virulence (Newman et al., 2007). For example, while bacterial LPS generally acts as an inducer of defenses (Zeidler et al., 2004), LPS of the symbiont Sinorhizobium meliloti suppresses defense responses in the host plant Medicago truncatula (Tellström et al., 2007). Furthermore, other bacterial exopolysaccharides also can suppress immune responses (Aslam et al., 2008).
There are other examples indicating that a strict discrimination between PAMPs and effectors cannot be maintained. CBEL is a cell wall glycoprotein PAMP that was identified in Phytophthora parasitica var nicotianae, the causal agent of tobacco Black Shank disease, which occurs widely in the genus Phytophthora (Mateos et al., 1997; Khatib et al., 2004). CBEL induces HR-like lesions and defense responses in tobacco (Nicotiana tabacum), but also in nonhost plants including Arabidopsis (Khatib et al., 2004). Compromised CBEL expression in P. parasitica var nicotianae demonstrates a role in cell wall polysaccharide deposition and adhesion to cellulose, although knockdown transformants do not display significantly reduced virulence (Gaulin et al., 2002). The Trichoderma viride ethylene-inducing xylanase (EIX) is a PAMP that likely contributes to the ability of the fungus to enter the host. In this case, the epitope that is recognized by plants consists of five surface-exposed amino acids that are not involved in enzyme activity. The finding that in the presumed PAMP Pep-13 the same amino acid residues that are required for eliciting plant defenses are also required for TGase activity (Nürnberger et al., 1994; Brunner et al., 2002) suggests that TGase activity also may contribute to virulence. In addition, Ax21 is characterized as a secreted peptide that is produced in a cell density–dependent manner and may play a role in quorum sensing and thereby virulence of the bacterium (Lee et al., 2006). The harpin HrpZ of the bacterial pathogen P. syringae represents another example, as it is a PAMP that appears to play a role in virulence by affecting host membrane integrity (Lee et al., 2001; Kvitko et al., 2007, 2009; Engelhardt et al., 2009).
Final examples of PAMPs that play a role in virulence are the PWL proteins of the fungus Magnaporthe oryzae, causal agent of rice blast that infects a large range of grass (Poaceae) species. Interestingly, the fungus secretes proteins that limit its host range, as strains that carry the PWL1 or -2 gene are avirulent on weeping lovegrass (Kang et al., 1995; Sweigard et al., 1995). The PWL gene family is large and ubiquitously represented throughout the M. oryzae population, and the encoded PWL proteins share significant homology. Interestingly, it was recently shown that PWLs are translocated to the cytoplasm of the invaded host cell and are likely to be involved in virulence. The proteins were found to be highly mobile in the symplast of the host tissue, as they moved to uninvaded neighboring cells where they may act to prime cells for fungal invasion, for instance through suppression of host defense responses (Khang et al., 2010; Valent and Khang, 2010).
THE AMBIGUOUS PRR-R PROTEIN DICHOTOMY
In accord with the current paradigm, PRRs are cell surface receptors that recognize PAMPs and are required for the first layer of active defense against invading pathogens. They are evolutionarily ancient receptors that may be conserved between species, exemplified by the homologous chitin receptors in rice and Arabidopsis (Kaku et al., 2006; Miya et al., 2007) and by the widespread occurrence of homologs of the FLS2 receptor for bacterial flagellin (Gómez-Gómez and Boller, 2000; Hann and Rathjen, 2007; Robatzek et al., 2007; Takai et al., 2008; Boller and Felix, 2009). By contrast, most R proteins typically are intracellular receptor proteins of the NB-LRR type that recognize effectors in the cytoplasm. R proteins are evolutionarily relatively young, and novel members continuously arise in the coevolutionary interaction between pathogens and their hosts. However, on the one hand, some of the currently recognized R proteins display the typical properties of PRR receptors, whereas on the other hand, PRRs may be relatively young as some PAMPs are only recognized by a narrow range of plant hosts (Felix and Boller, 2003; Zipfel et al., 2006). Furthermore, despite recognizing the same PAMP, different plant species may recognize different epitopes. For instance, distinct β-glucan cell wall fragments of fungi and oomycetes are recognized by different plant species (Côté et al., 2000; Klarzynski et al., 2000; Yamaguchi et al., 2000). Furthermore, flg15, a shortened version of the flagellin derived 22-amino acid peptide flg22 carrying the epitope, does not act as an elicitor in Arabidopsis or Nicotiana benthamiana, while it is fully active in tomato (Robatzek et al., 2007).
Typically, PAMPs are perceived by extracellular domains of plasma membrane receptors, and no intracellular PAMP receptors have yet been reported. Cell surface receptors that act as genuine PAMP receptors typically perceive pathogen molecules in a receptor-ligand fashion in which they physically interact with pathogen molecules (Chinchilla et al., 2006; Kaku et al., 2006; Zipfel et al., 2006). However, not all effectors are perceived inside host cells, and not all extracellular pathogen receptors perceive pathogen molecules through direct physical interactions. For example, the race-specific Cf-2 receptor from tomato monitors the presence of C. fulvum strains that produce the Avr2 effector by guarding the Rcr3 protease, which is targeted by Avr2 (Rooney et al., 2005; Shabab et al., 2008; van Esse et al., 2008). Similarly, direct as well as indirect recognition of pathogen effectors by intracellular pathogen receptors occurs (Dodds and Rathjen, 2010).
Cf-4–Mediated Recognition of C. fulvum Avr4 Homologs from Other Pathogenic Fungi
Prior to the discovery of Ecp6, the secreted race-specific chitin binding C. fulvum effector Avr4 was identified (Joosten et al., 1994). Three of the four disulfide bonds of Avr4 were found to encompass an invertebrate chitin binding domain, allowing it to specifically bind to chitin of fungal cell walls in planta (van den Burg et al., 2003, 2004, 2006). In this way, Avr4 contributes to fungal virulence by protecting the fungal hyphae against hydrolysis by host chitinases that are secreted into the tomato apoplast during infection (Joosten et al., 1995; van den Burg et al., 2006; van Esse et al., 2007).
Typical for a race-specific effector, initially no clear Avr4 homologs were reported in any other species. However, a query of the recently released publicly available genome sequence of Mycosphaerella fijiensis, phylogenetically related to C. fulvum and causal agent of the Black Sigatoka disease of banana, revealed the Avr4 homolog Mf-Avr4 (Stergiopoulos et al., 2010). The positioning and spacing of the six Cys residues that constitute the chitin binding domain of C. fulvum Avr4 is conserved, and Mf-Avr4 binds chitin and protects fungal hyphae against hydrolysis by plant chitinases in vitro. Additional Avr4 homologs were identified in several Cercospora species that also belong to the Mycosphaerellaceae family and that share the positioning and spacing of the six Cys residues required for chitin binding of C. fulvum Avr4 (Stergiopoulos et al., 2010).
Tomato genotypes that produce the C. fulvum (Cf)-4 resistance protein respond to Avr4 with an HR and are resistant to C. fulvum strains that secrete Avr4. Intriguingly, the Avr4 homolog of the banana pathogen M. fijiensis is recognized by the tomato Cf-4 resistance protein and triggers the activation of an HR in Cf-4 tomato plants. Mediated by various Cf-4–like proteins, recognition of C. fulvum Avr4 also occurs in the wild Solanum species S. peruvianum, S. chmielewskii, and S. parviflorum (Laugé et al., 2000; Kruijt et al., 2004). When coexpressed with Mf-Avr4 in N. benthamiana, these Cf-4–like proteins all trigger HR, confirming the occurrence of wide recognitional specificity for Avr4-like proteins within the Solanum genus (Stergiopoulos et al., 2010). Based on these findings Avr4 and its homologs can be classified as PAMPs, and based on this classification, Cf-4 qualifies as a PRR.
Interestingly, the Cercospora Avr4 homologs are neither recognized by Cf-4 nor by any of the Cf-4–like proteins. Although in these Cercospora homologs the Cys spacing is conserved, other amino acids that are essential for chitin binding are not (van den Burg et al., 2004; Stergiopoulos et al., 2010); therefore, it can be anticipated that the Cercospora Avr4s do not bind to chitin. This suggests that there is a strict correlation between the capacity of the various Avr4 homologs to bind chitin and their recognition by Cf-4 and that as a result of coevolution there is only recognition by tomato of Avr4 homologs that protect the pathogen against chitinases.
Recognition of C. fulvum Ecp2 Homologs
In addition to Ecp6, all C. fulvum strains produce the conserved extracellular protein 2 (Ecp2) (Stergiopoulos et al., 2007). Gene disruption revealed that Ecp2 is required for virulence as the C. fulvum mutants trigged a fast induction of pathogenesis-related proteins, caused leaf desiccation and senescence, accumulated much less biomass in susceptible plants, and produced very few conidia (Laugé et al., 1997). Interestingly, several tomato genotypes and lines of the wild currant tomato species Solanum pimpinellifolium were identified that recognize Ecp2 and thus presumably carry Cf-Ecp2 resistance genes (Laugé et al., 1998, 2000; Haanstra et al., 1999). Strikingly, specific HR-associated recognition of Ecp2 also occurs in several Nicotiana species that are nonhost to C. fulvum, such as N. tabacum, N. paniculata, N. undulata, and N. sylvestris (de Kock et al., 2004). Population analysis revealed quite some sequence variation in the Ecp2 gene. However, only four open reading frame mutations were identified and these do not affect recognition by the presumed Cf-Ecp2 resistance protein (Stergiopoulos et al., 2007).
Three genes encoding proteins that are highly homologous to C. fulvum Ecp2 were recently identified in the M. fijiensis genome, of which the most homologous was designated Mf-Ecp2 (Stergiopoulos et al., 2010). Interestingly, Mf-Ecp2 is also recognized by tomato plants that respond to C. fulvum Ecp2, as inoculation with recombinant potato virus X (PVX) expressing Mf-Ecp2 results in a systemic necrosis (Stergiopoulos et al., 2010). By contrast, the other two proteins Mf-Ecp2-1 and -2 do not trigger an HR in these tomato plants. Further data mining revealed that the M. graminicola genome also encodes three Ecp2 homologs (Stergiopoulos et al., 2010). In contrast with Avr4, Ecp2 homologs are not restricted to the Mycosphaerellaceae family, as they also occur in several fungal species outside this family, including plant pathogenic (Fusarium graminearum and Verticillium dahliae), animal pathogenic (Trichophyton equinum), and nonpathogenic (Aspergillus nidulans, Neurospora crassa, and Podospora anserina) species. Thus, like Ecp6 and Avr4, Ecp2 also qualifies as PAMP. As a consequence, the host proteins that mediate recognition of Ecp2 qualify as PRRs.
Tomato Ve1 as PRR
Tomato Cf proteins that provide resistance to C. fulvum strains that express the corresponding elicitors belong to the eLRR-RLP class of resistance proteins, which are cell surface receptors with extracellular LRRs that lack an obvious cytoplasmic signaling domain (Thomma et al., 2005; Wang et al., 2008, 2010). Tomato Eix2 is an RLP–type PRR that confers recognition of the PAMP EIX of the biocontrol fungus T. viride (Ron and Avni, 2004). The tomato RLP-encoding gene Ve1 has been characterized as a resistance gene that mediates race-specific resistance to race 1 strains of vascular fungal pathogens of the Verticillium genus (Fradin et al., 2009). Interestingly, race 1 resistance affects two distinct fungal species: V. dahliae and V. albo-atrum. This suggests that the yet unidentified elicitor of Ve1-mediated resistance is conserved between fungal species and is potentially a PAMP, which would in turn imply that that Ve1 is a PRR.
Three additional observations support the hypothesis that Ve1 is a PRR rather than an R protein, namely, the physiology of Ve1-mediated Verticillium resistance, the involvement of BAK1/SERK3 in Ve1 signaling, and the observation that functional Ve1 can be transferred across families. In resistant as well as susceptible plants, Verticillium enters root xylem vessels and sporulates, which results in colonization of stem vessels (Gold and Robb, 1995; Heinz et al., 1998; Chen et al., 2004). After a week, fungal elimination as a consequence of plant defense occurs. Whereas the pathogen is able to overcome this elimination in susceptible plants and extensive host colonization occurs, the fungus is not able to overcome elimination in resistant plants and remains present in the host at a low level (Gold and Robb, 1995; Heinz et al., 1998; Chen et al., 2004; van Esse et al., 2009). In accord with the paradigm that ETI quickly and strongly provides a robust type of resistance and PTI is a weak variant of ETI, Ve1-mediated resistance is reminiscent of PTI.
In several independent studies, the receptor-like kinase BAK1/SERK3 was identified as crucial factor in various PTI responses. In Arabidopsis and N. benthamiana, BAK1/SERK3 was found to interact with the flagellin receptor FLS2 (Chinchilla et al., 2007; Heese et al., 2007). In addition, BAK1 controls spreading necrosis after infection with bacterial and fungal pathogens in a brassinosteroid-independent manner, while silencing of BAK1 in N. benthamiana results in enhanced cell death upon infection with the oomycete pathogen Hyaloperonospora arabidopsidis (Heese et al., 2007; Kemmerling et al., 2007). Furthermore, silencing of BAK1 in N. benthamiana attenuates the response to the cold-shock protein and Phytophthora infestans INF1 (Heese et al., 2007). Recently, it was demonstrated that BAK1 is involved in Ve1-mediated Verticillium resistance of tomato (Fradin et al., 2009).
The transfer of race-specific R genes across species boundaries has been successful as long as recipient species are phylogenetically closely related. For example, transfer of the gene Rx that provides resistance to PVX, from potato to N. benthamiana, which both belong to the Solanaceae family, successfully provided PVX resistance (Bendahmane et al., 1999). However, interfamily transfer of race-specific R genes has met little success (Gust et al., 2010). By contrast, interfamily transfer of PAMP receptors appears to be more successful, as exemplified by the transfer of the PRRs EFR for bacterial EF-Tu and FLS2 for bacterial flagellin from Arabidopsis to N. benthamiana and tomato (Chinchilla et al., 2006; Zipfel et al., 2006; Lacombe et al., 2010). Intriguingly, preliminary results suggest that the tomato gene Ve1 might be transferred across family boundaries while remaining fully functional, as transgenic Arabidopsis expressing Ve1 appear to be resistant toward race 1 strains of V. dahliae and V. albo-atrum (E.F. Fradin and B.P.H.J. Thomma, unpublished data).
Kinases that carry an Arg (R) preceding the catalytic Asp (D) have been termed RD kinases, whereas non-RD kinases lack this Arg but carry a Cys or Gly instead (Johnson et al., 1996). The non-RD domain has been suggested to be a hallmark of kinases involved in innate immunity signaling (Dardick and Ronald, 2006). Despite the presence of relatively few non-RD kinases in the kinase families of yeast, fly, worm, human, Arabidopsis, and rice, 12 of 15 kinases known or predicted to function in PRR signaling fall into the non-RD class, including FLS2, EFR, and XA21 (Dardick and Ronald, 2006). It was hypothesized that the non-RD kinase motif has been recruited by plants and animals to mediate the activation of innate immune responses, which discriminates PRRs from other receptors (Dardick and Ronald, 2006; Ronald and Beutler, 2010). Nevertheless, in this respect, it is interesting to note that the receptor-like kinase BAK1/SERK3 that acts as a coreceptor in various PTI responses and has been implicated in Ve1 signaling does not belong to the class of non-RD kinases. Furthermore, the CERK1 receptors for chitin from Arabidopsis (Miya et al., 2007) and rice (Shimizu et al., 2010) also do not belong to this class (Dardick and Ronald, 2006).
Evasion of Host Recognition through PAMP Plasticity
Likely as a result of coevolution, C. fulvum strains have evolved that evade Cf-4–mediated recognition by producing Avr4 isoforms in which one disulfide bond is lost (Joosten et al., 1994). This results in a less compact overall structure of the Avr4 protein and prompt degradation by host proteases upon secretion into the apoplast (Joosten et al., 1997). However, once bound to fungal cell wall chitin, these instable Avr4 isoforms are stabilized and can still exert their intrinsic function for the pathogen (van den Burg et al., 2003). In this way, Cf-4–mediated recognition of mutant Avr4 isoforms is evaded without loss of intrinsic function, suggesting that both native and mutant Avr4 forms contribute to C. fulvum fitness (van den Burg et al., 2003; van Esse et al., 2007).
PAMPs are usually considered as invariant or highly constrained structures that are extremely difficult for microbes to alter because of fitness penalties. Nevertheless, PAMP plasticity has been observed, and pathogens apparently evolve to avoid recognition of their PAMPs. In mammalian pathogens, examples of LPS variants that perturb host recognition have been reported (Reife et al., 2006; Coats et al., 2007). Also, altered acylation compromises recognition of LPS by the corresponding TLR4 receptor (Montminy et al., 2006; Rebeil et al., 2006). A similar situation seems to occur in plants (Silipo et al., 2008). Furthermore, bacterial pathogens can suppress flagellin expression when colonizing their hosts (Akerley et al., 1995, Wolfgang et al., 2004; Shen and Higgins, 2006), and some bacteria, such as Agrobacterium spp, Rhizobium spp, and R. solanacearum, do not elicit flagellin-triggered immunity due to mutations in the epitope (Felix et al., 1999; Pfund et al., 2004). Similarly, a specific strain of X. campestris pv campestris does not cause disease symptoms due to a single amino acid difference in the flagellin sequence (Sun et al., 2006). Furthermore, a nonglycosylated flagellin mutant protein from P. syringae pv tabaci displays reduced HR-inducing capacity in tobacco and soybean (Glycine max) plants (Taguchi et al., 2003, 2006) and induces a weaker defense response in the nonhost plant Arabidopsis (Ishiga et al., 2005). Finally, as discussed above, whereas the sulfated 17–amino acid synthetic peptide (axYS22) derived from the N-terminal region of Xoo Ax21 displays elicitor activity, variants lacking sulfation were inactive (da Silva et al., 2004; Lee et al., 2009). Thus, although PAMPs are often thought to be invariant, it is evident that sequence variation and posttranslational modifications can modulate PAMP recognition.
THE AMBIGUOUS PTI-ETI DICHOTOMY
Although it is generally known that PTI and ETI share many signaling components, it has been proposed that immune responses in ETI occur more quickly, are more prolonged, and are more robust than those in PTI, suggesting that PTI is a weak variant of ETI (Tao et al., 2003; Jones and Dangl, 2006; Tsuda et al., 2009; Tsuda and Katagiri, 2010). Typically, ETI is associated with an HR and SAR, while PTI is not. However, the PAMPs CBEL, EIX, and harpins all induce HR in plants (Bailey et al., 1990; Wei et al., 1992; Khatib et al., 2004; Ron and Avni, 2004). Furthermore, it has been demonstrated that flg22 induces an HR in Arabidopsis (Naito et al., 2007, 2008), whereas flagellins from Pseudomonas avenae and distinct P. syringae pathovars activate HR in the nonhost plants rice and tobacco (Che et al., 2000; Taguchi et al., 2003; Hann and Rathjen, 2007). These data demonstrate that the occurrence of HR is not restricted to ETI but can also occur in PTI responses. Furthermore, it has been demonstrated that PAMP perception also results in SAR in Arabidopsis (Mishina and Zeier, 2007). Inoculation with nonhost strains of P. syringae, as well as application of flagellin and LPS, resulted in the activation of systemic resistance in Arabidopsis in the absence of necrosis. The activation of SAR was accompanied by the typical hallmarks of elevated SA levels, pathogenesis-related gene expression, and SAR marker gene expression and was impaired in typical SAR mutants.
Beside atypical examples of strong PTI responses, examples of weak ETI responses have been presented as well. In addition to the example of tomato Ve1-mediated Verticillium resistance mentioned above, resistance mediated by RPS4 that recognizes the P. syringae type III effector AvrRps4 is rather weak (Wirthmueller et al., 2007). Furthermore, it is well documented that the spectra of responses triggered by AvrRpm1 and AvrRpt2 in plants that carry the CC-NB-LRR–type R proteins RPM1 and RPS2, respectively, are very similar although the AvrRpm1-triggered responses are substantially faster (Ritter and Dangl, 1996; Tao et al., 2003). This illustrates that effector-triggered responses differ in speed and strength. Intriguingly, RPS4-mediated HR triggered by P. syringae AvrRps4 was recently found to depend on autophagy, whereas RPS2-mediated HR triggered by AvrRpt2 was not (Hofius et al., 2009). This demonstrates the existence of different mechanisms that culminate in HR in ETI triggered by different effectors.
In conclusion, it is questionable whether PTI and ETI are always distinct defense responses with PTI being a weak variant of ETI. Rather, it appears that ETI and PTI both can be robust or weak, depending on the specific interaction; different molecules activate different defense signaling pathways, depending on the trigger, the receptor, and possibly also environmental conditions.
CONCLUSION
Numerous examples, some of which are discussed in this review, illustrate that classifying a particular molecule as PAMP or effector, or as PRR or R protein, has become a nebulous exercise. Importantly, it should be realized that models are typically generalizations of real life situations based on single or few specific examples, and the field of PAMP signaling has mainly been driven by hallmark discoveries made for the Arabidopsis FLS2 (recognizing bacterial flagellin), EFR (recognizing bacterial EF-Tu), and CERK1 (recognizing fungal chitin receptors). Obviously, such models are important conceptual tools, yet they may lead to oversimplification of real life situations in nature. Cell surface pathogen receptors as well as cytoplasmic pathogen receptors both fall into the subclass of R proteins as originally described by Flor (1942). Whereas PRRs typically recognize conserved microbial signatures, NB-LRRs do not, which is the conceptual basis for the PTI-ETI dichotomy. This concept needs to be refined by accepting that some pathogens deploy evolutionarily ancient effectors that are instrumental for pathogenicity; thus, it became important for plants to evolve recognition of these molecules. Essentially, these effectors now act as PAMPs and thereby blur the PAMP-effector dichotomy. A further refinement is that PTI and ETI both can be robust or weak, depending on the specific interaction and that conserved microbial signature molecules can be modified to avoid recognition.
In addition to this, it is increasingly being recognized that activation of innate immunity in multicellular eukaryotic systems essentially boils down to recognition of danger signals (Matzinger, 2007; Boller and Felix, 2009). These danger signals are either molecules that are directly derived from the microbe (PAMPs and effectors) or represent microbial invasion-derived, damaged, or modified eukaryotic host structures that are not present or not released in noninfected organisms (Matzinger, 2002, 2007; Lotze et al., 2007; Boller and Felix, 2009).
Ultimately, plants sense the presence of microbial invaders by means of receptors that detect microbial structures or by receptors that directly or indirectly detect plant-manipulating activities of microbial effectors. In other words, microbe sensing and plant immune activation is determined by any type of plant receptor that recognizes appropriate ligands to activate defense. From the plant immunity point of view, the nature and intrinsic function of the ligand is not relevant as long as it timely and accurately betrays the potential microbial invader to the plant surveillance system. As a result of continuous coevolution between plant and pathogen, a wealth of plant perception systems for microbe-derived molecules has been shaped that reliably fulfills roles in mediating the establishment of plant immunity.
Acknowledgments
We thank Naoto Shibuya, Pierre de Wit, Peter van Esse, and Ronnie de Jonge for critically reading the manuscript. B.P.H.J.T. is supported by a Vidi grant of the Research Council for Earth and Life Sciences of the Netherlands Organization for Scientific Research (NWO). T.N. is supported by the German Research Foundation (SFB446, SFB766, GRK685, and SPP1212). M.H.A.J.J. is supported by the Technological Top Institute-Green Genetics. B.P.H.J.T. and T.N. are supported by ERA-NET Plant Genomics. B.P.H.J.T. and M.H.A.J.J. are supported by the Centre for BioSystems Genomics, which is part of the Netherlands Genomics Initiative/NWO.
References
- Akerley B.J., Cotter P.A., Miller J.F. (1995). Ectopic expression of the flagellar regulon alters development of the Bordetella-host interaction. Cell 80: 611–620 [DOI] [PubMed] [Google Scholar]
- Aslam S.N., et al. (2008). Bacterial polysaccharides suppress induced innate immunity by calcium chelation. Curr. Biol. 18: 1078–1083 [DOI] [PubMed] [Google Scholar]
- Ayers A.R., Ebel J., Finelli F., Berger N., Albersheim P. (1976). Host-pathogen interactions: IX. Quantitative assays of elicitor activity and characterization of the elicitor present in the extracellular medium of cultures of Phytophthora megasperma var. sojae. Plant Physiol. 57: 751–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey B.A., Dean J.F., Anderson J.D. (1990). An ethylene biosynthesis-inducing endoxylanase elicits electrolyte leakage and necrosis in Nicotiana tabacum cv xanthi leaves. Plant Physiol. 94: 1849–1854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendahmane A., Kanyuka K., Baulcombe D.C. (1999). The Rx gene from potato controls separate virus resistance and cell death responses. Plant Cell 11: 781–792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bent A.F., Mackey D. (2007). Elicitors, effectors, and R genes: The new paradigm and a lifetime supply of questions. Annu. Rev. Phytopathol. 45: 399–436 [DOI] [PubMed] [Google Scholar]
- Boller T. (1995). Chemoperception of microbial signals in plant cells. Annu. Rev. Plant Physiol. Plant Mol. Biol. 46: 189–214 [Google Scholar]
- Boller T., Felix G. (2009). A renaissance of elicitors: Perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu. Rev. Plant Biol. 60: 379–406 [DOI] [PubMed] [Google Scholar]
- Bolton M.D., et al. (2008). The novel Cladosporium fulvum lysin motif effector Ecp6 is a virulence factor with orthologues in other fungal species. Mol. Microbiol. 69: 119–136 [DOI] [PubMed] [Google Scholar]
- Brunner F., Rosahl S., Lee J., Rudd J.J., Geiler C., Kauppinen S., Rasmussen G., Scheel D., Nürnberger T. (2002). Pep-13, a plant defense-inducing pathogen-associated pattern from Phytophthora transglutaminases. EMBO J. 21: 6681–6688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Che F.S., Nakajima Y., Tanaka N., Iwano M., Yoshida T., Takayama S., Kadota I., Isogai A. (2000). Flagellin from an incompatible strain of Pseudomonas avenae induces a resistance response in cultured rice cells. J. Biol. Chem. 275: 32347–32356 [DOI] [PubMed] [Google Scholar]
- Chen P., Lee B., Robb J. (2004). Tolerance to a non-host isolate of Verticillium dahliae in tomato. Physiol. Mol. Plant Pathol. 64: 283–291 [Google Scholar]
- Chinchilla D., Bauer Z., Regenass M., Boller T., Felix G. (2006). The Arabidopsis receptor kinase FLS2 binds flg22 and determines the specificity of flagellin perception. Plant Cell 18: 465–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinchilla D., Zipfel C., Robatzek S., Kemmerling B., Nürnberger T., Jones J.D.G., Felix G., Boller T. (2007). A flagellin-induced complex of the receptor FLS2 and BAK1 initiates plant defence. Nature 448: 497–500 [DOI] [PubMed] [Google Scholar]
- Chisholm S.T., Coaker G., Day B., Staskawicz B.J. (2006). Host-microbe interactions: Shaping the evolution of the plant immune response. Cell 124: 803–814 [DOI] [PubMed] [Google Scholar]
- Coats S.R., Do C.T., Karimi-Naser L.M., Braham P.H., Darveau R.P. (2007). Antagonistic lipopolysaccharides block E. coli lipopolysaccharide function at human TLR4 via interaction with the human MD-2 lipopolysaccharide binding site. Cell. Microbiol. 9: 1191–1202 [DOI] [PubMed] [Google Scholar]
- Côté F., Roberts K.A., Hahn M.G. (2000). Identification of high-affinity binding sites for the hepta-beta-glucoside elicitor in membranes of the model legumes Medicago truncatula and Lotus japonicus. Planta 211: 596–605 [DOI] [PubMed] [Google Scholar]
- Dardick C., Ronald P. (2006). Plant and animal pathogen recognition receptors signal through non-RD kinases. PLoS Pathog. 2: e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Silva F.G., Shen Y., Dardick C., Burdman S., Yadav R.C., de Leon A.L., Ronald P.C. (2004). Bacterial genes involved in type I secretion and sulfation are required to elicit the rice Xa21-mediated innate immune response. Mol. Plant Microbe Interact. 17: 593–601 [DOI] [PubMed] [Google Scholar]
- de Jonge R., Thomma B.P.H.J. (2009). Fungal LysM effectors: extinguishers of host immunity? Trends Microbiol. 17: 151–157 [DOI] [PubMed] [Google Scholar]
- de Jonge R., van Esse H.P., Kombrink A., Shinya T., Desaki Y., Bours R., van der Krol S., Shibuya N., Joosten M.H., Thomma B.P. (2010). Conserved fungal LysM effector Ecp6 prevents chitin-triggered immunity in plants. Science 329: 953–955 [DOI] [PubMed] [Google Scholar]
- de Kock M.J.D., Iskandar H.M., Brandwagt B.F., Laugé R., de Wit P.J.G.M., Lindhout P. (2004). Recognition of Cladosporium fulvum Ecp2 elicitor by non-host Nicotiana spp. is mediated by a single dominant gene that is not homologous to known Cf-genes. Mol. Plant Pathol. 5: 397–408 [DOI] [PubMed] [Google Scholar]
- Dodds P.N., Rathjen J.P. (2010). Plant immunity: Towards an integrated view of plant-pathogen interactions. Nat. Rev. Genet. 11: 539–548 [DOI] [PubMed] [Google Scholar]
- Dow M., Newman M.-A., von Roepenack E. (2000). The induction and modulation of plant defense responses by bacterial lipopolysaccharides. Annu. Rev. Phytopathol. 38: 241–261 [DOI] [PubMed] [Google Scholar]
- Ebel J., Cosio E.G. (1994). Elicitors of plant defense responses. Int. Rev. Cytol. 148: 1–36 [Google Scholar]
- Erbs G., Silipo A., Aslam S., De Castro C., Liparoti V., Flagiello A., Pucci P., Lanzetta R., Parrilli M., Molinaro A., Newman M.A., Cooper R.M. (2008). Peptidoglycan and muropeptides from pathogens Agrobacterium and Xanthomonas elicit plant innate immunity: Structure and activity. Chem. Biol. 15: 438–448 [DOI] [PubMed] [Google Scholar]
- Engelhardt S., Lee J., Gäbler Y., Kemmerling B., Haapalainen M.L., Li C.M., Wei Z., Keller H., Joosten M., Taira S., Nürnberger T. (2009). Separable roles of the Pseudomonas syringae pv. phaseolicola accessory protein HrpZ1 in ion-conducting pore formation and activation of plant immunity. Plant J. 57: 706–717 [DOI] [PubMed] [Google Scholar]
- Felix G., Boller T. (2003). Molecular sensing of bacteria in plants. The highly conserved RNA-binding motif RNP-1 of bacterial cold shock proteins is recognized as an elicitor signal in tobacco. J. Biol. Chem. 278: 6201–6208 [DOI] [PubMed] [Google Scholar]
- Felix G., Duran J.D., Volko S., Boller T. (1999). Plants have a sensitive perception system for the most conserved domain of bacterial flagellin. Plant J. 18: 265–276 [DOI] [PubMed] [Google Scholar]
- Felix G., Regenass M., Boller T. (1993). Specific perception of subnanomolar concentrations of chitin fragments by tomato cells: Induction of extracellular alkalinization, changes in protein phosphorylation, and establishment of a refractory state. Plant J. 4: 307–316 [Google Scholar]
- Flor H.H. (1942). Inheritance of pathogenicity in Melampsora lini. Phytopathology 32: 653–669 [Google Scholar]
- Fradin E.F., Zhang Z., Juarez Ayala J.C., Castroverde C.D., Nazar R.N., Robb J., Liu C.-M., Thomma B.P.H.J. (2009). Genetic dissection of Verticillium wilt resistance mediated by tomato Ve1. Plant Physiol. 150: 320–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaulin E., Jauneau A., Villalba F., Rickauer M., Esquerré-Tugayé M.T., Bottin A. (2002). The CBEL glycoprotein of Phytophthora parasitica var-nicotianae is involved in cell wall deposition and adhesion to cellulosic substrates. J. Cell Sci. 115: 4565–4575 [DOI] [PubMed] [Google Scholar]
- Gijzen M., Nürnberger T. (2006). Nep1-like proteins from plant pathogens: Recruitment and diversification of the NPP1 domain across taxa. Phytochemistry 67: 1800–1807 [DOI] [PubMed] [Google Scholar]
- Gold J., Robb J. (1995). The role of the coating response in Craigella tomatoes infected with Verticillium dahliae, races 1 and 2. Physiol. Mol. Plant Pathol. 47: 141–157 [Google Scholar]
- Gómez-Gómez L., Boller T. (2000). FLS2: An LRR receptor-like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis. Mol. Cell 5: 1003–1011 [DOI] [PubMed] [Google Scholar]
- Gust A.A., Biswas R., Lenz H.D., Rauhut T., Ranf S., Kemmerling B., Götz F., Glawischnig E., Lee J., Felix G., Nürnberger T. (2007). Bacteria-derived peptidoglycans constitute pathogen-associated molecular patterns triggering innate immunity in Arabidopsis. J. Biol. Chem. 282: 32338–32348 [DOI] [PubMed] [Google Scholar]
- Gust A.A., Brunner F., Nürnberger T. (2010). Biotechnological concepts for improving plant innate immunity. Curr. Opin. Biotechnol. 21: 204–210 [DOI] [PubMed] [Google Scholar]
- Haanstra J.P.W., Laugé R., Meijer-Dekens F., Bonnema G., de Wit P.J.G.M., Lindhout P. (1999). The Cf-ECP2 gene is linked to, but not part of, the Cf-4/Cf-9 cluster on the short arm of chromosome 1 in tomato. Mol. Gen. Genet. 262: 839–845 [DOI] [PubMed] [Google Scholar]
- Haas B.J., et al. (2009). Genome sequence and comparative analysis of the Irish potato famine pathogen Phytophthora infestans. Nature 461: 393–398 [DOI] [PubMed] [Google Scholar]
- Haefele D.M., Lindow S.E. (1987). Flagellar motility confers epiphytic fitness advantages upon Pseudomonas syringae. Appl. Environ. Microbiol. 53: 2528–2533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hann D.R., Rathjen J.P. (2007). Early events in the pathogenicity of Pseudomonas syringae on Nicotiana benthamiana. Plant J. 49: 607–618 [DOI] [PubMed] [Google Scholar]
- Hattermann D.R., Ries S.M. (1989). Motility of Pseudomonas syringae pv. glycinea and its role in infection. Phytopathology 79: 284–289 [Google Scholar]
- Hauck P., Thilmony R., He S.Y. (2003). A Pseudomonas syringae type III effector suppresses cell wall-based extracellular defense in susceptible Arabidopsis plants. Proc. Natl. Acad. Sci. USA 100: 8577–8582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heese A., Hann D.R., Gimenez-Ibanez S., Jones A.M., He K., Li J., Schroeder J.I., Peck S.C., Rathjen J.P. (2007). The receptor-like kinase SERK3/BAK1 is a central regulator of innate immunity in plants. Proc. Natl. Acad. Sci. USA 104: 12217–12222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz R., Lee S.W., Saparno A., Nazar R.N., Robb J. (1998). Cyclical systemic colonization in Verticillium-infected tomato. Physiol. Mol. Plant Pathol. 52: 385–396 [Google Scholar]
- Hofius D., Schultz-Larsen T., Joensen J., Tsitsigiannis D.I., Petersen N.H., Mattsson O., Jørgensen L.B., Jones J.D., Mundy J., Petersen M. (2009). Autophagic components contribute to hypersensitive cell death in Arabidopsis. Cell 137: 773–783 [DOI] [PubMed] [Google Scholar]
- Hossain M.M., Shibata S., Aizawa S.-I., Tsuyumu S. (2005). Motility is an important determinant for pathogenesis of Erwinia carotovora subsp. carotovora. Physiol. Mol. Plant Pathol. 66: 134–143 [Google Scholar]
- Iizasa E., Mitsutomi M., Nagano Y. (2010). Direct binding of a plant LysM receptor-like kinase, LysM RLK1/CERK1, to chitin in vitro. J. Biol. Chem. 285: 2996–3004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishiga Y., Takeuchi K., Taguchi F., Inagaki Y., Toyota K., Shiraishi T., Ichinose Y. (2005). Defense responses of Arabidopsis thaliana inoculated with Pseudomonas syringae pv. tabaci wild type and defective mutants for flagellin (ΔfliC) and flagellin-glycosylation (Δorf1). J. Gen. Plant Pathol. 71: 302–307 [Google Scholar]
- Johnson L.N., Noble M.E., Owen D.J. (1996). Active and inactive protein kinases: Structural basis for regulation. Cell 85: 149–158 [DOI] [PubMed] [Google Scholar]
- Jones J.D.G., Dangl J.L. (2006). The plant immune system. Nature 444: 323–329 [DOI] [PubMed] [Google Scholar]
- Joosten M.H.A.J., Cozijnsen T.J., De Wit P.J.G.M. (1994). Host resistance to a fungal tomato pathogen lost by a single base-pair change in an avirulence gene. Nature 367: 384–386 [DOI] [PubMed] [Google Scholar]
- Joosten M.H.A.J., Verbakel H.M., Nettekoven M.E., Van Leeuwen J., Van der Vossen R.T.M., De Wit P.J.G.M. (1995). The phytopathogenic fungus Cladosporium fulvum is not sensitive to the chitinase and β-1,3-glucanase defence proteins of its host, tomato. Physiol. Mol. Plant Pathol. 46: 45–59 [Google Scholar]
- Joosten M.H.A.J., Vogelsang R., Cozijnsen T.J., Verberne M.C., De Wit P.J.G.M. (1997). The biotrophic fungus Cladosporium fulvum circumvents Cf-4-mediated resistance by producing unstable AVR4 elicitors. Plant Cell 9: 367–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaku H., Nishizawa Y., Ishii-Minami N., Akimoto-Tomiyama C., Dohmae N., Takio K., Minami E., Shibuya N. (2006). Plant cells recognize chitin fragments for defense signaling through a plasma membrane receptor. Proc. Natl. Acad. Sci. USA 103: 11086–11091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamoun S. (2006). A catalogue of the effector secretome of plant pathogenic oomycetes. Annu. Rev. Phytopathol. 44: 41–60 [DOI] [PubMed] [Google Scholar]
- Kang S., Sweigard J.A., Valent B. (1995). The PWL host specificity gene family in the blast fungus Magnaporthe grisea. Mol. Plant Microbe Interact. 8: 939–948 [DOI] [PubMed] [Google Scholar]
- Keen N.T. (1990). Gene-for-gene complementarity in plant-pathogen interactions. Annu. Rev. Genet. 24: 447–463 [DOI] [PubMed] [Google Scholar]
- Kemmerling B., et al. (2007). The BRI1-associated kinase 1, BAK1, has a brassinolide-independent role in plant cell-death control. Curr. Biol. 17: 1116–1122 [DOI] [PubMed] [Google Scholar]
- Khang C.H., Berruyer R., Giraldo M.C., Kankanala P., Park S.Y., Czymmek K., Kang S., Valent B. (2010). Translocation of Magnaporthe oryzae effectors into rice cells and their subsequent cell-to-cell movement. Plant Cell 22: 1388–1403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khatib M., Lafitte C., Esquerré-Tugayé M.-T., Bottin A., Rickauer M. (2004). The CBEL elicitor of Phytophthora parasitica var. nicotianae activates defence in Arabidopsis thaliana via three different signalling pathways. New Phytol. 162: 501–510 [Google Scholar]
- Klarzynski O., Plesse B., Joubert J.M., Yvin J.C., Kopp M., Kloareg B., Fritig B. (2000). Linear beta-1,3 glucans are elicitors of defense responses in tobacco. Plant Physiol. 124: 1027–1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruijt M., Brandwagt B.F., de Wit P.J.G.M. (2004). Rearrangements in the Cf-9 disease resistance gene cluster of wild tomato have resulted in three genes that mediate Avr9 responsiveness. Genetics 168: 1655–1663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kvitko B.H., Park D.H., Velásquez A.C., Wei C.F., Russell A.B., Martin G.B., Schneider D.J., Collmer A. (2009). Deletions in the repertoire of Pseudomonas syringae pv. tomato DC3000 type III secretion effector genes reveal functional overlap among effectors. PLoS Pathog. 5: e1000388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kvitko B.H., Ramos A.R., Morello J.E., Oh H.S., Collmer A. (2007). Identification of harpins in Pseudomonas syringae pv. tomato DC3000, which are functionally similar to HrpK1 in promoting translocation of type III secretion system effectors. J. Bacteriol. 189: 8059–8072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacombe S., Rougon-Cardoso A., Sherwood E., Peeters N., Dahlbeck D., van Esse H.P., Smoker M., Rallapalli G., Thomma B.P.H.J., Staskawicz B., Jones J.D., Zipfel C. (2010). Interfamily transfer of a plant pattern-recognition receptor confers broad-spectrum bacterial resistance. Nat. Biotechnol. 28: 365–369 [DOI] [PubMed] [Google Scholar]
- Laugé R., Goodwin P.H., de Wit P.J.G.M., Joosten M.H.A.J. (2000). Specific HR-associated recognition of secreted proteins from Cladosporium fulvum occurs in both host and non-host plants. Plant J. 23: 735–745 [DOI] [PubMed] [Google Scholar]
- Laugé R., Joosten M.H.A.J., Haanstra J.P., Goodwin P.H., Lindhout P., De Wit P.J.G.M. (1998). Successful search for a resistance gene in tomato targeted against a virulence factor of a fungal pathogen. Proc. Natl. Acad. Sci. USA 95: 9014–9018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laugé R., Joosten M.H.A.J., Van den Ackerveken G.F.J.M., Van den Broek H.W.J., De Wit P.J.G.M. (1997). The in planta-produced extracellular proteins ECP1 and ECP2 of Cladosporium fulvum are virulence factors. Mol. Plant Microbe Interact. 10: 725–734 [Google Scholar]
- Lee J., Klusener B., Tsiamis G., Stevens C., Neyt C., Tampakaki A.P., Panopoulos N.J., Nöller J., Weiler E.W., Cornelis G.R., Mansfield J.W., Nürnberger T. (2001). HrpZ(Psph) from the plant pathogen Pseudomonas syringae pv. phaseolicola binds to lipid bilayers and forms an ion-conducting pore in vitro. Proc. Natl. Acad. Sci. USA 98: 289–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S.W., Han S.W., Bartley L.E., Ronald P.C. (2006). From the Academy: Colloquium review. Unique characteristics of Xanthomonas oryzae pv. oryzae AvrXa21 and implications for plant innate immunity. Proc. Natl. Acad. Sci. USA 103: 18395–18400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S.W., Han S.W., Sririyanum M., Park C.J., Seo Y.S., Ronald P.C. (2009). A type I-secreted, sulfated peptide triggers XA21-mediated innate immunity. Science 326: 850–853 [DOI] [PubMed] [Google Scholar]
- Lotze M.T., Zeh H.J., Rubartelli A., Sparvero L.J., Amoscato A.A., Washburn N.R., Devera M.E., Liang X., Tör M., Billiar T. (2007). The grateful dead: Damage-associated molecular pattern molecules and reduction/oxidation regulate immunity. Immunol. Rev. 220: 60–81 [DOI] [PubMed] [Google Scholar]
- Mateos F.V., Rickauer M., Esquerré-Tugayé M.-T. (1997). Cloning and characterization of a cDNA encoding an elicitor of Phytophthora parasitica var. nicotianae that shows cellulose-binding and lectin-like activities. Mol. Plant Microbe Interact. 10: 1045–1053 [DOI] [PubMed] [Google Scholar]
- Matzinger P. (2002). The danger model: A renewed sense of self. Science 296: 301–305 [DOI] [PubMed] [Google Scholar]
- Matzinger P. (2007). Friendly and dangerous signals: Is the tissue in control? Nat. Immunol. 8: 11–13 [DOI] [PubMed] [Google Scholar]
- Medzhitov R., Janeway C.A., Jr. (1997). Innate immunity: The virtues of a nonclonal system of recognition. Cell 91: 295–298 [DOI] [PubMed] [Google Scholar]
- Mishina T.E., Zeier J. (2007). Pathogen-associated molecular pattern recognition rather than development of tissue necrosis contributes to bacterial induction of systemic acquired resistance in Arabidopsis. Plant J. 50: 500–513 [DOI] [PubMed] [Google Scholar]
- Miya A., Albert P., Shinya T., Desaki Y., Ichimura K., Shirasu K., Narusaka Y., Kawakami N., Kaku H., Shibuya N. (2007). CERK1, a LysM receptor kinase, is essential for chitin elicitor signaling in Arabidopsis. Proc. Natl. Acad. Sci. USA 104: 19613–19618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montminy S.W., et al. (2006). Virulence factors of Yersinia pestis are overcome by a strong lipopolysaccharide response. Nat. Immunol. 7: 1066–1073 [DOI] [PubMed] [Google Scholar]
- Naito K., Ishiga Y., Toyoda K., Shiraishi T., Ichinose Y. (2007). N-terminal domain including conserved flg22 is required for flagellin-induced hypersensitive cell death in Arabidopsis thaliana. J. Gen. Plant Pathol. 73: 281–285 [Google Scholar]
- Naito K., Taguchi F., Suzuki T., Inagaki Y., Toyoda K., Shiraishi T., Ichinose Y. (2008). Amino acid sequence of bacterial microbe-associated molecular pattern flg22 is required for virulence. Mol. Plant Microbe Interact. 21: 1165–1174 [DOI] [PubMed] [Google Scholar]
- Newman M.A., Dow J.M., Molinaro A., Parrilli M. (2007). Priming, induction and modulation of plant defence responses by bacterial lipopolysaccharides. J. Endotoxin Res. 13: 69–84 [DOI] [PubMed] [Google Scholar]
- Nürnberger T., Brunner F. (2002). Innate immunity in plants and animals: emerging parallels between the recognition of general elicitors and pathogen-associated molecular patterns. Curr. Opin. Plant Biol. 5: 318–324 [DOI] [PubMed] [Google Scholar]
- Nürnberger T., Nennstiel D., Jabs T., Sacks W.R., Hahlbrock K., Scheel D. (1994). High affinity binding of a fungal oligopeptide elicitor to parsley plasma membranes triggers multiple defense responses. Cell 78: 449–460 [DOI] [PubMed] [Google Scholar]
- Panopoulos N.J., Schroth M.N. (1974). Role of flagellar motility in the invasion of bean leaves by Pseudomonas phaseolicola. Phytopathology 64: 1389–1397 [Google Scholar]
- Petutschnig E.K., Jones A.M., Serazetdinova L., Lipka U., Lipka V. (2010). The lysin motif receptor-like kinase (LysM-RLK) CERK1 is a major chitin-binding protein in Arabidopsis thaliana and subject to chitin-induced phosphorylation. J. Biol. Chem. 285: 28902–28911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfund C., Tans-Kersten J., Dunning F.M., Alonso J.M., Ecker J.R., Allen C., Bent A.F. (2004). Flagellin is not a major defense elicitor in Ralstonia solanacearum cells or extracts applied to Arabidopsis thaliana. Mol. Plant Microbe Interact. 17: 696–706 [DOI] [PubMed] [Google Scholar]
- Rebeil R., Ernst R.K., Jarrett C.O., Adams K.N., Miller S.I., Hinnebusch B.J. (2006). Characterization of late acyltransferase genes of Yersinia pestis and their role in temperature-dependent lipid A variation. J. Bacteriol. 188: 1381–1388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reife R.A., Coats S.R., Al-Qutub M., Dixon D.M., Braham P.A., Billharz R.J., Howald W.N., Darveau R.P. (2006). Porphyromonas gingivalis lipopolysaccharide lipid A heterogeneity: Differential activities of tetra- and penta-acylated lipid A structures on E-selectin expression and TLR4 recognition. Cell. Microbiol. 8: 857–868 [DOI] [PubMed] [Google Scholar]
- Ritter C., Dangl J.L. (1996). Interference between two specific pathogen recognition events mediated by distinct plant disease resistance genes. Plant Cell 8: 251–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robatzek S., Bittel P., Chinchilla D., Köchner P., Felix G., Shiu S.H., Boller T. (2007). Molecular identification and characterization of the tomato flagellin receptor LeFLS2, an orthologue of Arabidopsis FLS2 exhibiting characteristically different perception specificities. Plant Mol. Biol. 64: 539–547 [DOI] [PubMed] [Google Scholar]
- Ron M., Avni A. (2004). The receptor for the fungal elicitor ethylene-inducing xylanase is a member of a resistance-like gene family in tomato. Plant Cell 16: 1604–1615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronald P.C., Beutler B. (2010). Plant and animal sensors of conserved microbial signatures. Science 330: 1061–1064 [DOI] [PubMed] [Google Scholar]
- Rooney H.C., Van’t Klooster J.W., van der Hoorn R.A.L., Joosten M.H.A.J., Jones J.D.G., de Wit P.J.G.M. (2005). Cladosporium Avr2 inhibits tomato Rcr3 protease required for Cf-2-dependent disease resistance. Science 308: 1783–1786 [DOI] [PubMed] [Google Scholar]
- Shabab M., Shindo T., Gu C., Kaschani F., Pansuriya T., Chintha R., Harzen A., Colby T., Kamoun S., van der Hoorn R.A. (2008). Fungal effector protein AVR2 targets diversifying defense-related cys proteases of tomato. Plant Cell 20: 1169–1183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen A., Higgins D.E. (2006). The MogR transcriptional repressor regulates nonhierarchal expression of flagellar motility genes and virulence in Listeria monocytogenes. PLoS Pathog. 2: e30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu T., Nakano T., Takamizawa D., Desaki Y., Ishii-Minami N., Nishizawa Y., Minami E., Okada K., Yamane H., Kaku H., Shibuya N. (2010). Two LysM receptor molecules, CEBiP and OsCERK1, cooperatively regulate chitin elicitor signaling in rice. Plant J. 64: 204–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silipo A., Sturiale L., Garozzo D., Erbs G., Jensen T.T., Lanzetta R., Dow J.M., Parrilli M., Newman M.A., Molinaro A. (2008). The acylation and phosphorylation pattern of lipid A from Xanthomonas campestris strongly influence its ability to trigger the innate immune response in Arabidopsis. ChemBioChem 9: 896–904 [DOI] [PubMed] [Google Scholar]
- Song W.Y., Wang G.L., Chen L.L., Kim H.S., Pi L.Y., Holsten T., Gardner J., Wang B., Zhai W.X., Zhu L.H., Fauquet C., Ronald P. (1995). A receptor kinase-like protein encoded by the rice disease resistance gene, Xa21. Science 270: 1804–1806 [DOI] [PubMed] [Google Scholar]
- Stergiopoulos I., De Kock M.J., Lindhout P., De Wit P.J.G.M. (2007). Allelic variation in the effector genes of the tomato pathogen Cladosporium fulvum reveals different modes of adaptive evolution. Mol. Plant Microbe Interact. 20: 1271–1283 [DOI] [PubMed] [Google Scholar]
- Stergiopoulos I., van den Burg H.A., Okmen B., Beenen H.G., van Liere S., Kema G.H., de Wit P.J.G.M. (2010). Tomato Cf resistance proteins mediate recognition of cognate homologous effectors from fungi pathogenic on dicots and monocots. Proc. Natl. Acad. Sci. USA 107: 7610–7615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun W., Dunning F.M., Pfund C., Weingarten R., Bent A.F. (2006). Within-species flagellin polymorphism in Xanthomonas campestris pv campestris and its impact on elicitation of Arabidopsis FLAGELLIN SENSING2-dependent defenses. Plant Cell 18: 764–779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweigard J.A., Carroll A.M., Kang S., Farrall L., Chumley F.G., Valent B. (1995). Identification, cloning, and characterization of PWL2, a gene for host species specificity in the rice blast fungus. Plant Cell 7: 1221–1233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taguchi F., Shimizu R., Inagaki Y., Toyoda K., Shiraishi T., Ichinose Y. (2003). Post-translational modification of flagellin determines the specificity of HR induction. Plant Cell Physiol. 44: 342–349 [DOI] [PubMed] [Google Scholar]
- Taguchi F., et al. (2006). Identification of glycosylation genes and glycosylated amino acids of flagellin in Pseudomonas syringae pv. tabaci. Cell. Microbiol. 8: 923–938 [DOI] [PubMed] [Google Scholar]
- Taguchi F., Yamamoto M., Ohnishi-Kameyama M., Iwaki M., Yoshida M., Ishii T., Konishi T., Ichinose Y. (2010). Defects in flagellin glycosylation affect the virulence of Pseudomonas syringae pv. tabaci 6605. Microbiology 156: 72–80 [DOI] [PubMed] [Google Scholar]
- Takai R., Isogai A., Takayama S., Che F.-S. (2008). Analysis of flagellin perception mediated by flg22 receptor OsFLS2 in rice. Mol. Plant Microbe Interact. 21: 1635–1642 [DOI] [PubMed] [Google Scholar]
- Tampakaki A.P., Skandalis N., Gazi A.D., Bastaki M.N., Sarris P.F., Charova S.N., Kokkinidis M., Panopoulos N.J. (2010). Playing the “Harp”: Evolution of our understanding of hrp/hrc genes. Annu. Rev. Phytopathol. 48: 347–370 [DOI] [PubMed] [Google Scholar]
- Tans-Kersten J., Brown D., Allen C. (2004). Swimming motility, a virulence trait of Ralstonia solanacearum, is regulated by FlhDC and the plant host environment. Mol. Plant Microbe Interact. 17: 686–695 [DOI] [PubMed] [Google Scholar]
- Tao Y., Xie Z., Chen W., Glazebrook J., Chang H.S., Han B., Zhu T., Zou G., Katagiri F. (2003). Quantitative nature of Arabidopsis responses during compatible and incompatible interactions with the bacterial pathogen Pseudomonas syringae. Plant Cell 15: 317–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tellström V., Usadel B., Thimm O., Stitt M., Küster H., Niehaus K. (2007). The lipopolysaccharide of Sinorhizobium meliloti suppresses defense-associated gene expression in cell cultures of the host plant Medicago truncatula. Plant Physiol. 143: 825–837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomma B.P.H.J., VAN Esse H.P., Crous P.W., DE Wit P.J. (2005). Cladosporium fulvum (syn. Passalora fulva), a highly specialized plant pathogen as a model for functional studies on plant pathogenic Mycosphaerellaceae. Mol. Plant Pathol. 6: 379–393 [DOI] [PubMed] [Google Scholar]
- Torto T.A., Li S., Styer A., Huitema E., Testa A., Gow N.A.R., van West P., Kamoun S. (2003). EST mining and functional expression assays identify extracellular effector proteins from the plant pathogen Phytophthora. Genome Res. 13: 1675–1685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuda K., Katagiri F. (2010). Comparing signaling mechanisms engaged in pattern-triggered and effector-triggered immunity. Curr. Opin. Plant Biol. 13: 459–465 [DOI] [PubMed] [Google Scholar]
- Tsuda K., Sato M., Stoddard T., Glazebrook J., Katagiri F. (2009). Network properties of robust immunity in plants. PLoS Genet. 5: e1000772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valent B., Khang C.H. (2010). Recent advances in rice blast effector research. Curr. Opin. Plant Biol. 13: 434–441 [DOI] [PubMed] [Google Scholar]
- van den Burg H.A., Harrison S.J., Joosten M.H.A.J., Vervoort J., de Wit P.J.G.M. (2006). Cladosporium fulvum Avr4 protects fungal cell walls against hydrolysis by plant chitinases accumulating during infection. Mol. Plant Microbe Interact. 19: 1420–1430 [DOI] [PubMed] [Google Scholar]
- van den Burg H.A., Spronk C.A., Boeren S., Kennedy M.A., Vissers J.P., Vuister G.W., de Wit P.J.G.M., Vervoort J. (2004). Binding of the AVR4 elicitor of Cladosporium fulvum to chitotriose units is facilitated by positive allosteric protein-protein interactions: The chitin-binding site of AVR4 represents a novel binding site on the folding scaffold shared between the invertebrate and the plant chitin-binding domain. J. Biol. Chem. 279: 16786–16796 [DOI] [PubMed] [Google Scholar]
- van den Burg H.A., Westerink N., Francoijs K.J., Roth R., Woestenenk E., Boeren S., de Wit P.J.G.M., Joosten M.H.A.J., Vervoort J. (2003). Natural disulfide bond-disrupted mutants of AVR4 of the tomato pathogen Cladosporium fulvum are sensitive to proteolysis, circumvent Cf-4-mediated resistance, but retain their chitin binding ability. J. Biol. Chem. 278: 27340–27346 [DOI] [PubMed] [Google Scholar]
- Van der Biezen E.A., Jones J.D. (1998). Plant disease-resistance proteins and the gene-for-gene concept. Trends Biochem. Sci. 23: 454–456 [DOI] [PubMed] [Google Scholar]
- van Esse H.P., Bolton M.D., Stergiopoulos I., de Wit P.J.G.M., Thomma B.P.H.J. (2007). The chitin-binding Cladosporium fulvum effector protein Avr4 is a virulence factor. Mol. Plant Microbe Interact. 20: 1092–1101 [DOI] [PubMed] [Google Scholar]
- van Esse H.P., Fradin E.F., de Groot P.J., de Wit P.J.G.M., Thomma B.P.H.J. (2009). Tomato transcriptional responses to a foliar and a vascular fungal pathogen are distinct. Mol. Plant Microbe Interact. 22: 245–258 [DOI] [PubMed] [Google Scholar]
- van Esse H.P., Van’t Klooster J.W., Bolton M.D., Yadeta K.A., van Baarlen P., Boeren S., Vervoort J., de Wit P.J.G.M., Thomma B.P.H.J. (2008). The Cladosporium fulvum virulence protein Avr2 inhibits host proteases required for basal defense. Plant Cell 20: 1948–1963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan J., Zhang X.-C., Neece D., Ramonell K.M., Clough S., Kim S.-Y., Stacey M.G., Stacey G. (2008). A LysM receptor-like kinase plays a critical role in chitin signaling and fungal resistance in Arabidopsis. Plant Cell 20: 471–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G., et al. (2008). A genome-wide functional investigation into the roles of receptor-like proteins in Arabidopsis. Plant Physiol. 147: 503–517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G., Fiers M., Ellendorff U., Wang Z., De Wit P.J.G.M., Angenent G., Thomma B.P.H.J. (2010). The diverse roles of extracellular leucine-rich repeat-containing receptor-like proteins in plants. Crit. Rev. Plant Sci. 29: 285–299 [Google Scholar]
- Wei Z.M., Laby R.J., Zumoff C.H., Bauer D.W., He S.Y., Collmer A., Beer S.V. (1992). Harpin, elicitor of the hypersensitive response produced by the plant pathogen Erwinia amylovora. Science 257: 85–88 [DOI] [PubMed] [Google Scholar]
- Wirthmueller L., Zhang Y., Jones J.D., Parker J.E. (2007). Nuclear accumulation of the Arabidopsis immune receptor RPS4 is necessary for triggering EDS1-dependent defense. Curr. Biol. 17: 2023–2029 [DOI] [PubMed] [Google Scholar]
- Wolfgang M.C., Jyot J., Goodman A.L., Ramphal R., Lory S. (2004). Pseudomonas aeruginosa regulates flagellin expression as part of a global response to airway fluid from cystic fibrosis patients. Proc. Natl. Acad. Sci. USA 101: 6664–6668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi T., Yamada A., Hong N., Ogawa T., Ishii T., Shibuya N. (2000). Differences in the recognition of glucan elicitor signals between rice and soybean: Beta-glucan fragments from the rice blast disease fungus Pyricularia oryzae that elicit phytoalexin biosynthesis in suspension-cultured rice cells. Plant Cell 12: 817–826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeidler D., Zähringer U., Gerber I., Dubery I., Hartung T., Bors W., Hutzler P., Durner J. (2004). Innate immunity in Arabidopsis thaliana: Lipopolysaccharides activate nitric oxide synthase (NOS) and induce defense genes. Proc. Natl. Acad. Sci. USA 101: 15811–15816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipfel C., Kunze G., Chinchilla D., Caniard A., Jones J.D., Boller T., Felix G. (2006). Perception of the bacterial PAMP EF-Tu by the receptor EFR restricts Agrobacterium-mediated transformation. Cell 125: 749–760 [DOI] [PubMed] [Google Scholar]
- Zipfel C., Robatzek S., Navarro L., Oakeley E.J., Jones J.D., Felix G., Boller T. (2004). Bacterial disease resistance in Arabidopsis through flagellin perception. Nature 428: 764–767 [DOI] [PubMed] [Google Scholar]