Abstract
Mechanosensitive TREK channels belong to the family of K2P channels, a family of widely distributed, well-modulated channels that uniquely have two similar or identical subunits, each with two TM1-P-TM2 motifs. Our goal is to build viable structural models of TREK channels, as representatives of K2P channels family. The structures available to be used as templates belong to the 2TM channels superfamily. These have low sequence similarity and different structural features: four symmetrically arranged subunits, each having one TM1-P-TM2 motif. Our model building strategy used two subunits of the template (KcsA) to build one subunit of the target (TREK-1). Our models of the closed channel were adjusted to differ substantially from those of the template, e.g., TM2 of the second repeat is near the axis of the pore whereas TM2 of the first repeat is far from the axis. Segments linking the two repeats and immediately following the last TM segment were modeled ab initio as α-helices based on helical periodicities of hydrophobic and hydrophilic residues, highly conserved and poorly conserved residues and statistically related positions from multiple sequence alignments. The models were further refined by two-fold symmetry-constrained MD simulations using a protocol we developed previously. We also built models of the open state and suggest a possible tension-activated gating mechanism characterized by helical motion with two-fold symmetry. Our models are consistent with deletion/truncation mutagenesis and thermodynamic analysis of gating described in the accompanying paper.
Key words: mechanosensitive ion channels, structure prediction, gating mechanism, proline hinge, mutual information, mutual information graph
Introduction
Two-pore-domain background potassium (K2P) channels form a distinct gene family of widely distributed background channels modulated by numerous physical and chemical factors that influence resting membrane potential and cell excitability.1. The family of mammalian K2P channels now include 15 members classified into six main structural classes2 based on sequence similarity and the stimuli to which they respond, as follows: (1) TWIK1, TWIK2 and KCNK7 channels (functional expression of KCNK7 has not yet been reported)-weak inward rectifiers; (2) TREK-1, TREK2 and TRAAK channels-mechano gated; (3) TASK1, TASK3 and TASK5 channels-acid inhibited; (4) THIK1 and THIK2 channels-halothane inhibited;3 (5) TALK1, TALK2 and TASK2 channels-alkaline-activated; and (6) TWIK-related spinal cord K+ (TRESK) channel, which is regulated by intracellular calcium. Despite low sequence similarity among the different classes and broad range of stimuli to which they respond, K2P channels share common structural features like two subunits per channel, each with two TM1-P-TM2 motifs. In one-pore-domain K+ (K1P) channels, four matching P loops are assembled in homo- or heterotetramers (all subunits have a similar P domain sequence, which contains the residues GYG or GFG), whereas in the dimeric K2P channels, the first and the second pore domains (P1 and P2) can have substantially different sequences. For example, many K2P channels have a F or L in the GXG motif (where X represents any amino acid) of the selectivity filter in the P2 domain instead of a Y.4,5
Therefore, in K2P channels, the pore is predicted to have a two-fold symmetry rather than the classical four-fold arrangement of other K+ channels. Although the selectivity of K2P channels for K+ over Na+ is high (permeability ratio (PNa/PK) < 0.03), these structural differences suggest a more varied permeation and gating compared with K1P channels.6,7
Members of the TREK subfamily, TREK-1, TREK-2 and TRAAK, are best known as background K+ channels exhibiting a low, weakly voltage-dependent, open probability that is strongly modulated by both chemical and physical stimuli.8 This subfamily is notable not only for their role in maintaining the resting potential and controlling membrane excitability, but also for contributing to diverse sensory transduction processes and metabolic regulation, including contributions to: mechanosensitivity,9–12 thermosensitivity,13,14 chemosensitivity,15 nociception16 and neuroprotection.17–19 Human TREK-1 is highly expressed in the brain, where it is particularly abundant in GABA (γ-aminobutyric acid)-ergic interneurons of the caudate nucleus and putamen.20 TREK-1 is also expressed in the prefrontal cortex, hippocampus, hypothalamus, midbrain serotonergic neurons of the dorsal raphé nucleus and sensory neurons of the dorsal root ganglia.21 TREK-1 is also found in peripheral tissues such as the gastrointestinal tract.22 TREK channels have also been identified as important drug targets for general anesthesia23 and potentially for treatment of depression.24
Effective drug design is hampered by lack of structural information and limited understanding of activation mechanism. Our goal is to build structural models of TREK-1 channel that can be used as a starting point for hypothesis-driven structural and functional experiments. If experimental tests confirm crucial aspects of the models, they can further be used as templates for modeling other channels in K2P family.
Predicting the structure of TREK channels is a challenging task, since available templates are distantly related KcsA channels, members of the voltage-gated channels subfamily. Therefore we used a complex approach combining homology-based and ab initio modeling incorporating additional constraints derived from sequence conservation patterns in multiple sequence alignments (MSA).
Results
Models of the closed, low-conductive and open states.
The first step in our modeling process was to use the KcsA crystal structure in the closed conformation to develop a homology model of TREK-1 (Fig. 1A). The alignment in Figure 1B was used to develop these models. KcsA and TREK-1 share only low sequence similarity, which makes alignment of some segments, especially the more peripheral TM1 segments, ambiguous when only two sequences are used. Much of this ambiguity was eliminated by choosing alignments in which highly conserved residues interact with other protein residues and align in both sequences, whereas poorly conserved residues are on the surfaces of the protein and interact primarily with either lipids in the TM region or water (Fig. 2).
Figure 1.
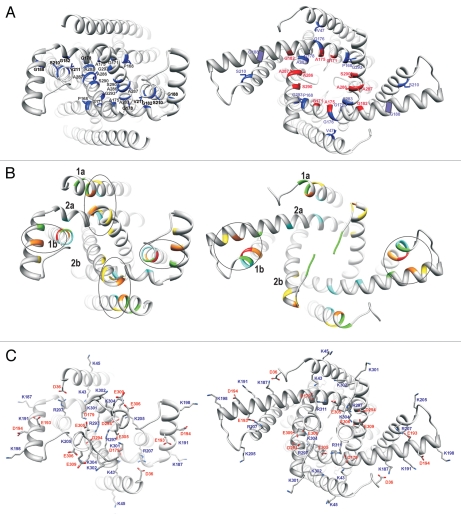
Modeling TREK-1 channel. (A) Schematic illustrating our approach to modeling TREK-1 channels: two KCSA subunits were used to model one TREK subunit; linker and C-terminal regions were modeled ab initio; helical segments are represented by cylinders. (B) Target-template sequence alignment used for modeling; identical residues are highlighted magenta, selectivity filter is highlighted orange; M1P1 loop was not included in the models; helical segments are underlined using the same color scheme as in (A). (C) Bottom view and (D) side view of structural models for the closed (left part), low-conductive (central part) and open (right part) conformation, in ribbon representation; helical segments are labeled and colored using the same color scheme as in (A and B).
Figure 2.
Residue conservation patterns from multiple sequence alignments, used as a modeling criteria. (A) Target-template sequence alignment in which residues are highlighted according to their degree of conservation, on a scale from red (highly conserved) to blue (not conserved). (B) Top view and (C) bottom view of the models for closed (left), low-conductive (center) and open (right) conformations, colored according to the same degree-of-conservation scheme.
The extracellular TM1-P loop of the first repeat is much longer in TREK-1 than in KcsA. The additional TREK-1 residues indicated in Figure 1B as an insert above the continuous sequence were not included in the model. Experimental studies showed that TREK-1 still forms functional channels when this segment is deleted (see Maksaev et al., pp. 34–42).
The segment linking the two repeats was modeled ab initio since there is no template for it in KcsA. The sequence pattern of this segment is consistent with a membrane surface amphipathic α-helix in which the hydrophobic face is poorly conserved and may interact with lipid alkyl chains while the polar face has positively charged sidechains that may interact with lipid head groups and residues conserved among TREK and TRAAK subfamilies that may interact with similarly conserved residues of other protein segments (see below). This pattern is also supported by a statistical analysis involving mutual information and graph theory discussed later. It was thus modeled in this way, with its highly conserved residues interacting with highly conserved residues near the N-terminus of TM1b. TM1b was selected for the postulated interactions with conserved residues of the linker because (a) the linker must be positioned near TM1b to connect the repeats, and (b) several conserved residues are on the “back” side of TM1b in our alignment where they would be exposed to lipid unless they interacted with the linker, and (c) the postulated interactions among similarly conserved residues of the two segments could be made in a complementary way that involved energetically favorable interactions among conserved hydrophobic residues and between conserved oppositely charged residues.
Some features of the initial KcsA-based model were not completely consistent with the TREK-1 sequence. TM2a has a highly conserved proline residue near the “hinge” region of other K+ channels where TM2 bends when the channel opens. Introduction of a proline in this region of the NaChBac channel stabilizes the open conformation.25 Thus, it seems likely that TM2a of TREK-1 may have a bent open-like conformation even when the channel is closed. The only residue on TM2b predicted to be exposed to lipid alkyl chains on the basis of mutability (poorly conserved blue in Fig. 2 and hydrophobic) is F269 near the N-terminus; whereas three (I161, L165 and I184) occur in TM2a. These patterns suggest that TM2b is more buried than TM2a and the TREK-1 structure deviates somewhat from the four-fold symmetry of KcsA. We adjusted our model to be consistent with these features while maintaining two-fold symmetry of the two subunits. In our closed conformation, TM2b is positioned nearer the central axis of the pore than TM2 of KcsA and TM2a is farther from the axis, in a position and conformation more like analogous segments of open K+ channel crystal structures (KVAP, MthK and KV1.2). This asymmetric collapse of the structure reduces the size of the central cavity and places side chains of the highly conserved L289 near the axis of the pore where they would block permeation of ions.
Channel opening was modeled in two ways. In the first approach the conformation of Repeat A was not altered much since it already had an open-like conformation and TM1b and TM2b were displaced radially outward, which confers approximate fourfold symmetry for the TM portion of the channel's backbone (low-conductive model of Fig. 1C and D). In this model most of the interactions among small residues at regions of close contact between helices are maintained. The difficulties with this type of model for the open conformation are that after MD simulations the radial in-plane expansion of the protein was ∼0.7 nm2 and after symmetry refinement it reduced to ∼0.1 nm2. Both of these values are much smaller than the ∼4 nm2 calculated from experimental measurements and the 0.75 nm2 cross-section area of the inner pore (∼0.6 × 1.2 nm at narrowest region). For comparison, the pore cross-section falls between ∼1 nm2 in the open structure of KcsA (pdb code 3f5w26) and ∼0.6 nm2 in KV1.2 (pdb code 2a79,27) with conductances ∼75 pS28 and ∼13 pS29 respectively, expected at 150 mM KCl concentration. Although the relationships between the pore geometry and conductance for such systems are rather complicated and the conductance is strongly influenced by charge at the channel's intracellular mouth,28 pore size in the above TREK model appears too small to allow a high rate of permeation. However, TREK-1 channels have two conductances, a small (∼60 pS) and a large (∼130 pS). Thus it is feasible that this model approximates the low-conductive state.
We thus developed another model for the open conformation with the inner portions of the TM helices located farther from the pore. This additional displacement required repositioning of the linker helix; it was modeled as a continuous extension of the TM2b helix (open model of Fig. 1C and D). The expansion area for this model after symmetry-restrained MD simulations was calculated to be ∼5.2 nm2, in close agreement with the value estimated from experimental analysis of dose-response curves (see Maksaev et al., pp. 34–42).
Due to insufficient information from template-based modeling alone, complementary strategies were used to derive additional constraints. Thus, several factors were considered in developing these models. The first was interactions among conserved residues. Functionally and/or structurally important sites or regions are typically composed of residues with similar degrees of conservation interacting among multiple segments. For example, the only residues conserved among all K+ channels are located in the selectivity filter where they interact with their counterparts of other subunits to determine the selectivity of the channel. These residues are not conserved between K+ channels and homologous channels with different selectivities. In contrast residues that are poorly conserved within closely related families or subfamilies tend to reside on the surfaces of proteins away from functionally important sites.30 In the analysis presented here, residues are classified into three categories: (1) highly conserved among all 2P sequences (red and orange in Fig. 2), (2) conserved among TREK and TRAAK mechanosensitive channels but not between these and other 2P families (yellow and green), and (3) poorly conserved among TREK and TRAAK sequences (cyan and blue). The outer half of the TM region that includes the K+ selectivity filter contains about 70% of residues that are well conserved among all 2P channels (red and orange residues of Fig. 2). In contrast, only less than 10% of residues of the inner pore or inner half of the TM region are conserved among all 2P families; however, many are conserved among mechanosensitive TREK and TRAAK sequences (yellow and green residues of Fig. 2). Supplementary Table 2 lists all of the interactions among residues of this region that are highly conserved at this level (red, orange and yellow residues of Fig. 2). All but two (M42 and W295) interact with at least one other equally conserved residue in the closed and/or open models. M42 may be conserved as an alternative initiation residue31 and W295 interacts with the cytoplasmic surface of the membrane, where it may serve as a lipid anchor.32 All poorly conserved residues of the transmembrane region (blue in Fig. 2) reside primarily on the outer surface of the protein.
The second factor was the relative positions of small residues. Small residues are frequently located at positions of closest contact between helices that do not cross at angles typical of “knobsinto-holes”33,34 or “ridges-into-grooves”35 helix packing patterns. In our closed model, residues S290 and G293 are near the crossing contact of the pore-lining TM2b helices; A175 and G178 of TM1a interact with A283, A286 and A287 of TM2b; P168 and G171 of TM2a are near TM2b of the other subunit; G182 of TM2a is near S210 and V211 of TM1b; and G188 of the linker is near S210 of TM1b (see blue residues of closed model in Fig. 3A). In open model G178 of TM2a packs next to A283 of TM2b, P168 of TM2a packs next to G293 of TM2b of the other subunit, G178 of TM2a is next to V47 of TM1a: and S210 of TM2a is near the linker helix (blue residues of open model in Fig. 3A). Energies required to transfer these types of small residues from an inaccessible environment inside a protein to an aqueous environment are relatively small.36 Thus, they are ideally suited to serve as residues that are relatively buried in a closed conformation but that become exposed to water inside the pore when the channel opens. G171, A175, G182, A286, A287 and S290 (red residues of Fig. 3A) of the TM2 segments have this property in our models.
Figure 3.
Ribbon representations of the inner pore region of closed (left) and open (right) models viewed from the inside. (A) Small residues located at positions of close contact between helices are colored blue. Small residues that become exposed in the lining of the pore when the channel opens are colored red. (B) High degree, high MI positions obtained from the MSA of TREK homologs, used as a modeling criteria. Residues with the highest degree values are colored according to the color spectrum scheme used in Supplementary Table 1, red for the highest score and cyan for the lowest. (C) Charged residues of the inner pore segments. Side chains are colored by element (gray = carbon, blue = nitrogen, red = oxygen, white = hydrogen). Positively charged resides are labeled in blue, negatively charged residues in red.
The third factor identified statistically related positions from our MSA of all 2P channels. To characterize evolutionary relations among residues within MSA columns, mutual information (MI) of pairs of MSA columns (j,k) was obtained. That MI was denoted as MI(j,k). For Ntotal (=1,557) positions in a MSA, a total of Ntotal(Ntotal − 1)/2 unique MI(j,k) values (=1,211,346 pairs) were calculated using Equation 1 (described in Materials and Methods). The distribution of MI(j,k) values is shown in Supplementary Figure S1. From the over one million MI(j,k) pairs, the leading 1,000 high MI pairs were used to compute a MI graph that represented the statistical association of the high MI pairs with each other. The degree (or connectivity) of the leading thirty-one positions is shown in the last column of Supplementary Table 1. Residues from TREK-1_human sequence that correspond to the MSA positions with high connectivity tend to form clusters at functionally important regions. In previous work on other membrane proteins such clusters differed somewhat among analyzed subfamilies.37 The analysis excludes residues, such as those of the selectivity filter, that are conserved among all sequences and surface residues where mutations can occur somewhat randomly in an uncoordinated manner. Twenty-one of the highest thirty one scoring residues are in the inner pore region near the cytoplasmic surface of the membrane in our models (Fig. 3B), six are in the deleted TM1a-Pa loop, three are in the outer TM portion of the protein and one is in the C-terminus cytoplasmic domain. The twenty-one interior residues are (in rank order for each segment, top six in bold) K43, V41, K45, W44 and V47 near the N-terminus of TM1a; G176 and G182 near the C-terminus of TM2a; E193, F196 and I189 on the conserved face of the helix linking the two repeats, S210, T211, R207, I208 and I209 near the N-terminus of TM1b and S300, V298, K302, G293, E306 and E305 near the C-terminus of TM2b. These residues form clusters in our models. The residue, corresponding to MSA position with the highest degree, S210 near the N-terminus of TM2b, interacts with a putative hinge region connecting the end of TM2a to the linking helix that changes conformation during gating. More precisely, in the closed conformation S210 interacts with I189, T211 with G182, R207 with I189 and I209 with F196; and in the open conformation S210 interacts with E193 and F196, T211 with I189 and R207 with E193 and I189. The N-terminus of TM1a interacts with the C-terminus of TM2a and C-terminus of TM2b and in the closed conformation; i.e., K43 with S300 and V47 with G176. The C-termini of TM2b also self associate in the closed conformation; i.e., G293, E305 and E306 self associate and K302 of one subunit interacts with E305 of the adjacent subunit. Thus, most residues associated with positions with high degree (obtained from MI graph) values in our model interact with other residues with high degree near the cytoplasmic surface of the membrane, where they are likely involved with mechanical activation of the channel.
A fourth factor was the location of charged residues; i.e., few isolated charges should be inaccessible to water, salt bridges and interactions of positively charged side chains with lipid head groups of the cytoplasmic surface are energetically favorable, permeation of inorganic cations through open channels can be assisted by negative charges near the pore residues and inhibition of permeation through closed channels can be impeded by positively charged residues near the pore. Our models have no charged residues throughout most of the transmembrane region. Charged residues of the inner pore region are illustrated in Figure 3C. K45, K187, K191, K198, and for the open conformation also K301 and K301 can interact with lipid head groups on the cytoplasmic surface. The following salt bridges form near the cytoplasmic surface in our models: K43-D179, K187-D36 (o), K191-D194, K205-D294, R207-D36 (c), R207-E193 (c), R297-D294, K301-E305 (c) and K304-E306, where (c) and (o) indicate only in the closed or open conformation. Negatively charged D179, D294, E305 and E306 are near the cytoplasmic entrance to the open pore where they may reduce the barrier to and/or increase the concentration of K+ ions. K205, R297 and K301 are near the axis of the closed pore, where they may impede permeation of K+ ions.
Post TM 2b cytoplasmic segment.
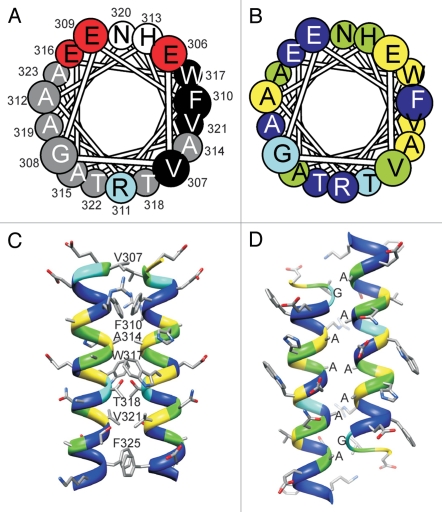
Segment 306–326 has a sequence consistent with formation of an amphipathic α-helix. This sequential pattern is unique to TREK and TRAAK subfamilies; sequences of other 2P channels cannot be aligned unambiguously with these segments and do not exhibit helical periodicities of hydrophobic and hydrophilic residues. Residues beyond this segment are not conserved among TREK and TRAAK subfamilies, and can be deleted in TREK-1 channels without affecting channel properties substantially (see Maksaev et al., pp. 34–42). The helical wheel representation of Figure 4A shows that the polar face (top) consists primarily of negatively charged residues, the hydrophobic face (right side) consists primarily of alkyl and aromatic residues, an “alanine” face (left side) consists primarily of small alanines and a glycine, and a “bottom” faces has two threonines flanking a positively charged arginine. The segment has five residue positions highly conserved (yellow) among TREK and TRAAK subfamilies; four on the hydrophobic face and one on the alanine face (Fig. 4B).
Figure 4.
Models of the post-TM C-terminal segment (residues 305–330). (A) Helical wheel representation in which residues are colored according to polarity: small hydrophobic (grey); large hydrophobic (black); negatively charged (red) and positively charged (blue). (B) Helical wheel representation in which residues are colored according to their degree of conservation, from yellow (highly conserved) to blue (not conserved). (C) “Parallel Dimer” model in which the highly conserved hydrophobic residues self associate; this arrangement was used in the models of the open and closed states. (D) “Antiparallel Dimer” model in which the small residues face pack next to each other; this arrangement was used in a tentative model of the inactivated state.
For closed, low-conductive and open conformations we modeled the two putative cytoplasmic helices to form a parallel dimer (dimeric coiled-coil) in which the axis of the dimer corresponds to the axis of the pore and the highly conserved hydrophobic residues self-associate (Fig. 4C). Similar types of cytoplasmic coiled-coils have been reported for other K+ (tetrameric coiled-coil in KcsA) and mechanosensitive (pentameric coiled-coil in MscL) channels, and our groups correctly modeled this type of interaction for MscL channels38,39 before the corrected conformation was published.40 These putative interactions remained relatively stable during MD simulations.
Although these models are consistent with the amphipathic nature of the cytoplasmic helices, they do not explain the presence of the alanine face. We have considered two possibilities, both related to inactivation mechanisms. TREK-1 channels do not usually inactivate when the patch is excised from the cell and the normal cytoplasmic environment is lost. Thus, inactivation appears to involve interactions with some cytoplasmic component, likely mediated by the C-terminus. If so, the cytoplasmic component may interact with the highly-conserved alanine faces of the putative C-terminus coiled-coils. We have not attempted to model this interaction since the cytoplasmic component is unknown. The second possibility is that tension increases within the segment linking the C-terminus of TM2B to the N-terminus of the coiled-coil when the channel opens, causing the cytoplasmic helices to eventually pull apart in a scissor-like motion to form an antiparallel dimer in which the “alanine” faces pack next to each other (Fig. 4D) when the channel inactivates. In our highly tentative models of this conformation, the highly-conserved hydrophobic faces interact with other highly-conserved residues of surrounding TM segments, the arginine on the “bottom” face extends into the pore where it salt-bridges to D294 and inhibits permeation through the inner pore, and the negatively-charged polar face is exposed to the cytoplasm. Thus, virtually all sequence characteristics of this segment are explained by this model. Preliminary MD simulations of this model indicated only marginal stability of the assembly; interactions among the alanines were maintained in only some symmetry-restrained simulations, but deviations from this motif were not large in any of the simulations.
Refinement of the models using symmetry-restrained MD simulations.
Although homology modeling is probably the most reliable structure prediction method,41 the accuracy of the resulting model depends on several factors: (a) accuracy of the template; (b) the level of sequence identity (above 30% typically yields a model with accuracy comparable to a low resolution crystal structure); (c) the accuracy of the target-template sequence alignment (the most important factor); and (d) the number of available templates.42 Given that our models were built based on a distant template and even certain regions were modeled in the absence of a template, they probably contain errors. Traditional MD simulations have limited capability for structure refinement of low-resolution models43,44 and these refinement procedures are applicable only to small to medium size proteins; therefore they cannot be used to refine TREK channels models. We have previously developed a protocol using symmetry-restrained MD simulations, named “symmetry annealing” (see Methods; reviewed in ref. 45), capable of substantially improving the accuracy of homology models in 80% of our benchmarking set. Two steps of symmetry annealing are sufficient to achieve maximum improvement and stability of the resulting models may reliably be used as a measure of success in the refinement process.
We initially evaluated the TREK models by measuring the stability of the models during 30 ns unrestrained MD simulations. All models show an RMSD value of less than 3 Å. These results are consistent with a reasonable level of accuracy since in our limited experience only structures with accuracy equivalent to a low-resolution x-ray structure exhibit this degree of stability.45 Surprisingly, the closed model was the least stable (RMSD values closed ∼2.8 Å, low-conductive ∼2.0 Å open ∼1.8 Å (Fig. 5B and black traces)).
Figure 5.
Refinement of structural models by symmetry-restrained MD simulations. (A) Simulation protocol: models were subject to 30 ns unrestrained MD simulation (black line); after 5, 10, 15, and respectively 20 ns, a symmetry annealing step was applied (blue and green thick lines) followed by 10 ns unrestrained simulation (blue and green thin lines); the model annealed after 15 ns was subject to a second step of symmetry annealing followed by unrestrained simulations (red lines). (B) RMSD plots of the Cα trace of each model, during the first 10 ns of unrestrained simulation, using as reference the models at the beginning of each simulation step. (C) Root-mean square fluctuations (RMSF) of each model, during the last step of unrestrained simulation (thin red lines in A).
Each TREK model was subject to structure refinement using a two-fold-symmetry annealing protocol applied after 5, 10, 15 and respectively 20 ns of unrestrained simulation (Fig. 5A). Following symmetry annealing, each structure was subject to 10 ns unrestrained MD simulation, to evaluate its stability. Results indicate that after the first step of symmetry annealing, all models become more stable (Fig. 5B and green and blue traces), which is an indicator of improved accuracy, the most improved model being the one of the closed state. A second step of annealing results only in marginal improvements (less than 0.5 Å decrease of RMSD, red traces in Fig. 5B), indicating that the procedure has reached its limits. However, after symmetry annealing all models have an RMSD value of less than 1.5 Å (equivalent to a middle-to-high-resolution x-ray structure; e.g., RMSD values from similar MD simulations of crystal structures of closed KcsA, NaK and MlotiK were ∼1.0 Å).45 This stability suggests that the models may be sufficiently accurate to be useful for understanding functional mechanism of TREK channels. It is somewhat surprising that the open model is the most stable (RMSD after second annealing ∼1.0 Å), since it is the least densely packed structure and deviates most from the closed template. As expected, root-mean-squared fluctuation measurements at each position indicate that the most stable regions are the TM helices (Fig. 5C), while the most flexible regions are the interconnecting segments, especially M1P1 loop and the ab-initio modeled linker between TM2a and TM1b.
Conclusions
We have developed structural models of TREK-1 channels in closed, low-conductive and open conformations, the least ambiguous of which are the closed and open models. These models are consistent with the following modeling criteria: (1) they are stable during MD simulations with RMSD values similar to those of crystal structures of K+ channels, (2) identical subunits have identical conformations, (3) almost all hydrophobic residues are in apolar environments while polar residues are in hydrophilic environments, (4) almost all charged residues form salt bridges, (5) numerous positively charged residues interact with cytoplasmic lipid head groups, (6) conserved residues interact with other comparably conserved residues, (7) residues from the human TREK-1 sequence that correspond to MSA positions with high degree of connectivity and high MI form spatial clusters, (8) regions of closest contact between helices that cross at atypical angles contain small residues, and (9) most residues that are buried in the closed conformation and exposed in the open conformation have intermediate polarities.
This family of K+ channels has numerous unique features. The tandem arrangement of two homologous but not identical repeats and two-fold symmetry allow for more variation in sequence, and hence more conformational freedom than in K+ channels made of four identical subunits which are expected to have identical average conformations. These differences are most apparent in our model of the closed conformation in which the inner pore has a collapsed conformation due to the location of the TM2b helix near the axis of the pore. A highly conserved proline 165 in TM2a is proposed to contribute to this asymmetry between the two repeats by constraining TM2a to an open-like configuration distant from the pore's axis in all conformations. The segment linking the two repeats is proposed to form a cytoplasmic membrane surface helix that changes conformation during gating. The cytoplasmic domain formed by the post TM2b segment of mechanosensitive TREK and TRAAK channels differ from other 2P channels in that they can form an amphipathic α-helix. In our models these form a parallel dimeric coiled-coil in closed and open conformations. Intersubunit interactions for the parallel interaction involve highly conserved hydrophobic residues. The inactivation mechanism is much more ambiguous and may involve interactions with cytoplasmic moieties not present in our models, and/or changes in the selectivity filter46 that we did not attempt to model. The model in which the cytoplasmic helices form an antiparallel dimer in the inactivated conformation is a rather speculative working hypothesis to provide a rationale for why the helix has alanines along one face. Although the models presented here are still tentative, they can be tested experimentally. One way is to introduce cysteine residues at positions predicted to be proximal in some conformations but distant in others. Distances between the residues can be analyzed in several ways; by oxidizing the cysteines to form disulfide bridges, by determining whether Cd2+ bridges can form between the sulfhydryl groups, by using reagents to cross-link the sulfhydryls or by attaching probes (e.g., EPR, FRET or LRET) to measure distances between the groups in the differing conformational states. Initial steps toward these types of experiments are described in the accompanying manuscript.
Materials and Methods
Multiple sequence alignment.
Sequences within K2P channels family were identified using Psi-Blast47,48 against RefSeq database.49 The multiple sequence alignment (MSA) of the resulting 235 sequences of K2P channels was generated using BioEdit50 then manually adjusted in the loop and terminal regions.
Calculation of mutabilities. The method by which mutabilities were calculated is described in Shrivastava et al.51 Calculations were performed for all 2P sequences of the initial alignment and for only the TREK and TRAAK subfamilies of this alignment. Mutabilities calculated from all sequences were used for the most highly conserved red and orange colored residues of Figure 2, mutabilities calculated for only the TREK and TRAAK subfamilies were used for all other residues.
Identification of statistically related MSA positions. The analysis for computing statistically related MSA positions is described in Fatakia et al.37 The independent probabilities of obtaining AAs x and y at positions j and k were represented as pj(x) and pk(y). The joint probability of AA pairs x and y at positions j and k was designated as pj,k(x,y). The MI of pairs of columns j and k was calculated using:
| Eqn. 1 |
Double counting was omitted because MI(j,k) = MI(k,j). Here NAA (= 20 + 1) indicates the twenty naturally occurring amino acid residues and the “-” symbol that exists as gap in the MSA. The symbol “X” from the MSA was not considered separately (there were four such instances) and was computed using the gap “-” instead.
MI graphs. To associate high MI(j,k) pairs with all other statistically significant high MI(j,k) pairs, we constructed a MI graph. The vertices of the MI graph represented positions from the MSA and the edges represented the statistically significant high MI pairs as demonstrated previously.37 From over one million MI pairs of MSA columns, the leading 800–1,000 MI pairs resulted in a consistent cohort of positions represented in the MSA (results not shown). Therefore, MI graph with leading 1,000 MI(j,k) pairs was constructed. The degree (connectivity) of the 31 leading vertices is reported in the last column of Supplementary Table 1.
Homology modeling.
The closed state model of the human TREK-1 channel (gb|AAD47569.1) was built using as template the K+ Channel KcsA at 2 Å resolution, pdb code 1k4c52 for each of the two repeats in one TREK subunit (Fig. 1A). Sequence alignment (Fig. 1B) was initially generated using Clustalx53 and refined by incorporating additional information from secondary structure prediction using JPred,54 Porter,55 Prof,56 Psipred57 and Sable58 and patterns of residue conservation. All initial models were generated using Modeller 9v1.59,60 Models were then adjusted manually to improve interactions among conserved residues and to bend TM2A just before a proline. The segment linking repeat A to repeat B was constructed as an α helix and then positioned and adjusted manually to connect the repeats while also optimizing interactions among conserved residues and placing the poorly-conserved hydrophobic face of the helix to interact with lipid alkyl chains at the inner surface of the membrane.
Two models of the open conformation were constructed from the closed conformation. The first was constructed by manually swinging only the TM2B and to a lesser extent TM1B helices radially outward to expand the inner portion of the pore. The TM region of this model was very similar to that of the “inactivated” model illustrated here. The second model was constructed by expanding the inner portion of the pore more by also swinging TM1A and TM2A radially outward slightly. These models were evaluated with MD simulations to determine which was more stable. The first model tended to return toward the closed conformation; whereas, the second remained stable as a highly open structure.
The cytoplasmic Post TM2B region was noted to have an amphipathic helical pattern. It was modeled to self-associate in two ways: as parallel and antiparallel dimers.
Molecular dynamics simulations.
The models for the open and closed states were used as starting structures for MD simulations and were processed according to the following protocol:
(a) Hydrogen atoms were added; ionizable residues were in their default protonation state.
(b) The structures were oriented so that the two-fold rotational symmetry axis coincided with z, with extracellular and selectivity filter side facing the positive direction. The channel was positioned along z coordinate to have the best match of the surface hydrophobicity pattern to the lipid membrane patch.
(c) Ions were positioned in the selectivity filters of HM the same way as in the corresponding template x-ray structures and with water molecules placed between the ions, since water is essential for ion conduction.61
(d) Channels were embedded into a patch of 1-palmitoyl-2-oleoyl-phosphatidylethanolamine (POPC) lipid bilayer fully hydrated with TIP3P water molecules.62 The lipid bilayer containing on average 228 molecules of POPC was pre-equilibrated in a solvated flexible simulation cell. The dimensions of the rectangular solvent box were chosen so that the minimum distance from the box boundaries to the protein was 25 Å. The simulation cell area was set at 112 × 116 Å2 for all the systems, thus leaving ∼50 Å (about 5–7 layers of lipid) between the copies of the channel in the periodic cells. To facilitate conformational rearrangements of channels in the membrane, the system was simulated with an anisotropic flexible cell maintaining 1 bar pressure in z direction and membrane tension of 40 dyne/cm in x−y plane (fixed x to y sizes ratio). Relatively high value of the tension was necessary to maintain an equilibrium area per POPC lipid matching the experimentally measured value of 68.3 Å2.63,64
(e) K+ and Cl− ions were added in order to maintain electroneutrality. Structure file was generated using the psfgen plugin in VMD.65 The total number of water molecules, lipids and ions contained in the systems are indicated in Supplementary Table 3.
Unrestrained MD simulations.
All MD simulations were performed in the NPT ensemble using NAMD2 package66 developed by the Theoretical and Computational Biophysics Group in the Beckman Institute for Advanced Science and Technology at the University of Illinois at Urbana-Champaign, with the CHARMM27 force field parameters67 including the grid-based CMAP correction68 for the φ ψ-angular dependence of the energy. This is especially important for α-helical proteins (as the systems simulated in this work), since CMAP not only yields significant improvements in the distribution of dihedral angles, but also corrects the tendency of CHARMM force field to distort α-helix structures to π-helix structures.69,70
The simulation systems were subject to a stepwise equilibration before the production phase, involving initial 1,000 steps of energy minimization, followed by 100 ps of gradual heating to 303 K with the protein constrained to the initial coordinates and 4 ns of simulation in which protein backbone was constrained but lipids were allowed to pack around the protein.
After the initial minimization, heating and equilibration stages, the production phase of each MD unrestrained simulation used a time step of 1 fs for the bonded interactions, with coordinates saved every 1 ps. The Langevin piston method71,72 was used to maintain a constant pressure of 1 atm. The temperature, set to 303 K, was controlled by using Langevin dynamics with a coupling coefficient of 1 ps−1. We used periodic boundary conditions and the particle mesh Ewald method68 with a real-space cutoff distance of 12 Å and a grid width of 0.97 Å. The switching distance for nonbonded electrostatics and van der Waals interactions was 10 Å and the integration time step was 4 and 2 fs respectively. Initial models of open, low-conductive and closed states were subject to 30 ns of unrestrained MD simulations.
Model refinement by symmetry annealing.
Structural models of the open, inactivated and closed states were refined by using two-fold symmetry restrained MD simulations (“ symmetry annealing”) according to a protocol we previously developed.45 During the symmetry annealing procedure all protein atoms were gradually driven towards the frequently (every 1 ps) updated two-fold symmetric average positions using harmonic restraints with spring constant slowly increasing from 0.001 to 10 kcal/mol/Å2. The rest of the system (lipids, water and ions) was unrestrained.
Simulation protocol.
The models were first subject to 30 ns of unrestrained MD simulations. During the unrestrained simulation stages all medium and protein atoms were free to move. Symmetry annealing was applied at different timesteps during the unrestrained simulation: after 5 ns, 10 ns, 15 ns and 20 ns (Fig. 5A) and each annealing step was followed by 10 ns unrestrained MD simulation to test the stability of the resulting model. Our previous experiments45 indicated that the accuracy of the model is well correlated with its stability, therefore we selected the most stable model (after annealing at 15 ns) and attempted further refinement by a second step of symmetry annealing.
Data analysis.
The size and shape of channels pore was estimated using the program HOLE.74
Protein secondary structure was estimated using the STRIDE algorithm75 embedded in VMD (http://www.ks.uiuc.edu/Research/vmd). RMSD relative to the starting structure, target RMSD structure, the symmetric average and RMSF were calculated using custom-written Tcl scripts in VMD.
Acknowledgements
This work was supported in part by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research. This study utilized the high-performance computational capabilities of the Biowulf Linux cluster at the National Institutes of Health (NIH), Bethesda, MD. The authors thank Drs. Stewart Durell, Yinon Shafrir, Sijung Yun (LCB, NCI) for fruitful discussions.
Abbreviations
- TM
transmembrane
- MD
molecular dynamics
- K2P
two-pore-domain background potassium
- P segment
pore-forming segment
- MSA
multiple sequence alignment
- MI
mutual information
Supplementary Material
References
- 1.Bayliss DA, Barrett PQ. Emerging roles for two-poredomain potassium channels and their potential therapeutic impact. Trends Pharmacol Sci. 2008;29:566–575. doi: 10.1016/j.tips.2008.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Honoré E. The neuronal background K2P channels: focus on TREK-1. Nat Rev Neurosci. 2007;8:251–261. doi: 10.1038/nrn2117. [DOI] [PubMed] [Google Scholar]
- 3.Rajan S, Wischmeyer E, Karschin C, Preisig-Müller R, Grzeschik KH, Daut J, et al. THIK-1 and THIK-2, a novel subfamily of tandem pore domain K+ channels. J Biol Chem. 2001;276:7302–7311. doi: 10.1074/jbc.M008985200. [DOI] [PubMed] [Google Scholar]
- 4.Lesage F, Lazdunski M. Molecular and functional properties of two-pore-domain potassium channels. Am J Physiol Renal Physiol. 2000;279:793–801. doi: 10.1152/ajprenal.2000.279.5.F793. [DOI] [PubMed] [Google Scholar]
- 5.Goldstein SA, Bockenhauer D, O'Kelly I, Zilberberg N. Potassium leak channels and the KCNK family of two-P-domain subunits. Nat Rev Neurosci. 2001;2:175–184. doi: 10.1038/35058574. [DOI] [PubMed] [Google Scholar]
- 6.Patel AJ, Honoré E. Properties and modulation of mammalian 2P domain K+ channels. Trends Neurosci. 2001;24:339–346. doi: 10.1016/s0166-2236(00)01810-5. [DOI] [PubMed] [Google Scholar]
- 7.O'Connell AD, Morton MJ, Hunter M. Two-pore domain K+ channels-molecular sensors. Biochim Biophys Acta. 2002;1566:152–161. doi: 10.1016/s0005-2736(02)00597-7. [DOI] [PubMed] [Google Scholar]
- 8.Lotshaw DP. Biophysical pharmacological and functional characteristics of cloned and native mammalian two-pore domain K+ channels. Cell Biochem Biophys. 2007;47:209–256. doi: 10.1007/s12013-007-0007-8. [DOI] [PubMed] [Google Scholar]
- 9.Bang H, Kim Y, Kim D. TREK-2, a new member of the mechanosensitive tandem-pore K+ channel family. J Biol Chem. 2000;275:17412–17419. doi: 10.1074/jbc.M000445200. [DOI] [PubMed] [Google Scholar]
- 10.Lesage F, Terrenoire C, Romey G, Lazdunski M. Human TREK2, a 2P domain mechanosensitive K+ channel with multiple regulations by polyunsaturated fatty acids, lysophospholipids and Gs, Gi and Gq protein-coupled receptors. J Biol Chem. 2000;275:28398–28405. doi: 10.1074/jbc.M002822200. [DOI] [PubMed] [Google Scholar]
- 11.Maingret F, Honoré E, Lazdunski M, Patel AJ. Molecular basis of the voltage-dependent gating of TREK-1, a mechanosensitive K+ channel. Biochem Biophys Res Commun. 2002;292:339–346. doi: 10.1006/bbrc.2002.6674. [DOI] [PubMed] [Google Scholar]
- 12.Patel AJ, Honoré E, Maingret F, Lesage F, Fink M, Duprat F, et al. A mammalian two pore domain mechano-gated S-like K+ channel. EMBO J. 1998;17:4283–4290. doi: 10.1093/emboj/17.15.4283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kang D, Choe C, Kim D. Thermosensitivity of the two-pore domain K+ channels TREK-2 and TRAAK. J Physiol. 2005;564:103–116. doi: 10.1113/jphysiol.2004.081059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maingret F, Lauritzen I, Patel AJ, Heurteaux C, Reyes R, Lesage F, et al. TREK-1 is a heat-activated background K+ channel. EMBO J. 2000;19:2483–2491. doi: 10.1093/emboj/19.11.2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maingret F, Patel AJ, Lesage F, Lazdunski M, Honoré E. Mechano- or acid stimulation, two interactive modes of activation of the TREK-1 potassium channel. J Biol Chem. 1999;274:26691–26696. doi: 10.1074/jbc.274.38.26691. [DOI] [PubMed] [Google Scholar]
- 16.Alloui A, Zimmermann K, Mamet J, Duprat F, Noël J, Chemin J, et al. TREK-1, a K+ channel involved in polymodal pain perception. EMBO J. 2006;25:2368–2376. doi: 10.1038/sj.emboj.7601116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chemin J, Patel A, Duprat F, Zanzouri M, Lazdunski M, Honoré E. Lysophosphatidic acid-operated K+ channels. J Biol Chem. 2005;280:4415–4421. doi: 10.1074/jbc.M408246200. [DOI] [PubMed] [Google Scholar]
- 18.Duprat F, Lesage F, Patel AJ, Fink M, Romey G, Lazdunski M. The neuroprotective agent riluzole activates the two P domain K+ channels TREK-1 and TRAAK. Mol Pharmacol. 2000;57:906–912. [PubMed] [Google Scholar]
- 19.Lauritzen I, Blondeau N, Heurteaux C, Widmann C, Romey G, Lazdunski M. Polyunsaturated fatty acids are potent neuroprotectors. EMBO J. 2000;19:1784–1793. doi: 10.1093/emboj/19.8.1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hervieu GJ, Cluderay JE, Gray CW, Green PJ, Ranson JL, Randall AD, et al. Distribution and expression of TREK-1, a two-pore-domain potassium channel, in the adult rat CNS. Neuroscience. 2001;103:899–919. doi: 10.1016/s0306-4522(01)00030-6. [DOI] [PubMed] [Google Scholar]
- 21.Medhurst AD, Rennie G, Chapman CG, Meadows H, Duckworth MD, Kelsell RE, et al. Distribution analysis of human two pore domain potassium channels in tissues of the central nervous system and periphery. Brain Res Mol Brain Res. 2001;86:101–114. doi: 10.1016/s0169-328x(00)00263-1. [DOI] [PubMed] [Google Scholar]
- 22.Talley EM, Solorzano G, Lei Q, Kim D, Bayliss DA. CNS distribution of members of the two-pore-domain (KCNK) potassium channel family. J Neurosci. 2001;21:7491–7505. doi: 10.1523/JNEUROSCI.21-19-07491.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Franks NP, Honoré E. The TREK K2P channels and their role in general anaesthesia and neuroprotection. Trends Pharmacol Sci. 2004;25:601–608. doi: 10.1016/j.tips.2004.09.003. [DOI] [PubMed] [Google Scholar]
- 24.Heurteaux C, Lucas G, Guy N, El Yacoubi M, Thümmler S, Peng XD, et al. Deletion of the background potassium channel TREK-1 results in a depression-resistant phenotype. Nat Neurosci. 2006;9:1134–1141. doi: 10.1038/nn1749. [DOI] [PubMed] [Google Scholar]
- 25.Zhao Y, Scheuer T, Catterall WA. Reversed voltagedependent gating of a bacterial sodium channel with proline substitutions in the S6 transmembrane segment. Proc Natl Acad Sci USA. 2004;101:17873–17878. doi: 10.1073/pnas.0408270101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cuello LG, Jogini V, Cortes DM, Perozo E. KcsA Potassium channel in the open-inactivated state with 32A opening at T112. (To be published) [Google Scholar]
- 27.Long SB, Campbell EB, Mackinnon R. Crystal structure of a mammalian voltage-dependent Shaker family K+ channel. Science. 2005;309:897–903. doi: 10.1126/science.1116269. [DOI] [PubMed] [Google Scholar]
- 28.Nimigean CM, Chappie JS, Miller C. Electrostatic tuning of ion conductance in potassium channels. Biochemistry. 2003;42:9263–9268. doi: 10.1021/bi0348720. [DOI] [PubMed] [Google Scholar]
- 29.Watanabe I, Zhu J, Sutachan JJ, Gottschalk A, Recio-Pinto E, Thornhill WB. The glycosylation state of KV1.2 potassium channels affects trafficking, gating and simulated action potentials. Brain Research. 2007;1144:1–18. doi: 10.1016/j.brainres.2007.01.092. [DOI] [PubMed] [Google Scholar]
- 30.Holm L, Rosenström P. Dali server: conservation mapping in 3D. Nucleic Acids Res. 2010;38:545–549. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Decker WK, Craigen WJ. The tissue-specific, alternatively spliced single ATG exon of the type 3 voltage-dependent anion channel gene does not create a truncated protein isoform in vivo. Mol Genet Metab. 2000;70:69–74. doi: 10.1006/mgme.2000.2987. [DOI] [PubMed] [Google Scholar]
- 32.Stopar D, Spruijt RB, Hemminga MA. Anchoring mechanisms of membrane-associated M13 major coat protein. Chem Phys Lipids. 2006;141:83–93. doi: 10.1016/j.chemphyslip.2006.02.023. [DOI] [PubMed] [Google Scholar]
- 33.Dunker AK, Jones TC. Proposed knobs-into-holes packing for several membrane proteins. Membr Biochem. 1978;2:1–16. doi: 10.3109/09687687809063855. [DOI] [PubMed] [Google Scholar]
- 34.Brown JH, Cohen C, Parry DA. Heptad breaks in alpha-helical coiled coils: stutters and stammers. Proteins. 1996;26:134–145. doi: 10.1002/(SICI)1097-0134(199610)26:2<134::AID-PROT3>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 35.Lee AG. Ca2+-ATPase structure in the E1 and E2 conformations: mechanism, helix-helix and helix-lipid interactions. Biochim Biophys Acta. 2002;1565:246–266. doi: 10.1016/s0005-2736(02)00573-4. [DOI] [PubMed] [Google Scholar]
- 36.Guy HR. Amino acid side-chain partition energies and distribution of residues in soluble proteins. Biophys J. 1985;47:61–70. doi: 10.1016/S0006-3495(85)83877-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fatakia SN, Costanzi S, Chow CC. Computing highly correlated positions using mutual information and graph theory for G protein-coupled receptors. PLoS One. 2009;4:4681. doi: 10.1371/journal.pone.0004681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sukharev S, Betanzos M, Chiang CS, Guy HR. The gating mechanism of the large mechanosensitive channel MscL. Nature. 2001;409:720–724. doi: 10.1038/35055559. [DOI] [PubMed] [Google Scholar]
- 39.Anishkin A, Gendel V, Sharifi NA, Chiang CS, Shirinian L, Guy HR, et al. On the conformation of the COOH-terminal domain of the large mechanosensitive channel MscL. J Gen Physiol. 2003;121:227–244. doi: 10.1085/jgp.20028768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Steinbacher S, Bass R, Strop P, Reesa DC. Structures of the prokaryotic mechanosensitive channels MscL and MscS. Current Topics in Membranes. 2007;58:1–24. [Google Scholar]
- 41.Xiang Z. Advances in homology protein structure modeling. Curr Protein Pept Sci. 2006;7:217–227. doi: 10.2174/138920306777452312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ginalski K. Comparative modeling for protein structure prediction. Curr Opin Struct Biol. 2006;16:172–177. doi: 10.1016/j.sbi.2006.02.003. [DOI] [PubMed] [Google Scholar]
- 43.Fan H, Mark AE. Refinement of homology-based protein structures by molecular dynamics simulation techniques. Protein Sci. 2004;13:211–220. doi: 10.1110/ps.03381404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen J, Brooks CL., III Can molecular dynamics simulations provide high-resolution refinement of protein structure? Proteins. 2007;67:922–930. doi: 10.1002/prot.21345. [DOI] [PubMed] [Google Scholar]
- 45.Anishkin A, Milac AL, Guy HR. Symmetry-restrained molecular dynamics simulations improve homology models of potassium channels. Proteins. 2010;78:932–949. doi: 10.1002/prot.22618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cordero-Morales JF, Cuello LG, Zhao Y, Jogini V, Cortes DM, Roux B, et al. Molecular determinants of gating at the potassium-channel selectivity filter. Nat Struct Mol Biol. 2006;13:311–318. doi: 10.1038/nsmb1069. [DOI] [PubMed] [Google Scholar]
- 47.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 48.Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pruitt KD, Tatusova T, Maglott DR. NCBI Reference Sequence (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2005;33:501–504. doi: 10.1093/nar/gki025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- 51.Shrivastava IH, Durell SR, Guy HR. A model of voltage gating developed using the KVAP channel crystal structure. Biophys J. 2004;87:2255–2270. doi: 10.1529/biophysj.104.040592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhou Y, Morais-Cabral JH, Kaufman A, Mackinnon R. Chemistry of ion coordination and hydration revealed by a K1 channel-Fab complex at 2.0A resolution. Nature. 2001;414:43–48. doi: 10.1038/35102009. [DOI] [PubMed] [Google Scholar]
- 53.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 54.Cole C, Barber JD, Barton GJ. The Jpred 3 secondary structure prediction server. Nucleic Acids Res. 2008;36:197–201. doi: 10.1093/nar/gkn238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pollastri G, McLysaght A. Porter: a new, accurate server for protein secondary structure prediction. Bioinformatics. 2005;21:1719–1720. doi: 10.1093/bioinformatics/bti203. [DOI] [PubMed] [Google Scholar]
- 56.Ouali M, King RD. Cascaded multiple classifiers for secondary structure prediction. Prot Sci. 2000;9:1162–1176. doi: 10.1110/ps.9.6.1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jones DT. Protein secondary structure prediction based on position-specific scoring matrices. J Mol Biol. 1999;292:195–202. doi: 10.1006/jmbi.1999.3091. [DOI] [PubMed] [Google Scholar]
- 58.Adamczak R, Porollo A, Meller J. Combining prediction of secondary structure and solvent accessibility in proteins. Proteins. 2005;59:467–475. doi: 10.1002/prot.20441. [DOI] [PubMed] [Google Scholar]
- 59.Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 60.Fiser A, Do RK, Sali A. Modeling of loops in protein structures. Protein Science. 2000;9:1753–1773. doi: 10.1110/ps.9.9.1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Khalili-Araghi F, Tajkhorshid E, Schulten K. Dynamics of K+ ion conduction through KV1.2. Biophys J. 2006;91:72–74. doi: 10.1529/biophysj.106.091926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of simple potential functions for simulating liquid water. J Chem Phys. 1983;79:926–935. [Google Scholar]
- 63.Gullingsrud J, Schulten K. Lipid bilayer pressure profiles and mechanosensitive channel gating. Biophys J. 2004;86:3496–3509. doi: 10.1529/biophysj.103.034322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kucerka N, Tristram-Nagle S, Nagle JF. Structure of fully hydrated fluid phase lipid bilayers with monoun-saturated chains. J Membr Biol. 2005;208:193–202. doi: 10.1007/s00232-005-7006-8. [DOI] [PubMed] [Google Scholar]
- 65.Humphrey W, Dalke A, Schulten K. VMD—Visual Molecular Dynamics. J Mol Graphics. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 66.Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, et al. Scalable molecular dynamics with NAMD. J Comput Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.MacKerell AD, Bashford D, Bellott M, Dunbrack RL, Evanseck JD, Field MJ, et al. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J Phys Chem B. 1998;102:3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 68.MacKerell AD. Empirical force fields for biological macromolecules: overview and issues. J Comput Chem. 2004;25:1584–1606. doi: 10.1002/jcc.20082. [DOI] [PubMed] [Google Scholar]
- 69.Freedberg DI, Venable RM, Rossi A, Bull TE, Pastor RW. Discriminating the helical forms of peptides by NMR and molecular dynamics simulation. J Am Chem Soc. 2004;126:10478–10484. doi: 10.1021/ja0484146. [DOI] [PubMed] [Google Scholar]
- 70.Buck M, Bouguet-Bonnet S, Pastor RW, MacKerell AD., Jr Importance of the CMAP correction to the CHARMM22 protein force field: dynamics of hen lysozyme. Biophys J. 2006;90:36–38. doi: 10.1529/biophysj.105.078154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Martyna GJ, Tobias DJ, Klein ML. Constant-pressure molecular-dynamics algorithms. J Chem Phys. 1994;101:4177–4189. [Google Scholar]
- 72.Feller SE, Zhang YH, Pastor RW, Brooks BR. Constant-pressure molecular-dynamics simulation—the Langevin piston method. J Chem Phys. 1995;103:4613–4621. [Google Scholar]
- 73.Darden T, York D, Pedersen L. Particle mesh Ewald—an N log(N) method for Ewald sums in large systems. J Chem Phys. 1993;98:10089–10092. [Google Scholar]
- 74.Smart OS, Goodfellow JM, Wallace BA. The Pore Dimensions of Gramicidin A. Biophys J. 993;65:2455–2460. doi: 10.1016/S0006-3495(93)81293-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Frishman D, Argos P. Knowledge-based secondary structure assignment. Proteins. 1995;23:566–579. doi: 10.1002/prot.340230412. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.