Abstract
In this work we describe the identification of a copper-inducible regulon in Mycobacterium tuberculosis (Mtb). Among the regulated genes was Rv0190/MT0200, a paralogue of the copper metalloregulatory repressor CsoR. The five-locus regulon, which includes a gene that encodes the copper-protective metallothionein MymT, was highly induced in wild type Mtb treated with copper, and highly expressed in an Rv0190/MT0200 mutant. Importantly, the Rv0190/MT0200 mutant was hyper-resistant to copper. The promoters of all five loci share a palindromic motif that was recognized by the gene product of Rv0190/MT0200. For this reason we named Rv0190/MT0200 RicR for regulated in copper repressor. Intriguingly, several of the RicR-regulated genes, including MymT, are unique to pathogenic Mycobacteria. The identification of a copper-responsive regulon specific to virulent mycobacterial species suggests copper homeostasis must be maintained during an infection. Alternatively, copper may provide a cue for the expression of genes unrelated to metal homeostasis, but nonetheless necessary for survival in a host.
Introduction
Mtb is the causative agent of tuberculosis and infects an estimated nine million people per year. One to two million people die from tuberculosis each year, making Mtb one of the most deadly infectious agents in the world (http://www.who.int/tb/publications/global_report/2009/update/en/index.html). Infection occurs upon inhalation of aerosolized droplets containing Mtb bacilli. Left untreated, most infections do not immediately lead to disease, but rather remain latent for many years [reviewed in (Saunders & Britton, 2007)]. Mtb has evolved to survive inside macrophages where it faces a multitude of stresses, including reactive nitrogen and oxygen intermediates (RNI, ROI), acidification of the phagosome, and iron limitation [reviewed in (Flynn & Chan, 2001, Pieters, 2008)].
While host-inflicted stresses are typically bactericidal, Mtb has developed ways to resist elimination by these defense mechanisms. Previous studies revealed that the Mtb proteasome is necessary for the bacteria to resist RNI toxicity in vitro and cause lethal infections in mice [reviewed in (Darwin, 2009)]. Proteasomes are barrel-shaped compartmentalized proteases that degrade proteins in a highly regulated manner [reviewed in (Cerda-Maira & Darwin, 2009)]. Mtb proteasome function requires Mpa (Mycobacterium proteasomal ATPase), a hexameric ATPase that unfolds and translocates substrates into the proteasome core (Darwin et al., 2005, Pearce et al., 2006, Striebel et al., 2010) and PafA (proteasome accessory factor A), which ligates the small post-translational modifier Pup (prokaryotic ubiquitin-like protein) to proteins, dooming them for degradation (Festa et al., 2007, Pearce et al., 2008, Striebel et al., 2009). Despite significant progress in characterizing the biochemistry of the Mtb proteasome system, it is not known how proteolysis is linked to Mtb resistance to RNI or virulence.
In an effort to begin to understand the link between Mtb proteasome function and pathogenesis, we sought to determine the role of the proteasome in transcriptional regulation. Compartmentalized proteases are often involved in regulating the stability of transcription factors, either directly or indirectly [reviewed in (Gottesman, 2003, Collins & Tansey, 2006)]. Notably, non-proteasomal, self-compartmentalized proteases have been implicated in virulence gene expression in bacterial pathogens [reviewed in (Butler et al., 2006)]. Based on these observations, we used pafA and mpa mutants to look for proteasomal degradation-dependent transcriptional changes that might reveal pathways important for pathogenesis. Under normal culture conditions, we observed significant changes in gene expression in less than 2% of the 4,009 predicted open reading frames (orfs) in pafA and mpa mutants compared to wild type (WT) Mtb. From this analysis, we identified a previously unrecognized copper responsive regulon. This regulon is controlled by RicR (GenBank accession number GU726749), a homologue of the Mtb copper-sensitive operon repressor, CsoR (Liu et al., 2007). Disruption of ricR resulted in the constitutive expression of the entire regulon and hyper-resistance of Mtb to copper. In contrast, CsoR regulated only a single operon that includes csoR and a putative copper transporter gene, ctpV (cation transporter P-type ATPase). Thus, our studies showed that defects in proteasome-dependent degradation result in transcriptional changes in Mtb. Furthermore, there appears to be at least two independent pathways to respond to copper-mediated stress, which may be critical during infection of a host.
Results
Transcriptional profiles of proteasome degradation-defective Mtb
We sought to identify genes that were regulated in a proteasome-dependent manner by comparing the transcriptomes of WT Mtb with pafA or mpa mutant strains (Table S1 for strain information). We did not analyze proteasome protease-disrupted Mtb because genetic or chemical disruption of protease activity results in a general growth defect (Gandotra et al., 2007, Darwin et al., 2003), which is not observed for mpa or pafA mutants (Darwin et al., 2003). Because PafA and Mpa are both involved in the Pup-proteasome degradation pathway, we hypothesized that the transcriptional profiles of pafA and mpa mutants would be similar. We also analyzed the transcriptome of a mutant with a transposon insertion in the penultimate codon of mpa (“mpa607”), which results in the synthesis of an Mpa allele with ATPase activity but unable to facilitate protein degradation (Darwin et al., 2003, Pearce et al., 2006). Therefore, we analyzed the mpa607 mutant to determine if it had a different transcriptional profile from an mpa transposon null mutant.
RNA for microarray analysis was harvested at mid-log [optical density absorbance at 580 nm (OD580) = 0.4] and early stationary (OD580 = 2.0) phases (see Experimental Procedures). Genes were considered significantly regulated if the change in transcript levels was greater than 2-fold after significance analysis of microarrays (SAM) analysis comparing all four transcriptomes (Kooperberg et al., 2002). Data from the mid-log cultures showed either minimal change (less than 2-fold) in the transcriptional profiles between strains, or were high variability among experiments (data not shown). The microarray results from early stationary phase were more consistent and therefore used for further studies and validation. We observed similar transcriptional profiles between all three mutants (pafA, mpa, mpa607) suggesting that PafA and Mpa do not have independent roles that lead to differing transcriptional responses under the conditions tested (Tables 1, 2, and S2; Geo Accession GSE23947).
Table 1.
Genes more than two-fold up-regulated in pafA, mpa, and mpa607 mutants as determined by microarray analysisa, sorted by locus.
| Locus | Gene Nameb | mpa607c | mpac | pafAc | Zur Regulond |
IdeR regulatede |
|---|---|---|---|---|---|---|
| Rv0106 | hypothetical protein | 2.27 | 3.25 | 4.21 | + | |
| Rv0280 | PPE FAMILY PROTEIN (ppe3) | 3.74 | 3.85 | 4.10 | + | + |
| Rv0281 | hypothetical protein | 1.97 | 1.94 | 2.46 | + | + |
| Rv0282 | hypothetical protein | 2.42 | 2.15 | 2.49 | + | + |
| Rv0282 | hypothetical protein | 1.94 | 2.00 | 2.62 | + | + |
| Rv0283 | POSSIBLE CONSERVED MEMBRANE PROTEIN | 1.80 | 2.06 | 2.34 | + | + |
| Rv0284 | POSSIBLE CONSERVED MEMBRANE PROTEIN | 2.25 | 2.33 | 2.51 | + | + |
| Rv0285 | PE FAMILY PROTEIN (pe5) | 1.82 | 1.91 | 2.19 | + | + |
| Rv0286 | PPE FAMILY PROTEIN (ppe4) | 1.56 | 1.85 | 2.16 | + | + |
| Rv0287 | ESAT-6 LIKE PROTEIN (esxG) | 1.73 | 1.61 | 2.10 | + | + |
| Rv0289 | hypothetical protein | 1.93 | 2.00 | 2.20 | + | + |
| Rv0290 | PROBABLE CONSERVED TRANSMEMBRANE PROTEIN |
1.68 | 1.94 | 1.96 | + | + |
| Rv0292 | PROBABLE CONSERVED TRANSMEMBRANE PROTEIN |
1.70 | 1.89 | 2.13 | + | + |
| Rv0885 | hypothetical protein | 1.92 | 1.75 | 2.49 | ||
| Rv2055c | Probable ribosomal protein S18 (rpsR2) | 2.63 | 2.65 | 3.81 | + | |
| Rv2056c | Probable ribosomal protein S14 (rpsN2) | 2.68 | 2.43 | 4.21 | + | |
| Rv2057c | Probable ribosomal protein L33 (rpmG1) | 2.94 | 2.91 | 4.20 | + | |
| Rv2058c | Probable 50S ribosomal protein L28 (rpmB2) | 4.15 | 3.47 | 6.12 | + | |
| Rv2059 | hypothetical protein | 1.66 | NaN | 2.17 | + | |
| Rv2359 | zur | 0.69 | 0.54 | 0.94 | ||
| Rv3074 | hypothetical protein | 2.57 | 1.76 | 1.87 |
Dye-flipped experiments were performed in duplicate for each mutant compared to WT. Statistical analysis was determined by SAM analysis between all conditions.
Functional annotations are taken from the Mtb strain H37Rv complete genome sequence (www.jcvi.org)
Numbers represent expression in the mutant strains relative to WT Mtb in 7H9 media grown to early stationary phase
Table 2.
Genes more than two-fold down-regulated in pafA, mpa, and mpa607 mutants as determined by microarray analysisa, sorted by locus.
| Locus | Gene Nameb | mpa607c | mpac | pafAc |
|---|---|---|---|---|
| Rv0140 | hypothetical protein | 0.40 | 0.56 | 0.48 |
| Rv0654 | PROBABLE DIOXYGENASE | 0.34 | 0.41 | 0.49 |
| Rv0765c | PROBABLE OXIDOREDUCTASE | 0.48 | 0.65 | 0.54 |
| Rv0766c | PROBABLE CYTOCHROME P450 123 (cyp123) | 0.41 | 0.47 | 0.54 |
| Rv0767c | hypothetical protein | 0.28 | 0.49 | 0.47 |
| Rv0768 | PROBABLE ALDEHYDE DEHYDROGENASE NAD DEPENDENT ALDA (aldA) | 0.38 | 0.48 | 0.46 |
| Rv0769 | PROBABLE DEHYDROGENASE/REDUCTASE | 0.47 | 0.53 | 0.51 |
| Rv0790c | hypothetical protein | 0.45 | 0.58 | 0.50 |
| Rv0791c | hypothetical protein | 0.39 | 0.52 | 0.39 |
| Rv0792c | PROBABLE TRANSCRIPTIONAL REGULATORY PROTEIN (PROBABLY GNTR-FAMILY) | 0.37 | 0.55 | 0.50 |
| Rv0793 | hypothetical protein | 0.47 | 0.67 | 0.56 |
| Rv0847 | PROBABLE LIPOPROTEIN (lpqS) | 0.24 | 0.52 | 0.33 |
| Rv0848 | POSSIBLE CYSTEINE SYNTHASE A (cysK2) | 0.32 | 0.56 | 0.42 |
| Rv0850 | PUTATIVE TRANSPOSASE (FRAGMENT) | 0.33 | 0.70 | 0.39 |
| Rv1169c | PE FAMILY PROTEIN (pe11) | 0.41 | 0.66 | 0.54 |
| Rv1218c | PROBABLE TETRONASIN-TRANSPORT ATP-BINDING PROTEIN ABC TRANSPORTER | 0.32 | 0.67 | 0.40 |
| Rv1285 | PROBABLE SULFATE ADENYLYLTRANSFERASE SUBUNIT 2 (cysD) | 0.24 | 0.41 | 0.35 |
| Rv1286 | PROBABLE BIFUNCTIONAL ENZYME CYSN/CYSC (cysN) | 0.30 | 0.54 | 0.37 |
| Rv1287 | hypothetical protein | 0.44 | 0.67 | 0.44 |
| Rv2466c | hypothetical protein | 0.34 | 0.49 | 0.45 |
| Rv2517c | hypothetical protein | 0.43 | 0.52 | 0.54 |
| Rv2641 | CADMIUM INDUCIBLE PROTEIN (cadI) | 0.47 | 0.63 | 0.50 |
| Rv2642 | POSSIBLE TRANSCRIPTIONAL REGULATORY PROTEIN (PROBABLY ARSR-FAMILY) | 0.39 | 0.53 | 0.46 |
| Rv2873 | CELL SURFACE LIPOPROTEIN (mpt83) | 0.46 | 0.52 | 0.54 |
| Rv2875 | MAJOR SECRETED IMMUNOGENIC PROTEIN (mpt70) | 0.40 | 0.42 | 0.41 |
| Rv2912c | PROBABLE TRANSCRIPTIONAL REGULATORY PROTEIN (PROBABLY TETR-FAMILY) | 0.26 | 0.42 | 0.25 |
| Rv2913c | POSSIBLE D-AMINO ACID AMINOHYDROLASE (D-AMINO ACID HYDROLASE) | 0.23 | 0.41 | 0.36 |
| Rv2962c | POSSIBLE GLYCOSYL TRANSFERASE | 0.41 | 0.64 | 0.51 |
| Rv2963 | PROBABLE INTEGRAL MEMBRANE PROTEIN | 0.25 | 0.60 | 0.38 |
| Rv2987c | PROBABLE 3-ISOPROPYLMALATE DEHYDRATASE (SMALL SUBUNIT) (leuD) | 0.56 | 0.40 | 0.43 |
| Rv3249c | POSSIBLE TRANSCRIPTIONAL REGULATORY PROTEIN (PROBABLY TETR-FAMILY) | 0.34 | 0.46 | 0.42 |
| Rv3250c | PROBABLE RUBREDOXIN (rubB) | 0.37 | 0.50 | 0.46 |
| Rv3251c | PROBABLE RUBREDOXIN (rubA) | 0.34 | 0.50 | 0.47 |
| Rv3252c | PROBABLE TRANSMEMBRANE ALKANE 1-MONOOXYGENASE (alkB) | 0.30 | 0.45 | 0.43 |
| Rv3269 | hypothetical protein | 0.40 | 0.65 | 0.59 |
| Rv3270 | PROBABLE METAL CATION-TRANSPORTING P-TYPE ATPASE C (ctpC) | 0.37 | 0.63 | 0.51 |
| Rv3463 | hypothetical protein | 0.37 | 0.53 | 0.48 |
| MT0196 | mymT | 0.36 | 0.71 | 0.47 |
Dye-flipped experiments were performed in duplicate for each mutant compared to WT. Statistical analysis was determined by SAM analysis between all conditions.
Functional annotations are taken from the Mtb strain H37Rv complete genome sequence (www.jcvi.org)
Numbers represent expression in the mutant strains relative to WT Mtb in 7H9 media grown to early stationary phase
In bold are genes with the palindrome.
Many genes in the Zur (zinc uptake regulator, Rv2359) regulon were up-regulated in the pafA and mpa mutants (Maciag et al., 2007) (Table 1). When zinc is low, Zur is released from its operators and gene expression is induced. The induced genes presumably aid the bacteria during zinc-limiting conditions. Among the Zur-regulated genes was the esx-3 (esat-6, region 3) operon, which is also controlled by IdeR (iron dependant repressor, Rv2711) (Rodriguez et al., 2002), and is important for iron uptake during iron-limiting conditions (Serafini et al., 2009, Siegrist et al., 2009). IdeR directly represses the esx-3 locus, but does not appear to regulate other Zur regulon genes (Maciag et al., 2007, Rodriguez et al., 2002). IdeR-dependent/Zur-independent genes (Gold et al., 2001) were not differentially expressed between the WT and mpa or pafA mutants, suggesting that the changes in esx-3 transcript levels were due to effects through Zur rather than IdeR. Notably, zur transcript levels were unchanged in all strains tested (Table 1, Geo Accession GSE23947).
We hypothesized that decreased intracellular levels of zinc in the pafA and mpa mutants resulted in the de-repression of the Zur regulon. To attempt to address this, we performed inductively coupled plasma dynamic reaction cell mass spectrometry (ICP-DRC-MS) to quantify intracellular zinc amounts. It is important to note that vanishingly small amounts of buffered or bio-available zinc are present within a cell; zinc-coordinating enzymes and ribosomes largely sequester zinc (Hitomi et al., 2001). The ICP-DRC-MS results indicated that total zinc concentrations in the WT and degradation-defective Mtb strains were relatively similar between samples and biological replicates (Fig. S1). It is notable that E. coli Zur can respond to less than femtomolar changes in free zinc, much like other zinc metalloregulatory proteins (Outten & O'Halloran, 2001, Ma et al., 2009c), and our analysis might not have been sensitive enough to detect small but significant alterations in the concentration of bio-available or rapidly exchanging zinc. Thus, it remains to be determined how disruption of proteasome function affects Zur-dependent transcription.
Identification of a regulatory element controlling promoters repressed in proteasome degradation-defective Mtb
Unlike the up-regulated genes, the down-regulated genes did not share a known regulatory mechanism. Most of these genes were unlinked and encoded hypothetical proteins (Table 2). Because the same transcriptional regulator typically controls co-regulated genes, we examined the putative promoter regions of all down-regulated genes in an attempt to identify a common regulator binding site. A palindrome, 5’-TACCC-N5-G/AGGTA-3’, was identified upstream of lpqS (putative lipoprotein; Rv0847) and Rv2963 (putative permease) (Fig. 1A). To determine if these palindromes were located near transcriptional start sites, we performed rapid amplification of 5’ complementary DNA ends (5’RACE) for both genes. The palindrome was between or overlapped tentative −10/−35 regions of each promoter, which strongly suggested the mechanism of transcriptional control was through the action of a repressor.
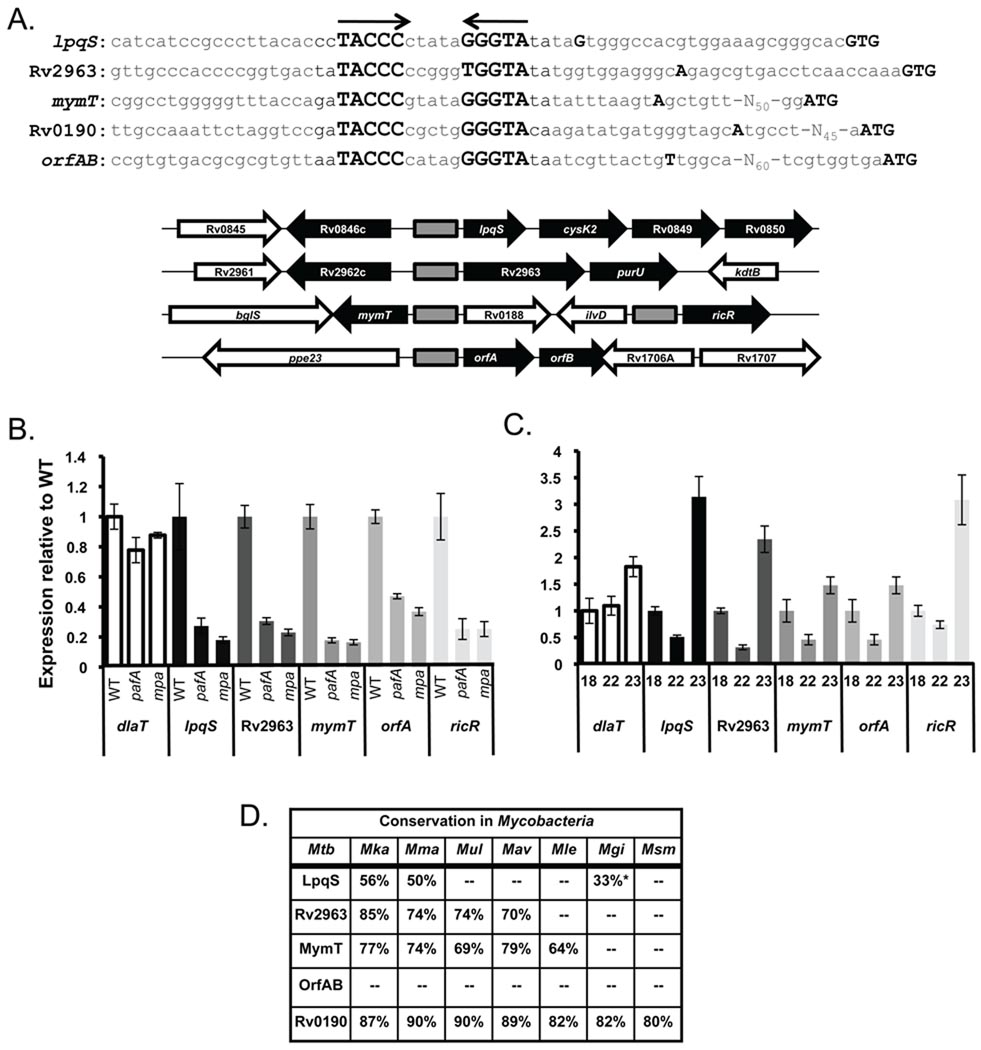
Fig. 1. Identification of a regulatory element in five loci repressed in proteasome degradation-defective Mtb.
(A) Alignment of the lpqS, Rv2963, mymT ricR, and orfAB promoters. The putative transcriptional start sites as determined by 5’RACE are indicated by double-underlines, and the presumed start codons are at the 3’ end of the sequence in capitals and in bold. The palindromic sequences are in bold and indicated by arrows. (Below) Genomic organization around each palindrome (gray boxes). Genes that were down-regulated in the pafA and mpa strains are in black arrows. (B) qRTPCR revealed that ricR orfA and mymT transcripts were decreased in pafA and mpa mutant strains when compared to WT Mtb. Transcript levels were normalized to rpoB (RNA polymerase β-subunit) for each gene in each condition or strain tested. Fold changes relative to WT Mtb are plotted on the y-axis. Experiments are representative of three biological replicates, each performed in triplicate. dlaT (dihydrolipoamide acyltransferase, Rv2215) was used as a negative control. Transcript levels of lpqS, Rv2963, mymT ricR, and orfAB were significantly lower in the pafA and mpa mutants (Student’s t-test, p < 0.05). (C) Complementation of the mpa mutation resulted in restoration of near WT levels of lpqS, Rv2963, mymT ricR, and orfA transcripts. Strains used for complementation were MHD18 (WT+pMV306, empty vector), MHD22 (mpa+pMV306), and MHD23 (mpa+pMV-mpa). The decreased levels in the mpa mutant were statistically significantly different from the WT and complemented strains. (D) Conservation of lpqS, Rv2963, mymT, orfAB and Rv0190 in other Mycobacteria. Mka = M. kansasii, Mma = M. marinum; Mul = M. ulcerans; Mav = M. avium, Mle = M. leprae, Mka = M. kansansii, Mgi = M. gilvum, Msm = M. smegmatis. The asterisk (*) signifies the homologue did not have the palindromic sequence found upstream of Mtb lpqS.
To determine if other promoters contained this regulatory element, we searched for this palindrome in the Mtb genome using the Search Pattern tool on GenoList (http://genodb.pasteur.fr). We performed a stringent search using the sequence 5’-TACCCNNNNNGGGTA-3’ and allowed for two mismatches. Three additional loci with this palindrome were identified: one upstream of a gene annotated as a "conserved hypothetical" (Rv0190, herein called ricR) and previously identified as a copper responsive gene (Ward et al., 2008); one upstream of mymT (encoding a copper metallothionein) (Gold et al., 2008); and the other in a region between ppe23 (Rv1706c) and Rv1706A (Fig. 1A). Upon closer examination, the palindrome near ppe23 was actually upstream of two small orfs: “orfA” and “orfB” were 186 bp and 165 bp, respectively, in length. Neither was annotated in H37Rv but orfB was annotated in the CDC1551 genome sequence as MT1746.1. A transcript corresponding to the orfAB operon was detected in Mtb H37Rv (Fig. S2).
5’RACE analysis suggested that the palindrome was between tentative −10/−35 sites of ricR, mymT and orfAB as observed for lpqS and Rv2963 (Fig. 1A). Upon further inspection of the microarray data, we found mymT was indeed down-regulated in the mpa and pafA mutants, but less than two-fold (Table 2). ricR and MT1746.1 were not identified in the microarray data (Geo Accession GSE23947), but it was possible that the changes in transcript levels were not consistent among experiments based on the statistical analysis of microarrays (SAM) analysis. Nonetheless, quantitative real time PCR (qRTPCR) analysis showed transcript levels of ricR, mymT, and orfAB were decreased in the pafA and mpa mutants as observed for lpqS and Rv2963 (Fig. 1B). Complementation of the mpa mutation resulted in the restoration of near WT expression of these genes (Fig. 1C).
lpqS, mymT and orfAB are only found in pathogenic mycobacteria, suggesting they play a role in host-pathogen interactions (Fig. 1D). LpqS homologues with greater than 50% identity are only found in three other sequenced species, M. kansasii, M. marinum and M. ulcerans. orfAB is present only in the Mtb complex, which includes M. bovis. Among Mycobacteria, Rv2963 is only found among pathogenic species although homologues with lower similarity are present in other bacterial genera. In contrast, RicR homologues are present in many bacterial species. Taken together, we identified five loci that share a palindrome in their promoters, suggesting they are controlled by a common regulatory mechanism.
Identification of a copper-inducible regulon
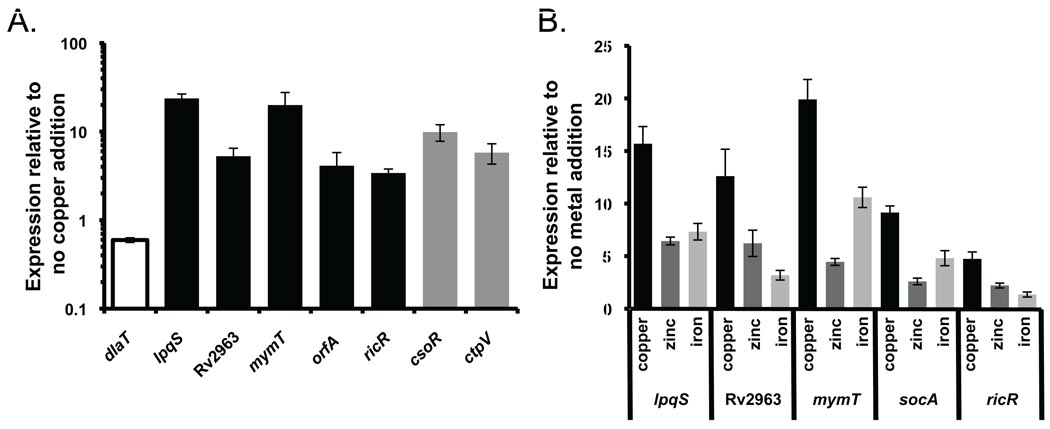
A previous report by Ward and co-workers identified numerous genes that are differentially regulated in Mtb treated with copper (Ward et al., 2008). Among these genes, Rv2963 and ricR were induced 15.7- and 8.5-fold, respectively (Ward et al., 2008). A gene downstream of lpqS, Rv0848, was also induced by copper (Ward et al., 2008). Metallothioneins are synthesized upon metal stress to bind excess zinc or copper (Sato & Bremner, 1993); consistent with this mymT is also highly induced by copper and other metals (Gold et al., 2008). Based on these observations, we decided to test if lpqS and orfAB, along with mymT, Rv2963 and ricR, were also copper inducible. After growing WT Mtb in Sauton’s minimal media to stationary phase (OD580 = 1.5) and treating with 500 µM copper sulfate (CuSO4) for four hours, we found that all five loci were induced by copper (Figure 2A). Because orfAB was not previously annotated, we now refer to this operon as socAB (small orf induced by copper A and B; GenBank accession number HM222605). As an additional control for copper induction, we measured the expression of two previously characterized copper-inducible genes, csoR and ctpV (Liu et al., 2007, Ward et al., 2008). As expected, copper induction was observed for both of these genes (Fig. 2A).
Fig. 2. Genes repressed in pafA and mpa mutants are copper inducible.
(A) Induction of genes in response to 500 µM CuSO4 for four hours. The genes repressed in the proteasome degradation-defective strains (in black) were all significantly induced in WT Mtb (H37Rv) in response to copper as shown by qRTPCR. csoR and ctpV are known copper-inducible genes that were used as positive controls. dlaT served as a negative control. (B) In WT Mtb, lpqS, Rv2963, mymT, ricR, and orfA were significantly induced by 500 µM CuSO4, 10-fold more iron (0.05% w/v ferric ammonium citrate) and 500 µM ZnSO4 when compared to untreated samples grown in Sauton’s minimal media. ricR transcripts appear to have the lowest response to the metals tested. Data are representative of two biological replicates, each performed in triplicate.
To test the metal specificity of this regulon, we treated WT Mtb with other biologically relevant metals (manganese, iron, cobalt, nickel, and zinc) or silver and measured the expression of all five genes. None of the genes was induced by manganese, cobalt, nickel and silver (data not shown). In contrast, all genes were induced to varying degrees by iron or zinc (Fig. 2B). Consistent with our data, mymT is induced by metals in addition to copper (Gold et al., 2008). While this increase was significant over untreated samples, it is unclear if iron or zinc specifically induced these promoters.
We hypothesized that lpqS, Rv2963, mymT, socAB, and ricR were down-regulated in the pafA and mpa mutants when compared to WT because these strains had lower intracellular copper concentrations. As with zinc, ICP-DRC-MS experiments showed that copper was not significantly reduced in any of the strains (Fig. S1). Thus, it remains unclear how defects in proteasome function affect the expression of these copper-inducible genes.
RicR is a copper responsive regulator
We hypothesized that the palindrome identified in the promoters of lpqS, Rv2963, mymT, ricR, and socAB was a repressor-binding site. Thus our next aim was to identify the repressor that controlled this regulon. We first tested CsoR as the possible repressor. The cso operon, consisting of csoR, Rv0968, ctpV and Rv0970, is de-repressed in the presence of copper as CsoR dissociates from its operator upon copper binding (Liu et al., 2007). However, it was not known if CsoR regulated other loci in Mtb. To determine whether or not CsoR was the regulator of the newly recognized regulon, we used microarrays to compare the transcriptomes of WT and csoR-deleted/disrupted (ΔcsoR∷hyg) Mtb (see Experimental Procedures). Because we were measuring the expression of genes physically unlinked to csoR, we were not concerned about polar effects caused by the hygromycin gene disrupting csoR.
In Sauton's minimal media without added copper, the expression of genes downstream of csoR (Rv0968, ctpV, Rv0970) in the ΔcsoR∷hyg mutant were slightly elevated compared to WT Mtb grown to stationary phase (OD580 ~1.5) (Tables 3 and S3, Geo Accession GSE23498). If we had introduced an in frame, unmarked deletion mutation in csoR, we would have expected to see a high level of expression of the cso operon due to the absence of its cognate repressor. However, because of the likely polar nature of the mutation we introduced into csoR, we did not observe this. We thus predicted that the addition of CuSO4 would not increase the expression of the cso operon in the ΔcsoR∷hyg mutant. Upon supplementation of the growth media with 500 µM CuSO4 and incubation for four hours, the cso operon was strongly induced in WT Mtb but not in the ΔcsoR∷hyg mutant, consistent with our prediction (Table 3). Interestingly, no other genes appeared to be differentially regulated between the WT and ΔcsoR∷hyg strains, suggesting CsoR only regulates its own promoter under the conditions tested. Importantly, lpqS, Rv2963, mymT, ricR, and socAB expression was not altered in the ΔcsoR∷hyg mutant, which was confirmed by qRTPCR (Fig. S3B). Thus, CsoR was not the regulator of the five copper-regulated loci that were repressed in the proteasome degradation-defective Mtb strains.
Table 3.
Genes differentially regulated in a ΔcsoR∷hyg mutant.
| locus | gene | Namec | Cu−a | Cu+b |
|---|---|---|---|---|
| Rv0967 | csoR | hypothetical protein | 0.09 | 0.02 |
| Rv0968 | hypothetical protein | 6.41 | 0.12 | |
| Rv0969 | ctpV | PROBABLE METAL CATION TRANSPORTER P-TYPE ATPASE CTPV |
4.55 | 0.14 |
| Rv0970 | PROBABLE CONSERVED INTEGRAL MEMBRANE PROTEIN | 1.66 | 0.21 |
Numbers represent average fold-change in the ΔcsoR∷hyg mutant compared to WT in the absence of copper. These data represents 2 dye-flip experiments after SAM analysis.
Numbers represent average fold-change in the ΔcsoR∷hyg mutant compared to WT in the presence of copper. These data represents 2 dye-flip experiments after SAM analysis.
Functional annotations are taken from the M. tuberculosis H37Rv complete genome sequence (www.jcvi.org)
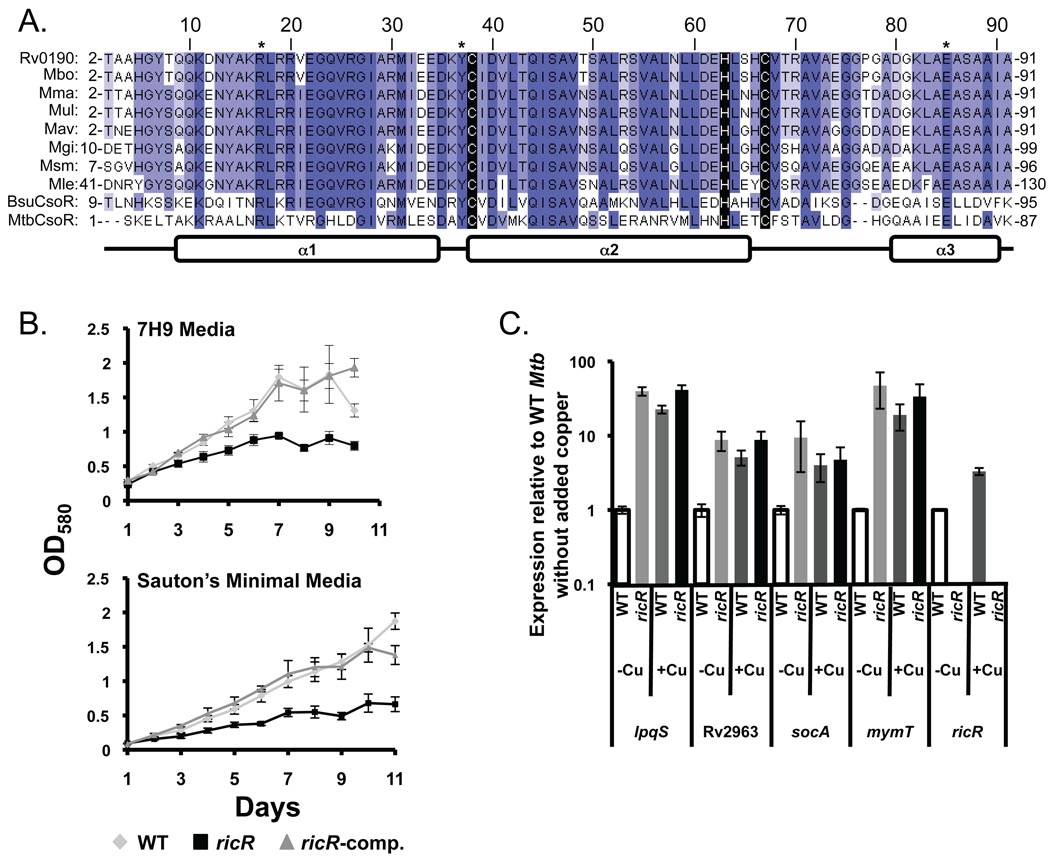
It was previously noted that RicR and CsoR are predicted to share the same overall structure and amino acids required for reduced copper [Cu(I)] coordination and allosteric regulation of DNA binding (Liu et al., 2007, Ma et al., 2009b) (Fig. 3A). The major difference between the two proteins is that Mtb CsoR has an extended C-terminal tail that is required for high affinity cso operator binding (Ma et al., 2009b). In fact, RicR is more similar to the only other well-characterized CsoR homologue, Bacillus subtilis CsoR (51% with BsuCsoR), which does not have the extended C-terminus found in Mtb CsoR (Smaldone & Helmann, 2007, Ma et al., 2009a). Furthermore, the BsuCsoR binding site is nearly identical to the palindrome identified upstream of lpqS.
Fig. 3. RicR is a copper-responsive regulator.
(A) Alignment of RicR (Rv0190/MT0200) homologues in other Mycobacteria along with MtbCsoR, and BsuCsoR. Gene identification: Mbo, Mb0196; Mma, MMAR_0433; Mul, MUL_1083; Mav, MAV_4988; Mgi, Mflv_0484; Msm, MSMEG_0230; Mle, ML2609. In black is conserved copper coordinating amino acids [cysteine (Cys) 38, histidine 63, Cys67] based on the crystal structure of MtbCsoR (Liu et al., 2007). The asterisk (*) indicates amino acids predicted to be important for DNA interactions in MtbCsoR (Ma et al., 2009a, Liu et al., 2007). Secondary structure prediction (alpha helices 1–3) was adapted from (Liu et al., 2007). Increasing blue intensity indicates increasing conservation of the highlighted amino acids. (B) Growth curves comparing the WT, ricR transposon mutant, and ricR-complemented strains in 7H9+ADN media (6 µM CuSO4) and Sauton’s minimal media (trace copper). OD580 was measured daily. These experiments represent three independent experiments, each done in duplicate. (C) lpqS, Rv2963, orfA, and mymT are highly expressed in the ricR mutant, comparable to levels in copper-induced WT bacteria. Cultures were grown in Sauton’s minimal media until to OD580 of ~0.6, after which RNA was harvested for analysis. For copper treatment, cells were treated for four hours with 500 µM CuSO4 prior to RNA harvest. ricR transcript was not detected (ND) in the ricR mutant due to disruption by the transposon insertion. qRTPCR was performed as described in the Experimental Procedures and Fig. 1. This is representative of two experiments, each done in triplicate.
To determine if RicR regulated the five gene copper-responsive regulon, we examined the expression profile of a previously uncharacterized ΦMycoMarT7 transposon mutant of ricR in Mtb strain CDC1551. We determined that the mutant had a growth defect under standard culture conditions (7H9 broth, Fig. 3B, top) as well as in Sauton’s minimal media without added copper (Fig. 3B, bottom); we thus harvested bacteria for qRTPCR analysis at an OD580 of ~0.6. If RicR were a repressor controlling its own expression as well as the transcription of lpqS, Rv2963, mymT, and socAB, a ricR mutation should result in copper-independent constitutive expression of these genes. Indeed, all genes were highly expressed in the ricR mutant compared to WT Mtb grown in Sauton's minimal media without added copper (Fig. 3C). Additionally, the expression of these genes in the ricR mutant was comparable to the expression observed in WT Mtb treated with 500 µM CuSO4 for four hours (Fig. 3C).
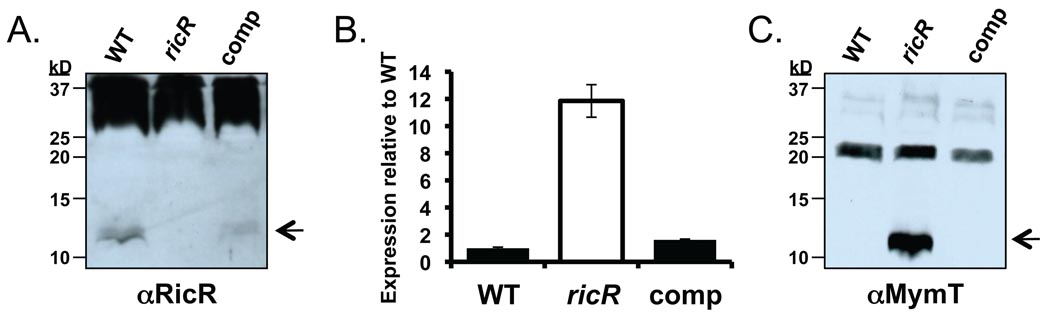
Finally, we complemented the ricR transposon mutation in single copy with ricR expressed from its native promoter. While RicR levels were partially restored (Fig. 4A), lpqS expression was repressed to WT levels (Fig. 4B). In addition, MymT levels, which drastically increased in the ricR mutant, were also restored to undetectable WT amounts in the complemented strain (Fig. 4C).
Fig. 4. Complementation of the ricR mutation.
(A) Immunoblot analysis shows that RicR levels were partially restored in the ricR-complemented strain grown in Sauton’s minimal media. Non-specific bands recognized by the RicR polyclonal antibodies served as loading controls. Data are representative of three biological replicates. (B) lpqS transcript levels at early stationary phase were highly expressed in the ricR mutant compared to WT Mtb and were restored to WT levels in the ricR-complemented strain. Bacteria were grown in Sauton’s minimal media for these experiments and this is representative of two biological replicates performed in triplicate. (C) Immunoblot analysis showed MymT levels were markedly increased in the ricR mutant and were restored to undetectable levels in the complemented strain grown in Sauton’s minimal media. Cross-reactive proteins served as the loading control.
To determine if RicR regulated other genes, we performed microarray analysis comparing the ricR transposon mutant to WT Mtb (Geo Accession GSE23498, Table S4). We grew cultures to an OD580 of ~0.6 in Sauton's minimal media containing no added copper. As expected, the most highly expressed genes in the ricR mutant were lpqS, Rv2963, mymT, and socAB (Table 4). ricR was also highly expressed, supporting the hypothesis that ricR is autoregulated. Transcription of ricR could be observed by microarray analysis because the oligonucleotide designed for the microarrays corresponded to sequence upstream of the transposon insertion, whereas the qRTPCR primers were designed to span the insertion site. Importantly, the difference in expression of all genes diminished between the WT and ricR strains upon treatment with 500 µM CuSO4 for four hours (Table 4). Taken together, these data supported the hypothesis that RicR is the copper-inducible repressor of this newly identified regulon.
Table 4.
Genes differentially regulated in a ricR mutant compared to WT Mtb with and without coppera.
| CDC1551 | H37Rv | gene | Name | Cu−a | Cu+b |
|---|---|---|---|---|---|
| MT0195 | Rv0186 | bglS | beta-glucosidase, putative | 15.94 | 3.29 |
| MT0196 | NA | mymT | hypothetical protein | 106.18 | 2.04 |
| MT0200 | Rv0190 | ricR |
homologue of copper sensitive operon repressor csoR |
4.74 | −1.50 |
| MT1746.1 | NA | socB | hypothetical protein | 16.22 | 2.62 |
| MT0868 | NA | hypothetical protein | 6.59 | 1.26 | |
| MT0869 | Rv0846c | copper-binding protein, putative | 13.53 | 1.83 | |
| MT0870 | Rv0847 | lpqS | hypothetical protein | 44.45 | 2.27 |
| MT0871 | Rv0848 | cysK2 | cysteine synthase/cystathionine beta-synthase family protein |
33.63 | 2.04 |
| MT0872 | Rv0849 | Probable conserved integral membrane transport protein |
27.26 | 2.35 | |
| MT0873 | Rv0850 | IS1606’, transposase | 10.58 | 2.28 | |
| MT3038 | Rv2962c | Rv2962c | UDP-glucoronosyl andUDP-glucosyltransferase family proteins |
13.34 | 1.95 |
| MT3039 | Rv2963 | hypothetical protein | 41.00 | 3.77 | |
| MT3041 | Rv2964 | purU | formyltetrahydrofolate deformylase | 15.23 | 5.95 |
| MT3041.1 | NA | hypothetical protein | 5.31 | 2.63 | |
| MT3159 | Rv3074 | hypothetical protein | 5.42 | 5.21 | |
| MT3885 | Rv3776 | hypothetical protein | 8.21 | 8.50 | |
| MT1422 | Rv1378c | hypothetical protein | 11.34 | 7.12 | |
| MT1511 | Rv1464 | aminotransferase, class V | 4.30 | 3.13 | |
| MT3753.2 | NA | hypothetical protein | 3.40 | 6.47 |
Numbers represent average fold-change over 2 dye-flip experiments after SAM analysis.
Numbers represent average fold-change over 3 dye-flip experiments after SAM analysis.
Functional annotations are taken from the Mtb strain H37Rv complete genome sequence (www.jcvi.org)
Genes with the palindromic sequence in their promoters are in bold.
RicR associates with the lpqS promoter palindrome
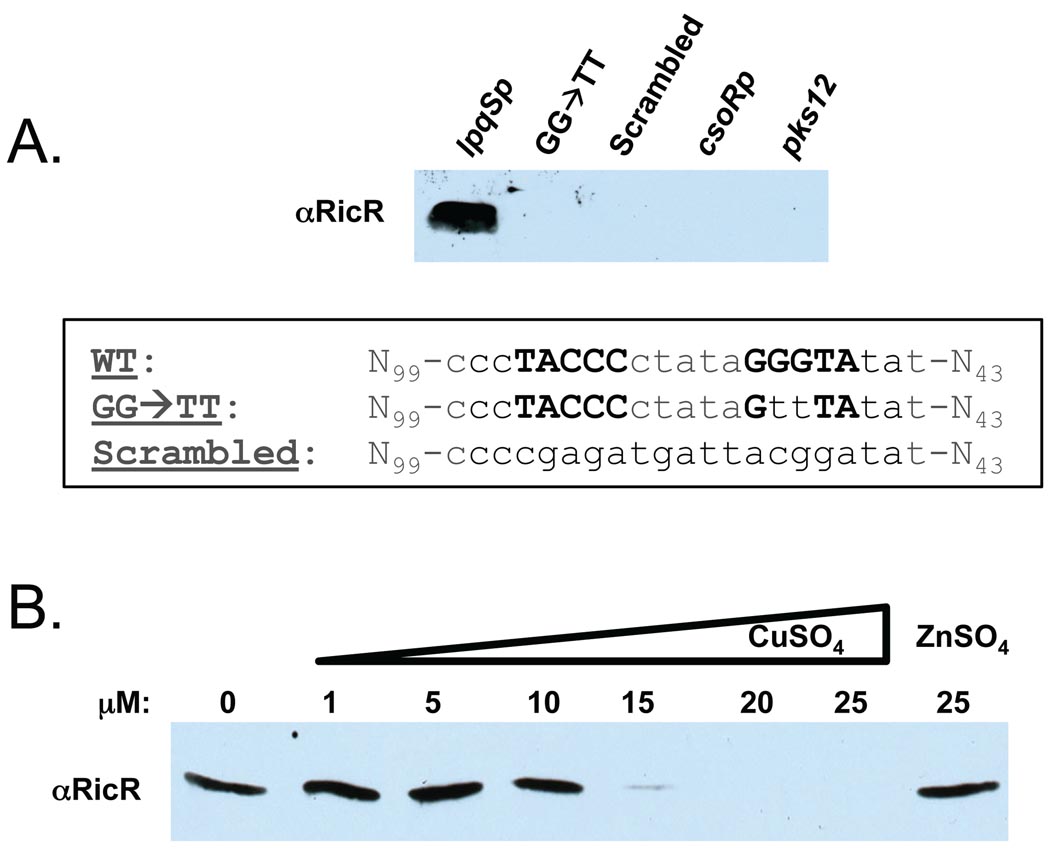
The next goal was to determine if RicR could associate with DNA encoding the palindrome identified in the lpqS, Rv2963, mymT, ricR and socAB promoters. Purified RicR-His6 could not be maintained in a soluble state upon dilution, and its use in gel-shift assays resulted in non-specific DNA binding (data not shown). As an alternative to using purified protein, we used lysates of E. coli producing untagged Mtb RicR. Furthermore, rather than using gel retardation, we examined the ability of biotinylated DNA encoding the lpqS promoter (lpqSp) to co-purify with RicR. RicR robustly co-purified with the lpqSp (Fig. 5A). Mutagenesis of a GGGTA interface (5’-TACCC-N5-GTTTA-3’) or scrambling of the palindrome sequence disrupted RicR association (Fig. 5A). RicR association appeared to be specific because it did not bind the csoR promoter (csoRp) or a randomly selected coding DNA sequence (pks12) (Fig. 5A). Finally, when copper was added to the reaction we observed a dose-dependent loss of DNA association, which did not occur in the presence of zinc (Fig. 5B). Although we cannot rule out the possibility that an E. coli protein bridged RicR to the lpqSp, it appears that RicR specifically bound to a palindrome found in the promoters of five loci, and dissociated from DNA in the presence of copper.
Fig. 5. RicR binds the lpqS promoter.
(A) DNA affinity chromatography revealed RicR associates with the lpqS promoter (lpqSp). Clarified lysates of E. coli expressing untagged ricR were incubated with biotinylated DNA (see Experimental Procedures for details). RicR-DNA interaction was disrupted upon palindrome mutagenesis (sequences indicated below). RicR did not bind the csoR promoter (csoRp) or intragenic DNA (pks12). (B) Copper disrupted the RicR-DNA interaction in a dose-dependent manner.
The ricR mutant is resistant to copper-mediated toxicity
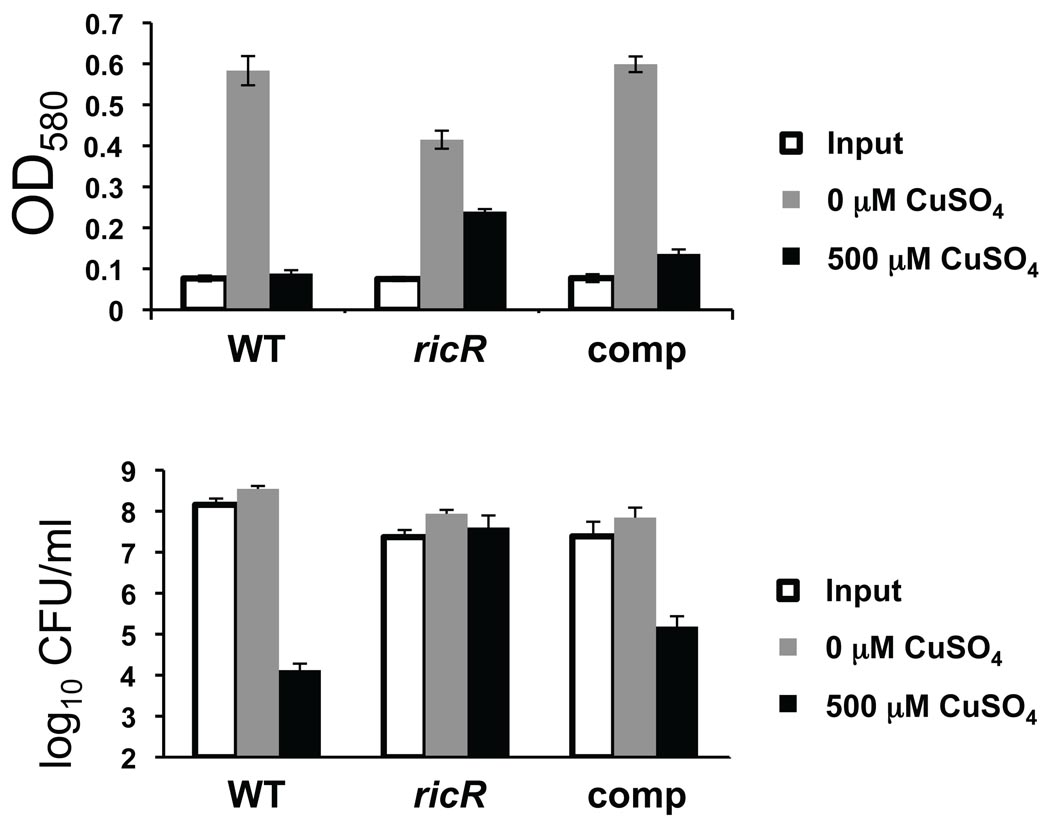
Because the RicR regulon was induced upon exposure to copper, we hypothesized that gene products of this regulon may be needed to combat toxicity related to excess copper levels. In support of this hypothesis, MymT is required for resistance to copper-mediated toxicity (Gold et al., 2008). Thus, a ricR mutant could be more resistant to copper toxicity when compared to WT Mtb due to the constitutive expression of mymT and the other RicR-regulated genes. We performed a copper sensitivity assay by treating WT, ricR, and ricR-complemented strains with CuSO4 in microtiter plates (see Experimental Procedures). After 10 days, WT and complemented strains grew well in the absence of copper, while the ricR mutant grew to a lower density (Fig. 6), consistent with the growth curve analysis (Fig. 3B). The phenotype of strains grown with copper were strikingly reversed: the WT and ricR-complemented bacteria were killed by copper in a dose-dependent manner whereas the ricR mutant was virtually unaffected by the presence of copper as determined by culture optical density or colony forming units (Fig. 6, Fig. S4,). These data strongly suggested that the constitutive expression of one or more genes in the RicR regulon protected the ricR mutant against copper toxicity.
Fig. 6. The ricR mutant is hyper-resistant to copper toxicity.
Copper sensitivity assay comparing the ability of WT, ricR and ricR-complemented Mtb strains to survive in 500 µM copper sulfate. The bottom panel shows colony-forming units (CFU) per ml from the same experiment. Data are representative of at least two experiments, each done in triplicate. CFU after 10 days in untreated versus copper-treated ricR strain were not statistically significantly different (Student’s t-test, p > 0.05), while the WT and complemented strains had significantly fewer CFU between copper-treated and untreated cultures.
Discussion
In this work, we identified a regulon controlled by the copper-sensing repressor RicR. We propose that under low copper conditions RicR represses gene expression, whereas exposure to excess copper results in RicR dissociation from DNA and the induction of lpqS, Rv2963, mymT, socAB and ricR. Consistent with this hypothesis, disruption of ricR resulted in the constitutive expression of all five loci in the absence of excess copper. Based on our copper sensitivity assays, we propose that the RicR regulon plays a protective role against copper toxicity. It is also possible that one or more of the regulated genes is not involved in counteracting the effects of copper toxicity, but uses copper as an in vivo cue to trigger gene expression. It is intriguing that RicR regulates several genes that are only found in pathogenic Mycobacteria, suggesting that these genes may be required for growth or survival in a specific niche in an animal host.
Of the genes that are controlled by RicR, mymT is the only one that has been previously characterized (Gold et al., 2008). MymT is a copper metallothionein that binds up to six Cu(I) ions. As would be predicted for a metallothionein gene, mymT is highly induced in the presence of copper (Fig. 2A, 3C) and appears to protect Mtb from the toxic effects of copper. However, a mymT null mutant is no less virulent than WT Mtb in a mouse model of infection (Gold et al., 2008). It is possible that all RicR-regulated genes need to be deleted or constitutively repressed in order to observe a measurable virulence phenotype, a hypothesis we are currently testing. Alternatively, a mouse model of infection may not be appropriate for examining the effects of copper on Mtb survival in vivo.
Because the RicR regulon is induced in copper, we propose that gene products in addition to MymT may also play a role in combating copper toxicity. Rv2963 is a putative permease that contains a carboxy(C)-terminus rich in histidines, which can potentially bind copper. LpqS is a probable lipoprotein that also has a histidine-rich region beginning 30 amino acids from its amino(N)-terminus. These histidines are predicted to remain after possible lipoprotein processing and lipidation (Sander et al., 2004). As a putative lipoprotein, LpqS may work with Rv2963 to export copper. That lipoproteins associate with permeases to export ligands has precedence in Mtb. LprG (Rv1411c) and Rv1410c transport ethidium bromide (Farrow & Rubin, 2008), while LppX (Rv2945c) is proposed to work with MmpL7 (Rv2942) to export phthiocerol dimycocerosate across the cell membrane (Camacho et al., 1999, Cox et al., 1999).
Divergently expressed from the lpqS promoter is Rv0846c, a predicted multi-copper oxidase whose expression is induced by copper (Ward et al., 2008). We also found that it is over-expressed in the ricR mutant (Table 4). It is possible that RicR bound to the lpqSp palindrome represses the divergently expressed Rv0846c, especially if RicR, like other metalloregulators, induces long-range changes in DNA structure (Iwig & Chivers, 2009). It was proposed that Rv0846c is chaperoned to the twin-arginine transport system to export copper (Ward et al., 2008); Rv0846c may function like E. coli CueO, a "cuproxidase" that oxidizes Cu(I) to the less toxic Cu(II) outside of the cell (Kosman, 2010).
In addition to LpqS and Rv2963, no function has been assigned to the previously unidentified socAB operon. Although socAB is transcribed (Fig. S2), it is unknown if the message is translated or if it is actually a small non-coding RNA. The role of socAB and the other RicR-regulated genes in protecting Mtb against copper toxicity is currently under investigation.
Despite the successful identification of the regulator of a new copper responsive regulon, we still do not understand why the RicR regulon is repressed in proteasome degradation-defective Mtb. The simplest model is that RicR is itself a proteasome substrate. The accumulation of RicR in the pafA and mpa mutants might result in the repression of the regulon under normal culture conditions. However, neither endogenous nor ectopically produced RicR levels were altered in proteasome degradation-defective strains under all conditions tested so far (data not shown). Another possibility is that the down-regulation of the RicR regulon in the mpa and pafA mutants is due to the accumulation of one or more copper-binding proteins that are normally proteasome substrates. This may result in the sequestration of available copper in the cell, mimicking copper-depleted conditions that result in RicR regulon repression. A similar explanation may account for the induction of the Zur regulon, where sequestered zinc would lead to higher gene expression. Ongoing studies are attempting to determine how proteasome activity affects these two metal-dependent regulons.
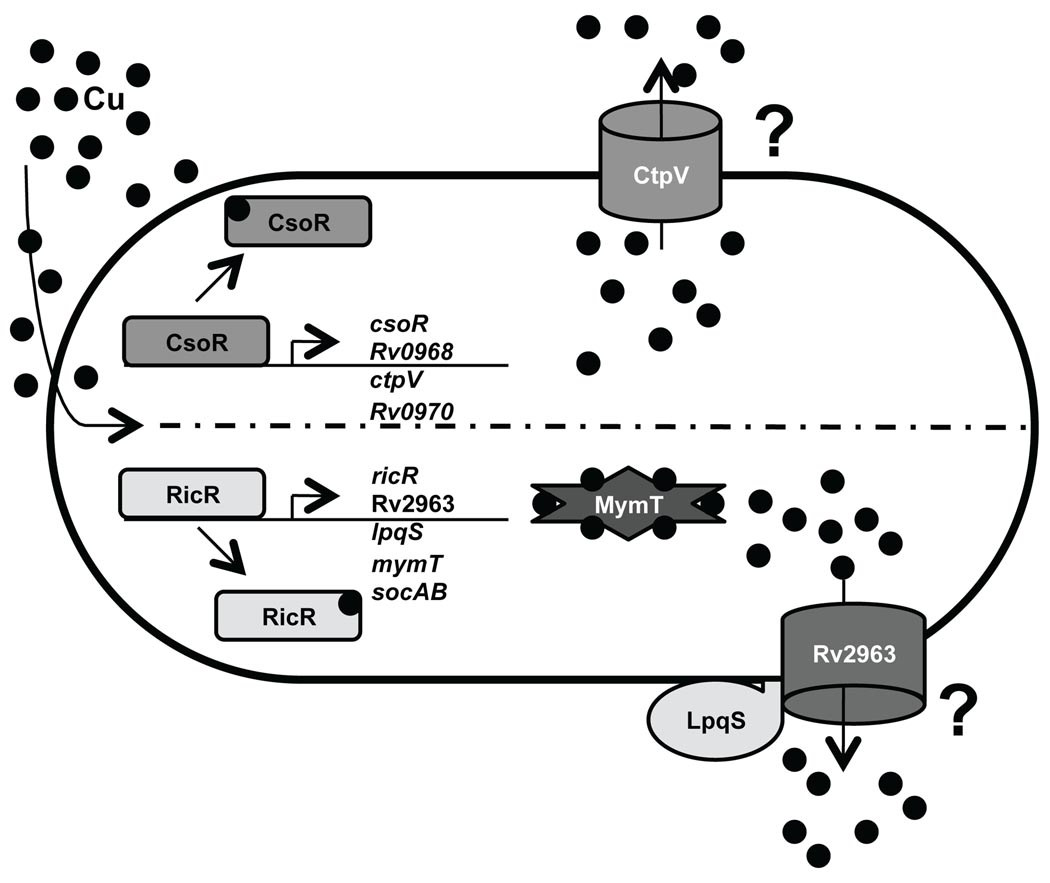
Prior to our work, CsoR was the only known copper responsive regulator in Mtb, controlling the expression of ctpV, which encodes a predicted membrane P-type ATPase. CtpV is proposed to be a copper efflux pump based on its regulation by CsoR and its similarity to other copper transporters (Liu et al., 2007). Supporting this hypothesis, a ctpV mutant is more susceptible to killing by high levels of copper than WT Mtb, presumably because the mutant cannot effectively export copper (Ward et al.). Importantly, the ctpV mutant is attenuated in an animal infection model, supporting the notion that the maintenance of copper homeostasis is important in vivo. Here, we present data that suggest the RicR regulon, which does not include ctpV, is also protective against high levels of copper in vitro. Taken together, we propose that Mtb has at least two mutually exclusive but parallel copper inducible pathways controlled by RicR and CsoR (Fig. 7). It remains to be determined if these two pathways represent a redundant response to copper, or if Mtb has evolved a graded response to copper toxicity; if so, this would predict that the Cu(I) binding affinities are different for RicR v. CsoR, a hypothesis currently under investigation (B. Keste, D. Giedroc, personal communication).
Fig. 7. Model for a dual copper response in Mtb.
Two parallel pathways appear to respond to copper in Mtb. (Top half) Copper induces the release of CsoR from its own promoter, resulting in the expression of ctpV, a predicted P-type ATPase metal transporter. (Bottom half) Copper also de-represses RicR from its own promoter as well as four other promoters. MymT binds up to six Cu(I) ions and may play a role in the sequestration or transport of excess copper. Rv2963 and LpqS are predicted to be membrane associated proteins and may play a role in copper export.
Little is understood about copper homeostasis during a bacterial infection. The exposure of microbes to excess copper could perhaps act as an innate immune defense, a hypothesis supported by a report suggesting that the mammalian ATP7A, a copper-transporting ATPase, provides some bactericidal activity in macrophages (White et al., 2009). Furthermore, copper levels are elevated in cultured macrophages infected with Mtb (Wagner et al., 2005). Copper has long been used as an anti-bacterial agent in laboratory and medical settings (Mikolay et al., 2010), thus perhaps it would not be surprising if animals have developed ways to mobilize cellular copper to battle invading microbes.
Experimental Procedures
Bacterial strains, growth conditions and primers
Bacterial strains, plasmids and primers used in this work are listed in Table S1. Mtb strains were grown in 7H9 broth (Difco) supplemented with 0.2% glycerol, 0.05% Tween-80, 0.5% bovine serum albumin, 0.2% dextrose and 0.085% sodium chloride (ADN), or Sauton’s minimal media (3.7 mM potassium phosphate, monobasic; 2.5 mM magnesium sulfate; 30 mM L-asparagine; 3.5 mM zinc sulfate; 9.5 mM citric acid; 6.0% glycerol; 0.005% ferric ammonium citrate; 0.05% Tween 80). Cultures were grown without agitation in 25, 75 or 125 cm2 vented flasks (Corning) in humidified incubators with 5% CO2 at 37°C. For growth on solid media, 7H11 agar (Difco) was supplemented with oleic acid, albumin, dextrose and catalase (OADC, BBL). When necessary, H37Rv strains were grown in 50 µg ml−1 kanamycin and/or 50 µg ml−1 hygromycin. CDC1551 strains were grown in 25µg ml−1 kanamycin and/or 25 µg ml−1 hygromycin. E. coli cultures were grown in Luria Bertani (LB) broth (Difco) on either a rolling drum or shaking at 37°C. LB agar was used for growth on solid medium. Selection was maintained with 200 µg ml−1 ampicillin, 100 µg ml−1 kanamycin, and 150 µg ml−1 hygromycin. Growth curves were performed exactly as described elsewhere (Darwin et al., 2003).
Primers were purchased from Invitrogen. All plasmids were sequenced by GENEWIZ, Inc.
Purification of CsoR and RicR for production of polyclonal antibodies
Purification of CsoR for antibody production was performed under denaturing conditions according to the manufacturer’s specifications (Qiagen). RicR was purified under native conditions, and dialyzed in 2 l of phosphate buffered-saline (PBS) at 4°C. Some protein was visibly insoluble after dialysis. Insoluble protein was recovered by centrifugation and washed twice MilliQ water. Protein preparations were about 80% pure and used as the immunogen for RicR antibody production. Polyclonal rabbit antibodies were produced by Covance (Denver, PA). DlaT and MymT antibodies were described previously (Tian et al., 2005, Gold et al., 2008). For immunoblotting analysis, equivalent cell numbers were harvested at the same phase of growth, i.e. when OD580 were the same. Bacteria were washed once in an equal volume of PBS with 0.05% Tween 20 and resuspended in 250 µl of lysis buffer (100 mM Tris-Cl, 100 mM potassium chloride, 1 mM EDTA, 5 mM magnesium chloride, pH 8.0). Cells were lysed by bead beating with zirconia beads three times for 30 s. Total cell lysate (150 µl) was mixed with 50 µl of 4× sodium dodecyl sulfate sample buffer and boiled for 10 min. Immunoblotting was performed as previously described (Harlow, 1988). Antibodies to RicR and CsoR were used at a dilution of 1:1,000. Horseradish peroxidase-conjugated goat anti-rabbit antibodies (Pierce) were used for chemiluminescent detection (ThermoScientific SuperSignal West Pico or Femto Chemiluminescent substrate).
Construction of the ΔcsoR∷hyg mutant
We made an allelic exchange plasmid to delete and disrupt csoR using a plasmid originally developed for specialized transduction (Bardarov et al., 2002). Briefly, approximately 700 bp of DNA upstream and downstream of csoR were amplified by PCR and cloned into pYUB854, flanking a hygromycin resistance cassette (see Table S1 for plasmids and primers). The plasmid was linearized with PacI and gel purified. 1 µg of linearized DNA was electroporated into Mtb (Hatfull, 2000). Transformants were inoculated onto 7H11+OADC and 50 µg ml−1 hygromycin. A no-DNA electroporation control was also inoculated onto the same media to check for spontaneous hygromycin resistance mutants. After three weeks, single colonies were inoculated into 200 µl of 7H9 + ADN in a 96-well plate for one week. The 200 µl starter cultures were used to inoculate a 5 ml cultures for further analysis. The deletion and replacement of csoR was confirmed by PCR and sequencing. Immunoblotting with antibodies raised to Mtb CsoR also demonstrated the absence of CsoR (data not shown).
RNA extraction
RNA was extracted from Mtb as follows: an equal volume of 4 M guanidinium isothiocyanate (GTC), 0.5% sodium N-lauryl sarcosine, 25 mM trisodium citrate) was added to cultures to arrest transcription, and cells were collected by centrifugation at 2,885 g. Bacterial were resuspended in 1 ml TRIzol reagent (Invitrogen) and lysed with zirconia silica beads (BioSpec Products) in a BioSpec Mini Bead Beater two times for 30 s, with 1 min cooling on ice in between. Samples were either stored at −80°C or used immediately. Samples were centrifuged for 5 min at 16,100 g at 4°C to pellet cellular debris. 300 µl of chloroform was added to PhaseLock tubes (Heavy 2 ml, 5Prime) after which the TRIzol samples were added. Tubes were vigorously inverted for 15 s and allowed to rest at RT for 5 min. Tubes were centrifuged for 10 min at 10,500 g at 4°C. The aqueous phases were added to microfuge tubes containing 270 µl isopropanol and 270 µl high salt solution (0.8 M trisodium citrate, 1.2 M NaCl) and vortexed for 5 s. After a 10 min incubation at RT, tubes were microcentrifuged at 16,100 g for 10 min at 4°C to pellet RNA. RNA pellets were washed with 1 ml of 75% ethanol and centrifuged at 7,500 g for 5 min at 4°C. Pellets were allowed to dry by air and resuspended in 50 µl of RNA storage buffer (Ambion). 2 µl were used for quantification and another 2 µl were added to 80% formamide/ 20% loading dye to check RNA integrity on an (1.6%) agarose gel. The purification process was repeated two times to obtain DNA-free RNA. RNA aliquots were stored at −80°C.
cDNA synthesis, qRTPCR, and 5’RACE
The Reverse Transcription System (Promega) was used to synthesize cDNA from purified RNA. 4 ng of random hexamers (Amersham or Invitrogen) were used to prime cDNA synthesis from 100 ng of RNA as a template. The cDNA equivalent of 1.9 ng of RNA was analyzed by qRTPCR using Platinum SYBR GREEN qPCR SuperMIx UDG (Invitrogen) in a MyIQ2 two-color real-time PCR detection system (Bio-Rad). Analysis was performed using the MyIQ2 software. Normalization for each primer pair was carried out using a standard curve with genomic DNA of known copy number. ΔΔCT analysis was performed using rpoB as a reference gene as previously described (Pearce et al., 2006).
5’RACE was performed as described by the manufacturer (Invitrogen). Briefly, 1 µg of RNA was used as template for cDNA production using a reverse primer 150–300 bp downstream of annotated translational start sites. A 3’ poly-C tail was added to cDNA by recombinant Tdt. The cDNA was then amplified using a nested reverse primer and a primer that anneals to the poly-C tail. Products were cloned and sequenced. Likely transcriptional start sites were selected based on clones that had the most sequence upstream of the start codon.
Microarray analysis
The microarray experiments comparing WT, pafA and mpa strains were grown in 7H9 media supplemented with ADN and RNA was harvested at early stationary phase. Microarrays comparing the ricR and csoR mutants with WT Mtb were performed using RNA harvested from cells grown in Sauton’s minimal media to OD580 ~0.6 or 1.5, respectively. 500 µM CuSO4 was added to the cultures for four hours for testing copper induction.
Labeled cDNA for microarray experiments were generated by adding 2 µg of total RNA in a mixture containing 6 µg of random hexamers (Invitrogen), 0.01 M dithiothreitol, an aminoallyl-deoxynucleoside triphosphate mixture containing 25 mM each dATP, dCTP, and dGTP, 15 mM dTTP, and 10 mM amino-allyl-dUTP (aa-dUTP) (Sigma), reaction buffer, and 400 units of SuperScript III reverse transcriptase (Invitrogen) at 42°C overnight. The RNA template was then hydrolyzed by adding sodium hydroxide and EDTA to a final concentration of 0.2 and 0.1 M, respectively, and incubated at 70°C for 15 min. Unincorporated aa-dUTP was removed with a Minelute column (Qiagen). The labeled cDNA was eluted with a phosphate elution buffer (4 mM potassium phosphate, pH 8.5, in ultrapure water), dried, and resuspended in 0.1 M sodium carbonate buffer (pH 9.0). To couple the amino-allyl cDNA with fluorescent labels, normal human serum-Cy3 or normal human serum-Cy5 (Amersham) was added at room temperature for 1 h. Uncoupled label was removed using the Qiagen Minelute column (Valencia, CA). A “dye-flipped” experiment was performed by reversing the coupling of cDNA of Cy3 and Cy5 per experimental pair to control for dye bias. All microarray hybridizations were performed with a minimum of two independent cultures per strain, resulting in at least two dye-flipped experiments per strain pairing.
Aminosilane-coated slides printed with a set of 4750 Mtb oligonucleotides representing all open reading frame sequences (www.jcvi.org) were pre-hybridized in 55 SSC (15 SSC: 0.15 M sodium chloride, 0.015 M sodium citrate; Invitrogen), 0.1% sodium dodecyl sulfate, and 1.0% bovine serum albumin at 42°C for 60 min. The slides were then washed at RT with distilled water, dipped in isopropanol, and allowed to dry. Equal volumes of the appropriate Cy3- and Cy5-labeled cDNA were combined, dried and then resuspended in a solution of 40% formamide, 5×SSC, and 0.1% dodecyl sulfate. Resuspended labeled cDNA were heated to 95°C prior to hybridization. The probe mixture was then added to the microarray slide and allowed to hybridize overnight at 42°C. Hybridized slides were washed sequentially in solutions of 15 SSC-0.2% SDS, 0.15 SSS-0.2% SDS, and 0.15 SSC at RT, then dried in air. All wash buffers were supplemented with 1 ml of 0.1 M DTT per liter of wash buffer. Slides were scanned with an Axon GenePix 4000 scanner and individual TIFF images from each channel were analyzed with TIGR Spotfinder (available at http://pfgrc.jcvi.org/index.php/bioinformatics.html). Microarray data were normalized by LOWESS normalization and with in-slide replicate analysis using the TM4 software MIDAS (available at http://pfgrc.jcvi.org/index.php/bioinformatics.html). We selected genes in which comparison of WT cells exposed to mutants being analyzed yielded log2 value |≥2.0| in all samples. Microarray data can be found at the NCBI Gene Expression Omnibus website (http://www.ncbi.nlm.nih.gov/geo/).
DNA affinity chromatography
DNA probes were made by amplifying sequences of interest with forward primers containing 5’ Biotin TEG modifications (Operon). 50 µl of Dynabeads M-280 Streptavidin (Invitrogen) were washed with 500 µl of 25 B&W buffer (2 M sodium chloride, 1 mM EDTA disodium salt, 10 mM tris hydroxymethyl, pH 7.5) and coupled to 1 µg of biotinylated probe in 15 B&W buffer (200 µl total volume). The bead-DNA mixture was incubated on a rotator at room temperature for 30 min. The coupled beads were then washed once with 500 µl of 15 B&W buffer followed by two washes with buffer A (20 mM Tris-Cl, pH 8.0; 100 mM sodium chloride; 1 mM EDTA disodium salt; 1 mM dithiothreitol (DTT); 10% glycerol). E. coli with the pET24b+RicR-Stop plasmid (no His6 tag) was grown to an optical density (A600) of 0.5 and allowed to grow for three additional hours without induction; leaky expression of ricR produced sufficient RicR for experiments. 50 ml of culture was aliquoted and cells were harvested and stored at −20°C until needed. 20 ml were used to prep 10 ml of lysate. Pellets were resuspended in 10 ml of buffer A and snap frozen in an ethanol dry ice bath. After thawing on ice, the cell suspension was sonicated twice for 30 sec (1 second on, 1 second off) with 10 min of icing in between. Lysates were clarified by centrifugation at 10,000 g for 30 min. DNA coupled Dynabeads were then added to 1 ml of clarified lysate and rotated for 30 min at room temperature. After incubation, tubes were centrifuged briefly to collect solution on the bottom of the tubes. The beads were washed in 1 ml buffer A, transferred to a new tube, and subsequently washed 3 more times with 1 ml buffer A. Protein interacting with the coupled DNA was eluted in 50 µl of elution buffer (1 M sodium chloride, 50 mM Tris pH7.5). 16.6 µl of 45 SDS-PAGE sample buffer was added to the elution and 15 µl was separated on a 15% SDS-PAGE gel for Coomassie blue staining or immunoblot analysis.
Copper sensitivity assay
Mtb starter cultures were grown in 25 cm2 flasks in Sauton’s minimal media to an optical density between A580 0.5 and 1.0. For each strain used, cultures were diluted to an A580 of 0.1 in Sauton’s minimal media. 194 µl of diluted cultures were aliquoted into 96 well plates. 6 µl of CuSO4 solution was added such that the final concentration of copper sulfate was 0, 100 or 500 µM. Each concentration for each strain was done in triplicate per experiment. OD580 readings were measured using a Molecular Devices plate spectrophotometer at day 0 and after 10 days of incubation at 37°C. Bacteria were inoculated onto 7H11 agar and incubated for 2–3 weeks for enumeration.
ICP-DRC-MS
10 ml cultures of WT, pafA, mpa or complemented strains were grown in 7H9 + ADN to an A580 of 2.0. 20 OD equivalents of bacteria were harvested by centrifugation and washed twice in 10 ml of 1 mM metal-free EDTA (EMD) to chelate all extracellular metal. Washed pellets were resuspended in 1 ml of 1 mM EDTA and transferred to centrifuge tubes. Cells were pelleted once more and resuspended in 100 µl of metal-free nitric acid (70%, Fisher). Cell suspensions were incubated overnight at 60°C and then cooled overnight. To neutralize the acid, 50 µl of metal free hydrogen peroxide was added. Finally, the volume was brought up to 3 ml with metal-free water. Total copper, zinc, and manganese analysis was performed via ICP-DRC-MS at Applied Speciation. Aliquots of each sample were introduced into a radio frequency plasma where energy-transfer processes cause desolvation, atomization and ionization. The ions are extracted from the plasma through a differentially-pumped vacuum interface and travel through a pressurized chamber (DRC) containing a specific reactive gas that preferentially reacts with interfering ions of the same target mass to charge ratios (m/z). A solid-state detector detects ions transmitted through the mass analyzer. Raw ICP-DRC-MS data was provided in µg/L. From this data, we determined the amount of metal in µg in the 3 mL preparations mentioned above. Because we used 20 OD bacteria equivalents, we were able to determine the µg/OD, and using the conversion of 1×107 colony forming units per 0.1 OD equivalent, we calculated the µg/CFU. Finally, we were able to convert the µg of metal into atoms using Avogadro’s number and the molecular weight of the specific metal ion.
Supplementary Material
Acknowledgements
We thank Kristin Burns, David Giedroc, Ben Gold and Victor Torres for critical review of this manuscript. We thank Jiri Zavadil, Michael S. Glickman and Hideki Makinoshima for preliminary microarray studies and technical support. We thank Naoko Tanese for helpful suggestions. We are grateful to Ben Gold and Carl Nathan for the gift of antibodies to MymT. This work was supported by a Center For AIDS Research (CFAR) Pilot Project grant (NIH S P30 A1027742-17) and NIH grants AI065437 and HL92774 awarded to K.H.D., AI15447 to S.N.P., and AI 30036, 37856, and 36973 awarded to W.R.B. D.S. was supported by T32AI007180-27. We are grateful to C.N. for early support of this study (1R01HL72718). K.H.D was supported by a Burroughs Wellcome Investigator in the Pathogenesis of Infectious Disease award.
References
- Bardarov S, Bardarov S, Jr, Pavelka MS, Jr, Sambandamurthy V, Larsen M, Tufariello J, Chan J, Hatfull G, Jacobs WR., Jr Specialized transduction: an efficient method for generating marked and unmarked targeted gene disruptions in Mycobacterium tuberculosis, M. bovis BCG and M. smegmatis. Microbiology. 2002;148:3007–3017. doi: 10.1099/00221287-148-10-3007. [DOI] [PubMed] [Google Scholar]
- Butler SM, Festa RA, Pearce MJ, Darwin KH. Self-compartmentalized bacterial proteases and pathogenesis. Mol Microbiol. 2006;60:553–562. doi: 10.1111/j.1365-2958.2006.05128.x. [DOI] [PubMed] [Google Scholar]
- Camacho LR, Ensergueix D, Perez E, Gicquel B, Guilhot C. Identification of a virulence gene cluster of Mycobacterium tuberculosis by signature-tagged transposon mutagenesis. Mol Microbiol. 1999;34:257–267. doi: 10.1046/j.1365-2958.1999.01593.x. [DOI] [PubMed] [Google Scholar]
- Cerda-Maira F, Darwin KH. The Mycobacterium tuberculosis proteasome: more than just a barrel-shaped protease. Microbes Infect. 2009;11:1150–1155. doi: 10.1016/j.micinf.2009.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins GA, Tansey WP. The proteasome: a utility tool for transcription? Curr Opin Genet Dev. 2006;16:197–202. doi: 10.1016/j.gde.2006.02.009. [DOI] [PubMed] [Google Scholar]
- Cox JS, Chen B, McNeil M, Jacobs WR., Jr Complex lipid determines tissue-specific replication of Mycobacterium tuberculosis in mice. Nature. 1999;402:79–83. doi: 10.1038/47042. [DOI] [PubMed] [Google Scholar]
- Darwin KH. Prokaryotic ubiquitin-like protein (Pup), proteasomes and pathogenesis. Nat Rev Microbiol. 2009;7:485–491. doi: 10.1038/nrmicro2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darwin KH, Ehrt S, Gutierrez-Ramos JC, Weich N, Nathan CF. The proteasome of Mycobacterium tuberculosis is required for resistance to nitric oxide. Science. 2003;302:1963–1966. doi: 10.1126/science.1091176. [DOI] [PubMed] [Google Scholar]
- Darwin KH, Lin G, Chen Z, Li H, Nathan CF. Characterization of a Mycobacterium tuberculosis proteasomal ATPase homologue. Mol Microbiol. 2005;55:561–571. doi: 10.1111/j.1365-2958.2004.04403.x. [DOI] [PubMed] [Google Scholar]
- Farrow MF, Rubin EJ. Function of a mycobacterial major facilitator superfamily pump requires a membrane-associated lipoprotein. J Bacteriol. 2008;190:1783–1791. doi: 10.1128/JB.01046-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Festa RA, Pearce MJ, Darwin KH. Characterization of the proteasome accessory factor (paf) operon in Mycobacterium tuberculosis. J Bacteriol. 2007;189:3044–3050. doi: 10.1128/JB.01597-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynn JL, Chan J. Immunology of tuberculosis. Annu Rev Immunol. 2001;19:93–129. doi: 10.1146/annurev.immunol.19.1.93. [DOI] [PubMed] [Google Scholar]
- Gandotra S, Schnappinger D, Monteleone M, Hillen W, Ehrt S. In vivo gene silencing identifies the Mycobacterium tuberculosis proteasome as essential for the bacteria to persist in mice. Nat Med. 2007;13:1515–1520. doi: 10.1038/nm1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gold B, Deng H, Bryk R, Vargas D, Eliezer D, Roberts J, Jiang X, Nathan C. Identification of a copper-binding metallothionein in pathogenic mycobacteria. Nat Chem Biol. 2008;4:609–616. doi: 10.1038/nchembio.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gold B, Rodriguez GM, Marras SA, Pentecost M, Smith I. The Mycobacterium tuberculosis IdeR is a dual functional regulator that controls transcription of genes involved in iron acquisition, iron storage and survival in macrophages. Mol Microbiol. 2001;42:851–865. doi: 10.1046/j.1365-2958.2001.02684.x. [DOI] [PubMed] [Google Scholar]
- Gottesman S. Proteolysis in bacterial regulatory circuits. Annu Rev Cell Dev Biol. 2003;19:565–587. doi: 10.1146/annurev.cellbio.19.110701.153228. [DOI] [PubMed] [Google Scholar]
- Harlow E, Lane DP. Antibodies: a laboratory manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1988. [Google Scholar]
- Hatfull GF, Jacobs WR. Molecular Genetics of Mycobacteria. ASM Press; 2000. [Google Scholar]
- Hitomi Y, Outten CE, O'Halloran TV. Extreme zinc-binding thermodynamics of the metal sensor/regulator protein, ZntR. J Am Chem Soc. 2001;123:8614–8615. doi: 10.1021/ja016146v. [DOI] [PubMed] [Google Scholar]
- Iwig JS, Chivers PT. DNA recognition and wrapping by Escherichia coli RcnR. J Mol Biol. 2009;393:514–526. doi: 10.1016/j.jmb.2009.08.038. [DOI] [PubMed] [Google Scholar]
- Kooperberg C, Sipione S, LeBlanc M, Strand AD, Cattaneo E, Olson JM. Evaluating test statistics to select interesting genes in microarray experiments. Hum Mol Genet. 2002;11:2223–2232. doi: 10.1093/hmg/11.19.2223. [DOI] [PubMed] [Google Scholar]
- Kosman DJ. Multicopper oxidases: a workshop on copper coordination chemistry, electron transfer, and metallophysiology. J Biol Inorg Chem. 2010;15:15–28. doi: 10.1007/s00775-009-0590-9. [DOI] [PubMed] [Google Scholar]
- Liu T, Ramesh A, Ma Z, Ward SK, Zhang L, George GN, Talaat AM, Sacchettini JC, Giedroc DP. CsoR is a novel Mycobacterium tuberculosis copper-sensing transcriptional regulator. Nat Chem Biol. 2007;3:60–68. doi: 10.1038/nchembio844. [DOI] [PubMed] [Google Scholar]
- Ma Z, Cowart DM, Scott RA, Giedroc DP. Molecular insights into the metal selectivity of the copper(I)-sensing repressor CsoR from Bacillus subtilis. Biochemistry. 2009a;48:3325–3334. doi: 10.1021/bi900115w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Z, Cowart DM, Ward BP, Arnold RJ, DiMarchi RD, Zhang L, George GN, Scott RA, Giedroc DP. Unnatural amino acid substitution as a probe of the allosteric coupling pathway in a mycobacterial Cu(I) sensor. J Am Chem Soc. 2009b;131:18044–18045. doi: 10.1021/ja908372b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Z, Jacobsen FE, Giedroc DP. Coordination chemistry of bacterial metal transport and sensing. Chem Rev. 2009c;109:4644–4681. doi: 10.1021/cr900077w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maciag A, Dainese E, Rodriguez GM, Milano A, Provvedi R, Pasca MR, Smith I, Palu G, Riccardi G, Manganelli R. Global analysis of the Mycobacterium tuberculosis zur (furB) regulon. J Bacteriol. 2007;189:730–740. doi: 10.1128/JB.01190-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikolay A, Huggett S, Tikana L, Grass G, Braun J, Nies DH. Survival of bacteria on metallic copper surfaces in a hospital trial. Appl Microbiol Biotechnol. 2010 doi: 10.1007/s00253-010-2640-1. [DOI] [PubMed] [Google Scholar]
- Outten CE, O'Halloran TV. Femtomolar sensitivity of metalloregulatory proteins controlling zinc homeostasis. Science. 2001;292:2488–2492. doi: 10.1126/science.1060331. [DOI] [PubMed] [Google Scholar]
- Pearce MJ, Arora P, Festa RA, Butler-Wu SM, Gokhale RS, Darwin KH. Identification of substrates of the Mycobacterium tuberculosis proteasome. EMBO J. 2006;25:5423–5432. doi: 10.1038/sj.emboj.7601405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce MJ, Mintseris J, Ferreyra J, Gygi SP, Darwin KH. Ubiquitin-like protein involved in the proteasome pathway of Mycobacterium tuberculosis. Science. 2008;322:1104–1107. doi: 10.1126/science.1163885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieters J. Mycobacterium tuberculosis and the macrophage: maintaining a balance. Cell Host Microbe. 2008;3:399–407. doi: 10.1016/j.chom.2008.05.006. [DOI] [PubMed] [Google Scholar]
- Rodriguez GM, Voskuil MI, Gold B, Schoolnik GK, Smith I. ideR, An essential gene in Mycobacterium tuberculosis: role of IdeR in iron-dependent gene expression, iron metabolism, and oxidative stress response. Infect Immun. 2002;70:3371–3381. doi: 10.1128/IAI.70.7.3371-3381.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sander P, Rezwan M, Walker B, Rampini SK, Kroppenstedt RM, Ehlers S, Keller C, Keeble JR, Hagemeier M, Colston MJ, Springer B, Bottger EC. Lipoprotein processing is required for virulence of Mycobacterium tuberculosis. Mol Microbiol. 2004;52:1543–1552. doi: 10.1111/j.1365-2958.2004.04041.x. [DOI] [PubMed] [Google Scholar]
- Sato M, Bremner I. Oxygen free radicals and metallothionein. Free Radic Biol Med. 1993;14:325–337. doi: 10.1016/0891-5849(93)90029-t. [DOI] [PubMed] [Google Scholar]
- Saunders BM, Britton WJ. Life and death in the granuloma: immunopathology of tuberculosis. Immunol Cell Biol. 2007;85:103–111. doi: 10.1038/sj.icb.7100027. [DOI] [PubMed] [Google Scholar]
- Serafini A, Boldrin F, Palu G, Manganelli R. Characterization of a Mycobacterium tuberculosis ESX-3 conditional mutant: essentiality and rescue by iron and zinc. J Bacteriol. 2009;191:6340–6344. doi: 10.1128/JB.00756-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegrist MS, Unnikrishnan M, McConnell MJ, Borowsky M, Cheng TY, Siddiqi N, Fortune SM, Moody DB, Rubin EJ. Mycobacterial Esx-3 is required for mycobactin-mediated iron acquisition. Proc Natl Acad Sci U S A. 2009;106:18792–18797. doi: 10.1073/pnas.0900589106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smaldone GT, Helmann JD. CsoR regulates the copper efflux operon copZA in Bacillus subtilis. Microbiology. 2007;153:4123–4128. doi: 10.1099/mic.0.2007/011742-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Striebel F, Hunkeler M, Summer H, Weber-Ban E. The mycobacterial Mpa-proteasome unfolds and degrades pupylated substrates by engaging Pup's N-terminus. EMBO J. 2010;29:1262–1271. doi: 10.1038/emboj.2010.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Striebel F, Imkamp F, Sutter M, Steiner M, Mamedov A, Weber-Ban E. Bacterial ubiquitin-like modifier Pup is deamidated and conjugated to substrates by distinct but homologous enzymes. Nat Struct Mol Biol. 2009;16:647–651. doi: 10.1038/nsmb.1597. [DOI] [PubMed] [Google Scholar]
- Tian J, Bryk R, Itoh M, Suematsu M, Nathan C. Variant tricarboxylic acid cycle in Mycobacterium tuberculosis: identification of alpha-ketoglutarate decarboxylase. Proc Natl Acad Sci U S A. 2005;102:10670–10675. doi: 10.1073/pnas.0501605102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner D, Maser J, Lai B, Cai Z, Barry CE, 3rd, Honer Zu, Bentrup K, Russell DG, Bermudez LE. Elemental analysis of Mycobacterium avium Mycobacterium tuberculosis-, and Mycobacterium smegmatis-containing phagosomes indicates pathogen-induced microenvironments within the host cell's endosomal system. J Immunol. 2005;174:1491–1500. doi: 10.4049/jimmunol.174.3.1491. [DOI] [PubMed] [Google Scholar]
- Ward SK, Abomoelak B, Hoye EA, Steinberg H, Talaat AM. CtpV: a putative copper exporter required for full virulence of Mycobacterium tuberculosis. Mol Microbiol. 77:1096–1110. doi: 10.1111/j.1365-2958.2010.07273.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward SK, Hoye EA, Talaat AM. The global responses of Mycobacterium tuberculosis to physiological levels of copper. J Bacteriol. 2008;190:2939–2946. doi: 10.1128/JB.01847-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White C, Lee J, Kambe T, Fritsche K, Petris MJ. A role for the ATP7A copper-transporting ATPase in macrophage bactericidal activity. J Biol Chem. 2009;284:33949–33956. doi: 10.1074/jbc.M109.070201. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.