Abstract
(−)-Gambogic acid (1), a biologically active “caged xanthone” from gamboge, the dried resin of Garcinia hanburyi, is of interest as a potential anticancer agent. The planar structure of (−)gambogic acid has been determined previously by analysis of its detailed NMR data and confirmed by single crystal X-ray diffraction, with the absolute configuration at C-13 deduced as R through a series of chemical degradations. Using (−)-morellic acid (2), an analogue of (−)gambogic acid, as a model compound, the 5R, 7S, 10aS, 13R, 27S absolute configuration of (−)gambogic acid was determined for the first time by comparison of physical and spectroscopic data, especially experimental and calculated electronic Circular Dichroism.
(−)-Gambogic acid (1) is a major constituent of gamboge, a resin exuded by Garcinia hanburyi Hook.f (Clusiaceae) used as a folk medicine to treat infections and tumors.1–5 This compound was isolated from G. hanburyi for the first time in 1949,6 and its planar structure, based on the same carbon skeleton as (−)-morellin, was established through several chemical reactions and NMR spectroscopy in 19657 and confirmed by X-ray diffraction analysis in 2001.8 The 1H and 13C NMR spectra of (−)-gambogic acid were assigned with the aid of HMBC and ROESY data,9 and its 13R absolute configuration determined by a series of chemical degradations,10 and supported by the later isolation of an epimer, (13S)-epigambogic acid.11
(−)-Gambogic acid has been found to be cytotoxic for various human cancer cell lines, such as BCG-823 gastric carcinoma,12 SMMC-7721 hepatoma,13 and SPC-A1 lung cancer14 cells, and has shown inhibitory activity in tumor-bearing mouse models.12–15 The mechanism of action of (−)-gambogic acid has been related to factors such as apoptosis induction,16 inhibition of human-topoisomerase-IIα17 and telomerase,18 and modulation of angiogenesis.19 The non-toxic dose of (−)-gambogic acid for Sprague-Dawley rats was established as 60 mg/kg, when administered by gavage once every other day for 13 weeks.20 This compound has been subjected to a Phase I clinical trial as an anticancer agent in the People’s Republic of China, with a dose regimen developed for subsequent Phase II testing.21
Previous studies have demonstrated that the bridged cage moiety of (−)-gambogic acid (1) plays a key role in mediating the cytotoxicity of this compound.22–24 However, the importance of the individual stereogenic centers present in the caged unit is unknown, and the CD spectrum of (−)-gambogic acid has not been reported. Even though the planar structure of (−)-gambogic acid was fully determined by X-ray diffraction analysis, and the absolute configuration of C-13 was deduced as R by a series of chemical degradations in a 1976 study, the absolute configuration at C-5, -7, -10a and -27 is still undetermined because insufficient data were provided by the X-ray crystallographic experiment.8 Several efforts at total synthesis of the caged part of (−)-gambogic acid have been successful,25–28 but they have not resolved the absolute configuration at C-5, -7, -10a and -27 of (−)-gambogic acid, as stated categorically in a recent review paper concerning caged xanthones.5
In our previous collaborative study using electronic circular dichroism (ECD), the absolute configuration at carbons 5, 7, 10a, and 27 of the structurally related caged xanthone, (−)-morellic acid, was determined.29 By comparison of the specific rotation, NMR, and CD spectra of (−)gambogic acid (1) with those of (−)-morellic acid (2), the absolute configuration of (−)-gambogic acid (1) has been determined in the present investigation.
The numbering system used for (−)-gambogic acid (1) and its analogues is not uniform in the literature.2–5 The compound has been numbered from the oxygen atom of a pyran ring linked to the xanthone core.9 (−)-Gambogic acid is a naturally prenylated xanthone, containing a 1,3,6-trihydroxyxanthone core, which may be numbered based on IUPAC provisional recommendations.30 It is recommended that (−)-gambogic acid is numbered in the same manner, based on a prenyl xanthone, 1,3,6-trihydroxy-2,4,5,7-tetraprenylxanthone.
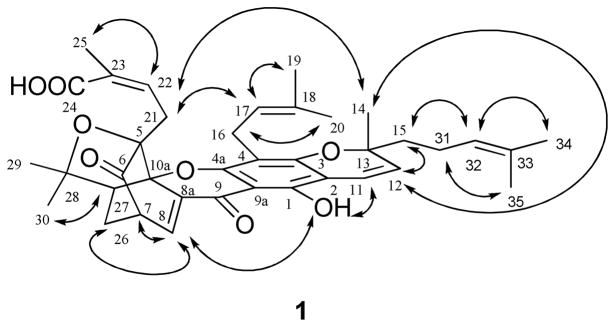
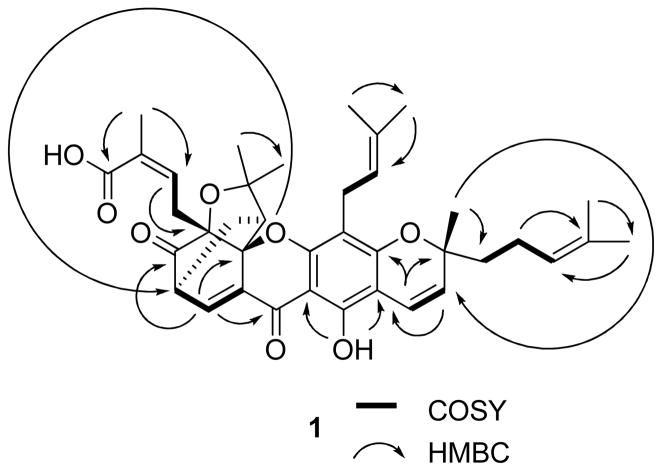
Although the 1H and 13C NMR signals of (−)-gambogic acid (1) were assigned by previous investigators,9 several carbons showed two signals each, indicating that the sample used earlier might be a mixture of two epimers. The 1H and 13C NMR spectra of our (−)-gambogic acid showed one signal for each proton and carbon, respectively, and all protons and carbons were assigned by analysis of its DEPT 90, DEPT 135, COSY, HMQC, and HMBC spectra (Table 1). A boat conformation for the 4-oxatricyclo[4.3.1.03,7]dec-2-one moiety of (−)-gambogic acid was proposed by Rao’s group when they attempted the total synthesis of morellin.31 This proposal was confirmed as result of the X-ray diffraction data of (−)-gambogic acid,8 which indicated an inverted boat conformation for the C-7, C-6, C-5, C-10a, C-26, and C-27 array, with C-27, -28, the oxygen between C-28 and C-5, and C-5 being essentially co-planar with the C-7, C-8, C-8a, and C-10a arrangement. In turn, C-27 lies below the plane defined by C-5, C-6, and C-26.8 Such a configurational diagram is shown in Figure 2, also indicating the NOESY correlations that were observed for 1.
Table 1.
1H and 13C NMR Spectroscopic Data of (−)-Gambogic Acid (1)
| position | δHa (J in Hz) | δCb |
|---|---|---|
| 1 | 157.3 C | |
| 2 | 102.7 C | |
| 3 | 161.5 C | |
| 4 | 107.6 C | |
| 4a | 157.6 C | |
| 5 | 84.0 C | |
| 6 | 203.2 C | |
| 7 | 3.48 (m) | 46.8 CH |
| 8 | 7.54 (d, 6.9) | 135.3 CH |
| 8a | 133.5 C | |
| 9 | 178.8 C | |
| 9a | 100.4 C | |
| 10a | 90.9 C | |
| 11 | 6.60 (d, 10.2) | 115.9 CH |
| 12 | 5.38 (d, 10.2) | 124.5 CH |
| 13 | 81.3 C | |
| 14 | 1.36 (s) | 27.7 CH3 |
| 15 | 1.76 (m) | 42.0 CH2 |
| 1.58 (m) | ||
| 16 | 3.27 (m) | 21.6 CH2 |
| 3.16 (m) | ||
| 17 | 5.04 (m) | 122.2 CH |
| 18 | 131.6 C | |
| 19 | 1.63 (s) | 25.6 CH3 |
| 20 | 1.70 (s) | 18.1 CH3 |
| 21 | 2.93 (m) | 29.3 CH2 |
| 22 | 6.10 (m) | 137.4 CH |
| 23 | 127.9 C | |
| 24 | 170.1 C | |
| 25 | 1.73 (s) | 20.8 CH3 |
| 26 | 1.40 (m) | 25.2 CH2 |
| 2.33 (m) | ||
| 27 | 2.52 (d, 9.3) | 49.0 CH |
| 28 | 83.8 C | |
| 29 | 1.68 (s) | 29.9 CH3 |
| 30 | 1.27 (s) | 28.9 CH3 |
| 31 | 2.03 (m) | 22.7 CH2 |
| 32 | 5.04 (m) | 123.8 CH |
| 33 | 131.8 C | |
| 34 | 1.61 (s) | 25.7 CH3 |
| 35 | 1.53 (s) | 17.6 CH3 |
| OH-1 | 12.80 (s) |
Data were measured in CDCl3 at 300 MHz. Chemical shifts (δ) are in ppm from TMS. J values are in Hz and omitted if the signals were overlapped as multiplets. s = singlet, d = doublet, t = triplet, m = multiplet.
Data were measured in CDCl3 at 75.5 MHz. Chemical shifts (δ) are in ppm from TMS.
Figure 2.
Selected NOESY correlations of (−)-gambogic acid (1).
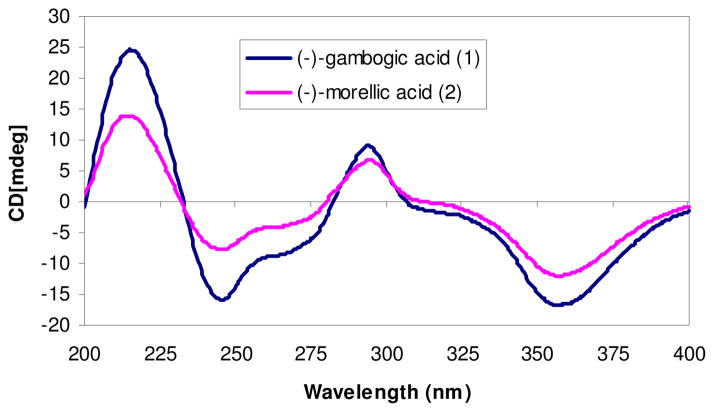
The NMR spectra for (−)-gambogic acid (1) and (−)-morellic acid (2) were measured under the same conditions. All protons and carbons of both compounds were assigned and confirmed by their 2D NMR spectra, and both compounds provided identical 1H and 13C NMR data at H-5, -7, -10a, and -27, and at C -5, -7, -10a, and -27. Further identical HMBC and NOESY correlation profiles of both (−)-gambogic acid and (−)-morellic acid indicated the same relative configurations for both compounds. Also, these two compounds exhibited well-matched CD spectra (Figure 3), displaying negative and positive Cotton effects (CEs) near 360 and 290 nm, respectively. They additionally displayed sequential negative and positive CEs at 246 and 215 nm, arising from exciton coupling of the α,β-unsaturated carbonyl and carboxylic acid chromophores. Critically, the absolute configuration of (−)-morellic acid (2) was defined unambiguously by a combination of experimental and theoretically calculated ECD spectra.29 Therefore, the absolute configuration of the caged unit of (−)-gambogic acid (1) may be defined unequivocally as 5R, 7S, 10aS, and 27S.
Figure 3.
CD spectra of (−)-gambogic acid (1) and (−)-morellic acid (2). The data were obtained in MeOH (0.017 mg/mL for both compounds) on the average of three scans corrected by subtracting a spectrum of the appropriate solution in the absence of the samples recorded under identical conditions. Each scan in the range 200–400 nm was obtained by taking points every 0.5 nm with an integration time of 0.5 sec and a 2 nm band width.
The NOESY correlations between H-7/H-8 and H-22, H-14/H-8, H-16, and H-21, H-16/H-22, and H-17/H-21 indicates the same orientation of the methyl group (C-14) as the prenyl goup linked at the C-5 position. This relative configuration of C-13 indicates a 13R absolute configuration, confirming the previous determination of the absolute configuration of C-13 by chemical degradations.10 Consequently, the absolute configuration of (−)-gambogic acid (1) was determined as 5R, 7S, 10aS, 13R, and 27S.
Both (−)-gambogic acid (1) and (−)-morellic acid (2) were evaluated for their cytotoxicity against the HT-29 human colon cancer cell line, using paclitaxel as positive control, and they were cytotoxic with ED50 values of 0.48 and 0.36 μM, respectively.
Experimental Section
General Experimental Procedures
Optical rotations were measured with a Perkin-Elmer 343 polarimeter. UV spectra were recorded on a Shimadzu UV-2401 PC UV-vis recording spectrophotometer. CD measurements were performed using a JASCO ORDM-401 instrument. IR spectra were recorded on a Nicolet 6700 FT-IR spectrometer. 1H and 13C NMR data, including DEPT, HMQC, HMBC, and COSY spectra, were recorded at room temperature on a Bruker Avance DPX-300 MHz spectrometer, and the NOESY or ROESY NMR spectra were recorded on a Bruker Avance DRX-600 NMR spectrometer, with TMS as internal standard for both the 300 and 600 MHz instruments. ESIMS and HRESIMS were recorded on a LCT-TOF mass spectrometer. (−)-Gambogic acid (1) was purchased from Sigma (G8171 5MG), and (−)-morellic acid (2) was isolated from Garcinia lateriflora.29
(−)-Gambogic acid (1)
Amorphous orange powder showing brown color under UV light at 365 nm; [α]20D – 714.1 (c 0.17, CHCl3); UV (MeOH) λmax (log ε) 290 (3.36), 362 (3.28) nm; CD (MeOH) λmax (Δε) 215 (+17.52), 245.5 (−10.82), 294 (+6.16), 355.5 (−11.45) nm; IR (dried film) νmax 2970, 2927, 1738, 1693, 1634, 1594, 1456, 1383, 1261, 1176, 756 cm−1; 1H and 13C NMR data, see Table 1; positive ESIMS m/z 651.4 [M + Na]+; positive HRESIMS found m/z 651.2931, calcd 651.2934 for C38H44O8Na.
Cytotoxicity Assay
Cytotoxicity of the samples was screened against HT-29 human colon cancer cells by a previously reported procedure, with paclitaxel was used as a positive control (ED50: 0.1 nM).32
Supplementary Material
Figure 1.
COSY and key HMBC correlations of (−)-gambogic acid (1).
Acknowledgments
This investigation was supported, in part, by grant P01 CA125066, funded by the National Cancer Institute, NIH, Bethesda, MD. We thank Jack Fowble, College of Pharmacy, and Dr. Kari Green-Church, Campus Chemical Instrument Center, The Ohio State University, for access to the NMR and MS spectrometers, respectively.
Footnotes
Dedicated to Dr. Koji Nakanishi of Columbia University for his pioneering work on bioactive natural products.
Supporting Information Available: MS and 1H, 13C, DEPT 90, DEPT 135, COSY, NOESY, HMQC, and HMBC NMR spectra of (−)-gambogic acid (1) and overlap profile of ROESY 2D NMR spectra of (−)-gambogic acid (1) and NOESY 2D NMR spectra of (−)-morellic acid (2). This information is available free-of-charge via the Internet at http://pubs.acs.org.
References and Notes
- 1.Asano J, Chiba K, Tada M, Yoshii T. Phytochemistry. 1996;41:815–820. doi: 10.1016/0031-9422(95)00682-6. [DOI] [PubMed] [Google Scholar]
- 2.Han QB, Wang YL, Yang L, Tso TF, Qiao CF, Song JZ, Xu LJ, Chen SL, Yang DJ, Xu HX. Chem Pharm Bull. 2006;54:265–267. doi: 10.1248/cpb.54.265. [DOI] [PubMed] [Google Scholar]
- 3.Reutrakul V, Anantachoke N, Pohmakotr M, Jaipetch T, Sophasan S, Yoosook C, Kasisit J, Napaswat C, Santisuk T, Tuchinda P. Planta Med. 2007;73:33–40. doi: 10.1055/s-2006-951748. [DOI] [PubMed] [Google Scholar]
- 4.Tao SJ, Guan SH, Wang W, Lu ZQ, Chen GT, Sha N, Yue QX, Liu X, Guo DA. J Nat Prod. 2009;72:117–124. doi: 10.1021/np800460b. [DOI] [PubMed] [Google Scholar]
- 5.Han Q-B, Xu H-X. Curr Med Chem. 2009;16:3775–3796. doi: 10.2174/092986709789104993. It is stated on page 3781 of this comprehensive review that the absolute configuration of (−)-gambogic acid and its analogues remained undetermined at the time of publication. [DOI] [PubMed] [Google Scholar]
- 6.Land M, Katz A. Pharm Acta Helv. 1949;24:387–401. [PubMed] [Google Scholar]
- 7.Ollis WD, Ramsay MVJ, Sutherland IO, Mongkolsuk S. Tetrahedron. 1965;21:1453–1470. [Google Scholar]
- 8.Weakley TJR, Cai SX, Zhang H-Z, Keana JFW. J Chem Cryst. 2001;31:501–505. On page 502 of this article, it is mentioned that the quality of the X-ray crystallographic data did not permit definition of the absolute configuration of (−)gambogic acid. [Google Scholar]
- 9.Lin LJ, Lin LZ, Pezzuto JM, Cordell GA, Ruangrungsi N. Magn Reson Chem. 1993;31:340–347. [Google Scholar]
- 10.Cardillo G, Merlini L. Tetrahedron Lett. 1967;27:2529–2530. [Google Scholar]
- 11.Han QB, Yang L, Liu Y, Wang YL, Qiao CF, Song JZ, Xu LJ, Yang DJ, Chen SL, Xu HX. Planta Med. 2006;72:281–284. doi: 10.1055/s-2005-916193. [DOI] [PubMed] [Google Scholar]
- 12.Liu W, Guo QL, You QD, Zhao L, Gu HY, Yuan ST. World J Gastroenterol. 2005;11:3655–3659. doi: 10.3748/wjg.v11.i24.3655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang Y, Yang L, You QD, Nie FF, Gu HY, Zhao L, Wang XT, Guo QL. Cancer Lett. 2007;256:259–266. doi: 10.1016/j.canlet.2007.06.014. [DOI] [PubMed] [Google Scholar]
- 14.Wu ZQ, Guo QL, You QD, Zhao L, Gu HY. Biol Pharm Bull. 2004;27:1769–1774. doi: 10.1248/bpb.27.1769. [DOI] [PubMed] [Google Scholar]
- 15.Zhao J, Qi Q, Yang Y, Gu HY, Lu N, Liu W, Wang W, Qiang L, Zhang LB, You QD, Guo QL. Eur J Pharmacol. 2008;589:127–131. doi: 10.1016/j.ejphar.2008.04.063. [DOI] [PubMed] [Google Scholar]
- 16.Pandey MK, Sung B, Ahn KS, Kunnumakkara AB, Chaturvedi MM, Aggrawal BB. Blood. 2007;110:3517–3525. [Google Scholar]
- 17.Qin YX, Meng LH, Hu CX, Duan WH, Zuo ZL, Lin LP, Zhang XW, Ding J. Mol Cancer Ther. 2007;6:2429–2440. doi: 10.1158/1535-7163.MCT-07-0147. [DOI] [PubMed] [Google Scholar]
- 18.Zhao Q, Yang Y, Yu J, You QD, Zeng S, Gu HY, Lu N, Qi Q, Liu W, Wang XT, Guo QL. Cancer Lett. 2008;262:223–231. doi: 10.1016/j.canlet.2007.12.002. [DOI] [PubMed] [Google Scholar]
- 19.Yi T, Yi Z, Cho SG, Luo J, Pandey MK, Aggrawal BB, Liu M. Cancer Res. 2008;68:1843–1850. doi: 10.1158/0008-5472.CAN-07-5944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Qi Q, You QD, Gu HY, Zhao L, Liu W, Lu N, Guo QL. J Ethnopharmacol. 2008;117:433–438. doi: 10.1016/j.jep.2008.02.027. [DOI] [PubMed] [Google Scholar]
- 21.Zhou ZT, Wang JW. Chin J N Drugs. 2007;16:79–83. [Google Scholar]
- 22.Zhang HZ, Kasibhatla S, Wang Y, Herich J, Guastella J, Tseng B, Drewe J, Cai SX. Bioorg Med Chem. 2004;12:309–317. doi: 10.1016/j.bmc.2003.11.013. [DOI] [PubMed] [Google Scholar]
- 23.Kuemmerle J, Jiang SC, Tseng B, Kasibhatla S, Drewe J, Cai SX. Bioorg Med Chem. 2008;16:4233–4241. doi: 10.1016/j.bmc.2008.02.084. [DOI] [PubMed] [Google Scholar]
- 24.Chantarasriwong O, Cho WC, Batova A, Chavasiri W, Moore C, Rheingold AL, Theodorakis EA. Org Biomol Chem. 2009;7:4886–4894. doi: 10.1039/b913496d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nicolaou KC, Sasmal PK, Xu H, Namoto K, Ritzen A. Angew Chem Int Ed. 2003;42:4225–4229. doi: 10.1002/anie.200351805. [DOI] [PubMed] [Google Scholar]
- 26.Nicolaou KC, Sasmal PK, Xu H. J Am Chem Soc. 2004;126:5493–5506. doi: 10.1021/ja040037+. [DOI] [PubMed] [Google Scholar]
- 27.Nicolaou KC, Xu H, Wartmann M. Angew Chem Int Ed. 2005;44:756–761. doi: 10.1002/anie.200462211. [DOI] [PubMed] [Google Scholar]
- 28.Hayden AE, Xu H, Nicolaou KC, Houk KN. Org Lett. 2006;8:2989–2992. doi: 10.1021/ol060917o. [DOI] [PubMed] [Google Scholar]
- 29.Ren Y, Lantvit DD, Carcache de Blanco EJ, Kardono LBS, Riswan S, Chai H, Cottrell CE, Farnsworth NR, Swanson SM, Ding Y, Li X-C, Marais JPJ, Ferreira D, Kinghorn AD. Tetrahedron. 2010;66:5311–5320. doi: 10.1016/j.tet.2010.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.El-Seedi HR, El-Ghorab DMH, El-Barbary MA, Zayed MF, Göransson U, Larsson S, Verpoorte R. Curr Med Chem. 2009;16:2581–2626. doi: 10.2174/092986709788682056. [DOI] [PubMed] [Google Scholar]
- 31.Raghavan S, Rao GSRS. Heterocycles. 1994;37:131–136. [Google Scholar]
- 32.Seo EK, Kim NC, Mi QW, Chai HB, Wall ME, Wani MC, Navarro HA, Burgess JP, Graham JG, Cabieses F, Tan GT, Farnsworth NR, Pezzuto JM, Kinghorn AD. J Nat Prod. 2001;64:1483–1485. doi: 10.1021/np0103158. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.