Nup133 links CENP-F, NudE/EL, and the dynein/dynactin complex to anchor centrosomes to the nuclear membrane.
Abstract
Centrosomes are closely associated with the nuclear envelope (NE) throughout the cell cycle and this association is maintained in prophase when they separate to establish the future mitotic spindle. At this stage, the kinetochore constituents CENP-F, NudE, NudEL, dynein, and dynactin accumulate at the NE. We demonstrate here that the N-terminal domain of the nuclear pore complex (NPC) protein Nup133, although largely dispensable for NPC assembly, is required for efficient anchoring of the dynein/dynactin complex to the NE in prophase. Nup133 exerts this function through an interaction network via CENP-F and NudE/EL. We show that this molecular chain is critical for maintaining centrosome association with the NE at mitotic entry and contributes to this process without interfering with the previously described RanBP2–BICD2-dependent pathway of centrosome anchoring. Finally, our study reveals that tethering of centrosomes to the nuclear surface at the G2/M transition contributes, along with other cellular mechanisms, to early stages of bipolar spindle assembly.
Introduction
In eukaryotic cells, the nuclear envelope (NE) separates the cytoplasm and the nucleus, where the chromosomes are confined. The NE is composed of a double membrane, the inner and outer nuclear membranes, which are connected at particular sites, the nuclear pore complexes (NPCs). These mediate all nucleocytoplasmic exchanges during interphase (Tran and Wente, 2006).
During cell division cytoplasmic microtubules nucleated from the centrosomes need to access mitotic chromosomes in order to establish a mitotic spindle and allow proper chromosome segregation in the two daughter cells. In vertebrates, this is achieved in prophase by nuclear envelope breakdown (NEBD), when the NE and its underlying lamina dismantle, whereas the NPC constituents—the nucleoporins—are dispersed into the mitotic cytoplasm in subcomplexes. At the end of mitosis, these soluble complexes and NE components are reused to form new NPCs and NEs within the two daughter cells (Antonin et al., 2008, Kutay and Hetzer, 2008).
A major player in both post-mitotic NPC reassembly and de novo assembly of NPCs during interphase is the evolutionarily conserved Nup107–160 complex, which is stably associated on both sides of the NPC and is composed of nine subunits in metazoans (Nup107, Nup160, Nup133, Nup96, Nup43, Nup85, Nup37, Sec13, and Seh1; Doucet et al., 2010; Wozniak et al., 2010). In addition, a fraction of the Nup107–160 complex localizes at spindle poles and proximal spindle fibers in prometaphase and at kinetochores from early prophase to late anaphase in mammalian cells, and throughout reconstituted spindles in Xenopus egg extracts (Belgareh et al., 2001; Harel et al., 2003; Loïodice et al., 2004; Orjalo et al., 2006). In the Xenopus in vitro system, immunodepletion of the entire Nup107–160 complex revealed its involvement at late stages of mitotic spindle assembly or stabilization (Orjalo et al., 2006). In mammalian cells, efficient depletion of this complex from kinetochores leads to an altered recruitment of a subset of kinetochore constituents and impairs mitotic progression (Zuccolo et al., 2007; Platani et al., 2009; Mishra et al., 2010; Wozniak et al., 2010).
A mitosis-specific interaction partner of the Nup107–160 complex is CENP-F (also called mitosin; Zuccolo et al., 2007), a large cell cycle–regulated protein that shows a very dynamic localization pattern: CENP-F is mainly found in the nucleus in G2, then binds to the NE at the G2/M transition before its accumulation at nascent kinetochores in early prophase, where it remains until anaphase onset. CENP-F subsequently localizes to the spindle midzone, and finally undergoes proteasome degradation in early G1 (Varis et al., 2006). Several studies, focusing on the mitotic role of CENP-F at kinetochores, showed that its depletion leads to a checkpoint-dependent mitotic delay, a phenotype likely underlying its requirement as an upstream recruitment factor for the kinetochore motor proteins dynein and CENP-E (Varis et al., 2006; Mao et al., 2010). Although CENP-F has also been implicated in centrosomal microtubule nucleation in mouse embryonic fibroblasts (Moynihan et al., 2009) and in timely G2/M progression (Hussein and Taylor, 2002), nothing was known so far about its specific function at the NE at the G2/M transition.
At the G2/M transition, the morphology of the NE is dramatically affected by mitotic microtubules, which are nucleated by the separating centrosomes that push on the nuclear surface and form pocket-like distortions of the NE, also termed prophase NE invaginations (PNEI; Robbins and Gonatas, 1964). At that stage, microtubules were found to facilitate NEBD by exerting pulling forces on the NE and thus contributing to its disruption, and by subsequently clearing the chromatin area from the NE membrane network (Beaudouin et al., 2002; Salina et al., 2002; Mühlhäusser and Kutay, 2007). The minus end–directed motor dynein and its regulatory complex dynactin, which interact with microtubules and are recruited to the NE specifically in early prophase, were shown to contribute to these processes (Busson et al., 1998; Salina et al., 2002; Mühlhäusser and Kutay, 2007). More recently, Ndel1 and Lis1, which associate with cytoplasmic dynein, were reported to localize at the NE in prophase cells and to contribute to dynein-dependent PNEI (Hebbar et al., 2008).
Although these studies suggested that NEBD and spindle formation are closely coordinated processes, how Ndel1 and in turn dynein/dynactin bind to the NE and how the localization of a subset of proteins at the NE in prophase may help to coordinate proper cell division remained unanswered. Recently, Splinter et al. (2010) demonstrated that RanBP2/Nup358, a nucleoporin that localizes at the cytoplasmic side of the NPC, contributes to dynein/dynactin recruitment and centrosome positioning, by recruiting BICD2 to NPCs in G2. We report here the characterization of a distinct prophase pathway, operating via the Nup133 subunit of the Nup107–160 nuclear pore subcomplex. We show that the N-terminal domain of Nup133 recruits CENP-F, which itself tethers NudE/NudEL to NPCs in prophase. The contribution of this molecular chain, along with the previously described RanBP2/Nup358-BICD2 pathway, in maintaining centrosome association with the NE at mitotic entry is further characterized.
Results
The N-terminal domain of hNup133 is largely dispensable for NPC assembly
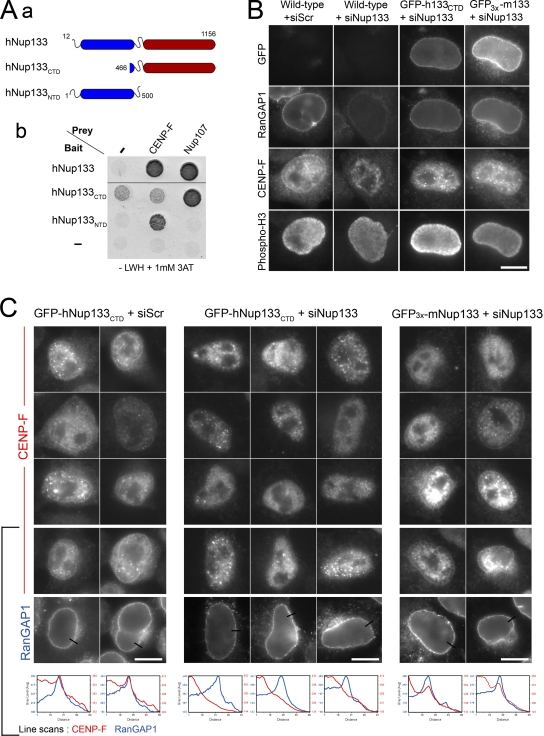
Structural studies have revealed that hNup133 is composed of an N-terminal domain with a β-propeller structure and an α-solenoid C-terminal domain, which is able to bind to hNup107 and to localize at NPCs (Berke et al., 2004; Boehmer et al., 2008; see also Fig. 2 A). Consistently, the C-terminal domain of hNup133 (aa 545–1156, subsequently referred as hNup133CTD) fused to GFP accumulated at the nuclear rim in interphase and colocalized with CENP-F and Hec1 at kinetochores in mitotic cells, like the full-length protein (Fig. 1 B and Fig. S1 A).
Figure 2.
The N-terminal domain of hNup133 tethers CENP-F at the NE in prophase. (Aa) Schematic representation of hNup133 constructs used in this study outlining its previously described β-propeller (N-terminal domain, NTD, blue) and α-solenoid (C-terminal domain, CTD, red) domains (adapted from Berke et al., 2004). (Ab) Yeast two-hybrid interactions between hNup133 (aa 12–1156), hNup133CTD (aa 466–1156) or hNup133NTD (aa 1–500), and hNup107 (aa 784–924), or CENP-F (aa 2644–3065) were analyzed as described in Materials and methods. Empty bait and prey vectors were used as negative controls (−). (B and C) Control HeLa cells (wild-type) or cell lines stably expressing GFP-hNup133CTD or GFP3x-mNup133 were transfected with a scramble siRNA (siScr) or with an siRNA duplex targeting the N-terminal domain of human Nup133. Cells were fixed after 3 d and processed for immunofluorescence using anti-RanGAP1, anti-CENP-F, and anti–phospho-H3 antibodies. Typical G2/M phospho-H3–positive cells are presented. Bars, 10 µm. In C, the anti-RanGAP1 signal is shown for cells on the bottom row. Line scans (with distances in pixels) measuring the intensity of CENP-F (red lines) and RanGAP1 (blue lines) reveal the peak of CENP-F that colocalizes with RanGAP1 at the NE in GFP-hNup133CTD treated with the scramble siRNA duplexes (left panels) or in GFP3x-mNup133 cells depleted for endogenous Nup133 (right panels), but not in Nup133-depleted GFP-hNup133CTD cells (middle panels). In some prophase cells, the bright intranuclear foci reflect the early recruitment of CENP-F at kinetochores.
Figure 1.
The N-terminal domain of hNup133 is largely dispensable for NPC assembly. (A) Control HeLa cells (WT) or cell lines stably expressing GFP-hNup133CTD or GFP3x-mNup133 were transfected with a scramble siRNA or with a hNup133 siRNA that does not target hNup133CTD and only mildly impacts mNup133 expression. After 3 d, whole-cell extracts were analyzed by Western blot using anti-hNup133 (which also recognizes mouse Nup133, albeit less efficiently than human Nup133) or anti-hNup107 antibodies. Anti–γ-tubulin was used as loading control. To allow quantifications, decreasing amounts of the reference sample were loaded (0.2-, 0.5-, and 1-fold equivalent). (B) Cells transfected with siRNAs as in A were fixed after 3 d and processed for immunofluorescence using anti-Nup107 and mAb414 antibodies. Bar, 20 µm.
To dissect the function of the N-terminal domain of hNup133, we established HeLa cell lines stably expressing RNAi-resistant GFP-hNup133CTD or full-length mouse GFP-Nup133. After depleting endogenous hNup133 with specific siRNAs (Fig. 1 A), these cells allowed us to test which functions of Nup133 specifically depend on the N-terminal β-propeller domain. Depletion of hNup133 is known to cause the degradation and mislocalization of the other members of the Nup107–160 complex, as well as a number of additional nucleoporins (Walther et al., 2003; Fig. 1, A and B). hNup107 was stabilized when depletion of endogenous hNup133 (reduced to ∼20% as compared with the level of hNup133 present in wild-type HeLa cells) was rescued with either GFP-hNup133CTD or GFP-mNup133 (Fig. 1 A). In addition, both Nup133CTD and full-length Nup133 restored the NE localization of Nup107 and the subset of nucleoporins recognized by the mAb414 antibody (Fig. 1 B). In some cells solely expressing GFP-hNup133CTD, the increased occurrence of cytoplasmic foci labeled by both the anti-Nup107 and mAb414 antibodies and likely corresponding to annulate lamellae may reflect a contribution of the N-terminal domain of Nup133 to some aspects of NPC biogenesis. Consistently, a membrane curvature-sensing domain (ALPS) located within the β-propeller domain of Nup133 (Drin et al., 2007) was recently demonstrated to be dispensable for post-mitotic NPC formation but required for the incorporation of the Nup107–160 complex into newly assembled NPCs during interphase (Doucet et al., 2010). Nevertheless, our study showed that the C-terminal domain of Nup133 is sufficient to assemble stable Nup107–160 complexes and that the N-terminal domain of Nup133 is largely dispensable for NPC assembly.
The N-terminal domain of hNup133 mediates the targeting of CENP-F to the NPC at the G2/M transition
We previously identified CENP-F as an interaction partner of hNup133 by yeast two-hybrid and immunoprecipitation experiments (Zuccolo et al., 2007). Further two-hybrid mapping of hNup133 domains showed that the N-terminal domain is necessary to interact with CENP-F whereas, as previously reported, the C-terminal domain mediates the interaction with hNup107 (Belgareh et al., 2001; Berke et al., 2004; Boehmer et al., 2008; Fig. 2 A). Moreover, a two-hybrid screen using the N-terminal domain of Nup133 (aa 1–500) as a bait identified the C-terminal domain of CENP-F as the only prey with very high confidence in the interaction (representing 25 out of 38 positive clones), indicating that this domain is sufficient to mediate interaction with CENP-F (Fig. 2 A).
We next asked whether the NE localization of CENP-F at the G2/M transition relies on the N-terminal domain of hNup133. Because CENP-F localizes at the NE only at the G2/M transition, we selected late G2 and prophase HeLa cells based on phospho-histone H3 staining. All control cells showed a distinct NE labeling of CENP-F (Fig. 2, B and C). In GFP-hNup133CTD cells treated with scramble siRNAs, CENP-F was localized at the NE in prophase, indicating that endogenous Nup133 in this cell line provides sufficient binding sites for CENP-F (Fig. 2 C). In contrast, knockdown of hNup133 abolished CENP-F localization to the NE (Fig. 2 B). CENP-F accumulation at the NE was largely rescued by full-length GFP-mNup133 but not by GFP-hNup133CTD (Fig. 2, B and C). Thus CENP-F localization to the NE in prophase requires the N-terminal domain of hNup133 (see Fig. 4 B).
Figure 4.
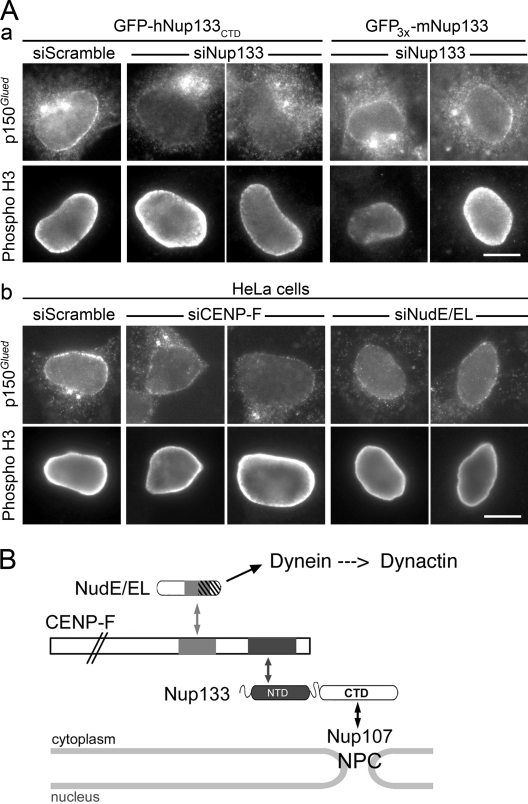
hNup133 contributes to dynactin anchoring at the NE at the G2/M transition via CENP-F and NudE/EL. (A) GFP-hNup133CTD or GFP3x-mNup133 cells (a) or wild-type HeLa cells (b) transfected with the indicated siRNA duplexes were processed for immunofluorescence using anti-p150Glued and anti–phospho-H3 antibodies. Bars, 10 µm. See also Fig. S3. (B) Schematic representation of the interaction networks connecting Nup133 to dynein/dynactin. Proteins are represented on approximate scale except for CENP-F. Boxes indicate the minimal domains involved in the interactions between Nup133 and CENP-F (black boxes; this paper and Zuccolo et al., 2007), CENP-F and NudE/EL (gray boxes), and between NudE/EL and dynein (dashed area overlapping with the CENP-F interaction domain; Liang et al., 2007; Stehman et al., 2007; Vergnolle and Taylor, 2007). Although not represented on this scheme, association of CENP-F with the pool of Nup133 localized on the nuclear side of NPCs cannot be excluded.
The effect of Nup133 depletion was specific to prophase NE localization of CENP-F because its localization at kinetochores was not affected under those conditions (Fig. 2, B and C and Fig. S1 A). Together with previous studies (Hussein and Taylor, 2002), we can conclude that the NPC targeting of CENP-F at the G2/M transition is not required for its accumulation at kinetochores, but likely has a separate function.
CENP-F anchors dynein/dynactin at the nuclear envelope via NudE/EL in prophase
Recent studies have demonstrated a role for CENP-F in indirectly recruiting dynein/dynactin to kinetochores via NudE and/or NudEL (also termed Ndel1), two highly related proteins that interact with both CENP-F and dynein (Liang et al., 2007; Stehman et al., 2007; Vergnolle and Taylor, 2007). Moreover, Ndel1, dynein, and dynactin accumulate at the NE in prophase cells (Busson et al., 1998; Salina et al., 2002; Hebbar et al., 2008). We therefore investigated whether NudE/EL could mediate the interaction of CENP-F with dynein/dynactin at the NE. As anticipated, a fraction of NudE/EL localized at the NE specifically at the G2/M transition, a stage at which NudE/EL already localizes at kinetochores in most cells (Fig. 3 and Fig. S2). Both NE and kinetochore targeting of NudE/EL were abolished by CENP-F knockdown in HeLa cells (Fig. 3 and Fig. S2). Furthermore, the NE localization of the dynactin subunit p150Glued was reduced in prophase cells expressing GFP-hNup133CTD as the only form of Nup133 (in which CENP-F is still expressed but no longer localized at the NE), as well as in CENP-F–depleted cells (Fig. 4 A and Fig. S3). Combined depletion of both NudE and NudEL (siNudE/EL) more severely reduced p150Glued localization at the NE in prophase (Fig. 4 Ab and Fig. S3). Consistent with previous studies (Guo et al., 2006), NudE/EL depletion also impaired the recruitment of p150Glued at the centrosome. In contrast, centrosomal localization of p150Glued was not significantly altered upon depletion of endogenous Nup133 in GFP-hNup133CTD cells or in CENP-F–depleted cells (Fig. 4 A and Fig. S3).
Figure 3.
CENP-F depletion impairs the NE localization of NudE/EL at the G2/M transition in HeLa cells. HeLa cells transfected with scramble or CENP-F siRNA duplexes were preextracted, fixed, and stained with anti-NudE/EL, anti-RanGAP1, and anti–phospho-H3 antibodies. Bars, 10 µm. See also Fig. S2.
Together, our results demonstrate that an interaction network starting from the N-terminal domain of Nup133 via CENP-F and NudE/EL contributes to the recruitment of dynein/dynactin represented by p150Glued to the NE in late G2/prophase HeLa cells (Fig. 4 B).
The Nup133–CENP-F–NudE/EL–dynein/dynactin network tethers the centrosomes to the nucleus in prophase
To understand the function of the Nup133-anchored dynein/dynactin network at the nuclear pore in prophase, we assayed mitotic progression by confocal time-lapse microscopy in cells where chromosomes (H2B-mCherry) and microtubule plus tips (EB3-GFP) were fluorescently labeled. The increased levels of EB3 and its accumulation around centrosomes in late G2/prophase cells (Ban et al., 2009) enabled us to identify cells about to enter mitosis.
Five different methods to disrupt the NPC–dynein/dynactin interaction, namely hNup133 depletion in the GFP-hNup133CTD cell line (n = 10 cells recorded at high resolution), transfection of siRNAs targeting CENP-F (n = 14) or NudE/EL (n = 6), or overexpression of dominant-negative dynactin inhibitors, CFP-p50/dynamitin (n = 10) and dsRed-p150cc1 (n = 9), all frequently caused centrosomes to detach from the nuclear periphery at the G2/M transition, albeit to various extents (Fig. 5, Videos 1–6, and unpublished data). Quantitative analysis of videos performed at lower resolution revealed a significant (>3 µm) detachment of centrosomes from the NE in 70% of CENP-F–depleted prophase cells, with more than 50% of the cells displaying at least one centrosome >3 µm apart from the NE for at least 5 min, a phenotype only rarely observed in cells treated with scrambled siRNAs (Fig. 5 C). In cells solely expressing GFP-hNup133CTD or depleted for CENP-F, detachment from the NE could sometimes be detected before centrosome separation (Videos 2 and 4) but became more pronounced once centrosomes started to move apart (Fig. 5, Ab–Ac and B; Videos 2–4). In contrast, unseparated centrosomes were more frequently observed far away from the NE, sometimes reaching cell borders, in NudE/EL-depleted cells (Fig. 5, Ad–d′; Video 5) or, consistent with a recent study by Splinter et al. (2010), in cells overexpressing CFP-p50/dynamitin (Fig. 5 Ae; Video 6). Analysis of cells recorded over 24 h (Fig. 5 Ad’) indicated that NudE/EL depletion induces either an earlier detachment of centrosomes from the NE or a delayed recruitment of EB3-GFP to centrosomes in prophase (a feature reminiscent of the decreased recruitment of p150Glued previously observed in these cells; Guo et al., 2006; Fig. 4 Ab and Fig. S3). Nevertheless, as observed in other conditions that disrupt the NPC–dynein/dynactin interaction (this paper and Splinter et al., 2010), centrosome movement away from the NE specifically occurred at the G2/M transition in NudE/EL-depleted cells (Fig. 5 Ad’).
Figure 5.
Interfering with Nup133-anchored dynein/dynactin impairs the tethering of centrosomes to the NE. (A) Time-lapse imaging of HeLa cells expressing EB3-GFP (green) and H2B-mCherry (red) and transfected with CENP-F (c and c′), NudE/EL siRNA duplexes (d and d′), or with a CFP-p50/dynamitin construct (e), or of cells stably expressing GFP-hNup133CTD treated with hNup133 siRNAs and subsequently transfected with plasmids encoding EB3-GFP and H2B-mCherry (b). Time (in min:sec) was set at 0:00 when centrosome splitting just became detectable. Bars, 10 µm. See also Videos 1–6. (B) Tracks representing centrosome (CTR) movements in a control and a CENP-F–depleted cell (see Aa, Ac′, and Videos 1 and 4). Trajectories before centrosome separation (red) and tracks of the separated centrosomes (blue and green) were superimposed on a schematic representation of the cell border and nuclear position (gray shading) at the beginning of the video. White and black dots indicate the positions of centrosomes at the beginning and at the end of the videos, respectively. (C) Analysis of the centrosome–NE distance over time in HeLa cells expressing EB3-GFP and H2B-mCherry and treated with scramble or CENP-F siRNA duplexes. For each cell entering mitosis, the maximum distance between the centrosomes and the NE reached during the G2/M transition was plotted over the time centrosomes spent >3 µm away from the NE. Each dot represents a single cell. The number of cell quantified is indicated.
We confirmed and further quantitated these phenotypes by measuring the centrosome–NE distance in fixed cells at various stages of the cell cycle (Fig. 6). In control cells centrosomes were rarely >2 µm away from the nuclear periphery in prophase. The mean centrosome–NE distance was significantly increased in prophase cells solely expressing GFP-hNup133CTD or depleted for CENP-F (Fig. 6, Aa–b and B). Consistent with our observations on live cells, this phenotype was even more pronounced upon depletion of NudE/EL (Fig. 6, Ab and B) or upon overexpression of CFP-p50 or DsRed-p150cc1 (Fig. 6, Ac and B; see also Splinter et al., 2010). In contrast, centrosome position was indistinguishable in S and early G2 between control cells and cells depleted for either CENP-F or NudE/EL (Fig. 6, Ab and B). Together, our data show that Nup133-mediated NE recruitment of dynein/dynactin is required to anchor centrosomes to the NE at the G2/M transition.
Figure 6.
The Nup133-anchored network tethers centrosomes to the NPCs specifically in prophase. (A) GFP3x-mNup133 or GFP-hNup133CTD cells treated for 3 d with scramble or hNup133 siRNAs (a), HeLa cells treated for 2 d with scramble, CENP-F, or NudE/EL siRNA duplexes (b), or HeLa cells transfected with a GFP-p50/dynamitin or a DsRec-p150cc1 construct (c) were processed for immunofluorescence using anti-pericentrin and anti–phospho-H3 antibodies. In b, cells were incubated before fixation with 40 µM BrdU for 3 h and anti-BrdU antibodies were further used. Bars, 10 µm. (B) Distances between centrosomes and the NE were measured on cells processed as in A. Prophase cells were identified by phospho-H3 staining and S/early G2 cells as BrdU-positive cells after a 3-h pulse with BrdU (G1 cells were not analyzed because centrioles were reported to be very mobile at that stage of the cell cycle; Piel et al., 2000). Distances are represented as box-plots using KaleidaGraph (see Materials and methods). The black and red bars indicate the median and mean values, respectively. The total number of cells quantified is indicated (n). ***, P < 10−5; **, P < 10−3; ns, P > 0.1 obtained using the Student’s t test.
Centrosome detachment from the NE in CENP-F or NudE/EL-depleted cells requires microtubules and Eg5 activity
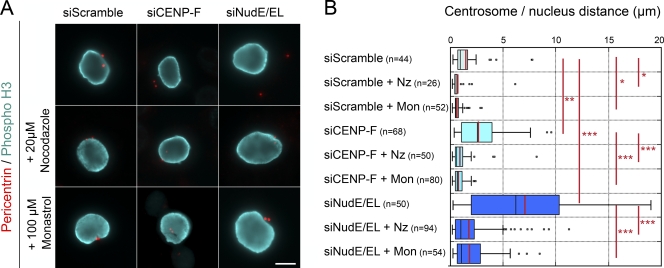
Tracking centrosomes in cells in which the Nup133-anchored dynein/dynactin pathway is impaired revealed that centrosomes moved in a directional manner with velocities around 0.4 µm/min, although peak velocities up to 0.8 µm/min were observed (Fig. 5 B), consistent with microtubule motor-mediated movements. In addition, centrosome movement appeared to be most pronounced after centrosome separation (Fig. 5, A and B), suggesting the implication of Eg5, a plus end–directed kinesin involved in centrosome separation (Tanenbaum et al., 2008; Splinter et al., 2010). Live-cell analysis revealed that acute depolymerization of microtubules by nocodazole at the onset of prophase microtubule nucleation as well as acute inhibition of Eg5 with the specific inhibitor monastrol (Mayer et al., 1999) largely abolished centrosome movement in CENP-F–depleted cells (Fig. S4). These results were confirmed quantitatively in fixed CENP-F–depleted prophase cells (Fig. 7, A and B). This quantitative analysis also revealed that centrosome detachment was strongly reduced, albeit not completely abolished, upon nocodazole or monastrol treatment of NudE/EL-depleted prophase cells (Fig. 7, A and B). Together, our data show that microtubules and Eg5 activity move centrosomes away from the nucleus in prophase cells if they are not tethered by the Nup133-anchored dynein/dynactin network.
Figure 7.
Centrosome movement away from the nuclear periphery requires microtubules and Eg5 activity. (A) HeLa cells transfected with scramble, CENP-F, or NudE/EL siRNA duplexes were either fixed (top row) or incubated with 20 µM nocodazole for 30 min or with 100 µM monastrol for 1 h before fixation. They were then stained with anti-pericentrin and anti–Phospho-H3 antibodies. Note that under those conditions, all phospho-H3–positive cells had entered prophase in the absence of microtubules or before Eg5 activation. All images arise from a single experimental dataset, although they were captured at different times using slightly different acquisition settings. Either a unique plane or maximum intensity projections of stacks are presented, as needed, depending on the locations of the centrosomes relative to the focal plane. Bar, 10 µm. (B) Distances between centrosomes and the NE, measured in phospho-H3–positive cells processed as above, are represented as box-plots using KaleidaGraph (see Materials and methods). The black and red bars indicate the median and mean values, respectively. The total number of cells quantified is indicated (n). ***, P < 10−5; **, P < 10−3; *, P < 0.05 obtained using the Student’s t test.
The Nup133–CENP-F–NudE/EL network tethers centrosomes to the NE in G2/prophase cells without interfering with the RanBP2–BICD2-dependent pathway
Because two distinct NPC-anchored networks (Nup133–CENP-F–NudE/EL, this study; and RanBP2-BICD2, Splinter et al., [2010]) contribute to dynein/dynactin recruitment at the NE and to centrosome positioning in G2/prophase cells, we next investigated whether there could be a cross talk between these two pathways. Unlike anticipated, BICD2 depletion in HeLa cells did not induce a significant detachment of the centrosomes from the NE (Fig. S5). Although this might reflect the existence of distinct modes of centrosome tethering among various cell lines, we could not exclude a nonoptimal depletion of BICD2 in HeLa cells (Fig. S5 B, see also the following paragraphs and Discussion). We thus decided to perform these studies in the U2OS cell line, used by Splinter et al. (2010), which has a more robust centrosome position adjacent to the nucleus (compare control cells in Fig. 6 B with those in Fig. 8 A) and sustains two successive rounds of siRNA depletion.
Figure 8.
Relationship between the Nup133–CENP-F–NudE/EL and RanBP2–BICD2 pathways in centrosome tethering to the NE in U2OS cells. (A) Distances between centrosomes and nuclear periphery, measured in phospho-H3–positive U2OS cells treated for 3 d with scramble, CENP-F, BICD2, a combination of CENP-F and BICD2, or NudE/EL siRNA duplexes. Distances are represented as box-plots using KaleidaGraph (see Materials and methods). The black and red bars indicate the median and mean values, respectively. The total number of cells quantified is indicated (n). ***, P < 10−5; **, P < 10−3; ns, P > 0.1 obtained using the Student’s t test. (B) Extracts from U2OS cells treated with the indicated siRNA duplexes were analyzed by Western blot using anti-BICD2 antibodies. Decreasing amounts of the reference sample (scramble siRNA) were loaded (1-, 0.5-, and 0.25-fold equivalent) and anti–γ-tubulin was used as loading control. (C and D) U2OS cells transfected with the indicated siRNA duplexes were preextracted, fixed, and stained with anti–CENP-F and anti-RanGAP1 (C) or anti-BICD2 (D), along with anti–phospho-H3 antibodies and DAPI. Bars, 10 µm.
Analysis of prophase U2OS cells revealed that CENP-F depletion leads to a mild detachment of the centrosome from the NE, similar to the one recorded in HeLa cells (Fig. 8 A). As already observed in HeLa cells (Figs. 6 and 7), NudE/EL depletion led to a more pronounced phenotype as compared with CENP-F or BICD2 depletion (Fig. 8 A). In this cell line, however, NudE/EL depletion phenocopied the phenotypes observed upon dynein heavy chain depletion or overexpression of CFP-p50 or DsRed-p150cc1 (Splinter et al., 2010 and Fig. 6 B). Somehow unexpectedly, the combined use of siRNAs targeting CENP-F and BICD2 did not lead to an additive or synergistic effect (Fig. 8 A). Although various cellular mechanisms may explain the lack of additive effect (see Discussion), we cannot formally rule out interference from multiple siRNA treatments done simultaneously as revealed by the slightly less efficient depletion of BICD2 under these conditions (Fig. 8, B–D). Immunofluorescence analyses further revealed that perturbation of the Nup133-dependent pathway does not significantly affect the localization of RanGAP1 whose NPC localization relies on RanBP2/Nup358 (Saitoh et al., 1997; Fig. 8 C, see also Fig. 2, B and C, Fig. 3 A, and Fig. S3). In addition, neither CENP-F nor NudE/EL depletion altered the NE localization of BICD2 (Fig. 8 D and Fig. S5). This indicates that the Nup133-anchored network does not interfere with the NE recruitment of RanBP2 or BICD2. Conversely, BICD2 depletion did not affect the recruitment of CENP-F at the NE (Fig. 8 C).
Together, these data reveal the lack of cross talk between the Nup133–CENP-F–NudE/EL and RanBP2–BICD2 pathways for tethering of centrosomes to the NE at the G2/M transition.
Centrosome tethering to the nucleus at the G2/M transition is required for timely establishment of a properly positioned mitotic spindle
CENP-F or NudE/EL RNAi, as well as overexpression of dynactin subunits, all impair kinetochore assembly and function. In contrast, as far as we can judge, the lack of the Nup133 N-terminal domain did not interfere with kinetochore assembly (Fig. S1). The GFP-hNup133CTD cell line thus provided a unique tool to specifically assess the consequences of centrosome detachment on spindle assembly and chromosome segregation. We assayed these functions in GFP-hNup133CTD cells with fluorescently labeled centromeres (GFP-CENP-A) and microtubules (mCherry-MAP4; Fig. 9 Aa and Video 7).
Figure 9.
Centrosome disconnection from the NE causes transiently aberrant spindle structures. (A) Cells stably expressing GFP-hNup133CTD and GFP-CENP-A (green) were transfected with a plasmid encoding the microtubule-binding protein mCherry-MAP4 (red) and with scramble (a) or hNup133 (b and c) siRNAs. Cells were imaged from prophase on. Top panels: mCherry-MAP4 signal; bottom panels: overlay of the mCherry-MAP4 and GFP-CENP-A + GFP-hNup133CTD signals (note that the GFP-hNup133CTD signal is hardly detectable over the GFP-CENP-A signal). Time is in min:sec. Times of NEBD, metaphase plate formation, and metaphase/anaphase transition are indicated. Bar, 10 µm. See also Videos 7–9. (B) Time spent from chromosome condensation to metaphase/anaphase transition for cells stably expressing GFP-hNup133CTD and GFP-CENP-A treated with scramble or hNup133 siRNA duplexes as indicated. Each dot represents a single cell, and the dashed line represents the average mitotic duration. Cells that display major mitotic defects (red dots) or minor chromosome segregation defects (one mis-segregated or lagging chromosome; black dots) are indicated. The number of cell quantified, the average mitotic duration, and standard deviation are indicated. ns, the difference between control and Nup133-depleted cells was not statistically significant as determined using the Student’s t test. (C) Analysis of centrosome localization (arrows) in prometaphase and metaphase U2OS cells treated with scramble or NudE/EL siRNA duplexes. Cells were fixed and then stained with anti-pericentrin (red), anti–phospho-H3 (green), and RanGAP1 (gray) antibodies. RanGAP1 that localizes at NPCs in interphase and on the mitotic spindle in mitosis was used to simultaneously assess NPC disassembly and spindle formation. Representative cells at various stages of prometaphase and metaphase are presented. The percentage of mitotic cells displaying the indicated phenotypes is indicated. Bar, 10 µm.
Time-lapse analysis of cells lacking the N-terminal domain of Nup133 revealed that spindle assembly initiated abnormally to various degrees depending on the position of centrosomes relative to the nucleus before NEBD (Fig. 9 A and Videos 8 and 9; see also Fig. 5 A and Video 2). When both centrosomes were on the same side and at similar distance from the nucleus, a horseshoe-shaped spindle-like structure was frequently observed (5 out of 18 cells; Fig. 9 Ab and Video 8; see also Fig. 5 Ab and Video 2). When one centrosome was closer to the NE than the other, microtubules nucleated by the proximal centrosome captured most chromosomes, leading to a transient formation of a monopolar spindle and sometimes an “isolated centrosome” (6 cells; Fig. 9 Ac and Video 9). More rarely, the two centrosomes were positioned on opposite sides of the nucleus, from where they effectively formed a bipolar spindle, despite the increased distance from the nuclear periphery at NEBD onset (1 cell; unpublished data). Despite these early defects, a bipolar spindle eventually formed after NEBD, and chromosomes aligned on the metaphase plate and were correctly segregated to the two daughter cells. Time-lapse analysis of a broader number of Nup133CTD cells stably expressing GFP-CENP-A and treated with either scrambled or Nup133 siRNA duplexes did not reveal any significant mitotic delay or chromosome segregation defect in Nup133-depleted cells as compared with control cells (Fig. 9 B). These data indicate that, unlike recently reported for cells with multipolar spindle poles (Ganem et al., 2009; Silkworth et al., 2009), the mild and transient detachment of centrosomes recorded in cells lacking the N-terminal domain of Nup133 does not, per se, lead to major alterations of cell cycle progression and chromosome missegregation (see Discussion).
Immunofluorescence analysis of mitotic NudE/EL-depleted U2OS cells revealed similar albeit more drastic mitotic defects: in most prometaphase cells, at least one centrosome was away from the chromosome mass, and major alteration of the mitotic spindles was also apparent upon RanGAP1 staining (Fig. 9 C). Nevertheless, 50% of the NudE/EL-depleted prometaphase/metaphase cells displayed a bipolar spindle (Fig. 9 C), indicating that even major alteration of centrosome positioning can be subsequently compensated by additional mechanisms to ensure bipolar spindle formation (see Discussion). Because of the additional contribution of NudE/EL to kinetochore functions (Liang et al., 2007; Stehman et al., 2007; Vergnolle and Taylor, 2007), the specific contribution of centrosome positioning to chromosome segregation could not be analyzed in these cells. However, the altered centrosome positioning in mitosis likely contributes, along with the reported kinetochore alterations, to the defects in metaphase chromosome alignment previously recorded upon NudE depletion (Vergnolle and Taylor, 2007).
Discussion
Nup133 is the base of a molecular network that tethers dynein/dynactin to the NE at the G2/M transition in mammalian cells
The Nup107–160 complex had previously been implicated at various stages of the cell cycle, notably spindle assembly, proper kinetochore function, cytokinesis, NPC reassembly at mitotic exit, and de novo NPC assembly in interphase (Doucet et al., 2010; Wozniak et al., 2010). This study uncovers an additional function, carried by a structurally defined domain of the Nup133 subunit, at the G2/M transition.
The CENP-F–NudE/EL–dynein/dynactin network, defined using biochemical and two-hybrid approaches had so far been shown to function at kinetochores in prometaphase (Liang et al., 2007; Stehman et al., 2007; Vergnolle and Taylor, 2007). We show here that the same molecular network is also used at the nuclear pore to tether centrosomes to the nucleus just before NEBD. The nucleoporin Nup133—structurally embedded in the NPC via its C-terminal α-solenoid domain (Boehmer et al., 2008)—anchors the C-terminal domain of CENP-F (residues 2644–3065; Zuccolo et al., 2007) via its N-terminal domain. CENP-F in turn uses a second, more central domain (residues 2122–2447; Vergnolle and Taylor, 2007) to bind to NudE/EL (Fig. 4 B). These nonoverlapping binding domains suggest that there is a direct molecular link from the NPC to NudE/EL, and in turn dynein/dynactin using bifunctional interactors. What could regulate the assembly of this link specifically at the onset of mitosis? Interestingly, both the N-terminal domain of hNup133 and the interacting C-terminal domain of CENP-F are specifically phosphorylated in mitosis (Zhu et al., 1995; Nousiainen et al., 2006; Glavy et al., 2007). It is therefore likely that, as recently reported for Nudel1 or p150Glued (Hebbar et al., 2008; Li et al., 2010), mitotic phosphorylation of Nup133, CENP-F, or both, temporally regulates the recruitment of CENP-F, and in turn NudE/EL and dynein/dynactin to the NPC at the G2/M transition.
Metazoans have evolved multiple, distinct cell cycle–specific pathways to tether the centrosome to the nucleus
Among the multiple functions of dynein and dynactin (Vallee et al., 2004), centrosome tethering to the NE appears to be an evolutionarily conserved function that relies, at least in Caenorhabditis elegans and mammalian cells, on dynein localization at the NE (Gönczy et al., 1999; Robinson et al., 1999; Malone et al., 2003; Zhang et al., 2009; Splinter et al., 2010; and this paper). The molecular details of this regulation are, however, different. In C. elegans early embryos, ZYG12, a divergent KASH domain protein anchored in the outer nuclear membrane by the inner nuclear membrane protein SUN-1, is responsible for dynein localization and centrosome tethering to the NE (Malone et al., 2003). However, ZYG12 is not present in C. elegans hyp7 cells in which a distinct KASH domain protein, UNC-83, contributes to nuclear migration by recruiting dynein to the NE through two distinct adaptor complexes (Fridolfsson et al., 2010). Similarly, mammals have apparently evolved distinct pathways to link dynein and/or centrosomes to the nucleus. In human dermal fibroblasts, constitutive centrosome–NE association was proposed to rely on emerin and microtubules (Salpingidou et al., 2007); however, this pathway does not significantly contribute to centrosome tethering in HeLa cells (unpublished data). More recently, SUN1/2 and the KASH domain proteins Syne/Nesprin-1/2 were shown to mediate the coupling between the nucleus and the centrosome in primary cultured neurons or glial cells (Zhang et al., 2009). Our data and the recent study from Splinter et al. (2010) reveal the existence of novel pathways for dynein/dynactin recruitment and centrosome positioning in G2/prophase cells that rely on nucleoporins rather than on SUN/KASH pairs. RanBP2/Nup358, a nucleoporin that localizes to the cytoplasmic side of the NPC, was demonstrated to contribute to this process by recruiting BICD2 to NPCs from early G2 on (Splinter et al., 2010). We show here that Nup133 anchors CENP-F that in turn recruits NudE/EL, recently shown to contribute to dynein localization at the NE in prophase cells (Hebbar et al., 2008). Because kinetochores were frequently positive for CENP-F or NudE/EL in cells that also exhibited NE localization for these proteins (Fig. 2, B and C, Fig. 3, and Fig. S2), we anticipate that this network might be recruited to NPCs later in G2 as compared with BICD2. At that stage, the extremely stable association of Nup133 with NPCs (Rabut et al., 2004) may be required to efficiently counteract the activity of Eg5 that pushes the two centrosomes away from the nucleus and apart from each other.
Importantly, perturbation of the Nup133-dependent pathway did not affect the NPC localization of BICD2 and conversely, BICD2 depletion did not affect CENP-F recruitment to the NE. This indicates that, as also observed at kinetochores (Mao et al., 2010) and at the NE in C. elegans hyp7 cells (Fridolfsson et al., 2010), two distinct pathways independently recruit dynein/dynactin to the NPC in G2/prophase cells. These two distinct NPC-anchored pathways are not redundant, as inhibition of either results in defects in centrosome–nucleus interactions. Although centrosome displacement was less pronounced for either case than for dynein/dynactin inhibition, combined RNAi depletion of CENP-F and BICD2 did not lead to a more pronounced phenotype as compared with each of the single depletions. Although this might possibly be attributed to the slightly less efficient depletion of BICD2 under these conditions (Fig. 8, B and C), it may also reflect the presence of counteracting forces mediated by kinesin-1 (which also interacts with BICD2; Splinter et al., 2010). Indeed, Splinter et al. (2010) previously reported that combined depletion of BICD2 and dynein leads to a less severe defect on centrosome positioning as compared with dynein depletion alone.
The specific recruitment of CENP-F and NudE/EL to the NE in prophase and the consequences of their depletion are clearly consistent with a role for these proteins at the NE. However, NudE/EL depletion leads to a more pronounced phenotype as compared with CENP-F–depleted cells, notably in U2OS cells in which it phenocopies dynein/dynactin inactivation (Figs. 5–8). These data suggest the participation of NudE/EL in another dynein/dynactin–dependent but CENP-F–independent process. Possible hypotheses include: (1) the destabilization of BICD2–dynein/dynactin interaction, (2) the contribution of NudE/EL to centrosomal functions, such as MT nucleation or anchoring (Guo et al., 2006; see also Fig. 4 B), (3) the implication in this process of an additional kinesin (in addition to kinesin-1 and Eg5; see for instance Tsai et al., 2010), or (4) alteration of the actin cytoskeleton. Indeed, Whitehead et al. (1996) previously reported movements of Eg5-immunoreactive centrosomes that frequently impinged on the plasma membrane upon cytochalasin-induced disassembly of actin fibers in prophase HeLa cells.
Finally, it is noteworthy that in C. elegans hyp7 cells a unique protein (the KASH protein UNC-83) anchors BICD1 and NUD2 (respectively homologous to mammalian BICD2 and NudE/EL) as well as kinesin-1 at the NE to coordinate nuclear migration (Fridolfsson et al., 2010). In human cells, the recruitment of these different actors via a complex multiprotein assembly (the NPC) may allow a more robust or more regulatable interaction between the nucleus and the cytoskeleton in prophase.
Tethering centrosomes to the nuclear surface cooperates with additional spindle positioning and assembly mechanisms in early stages of bipolar spindle assembly
The specific abrogation of NPC–centrosome tethering in HeLa cells transiently perturbed spindle positioning and assembly. However, most mispositioned or malformed spindles eventually recovered, apparently driven by additional mechanisms that also contribute to bipolar spindle formation. At NEBD, and concurrent with cell rounding, detached centrosomes moved back toward the chromosomes (Fig. 5 B). It was shown in Drosophila that cell rounding, driven by a mitosis-specific increase in cortical stiffness, also affects spindle positioning (Carreno et al., 2008; Kunda et al., 2008), and we therefore suggest that cell rounding contributes to rescue detached centrosomes by shortening the distance from cortex to chromosomes. In addition, NEBD releases mitotic kinases, spindle assembly factors, and RanGTP into the cytoplasm. This promotes microtubule growth and stabilizes K-fibers (Goodman and Zheng, 2006; Silverman-Gavrila and Wilde, 2006). The resulting microtubule-dependent forces are thus also likely to contribute to centrosome repositioning close to chromosomes. Aberrant spindles, including monopolars, usually evolved into normal bipolar structures, in a process likely involving microtubule self-assembly mechanisms similar to that used in cells recovering from monastrol treatment (Whitehead et al., 1996; Khodjakov et al., 2003).
Although NPC–centrosome tethering is therefore clearly not the only mechanism for early stages of spindle positioning and assembly in comparatively small HeLa cells that undergo strong cell rounding at mitosis entry, this mechanism may become crucial in larger cells such as oocytes or nonrounding cells in tissues, as well as under pathological situations when other pathways are impaired. In such cells, loss of NPC–centrosome tethering would be expected to lead to chromosome missegregation, a major source of aneuploidy. Aneuploidy has been linked to cancer progression in mammals (Chandhok and Pellman, 2009) and indeed mutations in the Nup133 β-propeller domain were identified in a global search for human breast cancers genes (Sjöblom et al., 2006). In the future, it will be important to test the hypothesis that mitotic defects caused by mutations within the Nup133 β-propeller domain contribute to tumorigenesis.
Materials and methods
Yeast two-hybrid
To construct the pB29-hNup133NTD plasmid, the N-terminal domain of hNup133 (aa 1–500) was cloned in an N-bait-LexA-C fusion vector optimized by Hybrigenics. The pB29-hNup133NTD construct was used to screen a random primed human breast tumor epithelial cell cDNA library (RP1) cloned into the pP6 plasmid using a high-throughput proprietary yeast two-hybrid based technology (Hybrigenics).
To investigate pairwise interactions by yeast two-hybrid, hNup133 (aa 12–1156; Belgareh et al., 2001) and hNup133CTD (aa 466–1156) cloned into the pLex12 vector and the pB29-hNup133NTD plasmid were transformed in L40ΔGal4 strain, while hNup107 (aa 784–924) and CENP-F (aa 2644–3065; Zuccolo et al., 2007) fused in frame with GAL4 AD in pP6 vectors (obtained from Hybrigenics) were transformed into the Y187 strain as described previously (Belgareh et al., 2001). Bait and prey strains were mated in rich medium, and diploids were grown on minimum medium lacking leucine, tryptophan, and histidine and containing 1 mM 3-amino-1,2,4-triazole (−LWH + 1 mM 3AT).
Cell culture and plasmid transfections
HeLa and U2OS cells were grown at 37°C in DME (Life Technologies) supplemented with 10% fetal calf serum, 1% l-glutamine, 100 µg/ml streptomycin, and 100 U/ml penicillin. Plasmid transfections were performed using Lipofectamine 2000 (Invitrogen). To establish HeLa stable cell lines, individual clones were isolated by G418 (0.5 mg/ml) or Hygromycin (0.4 mg/ml) selection.
The following plasmids were used in this study: The pEGFP-hNup133CTD plasmid (encoding aa 545–1156 of hNup133) was obtained by deletion of a SacI fragment from the pEGFP-hNup133 vector (Belgareh et al., 2001). To generate pEGFP3x-mNup133, an AflIII (blunt-ended)–NotI fragment was purified from the IMAGE Consortium (LLNL) cDNA clone (IMAGE: 1180761), obtained from the Resource Center of the German Human Genome Project, and subcloned in the pEGFP3x-C3 vector digested with SalI (blunt-ended) and NotI. The GFP-hNup133CTD and GFP3x-mNup133 fusions were subsequently inserted into the pIRES-neo vector (Takara Bio Inc.).
The pBOS-H2B-GFP-IRES-neo was constructed by inserting the NotI–XhoI (blunt-ended) fragment of pIRES-neo (Takara Bio Inc.) into the pBOS-H2B-GFP vector (BD) digested by NotI and AatII (blunt-ended). To generate the pBOS-H2B-mCherry-IRES-neo plasmid, the coding sequence of mCherry (Shaner et al., 2004) was amplified by PCR adding BamHI sites both at the 3′ and 5′ ends and inserted in pBOS-H2B-GFP-IRES-neo vector digested with BamHI. The pEGFP-CENP-A plasmid, in which full-length human CENP-A HA-tagged at its 3′ end is inserted into SmaI–XbaI sites of pEGFP-C1, was generously provided by K.F. Sullivan (University of California, Santa Cruz, Santa Cruz, CA). The EB3-GFP construct (pEGFP-N3-EB3; Stepanova et al., 2003) was a kind gift from N. Galjart (Erasmus University, Rotterdam, The Netherlands). The H2B-mCherry and GFP-CENP-A constructs were subsequently subcloned into the pIRES-hygro vector (Takara Bio Inc.).
MAP4 tagged with mCherry at its N terminus (Keppler and Ellenberg, 2009) was constructed by A. Keppler (EMBL, Heidelberg, Germany), the pEGFP-centrin1 plasmid (Piel et al., 2000) was provided by M. Piel and M. Bornens (Institut Curie, Paris, France), the DsRed p150-cc1 construct (DsRed-p150217–548; Quintyne and Schroer, 2002) was a kind gift of T. Schroer (Johns Hopkins University, Baltimrore, MD), and CFP-p50/dynactin was a kind gift from A. Popov and E. Karsenti (EMBL, Heidelberg, Germany).
Stable cell lines generated for this study are: GFP-hNup133CTD, GFP3x-mNup133FL, EB3-GFP + H2B-mCherry, GFP-centrin1, and GFP-hNup133CTD + mCherry-CENP-A. The EB3-GFP cell line was generously provided by Lucia Sironi (EMBL, Heidelberg, Germany).
siRNA transfections
The following siRNA duplexes (purchased from Eurogentec) were used in this study: scramble siRNA sequence (5′-CTGTGCAAGCCGTTGTGTA-3′), hNup133 (5′-AAGTCGATGACCAGCTGACCATT-3′; Walther et al., 2003), CENP-F (5′-CAGAATCTTAGTAGTCAAGTA-3′), NudE (5′-GGACCCAGCTCAAGTTTAATT-3′ and 5′-GGAAAGATCTGGCGATGACTT-3′; Vergnolle and Taylor, 2007), NudEL (5′-GCTAGGATATCAGCACTAATT-3′ and 5′-GGACCAAGCATCACGAAAATT-3′; Vergnolle and Taylor, 2007), and BICD2 (5′-GGUGGACUAUGAGGCUAUC-3′; BICD2#2; Splinter et al., 2010). siRNA duplexes were transfected in HeLa cells using HiPerfect reagent (QIAGEN) with 10 nM siRNA. siRNA-treated HeLa cells were analyzed 2 d after siRNA transfection, except Nup133-depleted cells, for which efficient depletion was achieved after 3–4 d of siRNA treatment (Walther et al., 2003). When combined with plasmid transfections, siRNA transfections were performed 12–24 h before plasmid transfections. U2OS cells were transfected as described previously (Splinter et al., 2010) using 20 nM siRNA and two successive rounds of siRNA depletion (during plating and 1 d after transfection) and analyzed 3 d after plating. For silencing both NudE and NudEL, a mixture of each of the four siRNA duplexes targeting these proteins was used.
Western blot analysis
For Western blot analysis, whole-cell lysates resuspended in SDS-PAGE sample buffer were separated on 4–12% SDS-PAGE gels and transferred to nitrocellulose filters. The resulting blots were saturated with TBS, 0.1% Tween, and 5% dried milk and probed with affinity-purified polyclonal rabbit anti-hNup133 (1:1,000; Belgareh et al., 2001) and polyclonal rabbit anti-hNup107 (1:2,000; Belgareh et al., 2001), polyclonal rabbit anti-BICD2 (1:500; Sigma-Aldrich), and monoclonal mouse anti–γ-tubulin (1:8,000; Abcam). Antibodies were detected with anti–rabbit or anti–mouse HRP (Jackson ImmunoResearch Laboratories) detected by enhanced chemiluminescence.
Immunofluorescence microscopy
Antibodies used in this study were: affinity-purified polyclonal rabbit anti-hNup107 (Belgareh et al., 2001), anti–phospho-H3 Ser10 (1:1,000; Millipore), IgG cut anti-NudE/EL (Stehman et al., 2007; 1:100), anti-BICD2 C-terminal (#2297; 1:250; generously provided by A. Akhmanova, Erasmus Medical Center, Rotterdam, The Netherlands), anti-pericentrin (1:500, Abcam), anti-CENP-E (Chan et al., 1998; generously provided by T. Yen, Fox Chase Cancer Center, Philadelphia, PA), monoclonal mouse mAb414 (1:400; Covance), anti-CENP-F (clone 11; 1:400; BD), anti-phospho-H3 Ser10 (1:2,000; Abcam), anti-p150Glued (1:250; BD), polyclonal goat anti-RanGAP1 (N-19; 1:200; Santa Cruz Biotechnology, Inc.), and polyclonal rat anti-BrdU (1:200; Harlan Sera-Lab).
Secondary antibodies were from Jackson ImmunoResearch Laboratories, Inc. (cy2, cy3, cy5, or AMCA-conjugated AffiPure IgGs) or Invitrogen (Alexa 488–conjugated IgGs), and were used at 1:500 to 1:1,000 dilutions.
For most immunofluorescence experiments, cells were washed in a solution containing 1% PFA + 0.01% glutaraldheyde, preextracted for 2 min in 0.5% Triton X-100 and then fixed for 20 min in 3% PFA. For BrdU incorporation, cells were incubated for 3 h with 40 µM BrdU and washed twice with phosphate-buffered saline before being processed for immunofluorescence using anti–phospho-H3 and anti-pericentrin antibodies. Cells were then postfixed for 15 min with 2% PFA, treated for 10 min with 4 N HCl, and immunostained with an anti-BrdU antibody. S and G2 cells were detected by BrdU staining and prophase cells by phospho-H3 signal.
Cells from Fig. 1 B were fixed for 20 min in 3% PFA followed by Triton X-100 permeabilization. For NudE/EL immunostaining (Fig. 3 and Fig. S2), cells were preextracted in 0.5% Triton X-100 in PHEM for 1 min and fixed in 4% PFA in PHEM for 20 min, followed by ice-cold methanol for 5 min. Widefield microscopic images were acquired using an upright motorized microscope (DMRA2; Leica). Acquisitions were performed using an oil immersion objective (100x PL APO HCX, 1.4 NA) and a high-sensitive cooled interlined CCD camera (CoolSnap HQ; Roper Industries). Rapid and precise Z-positioning was accomplished by a piezoelectric motor (LVDT; Physik Instrumente) mounted underneath the objective lens. Image stacks were acquired without camera binning, with a plane spacing of 0.2 µm. A unique plane is shown except for Figs. 6 A, 7 A (Nocodazole-treated cells), and 9 C, in which maximum intensity projections performed using MetaMorph software (Universal Imaging Corp.) are presented. For Fig. 3 and Fig. S2, confocal images were acquired with a scanning confocal microscope (LSM 510 NLO; Carl Zeiss, Inc.) using a 40x/1.3 Plan-Neofluar objective.
Live-cell imaging
For live-cell imaging, HeLa cells were seeded onto 35-mm glass dishes (Iwaki) or in LabTek chambers (Thermo Fisher Scientific) in Leibovitz’s CO2-independent medium without phenol red and imaged at 37°C. Prophase cells randomly chosen based on their typical EB3-GFP or mCherry-MAP4 staining were imaged every 30 s for up to 1 h (EB3-GFP/H2B-mCherry) or every 2–4 min for up to 3 h (mCherry-MAP4/GFP-CENP-A) with either a spinning disk confocal microscope (UltraVIEW ERS; PerkinElmer) mounted on an Axiovert200M body (Carl Zeiss, Inc.) and equipped with a 63x/1.4 NA objective (Carl Zeiss, Inc.) or a CSU22 spinning head (Yokagawa) mounted on a TE2000 microscope (Nikon) equipped with a 100x/1.4 NA objective and a Coolsnap HQ2 camera (Roper Industries). Maximum intensity projections of 15 Z-stack planes at a 2-µm interval for each time point are presented. Nocodazole (20 µM final concentration; Sigma-Aldrich) and monastrol (100 µM final concentration; Sigma-Aldrich) were added between two image acquisitions. For Fig. 5 Ad′ and C, time-lapse sequences were taken every 2.5 min for 48 h using a spinning disk microscope (PerkinElmer) as above with a 40x 1.30 NA oil objective (Carl Zeiss, Inc.). For Fig. 9 B, time-lapse sequences (20 Z-stack planes at a 1-µm interval every 4 min for 9 h) were acquired using a CSU22 spinning head (Yokagawa) mounted on a DMI6000 microscope (Leica) equipped with a Leica 100x/1.4 NA objective and a QuantEM 512SC camera (Photometrics). Microscopes were controlled either by MetaMorph software (Universal Imaging Corp.) or with UltraVIEW ERS software (PerkinElmer).
Image analysis
The mean distances between the NE and each of the two centrosomes were calculated on fixed cells using MetaMorph software in 2D on single planes or on maximum intensity projections of ∼20 Z-stacks at a 0.2-µm interval. Box-plots were generated using KaleidaGraph (Synergy Software): each box encloses 50% of the obtained distances centered on the median value. The bars extending from the top and bottom of each box mark the minimum and maximum values within the dataset falling within an acceptable range. Values falling outside of this range are displayed as an individual point. Unpaired Student’s t tests were used to evaluate differences in centrosome–NE distances between the various conditions.
Centrosome tracking on videos was performed with an ImageJ macro (http://weeman.inf.ethz.ch/ParticleTracker). For Fig. 5 C, the centrosome to NE distance was then calculated with a Matlab macro, taking as input the NE outline created with an ImageJ macro based on the H2B-mCherry signal. Quantifications and tracks were analyzed with Excel (Microsoft). Figures were assembled with Photoshop CS (Adobe).
Online supplemental material
Fig. S1 shows the kinetochore localization of Hec1, CENP-F, p150Glued, and Mad1 in mitotic GFP3x-mNup133 and GFP-hNup133CTD cells depleted for Nup133. Fig. S2 shows NudE/EL levels and localization in prophase HeLa cells treated with scramble, CENP-F, or NudE/EL siRNA duplexes. Fig. S3 shows the localization of p150Glued and RanGAP1 in prophase HeLa cells treated with scramble, CENP-F, or NudE/EL siRNA duplexes. Fig. S4 shows centrosome movements in CENP-F–depleted HeLa cells treated with nocodazole or monastrol. Fig. S5 shows the localization and expression of BICD2 and the centrosome to NE distances in HeLa cells treated with scramble, BICD2, or CENP-F siRNA duplexes. Videos 1–6 and 7–9 show the videos of cells presented in Fig. 5 A and Fig. 9 A, respectively. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201007118/DC1.
Acknowledgments
We are grateful to M. Gillard, A. Alves, and S. Chilaka for help with plasmid constructs and two-hybrid assays; to R. Tsien, K.F. Sullivan, N. Galjart, A. Popov, E. Karsenti, M. Piel, M. Bornens, T. Schroer, A. Keppler, M. Gillard, L. Sironi, T. Yen, V. Cordes, and A. Akhmanova for generously providing constructs, cell lines, or antibodies; to X. Baudin and the staffs of the Institut Jacques Monod (ImagoSeine) and Institut Curie Imaging Platforms for technical support; to J. Pines for suggesting monastrol experiments; to A. Akhmanova and M. Tanenbaum for advices during the revision process; and to M. Bornens, A. Paoletti, R. Karess, and members of our laboratories for valuable comments and critical reading of the manuscript.
This research was supported by the Centre National de la Recherche Scientifique, the Institut Curie, the Ligue Nationale Contre le Cancer (to V. Doye, équipe labelisée LIGUE), and the Agence Nationale de la recherche (ANR 07-BLAN-0063-01 to V. Doye). M. Zuccolo was supported by fellowships from the Ministère délégué à la Recherche et aux nouvelles Technologies and Boehringer Ingelheim Fonds.
Footnotes
Abbreviations used in this paper:
- CTD
- carboxy-terminal domain
- NE
- nuclear envelope
- NEBD
- nuclear envelope breakdown
- NPC
- nuclear pore complex
- NTD
- amino-terminal domain
References
- Antonin W., Ellenberg J., Dultz E. 2008. Nuclear pore complex assembly through the cell cycle: regulation and membrane organization. FEBS Lett. 582:2004–2016 10.1016/j.febslet.2008.02.067 [DOI] [PubMed] [Google Scholar]
- Ban R., Matsuzaki H., Akashi T., Sakashita G., Taniguchi H., Park S.Y., Tanaka H., Furukawa K., Urano T. 2009. Mitotic regulation of the stability of microtubule plus-end tracking protein EB3 by ubiquitin ligase SIAH-1 and Aurora mitotic kinases. J. Biol. Chem. 284:28367–28381 10.1074/jbc.M109.000273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaudouin J., Gerlich D., Daigle N., Eils R., Ellenberg J. 2002. Nuclear envelope breakdown proceeds by microtubule-induced tearing of the lamina. Cell. 108:83–96 10.1016/S0092-8674(01)00627-4 [DOI] [PubMed] [Google Scholar]
- Belgareh N., Rabut G., Baï S.W., van Overbeek M., Beaudouin J., Daigle N., Zatsepina O.V., Pasteau F., Labas V., Fromont-Racine M., et al. 2001. An evolutionarily conserved NPC subcomplex, which redistributes in part to kinetochores in mammalian cells. J. Cell Biol. 154:1147–1160 10.1083/jcb.200101081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berke I.C., Boehmer T., Blobel G., Schwartz T.U. 2004. Structural and functional analysis of Nup133 domains reveals modular building blocks of the nuclear pore complex. J. Cell Biol. 167:591–597 10.1083/jcb.200408109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehmer T., Jeudy S., Berke I.C., Schwartz T.U. 2008. Structural and functional studies of Nup107/Nup133 interaction and its implications for the architecture of the nuclear pore complex. Mol. Cell. 30:721–731 10.1016/j.molcel.2008.04.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busson S., Dujardin D., Moreau A., Dompierre J., De Mey J.R. 1998. Dynein and dynactin are localized to astral microtubules and at cortical sites in mitotic epithelial cells. Curr. Biol. 8:541–544 10.1016/S0960-9822(98)70208-8 [DOI] [PubMed] [Google Scholar]
- Carreno S., Kouranti I., Glusman E.S., Fuller M.T., Echard A., Payre F. 2008. Moesin and its activating kinase Slik are required for cortical stability and microtubule organization in mitotic cells. J. Cell Biol. 180:739–746 10.1083/jcb.200709161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan G.K., Schaar B.T., Yen T.J. 1998. Characterization of the kinetochore binding domain of CENP-E reveals interactions with the kinetochore proteins CENP-F and hBUBR1. J. Cell Biol. 143:49–63 10.1083/jcb.143.1.49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandhok N.S., Pellman D. 2009. A little CIN may cost a lot: revisiting aneuploidy and cancer. Curr. Opin. Genet. Dev. 19:74–81 10.1016/j.gde.2008.12.004 [DOI] [PubMed] [Google Scholar]
- Doucet C.M., Talamas J.A., Hetzer M.W. 2010. Cell cycle-dependent differences in nuclear pore complex assembly in metazoa. Cell. 141:1030–1041 10.1016/j.cell.2010.04.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drin G., Casella J.F., Gautier R., Boehmer T., Schwartz T.U., Antonny B. 2007. A general amphipathic alpha-helical motif for sensing membrane curvature. Nat. Struct. Mol. Biol. 14:138–146 10.1038/nsmb1194 [DOI] [PubMed] [Google Scholar]
- Fridolfsson H.N., Ly N., Meyerzon M., Starr D.A. 2010. UNC-83 coordinates kinesin-1 and dynein activities at the nuclear envelope during nuclear migration. Dev. Biol. 338:237–250 10.1016/j.ydbio.2009.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganem N.J., Godinho S.A., Pellman D. 2009. A mechanism linking extra centrosomes to chromosomal instability. Nature. 460:278–282 10.1038/nature08136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glavy J.S., Krutchinsky A.N., Cristea I.M., Berke I.C., Boehmer T., Blobel G., Chait B.T. 2007. Cell-cycle-dependent phosphorylation of the nuclear pore Nup107-160 subcomplex. Proc. Natl. Acad. Sci. USA. 104:3811–3816 10.1073/pnas.0700058104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gönczy P., Pichler S., Kirkham M., Hyman A.A. 1999. Cytoplasmic dynein is required for distinct aspects of MTOC positioning, including centrosome separation, in the one cell stage Caenorhabditis elegans embryo. J. Cell Biol. 147:135–150 10.1083/jcb.147.1.135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman B., Zheng Y. 2006. Mitotic spindle morphogenesis: Ran on the microtubule cytoskeleton and beyond. Biochem. Soc. Trans. 34:716–721 10.1042/BST0340716 [DOI] [PubMed] [Google Scholar]
- Guo J., Yang Z., Song W., Chen Q., Wang F., Zhang Q., Zhu X. 2006. Nudel contributes to microtubule anchoring at the mother centriole and is involved in both dynein-dependent and -independent centrosomal protein assembly. Mol. Biol. Cell. 17:680–689 10.1091/mbc.E05-04-0360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harel A., Orjalo A.V., Vincent T., Lachish-Zalait A., Vasu S., Shah S., Zimmerman E., Elbaum M., Forbes D.J. 2003. Removal of a single pore subcomplex results in vertebrate nuclei devoid of nuclear pores. Mol. Cell. 11:853–864 10.1016/S1097-2765(03)00116-3 [DOI] [PubMed] [Google Scholar]
- Hebbar S., Mesngon M.T., Guillotte A.M., Desai B., Ayala R., Smith D.S. 2008. Lis1 and Ndel1 influence the timing of nuclear envelope breakdown in neural stem cells. J. Cell Biol. 182:1063–1071 10.1083/jcb.200803071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussein D., Taylor S.S. 2002. Farnesylation of Cenp-F is required for G2/M progression and degradation after mitosis. J. Cell Sci. 115:3403–3414 [DOI] [PubMed] [Google Scholar]
- Keppler A., Ellenberg J. 2009. Chromophore-assisted laser inactivation of alpha- and gamma-tubulin SNAP-tag fusion proteins inside living cells. ACS Chem. Biol. 4:127–138 10.1021/cb800298u [DOI] [PubMed] [Google Scholar]
- Khodjakov A., Copenagle L., Gordon M.B., Compton D.A., Kapoor T.M. 2003. Minus-end capture of preformed kinetochore fibers contributes to spindle morphogenesis. J. Cell Biol. 160:671–683 10.1083/jcb.200208143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunda P., Pelling A.E., Liu T., Baum B. 2008. Moesin controls cortical rigidity, cell rounding, and spindle morphogenesis during mitosis. Curr. Biol. 18:91–101 10.1016/j.cub.2007.12.051 [DOI] [PubMed] [Google Scholar]
- Kutay U., Hetzer M.W. 2008. Reorganization of the nuclear envelope during open mitosis. Curr. Opin. Cell Biol. 20:669–677 10.1016/j.ceb.2008.09.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Liu X.S., Yang X., Song B., Wang Y., Liu X. 2010. Polo-like kinase 1 phosphorylation of p150Glued facilitates nuclear envelope breakdown during prophase. Proc. Natl. Acad. Sci. USA. 107:14633–14638 10.1073/pnas.1006615107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Y., Yu W., Li Y., Yu L., Zhang Q., Wang F., Yang Z., Du J., Huang Q., Yao X., Zhu X. 2007. Nudel modulates kinetochore association and function of cytoplasmic dynein in M phase. Mol. Biol. Cell. 18:2656–2666 10.1091/mbc.E06-04-0345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loïodice I., Alves A., Rabut G., Van Overbeek M., Ellenberg J., Sibarita J.B., Doye V. 2004. The entire Nup107-160 complex, including three new members, is targeted as one entity to kinetochores in mitosis. Mol. Biol. Cell. 15:3333–3344 10.1091/mbc.E03-12-0878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone C.J., Misner L., Le Bot N., Tsai M.C., Campbell J.M., Ahringer J., White J.G. 2003. The C. elegans hook protein, ZYG-12, mediates the essential attachment between the centrosome and nucleus. Cell. 115:825–836 10.1016/S0092-8674(03)00985-1 [DOI] [PubMed] [Google Scholar]
- Mao Y., Varma D., Vallee R. 2010. Emerging functions of force-producing kinetochore motors. Cell Cycle. 9:715–719 10.4161/cc.9.4.10763 [DOI] [PubMed] [Google Scholar]
- Mayer T.U., Kapoor T.M., Haggarty S.J., King R.W., Schreiber S.L., Mitchison T.J. 1999. Small molecule inhibitor of mitotic spindle bipolarity identified in a phenotype-based screen. Science. 286:971–974 10.1126/science.286.5441.971 [DOI] [PubMed] [Google Scholar]
- Mishra R.K., Chakraborty P., Arnaoutov A., Fontoura B.M., Dasso M. 2010. The Nup107-160 complex and gamma-TuRC regulate microtubule polymerization at kinetochores. Nat. Cell Biol. 12:164–169 10.1038/ncb2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moynihan K.L., Pooley R., Miller P.M., Kaverina I., Bader D.M. 2009. Murine CENP-F regulates centrosomal microtubule nucleation and interacts with Hook2 at the centrosome. Mol. Biol. Cell. 20:4790–4803 10.1091/mbc.E09-07-0560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mühlhäusser P., Kutay U. 2007. An in vitro nuclear disassembly system reveals a role for the RanGTPase system and microtubule-dependent steps in nuclear envelope breakdown. J. Cell Biol. 178:595–610 10.1083/jcb.200703002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nousiainen M., Silljé H.H., Sauer G., Nigg E.A., Körner R. 2006. Phosphoproteome analysis of the human mitotic spindle. Proc. Natl. Acad. Sci. USA. 103:5391–5396 10.1073/pnas.0507066103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orjalo A.V., Arnaoutov A., Shen Z., Boyarchuk Y., Zeitlin S.G., Fontoura B., Briggs S., Dasso M., Forbes D.J. 2006. The Nup107-160 nucleoporin complex is required for correct bipolar spindle assembly. Mol. Biol. Cell. 17:3806–3818 10.1091/mbc.E05-11-1061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piel M., Meyer P., Khodjakov A., Rieder C.L., Bornens M. 2000. The respective contributions of the mother and daughter centrioles to centrosome activity and behavior in vertebrate cells. J. Cell Biol. 149:317–330 10.1083/jcb.149.2.317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platani M., Santarella-Mellwig R., Posch M., Walczak R., Swedlow J.R., Mattaj I.W. 2009. The Nup107-160 nucleoporin complex promotes mitotic events via control of the localization state of the chromosome passenger complex. Mol. Biol. Cell. 20:5260–5275 10.1091/mbc.E09-05-0377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintyne N.J., Schroer T.A. 2002. Distinct cell cycle–dependent roles for dynactin and dynein at centrosomes. J. Cell. Biol. 159:245–254 10.1083/jcb.200203089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabut G., Doye V., Ellenberg J. 2004. Mapping the dynamic organization of the nuclear pore complex inside single living cells. Nat. Cell Biol. 6:1114–1121 10.1038/ncb1184 [DOI] [PubMed] [Google Scholar]
- Robbins E., Gonatas N.K. 1964. The ultrastructure of a mammalian cell during the mitotic cycle. J. Cell Biol. 21:429–463 10.1083/jcb.21.3.429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson J.T., Wojcik E.J., Sanders M.A., McGrail M., Hays T.S. 1999. Cytoplasmic dynein is required for the nuclear attachment and migration of centrosomes during mitosis in Drosophila. J. Cell Biol. 146:597–608 10.1083/jcb.146.3.597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitoh H., Pu R., Cavenagh M., Dasso M. 1997. RanBP2 associates with Ubc9p and a modified form of RanGAP1. Proc. Natl. Acad. Sci. USA. 94:3736–3741 10.1073/pnas.94.8.3736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salina D., Bodoor K., Eckley D.M., Schroer T.A., Rattner J.B., Burke B. 2002. Cytoplasmic dynein as a facilitator of nuclear envelope breakdown. Cell. 108:97–107 10.1016/S0092-8674(01)00628-6 [DOI] [PubMed] [Google Scholar]
- Salpingidou G., Smertenko A., Hausmanowa-Petrucewicz I., Hussey P.J., Hutchison C.J. 2007. A novel role for the nuclear membrane protein emerin in association of the centrosome to the outer nuclear membrane. J. Cell Biol. 178:897–904 10.1083/jcb.200702026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaner N.C., Campbell R.E., Steinbach P.A., Giepmans B.N., Palmer A.E., Tsien R.Y. 2004. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat. Biotechnol. 22:1567–1572 10.1038/nbt1037 [DOI] [PubMed] [Google Scholar]
- Silkworth W.T., Nardi I.K., Scholl L.M., Cimini D. 2009. Multipolar spindle pole coalescence is a major source of kinetochore mis-attachment and chromosome mis-segregation in cancer cells. PLoS ONE. 4:e6564 10.1371/journal.pone.0006564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman-Gavrila R.V., Wilde A. 2006. Ran is required before metaphase for spindle assembly and chromosome alignment and after metaphase for chromosome segregation and spindle midbody organization. Mol. Biol. Cell. 17:2069–2080 10.1091/mbc.E05-10-0991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjöblom T., Jones S., Wood L.D., Parsons D.W., Lin J., Barber T.D., Mandelker D., Leary R.J., Ptak J., Silliman N., et al. 2006. The consensus coding sequences of human breast and colorectal cancers. Science. 314:268–274 10.1126/science.1133427 [DOI] [PubMed] [Google Scholar]
- Splinter D., Tanenbaum M.E., Lindqvist A., Jaarsma D., Flotho A., Yu K.L., Grigoriev I., Engelsma D., Haasdijk E.D., Keijzer N., et al. 2010. Bicaudal D2, dynein, and kinesin-1 associate with nuclear pore complexes and regulate centrosome and nuclear positioning during mitotic entry. PLoS Biol. 8:e1000350 10.1371/journal.pbio.1000350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stehman S.A., Chen Y., McKenney R.J., Vallee R.B. 2007. NudE and NudEL are required for mitotic progression and are involved in dynein recruitment to kinetochores. J. Cell Biol. 178:583–594 10.1083/jcb.200610112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepanova T., Slemmer J., Hoogenraad C.C., Lansbergen G., Dortland B., De Zeeuw C.I., Grosveld F., van Cappellen G., Akhmanova A., Galjart N. 2003. Visualization of microtubule growth in cultured neurons via the use of EB3-GFP (end-binding protein 3-green fluorescent protein). J. Neurosci. 23:2655–2664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanenbaum M.E., Macůrek L., Galjart N., Medema R.H. 2008. Dynein, Lis1 and CLIP-170 counteract Eg5-dependent centrosome separation during bipolar spindle assembly. EMBO J. 27:3235–3245 10.1038/emboj.2008.242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran E.J., Wente S.R. 2006. Dynamic nuclear pore complexes: life on the edge. Cell. 125:1041–1053 10.1016/j.cell.2006.05.027 [DOI] [PubMed] [Google Scholar]
- Tsai J.W., Lian W.N., Kemal S., Kriegstein A.R., Vallee R.B. 2010. Kinesin 3 and cytoplasmic dynein mediate interkinetic nuclear migration in neural stem cells. Nat. Neurosci. 13:1463–1471 10.1038/nn.2665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallee R.B., Williams J.C., Varma D., Barnhart L.E. 2004. Dynein: An ancient motor protein involved in multiple modes of transport. J. Neurobiol. 58:189–200 10.1002/neu.10314 [DOI] [PubMed] [Google Scholar]
- Varis A., Salmela A.L., Kallio M.J. 2006. Cenp-F (mitosin) is more than a mitotic marker. Chromosoma. 115:288–295 10.1007/s00412-005-0046-0 [DOI] [PubMed] [Google Scholar]
- Vergnolle M.A., Taylor S.S. 2007. Cenp-F links kinetochores to Ndel1/Nde1/Lis1/dynein microtubule motor complexes. Curr. Biol. 17:1173–1179 10.1016/j.cub.2007.05.077 [DOI] [PubMed] [Google Scholar]
- Walther T.C., Alves A., Pickersgill H., Loïodice I., Hetzer M., Galy V., Hülsmann B.B., Köcher T., Wilm M., Allen T., et al. 2003. The conserved Nup107-160 complex is critical for nuclear pore complex assembly. Cell. 113:195–206 10.1016/S0092-8674(03)00235-6 [DOI] [PubMed] [Google Scholar]
- Whitehead C.M., Winkfein R.J., Rattner J.B. 1996. The relationship of HsEg5 and the actin cytoskeleton to centrosome separation. Cell Motil. Cytoskeleton. 35:298–308 [DOI] [PubMed] [Google Scholar]
- Wozniak R., Burke B., Doye V. 2010. Nuclear transport and the mitotic apparatus: an evolving relationship. Cell. Mol. Life Sci. 67:2215–2230 10.1007/s00018-010-0325-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Lei K., Yuan X., Wu X., Zhuang Y., Xu T., Xu R., Han M. 2009. SUN1/2 and Syne/Nesprin-1/2 complexes connect centrosome to the nucleus during neurogenesis and neuronal migration in mice. Neuron. 64:173–187 10.1016/j.neuron.2009.08.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X., Mancini M.A., Chang K.H., Liu C.Y., Chen C.F., Shan B., Jones D., Yang-Feng T.L., Lee W.H. 1995. Characterization of a novel 350-kilodalton nuclear phosphoprotein that is specifically involved in mitotic-phase progression. Mol. Cell. Biol. 15:5017–5029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuccolo M., Alves A., Galy V., Bolhy S., Formstecher E., Racine V., Sibarita J.B., Fukagawa T., Shiekhattar R., Yen T., Doye V. 2007. The human Nup107-160 nuclear pore subcomplex contributes to proper kinetochore functions. EMBO J. 26:1853–1864 10.1038/sj.emboj.7601642 [DOI] [PMC free article] [PubMed] [Google Scholar]