Abstract
Dendritic cells (DC) serve a key function in host defense, linking innate detection of microbes to the activation of pathogen-specific adaptive immune responses(1,2). Whether there is cell-intrinsic recognition of HIV-1 by host innate pattern-recognition receptors and subsequent coupling to antiviral T cell responses is not yet known(3). DC are largely resistant to infection with HIV-1(4), but facilitate infection of co-cultured T-helper cells through a process of trans-enhancement(5,6). We show here that, when DC resistance to infection is circumvented(7,8), HIV-1 induces DC maturation, an antiviral type I interferon response and activation of T cells. This innate response is dependent on the interaction of newly-synthesized HIV-1 capsid (CA) with cellular cyclophilin A (CypA) and the subsequent activation of the transcription factor IRF3. Because the peptidyl-prolyl isomerase CypA also interacts with CA to promote HIV-1 infectivity, our results suggest that CA conformation has evolved under opposing selective pressures for infectivity versus furtiveness. Thus, a cell intrinsic sensor for HIV-1 exists in DC and mediates an antiviral immune response, but it is not typically engaged due to absence of DC infection. The virulence of HIV-1 may be related to evasion of this response, whose manipulation may be necessary to generate an effective HIV-1 vaccine.
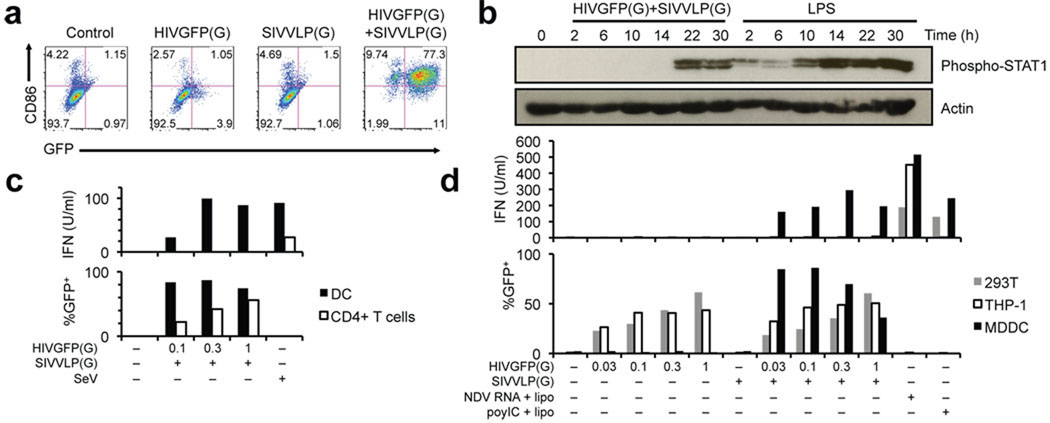
Exposure of monocyte-derived DC (MDDC) to GFP-encoding HIV-1 pseudotyped with VSV-G (HIVGFP(G)) (MOI 1–2, see Supplemental Table 1) resulted in little infection and absence of cell activation, as monitored by expression of CD86, CD80, CD38, and CD83 (Fig. 1a and Supplemental Fig. 2a). Likewise, VSV-G-pseudotyped SIVmac239 virus-like particles (SIVVLP(G)) had no effect DC activation. In contrast, co-infection of MDDC with HIVGFP(G) and SIVVLP(G), which provides Vpx-mediated relief of restriction to HIV-1 replication(8), resulted in GFP expression in more than 85% of the cells as well as up-regulation of CD86 and other activation markers after 48h (Fig. 1a and Supplemental Fig. 2a). Entry of both virions into the cytoplasm was required (Supplemental Fig. 2b), and activation occurred only beyond a variable threshold of infection (Supplemental Fig. 2c, 3a). A virus that expressed all accessory proteins and a CCR5-tropic replication competent virus also infected MDDC in the presence of SIVVLP(G) and induced expression of CD86 (Supplemental Fig. 3b, 4a). Expression of a Vpx-Vpr fusion protein in packaging cells rescued the ability of HIVGFP(G) to productively infect MDDC and to induce CD86 up-regulation, indicating that Vpx is the only SIVVLP component required for infection with HIV-1 (Supplemental Fig. 4b). Thus, MDDC carry an intact mechanism of activation following HIV-1 infection.
Figure 1.
Productive infection of MDDC with HIV-1 induces a type I interferon response. (a) GFP expression and CD86 surface expression in monocyte-derived dendritic cells (MDDC) at 48h after infection with HIVGFP(G) and SIVVLP(G), alone or in combination. (b) Immunoblot of phospho-STAT1 and total actin expression over time in MDDC infected with HIVGFP(G) and SIVVLP(G) or treated with LPS. (c) Type I interferon activity in the supernatant of MDDC or activated CD4+ T cells infected with SIVVLP(G) alone or in combination with dilutions of HIVGFP(G) or with Sendai virus (SeV). (d) Type I interferon activity in supernatants of MDDC, 293FT and THP-1 cells infected with SIVVLP(G) alone or in combination with dilutions of HIVGFP(G) or transfected with poly(I:C) or RNA from Newcastle Disease Virus (NDV)-infected cells.
As observed with MDDC, infection of primary peripheral blood CD11c+ DC with HIVGFP(G) did not result in detectable expression of GFP (Supplemental Fig. 4c). However, CD11c+ DC were infected with HIVGFP(G) in the presence of SIVVLP(G), resulting in up-regulation of CD86 in a proportion of cells similar to that observed after incubation with poly(I:C).
Genome-wide expression profiling demonstrated induction of a type I interferon response following co-infection, but not following infection with either HIV-1 or SIV particles (Supplemental Fig. 5a, 5b, 5c). After infection, expression of interferon-regulated genes was delayed as compared to the response to LPS (Supplemental Fig. 5d). Accordingly, STAT1 phosphorylation was present at 2h after LPS treatment, but only at 22h after infection (Fig. 1b). Type I interferon was produced by infected MDDC over the course of 48 hours (Supplemental Fig. 6a), but was not detected following infection of CD4+ T cells (Fig. 1c), 293T cells, and THP-1 cells (Fig. 1d), despite the ability of those cells to produce type I IFN after other viral innate stimuli. Blocking antibodies against IFNβ, but not IFNα, reduced expression of the activation markers on MDDC (Supplemental Fig. 6b, 6c). Further neutralization of type I and type III IFN did not improve the inhibition (data not shown). Together, these results suggest that CD86 induction is mainly due to the production of soluble type I IFNβ, in accordance with observations in murine DC(9).
Next, we sought to determine which step of the viral replication cycle is required for MDDC activation. Inhibitors of HIV-1 reverse transcriptase (AZT) and integrase (Raltegravir) inhibited transduction efficiency and MDDC activation only when added during the first 24 hours (Supplemental Fig. 7a) and had no effect on LPS- or poly(I:C)-induced CD86 up-regulation (data not shown). These results suggested that DC activation is induced after integration. There was no activation of MDDC after infection with an HIV-1-based vector devoid of viral protein-coding sequences (LKO1gfp) (Supplemental Fig. 7b). We therefore introduced mutations in the packaged HIVGFP genome, and evaluated activation of infected MDDC. Inactivation of Rev, required for nuclear export of unspliced viral RNA(10), and abrogation of Gag expression prevented MDDC activation (Fig. 2a), but mutation of the PTAP sequence in p6, required for viral budding(11), and treatment with HIV-1 protease inhibitors (Supplemental Fig. 7c) had no effect. In the absence of SIVVLP(G), intracellular CA from incoming viral particles failed to induce CD86 expression (Supplemental Fig. 8). These results suggested that newly synthesized GagPol is required for DC activation, which is consistent with the delayed induction of the type I interferon response. We next tested a panel of viruses with CA mutations(12) for ability to induce the innate response in MDDC. These mutants were defective for DC infection (Supplemental Fig. 9) and were thus partially rescued by co-transfection of wild type viral proteins in the packaging cells. The T54A/N57A and Q63A/Q67A mutants induced CD86 expression despite reduced infectivity compared to wild-type virus (Fig. 2b and Supplemental Fig. 10). In contrast, infection with the G89V mutant, which is compromised for CA binding to cyclophilin A (CypA), a peptidyl-prolyl isomerase required for optimal HIV-1 infectivity(13), resulted in substantially reduced CD86 expression at similar levels of infection.
Figure 2.
Dendritic cell activation requires cyclophilin A interaction with newly-synthesized HIV-1 capsid. (a) GFP and CD86 expression in MDDC infected with HIVGFP(G) or its mutants, ΔRev, ΔGag or PTAP− in the presence of SIVVLP(G). HIVGFP was rescued in all cases by co-expression of wild-type proteins in packaging cells. (b) Effect of CA mutations on proportion of GFP+ infected MDDC that express CD86. MDDC were infected with serially diluted wild-type (WT) pLaiΔEnv-GFP3(G) or CA mutants T54A/N57A, Q63A/Q67A or G89V, in the presence of SIVVLP(G). CA mutant infectivity was rescued by co-expression of wild-type proteins in packaging cells. * p < 0.026 (n=9). (c) Effect of Cyclosporin A and FK506 on expression of CD86 in MDDC infected with HIVGFP(G) and SIVVLP(G) or after treatment with LPS. CsA and FK506 target the calcineurin pathway but FK506 does not bind to CypA. (d) Expression of GFP, RFP and CD86 in HIV-infected cells following CypA knock-down by RNAi. MDDC were transduced with GFP-encoding control shRNA vector or a shRNA vector targeting the CypA-encoding PPIA, in the presence of SIVVLP(G), and subsequently challenged with HDVIRESRFP(G) or treated with LPS. Right panel: cells are gated on GFP+ populations shown in the left panels. Experiments were performed on a total of at least 6 donors, except (c) that was performed on 4 donors.
Treatment with Cyclosporin A (CsA), which disrupts the interaction between CypA and CA(14), prevented MDDC activation following infection with HIVGFP(G) and SIVVLP(G), but not following treatment with LPS (Fig. 2c). Because CsA also inhibits infection with HIV-1, we assessed its effect when administered at different times after infection of MDDC. When CsA was added as late as 12h following infection, it prevented up-regulation of CD86 despite highly efficient infection and expression of CA (Supplemental Fig. 11).
To study the role of CypA and other host genes in innate immune signaling following productive infection of MDDC with HIV-1, we employed an RNAi approach using shRNA lentiviral vectors (that also express GFP)(15) along with SIVVLP(G). Knockdown of PPIA markedly reduced expression of its product, CypA, and prevented CD86 up-regulation following infection with HIV-1 (HDVIRESRFP(G), encoding the reporter RFP), but not following treatment with LPS (Fig. 2d and Supplemental Fig. 12a). The interaction between CypA and newly synthesized CA is therefore essential for the innate response of MDDC to HIV-1.
Type I interferon responses following infection with multiple viruses requires the phosphorylation, dimerization, and nuclear translocation of IRF3(16). Productive infection of MDDC with HIV-1 resulted in CsA-sensitive nuclear accumulation of phosphorylated IRF3 (Fig. 3a). Knock-down of IRF3 in MDDC abrogated the induction of CD86 upon infection with HIV-1 and, as expected, following treatment with LPS or poly(I:C)), but not after treatment with curdlan, indicating that IRF3 knockdown did not lead to an intrinsic defect in CD86 expression (Fig. 3b and Supplemental Fig. 12b). IRF3 knockdown, as well as CypA knockdown, also increased the threshold at which virus induced CD86 and CD38 (Supplemental Fig. 12c, 12d).
Figure 3.
Dendritic cell activation by HIV-1 requires IRF-3. (a) Tubulin, Histone H3, IRF3 and Phosphor-Ser396-IRF3 expression in cytoplasmic and nuclear fractions of MDDC infected with SIVVLP(G) and dilutions of HIVGFP(G) in the presence or the absence of CsA. Cells were harvested at 8 hours after infection or after control treatment with poly(I:C). (b) GFP, RFP and CD86 expression in MDDC initially transduced with GFP-encoding control shRNA vector or a shRNA vector targeting IRF3 and subsequently challenged with HDVIRESRFP(G) or treated with poly(I:C) or Curdlan. Right panel: cells are gated on GFP+ transduced populations.
To determine if productive infection and subsequent activation of MDDC influences antiviral adaptive immunity, we first examined whether HIV-infected DC could activate HIV-1 Gag-specific CD4+ and CD8+ T cell clones. In the presence of MDDC incubated with HIV-1 alone, low levels of IFNγ were detected (17). In contrast, MDDC infected with HIVGFP(G) and SIVVLP(G) stimulated a high proportion of MHC class I and class II restricted T cell clones to produce IFNγ (Supplemental Fig. 13a). Maturation induced by unrelated TLR ligands coupled with abortive HIV infection was not sufficient for MDDC to potently stimulate HIV antigen-specific T cells (Supplemental Fig. 13b).
To directly measure the contribution of co-stimulation to T cell activation, we examined the polyclonal proliferation of naïve CD4+ T cells in response to infected DC in the presence of sub-optimal concentrations of anti-CD3 antibody(18–20). Under these conditions, T cells that were co-cultured with productively infected and activated MDDC proliferated through multiple cell cycles whereas T cells cultured with the abortively infected or uninfected MDDC had little proliferation (Fig. 4a and Supplemental Fig. 14). We next examined the effect of SCY, a non-immunosuppressive CsA analog(21,22) that, unlike CsA, does not have any direct effect on activation or proliferation of T cells(21) (Supplemental Fig. 15a). SCY inhibited DC activation induced by HIVGFP(G) similarly to CsA at similar levels of infection. DC treated with SCY or the RT inhibitor AZT at the time of HIVGFP(G) and SIVVLP(G) infection showed a reduced ability to induce proliferation (Fig. 4b and Supplemental Fig. 15b and 16), as did DC infected with the G89V CA mutant (Supplemental Fig. 17). These results are consistent with a requirement for interaction of newly-synthesized CA with CypA in the induction of DC co-stimulatory activity.
Figure 4.
Activation of T cells and inhibition of trans-enhancement by MDDC productively infected with HIV-1. (a) GFP and CD86 expression in control and HIV-1-infected DC (top) and CFSE dilution (bottom) in CFSE-labeled naïve CD4+ T cells cultured with the DC for four days in the presence of anti-CD3 antibody. (b) GFP and CD86 expression in DC (top) and CFSE dilution (bottom) in naïve CD4+ T cells cultured for four days with untreated DC or DC treated with 25 µM AZT or 1 µM SCY after infection. (c) Induction of a type I IFN-dependent antiviral state inhibits MDDC-dependent trans-enhancement. MDDC were infected with dilutions of HDVIRESRFP(G) and SIVVLP(G) in the presence or the absence of type I IFN neutralizing reagents. Activated CD4+ T cells and a CCR5-tropic HIV-1-GFP (R5-GFP) were added 2 days later. RFP and CD86 expression and GFP expression were measured in DC and CD4+ T cells, respectively. Trans-enhancement is indicated by the increase in GFP+ T cells in the presence or absence of MDDC in the top panel.
Trans-enhancement by MDDC of CD4+ T cell infection with a CCR5-tropic virus encoding GFP was inhibited if the DC were previously infected with HIV-1. The inhibition was relieved by neutralizing antibody against IFNβ, indicating that the innate response to HIV-1 in DC restricts infection of surrounding T cells (Figure 4c) and suggesting that activation of such response may also limit infection in vivo.
Our results show that, in contrast to CD4+ T cells, human dendritic cells have intrinsic machinery for responding to infection with HIV-1 and for activating antiviral defenses and adaptive immunity (Supplemental Fig. 1). However, they are unlikely to do so effectively in infected individuals because HIV-1 fails to replicate in DCs. HIV-2, which is not pandemic(23), encodes Vpx and has the potential to infect and activate MDDC in a CypA-dependent manner (Supplemental Fig. 18), which is consistent with the reported ability of HIV-2 CA to bind human CypA(24,25). The finding that newly-synthesized CA is required to induce DC activation through a pathway involving CypA and IRF3 implicates an intracellular viral protein, in addition to viral nucleic acids, among the type I interferon-inducing pathogen-associated molecular patterns (PAMPs)(26) and constitutes the first description of a cell-intrinsic recognition mechanism of retroviruses(26). It will be important to determine whether the mechanism described herein contributes to control of the viral load in individuals infected with HIV-2, as well as in HIV-1-infected long-term non-progressors or “elite controllers”(27). A better mechanistic understanding of this DC-intrinsic signaling pathway may also inform HIV vaccine development.
METHODS SUMMARY
Monocytes were isolated and incubated with GM-CSF and IL-4 to induce dendritic cell differentiation. Pseudotyped viruses and virus-like particles were produced by transient transfection of 293FT cells using TransIT-293 (Mirus). Infections were performed by incubating 105 MDDCs in 96 well U bottom plates in the presence of 8 µg/ml polybrene. Cell surface staining of activation markers was performed 48h after infection. shRNA vectors carrying GFP were transduced into fresh monocytes together with SIVVLP(G) and dendritic cell differentiation was induced. More than 90% of cells were routinely transduced and cells were challenged at day 4 with HDVIRESRFP(G) or other control PAMPs.
METHODS
Cells
GHOST(28) and 293FT (Invitrogen) cells were cultured in DMEM, 10% fetal bovine serum (FBS) (HyClone) and antibiotics. PBMC were isolated from IRB-approved buffy coats from normal donors. CD14+ cells were isolated by double positive selection with anti-human CD14 magnetic beads (Miltenyii). Purity was at least 99%. CD14+ cells were cultured in RPMI, 10% FBS, antibiotics and HEPES in the presence of recombinant human GM-CSF at 10 ng/ml and IL-4 at 50 ng/ml (eBioscience). Fresh media was added at day 3, and cells were stimulated or infected at day 4. To isolate CD11c+ DC, CD14-depleted PBMC were further depleted using biotin-labeled antibodies against CD3, CD16, CD19 and CD56 and streptavidin magnetic beads (Miltenyi). The negative fraction was stained and sorted on a FACSAria (BD Biosciences) as CD3−CD14−CD16− CD19−CD20−CD56−HLA-DR+CD11c+ (Supplemental Table 2). Purity was at least 98%. Total CD4+ T cells were isolated using human CD4 magnetic beads (Miltenyii). Naïve CD4+ T cells were further sorted as CD4+CD25−CD45RA+CD45RO−.
T cell clones
T cell clones were expanded as previously described (29). The clone DR4LI15 (LGLNKIVRMYSPTSI) was obtained from Nina Bhardwaj and the clones B81TL9 (TPQDLNTML) and B14DA9 (DRFYKTLRA) were obtained from Bruce Walker.
Reagents
LPS (lipopoylsaccharide) and poly(I:C) (polyriboinosinic:polyribocytidylic acid) were from Sigma and were used at 1 µg/ml and 10 µg/ml, respectively. Curdlan (CM-Curdlan) was form Wako and was used at 1 µg/ml. Cyclosporin A and FK506 were from Calbiochem. AZT (Zidovudine), RAL (Raltegravir), LPV (Lopinavir), SQV (Saquinavir), TPV (Tipranavir) were obtained through the NIH AIDS Research & Reference Reagent Program. SCY (SCYX00011867717) is similar to SCY-635 and was a generous gift from Scynexis.
Infection and stimulation
At day 4 of MDDC differentiation, cells were harvested, counted and resuspended in their own media at a concentration of one million/ml with 5 µg/ml polybrene and 100 µl were aliquoted in round bottom 96-well plates. For infection, 50 µl of media or SIVVLP(G) was first added. 100 µl of media or dilutions of various HIV-1-derived viral preparations were then added. SQV, TPV, AZT and RAL were added at 10 µM. Neutralizing anti-IFNα and anti-IFNβ were added at 20 µg/ml. B18R (eBioscience) was added at 0.2 µg/ml. IL28RA-Fc (R&D Systems) was added at 1 µg/ml. Neutralizing anti-IFNAR was added at 1 µg/ml. For shRNA-transduced DC, cells were harvested, counted and resuspended in fresh media containing GM-CSF, IL-4 and 1 µg/ml polybrene at a concentration of one million/ml. 100 µl were aliquoted in round bottom 96-well plate and 100 µl of media or virus was added.
Western blot analysis
Cells were lysed in 1% NP-40, 50 mM Tris pH 8, 120 mM NaCl, 4 mM EDTA, 50 mM NaF, 1 mM NA3VO4 and a protease inhibitor cocktail (Roche). Total lysates were resolved on SDS-PAGE, transferred to PVDF membranes and probed with primary antibodies and corresponding HRP-conjugated secondary antibodies (GE Healthcare).
Cytoplasmic and nuclear fractionation
MDDC were harvested at 24 hours after infection or treatment. 4×106 cells were washed once with room temperature PBS, gently pelleted and resuspended in 400 µl of cold cytoplasmic lysis buffer (CL buffer) containing 10 mM Hepes pH 7.9, 10 mM sodium potassium, 1.5 mM magnesium chloride, 1 mM sodium orthovanadate, 2 mM sodium pyrophosphate, 2 mM sodium β-glycerophosphate, 5 mM sodium fluoride, complete EDTA-free protease inhibitor cocktail (Roche) and phosphatase inhibitor cocktail (SIGMA P2850). Cells in cold CL buffer were immediately pelleted at 4°C, supernatant was discarded, 40 µl CL was added, and buffer and cells were gently resuspended by slow pipetting and soft flicking and left on ice for 15 minutes. 2.5 µl of 10% NP-40 was added and cells were lysed by gentle flicking. Nuclei were pelleted at 13,000 rpm for 5 minutes at 4°C. 40 µl of supernatant was harvested and saved as the cytoplasmic fraction, and remaining liquid was discarded. 40 µl of cold nuclear lysis buffer (NL buffer) containing 420 mM sodium chloride, 20 mM HEPES pH7.9, 1.5 mM magnesium chloride, 0.2 mM EDTA, 25% glycerol and protease and phosphatase inhibitors as in CL buffer was added. Nuclei were resuspended by vigorous flicking and incubated on ice for 15 minutes, with occasional flicking. Nuclei were vortexed for 10 seconds and sonicated for 10 minutes in a 4°C bath sonicator (30 seconds on, 30 seconds off). The nuclear lysate was cleared by centrifugation at 13,000rpm for 5 minutes at 4°C, and the resulting supernatant was saved as the nuclear extract. Western blot loading buffer with dithiothreitol was added to the cytoplasmic and nuclear extracts, and the samples were heated at 70°C for 15 minutes. 10 µl of each sample were run on a 7.5% SDS-PAGE gel and transferred to PVDF membrane (Roche). Membranes were blocked with 5% non-fat dry milk in TBS containing 0.1% Tween-20 (TBST) and probed with primary antibody overnight while rocking at 4°C, washed six times for 5 minutes with TBST, probed with secondary HRP-conjugated antibody (GE Healthcare) for one hour at room temperature, washed six times for 5 minutes in TBST, and incubated with ECL reagents (Pierce Pico or Pierce Femto). Chemiluminescence signal was visualized using Kodak film.
Quantitative Bioassay for Interferons
293FT and THP-1 were infected with HIVGFP(G) and SIVVLP(G) or transfected with polyI:C or total RNA from New Castle Disease (NDV)-infected A549 cells harvested in Trizol (Invitrogen) 8 hours after infection using lipofectamine 2000 (Invitrogen). NDV viral stock was produced by inoculating 10-day-old embryonated chicken eggs (Charles River). CD4+ T cells were expanded with 5 µg/ml Phytohemagglutinin-L (Sigma) and 10 U/ml human IL-2 for 4 days and infected with 100 HA units/ml of Sendai virus (Charles River) or infected with HIVGFP(G) and SIVVLP(G). Media was replaced after 24 hours and culture supernatants were harvested after another 24 hours. Cell culture supernatants were UV-irradiated to inactivate traces of Sendai virus. Supernatants were assayed for interferon activity using a recombinant COS-1 cell line, which carries a luciferase reporter containing multiple repeats of interferon-stimulated response element (ISRE). In brief, the reporter cells were exposed to cell culture supernatants for 8 hours to overnight, and assayed for luciferase activities, which were then translated to interferon activities by using a standard curve generated from a serial dilution of human interferon alpha 2a.
Microarray analysis
MDDC were infected with HIVGFP(G), SIVVLP(G), both or treated with LPS. Cells were harvested after 48h and a subset was analyzed by flow cytometry. RNA was prepared with TRIZOL and microarray data generation was done using standard protocols on Human Genome U133A 2.0 arrays (Affymetrix). Microarray analysis was performed using the Bioconductor package in R and Genespring GX10 (Agilent). Probes were filtered based on at least a 2-fold change in expression and p<0.05. Promoter analysis was performed using PRIMA(30) in EXPANDER(31). TLR Pathway analysis was performed using SPIKE(32).
qPCR
qPCR analysis was performed as described(33) using the standard curve method or the ΔCt method (for primer sets, see Table 3).
Plasmids
HIVGFP, which is env−vpu−vpr−vif−nef−, with GFP in place of nef, has already been described (34). HIVGFP ΔRev was generated by mutating the start codon of Rev. HIVGFP PTAP− was generated by mutating PTAP to LIRL. HIVGFP D52N is a point mutation of protease. HIVGFP ΔGag was generated by inserting a stop codon after 7 amino acids of Gag. VPX-VPR fusion protein was generated by fusing SIVmac251 Vpx with HIV-1 NL4-3 Vpr using the linker ANYAAAAAAADPS in pIRES2-EGFP (Clontech). LKO1gfp was generated by replacing the puroR open-reading frame in pLKO1puro(35) with the EGFP coding region. shRNAs were designed as described previously (Table 4), except that a partial mir30 sequence "CTGTGAAGCCACAGATGGG " was used for the loop. shRNAs were then cloned as described (35). T54A/N57A, Q63AQ67A, G89V and the parental vector pLaiΔEnv-GFP3 are env−nef−, with GFP in place of nef, and were previously described (12). HDVIRESRFP was described elsewhere(36). HIV-2 ROD9 Δenv GFP was generated from a HIV-2 Δenv construct(37) by inserting the GFP coding sequence in nef, thus disrupting nef. All plasmid DNA were prepared with Invitrogen HiPure plasmid kit. Plasmid DNA did not induce DC maturation, and viral producing cells were washed after DNA transfection.
Virus production
Viral particles were produced by transfection of 293FT cells with 3 µg DNA and 8 µl TransIT-293 (Mirus Bio); for shRNA vectors, we used 0.4 µg CMV-VSVG, 1 µg pCMV-ΔR8.91 and 1.6 µg shRNA; for SIVVLP(G), 0.4 µg CMV-VSVG and 2.6 µg pSIV3+ (38); for HIVGFP(G), 0.4 µg CMV-VSVG and 2.6 µg HIVGFP; for HIV2 ROD9 Δenv GFP(G), 0.4 µg CMV-VSVG and 2.6 µg HIV2 ROD9 Δenv GFP; for NL4-3-delta-EEGFP, 0.4 µg pCMV-VSVG and 2.6 µg pNL4-3-deltaE-EGFP(39). HIVGFP ΔRev, HIVGFP PTAP− were produced with 0.4 µg CMV-VSVG, 0.5 µg pCMV-ΔR8.91 and 2.1 µg HIV plasmid. HIVGFP ΔGag and CA mutants were produced with 0.4 µg CMV-VSVG, 1 µg pCMV-ΔR8.91 and 1.6 µg HIV plasmid. R5-GFP is NL4-3 encoding for the BAL envelope and GFP in nef (34,36). One day after transfection, media was removed, cells were washed out once, and fresh media was added. Viral supernatants were harvested one day later and filtered at 0.45 µM. In some experiments, p24 concentration was measured by p24 enzyme-linked immunosorbent assay (ELISA).
RNAi
Synthetic siRNA can be delivered in MDDC by electroporation, but this is highly and rapidly toxic and renders difficult the interpretation of dendritic cell activation, which can be altered by the presence of apoptotic or necrotic cells. In addition, we have not been able to achieve significant knock-down using synthetic siRNA with lipid-based reagents in MDDC. In fact, fluorescently-labeled siRNA appeared to be simply endocytosed with all the lipid-based reagents that we tested (data not shown). We thus utilized shRNA vectors. Using this method, we routinely obtained >90% transduction efficiency, alleviating the need for cell sorting or selection. Five million freshly isolated CD14+ cells were cultured in 5 ml of media containing GM-CSF, IL-4, and 1 µg/ml polybrene. One ml of SIVVLP(G) supernatant and 2.5 ml of shRNA vector supernatant were added to cells. At days 1 and 3, 2 ml of fresh media was added. At day 4, cells were transduced at more than 96% based on GFP expression and were used for further infections and stimulation as above.
HIV-specific T cell clone stimulation
48h after infection of MDDC, 105 rested HIV-specific T cell clones were added in the presence of GolgiStop (BD Biosciences). Where indicated, T cells were activated with 50 ng/ml PMA (Sigma) and 0.5 µg/ml ionomycin (Sigma). Cells were incubated for 6 hours and processed for intracellular staining(33).
Naïve T cell proliferation assay
48h after infection, naïve T cells were labeled with CFSE (eBioscience) as descried previously (18). DC were infected with dilutions of SIVVLP(G) and pLaiΔEnv-GFP3(G) WT (indicated as HIVGFP(G)) or G89V. AZT was added at the time of infection and SCY was added from 3 to 8 hours after infection. 48 hours after infection, half of the DC was processed for surface staining and cytometry. The other half was washed with media and resuspended in fresh media without cytokines. 20,000 T cells were mixed with DC at a DC to T ratio of 1:5 and 1:15. Cells were stimulated by dilutions of anti-CD3 (OKT3 hybridoma supernatant, approximately 1–100 ng/ml) in a total volume of 150 to 200 µl in round bottom 96-well plates. Cells were harvested and analyzed by flow cytometry at day 4 or day 5 postactivation.
Trans-enhancement
105 Day 4 MDDC were infected with dilutions of HDVIRESRFP(G) and SIVVLP(G) in 96 well round bottom plates in the presence of 5 µg/ml polybrene. Type I IFN neutralizing antibodies and recombinant proteins were maintained throughout the experiment in some samples. Media was replaced after 24 hours. Another 24 hours later, half of the cells were processed for surface staining and RFP and CD86 expression were measured by flow cytometry. The other half was mixed with a preparation of replication competent R5-GFP and 5×105 CD4+ T cells at day 4 post-activation with PHA-L (Sigma) and IL-2, in the absence of polybrene. GFP expression in CD4+ T cells was measured another 48 hours later.
Supplementary Material
ACKNOWLEDGMENTS
We thank members of the Littman laboratory for valuable discussions and critical reading of the manuscript, and Takeshi Egawa, Elke Kurz, Xing Gong, and Lina Kozyhaya for assistance with experiments. We thank Sam Hopkins at Scynexis for the generous gift of the SCY compound, Jiri Zavadil and the genomic core facility of NYU for performing the array studies, Michael Emerman, Masahiro Yamashita, and Wes Sundquist for HIV constructs and critical reading of the manuscript, Bruce Walker, Alicja Piechocka-Trocha, Douglas Kwon, Seanna M. Vine, Nina Bardhwaj and Elizabeth Miller for T cell clones and reagents, and Susan Schwab and Ruslan Medzhitov for critical reading of the manuscript. N.M. thanks Marc Sitbon, Naomi Taylor and Jean-Marie Blanchard for continuous support. The work was supported sequentially by EMBO and Cancer Research Institute fellowships (N.M.), by the Institut National de la Santé et de la Recherche Médicale (N.M.), by the Howard Hughes Medical Institute (D.R.L.), the Helen and Martin Kimmel Center for Biology and Medicine (D.R.L.), and National Institutes of Health grants AI33856 (D.R.L.), AI28900 (D.E.L.), U54AI57168 (D.E.L.) and R01AI065303 (D.U.).
Footnotes
AUTHOR CONTRIBUTIONS
N.M. and D.R.L. designed the study and wrote the manuscript. N.M. performed the experiments and analyses. B.H. provided technical help. D.U. provided expertise and contributed to experiments with human T cell proliferation assays. D.E.L. provided expertise in identifying the interferon response. D.E.L. and Y.W. designed the quantitative bioassay for interferons and Y.W. performed the assay. All authors discussed results and edited the manuscript.
DATA DEPOSITION
Microarray data has been deposited in the NCBI GEO database under the accession number GSE22589.
REFERENCES
- 1.Steinman RM, Hemmi H. Dendritic cells: translating innate to adaptive immunity. Current topics in microbiology and immunology. 2006;311:17–58. doi: 10.1007/3-540-32636-7_2. [DOI] [PubMed] [Google Scholar]
- 2.Takeuchi O, Akira S. Innate immunity to virus infection. Immunological reviews. 2009;227(1):75–86. doi: 10.1111/j.1600-065X.2008.00737.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stetson DB, Ko JS, Heidmann T, Medzhitov R. Trex1 prevents cell-intrinsic initiation of autoimmunity. Cell. 2008;134(4):587–598. doi: 10.1016/j.cell.2008.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Negre D, et al. Characterization of novel safe lentiviral vectors derived from simian immunodeficiency virus (SIVmac251) that efficiently transduce mature human dendritic cells. Gene therapy. 2000;7(19):1613–1623. doi: 10.1038/sj.gt.3301292. [DOI] [PubMed] [Google Scholar]
- 5.Cameron PU, et al. Dendritic cells exposed to human immunodeficiency virus type-1 transmit a vigorous cytopathic infection to CD4+ T cells. Science (New York, N.Y. 1992;257(5068):383–387. doi: 10.1126/science.1352913. [DOI] [PubMed] [Google Scholar]
- 6.Kwon DS, Gregorio G, Bitton N, Hendrickson WA, Littman DR. DC-SIGN-mediated internalization of HIV is required for trans-enhancement of T cell infection. Immunity. 2002;16(1):135–144. doi: 10.1016/s1074-7613(02)00259-5. [DOI] [PubMed] [Google Scholar]
- 7.Mangeot PE, et al. High levels of transduction of human dendritic cells with optimized SIV vectors. Mol Ther. 2002;5(3):283–290. doi: 10.1006/mthe.2002.0541. [DOI] [PubMed] [Google Scholar]
- 8.Goujon C, et al. With a little help from a friend: increasing HIV transduction of monocyte-derived dendritic cells with virion-like particles of SIV(MAC) Gene therapy. 2006;13(12):991–994. doi: 10.1038/sj.gt.3302753. [DOI] [PubMed] [Google Scholar]
- 9.Honda K, et al. Selective contribution of IFN-alpha/beta signaling to the maturation of dendritic cells induced by double-stranded RNA or viral infection. Proc Natl Acad Sci U S A. 2003;100(19):10872–10877. doi: 10.1073/pnas.1934678100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Malim MH, Hauber J, Fenrick R, Cullen BR. Immunodeficiency virus rev trans-activator modulates the expression of the viral regulatory genes. Nature. 1988;335(6186):181–183. doi: 10.1038/335181a0. [DOI] [PubMed] [Google Scholar]
- 11.Demirov DG, Orenstein JM, Freed EO. The late domain of human immunodeficiency virus type 1 p6 promotes virus release in a cell type-dependent manner. Journal of virology. 2002;76(1):105–117. doi: 10.1128/JVI.76.1.105-117.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yamashita M, Perez O, Hope TJ, Emerman M. Evidence for direct involvement of the capsid protein in HIV infection of nondividing cells. PLoS pathogens. 2007;3(10):1502–1510. doi: 10.1371/journal.ppat.0030156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yoo S, et al. Molecular recognition in the HIV-1 capsid/cyclophilin A complex. Journal of molecular biology. 1997;269(5):780–795. doi: 10.1006/jmbi.1997.1051. [DOI] [PubMed] [Google Scholar]
- 14.Luban J, Bossolt KL, Franke EK, Kalpana GV, Goff SP. Human immunodeficiency virus type 1 Gag protein binds to cyclophilins A and B. Cell. 1993;73(6):1067–1078. doi: 10.1016/0092-8674(93)90637-6. [DOI] [PubMed] [Google Scholar]
- 15.Boggiano C, Manel N, Littman DR. Dendritic cell-mediated trans-enhancement of human immunodeficiency virus type 1 infectivity is independent of DC-SIGN. Journal of virology. 2007;81(5):2519–2523. doi: 10.1128/JVI.01661-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sato M, et al. Distinct and essential roles of transcription factors IRF-3 and IRF-7 in response to viruses for IFN-alpha/beta gene induction. Immunity. 2000;13(4):539–548. doi: 10.1016/s1074-7613(00)00053-4. [DOI] [PubMed] [Google Scholar]
- 17.Moris A, et al. Dendritic cells and HIV-specific CD4+ T cells: HIV antigen presentation, T-cell activation, and viral transfer. Blood. 2006;108(5):1643–1651. doi: 10.1182/blood-2006-02-006361. [DOI] [PubMed] [Google Scholar]
- 18.Antons AK, Wang R, Kalams SA, Unutmaz D. Suppression of HIV-specific and allogeneic T cell activation by human regulatory T cells is dependent on the strength of signals. PLoS One. 2008;3(8):e2952. doi: 10.1371/journal.pone.0002952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gett AV, Sallusto F, Lanzavecchia A, Geginat J. T cell fitness determined by signal strength. Nature immunology. 2003;4(4):355–360. doi: 10.1038/ni908. [DOI] [PubMed] [Google Scholar]
- 20.Langenkamp A, et al. T cell priming by dendritic cells: thresholds for proliferation, differentiation and death and intraclonal functional diversification. Eur J Immunol. 2002;32(7):2046–2054. doi: 10.1002/1521-4141(200207)32:7<2046::AID-IMMU2046>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 21.Hopkins S, et al. SCY-635, a novel nonimmunosuppressive analog of cyclosporine that exhibits potent inhibition of hepatitis C virus RNA replication in vitro. Antimicrob Agents Chemother. 54(2):660–672. doi: 10.1128/AAC.00660-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chatterji U, et al. The isomerase active site of cyclophilin A is critical for hepatitis C virus replication. J Biol Chem. 2009;284(25):16998–17005. doi: 10.1074/jbc.M109.007625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.de Silva TI, Cotten M, Rowland-Jones SL. HIV-2: the forgotten AIDS virus. Trends Microbiol. 2008;16(12):588–595. doi: 10.1016/j.tim.2008.09.003. [DOI] [PubMed] [Google Scholar]
- 24.Neagu MR, et al. Potent inhibition of HIV-1 by TRIM5-cyclophilin fusion proteins engineered from human components. The Journal of clinical investigation. 2009;119(10):3035–3047. doi: 10.1172/JCI39354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Price AJ, et al. Active site remodeling switches HIV specificity of antiretroviral TRIMCyp. Nat Struct Mol Biol. 2009;16(10):1036–1042. doi: 10.1038/nsmb.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Janeway CA., Jr Approaching the asymptote? Evolution and revolution in immunology. Cold Spring Harb Symp Quant Biol. 1989;54(Pt 1):1–13. doi: 10.1101/sqb.1989.054.01.003. [DOI] [PubMed] [Google Scholar]
- 27.Kosmrlj A, et al. Effects of thymic selection of the T-cell repertoire on HLA class[thinsp]I-associated control of HIV infection. Nature. doi: 10.1038/nature08997. advance online publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
ADDITIONAL METHODS REFERENCES
- 28.Morner A, et al. Primary human immunodeficiency virus type 2 (HIV-2) isolates, like HIV-1 isolates, frequently use CCR5 but show promiscuity in coreceptor usage. Journal of virology. 1999;73(3):2343–2349. doi: 10.1128/jvi.73.3.2343-2349.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fonteneau JF, et al. Generation of high quantities of viral and tumor-specific human CD4+ and CD8+ T-cell clones using peptide pulsed mature dendritic cells. J Immunol Methods. 2001;258(1–2):111–126. doi: 10.1016/s0022-1759(01)00477-x. [DOI] [PubMed] [Google Scholar]
- 30.Elkon R, Linhart C, Sharan R, Shamir R, Shiloh Y. Genome-wide in silico identification of transcriptional regulators controlling the cell cycle in human cells. Genome research. 2003;13(5):773–780. doi: 10.1101/gr.947203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shamir R, et al. EXPANDER--an integrative program suite for microarray data analysis. BMC bioinformatics. 2005;6:232. doi: 10.1186/1471-2105-6-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Elkon R, et al. SPIKE--a database, visualization and analysis tool of cellular signaling pathways. BMC bioinformatics. 2008;9:110. doi: 10.1186/1471-2105-9-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Manel N, Unutmaz D, Littman DR. The differentiation of human T(H)-17 cells requires transforming growth factor-beta and induction of the nuclear receptor RORgammat. Nature immunology. 2008;9(6):641–649. doi: 10.1038/ni.1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Unutmaz D, KewalRamani VN, Marmon S, Littman DR. Cytokine signals are sufficient for HIV-1 infection of resting human T lymphocytes. The Journal of experimental medicine. 1999;189(11):1735–1746. doi: 10.1084/jem.189.11.1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moffat J, et al. A lentiviral RNAi library for human and mouse genes applied to an arrayed viral high-content screen. Cell. 2006;124(6):1283–1298. doi: 10.1016/j.cell.2006.01.040. [DOI] [PubMed] [Google Scholar]
- 36.Oswald-Richter K, et al. HIV infection of naturally occurring and genetically reprogrammed human regulatory T-cells. PLoS Biol. 2004;2(7):E198. doi: 10.1371/journal.pbio.0020198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Griffin SD, Allen JF, Lever AM. The major human immunodeficiency virus type 2 (HIV-2) packaging signal is present on all HIV-2 RNA species: cotranslational RNA encapsidation and limitation of Gag protein confer specificity. Journal of virology. 2001;75(24):12058–12069. doi: 10.1128/JVI.75.24.12058-12069.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mangeot PE, et al. Development of minimal lentivirus vectors derived from simian immunodeficiency virus (SIVmac251) and their use for gene transfer into human dendritic cells. Journal of virology. 2000;74(18):8307–8315. doi: 10.1128/jvi.74.18.8307-8315.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang H, et al. Novel single-cell-level phenotypic assay for residual drug susceptibility and reduced replication capacity of drug-resistant human immunodeficiency virus type 1. Journal of virology. 2004;78(4):1718–1729. doi: 10.1128/JVI.78.4.1718-1729.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.