Abstract
Histone H3 lysine-4 (H3K4) methylation is associated with transcribed genes in eukaryotes. In Drosophila and mammals, both di- and tri-methylation of H3K4 are associated with gene activation. In contrast to animals, in Arabidopsis H3K4 trimethylation, but not mono- or di-methylation of H3K4, has been implicated in transcriptional activation. H3K4 methylation is catalyzed by the H3K4 methyltransferase complexes known as COMPASS or COMPASS-like in yeast and mammals. Here, we report that Arabidopsis homologs of the COMPASS and COMPASS-like complex core components known as Ash2, RbBP5, and WDR5 in humans form a nuclear subcomplex during vegetative and reproductive development, which can associate with multiple putative H3K4 methyltransferases. Loss of function of ARABIDOPSIS Ash2 RELATIVE (ASH2R) causes a great decrease in genome-wide H3K4 trimethylation, but not in di- or mono-methylation. Knockdown of ASH2R or the RbBP5 homolog suppresses the expression of a crucial Arabidopsis floral repressor, FLOWERING LOCUS C (FLC), and FLC homologs resulting in accelerated floral transition. ASH2R binds to the chromatin of FLC and FLC homologs in vivo and is required for H3K4 trimethylation, but not for H3K4 dimethylation in these loci; overexpression of ASH2R causes elevated H3K4 trimethylation, but not H3K4 dimethylation, in its target genes FLC and FLC homologs, resulting in activation of these gene expression and consequent late flowering. These results strongly suggest that H3K4 trimethylation in FLC and its homologs can activate their expression, providing concrete evidence that H3K4 trimethylation accumulation can activate eukaryotic gene expression. Furthermore, our findings suggest that there are multiple COMPASS-like complexes in Arabidopsis and that these complexes deposit trimethyl but not di- or mono-methyl H3K4 in target genes to promote their expression, providing a molecular explanation for the observed coupling of H3K4 trimethylation (but not H3K4 dimethylation) with active gene expression in Arabidopsis.
Author Summary
Histones can be covalently modified and histone modifications regulate chromatin structure and gene transcription. One such modification is histone H3 lysine-4 (H3K4) methylation, which can be mono-, di-, or tri-methylated. In animals such as fruitfly and mammals, both di- and tri-methylation of H3K4 are associated with active gene expression. In contrast to animals, in the flowering plant Arabidopsis only H3K4 trimethylation has been implicated in gene transcriptional activation. H3K4 methylation is catalyzed by the H3K4 methyltransferase complexes known as COMPASS-like in mammals. Here, we report that COMPASS-like H3K4 methyltransferase complexes exist in Arabidopsis. Loss of function of a core complex protein causes a great decrease in Arabidopsis genome-wide H3K4 trimethylation, but not in di- or mono-methylation. Our analyses of several direct target genes of these COMPASS-like complexes show that they mediate deposition of trimethyl but not dimethyl H3K4 in these loci to activate their expression, providing concrete evidence for the notion that H3K4 trimethylation accumulation can activate eukaryotic gene expression. Furthermore, our findings provide a molecular explanation for the observed coupling of trimethylation but not dimethylation of H3K4 with active gene expression in Arabidopsis. In addition, we found that H3K4 trimethylation regulates leaf growth and development, flowering, and embryo development.
Introduction
Histone lysine methylation regulates chromatin structure and gene transcription in eukaryotes. Various lysine residues on histones can be methylated and the ε-amino group of lysines can be mono-, di- and tri-methylated. Lysine methylation is linked with transcriptional activation or repression depending on the particular residue that is methylated and the degree of methylation [1]. For instance, H3 lysine-27 trimethylation (H3K27me3) is exclusively involved in transcriptional repression, whereas H3K4 trimehtylation is associated with actively transcribed genes [2].
Recent genome-scale analyses of H3K4 methylation have revealed that different H3K4 methylation states are often associated with distinct transcription states in a gene. In the well-studied Saccharomyces cerevisiae, H3K4 trimethylation is a mark for actively transcribed genes, whereas mono- or di-methylation of H3K4 is not linked with active gene expression [3]. In Drosophila and mammals, both di- and tri-methylation of H3K4 are associated with gene activation [4], [5]. In contrast to animals, in the higher plant Arabidopsis thaliana, only H3K4 trimethylation, but not mono- or di-methylation of H3K4, is implicated in transcriptional activation [6].
H3K4 methylation is catalyzed by various methyltransferases. In Saccharomyces cerevisiae, the COMPASS (for Complex Proteins Associated with Set1) H3K4 methyltransferase complex catalyzes H3K4 methylation [7]. This complex contains an H3K4 methyltransferase called Set1, the only known H3K4 methyltransferase in yeast. By itself, Set1 is unable to catalyze H3K4 methylation, and requires other structural components in the complex for its catalytic activity [8], [9]. COMPASS-like complexes have been identified in mammals, and so far, five such complexes known as hSet1 and MLL1, MLL2, MLL3 and MLL4 have been biochemically purified [for a review, see [2]]. All of these complexes contain four core components including an H3K4 methyltransferase and three structural core components known as WDR5, Ash2 and RbBP5, homologs of the yeast SWD3, BRE2 and SWD1, respectively [2]. WDR5, Ash2 and RbBP5 together form a stable core subcomplex that provides a structural platform for H3K4 methylation [10]. The Ash2-RbBP5-WDR5 subcomplex interchangeably associates with different H3K4 methyltransferases such as hSet1, MLL1 and MLL2 to form different catalytic complexes, and is essential for both di- and tri-methylation of H3K4 [9], [10]. An in vitro reconstituted four-component mini-complex composed of WDR5, Ash2, RbBP5 and MLL1 methylates H3K4 specifically [9]. So far, COMPASS-like complexes have been identified in mammals, but still remain to be identified in other multicellular organisms such as plants.
H3K4 trimethylation typically occurs concomitantly with active gene transcription, and trimethyl H3K4 (H3K4me3) predominantly accumulates in the 5′ transcribed regions [2], [11]. In yeast, the RNA Polymerase II Associated Factor 1 complex (Paf1c) recruits the COMPASS complex to the initiating and early-elongating RNA Polymerase II (Pol II), resulting in H3K4 trimethylation in the 5′ genic regions [12]. A similar mechanism might exist in multicellular organisms as Paf1c appears to be evolutionarily conserved in animals and plants [13]–[15]. Although H3K4me3 is a chromatin mark for actively transcribed genes, whether it plays an active role in transcriptional activation remains unclear because the accumulation of H3K4me3 in the 5′ transcribed regions might merely result from active transcription by Pol II [16].
Recent studies have revealed a few components involved in H3K4 methylation in Arabidopsis. It has been shown that the Arabidopsis Paf1c is required for the accumulation of H3K4me3 in the 5′ regions of actively transcribed genes in Arabidopsis genome [15]. Bioinformatic and phylogenetic analyses reveal that there may be up to ten putative Arabidopsis H3K4 methyltransferases, among which are ARABIDOPSIS TRITHORAX1 (ATX1), ATX2, SET DOMAIN PROTEIN25 (SDG25; also known as ATXR7), SDG14 and SDG16 [17], [18]. Loss of ATX1 function causes a slight reduction in global H3K4me3 and accelerated developmental transition from a vegetative to a reproductive phase (i.e., flowering) [19]. In addition, atx1 mutation leads to floral organ abnormalities in a particular Arabidopsis ecotype [20]. Recently, it has been shown that loss of ATX2 or ATXR7 function causes moderately early flowering [21]–[23]. Besides these putative H3K4 methyltransferases, Arabidopsis has two homologs of the human COMPASS-like complex core component WDR5, namely WDR5a and WDR5b [24]. WDR5a binds histone H3 tails, is involved in H3K4 methylation in its target gene chromatin, and represses Arabidopsis flowering [24].
The timing of floral transition in Arabidopsis is genetically controlled by a network of flowering genes, among which FLC plays a central role. FLC, a MADS box transcriptional factor, is a key floral repressor that quantitatively inhibits the floral transition in Arabidopsis [for a review, see [25], [26]]. Besides FLC, there are five FLC homologs in Arabidopsis: FLOWERING LOCUS M (FLM)/MADS BOX AFFECTING FLOWERING 1 (MAF1) and MAF2-MAF5 [27], [28]. FLM, MAF2 and MAF4 moderately repress flowering [27]–[29], whereas the roles for MAF3 and MAF5 remain unclear [28].
FLC expression is under complex control and chromatin modification plays a critical role in FLC regulation which has become a model for understanding plant gene regulation by chromatin-based mechanisms [26], [30], [31]. FLC is expressed at low levels in many early-flowering Arabidopsis accessions because of the repression by ‘autonomous-pathway’ genes, among which are a few histone modifiers including CURLY LEAF (a putative H3K27 methyltransferase), HISTONE DEACETYLASE6, FLOWERING LOCUS D (FLD, a putative H3K4 demethylase) and several Type I and II arginine methyltransferases [32]–[36]. These proteins mediate ‘repressive’ histone modifications in FLC chromatin and repress FLC expression to promote flowering. In addition to repressive histone modifications, FLC chromatin can also be modified by H3K4 methylation. Recent studies have revealed that H3K4 trimethylation is linked with activation of FLC expression. H3K4me3 accumulates in the region around the transcription start site (TSS) of FLC including the 5′ end of transcribed region [13], [21]. This modification requires Paf1c and WDR5a, and in addition, ATX1 and ATXR7 are partly required [13], [15], [21], [22], [24], [37]. WDR5a can interact with ATX1, binds to FLC chromatin, is required for H3K4 trimethylation in FLC and for FLC activation, and has been proposed to act in the context of an H3K4 methyltransferase complex [24]. So far, although several components involved in H3K4 methylation have been characterized in Arabidopsis, it remains essentially unknown whether H3K4 methyltransferase complexes exist in Arabidopsis. In addition, although it has been well documented that H3K4me3 accumulation is associated with actively transcribed FLC chromatin, whether H3K4 trimethylation can activate FLC expression is unknown.
Here, we report that Arabidopsis homologs of the human COMPASS-like complex core components Ash2 and RbBP5, together with WDR5a, form a nuclear subcomplex for H3K4 methylation during vegetative and reproductive development. The subcomplex component WDR5a can associate with several putative H3K4 methyltransferases, suggesting that multiple COMPASS-like complexes exist in Arabidopsis. Loss of ASH2R function causes a great decrease in genome-wide H3K4 trimethylation, but not di- or mono-methylation. In addition, we found that the ASH2R subcomplex mediates H3K4 trimethylaiton, but not H3K4 dimethylation, in FLC and FLC homologs to activate their expression resulting in delayed flowering. Our findings suggest that the ASH2R-containing COMPASS-like complexes (ASH2R-COMPASS) deposit H3K4me3, but not H3K4me2 or H3K4me1 in Arabidopsis genome. Furthermore, we found that a null lesion in ASH2R causes arrested embryo development at globular stage and that ASH2R is also required for proper leaf growth and development, suggesting that the ASH2R-COMPASS-mediated H3K4 trimethylation plays important roles for multiple Arabidopsis developmental processes.
Results
Identification and Phylogenetic Analyses of Arabidopsis Homologs of the Human COMPASS-Like Complex Core Components, Ash2, RbBP5, and WDR5
Recently we have identified two homologs of the human WDR5, namely WDR5a and WDR5b in Arabidopsis genome [24]. To explore whether homologs of the other core COMPASS-like complex components Ash2 and RbBP5, exist in Arabidopsis, we searched the Arabidopsis protein database with the amino acid sequences of these proteins and identified a single homolog for each protein, namely ASH2R (At_1g51450) and RbBP5 LIKE (RBL; At_3g21060) (Figure S1 and Figure S2). The sequence similarity between ASH2R and the human Ash2 over the entire ASH2R is 46% (Figure S1). A recent phylogenetic analysis of Ash2 homologs from several animals and plants also indicates that ASH2R (also known as TRO) is a clear homolog of the human Ash2 [38]. We carried out phylogenetic analysis of RbBP5 homologs from representative animal and plant species. As shown in Figure S2, in each animal or plant species examined, there is only a single RbBP5 homolog; animal RbBP5 homologs form one clade, and plant homologs constitute another clade. These findings show that there are Ash2 and RbBP5 homologs in Arabidopsis.
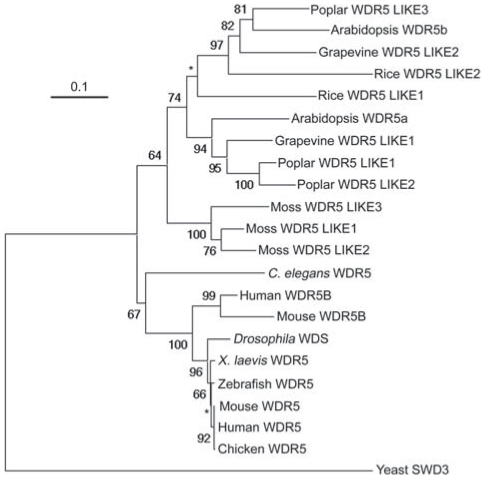
The evolutionary history of WDR5 homologs is complex. We found that there are multiple WDR5 homologs in land plants, for instance, three in the basal land plant moss, two in the monocot rice and three in the eudicot poplar, whereas in most animals, there is only a single WDR5 homolog (Figure 1). We further performed phylogenetic analysis of WDR5 homologs from representative species. The phylogenetic tree indicates that all WDR5 homologs from plants form a single clade with 64 bootstrap support, whereas all animal WDR5 homologs form another clade with 67 bootstrap support (Figure 1). There is no strong bootstrap support for an animal-plant clade. These results indicate that the animal WDR5 homologs might function like the human WDR5, but raise a question on biochemical functions of the multiple WDR5 homologs in plants.
Figure 1. Phylogenetic Tree for WDR5 Homologs from Representative Plant and Animal Species.
The tree of full-length WDR5 homologs was constructed by the Neighbor-Joining method using PHYLIP. Bootstrap values are shown for clades with a support greater than 60% (in 1000 sampling replicates). Star indicates a clade without a strong bootstrap support. The protein accession numbers are specified in Table S1.
RBL Promotes the Expression of FLC and FLC Homologs to Repress the Floral Transition
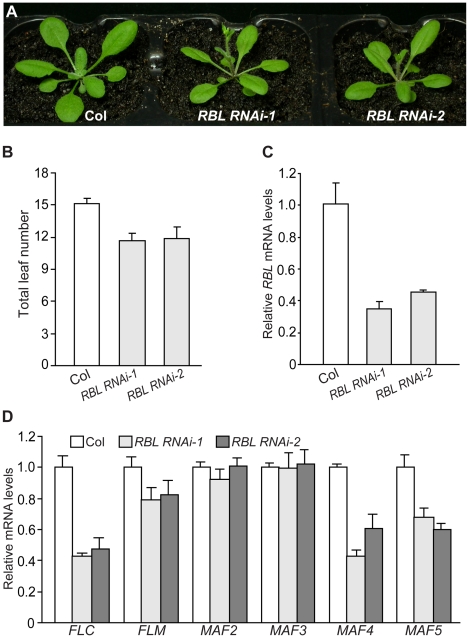
It was of great interest to determine biological functions of ASH2R and RBL. First, we employed a double-stranded RNA interference (dsRNAi) approach to knock down RBL expression in wildtype Col (knockout mutants in RBL were not publically available). Briefly, a 245-bp RBL-specific fragment from the 3′ transcribed region was used to create a dsRNAi cassette driven by the constitutive 35S promoter. Eight independent transgenic lines were generated, from which homozygous RBL RNAi-1 and RBL RNAi-2 lines with a single T-DNA locus, were identified. In long days (LD), five out of the eight lines flowered earlier than parental Col, but otherwise were normal; no noticeable phenotypes were observed in the remaining three lines (Figure 2A and 2B, and data not shown). RBL transcript levels were quantified in RBL RNAi-1 and -2 seedlings by real-time quantitative PCR, and indeed, RBL expression was knocked down compared to Col (Figure 2C). Together, these results show that RBL represses the floral transition.
Figure 2. RBL Represses the Floral Transition in Arabidopsis.
(A) Phenotypes of RBL RNAi lines grown in long days (LD). Col was used as the parental accession in RBL knockdown. (B) Flowering times of the RBL RNAi lines grown in LD. The total number of primary rosette and cauline leaves formed prior to bolting was counted, and 11–12 plants were scored for each line. Error bars indicate standard deviations (SD). (C) Relative RBL mRNA levels in seedlings of the RBL RNAi lines determined by real-time quantitative PCR. Relative expression to parental Col is presented. Bars indicate SD. (D) Relative mRNA levels of FLC and FLC homologs in RBL RNAi seedlings quantified by real-time PCR.
FLC plays a central role in floral repression in Arabidopsis, and as described in the Introduction, several components involved in H3K4 methylation promote FLC expression. Hence, it was of interest to examine whether FLC expression was suppressed upon RBL knockdown. FLC transcript levels were reduced in RBL RNAi seedlings compared to Col (Figure 2D). We further examined the expression of FLC homologs upon RBL knockdown, and found that MAF4 and MAF5 transcript levels were reduced, whereas levels of FLM, MAF2 and MAF3 transcripts in the RBL RNAi lines were similar to those in Col (Figure 2D). Together, these results suggest that RBL promotes the expression of FLC, MAF4 and MAF5, but not FLM, MAF2 or MAF3, to repress the floral transition.
ASH2R Represses Flowering and Is Essential for Seed Development
To explore the biological function of ASH2R, we identified an ash2r-1/+ heterozygous line from the SAIL collection [39], which carries an insertional T-DNA in the middle of ASH2R. We did not recover any ash2r-1 homozygotes from a large population of the selfed progeny of ash2r-1/+ heterozygotes. Subsequently, we found that in developing siliques from ash2r-1/+ plants, about one-fifth of the seeds (167 out of 788 seeds) were aborted white and brown seeds (Figure S3A), whereas in wild-type siliques only less than four thousandth of the seeds (3 out of 798) were aborted. Thus, we conclude that ash2r-1 homozygous seeds are not viable. Next, we examined ash2r-1 embryo development under a differential-interference-contrast microscope. Up to globular stage, ash2r-1 embryos developed similarly as wild-type ones, however, their further development was arrested (Figure S3B-S3H). When wild-type embryos reached heart stage (around 6 days after pollination; DAP), the ash2r-1 embryos were still at globular stage (Figure S3E and S3H). These findings are in line with a very recent study by Aquea et al. [38] showing that ASH2R/TRO is essential for Arabidopsis early embryogenesis.
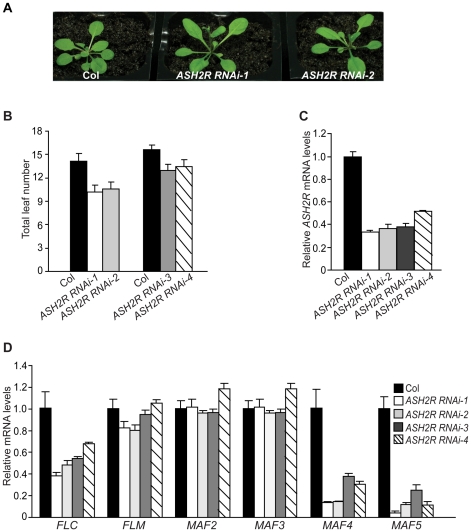
To explore the role of ASH2R in vegetative development, we exploited a dsRNAi approach to knock down ASH2R expression. A 223-bp ASH2R-specific fragment was used to create a dsRNAi cassette driven by the 35S promoter. Four independent homozygous transgenic lines with a single T-DNA locus, ASH2R RNAi-1 to -4, were created. These lines all flowered earlier than Col, but otherwise were normal in LD (Figure 3A, 3B). In addition, these lines set seeds normally. ASH2R expression was examined in ASH2R RNAi seedlings; indeed, it was knocked down and the ASH2R transcript levels in these lines were about 30–50% of those in Col (Figure 3C). Furthermore, we quantified the transcript levels of FLC and FLC homologs in these RNAi lines, and found that FLC, MAF4 and MAF5 expression was reduced, whereas FLM, MAF2 and MAF3 expression was not affected upon ASH2R knockdown (Figure 3D). Thus, ASH2R, like RBL, is required for the expression of FLC, MAF4 and MAF5 and represses Arabidopsis flowering.
Figure 3. ASH2R Knockdown Causes Early Flowering.
(A) Phenotypes of ASH2R RNAi lines grown in LD. (B) Flowering times of the ASH2R RNAi lines grown in LD. Total leaf number for each line (12 plants per line) was counted. Error bars indicate SD. (C) Relative ASH2R mRNA levels in ASH2R RNAi seedlings determined by real-time quantitative PCR. Relative expression to parental Col is presented. Bars indicate SD. (D) Relative mRNA levels of FLC and FLC homologs in ASH2R RNAi seedlings determined by real-time quantitative PCR. Bars indicate SD.
ASH2R Is Required for Normal Leaf Growth and Development
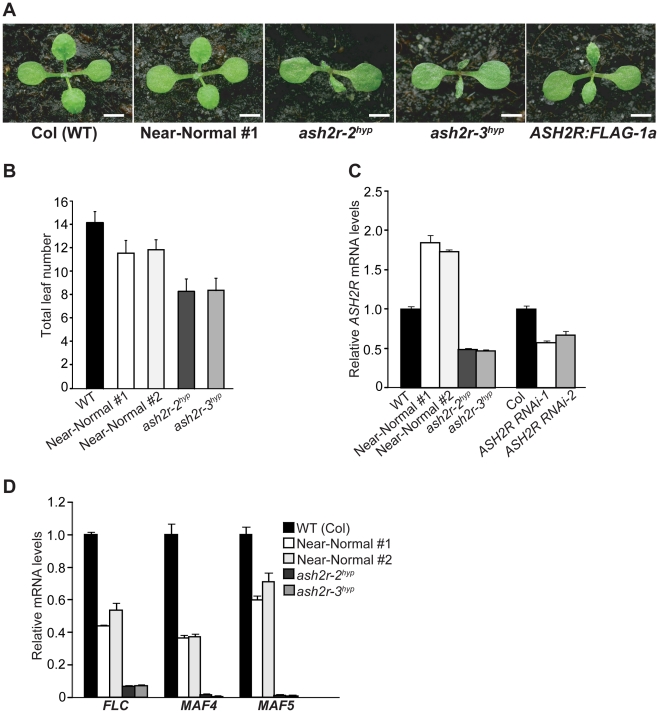
We further investigated the role for ASH2R in vegetative development using two mutant lines in which ASH2R function is severely disrupted as detailed below. In an effort to create glucocorticoid-inducible ASH2R expression lines, we unintentionally introduced two point mutations T136 to A and K187 to R, into the ASH2R coding sequence. Phylogenetic analysis suggests that T136 is conserved among Ash2 relatives in eudicots, whereas K187 appears not to be conserved (Figure S4B). The point-mutated ASH2R transgene was inserted downstream a synthetic promoter known as pOp6 (Figure S4A), which is responsive to glucocorticoid treatment [40]. In the absence of glucocorticoid induction, we identified five independent lines homozygous for the null ash2r-1 mutation and carrying the point-mutated ASH2R transgene, in which the transgene expression is leaky (see description below).
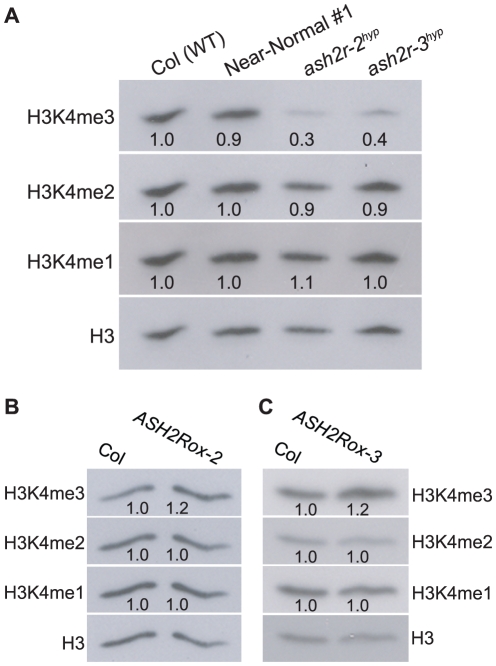
Individual T3 populations for these five lines were obtained (note that only transgenic progeny can grow into seedlings due to the embryo lethality of ash2r-1); subsequently, we examined a representative T3 population from each line phenotypically, and found that at seedling stage two lines (Near-Normal #1 and #2) developed like wildtype Col (WT), whereas the other three lines including ash2r-2 (hypomorphic; thereafter ash2r-2hyp) and ash2r-3 (hypomorphic; thereafter ash2r-3hyp) displayed leaf abnormalities (Figure 4A, and data not shown). Of note, all of the progeny within each examined population displayed nearly identical phenotypes that were stably inheritable. In terms of flowering time, the two near normal lines flowered moderately earlier than Col, whereas the other three mutant lines flowered much earlier than Col in LD (Figure 4B, and data not shown). Using real-time quantitative PCR, we further examined ASH2R transgene expression in shoot apices from 10-day (d) old seedlings, and found that the ASH2R transcript levels in both near-normal lines were higher than those in Col, whereas they were much lower in ash2r-2hyp and -3hyp compared to Col (Figure 4C). This suggests that the increased transcript levels of point-mutated ASH2R can partly ameliorate the partial functionality of point-mutated ASH2R. Interestingly, in all of these five lines the ASH2R transgene was expressed in the absence of the chemical inducers glucocorticoids; most likely this is because the strong 35S promoter located at the 3′ end of ASH2R may promote its expression (Figure S4A).
Figure 4. Characterization of the Hypomorphic ash2r Mutants.
(A) Phenotypes of the hypomorphic ash2r mutant seedlings (T3 generation) grown in LD. On the right is an ASH2R:FLAG-1a (T3) seedling with a leaf phenotype. Bars indicate 2.0 mm. (B) Flowering times of the hypomorphic ash2r mutants grown in LD. For each line, 12–15 plants were scored. Error bars indicate SD. (C) Relative mRNA levels of the ASH2R transgene in the shoot apices of 10-d-old seedlings of the indicated lines. The transcript levels were determined by real-time quantitative PCR. Relative expression to wildtype Col is presented. Bars indicate SD. (D) Relative mRNA levels of FLC, MAF4 and MAF5 in the shoot apices of 10-d-old seedlings of the indicated lines.
To confirm that the early-flowering phenotypes of ash2r-2hyp and -3hyp, like those of the ASH2R RNAi lines, were caused by reduced expression of FLC and its homologs, we quantified the transcript levels of FLC, MAF4 and MAF5 in shoot apices of these lines. Indeed, FLC expression was greatly reduced, and both MAF4 and MAF5 expression was nearly eliminated in ash2r-2hyp and -3hyp compared to Col (Figure 4D). Furthermore, we found that only about two-fifth of the siliques or silique-like structures of ash2r-2hyp and -3hyp bore seeds (typically only several viable seeds in each of these siliques) (Figure S5). As described earlier in the text, the ASH2R RNAi lines, unlike ash2r-2hyp and -3hyp, are normal except for early flowering as ASH2R expression is only partially suppressed in the RNAi lines. Together, these observations led us to conclude that both ash2r-2hyp and -3hyp are strong hypomorphic loss-of-function mutants, largely resulting from the point mutations of T136 to A and/or K187 to R and an expression reduction in the point-mutated ASH2R.
The role of ASH2R in leaf growth and development was further explored. We observed that all of the ash2r-2hyp and -3hyp mutants (T3) displayed leaf abnormalities: small, narrow, and sometimes curled leaf blades compared to wildtype Col (Figure 4A), suggesting that ASH2R is required for proper leaf growth and development. In an effort to create transgenic lines with the FLAG-tagged ASH2R transgene (ASH2R:FLAG), we identified one line named as ASH2R:FLAG-1a (T3 generation) homozygous for the null ash2r-1 mutation and carrying a single-locus ASH2R:FLAG fusion driven by the 35S promoter. The majority of ASH2R:FLAG-1a (T3) seedlings were near normal as Col, but about one-third of them displayed leaf phenotypes: typically small and narrow leaf blades as exemplified in Figure 4A. These phenotypes were due to the partial functionality of ASH2R:FLAG, not caused by the increased levels of ASH2R:FLAG because ASH2R overexpression did not give rise to any leaf phenotypes (see Figure 5A, next section). The leaf phenotype of ASH2R:FLAG-1a was similar to, but typically weaker than that of the strong hypomorphic ash2r-2hyp and -3hyp (Figure 4A). Together, these findings demonstrate that besides flowering repression and seed development, ASH2R is also required for proper leaf growth and development.
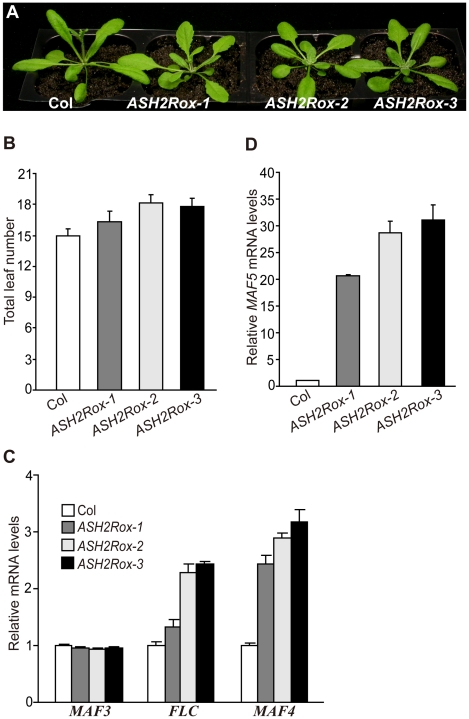
Figure 5. Overexpression of ASH2R Activates the Expression of FLC, MAF4, and MAF5 to Delay Flowering.
(A) Phenotypes of ASH2R overexpression lines grown in LD. (B) Flowering times of ASH2Rox lines grown in LD. Total leaf number at flowering for each line (11 plants per line) was counted. Error bars indicate SD. (C) Relative mRNA levels of FLC, MAF3 and MAF4 in ASH2Rox and Col seedlings determined by real-time quantitative PCR. Relative expression to parental Col is presented. Bars indicate SD. (D) Relative MAF5 mRNA levels in ASH2Rox and Col seedlings.
Gain of ASH2R Function Activates the Expression of Its Target Genes FLC, MAF4, and MAF5 to Delay Flowering
Loss of ASH2R function in vegetative development causes accelerated floral transition and leaf abnormalities. We sought to further explore the effects of gain of ASH2R function on Arabidopsis development. ASH2R coding region was overexpressed by the constitutive 35S promoter in Col. Eight independent transgenic lines were generated, among which three homozygous lines (T3 generation) with a single T-DNA locus, ASH2Rox-1 to -3 were further characterized. First, we confirmed that indeed ASH2R mRNA levels in these ASH2Rox lines were higher than those in parental Col (Figure S6). In opposite to the ASH2R RNAi lines, these three lines flowered later than Col (Figure 5A, 5B). In addition, we also observed that three out of the other five lines flowered later than Col (data not shown). Of note, except late flowering, these lines developed normally at vegetative phase (Figure 5A), and no noticeable phenotypes were observed in seed development. We further quantified transcript levels of FLC and its homologs in ASH2Rox seedlings. In opposite to ASH2R knockdown, both FLC and MAF4 expression was upregulated in the ASH2Rox lines, and surprisingly, MAF5 expression was significantly activated upon ASH2R overexpression (Figure 5C, 5D). Consistent with that ASH2R is not involved in the regulation of FLM, MAF2 and MAF3, their transcript levels in the ASH2Rox lines were similar to those in Col (Figure 5C and data not shown). Taken together, these results show that ASH2R plays an important role in activation of the expression of FLC, MAF4 and MAF5 to inhibit flowering. Interestingly, the significant activation of MAF5 expression upon ASH2R overexpression appears to have a limited effect on the floral repression. This could be attributed to that MAF5 may be a weak floral repressor.
The Spatial Expression Patterns of ASH2R, RBL, and WDR5a Overlap in Vegetative and Reproductive Development
It was of interest to examine whether the spatial expression patterns of ASH2R, RBL and WDR5a overlap in vegetative and reproductive phases. To uncover the ASH2R spatial pattern, a 1.5-kb 5′ promoter region plus part of the genomic coding region of ASH2R was translationally fused with the β-GLUCURONIDASE (GUS) gene. In addition, a 1.1-kb promoter plus 0.6-kb the genomic coding region of RBL was translationally fused with GUS. Both transgenes were introduced into Col, and using histochemical staining we found that in vegetative phase both ASH2R and RBL were strongly expressed in root tips, shoot apices and vascular tissues (Figure 6A). This pattern overlaps with those of WDR5a and FLC [21], [24].
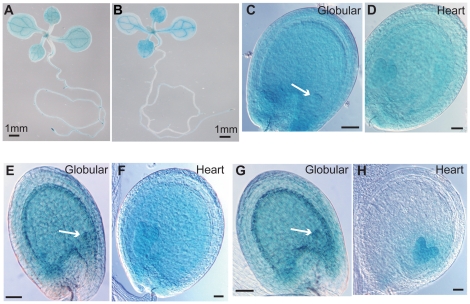
Figure 6. Spatial Expression Patterns of ASH2R, RBL, and WDR5a.
(A–B) ASH2R-GUS (A) and RBL-GUS (B) expression in 1-week-old seedlings. (C–D) ASH2R-GUS expression in a representative seed with globular embryo indicated by an arrow (C), and with heart embryo (D). (E–F) RBL-GUS expression in a representative seed with globular embryo (E), and with heart embryo (F). (G–H) WDR5a-GUS expression in a representative seed with globular embryo (G), and with heart embryo (H). (C–H) Bars = 50 µm.
Next, we analyzed the spatial patterns of ASH2R-GUS, RBL-GUS and WDR5a-GUS in seed development. ASH2R-GUS was strongly expressed in developing embryos and endosperms (Figure 6C, 6D), consistent with ASH2R playing an essential role in seed development. In addition, both WDR5a and RBL, like ASH2R, were expressed in developing embryos and endosperms at globular and heart stages (Figure 6E–6H). Together, these findings are consistent with the notion that ASH2R, RBL and WDR5a might act as part of a complex in vegetative and seed development.
ASH2R and RBL Form a Nuclear Subcomplex with WDR5a, but Not with WDR5b, in Vegetative and Reproductive Development
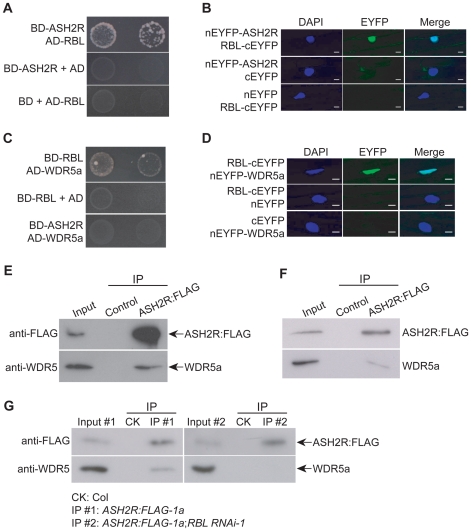
We further explored whether ASH2R, RBL and WDR5a form a stable complex. First, yeast two-hybrid assays were carried out to examine direct associations among these three proteins. In yeast, ASH2R interacted with RBL, not with WDR5a, whereas RBL interacted with WDR5a (Figure 7A and 7C). Next, to examine whether RBL associated with ASH2R and WDR5a in plant cells, we performed bimolecular fluorescence complementation (BiFC) experiments using non-fluorescent N-terminal and C-terminal EYFP (for Enhanced Yellow Fluorescent Protein) fragments (named as nEYFP and cEYFP, respectively) fused to the full-length WDR5a, ASH2R or RBL proteins. nEYFP-ASH2R and RBL-cEYFP were simultaneously expressed in onion epidermal cells, and fluorescence was observed in the nuclei of onion cells (Figure 7B), demonstrating that ASH2R associates with RBL. Similarly, we also found that RBL associated with WDR5a in the nuclei of onion cells (Figure 7D), consistent with that ASH2R, RBL and WDR5a form a complex.
Figure 7. ASH2R, RBL, and WDR5a Form a Nuclear Complex during Vegetative and Reproductive Development.
(A) Interaction of ASH2R with RBL in yeast. Full-length ASH2R and RBL proteins were respectively fused with the GAL4 DNA-binding domain (BD) and activation domain (AD). Yeast cells harboring these fusions, AD and/or BD, as indicated, were grown on the highly selective SD media lacking of Trp, Leu, His and adenine. (B) BiFC assay of nEYFP-ASH2R and RBL-cEYFP in onion epidermal cells. Plasmid pairs as indicated, were used to transiently co-transform onion epidermal cells by biolistic gene bombardment. Yellowish-green signal indicates the binding of nEYFP-ASH2R with RBL-cEYFP in the nuclei. Blue fluorescence from DAPI (4′,6-diamidino-2-phenylindole) staining indicates nuclei. Fluorescence signals were imaged using a laser scanning confocal microscope. Bar = 20 µm. (C) Interaction of WDR5a with RBL in yeast. Full-length WDR5a and RBL proteins were fused with the GAL4 BD and AD domains, respectively. (D) Physical interaction of WDR5a with RBL in onion epidermal cells as analyzed by BiFC. Bar = 20 µm. (E) Co-immunoprecipitation of ASH2R with WDR5a in 10-d-old seedlings. Total protein extracts from Col (WT; as a negative control) and ASH2R:FLAG-1a (T3) seedlings were immunoprecipitated with anti-FLAG agarose, and subsequently, the precipitates were analyzed by western blotting with anti-FLAG and anti-WDR5. The input was the ASH2R:FLAG-1a protein extract. (F) Co-immunoprecipitation of ASH2R with WDR5a in 5-DAP ASH2R:FLAG-1a siliques (T3). The input was the ASH2R:FLAG-1a protein extract from siliques. (G) Co-immunoprecipitation analysis of ASH2R with WDR5a upon RBL knockdown. Total protein extracts from the seedlings of Col (as a negative control), hemizygous ASH2R:FLAG-1a (from the crossing of ASH2R:FLAG-1a to Col), and doubly hemizygous RBL RNAi-1;ASH2R:FLAG-1a were immunoprecipitated with anti-FLAG agarose. Input #1 and Input #2 were the protein extracts of ASH2R:FLAG-1a and RBL RNAi-1;ASH2R:FLAG-1a, respectively.
Our phylogenetic analyses of the WDR5 homologs show that there are multiple WDR5 homologs in land plants (Figure 1). Besides WDR5a, Arabidopsis has another WDR5 homolog, WDR5b. We sought to determine whether WDR5b could form a complex with RBL and ASH2R. Using yeast two-hybrid approach, surprisingly, we found that WDR5b did not interact with either RBL or ASH2R in yeast (Figure S7). These results suggest that in Arabidopsis there may be only a single core subcomplex for H3K4 methylation, namely ASH2R-RBL-WDR5a.
We further performed co-immunoprecipitation experiments to determine if ASH2R, RBL and WDR5a form a complex in vivo using the ASH2R:FLAG-1a line (T3). Indeed, anti-FLAG specifically immunoprecipitated WDR5a from young seedlings and developing siliques (Figure 7E–7F). As described above, ASH2R physically associates with RBL which directly associates with WDR5a (Figure 7B and 7D), and there is no direct association between ASH2R and WDR5a (Figure 7C). Thus, ASH2R may form a complex with WDR5a via RBL. To determine whether RBL is required for the in vivo complex formation of ASH2R with WDR5a, we crossed the RBL RNAi-1 into the ASH2R:FLAG-1a line, and subsequently, the F1 seedlings were used for co-immunoprecipitation. Upon RBL knockdown, the anti-FLAG recognizing ASH2R:FLAG failed to pull down WDR5a in RBL RNAi-1;ASH2R:FLAG-1a seedlings (Figure 7G). Thus, RBL is required for the ASH2R-WDR5a complex formation. These results together led us to conclude that ASH2R, RBL and WDR5a form a complex during vegetative and reproductive development.
The physical association of RBL with ASH2R and WDR5a in the onion nuclei suggests that the ASH2R-RBL-WDR5a complex acts in the nucleus. Recently, it has been reported that a transiently expressed fusion protein of ASH2R with GFP (for Green Fluorescent Protein) in onion cells is localized into the nucleus [38]. Using an ASH2R-GFP transgenic line, we confirmed that the ASH2R fusion protein was indeed localized specifically in the nucleus (Figure S8). Together, these results are consistent with that the ASH2R subcomplex functions as a transcriptional regulator.
WDR5a Protein Interacts with Multiple Putative Arabidopsis H3K4 Methyltransferases
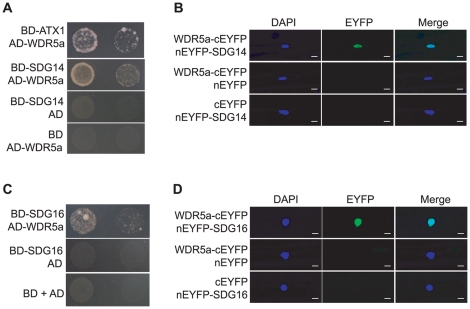
The human Ash2-RbBP5-WDR5 core subcomplex associates with MLL1, MLL2 and other H3K4 methyltransferases to form distinct catalytic complexes that deposit both di- and tri-methyl H3K4 [9], [10], [41], [42]. In Arabidopsis genome, there are may be up to ten putative H3K4 methyltransferases [17]. Recently we have found that WDR5a can associate with ATX1 [24]. Using yeast two-hybrid approach, we further explored whether WDR5a could associate with other putative H3K4 methyltransferases including SDG14 and SDG16. Full-length SDG14 and SDG16 proteins fused to the GAL4 DNA-Binding Domain (BD) were co-expressed with WDR5a fused to the GAL4 Activation Domain (AD) in yeast, and subsequently, we found that both SDG14 and SDG16 physically interacted with WDR5a (Figure 8A and 8C). Next, using BiFC we examined whether WDR5a could interact with SDG14 and SDG16 in plant cells. Upon simultaneous expression of WDR5a-cEYFP with nEYFP-SDG14 or nEYFP-SDG16 in onion epidermal cells, fluorescence was observed in the nuclei (Figure 8B and 8D). Hence, WDR5a indeed can physically interact with SDG14 and SDG16 in plant cells. In addition, we found that neither RBL nor ASH2R interacted with SDG14, SDG16 or ATX1 (Figure S9). Based on these results, we infer that the ASH2-RBL-WDR5a subcomplex associates with different H3K4 methyltransferases via WDR5a to form multiple functional COMPASS-like H3K4 methyltransferase complexes in Arabidopsis.
Figure 8. WDR5a Associates with Multiple Putative H3K4 Methyltransferases.
(A) Interaction of WDR5a with SDG14 in yeast. Full-length WDR5a and SDG14 proteins were respectively fused with the GAL4 AD and BD domains. Yeast cells harboring these fusions, AD and/or BD, as indicated, were grown on the highly selective SD media lacking of Trp, Leu, His and adenine. (B) Physical interaction of WDR5a-cEYFP with nEYFP-SDG14 in onion epidermal cells as analyzed by BiFC. Yellowish-green signal indicates the binding of WDR5a-cEYFP with nEYFP-SDG14 in the nuclei. Blue DAPI staining indicates nuclei. Bar = 20 µm. (C)Association of WDR5a with SDG16 in yeast. Full-length WDR5a and SDG16 proteins were respectively fused with the GAL4 AD and BD domains. (D) Physical interaction of WDR5a-cEYFP with nEYFP-SDG16 in onion epidermal cells analyzed by BiFC. Yellowish-green signal indicates the binding of WDR5a-cEYFP with nEYFP-SDG16 in the nuclei. Bar = 20 µm.
ASH2R Mediates Genome-Wide Trimethylation, but Not Di- or Mono-Methylation of H3K4
In yeast and mammal cells, Ash2-containing COMPASS and COMPASS-like complexes catalyze di- and tri-methylation of H3K4 [8], [9]; removal of BRE2 (the Ash2 homolog) function in yeast causes a great reduction in genome-wide H3K4me2 and H3K4me3 [8]. Recent studies reveal that the deficiency of MLL2 (the H3K4 methyltransferase of MLL2 COMPASS-like complex) in the mouse oocytes leads to a reduction in global H3K4me2 and H3K4me3 [42]. As described above, the Arabidopsis ASH2R core subcomplex can associate with multiple putative H3K4 methyltransferases to form catalytic COMPASS-like complexes. We sought to investigate the role of Arabidopsis ASH2R-COMPASS in H3K4 methylation. First, genome-wide H3K4 methylation was examined upon loss of ASH2R function. Total histones were extracted from seedlings of wildtype Col and the strong hypomorphic ash2r-2hyp and -3hyp, and levels of monomethyl H3K4 (H3K4me1), H3K4me2 and H3K4me3 were measured by western blotting with antibodies specifically recognizing these modifications. We found that H3K4me3, but surprisingly not H3K4me2, was strongly reduced upon loss of ASH2R function (Figure 9A). In addition, H3K4 monomethylation was not affected in ash2r-2hyp or -3hyp compared to the wildtype (Figure 9A). Next, we examined global H3K4 methylation in ASH2R-overexpression seedlings including ASH2Rox-2 and -3, and found that in opposite to loss of ASH2R function, H3K4me3 levels were increased in both ASH2Rox-2 and -3 lines, whereas levels of H3K4me1 and H3K4me2 remained unchanged compared to parental Col (Figure 9B, 9C). Together, these results suggest that in Arabidopsis, the ASH2R-COMPASS complexes are responsible for H3K4 trimethylation, but not for di- or mono-methylation of H3K4.
Figure 9. Analyses of Genome-Wide H3K4 Methylation upon Loss of ASH2R Function or ASH2R Overexpression.
(A) Loss of ASH2R function causes a strong reduction in global H3K4me3, but not H3K4me2 or H3K4me1. Total histones were extracted from 10-d-old seedlings of Col (WT) and the strong hypomorphic ash2r-2hyp and -3hyp, and subsequently were analyzed by western blotting. Total H3 in each sample serves as a loading control. The intensity of each protein band was quantified with the Quantity One software, and subsequently, was normalized to total H3. Values are fold changes of ash2r-2hyp or ash2r-3hyp over WT. (B–C) Analysis of levels of global H3K4me1, H3K4me2 and H3K4me3 in the ASH2R overexpression lines ASH2Rox-2 and -3. Total histones were extracted from 10-d-old seedlings of Col, ASH2Rox-2 and ASH2Rox-3, and subsequently, were analyzed by western blotting.
ASH2R Binds to the Chromatin of Its Target Genes and Mediates Tri-, but Not Di- Methylation of H3K4 in These Loci
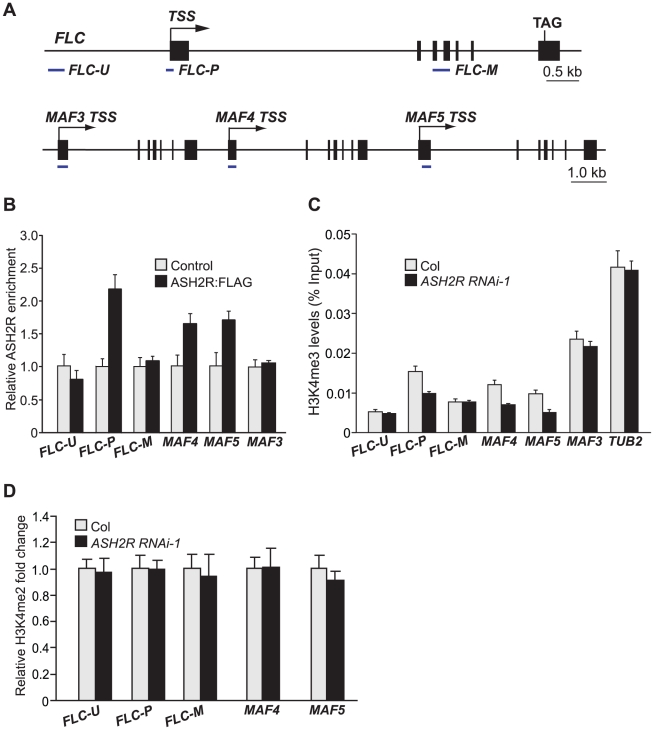
To investigate whether ASH2R directly interacted with its target the FLC locus to activate its expression, we performed chromatin immunoprecipitation (ChIP) with anti-FLAG using the wildtype-like seedlings of ASH2R:FLAG-1a (T3) in which FLC expression was only moderately suppressed (FLC transcript levels in ASH2R:FLAG-1a WT-like seedlings were about 60% of those in Col). ASH2R was enriched around the FLC TSS (FLC-P), but not in a distal region upstream of the TSS (FLC-U) or in the middle of FLC (FLC-M) (Figure 10B). Thus, we infer that the ASH2R core subcomplex directly binds to FLC chromatin to regulate its expression.
Figure 10. ASH2R Directly Mediates H3K4 Trimethylation in Its Target Genes FLC, MAF4, and MAF5.
(A) Schematic structures of FLC, MAF3, MAF4 and MAF5. The regions examined in ChIP experiments are indicated with solid lines under each region. The transcription start site (TSS) for each gene is indicated by an arrow, and filled boxes for exons. (B) ASH2R enrichment at the FLC, MAF4 and MAF5 loci. Anti-FLAG was used to immunoprecipitate target chromatin extracted from wildtype-like seedlings of ASH2R:FLAG-1a (T3) or Col (served as control). The amounts of immunoprecipitated genomic fragments were measured by real-time quantitative PCR, and subsequently normalized to TUBLIN2 (TUB2), an internal control. The fold enrichments of the ASH2R:FLAG line over control (Col) at the indicated regions are shown. Error bars indicate SD. (C) H3K4me3 levels in FLC, MAF4 and MAF5 in Col and ASH2R RNAi-1 seedlings. The amounts of immunoprecipitated genomic fragments were quantified, and subsequently normalized to the input DNA. For each amplicon, the percentage of input is shown. Error bars indicate SD. (D) Analysis of H3K4me2 in FLC, MAF4 and MAF5 in Col and ASH2R RNAi-1 seedlings.
Next, we investigated the effect of ASH2R knockdown on di- and tri-methylation of H3K4 in FLC chromatin in ASH2R RNAi seedlings by ChIP. Previously, it has been shown that H3K4 trimethylation predominantly occurs around the TSS in the actively transcribed FLC locus [13], [21]. We found that upon ASH2R knockdown, H3K4me3 levels were reduced in the FLC TSS region (Figure 10C), consistent with the ASH2R binding to this region. Furthermore, we found that levels of H3K4me2 in FLC chromatin remained unchanged upon ASH2R knockdown (Figure 10D). Hence, ASH2R is required for H3K4me3, but not for H3K4me2 in FLC chromatin, in line with that ASH2R mediates only genome-wide trimethylation, but not dimethylation of H3K4.
We further examined the association of ASH2R with the MAF4 and MAF5 loci, and found that ASH2R bound to the chromatin of 5′ transcribed regions of both MAF4 and MAF5, but not to the 5′ region of MAF3 that is located immediately upstream of MAF4 (Figure 10B), consistent with that ASH2R promotes the expression of MAF4 and MAF5, but not MAF3. Furthermore, ChIP assays show that levels of H3K4me3 in both MAF4 and MAF5 in their 5′ transcribed regions were strongly reduced, whereas H3K4me2 levels remained unchanged in both genes in ASH2R RNAi seedlings relative to Col (Figure 10C, 10D). Thus, ASH2R directly mediates H3K4 trimethylation not only in FLC, but also in MAF4 and MAF5.
Gain of ASH2R Function Causes Elevated H3K4 Trimethylation, but Not H3K4 Dimethylation, in Its Target Genes to Activate Their Expression
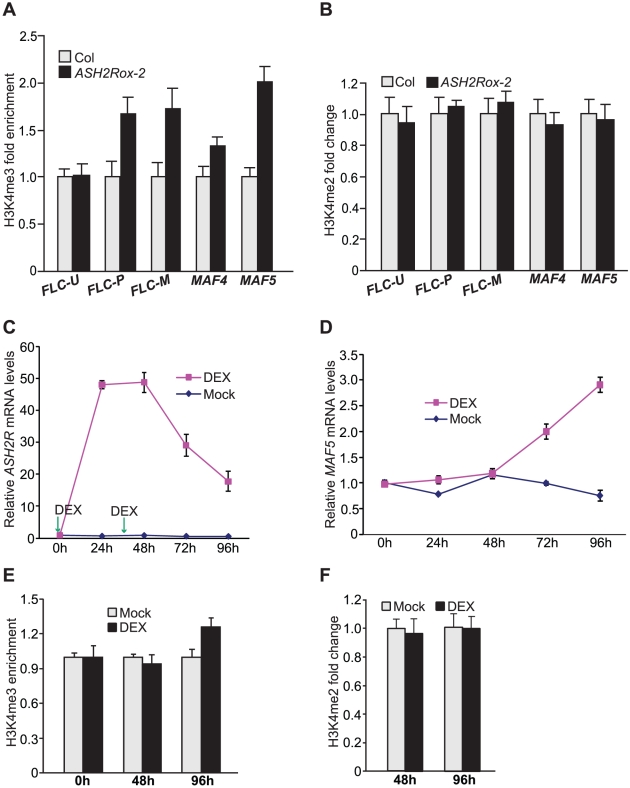
As described in Introduction, H3K4 trimethylation is a chromatin mark for actively transcribed eukaryotic genes, but whether H3K4me3 can cause transcriptional activation is unclear. We have found that ASH2R overexpression causes activation of FLC, MAF4 and MAF5 expression. Increased ASH2R expression is expected to increase the availability of ASH2R protein for assembly of ASH2R-COMPASS complexes. It was of great interest to determine whether ASH2R overexpression would cause elevated H3K4 trimethylation in FLC, MAF4 and MAF5, consequently leading to their activation. First, we performed ChIP experiments to examine tri-methyl H3K4 levels in these loci in Col and ASH2Rox-2 seedlings. At the FLC locus, levels of H3K4me3 in FLC-P, but not in FLC-U, were increased in ASH2Rox-2 relative to Col (Figure 11A). Interestingly, H3K4me3 levels in FLC-M were also increased upon elevated ASH2R expression (Figure 11A). In addition, we found that H3K4me3 levels in MAF4 were increased in ASH2Rox-2 seedlings relative to Col (Figure 11A). At the MAF5 locus, a strong increase of H3K4me3 was observed upon ASH2R overexpression (Figure 11A). Noteworthily, upon ASH2R overexpression in ASH2Rox-2 seedlings, a two-fold increase of H3K4me3 in MAF5 chromatin appears to cause an around 30-fold increase in MAF5 mRNA levels (Figure 5D and Figure 11A), indicating that at the MAF5 locus, H3K4 trimethylation state exerts a strong effect on its activation.
Figure 11. Gain of ASH2R Function Leads to Increased H3K4 Trimethylation in Its Direct Target Genes.
(A) Analysis of H3K4me3 in FLC, MAF4 and MAF5 in Col and ASH2Rox-2 seedlings. The amounts of genomic fragments immunoprecipitated with anti-H3K4me3 were quantified and subsequently normalized to TUB2. Relative fold changes of ASH2Rox-2 over Col at the indicated regions (as illustrated in Figure 10A) are shown. Error bars indicate SD. (B) Relative levels of H3K4me2 in FLC, MAF4 and MAF5 chromatin in Col and ASH2Rox-2 seedlings. (C) ASH2R expression in the pOp6-ASH2R;p35S-GR:LhG4 line (in the Col background) upon DEX treatments. Seedlings were treated with 10 µM DEX twice (at 0h and 36h as indicated by the arrows), and shoot apices with newly emerged leaves were harvested 0, 24, 48, 72 and 96 h after the initial treatment. DEX was omitted in the mock control. ASH2R transcript levels were quantified by real-time quantitative RT-PCR. Relative expression to 0h is presented; bars indicate SD. Note that the DEX treatments did not affect ASH2R expression in Col. (D) MAF5 expression in the shoot apices with newly emerged leaves of the pOp6-ASH2R;p35S-GR:LhG4 line upon DEX treatments as described in (C). Relative expression to 0h is presented; bars indicate SD. Note that the DEX treatments did not affect MAF5 expression in Col. (E) Analysis of H3K4me3 in MAF5 chromatin in the shoot apices with newly emerged leaves of the pOp6-ASH2R;p35S-GR:LhG4 line upon DEX-activated ASH2R induction. The DEX treatments were as described in (C). Relative fold changes of DEX-treated over mock samples at the indicated time points are shown. The examined MAF5 region is as illustrated in Figure 10A. Error bars indicate SD. (F) Analysis of H3K4me2 in MAF5 chromatin in the shoot apices with newly emerged leaves of the pOp6-ASH2R;p35S-GR:LhG4 line upon the DEX treatments as described in (C).
We further examined H3K4 dimethylation state in FLC, MAF4 and MAF5. Consistent with that the ASH2R core subcomplex is required for genome-wide H3K4 trimethylation, but not for H3K4 dimethylation, levels of H3K4me2 in these three loci remained unchanged in ASH2Rox-2 seedlings compared to Col (Figure 11B). Thus, consistent with ASH2R's role for H3K4 trimethylation, ASH2R overexpression leads to increased levels of H3K4me3, but not H3K4me2, in its target genes.
To further explore the role for ASH2R in the promotion of H3K4 trimethylation in its target genes, we carried out timed activations of ASH2R expression using a transgenic line (in the Col background) harboring a stringent two-component glucocorticoid-inducible transgene expression system that has been widely used for gene expression induction in plants [40], [43]. In this line (harboring pOp6-ASH2R;p35S-GR:LhG4), the expression of ASH2R transgene (free of mutations) was directed by the pOp6 promoter (Figure S10A), that is recognized by the trans-acting transcriptional activator GR:LhG4 (GR for Glucocorticoid Receptor). Upon binding of GR to glucocorticoids such as dexamethasone (DEX), GR:LhG4 is expected to specifically binds to pOp6 to turn on ASH2R expression. As noted earlier, we observed leaky expression of the point-mutated ASH2R transgene, likely caused by proximity of the 35S promoter driving GR:LhG4 expression; hence, we re-oriented p35S to maximize its distance to pOp6 (Figure S10A). Because ASH2R expression levels exert a much stronger effect on MAF5 expression than on FLC or MAF4 expression (Figure 5C, 5D), we followed MAF5 induction upon DEX-activated ASH2R expression as described next. We applied DEX to 7-d-old seedlings, and found that with a single application, MAF5 expression was slightly induced in 72 hour (h) (Figure S10C). Next, we applied DEX twice (the second application occurred 36h after the initial treatment); ASH2R expression was strongly activated upon DEX applications (still up to about 20 fold in 96h; see Figure 11C). Following ASH2R expression activation, MAF5 expression in shoot apices including newly emerged leaves, was induced to 2.0 fold over the mock in 72h and about 4.0 fold in 96h (Figure 11D). In addition, we found that MAF5 expression was not induced in the first pair of rosette leaves even though ASH2R expression was highly activated upon DEX applications (Figure S10D-S10E), suggesting that cell division activity is required for MAF5 induction. Interestingly, it took at least about 48h to induce MAF5 expression upon ASH2R expression activation (Figure 11D). Arabidopsis shoot apical meristematic cells divide once in 1–2 d [44]. It is likely that one to two rounds of cell division (or DNA replication) in the shoot apices may be required for MAF5 induction by ASH2R.
We performed ChIP experiments to examine H3K4 methylation state in MAF5 chromatin at 48h and 96h after the initial DEX application. Levels of H3K4me3, but not H3K4me2 in MAF5, increased in 96h, consistent with the induction of MAF5 expression (Figure 11E, 11F). Thus, the timed ASH2R induction causes increased H3K4me3 in MAF5 chromatin and turns on MAF5 expression. These findings together with the elevated levels of H3K4me3 in FLC, MAF4 and MAF5 upon ASH2R overexpression, strongly suggest that increased H3K4 trimethylation in these loci causes activation of their expression.
Discussion
In this study, we have found that ASH2R and RBL form a core subcomplex with WDR5a, but not WDR5b, which mediates genome-wide H3K4 trimethylation. The ASH2R core subcomplex, in association with H3K4 methyltransferases, mediates H3K4 trimethylation, but not H3K4 dimethylation in its target genes FLC, MAF4 and MAF5, resulting in activation of these gene expression, leading to late flowering. In short, our study provides strong evidence for that there are multiple ASH2R-COMPASS complexes in Arabidopsis, which deposit H3K4me3, but not H3K4me2 or H3K4me1, and that H3K4me3 accumulation can activate target gene expression in eukaryotes.
COMPASS-Like H3K4 Methyltransferase Complexes and Their Roles for Developmental Processes in Arabidopsis
The human COMPASS-like complexes contain three structural core components, one H3K4 methyltransferase and several non-conserved components [2]; the three core components together with the methyltransferase form a functional (catalytic) core complex for H3K4 methylation [10]. In this study, we have found that the Arabidopsis ASH2R, RBL and WDR5a, homologs of the three structural core components of the COMPASS-like complexes, form a core subcomplex during vegetative and reproductive development. Although there are two WDR5 homologs in Arabidopsis, our study suggests that Arabidopsis only has a single core subcomplex for H3K4 methylation, namely ASH2R-RBL-WDR5a. The biochemical and biological functions of WDR5b remain elusive. Our phylogenetic analyses show that in contrast to most animals, there are multiple WDR5 homologs in land plants. Biochemical functions of these proteins cannot be directly inferred simply based on the homology to WDR5, and have to be determined experimentally.
In Arabidopsis there are may be up to 10 putative H3K4 methyltransferases based on similarity of the catalytic SET domains to the known H3K4 methyltransferases in yeast and animals. We have found that WDR5a can associate with multiple putative H3K4 methyltransferases including ATX1, SDG14 and SDG16. Thus, we infer that multiple COMPASS-like H3K4 methyltransferase complexes exist in the higher plant Arabidopsis, for instance, the ASH2R-RBL-WDR5a-ATX1 complex. Of note, this ASH2R-ATX1 complex might contain other co-components beside the four-component catalytic core complex. These COMPASS-like complexes are expected to methylate H3K4 in various target genes and control multiple developmental processes including leaf growth and development, the floral transition and seed development.
The ASH2R-ATX1 COMPASS-like complex activates FLC expression to repress the floral transition in the rapid-cycling accession Col. Recently it has been shown that the ATX1 protein binds to FLC chromatin [21]; in this study, we have found that ASH2R, like ATX1, directly interacts with the FLC locus. These findings are consistent with that these two proteins are part of the ASH2R-ATX1 complex binding to FLC chromatin to activate FLC expression. Our genetic analyses show that loss-of-function ash2r mutants, similar to atx1, cause a great reduction in FLC expression and consequent early flowering. Furthermore, we have found that knockdown of either WDR5a or RBL expression, like ash2r and atx1, leads to reduced FLC expression and early flowering. These genetic findings are consistent with that WDR5a and RBL are part of the ASH2R-ATX1 COMPASS-like complex that activates FLC expression.
FLC is expressed at a level relatively low in the Col background (the wild-type accession used in this study and also commonly used in other Arabidopsis studies), compared to FRIGIDA (FRI)-containing accessions (note that FRI is mutated in Col) [45]. FRI, encoding a coiled-coil domain protein, upregulates the expression of FLC to a higher level that significantly delays flowering [45]. Previously, it has been shown that ATX1 is required for FRI-dependent FLC upregulation [21]. We have recently revealed that a functional FRI causes an increase in the amount of WDR5a protein bound to FLC chromatin, resulting in elevated H3K4 trimethylation in FLC [24]. WDR5a knockdown strongly suppresses FRI-dependent FLC upregulation, but not FLC upregulation/de-repression upon loss of FLD function (note that FLD, a putative H3K4 demethylase, represses FLC expression) [24]. Similarly, we have observed that ASH2R knockdown strongly suppresses FRI-dependent FLC upregulation, but not FLC de-repression in fld mutants (data not shown). Given the association of WDR5a with ATX1 and ASH2R (via RBL), these findings suggest that in the presence of a functional FRI, the ASH2R-WDR5a-ATX1 COMPASS-like complex is further enriched at FLC chromatin to upregulate FLC expression. Recently it has been reported that a histone methyltransferase known as EFS, that can methylate both H3K4 and H3K36 in vitro, is required for FRI recruitment to the FLC locus [46]. This recruitment is expected to cause the enrichment of ASH2R-WDR5a-ATX1 complex at FLC, which may function in concert with EFS leading to H3K4 and H3K36 methylation at FLC and FLC upregulation in the FRI background.
We and others have found that a null ASH2R/TRO lesion causes arrested embryogenesis at globular stage [see Figure S3 and [38]]; in addition, we found that ASH2R forms the core subcomplex with RBL and WDR5a in developing siliques and is required for genome-wide H3K4 trimethylation. These findings, together with the strong expression of ASH2R in developing seeds (Figure 6C, 6D), lead us to conclude that the ASH2R core subcomplex (presumably ASH2R-COMPASS) mediates H3K4 trimethylation in seed chromatin to control seed development. Interestingly, we noticed that knockdown of RBL or WDR5a, like ASH2R knockdown, does not disrupt seed development. There are two possible explanations. Firstly, the dsRNAi targeting RBL, ASH2R or WDR5a driven by the 35S promoter only partially suppresses target gene expression, and the remaining transcripts may be sufficient for proper embryogenesis. Secondly, WDR5a, ASH2R or RBL expression in the RNAi lines may not be affected in early embryogenesis because the 35S promoter has been shown to be inactive in the early seed development [47].
Role of ASH2R and ASH2R-COMPASS for H3K4 Trimethylation in Higher Plants
In yeast, the intact COMPASS complex is required for di- and tri-methylation of H3K4 [8]. Mammalian Ash2-COMPASS complexes are capable of catalyzing di- and tri-methylation of H3K4 [9], [41]; for instance, MLL2 deficiency in mouse oocytes causes a genome-wide reduction in H3K4me2 and H3K4me3 [42]. In mammals, both di- and tri-methylation of H3K4 are associated with actively transcribed genes [5], whereas recent genome-scale studies have revealed that only H3K4me3, but not H3K4me2, is correlated with active transcription and implicated in transcriptional activation in Arabidopsis [6]. Because H3K4me3 and H3K4me2 have distinct distribution patterns in Arabidopsis genome, they are expected to be deposited by different players. In this study, we have found that loss of ASH2R function causes a strong global reduction in H3K4me3, but not in H3K4me2 or H3K4me1 in Arabidopsis, whereas ASH2R overexpression leads to a genome-wide increase in H3K4me3, but not in H3K4me2 or H3K4me1. Furthermore, we have revealed that ASH2R knockdown causes a strong reduction in H3K4me3, but not in H3K4me2, whereas ASH2R overexpression leads to an increase in H3K4me3, but not in H3K4me2, in its direct target genes FLC, MAF4 and MAF5 to activate their expression. These findings further support the notion that the Arabidopsis ASH2R-COMPASS complexes specifically deposit H3K4me3 to promote target gene expression, providing a molecular explanation for the observed coupling of H3K4me3 (but not H3K4me2) with active gene expression in Arabidopsis. This indicates a difference in the role of Ash2-COMPASS in H3K4 methylation between Arabidopsis and yeast/mammals.
Both constitutive overexpression of ASH2R and the timed chemical induction of ASH2R expression give rise to increased H3K4me3 in its target genes such as MAF5, and target gene activation. Interestingly, overexpression of WDR5a and RBL by the constitutive 35S promoter, unlike ASH2R overexpression, did not give rise to any noticeable phenotypes (data not shown). Together, these observations indicate that the availability of ASH2R protein is a rate-limiting factor in Arabidopsis H3K4 trimethylation.
Transcriptional Activation by H3K4 Trimethylation
As noted in Introduction, H3K4 trimethylation is a chromatin mark for actively expressed genes in eukaryotic organisms that so far have been examined, but whether it can activate gene expression has been under much debate [see [16]] as the accumulation of H3K4me3 (predominantly in the 5′ transcribed regions) in actively transcribed chromatin may merely result from active transcription. In this study, we have found that the ASH2R core subcomplex, presumably ASH2R-COMPASS, binds to FLC, MAF4 and MAF5 chromatin, and that ASH2R knockdown leads to a reduction in H3K4me3 and suppresses the expression of these three genes. In addition, ASH2R overexpression causes increased H3K4me3 and activation of these gene expression. Furthermore, we have found that timed chemical induction of ASH2R expression causes increased H3K4 trimethylation in MAF5 (a direct target of ASH2R) in shoot apices and turns on its expression. These results strongly suggest that H3K4 trimethylation in FLC, MAF4 and MAF5 can activate their expression, providing concrete evidence for the notion that H3K4 trimethylation can activate eukaryotic gene expression. Ash2-COMPASS complexes are expected to be actively recruited to target gene chromatin to catalyze H3K4 trimethylation; subsequently, the evolutionarily conserved ATP-dependent chromatin remodeling factors such as NURF in human that recognizes and binds to H3K4me3 [11], may be recruited to target genes to actively mobilize/remodel nucleosomes, resulting in transcriptional activation or active gene expression.
Materials and Methods
Plant Materials and Growth Conditions
All of the Arabidopsis thaliana lines used in this study were in the Col background. The ash2r-1 allele (SAIL_851_H01) was isolated from the SAIL collection [39]. Plants were grown under cool white fluorescent lights in long days (16h light/8h dark).
Yeast Two-Hybrid Assay
The Matchmaker GAL4 Two-Hybrid System 3 (Clontech) was adapted for the yeast two-hybrid assay. The full-length coding sequences for the tested genes were cloned into the pGADT7 and/or pGBKT7 vectors (Clontech). The experiments were performed according to the manufacturer's instructions using the strain AH109 (Clontech). To test the interactions, yeast cells were spotted on the highly selective SD media lacking of leucine, tryptophan, histidine and adenine.
Bimolecular Fluorescence Complementation Analysis
The full-length coding sequences for WDR5a, ASH2R, RBL, SDG14 and SDG16 were translationally fused with either an N-terminal EYFP fragment in the pSAT1A-nEYFP-N1/pSAT1-nEYFP-C1 vectors and/or a C-terminal EYFP fragment in the pSAT1A-cEYFP-N1/pSAT1-cEYFP-C1-B vectors (www.bio.purdue.edu/people/faculty/gelvin/nsf/index.htm). Onion epidermal cells were transiently co-transformed by appropriate plasmid pairs using the Helium biolistic gene transformation system (Bio-Rad) following the manufacturer's instructions. Within 24–48 hrs after bombardment, YFP fluorescence was observed and imaged using a Zeiss LSM 5 EXCITER upright laser scanning confocal microscopy (Zeiss).
Whole-Mounted Clearing of Seeds
For the examination of seed development, seeds were cleared in 8∶1∶3 (W/V/V) chloral hydrate:glycerol: water for 1–2 hrs. The cleared seeds were examined using differential-interference-contrast optics on a Leica DM4500B microscope, and images were acquired with a Nikon DXM1200F digital camera.
Plasmid Constructions
To knock down RBL expression, two copies of a 245-bp RBL specific fragment (from +1853 to +2097 of the RBL cDNA; TSS as +1) were inserted into the pB7GWIWG2 vector [48] for hairpin RNA production. For ASH2R knockdown, two copies of a 223-bp ASH2R-specific fragment (from +1456 to +1678 of the ASH2 cDNA; TSS as +1), were inserted into the pB7GWIWG2 vector for hairpin RNA production. For overexpression of ASH2R and the rescue of ash2r-1 mutant, the full-length coding sequence for ASH2R was inserted downstream of the CaMV 35S promoter in the pMDC32 vector [49] via gateway technology (Invitrogen), resulting in the 35S-ASH2R construct. For ASH2R:FLAG construction, the full-length ASH2R coding sequence except the stop codon was first fused in frame with a FLAG tag (three copies), and subsequently the ASH2R-FLAG fragment was inserted downstream of the 35S promoter in the pMDC32 vector via gateway technology.
For ASH2R subcellular localization, the full-length ASH2R coding sequence except the stop codon was inserted between the 35S promoter and GFP in the pMDC85 vector [49]; the ASH2R coding sequence was in frame with the downstream GFP reporter gene. For ASH2R-GUS construction, an 1816-bp ASH2R genomic fragment (from −1537 to +279; A of the start codon as +1) including a 1537-bp native promoter and a 279-bp genomic coding region was inserted upstream of the GUS reporter gene in the pMDC162 vector [49]; the genomic coding sequence was in frame with GUS. To construct RBL-GUS, we inserted a 1665-bp RBL genomic fragment (from −1081 to +584; A of the start codon as +1) upstream of the GUS reporter gene in pMDC162; the genomic coding sequence of RBL was in frame with GUS.
Real-Time Quantitative RT-PCR
Total RNAs were extracted from aerial parts of 10-d-old seedlings or DEX-treated tissues as described previously [50]. The total RNAs were subsequently used as templates to synthesize cDNAs by reverse transcription. Real-time quantitative PCR was carried out on an ABI Prism 7900HT sequence detection system as previously described [24], [50]. Primers used to amplify the cDNAs of FLC, FLM, MAF2-MAF5 and TUB2 (At_5g62690) have been previously described [29], [50]. The primer pair, 5′-AGGAAGGGTACAAGGAAGGTGATG-3′ and 5′-AACGATATTTCACTGCCTGGTACAAC-3′, was used for ASH2R amplification; the primer pair, 5′-CGAAGATGAATTTGATTTGATACCTG-3′ and 5′-TGTCTCACCCATTTCTTCTGCTTGT-3′, was used to amplify RBL cDNAs. Each sample was quantified in triplicate and normalized to the endogenous control TUB2. Bars indicate standard deviations of triplicate measurements.
Histochemical β-Glucuronidase Staining
Plant tissues were fixed in 80% acetone at −20°C for 1 hr, washed by a staining buffer (5 mM EDTA pH 8.0, 0.05% Triton X-100, 2 mM potassium ferrocyanide, 2 mM potassium ferricyanide, 100 mM NaH2PO4 and 100 mM Na2HPO4), and incubated in the staining buffer with 0.5-mg 5-bromo-4-chloro-3-indolyl-β-d-glucuronic acid (X-Gluc) at 37°C for 4 to 12 hrs. The ASH2R-GUS seedlings were stained for 4 hrs, and other stainings were performed for 12 hrs. Seeds were cleared after staining for observation under a Leica DM4500B microscope.
Co-Immunoprecipitation
Immunoprecipitation experiments were performed as described previously [51] with minor modifications. Briefly, about 0.5-g 10-d-old seedlings were harvested and ground in liquid nitrogen, and subsequently, total proteins were extracted. 1.0-ml protein extracts were incubated with 60-µl slurry of anti-FLAG M2 affinity gel (Sigma, Cat#: A2220), and the immunoprecipitated proteins were washed three times and subsequently boiled in the SDS-PAGE loading buffer, followed by western blotting with anti-FLAG (Sigma, Cat#: A8592) or anti-WDR5 [Abcam, Cat#: ab75439 (Batch #: 639695); note that this antibody recognizes WDR5a; see [24]]. In the immunoprecipitation experiments with 5-DAP siliques, the samples were cross-linked with 1% formaldehyde for 30 minute before protein extraction as described previously [51].
Histone Extraction and Immunoblotting
Histone protein extraction and western analysis were performed as previously described [29]. Briefly, total histones were extracted from 10-d-old seedlings, separated in an SDS-PAGE gel, and transferred to a 0.2-µl nitrocellulose membrane (Bio-Rad). The protein blots were probed with anti-monomethyl histone H3K4 (Abcam, Cat#: ab8895), anti-dimethyl H3K4 (Millipore, Cat#: 07-030), anti-trimethyl H3K4 (Millipore, Cat#: 05-745R) and anti-histone H3 (Millipore, Cat#: 07-690).
ChIP-Quantitative PCR
ChIP experiments were performed with 10-d-old seedlings or DEX-treated shoot apices including newly emerged leaves as previously described [52]. Immunoprecipitations were carried out using Rabbit polyclonal anti-dimethyl H3K4 (Millipore, Cat#: 07-030), anti-trimethyl H3K4 (Millipore, Cat#: 05-745R) or anti-FLAG (Sigma, Cat#: A8592). The real-time quantitative PCR and the primers used to amplify FLC-U, FLC-P, FLC-M and TUB2 were described previously [29], [50]. The primer pair, 5′-CGGCGAGTTATGCAGACATCACA-3′ and 5′-GTGGCAGAGATGATGATAAGAGCG A-3′, was used to amplify MAF4; the primer pair, 5′-CAGGATCTCCGACCAGTTTATACAGAC-3′ and 5′-GAGGAGTTGTAGAGTTTGCCGGT-3′, was used to amplify MAF5. The MAF3 region was amplified by the primer pair: 5′-GTCTAGCCCAAAAGAAGAAGATAGAAACG-3′ and 5′-GGAGGCAGAGTCGTAGAGTTTTCC-3′. The relative fold changes are the averages of two independent biological repeats.
DEX Treatments
The initial treatments were carried out by applying 10 µM DEX plus 0.015% Silwet L-77 to 7-d old seedlings of Col and the pOp6-ASH2R;p35S-GR:LhG4 line grown in long days (the second treatment was carried out 36h after the initial treatment). In the mock control, DEX was omitted. The hour of initial DEX application was designated as 0h. Shoot apices including newly emerged leaves or the first pair of rosette leaves were collected at 0, 24, 48, 72 and 96h for RNA analyses. For ChIP experiments, shoot apices with newly emerged leaves were collected at 0, 48 and 96h.
Supporting Information
Alignment of Arabidopsis ASH2R (At ASH2R) with Homo sapiens Ash2 (Hs Ash2) and S. cerevisiae BRE2 (Sc BRE2). Numbers refer to amino acid residues. Identical residues among these proteins are shaded black, whereas similar residues are shaded gray.
(1.53 MB TIF)
Analysis of the Human RbBP5 Homologs in Representative Animal and Plant Species. (A) Alignment of Arabidopsis RBL (At RBL) with Homo sapiens RbBP5 (Hs RbBP5) and S. cerevisiae SWD1 (Sc SWD1). Numbers refer to amino acid residues. Identical residues among these proteins are shaded black, whereas similar residues are shaded gray. (B) Phylogenetic tree analysis of RbBP5 homologs from representative plant and animal species. The tree of full-length RbBP5 homologs was constructed by the Neighbor-Joining method using PHYLIP. Bootstrap values are shown for clades with support greater than 60% (in 1000 sampling replicates). The protein accession numbers are specified in Table S1.
(1.83 MB TIF)
ASH2R Is Essential for Embryo and Seed Development. (A) Wildtype (Col) and ash2r-1/+ heterozygote siliques. Portions of siliques were shown; arrows indicate aborted white and brown seeds. Shown on the right is an ash2r-1 (null) silique carrying the p35S-ASH2R transgene made of ASH2R CDS driven by the 35S promoter. (B–E) Wild-type seeds with globular embryo (B), with procambial embryo (C), with triangular embryo (D), and with heart embryo (E). (F, G and H) are ash2r-1 mutant seeds (with globular embryo) from the ash2r-1/+ siliques in which embryos with a wild-type phenotype were at procambial, triangular and heart stages, respectively. (B to H) Scale bars are 50 µm.
(4.09 MB TIF)
Analysis of the Point-Mutated ASH2R Transgene. (A) Schematic structure of the ASH2R transgene construct. Arrows indicate translation start sites. pOp6 is a synthetic promoter consists of six copies of the lac operator and the 35S minimal promoter. GR:LhG4 driven by the 35S promoter encodes a transcriptional activator whose active form can bind to pOp6. (B) Alignment of Arabidopsis ASH2R (At ASH2R) with Populus trichocarpa ASH2R (Pt ASH2R), Ricinus communis ASH2R (Rc ASH2R) and Vitis vinifera ASH2R (Vv ASH2R). Two missense point mutations introduced into the ASH2R transgene are indicated with stars. Numbers refer to amino acid residues. Identical residues among these proteins are shaded black, whereas similar residues are shaded gray.
(3.02 MB TIF)
Percentage of Seed-Bearing Siliques of Col, ash2r-2hyp, and ash2r-3hyp. Forty, sixty-six and eight-nine siliques or silique-like structures from main shoots were examined for Col, ash2r-2hyp and ash2r-3hyp, respectively.
(0.51 MB EPS)
Relative ASH2R mRNA Levels in the ASH2Rox Seedlings Determined by Real-Time Quantitative PCR. Relative expression to parental Col is presented. Bars indicate SD.
(0.50 MB EPS)
WDR5b Does Not Interact with RBL or ASH2R. Full-length WDR5b and the indicated proteins were fused with the GAL4-BD or AD domain. Yeast cells harboring these fusions, AD and/or BD, as indicated, were grown on SD media lacking Trp and Leu and selective SD media lacking of Trp, Leu, His and adenine. Note that AD-WDR5b was expressed in yeast as it could interact with an Arabidopsis protein in an unrelated yeast two-hybrid assay.
(8.00 MB TIF)
ASH2R Protein Subcellular Localization. The ASH2R-GFP fusion protein is localized in nuclei as indicated by the green fluorescence signals in Arabidopsis roots. Blue DAPI staining indicates nuclei.
(2.06 MB EPS)
RBL or ASH2R Does Not Interact with Putative H3K4 Methyltransferases. Yeast cells harboring the indicated fusions were grown on SD media lacking Trp and Leu, and selective SD media lacking of Trp, Leu, His and adenine.
(9.30 MB TIF)
Timed Activation of ASH2R Expression by DEX and Subsequent MAF5 Induction. (A) Schematic structure of the pOp6-ASH2R;p35S-GR:LhG4 construct. Arrows indicate translation start sites. (B) ASH2R expression in the pOp6-ASH2R;p35S-GR:LhG4 line upon DEX treatment. 7-d-old seedlings were treated with 10 µM DEX once (at 0h), and shoot apices with newly emerged leaves were harvested 0, 24, 48, 72 and 96 h after the DEX application. DEX was omitted in the mock control. ASH2R transcript levels were quantified by real-time quantitative RT-PCR. Relative expression to 0 h is presented; bars indicate SD. (C) MAF5 expression in the shoot apices with newly emerged leaves of the pOp6-ASH2R;p35S-GR:LhG4 line upon DEX treatment. The treatment was as described in (C). Relative expression to 0 h is presented; bars indicate SD. (D) ASH2R expression in the first pair of rosette leaves of the pOp6-ASH2R;p35S-GR:LhG4 line upon DEX treatment. Seedlings were treated with 10 µM DEX twice (at 0 h and 36 h), and rosette leaves were harvested 0, 24, 48, 72 and 96 h after the initial treatment. Relative expression to 0 h is presented; bars indicate SD. (E) MAF5 expression in the first pair of rosette leaves of the pOp6-ASH2R;p35S-GR:LhG4 line upon DEX treatment. The treatment was as described in (D). Relative expression to 0 h is presented; bars indicate SD.
(1.23 MB EPS)
List of WDR5 and RbBP5 Homologs from Representative Animal and Plant Species.
(0.04 MB DOC)
Acknowledgments
We thank Toshiro Ito for critically reading this manuscript, Stan Gevin for generously providing the BiFC vectors, and Flanders Interuniversity Institute for Biotechnology (Belgium) for providing the pB7GWIWG2 vector.
Footnotes
The authors have declared that no competing interests exist.
This work was supported by the Singapore Ministry of Education (AcRF Tier 2; MOE2009-T2-1-081) and by the Temasek Life Sciences Laboratory to YH. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Lan F, Nottke AC, Shi Y. Mechanisms involved in the regulation of histone lysine demethylases. Curr Opin Cell Biol. 2008;20:316–325. doi: 10.1016/j.ceb.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shilatifard A. Molecular implementation and physiological roles for histone H3 lysine 4 (H3K4) methylation. Curr Opin Cell Biol. 2008;20:341–348. doi: 10.1016/j.ceb.2008.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Santos-Rosa H, Schneider R, Bannister AJ, Sherriff J, Bernstein BE, et al. Active genes are tri-methylated at K4 of histone H3. Nature. 2002;419:407–411. doi: 10.1038/nature01080. [DOI] [PubMed] [Google Scholar]
- 4.Schubeler D, MacAlpine DM, Scalzo D, Wirbelauer C, Kooperberg C, et al. The histone modification pattern of active genes revealed through genome-wide chromatin analysis of a higher eukaryote. Genes Dev. 2004;18:1263–1271. doi: 10.1101/gad.1198204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bernstein BE, Kamal M, Lindblad-Toh K, Bekiranov S, Bailey DK, et al. Genomic maps and comparative analysis of histone modifications in human and mouse. Cell. 2005;120:169–181. doi: 10.1016/j.cell.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 6.Zhang X, Bernatavichute YV, Cokus S, Pellegrini M, Jacobsen SE. Genome-wide analysis of mono-, di- and trimethylation of histone H3 lysine 4 in Arabidopsis thaliana. Genome Biol. 2009;10:R62. doi: 10.1186/gb-2009-10-6-r62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Miller T, Krogan NJ, Dover J, Erdjument-Bromage H, Tempst P, et al. COMPASS: a complex of proteins associated with a trithorax-related SET domain protein. Proc Natl Acad Sci U S A. 2001;98:12902–12907. doi: 10.1073/pnas.231473398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schneider J, Wood A, Lee JS, Schuster R, Dueker J, et al. Molecular regulation of histone H3 trimethylation by COMPASS and the regulation of gene expression. Mol Cell. 2005;19:849–856. doi: 10.1016/j.molcel.2005.07.024. [DOI] [PubMed] [Google Scholar]
- 9.Dou Y, Milne TA, Ruthenburg AJ, Lee S, Lee JW, et al. Regulation of MLL1 H3K4 methyltransferase activity by its core components. Nat Struct Mol Biol. 2006;13:713–719. doi: 10.1038/nsmb1128. [DOI] [PubMed] [Google Scholar]
- 10.Ruthenburg AJ, Allis CD, Wysocka J. Methylation of lysine 4 on histone H3: intricacy of writing and reading a single epigenetic mark. Mol Cell. 2007;25:15–30. doi: 10.1016/j.molcel.2006.12.014. [DOI] [PubMed] [Google Scholar]
- 11.Sims RJ, Reinberg D. Histone H3 Lys 4 methylation: caught in a bind? Genes Dev. 2006;20:2779–2786. doi: 10.1101/gad.1468206. [DOI] [PubMed] [Google Scholar]
- 12.Ng HH, Robert F, Young RA, Struhl K. Targeted recruitment of Set1 histone methylase by elongating Pol II provides a localized mark and memory of recent transcriptional activity. Mol Cell. 2003;11:709–719. doi: 10.1016/s1097-2765(03)00092-3. [DOI] [PubMed] [Google Scholar]
- 13.He Y, Doyle MR, Amasino RM. PAF1-complex-mediated histone methylation of FLOWERING LOCUS C chromatin is required for the vernalization-responsive, winter-annual habit in Arabidopsis. Genes Dev. 2004;18:2774–2784. doi: 10.1101/gad.1244504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhu B, Mandal SS, Pham AD, Zheng Y, Erdjument-Bromage H, et al. The human PAF complex coordinates transcription with events downstream of RNA synthesis. Genes Dev. 2005;19:1668–1673. doi: 10.1101/gad.1292105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Oh S, Park S, van Nocker S. Genic and global functions for Paf1c in chromatin modification and gene expression in Arabidopsis. PLoS Genet. 2008;4:e1000077. doi: 10.1371/journal.pgen.1000077. doi: 10.1371/journal.pgen.1000077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Henikoff S. Nucleosome destabilization in the epigenetic regulation of gene expression. Nat Rev Genet. 2008;9:15–26. doi: 10.1038/nrg2206. [DOI] [PubMed] [Google Scholar]
- 17.Springer NM, Napoli CA, Selinger DA, Pandey R, Cone KC, et al. Comparative analysis of SET domain proteins in maize and Arabidopsis reveals multiple duplications preceding the divergence of monocots and dicots. Plant Physiol. 2003;132:907–925. doi: 10.1104/pp.102.013722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Baumbusch LO, Thorstensen T, Krauss V, Fischer A, Naumann K, et al. The Arabidopsis thaliana genome contains at least 29 active genes encoding SET domain proteins that can be assigned to four evolutionarily conserved classes. Nucleic Acids Res. 2001;29:4319–4333. doi: 10.1093/nar/29.21.4319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alvarez-Venegas R, Avramova Z. Methylation patterns of histone H3 Lys 4, Lys 9 and Lys 27 in transcriptionally active and inactive Arabidopsis genes and in atx1 mutants. Nucleic Acids Res. 2005;33:5199–5207. doi: 10.1093/nar/gki830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alvarez-Venegas R, Pien S, Sadder M, Witmer X, Grossniklaus U, et al. ATX-1, an Arabidopsis homolog of TRITHORAX, activates flower homeotic genes. Curr Biol. 2003;13:627–637. doi: 10.1016/s0960-9822(03)00243-4. [DOI] [PubMed] [Google Scholar]
- 21.Pien S, Fleury D, Mylne JS, Crevillen P, Inze D, et al. ARABIDOPSIS TRITHORAX1 dynamically regulates FLOWERING LOCUS C activation via histone H3 lysine-4 trimethylation. Plant Cell. 2008;20:580–588. doi: 10.1105/tpc.108.058172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tamada Y, Yun JY, Woo SC, Amasino RM. ARABIDOPSIS TRITHORAX-RELATED7 is required for methylation of lysine 4 of histone H3 and for transcriptional activation of FLOWERING LOCUS C. Plant Cell. 2009;21:3257–3269. doi: 10.1105/tpc.109.070060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Berr A, Xu L, Gao J, Cognat V, Steinmetz A, et al. SET DOMAIN GROUP25 encodes a histone methyltransferase and is involved in FLOWERING LOCUS C activation and repression of flowering. Plant Physiol. 2009;151:1476–1485. doi: 10.1104/pp.109.143941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jiang D, Gu X, He Y. Establishment of the winter-annual growth habit via FRIGIDA-mediated histone methylation at FLOWERING LOCUS C in Arabidopsis. Plant Cell. 2009;21:1733–1746. doi: 10.1105/tpc.109.067967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Michaels SD. Flowering time regulation produces much fruit. Curr Opin Plant Biol. 2009;12:75–80. doi: 10.1016/j.pbi.2008.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Baurle I, Dean C. The timing of developmental transitions in plants. Cell. 2006;125:655–664. doi: 10.1016/j.cell.2006.05.005. [DOI] [PubMed] [Google Scholar]
- 27.Scortecci KC, Michaels SD, Amasino RM. Identification of a MADS-box gene, FLOWERING LOCUS M, that represses flowering. Plant J. 2001;26:229–236. doi: 10.1046/j.1365-313x.2001.01024.x. [DOI] [PubMed] [Google Scholar]
- 28.Ratcliffe OJ, Kumimoto RW, Wong BJ, Riechmann JL. Analysis of the Arabidopsis MADS AFFECTING FLOWERING gene family: MAF2 prevents vernalization by short periods of cold. Plant Cell. 2003;15:1159–1169. doi: 10.1105/tpc.009506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gu X, Jiang D, Wang Y, Bachmair A, He Y. Repression of the floral transition via histone H2B monoubiquitination. Plant J. 2009;57:522–533. doi: 10.1111/j.1365-313X.2008.03709.x. [DOI] [PubMed] [Google Scholar]
- 30.Sung S, Amasino RM. REMEMBERING WINTER: Toward a molecular understanding of vernalization. Annu Rev Plant Biol. 2005;56:491–508. doi: 10.1146/annurev.arplant.56.032604.144307. [DOI] [PubMed] [Google Scholar]
- 31.He Y. Control of the transition to flowering by chromatin modifications. Molecular Plant. 2009;2:554–564. doi: 10.1093/mp/ssp005. [DOI] [PubMed] [Google Scholar]
- 32.Schubert D, Primavesi L, Bishopp A, Roberts G, Doonan J, et al. Silencing by plant Polycomb-group genes requires dispersed trimethylation of histone H3 at lysine 27. Embo J. 2006;25:4638–4649. doi: 10.1038/sj.emboj.7601311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu K, Zhang L, Zhou C, Yu CW, Chaikam V. HDA6 is required for jasmonate response, senescence and flowering in Arabidopsis. J Exp Bot. 2008;59:225–234. doi: 10.1093/jxb/erm300. [DOI] [PubMed] [Google Scholar]
- 34.Schmitz RJ, Sung S, Amasino RM. Histone arginine methylation is required for vernalization-induced epigenetic silencing of FLC in winter-annual Arabidopsis thaliana. Proc Natl Acad Sci U S A. 2008;105:411–416. doi: 10.1073/pnas.0710423104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang X, Zhang Y, Ma Q, Zhang Z, Xue Y, et al. SKB1-mediated symmetric dimethylation of histone H4R3 controls flowering time in Arabidopsis. Embo J. 2007;26:1934–1941. doi: 10.1038/sj.emboj.7601647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu C, Lu F, Cui X, Cao X. Histone methylation in higher plants. Annu Rev Plant Biol. 2010;61:395–420. doi: 10.1146/annurev.arplant.043008.091939. [DOI] [PubMed] [Google Scholar]
- 37.Yu X, Michaels SD. The Arabidopsis Paf1c complex component CDC73 participates in the modification of FLC chromatin. Plant Physiol: 2010;153:1074–1084. doi: 10.1104/pp.110.158386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Aquea F, Johnston AJ, Canon P, Grossniklaus U, Arce-Johnson P. TRAUCO, a Trithorax-group gene homologue, is required for early embryogenesis in Arabidopsis thaliana. J Exp Bot. 2010;61:1215–1224. doi: 10.1093/jxb/erp396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sessions A, Burke E, Presting G, Aux G, McElver J, et al. A high-throughput Arabidopsis reverse genetics system. Plant Cell. 2002;14:2985–2994. doi: 10.1105/tpc.004630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Craft J, Samalova M, Baroux C, Townley H, Martinez A, et al. New pOp/LhG4 vectors for stringent glucocorticoid-dependent transgene expression in Arabidopsis. Plant J. 2005;41:899–918. doi: 10.1111/j.1365-313X.2005.02342.x. [DOI] [PubMed] [Google Scholar]
- 41.Patel A, Vought VE, Dharmarajan V, Cosgrove MS. A conserved arginine-containing motif crucial for the assembly and enzymatic activity of the Mixed Lineage Leukemia Protein-1 core complex. J Biol Chem. 2008;283:32162–32175. doi: 10.1074/jbc.M806317200. [DOI] [PubMed] [Google Scholar]
- 42.Andreu-Vieyra CV, Chen R, Agno JE, Glaser S, Anastassiadis K, et al. MLL2 is required in oocytes for bulk Histone 3 lysine 4 trimethylation and transcriptional silencing. PLoS Biol. 2010;8:e1000453. doi: 10.1371/journal.pbio.1000453. doi: 10.1371/journal.pbio.1000453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sun B, Xu Y, Ng KH, Ito T. A timing mechanism for stem cell maintenance and differentiation in the Arabidopsis floral meristem. Genes Dev. 2009;23:1791–1804. doi: 10.1101/gad.1800409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Reddy GV, Meyerowitz EM. Stem-cell homeostasis and growth dynamics can be uncoupled in the Arabidopsis shoot apex. Science. 2005;310:663–667. doi: 10.1126/science.1116261. [DOI] [PubMed] [Google Scholar]
- 45.Johanson U, West J, Lister C, Michaels S, Amasino R, et al. Molecular analysis of FRIGIDA, a major determinant of natural variation in Arabidopsis flowering time. Science. 2000;290:344–347. doi: 10.1126/science.290.5490.344. [DOI] [PubMed] [Google Scholar]
- 46.Ko JH, Mitina I, Tamada Y, Hyun Y, Choi Y, et al. Growth habit determination by the balance of histone methylation activities in Arabidopsis. Embo J. 29:3208–3215. doi: 10.1038/emboj.2010.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kwong RW, Bui AQ, Lee H, Kwong LW, Fischer RL, et al. LEAFY COTYLEDON1-LIKE defines a class of regulators essential for embryo development. Plant Cell. 2003;15:5–18. doi: 10.1105/tpc.006973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Karimi M, De Meyer B, Hilson P. Modular cloning in plant cells. Trends Plant Sci. 2005;10:103–105. doi: 10.1016/j.tplants.2005.01.008. [DOI] [PubMed] [Google Scholar]
- 49.Curtis MD, Grossniklaus U. A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 2003;133:462–469. doi: 10.1104/pp.103.027979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jiang D, Yang W, He Y, Amasino RM. Arabidopsis relatives of the human Lysine-Specific Demethylase1 repress the expression of FWA and FLOWERING LOCUS C and thus promote the floral transition. Plant Cell. 2007;19:2975–2987. doi: 10.1105/tpc.107.052373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wood CC, Robertson M, Tanner G, Peacock WJ, Dennis ES, et al. The Arabidopsis thaliana vernalization response requires a Polycomb-like protein complex that also includes VERNALIZATION INSENSITIVE 3. Proc Natl Acad Sci U S A. 2006;103:14631–14636. doi: 10.1073/pnas.0606385103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Johnson L, Cao X, Jacobsen S. Interplay between two epigenetic marks: DNA methylation and histone H3 lysine 9 methylation. Curr Biol. 2002;12:1360–1367. doi: 10.1016/s0960-9822(02)00976-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Alignment of Arabidopsis ASH2R (At ASH2R) with Homo sapiens Ash2 (Hs Ash2) and S. cerevisiae BRE2 (Sc BRE2). Numbers refer to amino acid residues. Identical residues among these proteins are shaded black, whereas similar residues are shaded gray.
(1.53 MB TIF)
Analysis of the Human RbBP5 Homologs in Representative Animal and Plant Species. (A) Alignment of Arabidopsis RBL (At RBL) with Homo sapiens RbBP5 (Hs RbBP5) and S. cerevisiae SWD1 (Sc SWD1). Numbers refer to amino acid residues. Identical residues among these proteins are shaded black, whereas similar residues are shaded gray. (B) Phylogenetic tree analysis of RbBP5 homologs from representative plant and animal species. The tree of full-length RbBP5 homologs was constructed by the Neighbor-Joining method using PHYLIP. Bootstrap values are shown for clades with support greater than 60% (in 1000 sampling replicates). The protein accession numbers are specified in Table S1.
(1.83 MB TIF)
ASH2R Is Essential for Embryo and Seed Development. (A) Wildtype (Col) and ash2r-1/+ heterozygote siliques. Portions of siliques were shown; arrows indicate aborted white and brown seeds. Shown on the right is an ash2r-1 (null) silique carrying the p35S-ASH2R transgene made of ASH2R CDS driven by the 35S promoter. (B–E) Wild-type seeds with globular embryo (B), with procambial embryo (C), with triangular embryo (D), and with heart embryo (E). (F, G and H) are ash2r-1 mutant seeds (with globular embryo) from the ash2r-1/+ siliques in which embryos with a wild-type phenotype were at procambial, triangular and heart stages, respectively. (B to H) Scale bars are 50 µm.
(4.09 MB TIF)
Analysis of the Point-Mutated ASH2R Transgene. (A) Schematic structure of the ASH2R transgene construct. Arrows indicate translation start sites. pOp6 is a synthetic promoter consists of six copies of the lac operator and the 35S minimal promoter. GR:LhG4 driven by the 35S promoter encodes a transcriptional activator whose active form can bind to pOp6. (B) Alignment of Arabidopsis ASH2R (At ASH2R) with Populus trichocarpa ASH2R (Pt ASH2R), Ricinus communis ASH2R (Rc ASH2R) and Vitis vinifera ASH2R (Vv ASH2R). Two missense point mutations introduced into the ASH2R transgene are indicated with stars. Numbers refer to amino acid residues. Identical residues among these proteins are shaded black, whereas similar residues are shaded gray.
(3.02 MB TIF)
Percentage of Seed-Bearing Siliques of Col, ash2r-2hyp, and ash2r-3hyp. Forty, sixty-six and eight-nine siliques or silique-like structures from main shoots were examined for Col, ash2r-2hyp and ash2r-3hyp, respectively.
(0.51 MB EPS)
Relative ASH2R mRNA Levels in the ASH2Rox Seedlings Determined by Real-Time Quantitative PCR. Relative expression to parental Col is presented. Bars indicate SD.
(0.50 MB EPS)
WDR5b Does Not Interact with RBL or ASH2R. Full-length WDR5b and the indicated proteins were fused with the GAL4-BD or AD domain. Yeast cells harboring these fusions, AD and/or BD, as indicated, were grown on SD media lacking Trp and Leu and selective SD media lacking of Trp, Leu, His and adenine. Note that AD-WDR5b was expressed in yeast as it could interact with an Arabidopsis protein in an unrelated yeast two-hybrid assay.
(8.00 MB TIF)
ASH2R Protein Subcellular Localization. The ASH2R-GFP fusion protein is localized in nuclei as indicated by the green fluorescence signals in Arabidopsis roots. Blue DAPI staining indicates nuclei.
(2.06 MB EPS)
RBL or ASH2R Does Not Interact with Putative H3K4 Methyltransferases. Yeast cells harboring the indicated fusions were grown on SD media lacking Trp and Leu, and selective SD media lacking of Trp, Leu, His and adenine.
(9.30 MB TIF)
Timed Activation of ASH2R Expression by DEX and Subsequent MAF5 Induction. (A) Schematic structure of the pOp6-ASH2R;p35S-GR:LhG4 construct. Arrows indicate translation start sites. (B) ASH2R expression in the pOp6-ASH2R;p35S-GR:LhG4 line upon DEX treatment. 7-d-old seedlings were treated with 10 µM DEX once (at 0h), and shoot apices with newly emerged leaves were harvested 0, 24, 48, 72 and 96 h after the DEX application. DEX was omitted in the mock control. ASH2R transcript levels were quantified by real-time quantitative RT-PCR. Relative expression to 0 h is presented; bars indicate SD. (C) MAF5 expression in the shoot apices with newly emerged leaves of the pOp6-ASH2R;p35S-GR:LhG4 line upon DEX treatment. The treatment was as described in (C). Relative expression to 0 h is presented; bars indicate SD. (D) ASH2R expression in the first pair of rosette leaves of the pOp6-ASH2R;p35S-GR:LhG4 line upon DEX treatment. Seedlings were treated with 10 µM DEX twice (at 0 h and 36 h), and rosette leaves were harvested 0, 24, 48, 72 and 96 h after the initial treatment. Relative expression to 0 h is presented; bars indicate SD. (E) MAF5 expression in the first pair of rosette leaves of the pOp6-ASH2R;p35S-GR:LhG4 line upon DEX treatment. The treatment was as described in (D). Relative expression to 0 h is presented; bars indicate SD.
(1.23 MB EPS)
List of WDR5 and RbBP5 Homologs from Representative Animal and Plant Species.
(0.04 MB DOC)