Abstract
MicroRNAs (miRNAs) are initially expressed as long transcripts that are processed in the nucleus to yield ∼65-nucleotide (nt) RNA hairpin intermediates, termed pre-miRNAs, that are exported to the cytoplasm for additional processing to yield mature, ∼22-nt miRNAs. Here, we demonstrate that human pre-miRNA nuclear export, and miRNA function, are dependent on Exportin-5. Exportin-5 can bind pre-miRNAs specifically in vitro, but only in the presence of the Ran-GTP cofactor. Short hairpin RNAs, artificial pre-miRNA analogs used to express small interfering RNAs, also depend on Exportin-5 for nuclear export. Together, these findings define an additional cellular cofactor required for miRNA biogenesis and function.
Keywords: MicroRNA, nuclear export, RNA interference, siRNA
MicroRNAs (miRNAs) are ∼22-nucleotide (nt) noncoding RNAs observed in a wide range of eukaryotes. Over 200 genomically encoded miRNAs have been identified in several different species (Ambros 2003). Although few animal miRNAs have been assigned a function to date, the Caenorhabditis elegans let-7 and lin-4 miRNAs and the miR-14 and bantam miRNAs found in Drosophila all repress the expression of mRNAs bearing partially complementary target sites (Ambros 2003). In C. elegans, let-7 and lin-4 expression is developmentally regulated, and loss of let-7 or lin-4 function results in disruption of normal larval development due to the inappropriate expression of their mRNA targets (Lee et al. 1993; Reinhart et al. 2000). Consistent with a similar role for miRNAs in vertebrate development, many murine miRNAs are also expressed in a developmentally regulated or tissue-specific manner (Lagos-Quintana et al. 2002; Houbaviy et al. 2003).
The miRNAs are initially expressed as part of an imperfect RNA hairpin of ∼80 nt in length that in turn forms part of a longer initial transcript termed a primary miRNA (pri-miRNA; Lee et al. 2002). The first step in miRNA biogenesis is the nuclear excision of the upper part of this RNA hairpin to give the ∼65-nt pre-miRNA intermediate (Lee et al. 2002; Zeng and Cullen 2003). This processing step is performed by human RNAse III, also called `Drosha' (Lee et al. 2003). The pre-miRNA intermediate, which in the case of human miR-30 consists of a 63-nt hairpin bearing a 2-nt 3′ overhang, is then exported to the cytoplasm by a currently unknown mechanism. Once there, the pre-miRNA is processed by a second RNAse III family member called `Dicer' to give the mature ∼22-nt miRNA (Grishok et al. 2001; Hutvágner et al. 2001; Ketting et al. 2001). The miRNA is then incorporated into the RNA-induced silencing complex (RISC), where it functions to guide RISC to appropriate mRNA targets (Hammond et al. 2000; Martinez et al. 2002; Mourelatos et al. 2002; Schwarz et al. 2002).
In addition to miRNAs, cells can also generate similar ∼22-nt noncoding RNAs called small interfering RNAs (siRNA), by Dicer processing of long double-stranded RNAs (dsRNAs; Zamore et al. 2000; Bernstein et al. 2001). siRNAs, which can also be introduced into cells by transfection of synthetic siRNA duplexes (Elbashir et al. 2001), can program RISC to cleave mRNAs bearing perfectly complementary target sites. This process, termed RNA interference (RNAi; Fire et al. 1998), has emerged as a powerful technique for the analysis of gene function in both invertebrate and vertebrate cells. However, because RNAi induced by artificial siRNA duplexes gives only a transient repression of the target gene, efforts have been made to derive vectors able to produce siRNAs constitutively. The most prevalent technique for stable siRNA expression involves transcription of a short hairpin RNA (shRNA) using an RNA polymerase III (pol III)-dependent promoter (Brummelkamp et al. 2002; Paddison et al. 2002). The shRNAs that have been described generally feature perfect 19-29-base pair (bp) stems with small terminal loops and a 2-nt 3′ overhang, consisting of two “U” residues, that results from termination of pol III transcription. Therefore, the structure of shRNAs is closely analogous to the structure of the miR-30 pre-miRNA, which also forms an RNA hairpin bearing a 2-nt 3′ overhang.
Several members of the karyopherin family of nucleocytoplasmic transport factors have been shown to play a role in the nuclear export of noncoding RNAs, including tRNAs, snRNAs, and rRNAs (Lei and Silver 2002). A defining characteristic of karyopherins that function in nuclear export is that nuclear cargo binding requires the GTP-bound form of the Ran GTPase, whereas cytoplasmic hydrolysis of Ran-GTP to Ran-GDP induces cargo release. The karyopherin Exportin-5 (Exp5; Brownawell and Macara 2002) mediates the nuclear export of adenovirus VA1, a highly structured 160-nt noncoding RNA, and can also function as a secondary receptor for tRNA nuclear export (Gwizdek et al. 2003; Bohnsack et al. 2002; Calado et al. 2002). In the case of VA1, Exp5 binding requires a terminal dsRNA “mini-helix” of >14 bp bearing a base-paired 5′ end and a 3′ extension of ≥3 nt (Gwizdek et al. 2003). As these structural determinants are shared with both pre-miRNAs and shRNAs, we asked whether Exp5 might also be required for nuclear export of these small noncoding RNAs. Here, we demonstrate that Exp5 is indeed required for the nuclear export and, hence biological activity, of pre-miRNAs and shRNAs but not for synthetic siRNA function. Moreover, we show that Exp5 binds the miR-30 pre-miRNA precursor specifically in vitro, but only in the presence of Ran-GTP. These results identify a major cellular export cargo for Exp5 and define a novel cofactor for miRNA biogenesis and function.
Results and Discussion
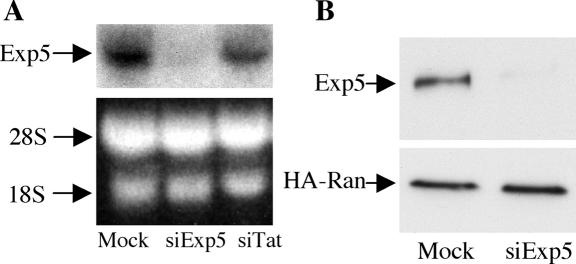
To test whether Exp5 is indeed involved in pre-miRNA nuclear export, we first identified a synthetic siRNA able to effectively knock-down Exp5 expression in human cells. An siRNA duplex targeted to residues 339-360 of the human Exp5 open reading frame (ORF) was transfected into 293T cells at time 0 and again 36 h and 60 h later. Exp5 gene expression was analyzed an additional 36 h later. As shown in Figure 1, this treatment dramatically and specifically reduced the endogenous level of expression of both Exp5 mRNA and protein.
Figure 1.
Knockdown of endogenous human Exp5 mRNA and protein expression by RNA interference. (A) 293T cells were transfected at 0, 36, and 60 h with the siExp5 RNA duplex, with the siTat duplex as a negative control, or mock-transfected. At 96 h, Exp5 mRNA expression levels were determined by Northern analysis. Ribosomal RNA served as a loading control. (B) Similar to panel A, except that 293T cells were cotransfected with pBC12/MS-HA-Ran, which expresses an HA-tagged Ran protein, at 60 h. Western analysis was performed at 96 h using a rabbit polyclonal anti-Exp5 antiserum or an HA-specific mouse monoclonal antibody.
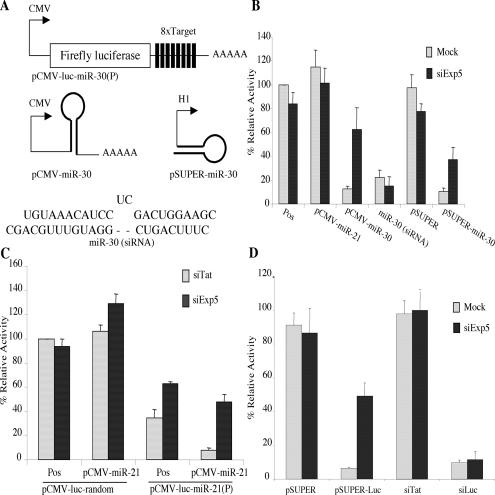
We previously described a firefly luciferase (luc)-based indicator plasmid, termed pCMV-luc-miR-30(P), that contains eight target sites for the human miR-30 miRNA inserted into the 3′ untranslated region (Fig. 2A; Zeng et al. 2003). As a result, luc expression from this plasmid is potently reduced by expression of miR-30. We also previously described a plasmid, pCMV-miR-30, that expresses authentic human miR-30 as a long mRNA similar to a pri-miRNA (Fig. 2A; Zeng et al. 2002). As in the case of endogenous miR-30, expression of miR-30 by pCMV-miR-30 requires nuclear excision of the pre-miRNA intermediate, followed by nuclear export and Dicer processing (Zeng and Cullen 2003). A second miR-30 expression plasmid, termed pSuper-miR-30, uses the pol III-dependent H1 promoter to express a 63-nt RNA hairpin that is identical to the authentic pre-miR-30 intermediate except that the 2-nt 3′ overhang has been changed from 5′-GC-3′ to 5′-UU-3′ (Fig. 2A). Finally, we can also introduce miR-30 into cells by direct transfection of a synthetic ∼22-nt RNA duplex consisting of the miR-30 miRNA annealed to the miR-30 antisense miRNA (Fig. 2A).
Figure 2.
Loss of Exp5 expression specifically relieves inhibition of gene expression caused by pre-miRNAs or shRNAs. (A) Schematic representation of the pCMV-luc-miR-30(P) indicator plasmid, and of the different miR-30 RNA variants used. (B) At 60 h, coincident with the third siExp5 or mock transfection, 293T cells were cotransfected with the pCMV-luc-miR-30(P) indicator plasmid, a Renilla luciferase internal control plasmid, the indicated pBC12/CMV- or pSuper-derived control or miRNA expression plasmids, or the synthetic miR-30(siRNA) RNA duplex. Firefly and Renilla luciferase expression levels were determined at 96 h and adjusted for minor variations in the Renilla internal control. Pos (Positive control) refers to mock- or siExp5-transfected cultures that had been cotransfected with pCMV-luc-miR-30(P) and the Renilla and pBC12/CMV control plasmids. Data are presented relative to the firefly luciferase activity detected in the mock-transfected positive control culture, which was set at 100. Average of three experiments with standard deviation indicated. (C) These data were generated as described in panel B except that the indicator plasmids pCMV-luc-miR-21(P) and pCMV-luc-random were used. In addition, this panel used cells transfected with the siTat duplex as the control for siExp5. (D) These data were obtained as described in B, using the pCMV-luc-miR-30(P) indicator plasmid, and are presented as shown in B.
If Exp5 is essential for pre-miRNA export, but not for miRNA function per se, then RNAi of Exp5 should relieve the inhibition of pCMV-luc-miR-30(P)-derived luc enzyme expression caused by pCMV-miR-30 and pSuper-miR-30, both of which produce nuclear pre-miR-30 intermediates, but should not relieve inhibition caused by the synthetic miR-30 RNA duplex. As shown in Figure 2B, that is exactly what is observed. Specifically, knockdown of Exp5 expression had no enhancing effect on the level of luc expression observed in the absence of miR-30, or when miR-30 was introduced as part of a synthetic RNA duplex, but it markedly enhanced the level of luc expression seen in cells cotransfected with the pCMV-luc-miR-30(P) indicator and either pCMV-miR-30 or pSuper-miR-30.
The pCMV-luc-miR-21(P) indicator plasmid (Zeng et al. 2003) is identical to pCMV-luc-miR-30(P) (Fig. 2A) except that it contains eight target sites for the human miR-21 miRNA. The control indicator plasmid pCMV-luc-random instead contains eight insertions of a random sequence of similar size. As shown in Figure 2C, luc expression from the control pCMV-luc-random indicator plasmid was not significantly affected by overexpression of miR-21, by cotransfection of the pCMV-miR-21 expression plasmid, or by transfection of cells with siExp5. As previously reported (Zeng et al. 2003), the pCMV-lucmiR-21(P) indicator plasmid gives rise to a significantly lower level of luc activity in transfected 293T cells compared to pCMV-luc-random, due to the action of endogenous miR-21. The miR-21 miRNA, unlike miR-30, is expressed at readily detectable levels in 293T cells (Zeng et al. 2003). Overexpression of miR-21, by cotransfection of pCMV-miR-21, results in a further, profound inhibition of luc expression. Importantly, the level of luc activity encoded by pCMV-luc-miR-21(P) was significantly increased in siExp5-transfected cells, in both the presence and absence of overexpressed miR-21. These data argue that the inhibitory effect of both exogenous and endogenous miR-21 is dependent on the biological activity of the Exp5 nuclear export factor.
Finally, we asked whether Exp5 is also required for RNA interference by an shRNA. In fact, knockdown of Exp5 expression did not rescue luc expression that had been inhibited by a synthetic, luc-specific siRNA but did enhance luc expression that had been inhibited when that same siRNA was initially transcribed as an shRNA (Fig. 2D).
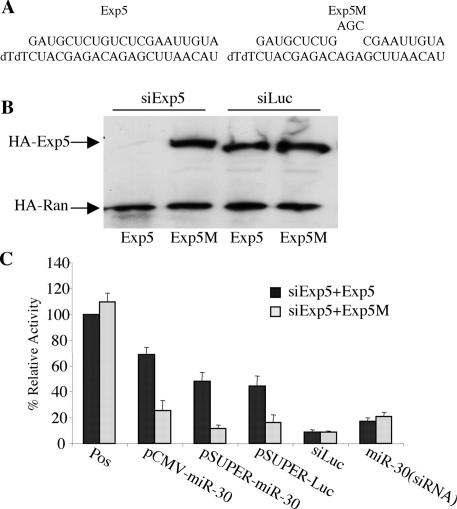
Given recent data suggesting that RNA interference can sometimes exert a nonspecific inhibitory effect (Bridge et al. 2003), we wished to confirm that the inhibition in miRNA or shRNA function shown in Figure 2 was specific. We therefore constructed expression plasmids encoding influenza hemagglutinin (HA)-tagged forms of either wild-type human Exp5 or an Exp5 mutant (Exp5M) that contains a 3-nt mutation in the center of the siRNA target (Fig. 3A). This mutation is predicted to inhibit RNAi produced by this siRNA but does not change the Exp5 amino acid sequence. Cotransfection of 293T cells with the Exp5-specific siRNA gave no detectable Exp5 protein expression in cells transfected with the wild-type Exp5 sequence, but resulted in readily detectable levels in cells transfected with the mutated Exp5M gene (Fig. 3B). Knockdown of wild-type Exp5 expression was specific, as expression of an HA-tagged control protein was unaffected.
Figure 3.
RNAi of Exp5 exerts a specific phenotype. (A) Sequence of the Exp5 siRNA (lower strand) and of the mRNA target (upper strand) in wild-type Exp5 and in the Exp5M mutant. (B) Coincident with the third siRNA transfection, here using either siExp5 or siLuc as a control, cells were cotransfected with plasmids expressing HA-tagged versions of wild-type or mutant Exp5 or Ran. A further 36 h later, HA-tagged protein levels were determined by Western analysis. (C) This transfection experiment was performed as described in Fig. 2B, except that at 60 h the 293T cells were also cotransfected with plasmids expressing wild-type Exp5 or the Exp5M mutant. All cultures were treated with the siExp5 RNA duplex. The Pos (Positive control) culture here refers to cells transfected with the pCMV-luc-miR-30(P) indicator, the Renilla and pBC12/CMV control plasmids, and the siExp5 RNA duplex. Data are given relative to the Pos culture cotransfected with the wild-type Exp5 expression plasmid, which was set at 100.
If the Exp5-specific siRNA is indeed blocking the biological activity of pre-miRNAs and shRNAs by knocking down Exp5 expression, then cotransfection of cells with a vector expressing the Exp5M mutant should rescue this activity. In fact, expression of Exp5M did restore the inhibition of luc expression induced by pCMV-miR-30, pSuper-miR-30, and pSuper-luc (Fig. 3C). In contrast, pCMV-Exp5M did not reduce the level of luc activity seen in the absence of any overexpressed pre-miRNA or shRNA and also did not further lower the level of luc activity seen in cells cotransfected with the synthetic miR-30 or luc siRNA duplex. Similarly, overexpression of Exportin-t or Crm1, two karyopherins involved in the export of other noncoding RNAs, also had no significant effect (data not shown). We conclude that the Exp5-specific siRNA reverses the ability of pre-miRNA and shRNA expression plasmids to inhibit target mRNA expression (Fig. 2) by selectively inhibiting Exp5 expression.
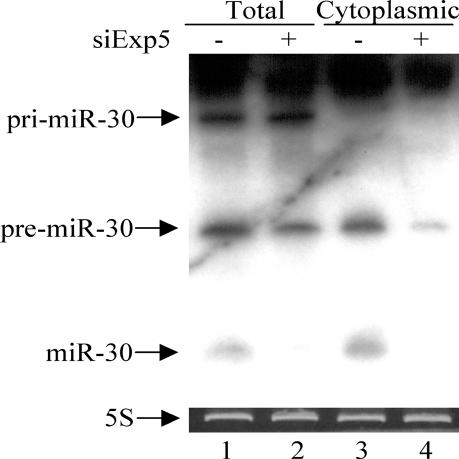
If Exp5 is required for pre-miRNA nuclear export and processing, then knockdown of Exp5 expression should result in a significant drop in the level of expression of the mature miRNA and possibly also an increase in the level of expression of the pre-miRNA intermediate. As shown in Figure 4, knockdown of Exp5 expression had little effect on the level of expression of the initial pri-miR-30 transcript or of the pre-miR-30 intermediate, as assessed by Northern analysis of total cellular RNA, but did significantly reduce the level of expression of the mature miR-30 miRNA (Fig. 4, cf. lanes 1 and 2). A more dramatic Exp5 phenotype was observed upon analysis of cytoplasmic RNA. Specifically, there was a clear reduction in the expression of both pre-miR-30 and mature miR-30 in the cytoplasm of cells expressing reduced levels of Exp5 (Fig. 4, cf. lanes 3 and 4). No pri-miR-30 was detected in the cytoplasm of pCMV-miR-30-transfected cells regardless of the level of Exp5 expression. Although these data are clearly consistent with the hypothesis that Exp5 is critical for the nuclear export, and hence cytoplasmic processing, of pre-miRNAs, it is interesting that no accumulation of the pre-miRNA intermediate was detected. We note that Exp5 binding to the VA1 noncoding RNA is highly sensitive to minor changes at the base of the terminal RNA helix, for example being disrupted by a 5′ overhang (Gwizdek et al. 2003; data not shown). Therefore, we hypothesize that Exp5 may bind to the base of the RNA mini-helix found in both VA1 and pre-miRNAs and thereby not only activate their nuclear export but also protect these short noncoding RNAs from exonucleolytic digestion. Loss of Exp5 expression would therefore potentially lead not only to the nuclear retention of pre-miRNAs but also to their nuclear degradation.
Figure 4.
Analysis of miR-30 RNA expression. 293T cells were transfected with siExp5 or mock-transfected. At 60 h, coincident with the third siExp5 or mock transfection, cells were also cotransfected with pCMV-miR-30; 36 h later, total and cytoplasmic RNA fractions were isolated and subjected to Northern analysis. The location of the pri-miR-30, pre-miR-30, and mature miR-30 RNAs is indicated. 5S rRNA served as a loading control.
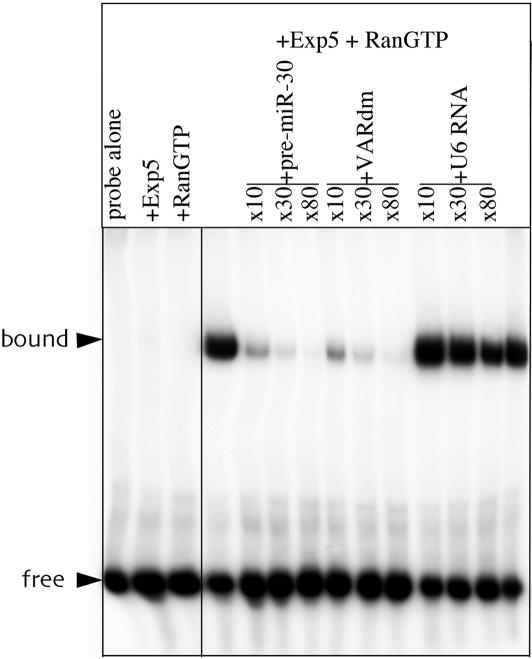
Exp5 is a member of the karyopherin family of nucleocytoplasmic factors. A defining characteristic of karyopherins that function in nuclear export is that cargo binding requires the GTP-bound form of the Ran GTPase (Lei and Silver 2002). To test whether Exp5 is indeed able to bind the 63-nt pre-miR-30 RNA in a Ran-GTP dependent manner, we used T7 RNA polymerase to transcribe a radiolabeled 63-nt RNA probe that is identical to the pre-miR-30 RNA except that the first nucleotide was changed from U to G to accommodate the requirements of this polymerase. To maintain the integrity of the pre-miR-30 RNA stem, this also necessitated a change in residue 61, at the 3′ end of the pre-miRNA stem, from A to C. This RNA probe was incubated with purified, recombinant His-tagged human Exp5 and/or recombinant Ran-GTP. As determined by gel-shift analysis, the pre-miR-30 probe did not bind either Exp5 or Ran-GTP alone but formed a readily detectable complex when incubated with both proteins simultaneously (Fig. 5).
Figure 5.
Specific binding of pre-miR-30 by Exp5 in the presence of Ran-GTP. A 32P-labeled pre-miR-30 RNA probe was incubated with recombinant Exp5-His and/or recombinant Ran-GTP in the presence or absence of the indicated fold excess of various unlabeled competitor RNAs. Protein:RNA complexes were detected by nondenaturing gel electrophoresis and autoradiography.
To show that formation of this RNA-protein complex was specific, we next asked whether unlabeled RNA competitors would block complex formation if added in excess. As shown in Figure 5, formation of the pre-miR-30/Exp5/Ran-GTP complex was indeed inhibited when the unlabeled competitor used was either the pre-miR-30 RNA itself or VARdm, a derivative of adenovirus VA1 RNA previously shown to bind Exp5 with high affinity (Gwizdek et al. 2003). In contrast, an irrelevant RNA competitor, the moderately structured, 104-nt U6 snRNA, failed to affect complex formation. We therefore conclude that Exp5 binds the pre-miR-30 miRNA precursor specifically and in a Ran-GTP-dependent manner.
In this paper, we have demonstrated that the nuclear export, and hence biological activity, of the human miR-30 pre-miRNA is dependent on the Exp5 nuclear export factor. The miR-30 pre-miRNA, the only pre-miRNA whose structure is currently known, consists of an imperfect 63-nt RNA hairpin bearing a 2-nt 3′ extension generated by nuclear processing by Drosha (Lee et al. 2003). As other pre-miRNAs also require Drosha for nuclear excision from their cognate pri-miRNA (Lee et al. 2003), it seems probable that other pre-miRNAs will also consist of small RNA hairpins bearing 2-nt 3′ overhangs and therefore will also be able to bind Exp5 specifically. The finding that the inhibitory activity of both endogenous and overexpressed human miR-21 is also dependent on Exp5 (Fig. 2C) is consistent with this hypothesis.
Although evidence documenting the structure of pre-miR-30 has only recently been reported, we note that the most prevalent design for artificial shRNAs also consists of an RNA hairpin bearing a 2-nt overhang (Brummelkamp et al. 2002; Paddison et al. 2002). Our data suggest that this structural similarity to pre-miRNAs allows artificial shRNAs to also recruit Exp5 for their nuclear export (Fig. 2D). Evidence obtained with the adenovirus VA1 RNA suggests that Exp5 binding requires a >14-bp RNA stem with a base-paired 5′ end and a short 3′ overhang (Gwizdek et al. 2003). Therefore, both pre-miR-30 and the luc shRNA would appear to be equivalently good substrates for Exp5 binding. However, we note that Kawasaki and Taira (2003) reported that shRNAs bearing artificial terminal loops are exported from the nucleus less effectively than shRNAs bearing terminal loops derived from miRNAs, and thus there may be room to optimize shRNA design for enhanced Exp5 recruitment. We also note that two groups have reported the accumulation of pre-let-7 miRNA precursors at different stages during fruit fly and sea urchin development (Pasquinelli et al. 2000; Hutvágner et al. 2001). Although this could represent a defect in RNA processing per se, it is also possible that it is the nuclear export of the prelet-7 miRNA precursor, and hence access to cytoplasmic Dicer, that is developmentally regulated.
In addition to defining a cellular factor that plays a critical role in miRNA biogenesis and function, the present study also identifies the first RNA cargo of cellular origin that is dependent on Exp5 for nuclear export. Given evidence that individual miRNAs may be expressed at up to 50,000 copies per cell in some organisms (Lim et al. 2003), it appears possible that pre-miRNAs may represent the primary cellular export cargo for Exp5. We also note that it was recently reported that loss-of-function mutations of HASTY, the Arabidopsis homolog of Exp5, severely disrupt the normal development of this plant species (Bollman et al. 2003). Although the mechanism underlying this phenotype was not determined, our data suggest that this likely results from a global inhibition in plant miRNA nuclear export and function.
Materials and methods
Plasmids and siRNAs
The parental expression plasmid pBC12/CMV and its derivatives pCMV-luc-miR-30(P), pCMV-luc-miR-21(P), pCMV-luc-random (Zeng et al. 2003), pCMV-miR-30, pCMV-miR-21 (Zeng and Cullen 2003), pBC12/MS2-HA-Ran (Yi et al. 2002), and pSuper (Brummelkamp et al. 2002) have been described. pSuper-miR-30 was derived by insertion of a DNA oligonucleotide, encoding the first 61 nt of the 63-nt pre-miR-30 RNA linked 3′ to the sequence 5′-TTTTT-3′, between the unique BglII and HindIII sites in pSuper. pSuper-Luc was designed as described (Brummelkamp et al. 2003) and is predicted to transcribe an shRNA consisting of a 27-bp RNA stem in which the 3′ arm is complementary to residues 155-181 of GL2 luciferase (Elbashir et al. 2001). pCMV-HA-Exp5 was constructed by inserting the Exp5 ORF (Bohnsack et al. 2002) into a pBC12/CMV derivative encoding an amino-terminal HA tag. pCMV-Exp5M was generated from pCMV-HA-Exp5 by PCR-mediated mutagenesis.
All synthetic RNAs were obtained from Dharmacon. The siTat and siLuc siRNA duplexes have been described (Elbashir et al. 2001; Coburn and Cullen 2002). The miR-30(siRNA) duplex was obtained by annealing synthetic RNAs identical in sequence to human miR-30 and miR-30as (Zeng et al. 2002). The 22-bp siExp5 RNA duplex was designed to target residues 339-360 of the human Exp5 ORF (Fig. 3).
Transfection and luciferase assays
All transfections were performed on human 293T cells cultured in 24-well tissue culture plates. Lipofectamine 2000 (Invitrogen) was used for both siRNA and plasmid DNA transfections, using the manufacturer's protocols. RNAi for Exp5 was routinely performed by transfection of 100 nM of the siExp5 RNA duplex at 0 h and again at ∼36 h. A further 24 h later, cells were again transfected with siExp5 together with relevant indicator and reporter plasmids. At this stage, cells were transfected with 10 ng of a reporter plasmid and 1 ng of the pRL-CMV Renilla luciferase internal control plasmid (Promega). In addition, cells were cotransfected with 50 ng of pCMV-miR-30, pCMV-miR-21, or pBC12/CMV; 10 ng of pSuper-miR-30, pSuper-luc, or pSuper; or 100 nM of siTat, siLuc, or miR-30(siRNA). After a final 36-h period, cells were lysed and assayed for both firefly and Renilla luciferase activity. Where indicated, cells were mock-transfected rather than transfected with siExp5.
RNA and protein expression analysis
For analysis of miR-30 RNA expression, total and cytoplasmic RNA fractions were isolated as described (Yi et al. 2002). Equal amounts of RNA were separated by electrophoresis through a 15% TBE-Urea gel (Bio-Rad), transferred to Hybond-N nylon membranes, and probed using a 63-nt end-labeled DNA probe complementary to full-length pre-miR-30. For RNA analysis of Exp5 mRNA expression, total RNA was isolated and Northern analyses performed using a nick-translated, 32P-labeled Exp5 probe.
For analysis of endogenous Exp5 protein expression levels, 293T cells were cotransfected with pBC12/MS2-HA-Ran coincident with the third siExp5 transfection. Cells were lysed 36 h later, and extracts were subjected to Western analysis using a 1:350 dilution of an affinity-purified rabbit polyclonal antiserum raised against the full-length recombinant Exp5 protein (Gwizdek et al. 2003). For analysis of HA-tagged protein expression, 293T cells were cotransfected with pCMV-HA-Exp5, pCMVHA-Exp5M, and/or pBC12/MS2-HA-Ran coincident with the third siExp5 transfection, and cells were lysed 36 h later. HA-tagged protein expression levels were determined by Western analysis using a monoclonal mouse anti-HA antibody (Covance). Horseradish peroxidase-conjugated secondary antibodies (Amersham) and enhanced chemiluminescence followed by autoradiography were used to detect reactive proteins.
Electrophoretic mobility shift assay
A [32P]-labeled pre-miR-30 RNA probe, and unlabeled competitor RNAs, were synthesized using Riboscribe T7 probe synthesis and Ampliscribe T7 transcription kits (Epicentre), respectively. Recombinant His-tagged full-length Exp5 (15 nM; Gwizdek et al. 2003) was incubated with 0.5 μM of recombinant Ran(Q69L)-GTP plus 3 nM of the [32P]-pre-miR-30 probe, and varying concentrations of competitor RNAs, in a total volume of 20 μL at 25°C. The binding buffer contained 20 mM HEPES, pH 7.3, 150 mM potassium acetate, 2 mM magnesium acetate, 0.1% Tween-20, 7 mM 2-mercaptoethanol, 1.5 μg/mL poly dIdC, and 0.2% BSA. After 45 min, 0.6 μg of heparin was added to the reaction and incubated for another 5 min. Samples were then analyzed by electrophoresis on a pre-run 5% nondenaturing polyacrylamide gel at 4°C, followed by autoradiography.
Acknowledgments
The authors thank Dirk Görlich (ZMBH, Heidelberg, Germany), James Dahlberg (University of Wisconsin, Madison), and Carole Gwidzek and Catherine Dargemont (Institut Jacques Monod, CNRS, France) for reagents used in this work. This work was supported by the Howard Hughes Medical Institute (B.R.C.) and by grant R01 GM50526 (I.G.M.) from the NIH, DHHS.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article published online ahead of print. Article and publication date are at http://www.genesdev.org/cgi/doi/10.1101/gad.1158803.
References
- Ambros V. 2003. MicroRNA pathways in flies and worms: Growth, death, fat, stress, and timing. Cell 113: 673-676. [DOI] [PubMed] [Google Scholar]
- Bernstein E., Caudy, A.A., Hammond, S.M., and Hannon, G.J. 2001. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 409: 363-366. [DOI] [PubMed] [Google Scholar]
- Bohnsack M.T., Regener, K., Schwappach, B., Saffrich, R., Paraskeva, E., Hartmann, E., and Görlich, D. 2002. Exp5 exports eEF1A via tRNA from nuclei and synergizes with other transport pathways to confine translation to the cytoplasm. EMBO J. 21: 6205-6215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollman K.M., Aukerman, M.J., Park, M.-Y., Hunter, C., Berardini, T.Z., and Poethig, R.S. 2003. HASTY, the Arabidopsis ortholog of exportin 5/MSN5, regulates phase change and morphogenesis. Development 130: 1493-1504. [DOI] [PubMed] [Google Scholar]
- Bridge A.J., Pebernard, S., Ducraux, A., Nicoulaz, A.-L., and Iggo, R. 2003. Induction of an interferon response by RNAi vectors in mammalian cells. Nat. Genet. 34: 263-264. [DOI] [PubMed] [Google Scholar]
- Brownawell A.M. and Macara, I.G. 2002. Exportin-5, a novel karyopherin, mediates nuclear export of double-stranded RNA binding proteins. J. Cell Biol. 156: 53-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brummelkamp T.R., Bernards, R., and Agami, R. 2002. A system for stable expression of short interfering RNAs in mammalian cells. Science 296: 550-553. [DOI] [PubMed] [Google Scholar]
- Calado A., Treichel, N., Müller, E.-C., Otto, A., and Kutay, U. 2002. Exportin-5-mediated nuclear export of eukaryotic elongation factor 1A and tRNA. EMBO J. 21: 6216-6224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coburn G.A. and Cullen, B.R. 2002. Potent and specific inhibition of human immunodeficiency virus type 1 replication by RNA interference. J. Virol. 76: 9225-9231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbashir S.M., Harborth, J., Lendeckel, W., Yalcin, A., Weber, K., and Tuschl, T. 2001. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 411: 494-498. [DOI] [PubMed] [Google Scholar]
- Fire A., Xu, S., Montgomery, M.K., Kostas, S.A., Driver, S.E., and Mello, C.C. 1998. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391: 806-811. [DOI] [PubMed] [Google Scholar]
- Grishok A., Pasquinelli, A.E., Conte, D., Li, N., Parrish, S., Ha, I., Baillie, D.L., Fire, A., Ruvkun, G., and Mello, C.C. 2001. Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell 106: 23-34. [DOI] [PubMed] [Google Scholar]
- Gwizdek C., Ossarch-Nazari, B., Brownawell, A.M., Doglio, A., Bertrand, E., Macara, I.G., and Dargemont, C. 2003. Exportin-5 mediates nuclear export of minihelix-containing RNAs. J. Biol. Chem. 278: 5505-5508. [DOI] [PubMed] [Google Scholar]
- Hammond S.M., Bernstein, E., Beach, D., and Hannon, G.J. 2000. An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature 404: 293-295. [DOI] [PubMed] [Google Scholar]
- Houbaviy H., Murray, M.F., and Sharp, P.A. 2003. Embryonic stem cell specific microRNAs. Dev. Cell 5: 351-358. [DOI] [PubMed] [Google Scholar]
- Hutvágner G., McLachlan, J., Pasquinelli, A.E., Bálint, É., Tuschl, T., and Zamore, P.D. 2001. A cellular function for the RNA-interference enzyme dicer in the maturation of the let-7 small temporal RNA. Science 293: 834-838. [DOI] [PubMed] [Google Scholar]
- Kawasaka H. and Taira, K. 2003. Short hairpin type of dsRNAs that are controlled by tRNAVal promoter significantly induce RNAi-mediated gene silencing in the cytoplasm of human cells. Nucleic Acids Res. 31: 700-707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ketting R.F., Fischer, S.E., Bernstein, E., Sijen, T., Hannon, G.J., and Pasterk, R.H. 2001. Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans. Genes & Dev. 15: 2654-2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagos-Quintana M., Rauhut, R., Yalcin, A., Meyer, J., Lendeckel, W., and Tuschl, T. 2002. Identification of tissue-specific microRNAs from mouse. Curr. Biol. 12: 735-739. [DOI] [PubMed] [Google Scholar]
- Lee R.C., Feinbaum, R., and Ambros, V. 1993. The heterochronic gene lin-4 of C. elegans encodes two small RNA's with antisense complementarity to lin-14. Cell 75: 843-854. [DOI] [PubMed] [Google Scholar]
- Lee Y., Jeon, K., Lee, J.-T., Kim, S., and Kim, V.N. 2002. MicroRNA maturation: Stepwise processing and subcellular localization. EMBO J. 21: 4663-4670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y., Ahn, C., Han, J., Choi, H., Kim, J., Yim, J., Lee, J., Provost, P., Radmark, O., Kim, S., et al. 2003. The nuclear RNase III drosha initiates microRNA processing. Nature 425: 415-419. [DOI] [PubMed] [Google Scholar]
- Lei E.P. and Silver, P.A. 2002. Protein and RNA export from the nucleus. Dev. Cell 2: 261-272. [DOI] [PubMed] [Google Scholar]
- Lim L.P., Lau, N.C., Weinstein, E.G., Abdelhakim, A., Yekta, S., Rhoades, M.W., Burge, C.B., and Bartel, D.P. 2003. The microRNAs of Caenorhabditis elegans. Genes & Dev. 17: 991-1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez J., Patkaniowska, A., Urlaub, H., Lührmann, R., and Tuschl, T. 2002. Single-stranded antisense siRNAs guide target RNA cleavage in RNAi. Cell 110: 563-574. [DOI] [PubMed] [Google Scholar]
- Mourelatos Z., Dostie, J., Paushkin, S., Sharma, A., Charroux, B., Abel, L., Rappsilber, J., Mann, M., and Dreyfuss, G. 2002. miRNPs: A novel class of ribonucleoproteins containing numerous microRNAs. Genes & Dev. 16: 720-728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paddison P.J., Caudy, A.A., Bernstein, E., Hannon, G.J., and Conklin, D.S. 2002. Short hairpin RNAs (shRNAs) induce sequence-specific silencing in mammalian cells. Genes & Dev. 16: 948-958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasquinelli A.E., Reinhart, B.J., Slack, F., Martindale, M.Q., Kuroda, M.I., Maller, B., Hayward, D.C., Ball, E.E., Degnan, B., Müller, P., et al. 2000. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature 408: 86-89. [DOI] [PubMed] [Google Scholar]
- Reinhart B.J., Slack, F.J., Basson, M., Pasquinelli, A.E., Bettinger, J.C., Rougvie, A.E., Horvitz, H.R., and Ruvkun, G. 2000. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature 403: 901-906. [DOI] [PubMed] [Google Scholar]
- Schwarz D.S., Hutvágner, G., Haley, B., and Zamore, P.D. 2002. Evidence that siRNAs function as guides, not primers, in the Drosophila and human RNAi pathways. Mol. Cell 10: 537-548. [DOI] [PubMed] [Google Scholar]
- Yi R., Bogerd, H.P., Wiegand, H.L., and Cullen, B.R. 2002. Both Ran and importins have the ability to function as nuclear mRNA export factors. RNA 8: 180-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamore P.D., Tuschl, T., Sharp, P.A., and Bartel, D.P. 2000. RNAi: Double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell 101: 25-33. [DOI] [PubMed] [Google Scholar]
- Zeng Y. and Cullen, B.R. 2003. Sequence requirements for micro RNA processing and function in human cells. RNA 9: 112-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Y., Wagner, E.J., and Cullen, B.R. 2002. Both natural and designed micro RNAs can inhibit the expression of cognate mRNAs when expressed in human cells. Mol. Cell 9: 1327-1333. [DOI] [PubMed] [Google Scholar]
- Zeng Y., Yi, R., and Cullen, B.R. 2003. MicroRNAs and small interfering RNAs can inhibit mRNA expression by similar mechanisms. Proc. Natl. Acad. Sci. 100: 9779-9784. [DOI] [PMC free article] [PubMed] [Google Scholar]