Abstract
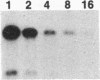
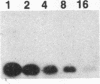
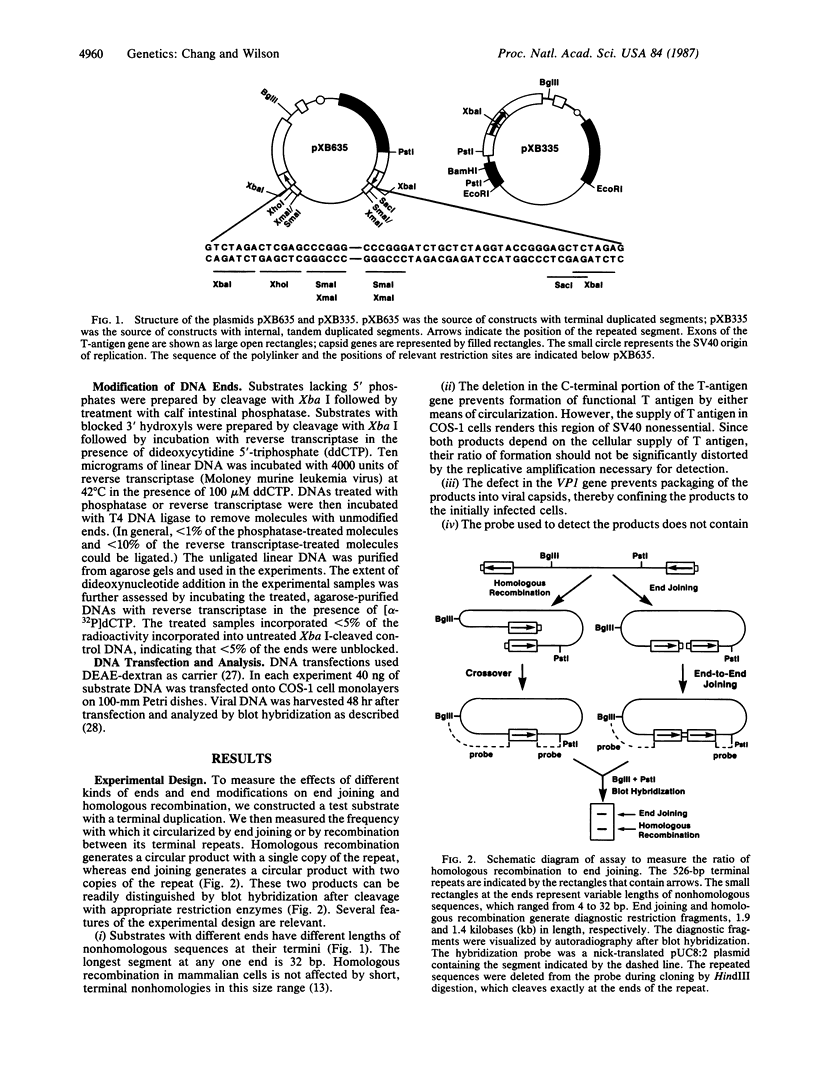
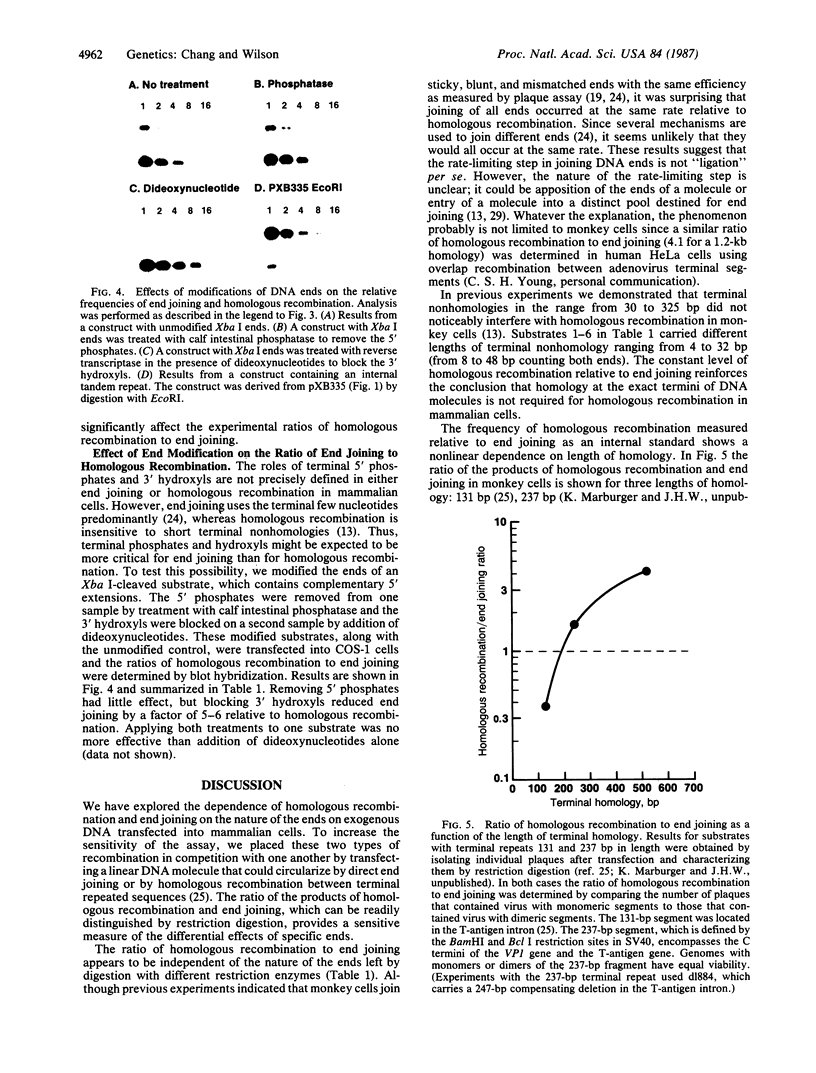
In animal cells, exogenous DNA recombines into random chromosomal sites much more frequently than it recombines into homologous sites. Free DNA ends are "recombinogenic" in both processes. To test the effects of specific ends on analogous extrachromosomal processes, we constructed a linear genome of simian virus 40 with terminal repeated sequences. After transfection into monkey cells, the model substrate can circularize by end joining (analogous to random integration) or by homologous recombination between its terminal repeats (analogous to targeted recombination). Since the two types of recombination are in competition with one another, the ratio of homologous-recombination to end-join products is a sensitive indicator of the differential effects of specific ends. Substrates with blunt ends, complementary sticky ends, or mismatched ends generated the same ratio of homologous-recombination to end-join products. However, addition of dideoxynucleotides to the 3' hydroxyls of the substrate decreased the frequency of end joining by a factor of 5-6 relative to homologous recombination. Thus, the frequency of end joining can be decreased relative to that of homologous recombination by modification of the ends of the input DNA. These results suggest an approach to altering the ratio of random to targeted integration in mammalian cells.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ayares D., Chekuri L., Song K. Y., Kucherlapati R. Sequence homology requirements for intermolecular recombination in mammalian cells. Proc Natl Acad Sci U S A. 1986 Jul;83(14):5199–5203. doi: 10.1073/pnas.83.14.5199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarti S., Joffe S., Seidman M. M. Recombination and deletion of sequences in shuttle vector plasmids in mammalian cells. Mol Cell Biol. 1985 Sep;5(9):2265–2271. doi: 10.1128/mcb.5.9.2265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang X. B., Wilson J. H. Formation of deletions after initiation of simian virus 40 replication: influence of packaging limit of the capsid. J Virol. 1986 May;58(2):393–401. doi: 10.1128/jvi.58.2.393-401.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folger K. R., Wong E. A., Wahl G., Capecchi M. R. Patterns of integration of DNA microinjected into cultured mammalian cells: evidence for homologous recombination between injected plasmid DNA molecules. Mol Cell Biol. 1982 Nov;2(11):1372–1387. doi: 10.1128/mcb.2.11.1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folger K., Thomas K., Capecchi M. R. Analysis of homologous recombination in cultured mammalian cells. Cold Spring Harb Symp Quant Biol. 1984;49:123–138. doi: 10.1101/sqb.1984.049.01.016. [DOI] [PubMed] [Google Scholar]
- Gluzman Y. SV40-transformed simian cells support the replication of early SV40 mutants. Cell. 1981 Jan;23(1):175–182. doi: 10.1016/0092-8674(81)90282-8. [DOI] [PubMed] [Google Scholar]
- Hasson J. F., Mougneau E., Cuzin F., Yaniv M. Simian virus 40 illegitimate recombination occurs near short direct repeats. J Mol Biol. 1984 Jul 25;177(1):53–68. doi: 10.1016/0022-2836(84)90057-3. [DOI] [PubMed] [Google Scholar]
- Hinnen A., Hicks J. B., Fink G. R. Transformation of yeast. Proc Natl Acad Sci U S A. 1978 Apr;75(4):1929–1933. doi: 10.1073/pnas.75.4.1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato S., Anderson R. A., Camerini-Otero R. D. Foreign DNA introduced by calcium phosphate is integrated into repetitive DNA elements of the mouse L cell genome. Mol Cell Biol. 1986 May;6(5):1787–1795. doi: 10.1128/mcb.6.5.1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopchick J. J., Stacey D. W. Differences in intracellular DNA ligation after microinjection and transfection. Mol Cell Biol. 1984 Feb;4(2):240–246. doi: 10.1128/mcb.4.2.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin F. L., Sperle K., Sternberg N. Homologous recombination in mouse L cells. Cold Spring Harb Symp Quant Biol. 1984;49:139–149. doi: 10.1101/sqb.1984.049.01.017. [DOI] [PubMed] [Google Scholar]
- Lin F. L., Sperle K., Sternberg N. Model for homologous recombination during transfer of DNA into mouse L cells: role for DNA ends in the recombination process. Mol Cell Biol. 1984 Jun;4(6):1020–1034. doi: 10.1128/mcb.4.6.1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin F. L., Sperle K., Sternberg N. Recombination in mouse L cells between DNA introduced into cells and homologous chromosomal sequences. Proc Natl Acad Sci U S A. 1985 Mar;82(5):1391–1395. doi: 10.1073/pnas.82.5.1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meselson M. S., Radding C. M. A general model for genetic recombination. Proc Natl Acad Sci U S A. 1975 Jan;72(1):358–361. doi: 10.1073/pnas.72.1.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller C. K., Temin H. M. High-efficiency ligation and recombination of DNA fragments by vertebrate cells. Science. 1983 May 6;220(4597):606–609. doi: 10.1126/science.6301012. [DOI] [PubMed] [Google Scholar]
- Roth D. B., Porter T. N., Wilson J. H. Mechanisms of nonhomologous recombination in mammalian cells. Mol Cell Biol. 1985 Oct;5(10):2599–2607. doi: 10.1128/mcb.5.10.2599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth D. B., Wilson J. H. Nonhomologous recombination in mammalian cells: role for short sequence homologies in the joining reaction. Mol Cell Biol. 1986 Dec;6(12):4295–4304. doi: 10.1128/mcb.6.12.4295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth D. B., Wilson J. H. Relative rates of homologous and nonhomologous recombination in transfected DNA. Proc Natl Acad Sci U S A. 1985 May;82(10):3355–3359. doi: 10.1073/pnas.82.10.3355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothstein R. J. One-step gene disruption in yeast. Methods Enzymol. 1983;101:202–211. doi: 10.1016/0076-6879(83)01015-0. [DOI] [PubMed] [Google Scholar]
- Rubnitz J., Subramani S. The minimum amount of homology required for homologous recombination in mammalian cells. Mol Cell Biol. 1984 Nov;4(11):2253–2258. doi: 10.1128/mcb.4.11.2253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherer S., Davis R. W. Replacement of chromosome segments with altered DNA sequences constructed in vitro. Proc Natl Acad Sci U S A. 1979 Oct;76(10):4951–4955. doi: 10.1073/pnas.76.10.4951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith A. J., Berg P. Homologous recombination between defective neo genes in mouse 3T6 cells. Cold Spring Harb Symp Quant Biol. 1984;49:171–181. doi: 10.1101/sqb.1984.049.01.020. [DOI] [PubMed] [Google Scholar]
- Smithies O., Gregg R. G., Boggs S. S., Koralewski M. A., Kucherlapati R. S. Insertion of DNA sequences into the human chromosomal beta-globin locus by homologous recombination. Nature. 1985 Sep 19;317(6034):230–234. doi: 10.1038/317230a0. [DOI] [PubMed] [Google Scholar]
- Smithies O., Koralewski M. A., Song K. Y., Kucherlapati R. S. Homologous recombination with DNA introduced into mammalian cells. Cold Spring Harb Symp Quant Biol. 1984;49:161–170. doi: 10.1101/sqb.1984.049.01.019. [DOI] [PubMed] [Google Scholar]
- Stringer J. R. DNA sequence homology and chromosomal deletion at a site of SV40 DNA integration. Nature. 1982 Mar 25;296(5855):363–366. doi: 10.1038/296363a0. [DOI] [PubMed] [Google Scholar]
- Szostak J. W., Orr-Weaver T. L., Rothstein R. J., Stahl F. W. The double-strand-break repair model for recombination. Cell. 1983 May;33(1):25–35. doi: 10.1016/0092-8674(83)90331-8. [DOI] [PubMed] [Google Scholar]
- Thomas K. R., Folger K. R., Capecchi M. R. High frequency targeting of genes to specific sites in the mammalian genome. Cell. 1986 Feb 14;44(3):419–428. doi: 10.1016/0092-8674(86)90463-0. [DOI] [PubMed] [Google Scholar]
- Wake C. T., Gudewicz T., Porter T., White A., Wilson J. H. How damaged is the biologically active subpopulation of transfected DNA? Mol Cell Biol. 1984 Mar;4(3):387–398. doi: 10.1128/mcb.4.3.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wake C. T., Vernaleone F., Wilson J. H. Topological requirements for homologous recombination among DNA molecules transfected into mammalian cells. Mol Cell Biol. 1985 Aug;5(8):2080–2089. doi: 10.1128/mcb.5.8.2080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson J. H., Berget P. B., Pipas J. M. Somatic cells efficiently join unrelated DNA segments end-to-end. Mol Cell Biol. 1982 Oct;2(10):1258–1269. doi: 10.1128/mcb.2.10.1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson J. H. Genetic analysis of host range mutant viruses suggests an uncoating defect in simian virus 40-resistant monkey cells. Proc Natl Acad Sci U S A. 1977 Aug;74(8):3503–3507. doi: 10.1073/pnas.74.8.3503. [DOI] [PMC free article] [PubMed] [Google Scholar]