Abstract
Bacterial type III secretion systems deliver protein virulence factors to host cells. Here we characterize the interaction between HrpB2, a small protein secreted by the Xanthomonas citri subsp. citri type III secretion system, and the cytosolic domain of the inner membrane protein HrcU, a paralog of the flagellar protein FlhB. We show that a recombinant fragment corresponding to the C-terminal cytosolic domain of HrcU produced in E. coli suffers cleavage within a conserved Asn264-Pro265-Thr266-His267 (NPTH) sequence. A recombinant HrcU cytosolic domain with N264A, P265A, T266A mutations at the cleavage site (HrcUAAAH) was not cleaved and interacted with HrpB2. Furthermore, a polypeptide corresponding to the sequence following the NPTH cleavage site also interacted with HrpB2 indicating that the site for interaction is located after the NPTH site. Non-polar deletion mutants of the hrcU and hrpB2 genes resulted in a total loss of pathogenicity in susceptible citrus plants and disease symptoms could be recovered by expression of HrpB2 and HrcU from extrachromossomal plasmids. Complementation of the ΔhrcU mutant with HrcUAAAH produced canker lesions similar to those observed when complemented with wild-type HrcU. HrpB2 secretion however, was significantly reduced in the ΔhrcU mutant complemented with HrcUAAAH, suggesting that an intact and cleavable NPTH site in HrcU is necessary for total functionally of T3SS in X. citri subsp. citri. Complementation of the ΔhrpB2 X. citri subsp. citri strain with a series of hrpB2 gene mutants revealed that the highly conserved HrpB2 C-terminus is essential for T3SS-dependent development of citrus canker symptoms in planta.
Introduction
Many Gram-negative bacterial pathogens produce proteinaceous pathogenic factors that are secreted and injected into the host cell via the type III secretion system (T3SS) during the infective process [1], [2], [3]. A great deal of focus has been aimed at understanding the T3SS of phytopathogenic Xanthomonas species that infect a wide variety of plant hosts, many of which are of great economic importance [1], [4], [5], [6], [7], [8], [9], [10], [11], [12], [13], [14], [15]. The phytopathogen Xanthomonas citri subsp. citri (Xanthomonas axonopodis pv citri strain 306; Xac) is the causal agent of citrus canker, a disease that threatens citrus crops world-wide [16]. The Xac hrp locus (hrp: “hypersensitive response and pathogenicity”) encompasses a group of 25 genes that code for a T3SS. Some products encoded by these genes are conserved in all T3SS, including core flagellar secretory components, while others are proteins of unknown function but whose homologs are essential for T3SS function in other Xanthomonas species [5], [7], [17].
The Xac T3SS is required for the development of disease symptoms in susceptible citrus plants as well as for the hypersensitive response (HR) in resistant plants [17], [18]. Deletions in the hrpB and hrpD operons and deletions of the hrpF gene in Xac failed to produce canker in citrus plants or hypersensitive response (HR) in cotton [17]. Furthermore, a specific T3SS substrate, PthA (a member of the AvrBs3 family), has been shown to contribute significantly to T3SS-dependent development of disease symptoms by Xac in citrus and the introduction of the pthA gene into strains of X. phaseoli and X. campestris pv. malvacearum (neither pathogenic in citrus) resulted in the elicitation of HR in their respective hosts, bean and cotton [18]. A PthA homolog coded by the hssB3.0 gene was found to be required for virulence of Xac KC21 on Citrus grandis cultivars [19]. Other possible T3SS-related factors have been identified in the Xac genome by bioinformatics analysis [7] but have not been studied at the genetic or protein level.
We have previously identified protein-protein interactions involving components, substrates and regulators of the T3SS of Xac strain 306 [5] whose genome has been sequenced [7]. One of the interactions identified was that involving HrpB2 and HrcU. HrpB2 is a small protein found associated with the T3SS of only a few phytopathogenic bacteria (Xanthomonas spp., Ralstonia solanacearum, Acidovorax avenae) and of Burkholderia spp that can infect animals and plants. In Xanthomonas campestris pv. vesicatoria (Xcv), HrpB2 is secreted and is essential for the secretion of the AvrBs3 virulence protein by the T3SS [20] and has been shown to interact with HpaC, a protein required for the efficient secretion of other effectors proteins [21]. These observations have led to the suggestion that HrpB2 may play a role in controlling the hierarchy of a stepwise secretion process [20], [21].
HrcU homologs are found in all known T3SSs and flagellar systems and are made up of an N-terminal domain containing several transmembrane helices and a cytoplasmic C-terminal domain. In Xanthomonas campestris pv. glycines 8ra, the HrcU homolog is not required for HR induction on non-host plants, pepper and tomato, or for the multiplication of bacteria in the host plant, but was required for the pathogenic symptoms on soybean [22]. On the other hand, insertion mutagenesis in the Xcv hrpC operon, which codes for both HrcU and HrcV, resulted in nonpathogenic mutants that exhibited significantly reduced growth in pepper leaves and lost the ability to induce HR in resistant host plants and in non-hosts [23].
HrcU is a paralog of the flagellar protein FlhB. The 173-residue C-terminal domain of FlhB from Salmonella is specifically cleaved between Asn-269 and Pro-270 within a NPTH motif [24] via an autocatalytic process [25]. This NPTH motif is conserved in all FlhB homologs, including those found in T3SS of animal and plant pathogens and a similar cleavage has been observed in the homolog YscU from the T3SS of Yersinia pseudotuberculosis [26]. In flagellar systems, mutations that abolish cleavage in FlhB also abolish the secretion of flagellin and other late export extracellular components but not early export proteins such as FlgD [27]. Cleavage of YscU does not however seem to be essential for the secretion of virulence factors by the Yersinia T3SS [26] and thus appears to discriminate between translocator and effector proteins [28]. Substitutions of N263 abolish autocleavage of YscU while P264 and H266 showed partial cleavage [29]. Structural studies of YcsU [29] and its homologs EscU from enteropatogenic E. coli, SpaS from Salmonella typhimurium [30] and Spa40 from Shigella flexneri [31] reported similar structural and functional data.
In this report, we have characterized the interaction between HrpB2 and the C-terminal domain of HrcU of Xac using purified recombinant proteins. We show that when expressed in E. coli, HrcUXAC suffers a cleavage within the NPTH motif in a manner similar to that observed for the HrcU homologs FlhB and YscU and that the HrpB2XAC binding site on HrcUXAC corresponds to the region C-terminal to the cleavage site. Deletion mutations in the hrcU and hrpB2 genes (ΔhrcU and ΔhrpB2) resulted in a total loss of virulence in planta and pathogenicity could be regained by the expression of HrcUXAC and HrpB2XAC from extrachromosomal plasmids. Furthermore, citrus canker symptoms could be observed in infections of the ΔhrcU mutant expressing a HrcUXAC variant in which the NPTH site has was abolished. We also show that HrpB2XAC is secreted in a manner that depends on HrcUXAC but is only partly dependent on HrcUXAC cleavage. Expression of HrpB2XAC variants in a ΔhrpB2 background showed that the last seven amino acids are essential for HrpB2XAC function in the development of canker disease symptoms.
Results
Expression of the cytosolic domain of HrcUXAC (HrcUXAC_207–357) in E. coli produces a 7 kDa polypeptide
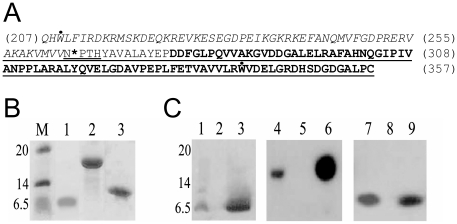
The C-terminal domain of HrcU corresponding to residues 207–357 (HrcUXAC_207–357, sequence shown in Fig. 1A) was expressed in E. coli BL21(DE3) cells. The expression of the recombinant protein was expected to produce a 158 residue, 17 kDa polypeptide. However, SDS-PAGE analysis failed to detect a 17 kDa fragment but instead a 7 kDa fragment appeared in Coomassie-stained gels after induction with IPTG (data not shown). This fragment was subsequently purified (Fig. 1B, lane 1). This result was obtained after expression in a variety of different E. coli strains including BL21(DE3), BL21(DE3)RP, BL21(DE3)RIL, BL21(DE3)pLysS, BL21(DE3)CY, BL21(DE3)SI and BL21(DE3)Star (data not shown).
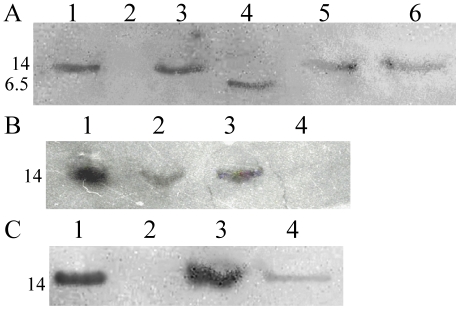
Figure 1. Expression of HrcUXAC C-terminal fragments.
A) Primary sequence of the C-terminal domain (residues 207-357) of HrcUXAC. Residues 207-264 are in italic and residues 277-357 are shown in bold. The underlined sequence was shown to interact with HrpB2XAC in yeast two-hybrid assays [5]. In HrcUXAC_207-264 and HrcUXAC_207-357(AAAH), residues Q207 and H208 were replaced with Met and Asp residues respectively. The highly conserved NPTH sequence is double-underlined and the cleavage site between N264 and P265 is indicated with an asterisk. The two tryptophan (W209 and W340) residues are indicated with a dot above their letter symbols. B) Coomassie-stained SDS-PAGE of purified recombinant HrcU fragments. Purified HrcUXAC_208-264 (lane 1), HrcUXAC_207-357AAAH (lane 2) and HrcUXAC_His277-357 (lane 3). Molecular mass markers (M) are shown on the left with masses in kDa. C) Western blots of purified HrcUXAC fragments (lanes 3, 6, 9) and of E. coli cell lysates before (lanes 2, 5, 8) and after (lanes 1, 4, 7) expression using the polyclonal antiserum raised against HrcUXAC_207-357AAAH. HrcUXAC_207-357 (lanes 1-3), HrcUXAC_207-357AAAH (lanes 4-6), HrcUXAC_His277-357 (lanes 7-9).
Mutation of the conserved NPTH site results in the production of a full-length 17 kDa polypeptide
HrcU orthologs and paralogs all possess a conserved Asn-Pro-Thr-His (NPTH) sequence (residues 264–267 in HrcUXAC, Fig. 1A) which has been shown to be a site of auto-cleavage in the flagellar protein FlhB [24], [25]. To test the hypothesis that a similar cleavage was occurring in HrcUXAC_207–357, we mutated residues 264–266 to alanine and expressed the polypeptide in E. coli. As shown in Figure 1B (lane 2), expression and purification of HrcUXAC_207–357(AAAH) produced a protein of the expected size (17 kDa).
To test whether the 7 kDa fragment was in fact derived from HrcUXAC_207–357 we used purified HrcUXAC_207–357(AAAH) to obtain polyclonal antiserum against the HrcUXAC C-terminal domain. Western blot assays against lysates of E. coli cultures obtained before and after IPTG-induced expression of HrcUXAC_207–357 and HrcUXAC_207–357(AAAH) showed that the antibody recognizes the 17 kDa HrcUXAC_207–357(AAAH) fragment (Fig. 1C, lanes 4 and 5) as well as the 7 kDa fragment (Fig. 1C, lanes 1 and 2). The antibody also recognized the purified 7 kDa fragment (Fig. 1C, lane 3). These results indicate that the purified 7 kDa fragment obtained after HrcU207–357 expression is in fact derived from HrcUXAC.
N-terminal sequencing by Edman degradation of the 7 kDa fragment was consistent with the N-terminus beginning at position 207 (XXXLFIRDKR), indicating that the initiation Met residue was indeed retained. The mass of the purified 7 kDa fragment determined by MALDI-ToF analysis was very close to the mass expected from the N-terminal fragment (6911 Da with retention of the initiation methionine) produced from cleavage between residues Ans264 and Pro265 of the NPTH sequence within HrcUXAC_207–357. The above results thus allow us to designate the name HrcUXAC_207–264 to the 7 kDa polypeptide that was detected and purified after expression of HrcU207–357.
Cleavage of HrcUXAC_207–357 between residues Asn264 and Pro265 would be expected to produce two fragments, one N-terminal fragment beginning at residue 207 and ending at residue 264 (6911 Da) and one C-terminal fragment corresponding to residues 265–357 (9931 Da). As mentioned above, only a 7 kDa fragment could be observed to be induced in Coomassie-stained gels (data not shown). While no 10 kDa fragment was observed to be induced in Coomassie-stained gels, a faint band could be observed above the 7 kDa band in the Western blot of E. coli lysates after induction of HrcU207–357 expression with IPTG (Fig. 1C, lane 1). Therefore, the evidence so far is consistent with the cleavage at residue 264 possibly followed by a degradation of a significant fraction of the 10 kDa fragment in E. coli.
Interactions between HrpB2XAC and fragments derived from the cytosolic C-terminal domain of HrcUXAC
We have previously shown that HrpB2XAC interacts with fragments derived from the C-terminal domain of HrcUXAC in yeast two-hybrid assays [5]. In that study, the smallest HrcUXAC fragment observed to interact corresponded to residues 256 to 357 (underlined sequence in Fig. 1A). It was therefore not clear whether HrcUXAC sequences before or after the conserved NPTH site (or both) were necessary for interaction with HrpB2. We therefore expressed and purified recombinant HrpB2XAC to perform in vitro interaction assays with HrcUXAC_207–264 and HrcUXAC_207–357(AAAH). We also expressed and purified an HrcUXAC fragment corresponding to residues 277–357 with an N-terminal His-tag fusion (HrcUXAC_His277–357) (Figure 1B, lane 3). This fragment is recognized by polyclonal anti-HrcUXAC antibodies both in E. coli lysates and after purification (Figure 1C, lanes 7, 8 and 9) and its estimated mass determined by MALDI-ToF spectrometry corresponds well with the expected mass of a fragment in which the initiation methionine has been retained (data not shown).
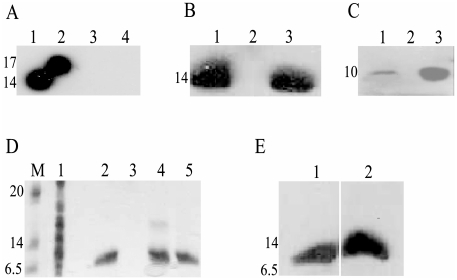
Figure 2A shows the results of Far-Western blot analysis of the interaction between HrpB2XAC and HrcUXAC_207–357(AAAH) using polyclonal antibodies raised against HrpB2XAC. HrpB2XAC bound to immobilized HrcUXAC_207–357(AAAH) (lane 2) but not to an immobilized recombinant C-terminal chicken α-tropomyosin fragment used as a negative control (lane 3). Similar experiments using immobilized HrcUXAC_207–264 failed to detect an interaction (data not shown). Figure 2B shows that Far-Western assays using immobilized cell lysates obtained before (lane 2) and after induction (lane 1) of HrpB2XAC expression as well as purified HrpB2XAC (lane 3). After incubation of the membranes with HrcUXAC_207–357(AAAH), bound HrcUXAC_207–357(AAAH) could be detected with polyclonal anti-HrcUXAC antibodies. Again, no interactions could be detected in similar experiments in which membranes were incubated with HrcUXAC_207-264 (data not shown).
Figure 2. Interaction of HrpB2XAC with HrcUXAC_207-357AAAH and with HrcUXAC_His277-357.
A) Far-Western blot assays demonstrating the HrpB2XAC interaction with immobilized HrcUXAC_207-357AAAH. The following purified proteins were separated by SDS-PAGE and transferred to a nitrocellulose membrane: Lane 1: HrpB2XAC. Lanes 2 and 4: HrcUXAC_207-357AAAH. Lane 3: chicken muscle tropomyosin fragment Tm143-284 [60]. Nitrocellulose strips corresponding to lanes 2 and 3 were incubated with HrpB2 followed by washing to remove unbound proteins. Nitrocellulose strips corresponding to lanes 1 to 4 were then incubated with polyclonal antiserum raised against HrpB2XAC. The strips were rejoined and revealed using anti-mouse IgG conjugated with horseradish peroxidase. B) Far-Western Blot assays demonstrating the HrcUXAC_207-357AAAH interaction with immobilized HrpB2XAC. The following samples were separated by SDS-PAGE and transferred to a nitrocellulose membrane: Lysates of E. coli cells after (lane 1) and before (lane 2) expression of HrpB2XAC and purified HrpB2XAC (lane 3). The nitrocelulose membrane was incubated with HrcUXAC_207-357AAAH following by incubation with polyclonal antiserum raised against HrcUXAC_207-357AAAH and revealed using protein A conjugated with horseradish peroxidase. C) Far-Western Blot assays demonstrating the HrpB2XAC interaction with immobilized HrcUXAC_His277-357. The following samples were separated by SDS-PAGE and transferred to a nitrocellulose membrane: Lysates of E. coli cells after (lane 1) and before (lane 2) expression of HrcUXAC_His277-357 and purified HrcUXAC_His277-357 (lane 3). The nitrocellulose membrane was incubated with HrpB2 following by incubation with polyclonal antiserum raised against HrpB2XAC and revealed as described in part A. D) Pull-down assay demonstrating the interaction of HrpB2XAC with HrcUXAC_His277-357 immobilized on Ni2+-chelating resin. HrcUXAC_His277-357 (lane 2), HrpB2XAC (lane 3), a HrpB2XAC plus HrcUXAC_His277-357 mixture (lane 4) and a mixture of HrcUXAC_His277-357 with an E. coli BL21(DE3) cell lysate (lane 5) were applied to a Ni2+-chelating resin, washed with buffer containing 25 mM imidazole and bound proteins were eluted by washing with 500 mM imidazole. Eluted proteins were separated by SDS-PAGE and visualized by Coomassie brilliant blue staining. Lane 1 shows the contents of the E. coli lysate employed in lane 5. Molecular mass markers (M) are shown to the left in kilodaltons. E) Since HrpB2XAC and HrcUXAC_His277-357 are not easily separated by SDS-PAGE, the presence of both HrpB2XAC and HrcUXAC_His277-357 in the bound fraction shown in lane 4 of Figure 2D was demonstrated by Western blot using polyclonal antisera raised against HrcUXAC_207-357AAAH (lane 1) and against HrpB2XAC (lane 2). The masses of molecular weight markers (in kDa) are indicated to the left of parts A-E.
HrcUXAC_His277–357 corresponds to a fragment that begins 10 residues after the conserved NPTH site. Binding of HrcUXAC_His277–357 to HrpB2XAC was demonstrated in Far-Western experiments using E. coli lysates obtained after induction of expression of HrpB2XAC as well as purified HrpB2XAC. These samples were submitted to SDS-PAGE, transferred to nitrocellulose membranes, overlayed with HrcUXAC_His277–357 and bound HrcUXAC_His277–357 was detected using anti-HrcUXAC antibodies (Fig. 2C). This interaction was further demonstrated by immobilizing HrcUXAC_His277–357 on a Ni2+-chelating resin and testing whether it could retain HrpB2XAC (Fig. 2D and 2E). While purified HrpB2XAC did not interact with the Ni2+-chelating resin on its own (Fig. 2D, lane 3), it was retained by HrcUXAC_His277–357 bound to the column (Fig. 2D, lane 4 and Fig. 2E, lane 2). Since HrpB2XAC (14 kDa) and HrcUXAC_His277–357 (10 kDa) have similar mobility in SDS-PAGE, we detected the individual components of the complex using HrcUXAC-specific and HrpB2XAC-specific antisera (Fig. 2E, lanes 1 and 2 respectively).
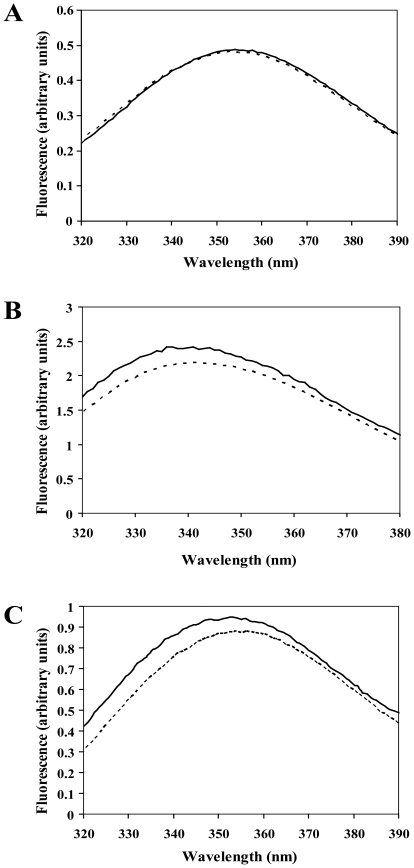
The specific interaction between HrpB2XAC and the region C-terminal to the HrcUXAC NPTH site was further demonstrated in fluorescence perturbation assays. The HrpB2XAC protein does not possess any tryptophan residues. On the other hand, the C-terminal cytosolic domain of HrcUXAC has two tryptophans, one at position 209, before the NPTH site, and the other at position 340, after the NPTH site (Fig. 1A). We therefore used the intrinsic fluorescence of purified HrcUXAC_207–357(AAAH), HrcUXAC_207–264 and HrcUXAC_His277–357 as probes to detect interactions with HrpB2. Figure 3 shows that the fluorescence of HrcUXAC_207–357(AAAH) and HrcUXAC_His277–357 is perturbed by the addition of HrpB2XAC (Fig. 3B and 3C) while the fluorescence of HrcUXAC_207–264 remains unchanged (Fig. 3A). The addition of HrpB2 caused slight blue-shifts in the emission spectra of both HrcUXAC_207–357(AAAH) and HrcUXAC_His277–357 as well as a small increase in intensity. These results confirm that the site of HrpB2XAC interaction on HrcUXAC corresponds to the sequence C-terminal to the NPTH cleavage site.
Figure 3. HrpB2XAC induced changes in HrcUXAC fluorescence.
Fluorescence emission spectra of HrcUXAC_207-264 (A), HrcUXAC_207-357AAAH (B) and HrcUXAC_His277-357 (C) in the absence (dotted lines) and presence (solid lines) of HrpB2. All proteins (2 µM) were dissolved in 5 mM sodium acetate (pH 6.0). Spectra were recorded at 25°C using an excitation wavelength of 280 nm.
The NPTH cleavage site is not required for the development of canker symptoms
To study the contribution of HrcUXAC and its NPTH site to Xac pathogenicity we employed an allelic exchange protocol to the produce the ΔhrcU Xac strain containing in-frame deletions of hrcU codons 14-347 (Table 1). We also produced plasmids containing the hrcU open reading frame plus 1 kb upstream sequences that contain the promoter region (pUFR047_hrcU; Table 2 ). Furthermore, we introduced mutations in this plasmid that change the NPTH site to AAAH (pUFR047_hrcUAAAH).
Table 1. Strains used in this study.
| Strains | Relevant characteristics | Source |
| Bacterial Strains: | ||
| E. coli DH10B | Recipient for cloning experiments | [56] |
| E. coli BL21(DE3) | IPTG-inducible T7 RNA polymerase | [58] |
| E. coli BL21(DE3) (RIL) | IPTG-inducible T7 RNA polymerase | [59] |
| Xac strain 306 | Template for PCR-based cloning | [7] |
| Xac ΔhrcU | Xac strain carrying deletion of hrcU gene (codons 14–347) | This study |
| Xac ΔhrpB2 | Xac strain carrying deletion of hrpB2 gene (codons 10–119) | This study |
| Xac ΔhrcU+pUFR047 _hrcU | Xac ΔhrcU carrying pUFR047_hrcU | This study |
| Xac ΔhrcU+pUFR047_hrcUAAAH | Xac ΔhrcU carrying pUFR047_hrcUAAAH | This study |
| Xac ΔhrpB2+pUFR047_hrpB2 | Xac ΔhrpB2 carrying pUFR047_hrpB2 | This study |
| Xac ΔhrpB2+pUFR047_hrpB21-56 | Xac ΔhrpB2 carrying pUFR047_hrpB21-56 | This study |
| Xac ΔhrpB2+pUFR047_hrpB21-123 | Xac ΔhrpB2 carrying pUFR047_hrpB21-123 | This study |
| Xac ΔhrpB2+pUFR047_hrpB2LQGPR | Xac ΔhrpB2 carrying pUFR047_hrpB2LQGPR | This study |
| Xac ΔhrpB2+pUFR047_hrpB2T125A | Xac ΔhrpB2 carrying pUFR047_hrpB2T125A | This study |
| Xac ΔhrpB2+pUFR047_hrpB2L126A | Xac ΔhrpB2 carrying pUFR047_hrpB2L126A | This study |
| Xac ΔhrpB2+pUFR047_hrpB2V127A | Xac ΔhrpB2 carrying pUFR047_hrpB2V127A | This study |
| Xac ΔhrpB2+pUFR047_hrpB2K128A | Xac ΔhrpB2 carrying pUFR047_hrpB2K128A | This study |
| Xac ΔhrpB2+pUFR047_hrpB2N129A | Xac ΔhrpB2 carrying pUFR047_hrpB2N129A | This study |
| Xac ΔhrpB2+pUFR047_hrpB2Q130A | Xac ΔhrpB2 carrying pUFR047_hrpB2Q130A | This study |
*See Table 2 for plasmid construction details.
Table 2. Plasmids used in this study.
| Plasmids | Relevant characteristics | Source |
| pET-11d | T7 RNA polymerase - based expression vector | [57] |
| pET-3a | T7 RNA polymerase - based expression vector | [57] |
| pET-28a (+) | T7 RNA polymerase - based expression vector | Novagen |
| pU1 | pET-11d based vector expressing HrcUXAC_207-357 | This study |
| pU2 | pET-11d based vector expressing HrcUXAC_207-357(AAAH) | This study |
| pU3 | pET-28a(+) based vector expressing HrcUXAC_His277-357 | This study |
| pB2 | pET-3a based vector expressing HrpB2XAC | This study |
| pET-Tmy143-284 | pET-3a based vector expressing chicken alpha tropomyosin | [60] |
| pNPTS138 | Suicide vector, Kmr/SacB | Dickon Alley* |
| pUFR047 | Wide host range vector, Gmr | [63] |
| pBBR1MCS-5 | Wide host range vector, Gmr | [62] |
| pNPTS138_ΔhrcU | Suicide vector carrying internal truncation of Xac hrcU gene | This study |
| pNPTS138_ΔhrpB2 | Suicide vector carrying internal truncation of Xac hrpB2 gene | This study |
| pBBR_hrcU | pBBR1MCS-5 vector carrying Xac hrcU gene | This study |
| pBBR_hrcUAAAH | pBBR1MCS-5 vector carrying Xac hrcU gene with mutations that change NPTH motif to AAAH | This study |
| pUFR047_hrcU | pUFR047 based vector for expression of HrcUXAC in Xac | This study |
| pUFR047_hrcUAAAH | pUFR047 based vector for expression of HrcUXAC_AAAH in Xac | This study |
| pBBR_hrpB2 | pBBR1MCS-5 vector carrying Xac hrpB2 gene | This study |
| pUFR047_hrpB2 | pUFR047 based vector for expression of HrpB2XAC in Xac | This study |
| pUFR047_hrpB21 -56 | pUFR047 based vector for expression of HrpB2XAC_1-56 in Xac | This study |
| pUFR047_hrpB21 -123 | pUFR047 based vector for expression of HrpB2XAC_1-123 in Xac | This study |
| pUFR047_hrpB2LQGPR | pUFR047 based vector for expression of HrpB2XAC_LQGPR in Xac | This study |
| pUFR047_hrpB2T125A | pUFR047 based vector for expression of HrpB2XAC_T125A in Xac | This study |
| pUFR047_hrpB2L126A | pUFR047 based vector for expression of HrpB2XAC_L126A in Xac | This study |
| pUFR047_hrpB2V127A | pUFR047 based vector for expression of HrpB2XAC_V127A in Xac | This study |
| pUFR047_hrpB2K128A | pUFR047 based vector for expression of HrpB2XAC_K128A in Xac | This study |
| pUFR047_hrpB2N129A | pUFR047 based vector for expression of HrpB2XAC_N129A in Xac | This study |
| pUFR047_hrpB2Q306A | pUFR047 based vector for expression of HrpB2XAC_Q130A in Xac | This study |
*unpublished.
Figure 4A shows the results of inoculation of sweet orange leaf tissue with the Xac wild-type, ΔhrcU, ΔhrcU+pUFR047_hrcU and ΔhrcU+pUFR047_hrcUAAAH strains 15 days after infection. While infection with the wild-type strain showed clear disease symptoms including water-soaking, hyperplasy and necrosis, the ΔhrcU strain failed to produce any disease symptoms in the susceptible citrus host. This result is consistent with the absolute requirement for HrcU homologs for the functioning of all T3SS systems [22], [32]. The phenotype of the ΔhrcU strain could be reverted by the expression of wild-type HrcU coded by the pUFR047_hrcU plasmid or by expression of the HrcUXAC_AAAH coded by the pUFR047_hrcUAAAH plasmid (Fig. 4A). In both cases, canker symptoms were less severe than those observed using the wild-type strain. It is not clear why the reversion of disease symptoms was attenuated in these experiments. We note that the native upstream promoter regions contained within these plasmids contain PIP (plant-inducible promoter) boxes [7] that have been shown to be recognized by the HrpX transcription factor that controls hrp expression in Xcv [33], [34], [35].
Figure 4. HrcU and HrpB2 contribute to Xac pathogenicity during infection of Citrus sinensis.
Macroscopic symptoms 15 days after inoculation on the abaxial surface of leafs with ΔhrcU (A) and ΔhrpB2 (B-D) mutants. The following strains were used: Xac wild-type (WT), ΔhrcU, ΔhrcU+pUFR047_hrcU (ΔhrcU+hrcU), ΔhrcU+pUFR047_hrcUAAAH (ΔhrcU+hrcUAAAH), ΔhrpB2, ΔhrpB2+pUFR047_hrpB2 (ΔhrpB2+hrpB2), ΔhrpB2+pUFR047_hrpB21-56 (ΔhrpB2+hrpB21-56), ΔhrpB2+pUFR047_hrpB21-123 (ΔhrpB2+hrpB21-123), ΔhrpB2+pUFR047_hrpB2LQGPR (ΔhrpB2+hrpB2LQGPR), ΔhrpB2+pUFR047_hrpB2T125A (ΔhrpB2+hrpB2T125A), ΔhrpB2+pUFR047_hrpB2L126A (ΔhrpB2+hrpB2L126A), ΔhrpB2+pUFR047_hrpB2V127A (ΔhrpB2+hrpB2V127A), ΔhrpB2+pUFR047_hrpB2K128A (ΔhrpB2+hrpB2K128A), ΔhrpB2+pUFR047_hrpB2N129A (ΔhrpB2+hrpB2N129A) and ΔhrpB2+pUFR047_hrpB2L130A (ΔhrpB2+hrpB2L130A).
The HrpB2XAC C-terminal region is required to elicit citrus canker symptoms
To study the contribution of HrpB2XAC to Xac pathogenicity, the allelic exchange protocol was used to produce the ΔhrpB2 strain with an in-frame deletion of hrpB2 codons 10-119 (Table 1). We also produced plasmid pUFR047_hrpB2 (Table 2) which codes for the wild-type HrpB2XAC protein plus a 1 kb upstream region that includes the hrpB1 gene between the promoter and hrpB2. Figure 4B shows that the ΔhrpB2 strain was unable to elicit disease symptoms and that the virulence of the mutant strain was fully restored by transformation with pUFR047_hrpB2.
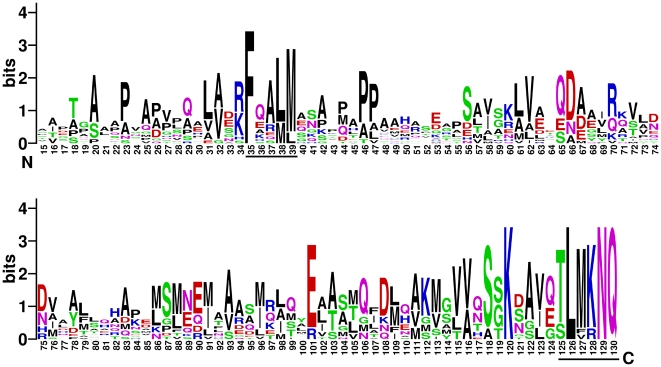
Multiple sequence alignment analysis of HrpB2 proteins from Xanthomonas, Burkholderia, Acidovorax and Ralstonia species (Figure 5) indicates that there are two regions of sequence conservation in an otherwise variable protein family: i) a five residue motif which we name FQALM that corresponds to positions 35–39 of HrpB2XAC and ii) the last six amino acids of the protein (HrpB2XAC residues 125–130) which we name the TLMKNQ motif (in Xac the methionine residue is substituted with a valine).
Figure 5. Graphical Representation of the multiple sequence alignment of the HrpB2 protein family.
The Pfam database [64] lists 61 sequences in this group (PF09487) from Xanthomonas (13 sequences), Burkholderia (43 sequences), Ralstonia (3 sequences), and Acidovorax (2 sequences) species. However, after removal of all sequences with greater that 95% identity, only 16 remain. These 16 sequences were used to generate this representation using the WebLogo server (http://weblogo.berkeley.edu/) [65] in which the height of the residue symbol indicates the degree of conservation (the representation obtained using all 61 sequences is highly similar). Numbers refer to residue positions in HrpB2XAC. The FQALM and TLMKNQ motifs are underlined.
In order to determine whether either or both of these motifs is important for HrpB2XAC function in the elicitation of citrus canker symptoms, we expressed HrpB2XAC fragments or full-length HrpB2XAC variants (Table 1) in the ΔhrpB2 strain. To test the importance of the FQALM motif we mutated these residues to LQGPR and expressed the mutant protein (HrpB2XAC_LQGPR) in the ΔhrpB2 strain using the pUFR047_hrpB2LQGPR plasmid. The ΔhrpB2+pUFR047_hrpB2LQGPR strain was able to cause citrus canker symptoms in a manner indistinguishable from the wild-type Xac strain (Fig. 4B). Therefore, the FQALM motif does not seem to be essential for HrpB2 function. When the ΔhrpB2 strain was transformed with plasmids pUFR047_hrpB21-56 and pUFR047_hrpB21-123, leading to the expression of HrpB2XAC_1-56 and HrpB2XAC_1-123 respectively, neither of the resulting strains were able to induce citrus canker symptoms in orange leaves (Fig. 4C). These results suggested that the C-terminal region of HrpB2XAC which contains the conserved TLMKNQ motif is important for HrpB2 function. To test the importance of each residue in this motif, six hrpB2 XAC mutants in which each of these residues were changed to alanine were expressed the ΔhrpB2 strain. The results showed while the Xac strain expressing HrpB2XAC_T125A was not able produce canker symptoms, the strains expressing HrpB2XAC_V127A, HrpB2XAC_K128A, HrpB2XAC_N129A and HrpB2XAC_Q130A produced canker symptoms to the same extent as wild-type Xac. Furthermore, ΔhrpB2 cells expressing HrpB2XAC_L126A produced attenuated citrus canker symptoms when compared to the same cells containing the plasmid that expresses wild-type HrpB2XAC (Fig. 4D). These results point to the importance of the TLMKNQ motif, and especially to the first residue of this motif (T125), in the role of HrpB2XAC in the development disease in citrus plants.
HrpB2XAC is secreted by Xac in liquid media
Rossier et al. [20], showed that in Xcv HrpB2 is secreted in a T3SS-dependent manner. In that study, a mutant Xcv strain with constitutive expression of the hrp locus (due to a constitutively activated HrpG mutation) was used. No such mutant Xac strain has yet been isolated or produced. Expression of hrp genes in Xcv is dependent on unknown plant signals and is controlled by specific promoters with PIP boxes [33], [34], [35]. In Ralstonia solanacearum, hrp expression is dependent on contact with an unidentified component derived from the host cell wall [36], [37], [38] and passion fruit leaf extracts have been shown to modify the proteome of X. axonopodis pv. passiflorae [39]. We therefore grew liquid Xac cultures in the presence of extracts derived from sweet orange (C. sinensis) leaves. Proteins in the secreted fraction were separated by SDS-PAGE and probed for HrpB2XAC by Western blot analysis using anti-HrpB2XAC antiserum. We found that HrpB2XAC could be observed in the secreted fraction of wild-type cells (Fig. 6A, lane 1). As expected, secretion of HrpB2XAC was abolished in the ΔhrpB2 mutants and complementation with pUFR047_hrpB2 restored HrpB2XAC secretion (Fig. 6A, lanes 2 and 3, respectively). We did not detect HrpB2XAC in the cellular fractions (data not shown) but note that the secreted fraction was concentrated 60-fold in relation to the cellular fraction (see Experimental Procedures).
Figure 6. HrpB2XAC is secreted by Xac.
Liquid cultures of Xac were grown as described in Materials and Methods. Secreted fractions were concentrated and separated by SDS-PAGE 18% and proteins were transferred to nitrocellulose membranes. HrpB2XAC was detected using anti-HrpB2XAC antiserum and revealed using anti-mouse IgG conjugated with horseradish peroxidase. (A) Lane 1: Xac wild-type, lane 2: Xac ΔhrpB2, lane 3: Xac ΔhrpB2+pUFR047_hrpB2, lane 4: Xac ΔhrpB2+pUFR047_hrpB21-56, lane 5: Xac ΔhrpB2+pUFR047_hrpB21-123, lane 6: Xac ΔhrpB2+pUFR047_hrpB2LQGPR. (B) Lane 1: Xac wild-type, lane 2: Xac ΔhrpB2+pUFR047_hrpB2T125A, lane 3: Xac ΔhrpB2+pUFR047_hrpB2Q130A, lane 4: Xac ΔhrpB2. (C) Lane 1: Xac wild-type, lane 2: Xac ΔhrcU, lane 3: Xac ΔhrcU+pUFR047_hrcU and lane 4: Xac ΔhrcU+pUFR047_hrcUAAAH.
We then asked whether the HrpB2XAC mutants described above were secreted when expressed in the ΔhrpB2 strain. Figure 6A (lanes 4, 5 and 6) shows that HrpB21-56 (5.7 kDa), HrpB21-123 (13 kDa) and HrpB2XAC_LQGPR were all observed in Xac culture supernatants. Furthermore, all six mutants carrying alanines at each position of the TLMKNQ motif could be detected in Xac culture supernatants (Fig. 6B, lanes 2 and 3 and data not shown). Finally, we observed that HrpB2XAC secretion was abolished in the ΔhrcU mutant (Fig. 6C, lane 2). Complementation of the ΔhrcU mutant with pUFR047_hrcU restored HrpB2XAC secretion to wild-type levels (Fig. 6C, lane 3). Interestingly, complementation of the ΔhrcU mutant with pUFR047_hrcUAAAH resulted in significantly reduced levels of HrpB2XAC secretion (Fig. 6, lane 4). This difference in levels of HrpB2XAC secretion may, therefore, be due to the inability of the HrcUXAC_AAAH protein to undergo the self-cleavage reaction. Apparently, only minimal amounts of HrpB2XAC are necessary to elicit citrus canker symptoms during the infection process.
HrcUXAC is not required for Xac survival in planta
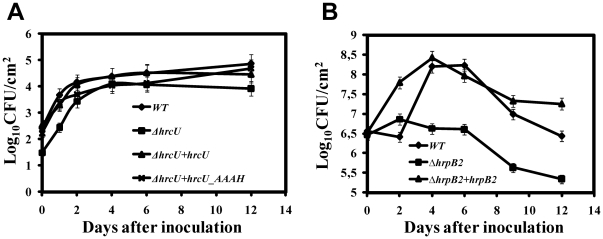
In order to determine whether HrcUXAC and its NPTH site were necessary for Xac survival in inoculated host leafs, we inoculated Citrus leafs with Xac bacterial suspensions and accompanied bacterial numbers during a 12 day period after infection. Figure 7A shows that wild-type Xac, ΔhrcU, ΔhrcU+pUFR047_hrcU, ΔhrcU+pUFR047_hrcUAAAH strains all presented similar growth curves. This suggests that HrcUXAC is not absolutely required for bacterial survival in planta, in spite of the fact that the ΔhrcU does not produce canker disease symptoms. In contrast, the ΔhrpB2 strain presented significantly reduced survival when compared to the wild-type and ΔhrpB2+pUFR047_hrpB2 strains (Fig. 7B).
Figure 7. Number of colony-forming units (CFU) of Xac strains per cm2 of leaf tissue during the first twelve days after inoculation.
The abaxial surface of young leaves was pricked by using insect pins whose tips were previously immersed in the bacterial suspension for Xac hrcU mutant strains (A) or by infiltration into leaves with needleless syringes for Xac hrpB2 mutant strains (B). Discs of infected leaves were excised, homogenized and cultured quantitatively by incubation on agar plates. The assays were performed in triplicate and error bars represent the standard deviation of the data. Differences in the initial bacterial populations are due to differences in the inoculation protocols. (A) Xac wild-type (diamonds), ΔhrcU (squares), ΔhrcU+pUFR047_hrcU (triangles), ΔhrcU+pUFR047_hrcUAAAH (crosses). (B) Xac wild-type (diamonds), ΔhrpB2 (squares), ΔhrpB2+pUFR047_hrpB2 (triangles).
Discussion
In this study we constructed non-polar knock-out mutants for the hrcU and hrpB2 genes and show that they completely abolish pathogenicity of Xac in sweet orange. Complementation of the ΔhrcU strain with plasmids pUFR047_hrcU or pUFR047_hrcUAAAH recovered the capacity to induce disease symptoms. We also demonstrated that HrpB2XAC is secreted to the extracellular space in a HrcUXAC-dependent manner by the Xac T3SS. HrpB2XAC secretion was abolished in the ΔhrcU knockout and restored in the ΔhrcU+pUFR047_hrcU and ΔhrcU+pUFR047_hrcUAAAH strains, but the amount of HrpB2XAC secreted by the ΔhrcU+pUFR047_hrcUAAAH strain was reduced with respect to that observed for the wild-type and ΔhrcU+pUFR047_hrcU strains (Figure 6C). This result suggests that while HrcUXAC cleavage may not be absolutely necessary for the proper functioning of the Xac T3SS, it may contribute to the efficiency by which it carries out its tasks.
In this report we have shown that HrcUXAC expressed in E. coli suffers proteolysis at a highly conserved NPTH site in a manner similar to that already described for its paralog FlhB of the flagellar system [24] and its orthologs YscU, EscU and SpaS from the T3SSs of Yersina [26], [40], E. coli and Salmonella [30] respectively. This, and a similar report for the HrcU protein from Xanthomonas campestris pv. vesicatoria [21], are the first observations of NPTH-dependent cleavage of a FlhB homolog from the T3SS of a plant pathogen. Ferris et al. [25] have shown that FlhB cleavage at the NPTH site is an autocatalytic process; that is, FlhB catalyzes its own hydrolysis at this site. Furthermore, a series of crystal structures of the C-terminal domains of the FlhB homologs EscU and SpaS from the E. coli and Salmonella T3SSs [30], and YscU from Yersinia enterocolitica [29] have recently provided information regarding the mechanism and conformational changes associated with self-cleavage.
We also show that the HrcUXAC C-terminal fragment that is released upon HrcUXAC self-cleavage interacts with HrpB2XAC, whose only known homologs are found in the phytopathogens Xanthomonas spp., Ralstonia solanacearum, and Acidovorax avenae, as well as Burkholderia spp that infect both animals and plants. Our results show that HrpB2XAC does not interact specifically with the site of HrcUXAC cleavage since it could bind to HrcUXAC_207-357(AAAH) and to HrcUHis277-357, a fragment that begins 10 residues after the NPTH site. Unfortunately we were not able to detect HrcUXAC in the wild-type or complemented mutant strains using the anti-HrcUXAC polyclonal antibodies in this study (data not shown) and so could not determine relative levels of HrcUXAC in the Xac strains nor have we so far been able to determine whether HrcUXAC is in fact cleaved at the NPTH site in Xac cells. However, during the preparation of this work, HrcU cleavage was observed in Xcv [21].
In order to understand HrcU function it is useful to recall what we know about the functioning of HrcU homologs. Mutants that inhibit cleavage at the NPTH site of HrcU homologs exhibit defects in the secretion of specific substrates. For example, in FlhB, mutations at this site inhibit the export of “late” flagellar proteins, while normal levels of early substrates, including hook protein FlgE, are secreted [27]. Also, a Y. enterocolitica ΔyscU strain expressing YscUN263A, in which the conserved Asn residue of the NPTH sequence was mutated to Ala, produced longer needles, exported reduced amounts of YscP (a FliK homolog, see below) and did not export the translocator proteins LcrV, YopB and YopD. The first two defects could be compensated by overexpression of YscP (see below) while export of LcrV, YopB and YopD was absolutely dependent on a cleavable NPTH site [28].
The cleaved FlhB C-terminal fragment binds to both early and late flagellar export substrates (FlgD, FliC). Furthermore, the product of the fliK gene, FliK or flagellar hook-length control protein, binds to the self-cleavage C-terminal fragment of FlhB [24] and during flagellar assembly FliK is itself secreted subsequent to hook protein subunit secretion [41], [42]. Also, fliK mutants do not secrete late substrates but do secrete excessive amounts of hook protein (FlgE), resulting in the production of characteristic polyhooks [43]. This phenotype can be reverted by single amino acid substitutions in FlhB, almost all of which map to the C-terminal self-cleavage fragment [27], [43], [44], [45]. Thus, in the flagellar system, FlhB and FliK act together to control substrate switching from early to late substrates, though the molecular mechanism by which this is achieved is not fully understood [41], [46].
In the animal pathogens Yersinia, Salmonella and Shigella, the formation of needle complexes and subsequent secretion of virulence factors by T3SSs are controlled by an interplay between FlhB and FliK homologs. In these systems, mutations in the FliK homologs YscP [47], [48], InvJ [49] or Spa32 [50] result in the formation of needles of variable length and compromised virulence factor secretion. In the case of Yersinia, the phenotypes can be reverted by mutations in the cytosolic domains of the FlhB homolog YscU [47]. Futhermore, YscP, InvJ and Spa32 are secreted during T3SS assembly [50], [51], [52] in a manner similar to the secretion of FliK in the flagellar system. Finally, in Yersinia, YscP secretion appears to be coupled to the secretion of another small protein (YscO) that binds preferentially to the uncleaved form of YscU [53].
Few YscP homologs from non-flagellar T3SS have been identified in plant-associated bacteria: HrpP from Pseudomonas syringae, RspP from P. fluorescens, HpaP from R. solanacearum [54] and the HpaP/HpaC proteins coded by the hrp gene clusters of Xanthomonas spp (for example HpaC in Xcv and HpaP in Xac). During the preparation of this work Lorenz et al. [21] published a study on the HpaC and HrpB2 proteins from Xcv. They found that: 1) amino acids 10 to 25 of HrpB2 are crucial for its efficient secretion and function and that HrpB2 is necessary for the secretion of effectors and of extracellular components of the secretion apparatus, 2) HrpB2 and HpaC interact with each other and both also interact with the C-terminal domain of HrcU and 3) HrpB2 secretion is suppressed by HpaC. They therefore speculated that HpaC acts to control the switch between the secretion of early to late T3SS substrates (see also reference [20]) and that HpaC binding to HrcU specifically inhibits HrpB2 binding and secretion [21]. While HpaC from Xcv has been shown to be necessary for the secretion of both T3SS effector and translocon proteins, it is not required for the export of the Hrp pilus protein HrpE [55]. In this sense, the hpaC mutant phenotype in Xcv is similar to that observed for yscP, invJ and spa32 mutants (see above). However, HpaC itself is not secreted by Xcv and Hrp pilus formation was not affected in hpaC mutant strains (different from that observed for yscP, invJ and spa32 mutants as described above) [55]. On the other hand, HrpB2 binds to HrcU and, like FliK and YscP, HrpB2 is itself secreted. Since both HrpB2 and HpaC bind to the C-terminal domain of HrcU, the accumulated evidence so far is not clear as to which (if either) HpaC-HrcU or HrpB2-HrcU complexes carry out molecular functions in the Xanthomonas T3SS that are orthologous to those of YscU-YscP and FlhB-FliK described above.
One interesting observation from our study was that while both ΔhrcU and ΔhrpB2 knockout strains do not induce citrus canker symptoms, only the latter presents a significant reduction in survival in the host tissue. The ΔhrcU mutant survives as well as the wild type strain in the host tissue, but does not detectably secrete HrpB2. A similar phenomena has been observed in X. campestris pv. glycines 8ra where HrcU is required for pathogenicity in its natural soybean host but is not required for multiplication in the host plant nor is it required for the induction of HR in non-hosts [22]. The molecular basis for the differences in the ΔhrcU and ΔhrpB2 phenotypes in Xac is not yet clear. One possibility is that the ΔhrcU mutant fails to secrete effector(s) that trigger specific host defense mechanisms resulting in the bacterial survival. Another possibility is that intracellular HrpB2 may contribute to Xac survival while extracelular HrpB2 contributes to citrus canker symptom development.
In Xcv, deletion of HrpB2 residues 10-25 impaired protein secretion and disease symptom formation, which led to the conclusion that secretion is required for function [21]. We demonstrated that while ΔhrpB2+pUFR047_hrpB21-56, ΔhrpB2+ pUFR047_hrpB21-123 and ΔhrpB2+pUFR047_hrpB2T125A strains are not able to cause citrus canker, the truncated HrpB2XAC polypeptides and HrpB2XAC single amino acid substitution mutants are all however secreted to the extracellular space. Therefore, HrpB2XAC secretion, per se, is not sufficient for HrpB2XAC function. Apparently, the conserved C-terminal region of the protein, more specifically residue T125 in the conserved TLMKNQ motif, is especially important for HrpB2XAC-dependent pathogenicity.
Important unanswered questions remain regarding HrpB2 function at the molecular level. Further studies are needed to determine whether HrpB2 exercises a role in substrate switching or as a minor structural component of the T3SS pilus (as do hook-filament junction and capping proteins in bacterial flagella) or carries out other, as yet not contemplated, functions and also whether these functions are effected within the bacterial cell or in the exterior subsequent to its secretion (or both).
Materials and Methods
Construction vectors for the expression of HrcUXAC_207-357, HrcUXAC_His277-357, HrcUXAC_207-357(AAAH) and HrpB2XAC in E. coli
E. coli strains and plasmids are described in Table 1 and Table 2, respectively. E. coli cells were cultivated at 37°C in 2xYT media [56]. When necessary, the appropriate antibiotics were added at the following final concentrations: ampicillin 200 µg/ml, kanamycin 50 µg/ml and chloramphenicol 200 µg/ml. Synthetic oligonucleotide primers (Table 3) for polymerase chain reactions (PCR) were designed containing restriction sites useful for cloning (see below). PCR products were purified from agarose gels using the QIAquick Gel Extraction Kit (Qiagen). To produce a vector for the expression of HrcUXAC_207-357, the DNA sequence coding residues 207-357 of the hrcU gene was amplified from genomic Xac DNA using the oligonucleotides F-U207-357 and R-U207-357 (Table 3). The PCR product was digested with endonucleases NcoI and HindIII and inserted into the expression vector pET-11d [57], previously digested with the same enzymes to produce plasmid pU1. Primers F-UAAAH and R-UAAAH (Table 3) were used in PCR with pU1 as template in order to change the codons for residues 264-266 to alanine codons using the QuikChange Site-Directed Mutageneis Kit (Stratagene). The resulting recombinant plasmid (pU2) directs the expression of HrcUXAC_207-357(AAAH). Note that in both recombinant HrcUXAC_207-357 (through which HrcUXAC_207-264 is purified, see below) and HrcUXAC_207-357(AAAH), residues Gln207 and His208 have been mutated to Met and Asp residues, respectively, due to the introduction of restriction sites used in the cloning protocol. To produce a vector for the expression of HrcUXAC_His277-357, the sequence coding for HrcU residues 277-357 was amplified using primers F-UHis277-357 and R-UHis277-357 (Table 3). This product was digested with NdeI and HindIII and ligated into the expression vector pET-28a (Novagen) previously digested with the same enzymes to produce the recombinant plasmid pU3. To produce a vector for the expression of full-length HrpB2XAC, the expression vector pET-3a (Studier et al., 1990) was digested with HindIII, filled in with the Klenow fragment of E. coli DNA polymerase I and then digested with NdeI. Primers F-B2 and R-B2 were used in a PCR with Xac genomic DNA, the product was treated with Klenow fragment and polynucleotide kinase to produce blunt ends, digested with NdeI and then ligated into the pET-3a vector described above to produce the recombinant plasmid pB2. The accession numbers for the complete Xac genome sequence and the HrpB2XAC and HrcUXAC protein sequences are NC_003919, NP_640763 and NP_640761, respectively.
Table 3. Oligonucleotides used in this study.
| Oligonucleotides | Sequence |
| F-U207-357 | 5′ CATCCCATGGACTGGCTGTTCATCCGGGAC 3′ |
| R-U207-357 | 5′ CCCAAGCTTCTCGAGGCTCGCACGCGATCTCCTAG 3′ |
| F-UAAAH | 5′ GTGATGGTGGTCGCCGCGGCCCATTACGCGGTGGCAC |
| R-UAAAH | 5′ GTGCCACCGCGTAATGGGCCGCGGCGACCACCATCAC 3′ |
| F-UHis277-357 | 5′ TAAATTGCTCATATGGATGACTTCGGCCTA 3′ |
| R-UHis277-357 | 5′ TAAATTGCTCCATGGATGACTTCGGCCTA 3′ |
| F-B2 | 5′ CGGAATTCCATATGACGCTCATTCCTCCTGTC 3′ |
| R-B2 | 5′ CCGCTCGAGCTATTGGTTCTTGACCAGTGTCTG 3′ |
| F1-U | 5′ TCGGGACTAAAGCTTGCATCAACT TGATCT 3′ |
| R1-U | 5′ GGAATTACCATATGCAGTTTCTTCTCGGTCGGCTTCTC 3′ |
| F2-U | 5′ GGAATTACCATATGCACAGCGACGGCGATGGAGCT 3′ |
| R2-U | 5′ TTTGAACTTGCTAGCTGATCGGTGCCGCTG 3′ |
| R-compU | 5′ ATTTTAAGCTTGTCGACCTAGCATGGCAGAGCTCC 3′ |
| F1-B2 | 5′ CACTACAAGCTTAAGCAACCAGCAAGGGGA 3′ |
| R1-B2 | 5′ GGAATTACCATATGAATCGCTTGGACAGGAGGAAT 3′ |
| F2-B2 | 5′ GGAATTACCATATGAAGAACGCCGTGCAGACACTG 3′ |
| R2-B2 | 5′ AACATTAAATCTAGAGTCGACTGGTTCGCATGCAGGCCGAGC 3′ |
| R-compB2 | 5′ AATTTAAGCTTGTCGACCTATTGGTTCTTGACCAGTGTC3′ |
| F-M57 | 5′GCAGCGAGTGGGCAACCCGAGCTAGATGAGCCGCGTGGTCGATGTGC3′ |
| R-M57 | 5′GCACATCGACCACGCGGCTCATCTAGCTCGGGTTGCCCACTCGCTGC3′ |
| F-Q124 | 5′GCAATCGGGAAAGAACGCAGTGTAGACACTGGTCAAGAATCAATAG3′ |
| R-Q124 | 5′CTATTGATTCTTGACCAGTGTCTACACTGCGTTCTTTCCCGATTGC3′ |
| F-FQALM | 5′CGCTAGTGAATCGCTTACAAGGGCCGAGGCAGTCCTCTAGC3′ |
| R-FQALM | 5′GCTAGAGGACTGCCTCGGCCCTTGTAAGCGATTCACTAGCG3′ |
| F-T125A | 5′GGAAAGAACGCAGTGCAGGCACTGGTCAAGAATCAATAG3′ |
| R-T125A | 5′CTATTGATTCTTGACCAGTGCCTGCACTGCGTTCTTTCC3′ |
| F-L126A | 5′GAAAGAATGCAGTGCAGACAGCGGTCAAGAACCAATAGGT3′ |
| R-L126A | 5′ACCTATTGGTTCTTGACCGCTGTCTGCACTGCATTCTTTC3′ |
| F-V127A | 5′AGAACGCAGTGCAGACACTGGCCAAGAATCAATAGGTCGAC3′ |
| R-V127A | 5′GTCGACCTATTGATTCTTGGCCAGTGTCTGCACTGCGTTCT3′ |
| F-K128A | 5′GCCGTGCAGACACTGGTAGCAAACCAATAGGTCGACCTCGA3′ |
| R-K128A | 5′TCGAGGTCGACCTATTGGTTTGCTACCAGTGTCTGCACGGC3′ |
| F-N129A | 5′CCGTGCAGACACTAGTCAAGCGCCAATAGGTCGACCTCGAGGG3′ |
| R-N129A | 5′CCCTCGAGGTCGACCTATTGGCGCTTGACTAGTGTCTGCACGG3′ |
| F-Q130A | 5′TGCAGACACTGGTCAAGAACGCATAGGTCGACCTCGAGGGGGG3′ |
| R-Q130A | 5′CCCCCCTCGAGGTCGACCTATGCGTTCTTGACCAGTGTCTGCA3′ |
Expression and purification of recombinant HrpB2XAC and HrcUXAC fragments
Plasmid constructs pU1, pU2, and pB2 were used to transform E. coli strain BL21(DE3) [58] and pU3 was used to transform BL21(DE3)RIL cells [59]. The synthesis of recombinant proteins was induced by the addition of 0.4 mM isopropyl-β-D-thiogalactopyranoside when cultures grown at 37°C attained an optical density of 0.8 at 600 nm. After three more hours of growth, cells were collected by centrifugation at 4500 x g for 15 min at 4°C and ressuspended in 20 ml/l of culture of 25 mM Tris-HCl (pH 8.0) for HrcUXAC fragments, and 5 mM sodium acetate (pH 6.0) for HrpB2XAC. Cells were lysed by passage through a French pressure cell followed by centrifugation at 37000 x g for 1 hour at 4°C. Expression of HrcUXAC_207-357 led to the production of a 7 kDa polypeptide, not 17 kDa expected from the size of the protein coded by the gene fragment in the pU1 vector (see Results). This polypeptide was purified from the soluble fraction of the bacterial lysate by Q-Sepharose (Amersham Bioscience) anion-exchange chromatography (25 mM Tris-HCl (pH 8.0), 14 mM β-mercaptoethanol) using a 0-300 mM NaCl gradient, followed by Superdex G-75 (Amersham Bioscience) size exclusion chromatography (25 mM Tris-HCl (pH 8.0), 100 mM NaCl, 14 mM β-mercaptoethanol). HrcUXAC_207-357(AAAH) and HrpB2XAC recombinant proteins were recovered from the insoluble fraction of the bacterial lysate by solubilizing in 25 mM Tris-HCl (pH 8.0), 14 mM β-mercaptoethanol, 8 M urea for HrcUXAC_207-357(AAAH) or 5 mM sodium acetate (pH 6.0), 14 mM β-mercaptoethanol, 8 M urea for HrpB2XAC. HrcUXAC_207-357(AAAH) was purified by Q-Sepharose anion-exchange chromatography, using the solubilization buffer (above) and a 0-300 mM NaCl gradient followed by Superdex G-75 size exclusion chromatography using 25 mM Tris-HCl (pH 8.0), 100 mM NaCl, 14 mM β-mercaptoethanol, 8 M urea. HrpB2XAC was purified by passing the protein mixture through a Q-sepharose column equilibrated with 5 mM sodium acetate (pH 6.0), 14 mM β-mercaptoethanol, 8 M urea. HrpB2XAC does not bind to this column under these conditions. The unbound fraction containing HrpB2XAC was concentrated using an 10 kDa Amicon filter (Millipore) and separated by Superdex G-75 size exclusion chromatography using 5 mM sodium acetate (pH 6.0), 100 mM NaCl, 14 mM β-mercaptoethanol, 8 M urea. HrcUXAC_207-357(AAAH) and HrpB2XAC were refolded by dialyses against 25 mM Tris-HCl (pH 8.0), 14 mM β-mercaptoethanol for HrcUXAC_207-357(AAAH), or 5 mM sodium acetate (pH 6.0), 14 mM β-mercaptoethanol for HrpB2XAC containing successively reduced amounts of urea: 6 M, 4 M, 2 M, 0 M. HrcUHis277-357 was purified from the insoluble fraction of the bacterial lysate by solubilizing in 25 mM Tris-HCl (pH 8.0), 10 mM imidazole, 100 mM NaCl, 2 mM β-mercaptoethanol, 8 M urea. The protein mixture was applied to a Ni2+-chelating Sepharose column equilibrated with the same buffer and eluted using a 25-500 mM imidazole gradient. HrcUXAC_His277-357 fractions were pooled and the protein was refolded by successive dialyses against 25 mM Tris-HCl (pH 8.0), 14 mM β-mercaptoethanol containing 6 M, 4 M, 2 M and 0 M urea.
Production of polyclonal antibodies against HrcUXAC207-357(AAAH) and HrpB2XAC proteins
Swiss Webster mice were immunized with four injections, separated by one week intervals, of 10 µg soluble HrpB2XAC. New Zealand white rabbits were immunized with HrcUXAC_207-357(AAAH) using four 200 µg injections separated by one week intervals. In both cases, the antigens were diluted with one volume of complete Freund's adjuvant (Sigma) for the first immunization and one volume of incomplete Freund's adjuvant (Sigma) for the remaining immunizations. Blood was collected and incubated for 1 hr at 37°C and the serum was recovered by centrifugation at 5000 x g for 15 min at room temperature, aliquoted and stored at -20°C. Before use, antiserum aliquots were incubated with an E. coli lysate as described [56].
Edman degradation N-terminal sequencing
The N-terminus of the 7 kDa polypeptide purified after the expression of HrcUXAC_207-357 was lyophilized and dissolved in ultrapure water. N-terminal sequencing was carried out by Edman degradation using a PPSQ/23 sequencer (Shimadzu Corporation, Tokyo).
Mass spectrometry experiments
Purified proteins were analyzed by Matrix Assisted Laser Desorption Ionization (MALDI) Time of Flight (TOF) Mass Spectrometry (MS) using an Ettan MALDI-TOF Pro system (Amersham Biosciences). All MALDI-TOF MS spectra were externally calibrated using a cytochrome C standard (12327 Da). Protein mass was identified in linear mode with positive ionization at 20 kV. The samples were mixed with an equal volume of sinapinic acid matrix dissolved in 50% acetonitrile, 0.5% of trifluoroacetic acid. A 0.5 µl aliquot was loaded onto stainless steel MALDI slides for analysis. Spectra were analyzed using the Ettan Maldi-Tof Pro v2.0 software package.
Western blot assays
Samples were separated by SDS-PAGE (18% acrylamide) and electroblotted onto a nitrocellulose membrane. The membrane was colored with Ponceau red to identify the positions of specific proteins and then blocked for 2 h with 10 mM Tris-HCl (pH 7.5), 150 mM NaCl, 0.1% Tween 20, 0.1% Triton (TBS-TT) and 5% non-fat dry milk. The membranes were probed for 2 h with the appropriate polyclonal antiserum in 5-10 ml of the above blocking buffer (1∶3000 dilution for anti-HrcUXAC antibody and 1∶20000 dilution for the anti-HrpB2XAC antibody) and then washed four times for 15 min with TBS-TT. The anti-HrpB2XAC antibody was detected using an anti-mouse IgG conjugated with horseradish peroxidase (Sigma) at a dilution of 1∶6000. The anti-HrcUXAC antibody was detected using protein A conjugated with horseradish peroxidase (Sigma) at a dilution of 1∶30000. The membranes were incubated for 2 h with the protein-A or anti-IgG conjugates in 5-10 ml of blocking buffer following by washing with TBS-TT. Reactive bands were detected using the ECL AdvanceTM Western Blotting Detection Kit (GE Heathcare-Amersham) according to the manufacturer's instructions.
Far-Western assays
Far-Western blot assays were carried out to detect specific protein-protein interactions. Approximately 15 µg of purified recombinant protein or lysates from E. coli cells was separated by SDS-PAGE (18% acrylamide) and electroblotted onto nitrocellulose membranes. The membrane was blocked for 2 h with TBS-TT plus 5% nonfat dry milk followed by 14 h incubation with 50 µg/ml of a second purified recombinant protein (indicated in the figure legends) at 4°C. Unbound proteins were removed by washing the membranes four times for 15 min with TBS-TT. Bound proteins were then detected as described for the Western blot assays (above). In some cases, negative control experiments were performed using a polypeptide derived from residues 143-284 of chicken muscle α-tropomyosin [60].
His-tag pulldown assays
HrcUXAC_His277-357, HrpB2XAC and an E. coli lysate were dialyzed at 4°C against 25 mM Tris-HCl, 100 mM NaCl, 2 mM β-mercaptoethanol, 10 mM imidazole (pH 8.0). A mixture of HrcUHis277-357 (30 µM) and HrpB2 (30 µM) was added to a 0.25-ml aliquot of Ni2+-chelating Sepharose resin (Amersham Bioscience) equilibrated in the above buffer at room temperature. In control experiments, the resin was mixed with only HrcUXAC_His277-357 or HrpB2 or with a mixture of HrcUHis277-357 and a lysate derived from 10 ml of E. coli BL21(DE3) culture (OD600 = 0.8). The mixtures were washed four times with 1 ml of 25 mM Tris-HCl, 100 mM NaCl, 2 mM 2-β-mercaptoethanol, 25 mM imidazole (pH 8.0). Bound proteins were released by washing with 50 µl of 25 mM Tris-HCl, 100 mM NaCl, 2 mM 2-mercaptoethanol, 500 mM imidazole (pH 8.0). Samples were then analyzed by SDS-PAGE and Western blot assay.
Fluorescence experiments
HrcUXAC_207-357(AAAH) and HrpB2XAC (both 2 µM) were dissolved in 5 mM sodium acetate pH 6.0 at 25°C. Fluorescence emission spectra were obtained using an AVIV (Lakewood, NJ) ATF 105 Automated Titrating Differential/Ratio spectrofluorometer and were collected between 320 and 400 nm using an excitation wavelength of 280 nm and excitation and emission bandwidths of 2 nm and 5 nm respectively.
Production of Xac genes knockouts
Deletion strains were constructed using the suicide vector pNPTS138 (Alley Dickon, unpublished) by allelic exchange as described [61]. DNA fragments (1 kb) flanking each side of the Xac hrpB2 and hrcU genes were amplified by PCR using oligonucleotides listed in Table 3. For hrcU, primer pairs F1-U + R1-U and F2-U + R2-U were used. For hrpB2, primer pairs F1-B2 + R1-B2 and F2-B2 + R2-B2 were used. The products were digested with endonuclease NdeI and specific pairs were joined together with T4 DNA ligase (New England Biolabs). The resulting fragments were cloned into pNPTS138 generating pNPTS138-ΔhrcU using HindIII and NheI and pNPTS138-ΔhrpB2 by using HindIII and SalI. These plasmids were introduced by electroporation into Xac strain 306. Kanamycin and ampicillin-resistant colonies were selected and grown on plates containing 5% sucrose and ampicillin. Sucrose-sensitive and kanamycin- and ampicilin-resistant colonies were selected and used to inoculate 10 ml of 2xYT-ampicilin medium, which was incubated overnight with agitation at 28°C. A 100 µl aliquot of this culture was plated without dilution on 2xYT agar plates containing 200mg/L ampicillin. The resulting colonies were transferred in replica on two plates: one containing kanamycin and ampicillin and another containing sucrose and ampicilin. Clones that were simultaneously kanamycin-sensitive and sucrose-resistant were selected, and the deletion was confirmed by PCR.
Production of expression vectors for complementation of ΔhrcU and ΔhrpB2 in Xac
A fragment containing the hrcU gene plus 1 kb upstream sequences was amplified by PCR using primers F1-U and R-compU (Table 3). This fragment contains the complete HrcU open reading frame as well as its native promoter. After digestion with HindIII and SalI, this fragment was cloned into the HindIII-SalI sites of pBBR1MCS-5 [62], resulting in pBBR_hrcU (Table 2). To construct pBBR_hrcUAAAH (Table 2), primers F-UAAAH and R-UAAAH (see Table 3) were used in a PCR amplification with pBBR_hrcU as template to change the codons for residues 264-266 (NPT) to alanine codons using the QuikChange Site-Directed Mutagenesis Kit (Stratagene). The mutation was confirmed by sequencing. The HindIII/SalI fragments of pBBR_hrcU and pBBR_hrcUAAAH, which contain the complete HrcUXAC open reading frame as well as its native promoter, were cloned into the same sites of pUFR047, a broad-host range vector carrying a gentamycin resistance gene [63], generating constructs pUFR047_hrcU and pUFR047_hrcUAAAH (Table 2). These plasmids were used to transform the ΔhrcU mutant strain by electroporation followed by selection on LB plates with 10 µg/ml gentamycin and 200 µg/ml ampicillin.
A fragment containing the hrpB2 gene plus 1 kb upstream sequences was amplified by PCR using primers F1-B2 and R-compB2 (Table 3), digested with HindIII and cloned into the HindIII site of pUFR047. The resulting construct, pUFR047_hrpB2 (Table 2) was used to transform the Xac ΔhrpB2 mutant strain by electroporation. Transformed colonies were selected on LB/gentamycin/ampicilin plates to produce strain ΔhrpB2+pUFR047_hrpB2 (Table 1). To produce hrpB2 gene mutants for expression in Xac, the HindIII/SalI fragment of the PCR product above was cloned between the HindIII and SalI sites of pBBR1MCS-5 generating the construct pBBR_hrpB2 (Table 2) which was then used as a template to produce mutants using the QuikChange Site-Directed Mutagenesis Kit (Stratagene). Primers F-M57 and R-M57, F-Q124 and R-Q124 (Table 3) were used to change the codons 171 and 372 to stop codons; primers F-FQALM and R-FQALM (Table 3) were used to change the codons for the FQALM motif (residues 35-39) to LQGPR codons and finally, primers pairs F-T125A and R-T125A, F-L126A and R-L126A, F-V127A and R-V127A, F-K128A and R-K128A, F-N129A and R-N129A and F-Q130A and R-Q130A (Table 3) were used to change the respective codons to alanine codons. The HindIII/SalI fragments from all these pBBR_hrpB2 derived constructs were cloned between the same sites of pUFR047 generating the constructions pUFR047_hrpB21-56, pUFR047_hrpB21-123, pUFR047_hrpB2T125A, pUFR047_hrpB2L126A, pUFR047_hrpB2V127A, pUFR047_hrpB2K128A, pUFR047_hrpB2N129A, and pUFR047_hrpB2Q130A (Table 2). All the mutations were confirmed by sequencing. Theses constructions were used to transform the Xac ΔhrpB2 strain by electroporation (Table 1).
Plant bioassays
Highly susceptible Navel sweet orange (Citrus sinensis (L.) Osbeck) plants were grown under greenhouse conditions and maintained at 28°C with daylight for virulence assays. To visually monitor the development of citrus canker symptoms, Xac 306 and mutant strains were grown overnight at 30°C and adjusted to an optical density of 0.3 at 600 nm in 2xYT culture medium. The suspensions were hand-infiltrated with a 1-ml syringe with needle into the abaxial surface of attached leaves. To monitor bacterial growth in planta, Xac strains were grown overnight at 30°C and adjusted to an optical density at 600 nm (OD 600) of 0.5 in NB culture medium (8g of nutrient broth liter-1, 5 g of NaCl liter−1, pH 7). The abaxial surface of young leaves was pricked by using pins whose tips were previously immersed in the bacterial suspension for Xac hrcU mutant strains (Fig. 7A) or by infiltration into leaves with needleless syringes for Xac hrpB2 mutant strains (Fig. 7B). In both cases, leaf disks (0.8 cm2) from infected plants were removed with a cork borer during a 12 day period post-inoculation, macerated in 0.85% NaCl with a mortar and pestle. Different dilutions were spread on LB plates with the appropriate antibiotics and the bacterial population was determined by counting colonies after a 2-day incubation period at 28-30°C. Experiments were performed in triplicate.
Preparation of orange leaf extracts
Sweet orange leaf extracts were prepared as described previously for passion fruit leaf extracts [39]. Leaves were washed extensively with sterile water. Midribs were excluded and 1 g of tissue was mixed with liquid nitrogen and pulverized to form a fine powder. One-hundred milliliters of MM medium [39] plus carbenicillin 100 µg/ml, pH 7.4 were added and the mixture was macerated followed by centrifugation at 5000 x g for 15 min at 4°C. The supernatant was recovered and passed through 0.45 µm and 0.22 µm filters (Millipore) and stored at -80°C.
HrpB2 secretion by Xac
Xac 306 cells were cultivated at 30°C in MM medium (pH 5.4) plus 100 µg/ml carbenicillin containing sweet orange leaf extract (extract derived from 1 g of leaf tissue per litre of MM medium). Xac cultures (50 mL) were grown for 24 h to an OD 600 = 0.3 after which cells were collected by centrifugation and resuspended in 3 ml of urea-SB: 8 M urea, 10% glycerol, 52 mM Tris-HCl (pH 6.8), 2% SDS, 0.1% bromphenol blue, 140 mM 2-mercaptoethanol. The extracellular (secreted) fraction from a 50 ml culture was passed through a low protein-binding filter 0.45 µm (Millipore). Proteins in the filtrates were precipitated by adding 10% trichloroacetic acid and freezing at −20°C for 12 h followed by centrifugation at 12000 x g (4°C). The precipitate was washed twice with cold acetone and resuspended in 50 µl urea-SB. Note that the above procedure produces a secreted fraction that is derived from 60 times as many bacterial cells per unit volume as the cellular fraction. Equal volumes of cellular and secreted protein fractions were separated by SDS-PAGE (18% acrylamide) and transferred onto the nitrocellulose membrane. HrpB2 was detected by Western blot using anti-HrpB2XAC antibodies (above).
Acknowledgments
We thank Fernando Corrêa for the help in the fluorescence measurements, Izaura Nobuko Toma and Paolo di Mascio for MALDI-TOF MS analyses, Izaura Yoshico Hirata for Edman degradation analyses, Ângela Mika Katsuyama for the clone for expression of HrpB2, and Marcos C. Alegria for helpful discussions.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was supported by research grants from the Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP grant # 2005/59243-3) and the Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) to C.S.F. and a graduate student scholarship to P.A.C. from FAPESP. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Blocker A, Komoriya K, Aizawa S. Type III secretion systems and bacterial flagella: insights into their function from structural similarities. Proc Natl Acad Sci U S A. 2003;100:3027–3030. doi: 10.1073/pnas.0535335100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.He SY, Nomura K, Whittam TS. Type III protein secretion mechanism in mammalian and plant pathogens. Biochim Biophys Acta. 2004;1694:181–206. doi: 10.1016/j.bbamcr.2004.03.011. [DOI] [PubMed] [Google Scholar]
- 3.Buttner D, He SY. Type III protein secretion in plant pathogenic bacteria. Plant Physiol. 2009;150:1656–1664. doi: 10.1104/pp.109.139089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rong W, Feng F, Zhou J, He C. Effector-triggered innate immunity contributes Arabidopsis resistance to Xanthomonas campestris. Mol Plant Pathol. 2010;11:783–793. doi: 10.1111/j.1364-3703.2010.00642.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alegria MC, Docena C, Khater L, Ramos CH, da Silva AC, et al. New protein-protein interactions identified for the regulatory and structural components and substrates of the type III Secretion system of the phytopathogen Xanthomonas axonopodis Pathovar citri. J Bacteriol. 2004;186:6186–6197. doi: 10.1128/JB.186.18.6186-6197.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buttner D, Bonas U. Who comes first? How plant pathogenic bacteria orchestrate type III secretion. Curr Opin Microbiol. 2006;9:193–200. doi: 10.1016/j.mib.2006.02.006. [DOI] [PubMed] [Google Scholar]
- 7.da Silva AC, Ferro JA, Reinach FC, Farah CS, Furlan LR, et al. Comparison of the genomes of two Xanthomonas pathogens with differing host specificities. Nature. 2002;417:459–463. doi: 10.1038/417459a. [DOI] [PubMed] [Google Scholar]
- 8.Kay S, Bonas U. How Xanthomonas type III effectors manipulate the host plant. Curr Opin Microbiol. 2009;12:37–43. doi: 10.1016/j.mib.2008.12.006. [DOI] [PubMed] [Google Scholar]
- 9.Van Gijsegem F, Gough C, Zischek C, Niqueux E, Arlat M, et al. The hrp gene locus of Pseudomonas solanacearum, which controls the production of a type III secretion system, encodes eight proteins related to components of the bacterial flagellar biogenesis complex. Mol Microbiol. 1995;15:1095–1114. doi: 10.1111/j.1365-2958.1995.tb02284.x. [DOI] [PubMed] [Google Scholar]
- 10.Furutani A, Takaoka M, Sanada H, Noguchi Y, Oku T, et al. Identification of novel type III secretion effectors in Xanthomonas oryzae pv. oryzae. Mol Plant Microbe Interact. 2009;22:96–106. doi: 10.1094/MPMI-22-1-0096. [DOI] [PubMed] [Google Scholar]
- 11.Marguerettaz M, Pieretti I, Gayral P, Puig J, Brin C, et al. Genomic and evolutionary features of the SPI-1 type III secretion system that is present in Xanthomonas albilineans but is not essential for xylem colonization and symptom development of sugarcane leaf scald. Mol Plant Microbe Interact. 2010;24:246–259. doi: 10.1094/MPMI-08-10-0188. [DOI] [PubMed] [Google Scholar]
- 12.Szczesny R, Buttner D, Escolar L, Schulze S, Seiferth A, et al. Suppression of the AvrBs1-specific hypersensitive response by the YopJ effector homolog AvrBsT from Xanthomonas depends on a SNF1-related kinase. New Phytol. 2010;187:1058–1074. doi: 10.1111/j.1469-8137.2010.03346.x. [DOI] [PubMed] [Google Scholar]
- 13.Song C, Yang B. Mutagenesis of 18 type III effectors reveals virulence function of XopZ(PXO99) in Xanthomonas oryzae pv. oryzae. Mol Plant Microbe Interact. 2010;23:893–902. doi: 10.1094/MPMI-23-7-0893. [DOI] [PubMed] [Google Scholar]
- 14.Moreira LM, Almeida NF, Jr, Potnis N, Digiampietri LA, Adi SS, et al. Novel insights into the genomic basis of citrus canker based on the genome sequences of two strains of Xanthomonas fuscans subsp. aurantifolii. BMC Genomics. 2010;11:238. doi: 10.1186/1471-2164-11-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pieretti I, Royer M, Barbe V, Carrere S, Koebnik R, et al. The complete genome sequence of Xanthomonas albilineans provides new insights into the reductive genome evolution of the xylem-limited Xanthomonadaceae. BMC Genomics. 2009;10:616. doi: 10.1186/1471-2164-10-616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brunings AM, Gabriel DW. Xanthomonas citri: breaking the surface. Mol Plant Pathol. 2003;4:141–157. doi: 10.1046/j.1364-3703.2003.00163.x. [DOI] [PubMed] [Google Scholar]
- 17.Dunger G, Arabolaza AL, Gottig N, Orellano EG, Ottado J. Participation of Xanthomonas axonopodis pv. citri hrp cluster in citrus canker and nonhost plant responses. Plant Pathology. 2005;54:8. [Google Scholar]
- 18.Swarup S, Yang Y, Kingsley MT, Gabriel DW. An Xanthomonas citri pathogenicity gene, pthA, pleiotropically encodes gratuitous avirulence on nonhosts. Mol Plant Microbe Interact. 1992;5:204–213. doi: 10.1094/mpmi-5-204. [DOI] [PubMed] [Google Scholar]
- 19.Hiroshi S, Takashi F, Hiromichi I, Sinji T, Katsumi O. A pthA homolog from Xanthomonas axonopodis pv. citri responsible for host-specific suppression of virulence. Journal of Bacteriology. 2007;189:3271–3279. doi: 10.1128/JB.01790-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rossier O, Van den Ackerveken G, Bonas U. HrpB2 and HrpF from Xanthomonas are type III-secreted proteins and essential for pathogenicity and recognition by the host plant. Mol Microbiol. 2000;38:828–838. doi: 10.1046/j.1365-2958.2000.02173.x. [DOI] [PubMed] [Google Scholar]
- 21.Lorenz C, Schulz S, Wolsch T, Rossier O, Bonas U, et al. HpaC controls substrate specificity of the Xanthomonas type III secretion system. PLoS Pathog. 2008;4:e1000094. doi: 10.1371/journal.ppat.1000094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oh C, Heu S, Yoo JY, Cho Y. An hrcU-homologous gene mutant of Xanthomonas campestris pv. glycines 8ra that lost pathogenicity on the host plant but was able to elicit the hypersensitive response on nonhosts. Mol Plant Microbe Interact. 1999;12:633–639. doi: 10.1094/MPMI.1999.12.7.633. [DOI] [PubMed] [Google Scholar]
- 23.Bonas U, Shulte R, Fenselau S, Minsavage GV, Staskawicz BJ, et al. Isolation of a gene cluster from Xanthomonas campestris pv. vesicatoria that determines pathogenicity and the hypersensitive response on pepper and tomato. Molecular Plant-Microbe Interactions. 1991;15:109–119. [Google Scholar]
- 24.Minamino T, Macnab RM. Domain structure of Salmonella FlhB, a flagellar export component responsible for substrate specificity switching. J Bacteriol. 2000;182:4906–4914. doi: 10.1128/jb.182.17.4906-4914.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ferris HU, Furukawa Y, Minamino T, Kroetz MB, Kihara M, et al. FlhB regulates ordered export of flagellar components via autocleavage mechanism. J Biol Chem. 2005;280:41236–41242. doi: 10.1074/jbc.M509438200. [DOI] [PubMed] [Google Scholar]
- 26.Lavander M, Sundberg L, Edqvist PJ, Lloyd SA, Wolf-Watz H, et al. Proteolytic cleavage of the FlhB homologue YscU of Yersinia pseudotuberculosis is essential for bacterial survival but not for type III secretion. J Bacteriol. 2002;184:4500–4509. doi: 10.1128/JB.184.16.4500-4509.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fraser GM, Hirano T, Ferris HU, Devgan LL, Kihara M, et al. Substrate specificity of type III flagellar protein export in Salmonella is controlled by subdomain interactions in FlhB. Mol Microbiol. 2003;48:1043–1057. doi: 10.1046/j.1365-2958.2003.03487.x. [DOI] [PubMed] [Google Scholar]
- 28.Sorg I, Wagner S, Amstutz M, Muller SA, Broz P, et al. YscU recognizes translocators as export substrates of the Yersinia injectisome. EMBO J. 2007;26:3015–3024. doi: 10.1038/sj.emboj.7601731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wiesand U, Sorg I, Amstutz M, Wagner S, van den Heuvel J, et al. Structure of the type III secretion recognition protein YscU from Yersinia enterocolitica. J Mol Biol. 2009;385:854–866. doi: 10.1016/j.jmb.2008.10.034. [DOI] [PubMed] [Google Scholar]
- 30.Zarivach R, Deng W, Vuckovic M, Felise HB, Nguyen HV, et al. Structural analysis of the essential self-cleaving type III secretion proteins EscU and SpaS. Nature. 2008;453:124–127. doi: 10.1038/nature06832. [DOI] [PubMed] [Google Scholar]
- 31.Deane JE, Graham SC, Mitchell EP, Flot D, Johnson S, et al. Crystal structure of Spa40, the specificity switch for the Shigella flexneri type III secretion system. Mol Microbiol. 2008;69:267–276. doi: 10.1111/j.1365-2958.2008.06293.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Allaoui A, Woestyn S, Sluiters C, Cornelis GR. YscU, a Yersinia enterocolitica inner membrane protein involved in Yop secretion. J Bacteriol. 1994;176:4534–4542. doi: 10.1128/jb.176.15.4534-4542.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Astua-Monge G, Freitas-Astua J, Bacocina G, Roncoletta J, Carvalho SA, et al. Expression profiling of virulence and pathogenicity genes of Xanthomonas axonopodis pv. citri. J Bacteriol. 2005;187:1201–1205. doi: 10.1128/JB.187.3.1201-1205.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fenselau S, Bonas U. Sequence and expression analysis of the hrpB pathogenicity operon of Xanthomonas campestris pv. vesicatoria which encodes eight proteins with similarity to components of the Hrp, Ysc, Spa, and Fli secretion systems. Mol Plant Microbe Interact. 1995;8:845–854. doi: 10.1094/mpmi-8-0845. [DOI] [PubMed] [Google Scholar]
- 35.Koebnik R, Kruger A, Thieme F, Urban A, Bonas U. Specific binding of the Xanthomonas campestris pv. vesicatoria AraC-type transcriptional activator HrpX to plant-inducible promoter boxes. J Bacteriol. 2006;188:7652–7660. doi: 10.1128/JB.00795-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Aldon D, Brito B, Boucher C, Genin S. A bacterial sensor of plant cell contact controls the transcriptional induction of Ralstonia solanacearum pathogenicity genes. EMBO J. 2000;19:2304–2314. doi: 10.1093/emboj/19.10.2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brito B, Aldon D, Barberis P, Boucher C, Genin S. A signal transfer system through three compartments transduces the plant cell contact-dependent signal controlling Ralstonia solanacearum hrp genes. Mol Plant Microbe Interact. 2002;15:109–119. doi: 10.1094/MPMI.2002.15.2.109. [DOI] [PubMed] [Google Scholar]
- 38.Marenda M, Brito B, Callard D, Genin S, Barberis P, et al. PrhA controls a novel regulatory pathway required for the specific induction of Ralstonia solanacearum hrp genes in the presence of plant cells. Mol Microbiol. 1998;27:437–453. doi: 10.1046/j.1365-2958.1998.00692.x. [DOI] [PubMed] [Google Scholar]
- 39.Tahara ST, Mehta A, Rosato YB. Proteins induced by Xanthomonas axonopodis pv. passiflorae with leaf extract of the host plant (Passiflorae edulis). Proteomics. 2003;3:95–102. doi: 10.1002/pmic.200390014. [DOI] [PubMed] [Google Scholar]
- 40.Riordan KE, Sorg JA, Berube BJ, Schneewind O. Impassable YscP substrates and their impact on the Yersinia enterocolitica type III secretion pathway. J Bacteriol. 2008;190:6204–6216. doi: 10.1128/JB.00467-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Macnab RM. How bacteria assemble flagella. Annu Rev Microbiol. 2003;57:77–100. doi: 10.1146/annurev.micro.57.030502.090832. [DOI] [PubMed] [Google Scholar]
- 42.Minamino T, Gonzalez-Pedrajo B, Yamaguchi K, Aizawa SI, Macnab RM. FliK, the protein responsible for flagellar hook length control in Salmonella, is exported during hook assembly. Mol Microbiol. 1999;34:295–304. doi: 10.1046/j.1365-2958.1999.01597.x. [DOI] [PubMed] [Google Scholar]
- 43.Williams AW, Yamaguchi S, Togashi F, Aizawa SI, Kawagishi I, et al. Mutations in fliK and flhB affecting flagellar hook and filament assembly in Salmonella typhimurium. J Bacteriol. 1996;178:2960–2970. doi: 10.1128/jb.178.10.2960-2970.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kutsukake K, Minamino T, Yokoseki T. Isolation and characterization of FliK-independent flagellation mutants from Salmonella typhimurium. J Bacteriol. 1994;176:7625–7629. doi: 10.1128/jb.176.24.7625-7629.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Suzuki T, Iino T. Role of the flaR gene in flagellar hook formation in Salmonella spp. J Bacteriol. 1981;148:973–979. doi: 10.1128/jb.148.3.973-979.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Macnab RM. The bacterial flagellum: reversible rotary propellor and type III export apparatus. J Bacteriol. 1999;181:7149–7153. doi: 10.1128/jb.181.23.7149-7153.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Edqvist PJ, Olsson J, Lavander M, Sundberg L, Forsberg A, et al. YscP and YscU regulate substrate specificity of the Yersinia type III secretion system. J Bacteriol. 2003;185:2259–2266. doi: 10.1128/JB.185.7.2259-2266.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Journet L, Agrain C, Broz P, Cornelis GR. The needle length of bacterial injectisomes is determined by a molecular ruler. Science. 2003;302:1757–1760. doi: 10.1126/science.1091422. [DOI] [PubMed] [Google Scholar]
- 49.Kubori T, Sukhan A, Aizawa SI, Galan JE. Molecular characterization and assembly of the needle complex of the Salmonella typhimurium type III protein secretion system. Proc Natl Acad Sci U S A. 2000;97:10225–10230. doi: 10.1073/pnas.170128997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tamano K, Katayama E, Toyotome T, Sasakawa C. Shigella Spa32 is an essential secretory protein for functional type III secretion machinery and uniformity of its needle length. J Bacteriol. 2002;184:1244–1252. doi: 10.1128/JB.184.5.1244-1252.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Collazo CM, Zierler MK, Galan JE. Functional analysis of the Salmonella typhimurium invasion genes invl and invJ and identification of a target of the protein secretion apparatus encoded in the inv locus. Mol Microbiol. 1995;15:25–38. doi: 10.1111/j.1365-2958.1995.tb02218.x. [DOI] [PubMed] [Google Scholar]
- 52.Payne PL, Straley SC. YscP of Yersinia pestis is a secreted component of the Yop secretion system. J Bacteriol. 1999;181:2852–2862. doi: 10.1128/jb.181.9.2852-2862.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Riordan KE, Schneewind O. YscU cleavage and the assembly of Yersinia type III secretion machine complexes. Mol Microbiol. 2008;68:1485–1501. doi: 10.1111/j.1365-2958.2008.06247.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Agrain C, Callebaut I, Journet L, Sorg I, Paroz C, et al. Characterization of a Type III secretion substrate specificity switch (T3S4) domain in YscP from Yersinia enterocolitica. Mol Microbiol. 2005;56:54–67. doi: 10.1111/j.1365-2958.2005.04534.x. [DOI] [PubMed] [Google Scholar]
- 55.Buttner D, Lorenz C, Weber E, Bonas U. Targeting of two effector protein classes to the type III secretion system by a HpaC- and HpaB-dependent protein complex from Xanthomonas campestris pv. vesicatoria. Mol Microbiol. 2006;59:513–527. doi: 10.1111/j.1365-2958.2005.04924.x. [DOI] [PubMed] [Google Scholar]
- 56.Sambrook J, Russel DW. New York: Cold Spring Harbour Press; 2000. Molecular Cloning: A Laboratory Manual. [Google Scholar]
- 57.Studier FW, Rosenberg AH, Dunn JJ, Dubendorff JW. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- 58.Studier FW, Moffatt BA. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J Mol Biol. 1986;189:113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- 59.Kleber-Janke T, Becker WM. Use of modified BL21(DE3) Escherichia coli cells for high-level expression of recombinant peanut allergens affected by poor codon usage. Protein Expr Purif. 2000;19:419–424. doi: 10.1006/prep.2000.1265. [DOI] [PubMed] [Google Scholar]
- 60.Paulucci AA, Hicks L, Machado A, Miranda MT, Kay CM, et al. Specific sequences determine the stability and cooperativity of folding of the C-terminal half of tropomyosin. J Biol Chem. 2002;277:39574–39584. doi: 10.1074/jbc.M204749200. [DOI] [PubMed] [Google Scholar]
- 61.Guzzo CR, Salinas RK, Andrade MO, Farah CS. PILZ protein structure and interactions with PILB and the FIMX EAL domain: implications for control of type IV pilus biogenesis. J Mol Biol. 2009;393:848–866. doi: 10.1016/j.jmb.2009.07.065. [DOI] [PubMed] [Google Scholar]
- 62.Kovach ME, Elzer PH, Hill DS, Robertson GT, Farris MA, et al. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene. 1995;166:175–176. doi: 10.1016/0378-1119(95)00584-1. [DOI] [PubMed] [Google Scholar]
- 63.De Feyter R, Yang Y, Gabriel DW. Gene-for-genes interactions between cotton R genes and Xanthomonas campestris pv. malvacearum avr genes. Mol Plant Microbe Interact. 1993;6:225–237. doi: 10.1094/mpmi-6-225. [DOI] [PubMed] [Google Scholar]
- 64.Finn RD, Mistry J, Tate J, Coggill P, Heger A, et al. The Pfam protein families database. Nucleic Acids Res. 2010;38:D211–222. doi: 10.1093/nar/gkp985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]