Abstract
Background Aims
Bone marrow-derived progenitor cells are under investigation for cardiovascular repair, but may be altered by disease. Our purpose was to identify differences in gene expression in CD133+ cells of patients with coronary artery disease (CAD) and healthy controls, and to determine whether exercise modifies gene expression.
Methods
CD133+ cells were flow-sorted from 10 CAD patients and 4 controls, and total RNA was isolated for microarray-based gene expression profiling. Genes that were found to be differentially regulated in patients were further analyzed to investigate if exercise has any normalizing effect on CD133+ cells in CAD patients following 3 months of an exercise program.
Results
Improvement in effort tolerance and increase in the number of CD133+ cells was observed in CAD patients after 3 months of exercise. Gene expression analysis of the CD133+ cells identified 82 differentially expressed genes (2-fold cutoff, 25% false-discovery rate and present calls) in patients compared with controls, of which 59 were found to be up-regulated and 23 down-regulated. These genes were found to be involved in carbohydrate metabolism, cell cycle, cellular development and signaling, and molecular transport. Following completion of the exercise program, gene expression patterns resembled those of controls in 7 of 10 patients.
Conclusion
The present study finds alterations in gene expression of bone marrow-derived CD133+ progenitor cells in CAD patients, which in part may be normalized by exercise.
Keywords: progenitor cells, gene expression, coronary artery disease, exercise, vascular repair
Introduction
It is widely believed that dysfunctional endothelium, associated with reduced nitric oxide bioactivity, induces a variety of proinflammatory and proliferative responses contributing to progression of atherosclerosis and clinical expression of disease1. Bone marrow-derived progenitor cells, first described in 1997 as CD34+ cells, are believed to play an important role in endothelial repair, as demonstrated in animal models of induced vascular injury2. Markers VEGFR-2 (KDR) and CD133 were subsequently reported to define a population of particular relevance to vascular repair, as VEGFR-2 is expressed by endothelial cells and CD133 is only expressed on immature hematopoietic stem cells3. These CD133+/VEGFR-2+ cells, though rare (<0.01% of mononuclear cells), circulate in peripheral blood and have the capacity to differentiate into cells with endothelial characteristics4. Several groups have reported that bone marrow derived progenitor cells with endothelial differentiation potential are reduced in patients with coronary artery disease (CAD)5–7. We undertook the current study to test the hypothesis that in patients with CAD, CD133+ bone marrow-derived progenitor cells may have altered gene expression as a result of comorbidities of atherosclerosis and that exercise, previously reported to increase bone marrow-derived progenitor cells in the circulation with improved endothelial characteristics,8–13 might favorably affect gene expression towards that seen in healthy subjects. To this end, we examined the global gene expression profiles of circulating CD133+ endothelial progenitor cells (EPCs) in CAD patients who participated in a 3 month exercise program. We report here for the first time that gene expression is significantly altered in CD133+ cells from CAD patients and demonstrate the reversal of some of these altered genes towards normal expression by exercise.
Methods
CAD patients enrolled in a cardiac rehabilitation program consisting of 36 sessions of 60 minutes each, spaced over approximately 3 months. Medical management was stable for at least a month prior to program participation and all patients were maintained on medications throughout the study as prescribed without change in preparation or dose. Cardiovascular examination and baseline testing were performed after an overnight fast (except for water) with blood samples drawn prior to exercise. Symptom-limited treadmill exercise testing was performed using the modified Bruce protocol14 (3 minute stages at increments of 1.7, 1.7, 1.7, 2.5, 3.4 miles per hour and 0%, 0.5%, 10%, 12% and 14% inclination, respectively). Healthy subjects were recruited for the study to serve as controls for baseline analyses. This protocol was approved by the institutional review boards of the National Heart, Lung, and Blood Institute and Suburban Hospital in Bethesda, Maryland (location of the cardiac rehabilitation program), and all participants provided written, informed consent.
Flow Cytometry Characterization of Peripheral Blood Mononuclear Cells
Flow cytometry analysis was performed on buffy coat cells isolated from EDTA-anticoagulated peripheral blood, with mononuclear cells isolated by density gradient centrifugation, as previously reported15. Cells were stained with CD133-PE (Miltenyi Biotech, Auburn, CA), and with CD3-PECy5, and CD19-PECy5 (BD Biosciences) for exclusion of lymphocytes. All cells were stained with either 7AAD (BD Biosciences) or Live/Dead (BD Biosciences, San Jose, CA) to allow exclusion of dead cells. Combinations of isotype controls were used as negative controls based on the species and IgG subclass of each antibody. Data were gated on mononuclear cell population during data acquisition, and 200,000 to 500,000 events were collected in the gated region for each sample of cells. The CYAN High-Performance Flow Cytometer with Summit Software (Dako Cytomation, Ft Collins, CO) was used for data acquisition and FCSExpress (De Novo Software, Thornhill, ONT, Canada) for analysis. (Supplemental Figure 1 depicts the characterization of CD 133 + cells by flow cytometry). Cell populations were expressed as the number of circulating cells per volume of peripheral blood, based on the percent of mononuclear cells counted during analysis and the total nucleated white cell count.
Isolation of CD133+ RNA
CD133+ cells were isolated from peripheral blood using fluorescence-activated cell sorting with a MoFlo cytometer (Dako Cytomation) and collected in lysis buffer containing guanidinium thiocyanate (Ambion Inc, Austin, Texas), mixed with ethanol and applied to a silica-based filter that selectively binds RNA. Genomic DNA was removed by DNase treatment. The concentration of the isolated RNA was determined with the Nanodrop ND-1000 spectrophotometer (Nanodrop Technologies, Wilmington, Delaware). Quality and integrity of the total RNA isolated were assessed on the Agilent 2100 bioanalyzer (Agilent Technologies, Palo Alto, California).
Amplification of RNA for Gene Expression Profiling
T7-based RNA amplification was performed on 5 ng of the isolated total RNA with the Riboamp OA 2-round amplification kit as suggested by the manufacturer (Arcturus, Mountain View, California). Briefly, total RNA was incubated with oligo dT/T7 primers and reverse-transcribed into double-stranded cDNA. In vitro transcription of the purified cDNA was performed with T7 RNA polymerase at 42°C for 6 hours. The amplified RNA was purified and subjected to a second round of amplification and biotin labeling with Affymetrix's IVT labeling kit according to the manufacturer's directions (Affymetrix, Santa Clara, California). The yield and integrity of the biotin-labeled cRNA were determined with the Nanodrop ND-1000 spectrophotometer and the Agilent 2100 bioanalyzer. Twenty micrograms of biotin-labeled RNA was fragmented to ~200-bp size by incubation in fragmentation buffer containing 200 mmol/L Tris-acetate pH 8.2, 500 mmol/L potassium acetate, and 500 mmol/L magnesium acetate for 35 minutes at 94°C before hybridization. Fragmented RNA was assessed for relative length on Agilent 2100 bioanalyzer and hybridized to Affymetrix Human Genome (HG) U133 Plus 2.0 chips for 16 hours, washed, stained on an Affymetrix fluidics station, and scanned with Affymetrix GeneChip scanner.
Microarray Data Processing and Statistical Analysis
Affymetrix GeneChip operating software version 1.4 was used to calculate MAS5 signal intensity and the percent present calls on the hybridized Affymetrix chip. The signal-intensity values obtained for probe sets present in the microarrays were transformed with an adaptive variance-stabilizing, quantile-normalizing transformation termed “S10”, (P.J. Munson, GeneLogic Workshop of Low Level Analysis of Affymetrix GeneChip Data, 2001, software available at http://abs.cit.nih.gov/geneexpression.html).Transformed data from all the chips were subjected to a principal component analysis to detect outliers.
JMP statistical software package 7.0 (SAS Institute, Cary, NC) was used in microarray data analysis. Differentially regulated genes in CAD were identified by comparing the gene expression profiles of CD133+ cells between healthy controls and CAD patients before exercise using the criterions of 2-fold change, 25% false discovery rate (FDR). The expression levels of selected up- and down-altered genes for each CAD patient between baseline and exercise program completion were compared separately using paired one-sided t-test. Ingenuity pathway analysis program (Ingenuity Systems, Redwood, CA) was used for following-up functional characterization of the selected genes. Microarray data are available for public access with accession number GSE 18608 at the National Center for Biotechnology Information gene expression and hybridization array data repository Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo).
Wilcoxon signed-rank test was used to estimate significance level of improvements in some clinical measurements in follow-up relative to baseline. Data are reported as mean ± standard deviation, unless otherwise indicated. Spearman's rank correlation method was used to estimate the significance of association between 1) baseline expression of the selected genes and baseline clinical measurements of 10 CAD patients, and 2) expression of genes and clinical measurements prior to and following exercise program participation by the patients.
Benjamini-Hochberg16 method was used to correct for multiple testing for the significance of correlation.
Validation of Gene Expression Data by Quantitative Polymerase Chain Reaction
First-strand cDNA was synthesized with 500ng of amplified RNA and random primers in a 20-μL reverse-transcriptase reaction mixture using Invitrogen's Superscript cDNA synthesis kit (Invitrogen, Carlsbad, Calif) according to the manufacturer's directions. Quantitative real-time polymerase chain reaction assays were performed with the use of gene-specific double–fluorescently labeled probes in a 7900 Sequence Detector (PE Applied Biosystems, Norwalk, Conn). Probes and primers were obtained from Applied Biosystems. In brief, polymerase chain reaction amplification was performed in a 384 well plate with a 20 μL reaction mixture containing 300 nm of each primer, 200 nm of probe, 200 nm of dNTP in 1× real-time polymerase chain reaction buffer and passive reference (ROX) fluorochrome. The thermal cycling conditions were 2 minutes at 50°C and 10 minutes at 95°C, followed by 40 cycles of 15 second denaturation at 95°C and 1 minute of annealing and extension at 60°C. Samples were analyzed in duplicate, and the cycle threshold values obtained were normalized to the housekeeping gene ß-actin. The comparative cycle threshold method, which compares the differences in cycle threshold values between groups, was used to achieve the relative fold change in gene expression between CAD patients and normal healthy subjects.
Results
Clinical Evaluation
Twenty-five CAD patients initially consented for protocol participation and completed the 3 month exercise program. For the group, fitness as measured by treadmill exercise duration was significantly improved following completion of the program, from 485 ± 159 to 555 ± 148 seconds (P=0.0002). Numbers of CD133+ cells in blood increased from 873 ± 556 to 1666 ± 1747 cells/mL (P=0.0384) following the exercise program.
Global Gene Expression Analysis of CD133 + cells
Total RNA isolated from CD133+ cells derived from baseline and 3 month samples from patients was processed for microarray analysis. Upon hybridization to gene chips, 13 patients' baseline samples passed our gene chip quality control metrics with scale factor set at less than 10 and % present calls ≥ 20. In these 13 patients, 3 of the samples showed a slight increase in scale factor in their post-exercise sample and were excluded for analysis. Thus, data analysis and mining were performed with paired samples from 10 patients (6 males, 4 females; age range 30–76 years) selected with the above mentioned quality control filters (Table 1). Similar analyses were performed on samples were obtained from 4 healthy volunteers (3 males, 1 female; age range 50–64 years). Clinical observations and measurements performed prior to and at the completion of the 3 month exercise program were obtained from these subjects (Table 2).
Table 1.
Characteristics of CAD Patients (N = 10)
| Parameter | |
|---|---|
| Age (average, range) | 62.9 ± 13, 30–76 |
| Sex Distribution (% male) | 60 |
| Diseased Vessels (average, range) | 2, 1–4 |
| Ejection Fraction (%) | 61 ± 6 |
| Myocardial Infarction (% of patients) | 40 |
| Revascularization (% of patients) | 100 |
| Coronary artery bypass grafting (% of patients) | 20 |
| Coronary Stent/Angioplasty (% of patients) | 80 |
| Risk Factor History | |
| Hypertension | 6 |
| treated | 6 |
| Dyslipidemia | 9 |
| treated | 9 |
| Diabetes | 4 |
| treated | 4 |
| Cigarette Smoking (current) | 0 |
| Medications | |
| Antihypertensive | 6 |
| ace inhibitor | 4 |
| angiotensin receptor blocker | 1 |
| beta blocker | 4 |
| calcium channel blocker | 1 |
| diuretic | 1 |
| Antilipidemic | 9 |
| atorvastatin | 3 |
| ezetimibe | 1 |
| pravastatin | 1 |
| rosuvastatin | 3 |
| simvastatin | 2 |
| Antidiabetic | 4 |
| insulin | 1 |
| metformin | 3 |
| rosiglitazone | 2 |
Table 2.
Testing data for CAD patients at baseline and following completion of exercise program
| Parameter | Baseline (N = 10) | Completion (N =10) |
|---|---|---|
| Vital Signs | ||
| Body Mass Index (kg/m2) | 27.9 ± 5.9 | 27.9 ± 5.9 |
| Systolic Blood Pressure (mmHg) | 124 ± 15 | 124 ± 13 |
| Diastolic Blood Pressure (mmHg) | 76 ± 10 | 73 ± 10 |
| Resting Heart Rate (beats/min) | 74 ± 15 | 73 ± 10 |
| Lab Values | ||
| Total Cholesterol (mg/dL) | 128 ± 24 | 130 ± 22 |
| Low-density Lipoprotein Cholesterol (mg/dL) | 72 ± 26 | 72 ± 21 |
| High-density Lipoprotein Cholesterol (mg/dL) | 45 ± 7 | 45 ± 7 |
| Triglycerides (mg/dL) | 127 ± 51 | 122 ± 49 |
| C-Reactive Protein (mg/dL) | 2.0 ± 3.5 | 2.3 ± 3.4 |
| Fasting Glucose (mg/dL) | 113 ± 28 | 117 ± 28 |
| Hemoglobin (g/uL) | 13.9 ± 1.3 | 13.9 ± 1.2 |
| White Blood Cells (K/uL) | 6.0 ± 1.7 | 6.2 ± 1.2 |
| Neutrophils (%) | 57.6 ± 8.0 | 59.2 ± 7.3 |
| Lymphocytes (%) | 29.7 ± 8.6 | 28.4 ± 7.5 |
| Monocytes (%) | 8.3 ± 1.9 | 8.3 ± 1.9 |
| Exercise Data | ||
| Exercise Duration (sec) | 478 ± 174 | 526 ± 168 |
| CD133 + (cells/mL) | 873 ± 556 | 1666 ± 1747 |
Identification of Highly Abundant Transcripts in CD 133+ cells
In an effort to determine transcripts that are highly abundant in CD133+ cells from our study, positively detected transcripts from healthy volunteers were ranked in order of their abundance from high to low based on their log-transformed median signal intensity. The top 500 ranking probe sets, representing 351 genes (Supplemental Table 1) identified to be of high abundance in our CD133+ cells, included annexin, adducin, calmodulin, cathepsin, chemokines, transcription factors, histones, interleukins, splicing factors and some universally expressed proteins such as actin, ferritin, ribosomal proteins, myosin light chain, and microglobulin. Ingenuity Pathway analysis of these CD133+ abundant genes indicated them to be involved in multiple cellular functions especially protein synthesis, post-transcriptional modification and cell death as shown in Figure 1.
Figure 1.
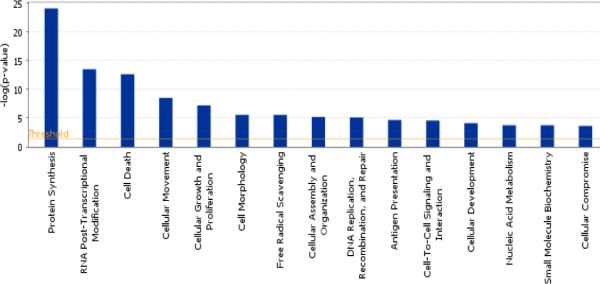
Distribution of some top molecular and cellular functions associated with the top 500 abundant transcripts (367 genes) in the expressions of 4 healthy controls. X axis shows the pathways and the Y axis denotes the P value for each of the pathways represented.
Differential CD133+ Gene Expression in Coronary Artery Disease
To establish a set of reference genes which can be used to measure for any improvement in expression of CD133+ from baseline to post-exercise, we first compared the CD133+ cell gene expression profiles in 10 subjects with CAD and 4 healthy controls - Using statistical filters of 25% FDR, fold change greater than 2 and present calls, we identified 105 differentially expressed probe sets representing 82 unique genes (59 up-regulated and 23 down-regulated genes, respectively, in Table 3). Figures 2A & B show the hierarchical clustering for healthy controls and for CAD patients at baseline and following 3 months of exercise program participation. Molecular and cellular functions of the 59 up-regulated genes were classified by Ingenuity Pathway analysis and these genes were found to be involved in multiple cellular functions especially carbohydrate metabolism and molecular transport (Figure 3). The 23 down-regulated genes were found to be involved in cellular function and maintenance, cell cycle, cellular development and cell signaling. Genes that are of potential biological relevance to our study include HOXA9, TSC22D1, RIPK1, NARF, ANAPC13, HAT1.
Table 3.
List of genes differentially regulated in CD133+ cells between coronary artery disease patients and healthy controls. FC stands for fold change of gene expression in patients over healthy controls in signal intensity. A negative value indicates down-regulation; positive value indicates up-regulation.
| Probeset ID | Gene Symbol | Gene Name | FC |
|---|---|---|---|
| 209905_at | HOXA9 | homeobox A9 | 19.04 |
| 215111_s_at | TSC22D1 | TSC22 domain family, member 1 | 11.00 |
| 203616_at | POLB | polymerase (DNA directed), beta | 4.72 |
| 234295_at | DBR1 | debranching enzyme homolog 1 | 3.88 |
| 221740_x_at | LRRC37A2 | leucine rich repeat containing 37 A2 | 3.64 |
| 213313_at | RABGAP1 | RAB GTPase activating protein 1 | 3.62 |
| 201641_at | BST2 | bone marrow stromal cell antigen 2 | 3.43 |
| 225674_at | BCAP29 | B-cell receptor-associated protein 29 | 3.38 |
| 1554423_a_at | FBXO7 | F-box protein 7 | 3.20 |
| 224786_at | SCOC | short coiled-coil protein | 3.19 |
| 202293_at | STAG1 | stromal antigen 1 | 3.18 |
| 202282_at | HSD17B10 | hydroxysteroid 17b dehydrogenase 10 | 3.08 |
| 208991_at | STAT3 signal transducer and activator of transcription 3 | 3.03 | |
| 221458_at | HTR1F5-hydroxytryptamine (serotonin) receptor 1F | 2.96 | |
| 213518_at | PRKCI | protein kinase C, iota | 2.96 |
| 206495_s_at | HINFP | histone H4 transcription factor | 2.93 |
| 37966_at | PARVB | parvin, beta | 2.92 |
| 225922_at | FNIP2 | folliculin interacting protein 2 | 2.92 |
| 222516_at | AP3M1 | adaptor-related protein complex 3, mu 1 subunit | 2.88 |
| 209786_at | HMGN4 | high mobility group nucleosomal binding domain 4 | 2.84 |
| 228899_at | LOC100132884 | hypothetical protein LOC100132884 | 2.83 |
| 203680_at | PRKAR2B | protein kinase, regulatory, type II, beta | 2.80 |
| 203138_at | HAT1 | histone acetyltransferase 1 | 2.77 |
| 225892_at | IREB2 | iron-responsive element binding protein 2 | 2.75 |
| 227942_s_at | CRIPT | cysteine-rich PDZ-binding protein | 2.73 |
| 218408_at | TIMM10 | translocase of inner mitochondrial membrane 10 | 2.71 |
| 200853_at | H2AFZH2A | histone family, member Z | 2.68 |
| 219862_s_at | NARF | nuclear prelamin A recognition factor | 2.66 |
| 235244_at | CCDC58 | coiled-coil domain containing 58 | 2.65 |
| 209001_s_at | ANAPC13 | anaphase promoting complex subunit 13 | 2.65 |
| 200820_at | PSMD8 | proteasome (prosome, macropain) 26S subunit, 8 | 2.63 |
| 228667_at | AGPAT4 | 1-acylglycerol-3-phosphate O-acyltransferase 4 | 2.63 |
| 226551_at | RIPK1 receptor (TNFRSF)-interacting serine-threonine kinase 1 | 2.59 | |
| 222702_x_at | CRIPT | cysteine-rich PDZ-binding protein | 2.58 |
| 203136_at | RABAC1 | Rab acceptor 1 (prenylated) | 2.58 |
| 234984_at | NEDD11 | neural precursor cell expressed, developmentally down-regulated | 2.56 |
| 224448_s_at | C6ORF125 | chromosome 6 open reading frame 125 | 2.53 |
| 218214_at | C12ORF44 | chromosome 12 open reading frame 44 | 2.52 |
| 224914_s_at | SARNP | SAP domain containing ribonucleoprotein | 2.48 |
| 232024_at | GIMAP2 | GTPase, IMAP family member 2 | 2.44 |
| 227207_x_at | ZNF213 | zinc finger protein 213 | 2.42 |
| 217803_at | GOLPH3 | golgi phosphoprotein 3 (coat-protein) | 2.41 |
| 200075_s_at | GUK1 | guanylate kinase 1 | 2.39 |
| 210460_s_at | PSMD4 | proteasome macropain 26S subunit, non-ATPase 4 | 2.38 |
| 226515_at | CCDC127 | coiled-coil domain containing 127 | 2.38 |
| 212936_at | FAM172A | family with sequence similarity 172, member A | 2.37 |
| 203106_s_at | VPS41 | vacuolar protein sorting 41 homolog (S. cerevisiae) | 2.36 |
| 228167_at | KLHL6 | kelch-like 6 (Drosophila) | 2.29 |
| 224364_s_at | PPIL3 | peptidylprolyl isomerase (cyclophilin)-like 3 | 2.28 |
| 203467_at | PMM1 | phosphomannomutase 1 | 2.28 |
| 223215_s_at | JKAMP | JNK1/MAPK8-associated membrane protein | 2.22 |
| 224817_at | SH3PXD2A | SH3 and PX domains 2A | 2.21 |
| 1557132_at | WDR17 | WD repeat domain 17 | 2.21 |
| 202204_s_at | AMFR | autocrine motility factor receptor | 2.20 |
| 226450_at | INSR | insulin receptor | 2.17 |
| 1554479_a_at | CARD8 | caspase recruitment domain family, member 8 | 2.15 |
| 213738_s_at | ATP5A1 | ATP synthase, H+ transporting, alpha subunit 1, | 2.10 |
| 214527_s_at | PQBP1 | polyglutamine binding protein 1 | 2.09 |
| 217317_s_at | HERC2P2 | hect domain and RLD 2 pseudogene 2 | 2.02 |
| 217120_s_at | LOC728142 | hypothetical LOC728142 | −2.00 |
| 231825_x_at | ATF7IP | activating transcription factor 7 interacting protein | −2.07 |
| 235060_at | LOC100190986 | hypothetical LOC100190986 | −2.18 |
| 242224_at | GPATCH2 | G patch domain containing 2 | −2.22 |
| 235623_at | ELP2 | elongation protein 2 homolog (S. cerevisiae) | −2.29 |
| 206323_x_at | OPHN1 | oligophrenin 1 | −2.37 |
| 1555024_at | ADAM22 | ADAM metallopeptidase domain 22 | −2.37 |
| 213065_at | ZFC3H1 | zinc finger, C3H1-type containing | −2.41 |
| 200772_x_at | PTMA (includes EG:5757) | prothymosin, alpha | −2.43 |
| 232689_at | LOC284561 | hypothetical protein LOC284561 | −2.47 |
| 214969_at | MAP3K9 | mitogen-activated protein kinase kinase kinase 9 | −2.53 |
| 208913_at | GGA2 golgi associated, gamma adaptin ARF binding protein 2 | −2.53 | |
| 202864_s_at | SP100 | SP100 nuclear antigen | −2.56 |
| 244641_at | C7ORF30 | chromosome 7 open reading frame 30 | −2.60 |
| 208191_x_at | PSG4 | pregnancy specific beta-1-glycoprotein 4 | −2.67 |
| 203181_x_at | SRPK2 | SFRS protein kinase 2 | −2.71 |
| 208549_x_at | PTMAP7 | prothymosin, alpha pseudogene 7 | −2.73 |
| 204602_at | DKK1 | dickkopf homolog 1 (Xenopus laevis) | −2.75 |
| 213093_at | PRKCA | protein kinase C, alpha | −2.94 |
| 211921_x_at | PTMA (includes EG:5757) | prothymosin, alpha | −3.01 |
| 1554696_s_at | TYMS | thymidylate synthetase | −3.07 |
| 230742_at | RBM6 | RNA binding motif protein 6 | −3.47 |
| 229272_at | FNBP4 | formin binding protein 4 | −4.55 |
Figure 2A.
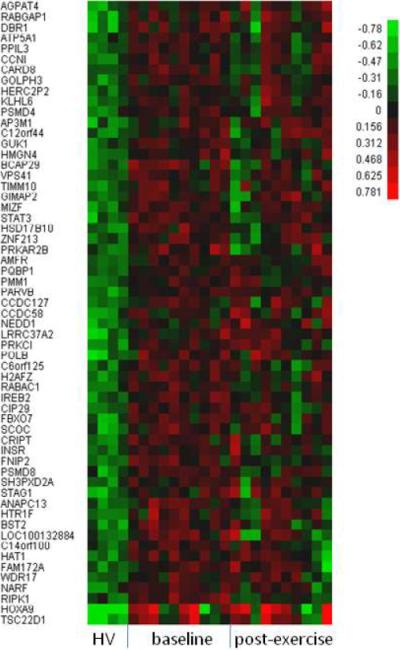
Hierarchical clustering of 59 up-regulated genes selected with 2-fold change, 25%FDR and present calls from the comparison of CD133+ gene expression between 10 CAD patients at baseline versus 4 healthy controls (HC). The values are in log10 scale.
Figure 2B.
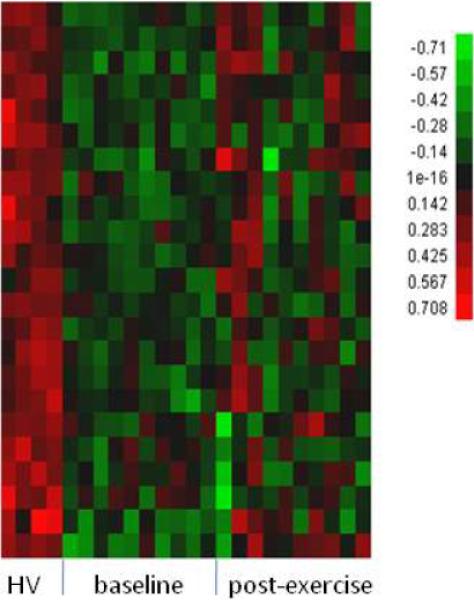
Hierarchical clustering of 23 down-regulated genes selected with 2-fold change, 25%FDR and present calls from the comparison of CD133+ gene expression between 10 CAD patients at baseline versus 4 healthy controls (HC). The values are in log10 scale.
Figure 3.
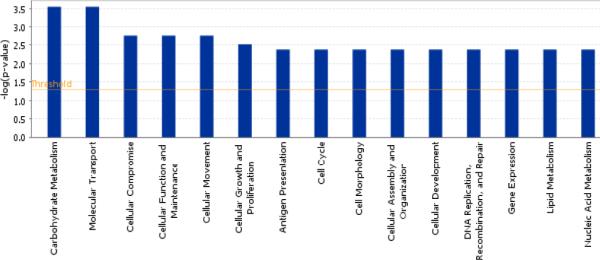
Distribution of some top molecular and cellular functions associated with the 82 genes selected from baseline CAD (n=10) Vs healthy control (n=4). X axis shows the pathways and the Y axis denotes the P value for each of the pathways represented.
Further validation of a few selected genes from Table 3 was performed by quantitative real-time reverse-transcription polymerase chain reaction (TaqMan) studies on ABI 7900. Results are represented as fold change comparing CAD patients to controls subjects. TaqMan fold changes were comparable and showed the same trend as the microarray data, as shown in Table 4.
Table 4.
Validation of few selected genes from microarray data by quantitative polymerase chain reaction (QPCR)
| Gene Symbol | Gene Title | Fold change CAD Vs HC | P value* | |
|---|---|---|---|---|
| QPCR | MA | |||
| ATP5A1 | ATPsynthase, H+transporting alpha1 | 2.53 | 2.1 | P< 0.005 |
| BST2 | Bone Marrow Stromal Cell Antigen 2 | 1.74 | 3.43 | P< 0.001 |
| HAT1 | Histone Acetyl Transferase1 | 2.25 | 2.77 | P< 0.005 |
| HOXA9 | Homeobox A9 | 10.55 | 19.04 | P< 0.001 |
| PTMA | Prothymosin alpha | −3.81 | −3.01 | P< 0.05 |
| TYMS | Thymidylate Synthetase | −2.21 | −3.07 | P< 0.005 |
• P values were calculated on the QPCR data.
• MA - Microarray
Effect of Exercise on the altered genes in CAD
We then examined the effect of exercise on these 59 up-regulated and 23 down regulated genes in the CAD patients. By paired one-sided t-test analysis, 7 out of 10 patients showed significant response to exercise (p<0.05) for the up-regulated genes on average as evidenced by the level of expression reaching levels approximating those in the healthy subjects. Similarly, 5 CAD patients showed significant response to exercise (p<0.05) for the down-regulated genes on average reaching levels approximating those in the healthy subjects. 4 CAD patients showed significant changes (p<0.01) in both the up and down regulated genes. Two patients showed no response to this exercise in either up- or down-regulated genes, on average. However, when individual genes from the set of 82 genes were examined for the group as a whole, all the genes on average showed at least 2-fold change from baseline measurements.
Correlation analysis of these 82 differentially expressed genes with FDR < 20% to clinical measurements showed strong association, as determined by FDR<20%, between the baseline gene expression for GUK1, HOXA9, NARF, PRKC1, SP100, STAG1 and TCS22D1 to at least one of the categories such as patient age, CD133 cell number, white blood cell count, C-reactive protein and triglyceride levels. These genes and the associated clinical categories are listed in Supplemental Table 2. Similarly, correlation analysis on the changes in gene expression at the completion of exercise program showed strong correlation of 15 genes to changes in glucose, C-reactive protein, total cholesterol, low density lipoprotein cholesterol, triglycerides and heart rate (Supplementary Table 3). In this small set of samples from patients we did not observe statistically significant correlation between changes in gene expression and CD 133 cells following the completion of the 3 month exercise program.
Discussion
There is considerable basic research and clinical interest in the relevance of bone marrow-derived cells to vascular homeostasis and repair. Initial reports focused on hematopoietic progenitor cells with CD34 marker, but subsequently included a population of immature progenitor cells indentified by the CD133 marker. The relevance of these progenitor cells in CAD was suggested by Werner et al17, who reported an inverse relation between CD34+ and CD133+ cells and subsequent cardiac events at 12 months in a series of 519 patients with disease confirmed at coronary angiography. Currently, the common clinical concept claims a protective role of bone marrow-derived progenitor cells, which may be compromised in CAD due to reduced number and impaired function18,19. Considering these reported findings, we undertook the current study with the aim of characterizing CD133+ cells from CAD patients at the genomic level.
Our analysis showed significant differences in expression of several genes in CD133+ cells from CAD patients that may be important to survival and differentiation potential of cells compared with gene expression in CD133+ cells isolated from healthy controls. Thus we identified 82 differentially expressed genes in CAD patients compared with controls, of which 59 genes, such as HOX A920,TSC22D121, RIPK1, NARF, ANAPC1322 and HAT23, were found to be significantly up-regulated, and 23 genes, such as MAP3K24–26, PTMA27–29, FNBP4 and RBM6, were found to be down-regulated. By Ingenuity Pathway analysis, these genes were found to be involved in multiple cellular functions, especially carbohydrate metabolism and molecular transport. After 3 months of rehabilitation associated with significant improvement in effort tolerance and increase in the number of CD133+ cells, gene expression patterns of the CD133+ cells resembled those of controls in 7 out of 10 patients in our study. In 4 CAD patients, exercise reversed expression levels towards those measured in controls in both up- and down-regulated genes determined at baseline. Correlation analysis identified only SP 100 expression to have statistically significant association to baseline CD133 cell number. The relevance of SP 100 gene product to stem cells of vascular endothelium and cardio vascular disease remains unknown.
To identify transcripts that are highly abundant in CD133+ cells from our study, positively detected transcripts from healthy controls were ranked in order of their abundance from high to low based on their log-transformed median signal intensity. The top 500 ranking probe sets representing 351 genes identified to be of high abundance in CD133+ cells by Ingenuity pathway analysis at p<0.05 indicated them to be involved in multiple cellular functions such as protein synthesis, post-transcriptional modification and cell death, and this list provides a novel expanded transcriptome of CD 133+ cells.
A comparative analysis of previously published 257 CD133+ highly abundant transcripts from cord blood and peripheral blood30–32 and CD133+ transcripts from the current study showed overlap of 42 highly expressed transcripts such as growth factor binding proteins, nucleosporin, transcription factors, sideroflexin, anaphase promoting complex, and map kinases. The non-overlapping genes included several transcripts with unknown functions and several zinc finger proteins. It should be noted that the methodology to sort the CD133+ cells and RNA processing for gene chip hybridization differ considerably among these studies.
Of particular interest among these gene expression modulations is the significant alteration in the expression of HOXA9 and histone acetyl transferase (HAT1) genes in our study. It is believed that the interplay between HAT, histone deacetyalse (HDAC) and HOXA9 genes determines the endothelial differentiation of progenitor cells23. HOXA9 has been shown to be crucial for hematopoietic stem cell development and stem cell expansion33. HOXA9 acts as a homeobox transcription factor expressed in mature endothelial cells and its expression is downregulated by inflammatory signals. HOXA9 overexpression is associated with an increase in the number of circulating EPCs while in HOXA deficient mice, there was a decrease in the number of EPCs and impaired postnatal neovasculaization23,34. Studies on hypertensive patients with significant endothelial injury by Pirro et al35 revealed decrease in the number of peripheral EPCs and significantly lowered expression of HOXA9 gene in those cells. HAT1 is involved in fundamental processes that regulate the expression of multiple genes that play an important role in the multifactorial processes that lead to atherosclerosis and re stenosis36,37. It is hypothesized that HOXA9 functions by modulating downstream target genes that contributes in physiological conditions to endothelial commitment of EPCs, postnatal neovascularization and injured endothelium repair20. The relevance of expression of these genes in CAD patients at baseline or follow up is unclear as we observed no association with CD133 cell numbers and we did not measure the endothelial function in our patients.
Our findings may have clinical implications, as CD133+ cells may have limited capacity to repair vascular disease in patients with CAD and thus compromise cell-based treatment approaches, including autologous administration of CD133+ cells isolated from the circulation and injected in large numbers into the heart (17). Although CD34+ selected cells or nonselected bone marrow mononuclear cells are more commonly used for this purpose, at least 4 trials using CD133+ cells are registered in www.ClinicalTrials.gov. We also report that exercise might reverse some of the gene expression patterns towards levels seen in cells from healthy subjects, thus improving their potential for vascular repair or new vessel growth. Further studies are needed to determine whether changes in CD133+ gene expression patterns towards that seen in healthy subjects are accompanied by changes in phenotype and differentiation potential that may be relevant to cardiovascular repair.
Supplementary Material
Acknowledgement
This research was supported by the Intramural Research Program of the NIH, National Heart, Lung and Blood Institute. We would like to acknowledge Ms. Kimberly Woodhouse for her technical assistance in performing the QPCR assays.
References
- 1.Libby P, Willerson JT, Braunwald E. C-reactive protein and coronary heart disease. N Engl J Med. 2004;351:295–298. author reply 295-298. [PubMed] [Google Scholar]
- 2.Gill M, Dias S, Hattori K, Rivera ML, Hicklin D, Witte L, Girardi L, Yurt R, Himel H, Rafii S. Vascular trauma induces rapid but transient mobilization of VEGFR2(+)AC133(+) endothelial precursor cells. Circ Res. 2001;88:167–174. doi: 10.1161/01.res.88.2.167. [DOI] [PubMed] [Google Scholar]
- 3.Peichev M, Naiyer AJ, Pereira D, Zhu Z, Lane WJ, Williams M, Oz MC, Hicklin DJ, Witte L, Moore MA, Rafii S. Expression of VEGFR-2 and AC133 by circulating human CD34(+) cells identifies a population of functional endothelial precursors. Blood. 2000;95:952–958. [PubMed] [Google Scholar]
- 4.Gehling UM, Ergun S, Schumacher U, Wagener C, Pantel K, Otte M, Schuch G, Schafhausen P, Mende T, Kilic N, Kluge K, Schafer B, Hossfeld DK, Fiedler W. In vitro differentiation of endothelial cells from AC133-positive progenitor cells. Blood. 2000;95:3106–3112. [PubMed] [Google Scholar]
- 5.Heeschen C, Lehmann R, Honold J, Assmus B, Aicher A, Walter DH, Martin H, Zeiher AM, Dimmeler S. Profoundly reduced neovascularization capacity of bone marrow mononuclear cells derived from patients with chronic ischemic heart disease. Circulation. 2004;109:1615–1622. doi: 10.1161/01.CIR.0000124476.32871.E3. [DOI] [PubMed] [Google Scholar]
- 6.Scheubel RJ, Zorn H, Silber RE, Kuss O, Morawietz H, Holtz J, Simm A. Age-dependent depression in circulating endothelial progenitor cells in patients undergoing coronary artery bypass grafting. J Am Coll Cardiol. 2003;42:2073–2080. doi: 10.1016/j.jacc.2003.07.025. [DOI] [PubMed] [Google Scholar]
- 7.Vasa M, Fichtlscherer S, Aicher A, Adler K, Urbich C, Martin H, Zeiher AM, Dimmeler S. Number and migratory activity of circulating endothelial progenitor cells inversely correlate with risk factors for coronary artery disease. Circ Res. 2001;89:E1–7. doi: 10.1161/hh1301.093953. [DOI] [PubMed] [Google Scholar]
- 8.Aicher A, Heeschen C, Mildner-Rihm C, Urbich C, Ihling C, Technau-Ihling K, Zeiher AM, Dimmeler S. Essential role of endothelial nitric oxide synthase for mobilization of stem and progenitor cells. Nat Med. 2003;9:1370–1376. doi: 10.1038/nm948. [DOI] [PubMed] [Google Scholar]
- 9.Laufs U, Werner N, Link A, Endres M, Wassmann S, Jurgens K, Miche E, Bohm M, Nickenig G. Physical training increases endothelial progenitor cells, inhibits neointima formation, and enhances angiogenesis. Circulation. 2004;109:220–226. doi: 10.1161/01.CIR.0000109141.48980.37. [DOI] [PubMed] [Google Scholar]
- 10.Paul JD, Powell TM, Thompson M, Benjamin M, Rodrigo M, Carlow A, Annavajjhala V, Shiva S, Dejam A, Gladwin MT, McCoy JP, Zalos G, Press B, Murphy M, Hill JM, Csako G, Waclawiw MA, Cannon RO., 3rd Endothelial progenitor cell mobilization and increased intravascular nitric oxide in patients undergoing cardiac rehabilitation. J Cardiopulm Rehabil Prev. 2007;27:65–73. doi: 10.1097/01.HCR.0000265031.10145.50. [DOI] [PubMed] [Google Scholar]
- 11.Rehman J, Li J, Parvathaneni L, Karlsson G, Panchal VR, Temm CJ, Mahenthiran J, March KL. Exercise acutely increases circulating endothelial progenitor cells and monocyte-/macrophage-derived angiogenic cells. J Am Coll Cardiol. 2004;43:2314–2318. doi: 10.1016/j.jacc.2004.02.049. [DOI] [PubMed] [Google Scholar]
- 12.Sandri M, Adams V, Gielen S, Linke A, Lenk K, Krankel N, Lenz D, Erbs S, Scheinert D, Mohr FW, Schuler G, Hambrecht R. Effects of exercise and ischemia on mobilization and functional activation of blood-derived progenitor cells in patients with ischemic syndromes: results of 3 randomized studies. Circulation. 2005;111:3391–3399. doi: 10.1161/CIRCULATIONAHA.104.527135. [DOI] [PubMed] [Google Scholar]
- 13.Steiner S, Niessner A, Ziegler S, Richter B, Seidinger D, Pleiner J, Penka M, Wolzt M, Huber K, Wojta J, Minar E, Kopp CW. Endurance training increases the number of endothelial progenitor cells in patients with cardiovascular risk and coronary artery disease. Atherosclerosis. 2005;181:305–310. doi: 10.1016/j.atherosclerosis.2005.01.006. [DOI] [PubMed] [Google Scholar]
- 14.Hsia D, Casaburi R, Pradhan A, Torres E, Porszasz J. Physiological responses to linear treadmill and cycle ergometer exercise in COPD. Eur Respir J. 2009;34:605–615. doi: 10.1183/09031936.00069408. [DOI] [PubMed] [Google Scholar]
- 15.Desai A, Glaser A, Liu D, Raghavachari N, Blum A, Zalos G, Lippincott M, McCoy JP, Munson PJ, Solomon MA, Danner RL, Cannon RO., 3rd Microarray-based characterization of a colony assay used to investigate endothelial progenitor cells and relevance to endothelial function in humans. Arterioscler Thromb Vasc Biol. 2009;29:121–127. doi: 10.1161/ATVBAHA.108.174573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J.R. Statist.Soc.B. 1995;57:289–300. [Google Scholar]
- 17.Werner N, Kosiol S, Schiegl T, Ahlers P, Walenta K, Link A, Bohm M, Nickenig G. Circulating endothelial progenitor cells and cardiovascular outcomes. N Engl J Med. 2005;353:999–1007. doi: 10.1056/NEJMoa043814. [DOI] [PubMed] [Google Scholar]
- 18.Leor J, Marber M. Endothelial progenitors: a new Tower of Babel? J Am Coll Cardiol. 2006;48:1588–1590. doi: 10.1016/j.jacc.2006.07.032. [DOI] [PubMed] [Google Scholar]
- 19.Schmidt-Lucke C, Rossig L, Fichtlscherer S, Vasa M, Britten M, Kamper U, Dimmeler S, Zeiher AM. Reduced number of circulating endothelial progenitor cells predicts future cardiovascular events: proof of concept for the clinical importance of endogenous vascular repair. Circulation. 2005;111:2981–2987. doi: 10.1161/CIRCULATIONAHA.104.504340. [DOI] [PubMed] [Google Scholar]
- 20.Bruhl T, Urbich C, Aicher D, Acker-Palmer A, Zeiher AM, Dimmeler S. Homeobox A9 transcriptionally regulates the EphB4 receptor to modulate endothelial cell migration and tube formation. Circ Res. 2004;94:743–751. doi: 10.1161/01.RES.0000120861.27064.09. [DOI] [PubMed] [Google Scholar]
- 21.Zhang J, Oh KH, Xu H, Margetts PJ. Vascular endothelial growth factor expression in peritoneal mesothelial cells undergoing transdifferentiation. Perit Dial Int. 2008;28:497–504. [PubMed] [Google Scholar]
- 22.Miller F, Kentsis A, Osman R, Pan ZQ. Inactivation of VHL by tumorigenic mutations that disrupt dynamic coupling of the pVHL.hypoxia-inducible transcription factor-1alpha complex. J Biol Chem. 2005;280:7985–7996. doi: 10.1074/jbc.M413160200. [DOI] [PubMed] [Google Scholar]
- 23.Rossig L, Urbich C, Bruhl T, Dernbach E, Heeschen C, Chavakis E, Sasaki K, Aicher D, Diehl F, Seeger F, Potente M, Aicher A, Zanetta L, Dejana E, Zeiher AM, Dimmeler S. Histone deacetylase activity is essential for the expression of HoxA9 and for endothelial commitment of progenitor cells. J Exp Med. 2005;201:1825–1835. doi: 10.1084/jem.20042097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Song MB, Yu XJ, Zhu GX, Chen JF, Zhao G, Huang L. Transfection of HGF gene enhances endothelial progenitor cell (EPC) function and improves EPC transplant efficiency for balloon-induced arterial injury in hypercholesterolemic rats. Vascul Pharmacol. 2009;51:205–213. doi: 10.1016/j.vph.2009.06.009. [DOI] [PubMed] [Google Scholar]
- 25.Zakkar M, Chaudhury H, Sandvik G, Enesa K, Luong le A, Cuhlmann S, Mason JC, Krams R, Clark AR, Haskard DO, Evans PC. Increased endothelial mitogen-activated protein kinase phosphatase-1 expression suppresses proinflammatory activation at sites that are resistant to atherosclerosis. Circ Res. 2008;103:726–732. doi: 10.1161/CIRCRESAHA.108.183913. [DOI] [PubMed] [Google Scholar]
- 26.Zakkar M, Van der Heiden K, Luong le A, Chaudhury H, Cuhlmann S, Hamdulay SS, Krams R, Edirisinghe I, Rahman I, Carlsen H, Haskard DO, Mason JC, Evans PC. Activation of Nrf2 in endothelial cells protects arteries from exhibiting a proinflammatory state. Arterioscler Thromb Vasc Biol. 2009;29:1851–1857. doi: 10.1161/ATVBAHA.109.193375. [DOI] [PubMed] [Google Scholar]
- 27.Grunberg E, Eckert K, Maurer HR. Prothymosin alpha1 antagonizes the inhibitory effects of transforming growth factor-beta1 on the adhesion of peripheral blood lymphocytes to human umbilical vein endothelial cells. Int J Mol Med. 1998;1:741–746. doi: 10.3892/ijmm.1.4.741. [DOI] [PubMed] [Google Scholar]
- 28.Grunberg E, Eckert K, Maurer HR, Immenschuh P, Kreuser ED. Prothymosin alpha1 effects on IL-2-induced expression of LFA-1 on lymphocytes and their adhesion to human umbilical vein endothelial cells. J Interferon Cytokine Res. 1997;17:159–165. doi: 10.1089/jir.1997.17.159. [DOI] [PubMed] [Google Scholar]
- 29.Roland I, Minet E, Ernest I, Pascal T, Michel G, Remacle J, Michiels C. Identification of hypoxia-responsive messengers expressed in human microvascular endothelial cells using differential display RT-PCR. Eur J Biochem. 2000;267:3567–3574. doi: 10.1046/j.1432-1327.2000.01385.x. [DOI] [PubMed] [Google Scholar]
- 30.Ivanova NB, Dimos JT, Schaniel C, Hackney JA, Moore KA, Lemischka IR. A stem cell molecular signature. Science. 2002;298:601–604. doi: 10.1126/science.1073823. [DOI] [PubMed] [Google Scholar]
- 31.Ramalho-Santos M, Yoon S, Matsuzaki Y, Mulligan RC, Melton DA. “Stemness”: transcriptional profiling of embryonic and adult stem cells. Science. 2002;298:597–600. doi: 10.1126/science.1072530. [DOI] [PubMed] [Google Scholar]
- 32.Toren A, Bielorai B, Jacob-Hirsch J, Fisher T, Kreiser D, Moran O, Zeligson S, Givol D, Yitzhaky A, Itskovitz-Eldor J, Kventsel I, Rosenthal E, Amariglio N, Rechavi G. CD133-positive hematopoietic stem cell “stemness” genes contain many genes mutated or abnormally expressed in leukemia. Stem Cells. 2005;23:1142–1153. doi: 10.1634/stemcells.2004-0317. [DOI] [PubMed] [Google Scholar]
- 33.Panepucci RA, Oliveira LH, Zanette DL, Carrara RD, Araujo AG, Orellana MD, Palma PV, Menezes CC, Covas DT, Zago MA. Increased Levels of NOTCH1, NF-kappaB, and Other Interconnected Transcription Factors Characterize Primitive Sets of Hematopoietic Stem Cells. Stem Cells Dev. 2010;19:321–32. doi: 10.1089/scd.2008.0397. [DOI] [PubMed] [Google Scholar]
- 34.Thorsteinsdottir U, Mamo A, Kroon E, Jerome L, Bijl J, Lawrence HJ, Humphries K, Sauvageau G. Overexpression of the myeloid leukemia-associated Hoxa9 gene in bone marrow cells induces stem cell expansion. Blood. 2002;99:121–129. doi: 10.1182/blood.v99.1.121. [DOI] [PubMed] [Google Scholar]
- 35.Pirro M, Schillaci G, Menecali C, Bagaglia F, Paltriccia R, Vaudo G, Mannarino MR, Mannarino E. Reduced number of circulating endothelial progenitors and HOXA9 expression in CD34+ cells of hypertensive patients. J Hypertens. 2007;25:2093–2099. doi: 10.1097/HJH.0b013e32828e506d. [DOI] [PubMed] [Google Scholar]
- 36.Pons D, de Vries FR, van den Elsen PJ, Heijmans BT, Quax PH, Jukema JW. Epigenetic histone acetylation modifiers in vascular remodelling: new targets for therapy in cardiovascular disease. Eur Heart J. 2009;30:266–277. doi: 10.1093/eurheartj/ehn603. [DOI] [PubMed] [Google Scholar]
- 37.Pons D, Jukema JW. Epigenetic histone acetylation modifiers in vascular remodelling - new targets for therapy in cardiovascular disease. Neth Heart J. 2008;16:30–32. doi: 10.1007/BF03086114. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


