Abstract
Microbes in nature frequently function as members of complex multitaxon communities, but the structural organization of these communities at the micrometer level is poorly understood because of limitations in labeling and imaging technology. We report here a combinatorial labeling strategy coupled with spectral image acquisition and analysis that greatly expands the number of fluorescent signatures distinguishable in a single image. As an imaging proof of principle, we first demonstrated visualization of Escherichia coli labeled by fluorescence in situ hybridization (FISH) with 28 different binary combinations of eight fluorophores. As a biological proof of principle, we then applied this Combinatorial Labeling and Spectral Imaging FISH (CLASI-FISH) strategy using genus- and family-specific probes to visualize simultaneously and differentiate 15 different phylotypes in an artificial mixture of laboratory-grown microbes. We then illustrated the utility of our method for the structural analysis of a natural microbial community, namely, human dental plaque, a microbial biofilm. We demonstrate that 15 taxa in the plaque community can be imaged simultaneously and analyzed and that this community was dominated by early colonizers, including species of Streptococcus, Prevotella, Actinomyces, and Veillonella. Proximity analysis was used to determine the frequency of inter- and intrataxon cell-to-cell associations which revealed statistically significant intertaxon pairings. Cells of the genera Prevotella and Actinomyces showed the most interspecies associations, suggesting a central role for these genera in establishing and maintaining biofilm complexity. The results provide an initial systems-level structural analysis of biofilm organization.
Keywords: cell biology, fluorescence, microbial diversity, oral biofilm
The first direct observation of bacteria is credited to Antony van Leeuwenhoek, who in the 17th century produced drawings with remarkable realism of several microbes that he had viewed in a sample of his own dental plaque (1). In hindsight, van Leeuwenhoek's discovery clearly revealed microbes living not as isolated cells but rather as members of a complex community of morphologically distinct organisms. Modern methods of microbial imaging no longer rely solely on phenotypic characteristics, which provide only limited usefulness for taxonomic classification. An important advance was the introduction of in situ hybridization with oligonucleotide probes targeted to ribosomal RNA for the taxonomic identification and visualization of microbes in situ (2, 3). However, to date, limitations in current technologies have prevented a full systems-level study of microbial community organization.
In principle, fluorescence in situ hybridization (FISH) probes could be designed with rRNA sequence specificity for nearly any microbial phylotype or taxon. In practice, the use of bandpass filters in fluorescence image acquisition and the excitation crosstalk and emission bleed-through of available organic fluorochromes limit the number of fluorophores that can be differentiated simultaneously (4). This limitation restricts routine FISH technology to the identification of only one or a few types of microbes in any single experiment. Thus, although it is possible in many cases to identify with high certainty a few microbial taxa within the context of an entire community, the simultaneous microscopic identification of more than a handful of taxa in a single sample has yet to be achieved.
Microbial communities can be considered as networks of interconnected organisms, often growing in biofilms attached to a substratum and surrounded by a pericellular matrix (5). The complexity of microbial biofilms varies from simple single-species populations to extraordinarily species-rich communities as in the case of soil particle biofilms or those associated with the epithelial surfaces of the human body (6, 7). Environmental metagenomics (8) and microbial coculture experiments (9) have suggested a number of specific molecular interactions between different phylotypes of microbes that make up communities. These interactions include communication via soluble factors (10), syntrophic metabolism in which the enzymatic breakdown of complex biopolymers is carried out between two or more cooperating organisms (11), and specific structural associations mediated by cell-surface receptors and ligands (12). Molecular methods provide an exhaustive list of potential functions that are present in a community but provide little information on how these metabolic functions are segmented in space and how different organisms might cooperate. Analogous to protein complexes in cellular systems biology, the observation of physical cell–cell interactions between organisms has been suggested as strong indication of functional interaction in the community (13). Therefore, for a comprehensive understanding of microbial ecosystem function, a systems-level analysis of the cellular architecture of microbial communities is crucial.
Human dental plaque is a well-studied and highly diverse microbial community that serves as a model for biofilm biology in general (14). More than 600 species of microbes have been identified as resident in the human oral cavity by traditional molecular methods (15), and recent studies suggest far greater diversity (16). A small subset of oral microbes such as species of the genera Streptococcus and Actinomyces are able to colonize the saliva-coated surface of teeth after tooth brushing (17). Once the tooth surface is initially colonized, an ecological succession is thought to occur as other species including those of the genera Veillonella, Fusobacterium, and others bind to these founder organisms. Coadhesion and coaggregation of genetically distinct oral microbes, in addition to growth of already adherent cells, further contributes to biofilm maturation and ultimately may lead to a highly complex and species-rich microbial community (18). Biochemical mediators of species-specific cell-to-cell adhesion, the cell-surface adhesins and receptors, have been elucidated by means of in vitro biochemical and genetic studies (e.g., refs. 19–21), leading to a general hypothesis for the structure of dental plaque (22). In addition, microscopic imaging studies using FISH or immunofluorescence to identify specific microbial taxa have confirmed some of the interspecies spatial relationships in dental plaque biofilms, but only for a few species or taxonomic groups in any single assay (e.g., 23–27). Thus, the system-level taxonomic spatial structure of dental plaque remains to be determined directly.
Recent advances in fluorescence spectral image acquisition and the application of linear unmixing algorithms to spectrally recorded image data allow the unambiguous identification of fluorophores with overlapping spectra, even within the same pixel in a digitally recorded image (28, 29). Here we report the development of a biological labeling and fluorescence image analysis strategy, which we call “Combinatorial Labeling and Spectral Imaging” (CLASI) (Fig. 1), and its application to the simultaneous identification of tens to potentially hundreds of microbial taxa in a single microscopic image. Our strategy entails the labeling by FISH of a given type of microorganism with two or more fluorophores selected from a library of fluorophores chosen to maximize spectral discrimination. With this combinatorial labeling approach we greatly expand the number of different kinds of microbes distinguishable in a single field of view.
Fig. 1.
Overview of the CLASI-FISH method. (A) Schematic diagram of the combinatorial labeling approach. To a sample of fixed microbes, fluorophore-conjugated oligonucleotide probes are added so that each kind of microbe is labeled with exactly two different fluorophores. (B) Image processing and analysis flowchart. Linear unmixing is applied to raw spectrally acquired images, cells are segmented from background, and then each particle is analyzed for its binary fluorophore composition. The binary label restriction and averaging the unmixing result over all pixels within a segmented object result in less noisy data and a more robust analysis.
Results
Imaging Proof of Principle.
We first sought to establish that microbe-sized objects labeled with binary combinations of fluorophores with highly overlapping emission spectra could be distinguished in spectrally acquired microscope images. Escherichia coli were used as test objects for CLASI-FISH. Eight versions of a FISH probe with the same oligonucleotide sequence, the Eub338 probe (30), targeting a conserved region of the bacterial 16S ribosomal RNA and confirmed to be present in E. coli, were custom synthesized (Materials and Methods). Each version had a different fluorophore covalently conjugated to its 5′ end. These eight probes were used to label separate aliquots of paraformaldehyde-fixed E. coli cells in FISH reactions so that 28 populations of microbes were created, each population labeled with a unique binary combination of probes, hereafter referred to as a ”label type.” After FISH, the separately labeled E. coli populations were combined to create a mixture of the 28 different label types.
Specimens were imaged with a laser scanning confocal microscope equipped with a 32-anode spectral detector (Materials and Methods). For quantitative analysis, to maximize both fluorescence signal within pixels and number of cells in a single field of view, specimens were imaged with a low-magnification, high numerical aperture (NA) objective lens (20×/0.8 NA) (Fig. 2 A and B). For ease of visualization, we also present a high-magnification image acquired with a 100×/1.4 NA objective (Fig. 2 C and D). Separate spectral image acquisitions were made of each field of view of labeled E. coli, with each image acquired using a different laser line available on the microscope system for fluorophore excitation.
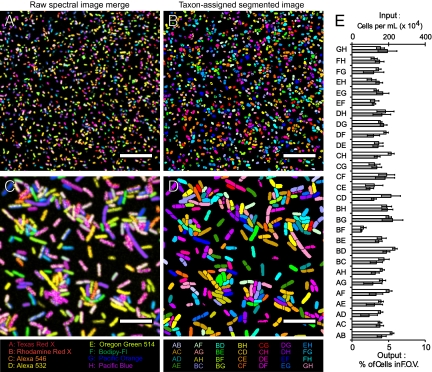
Fig. 2.
CLASI-FISH proof of principle with E. coli. (A–D) Details from images of a mixture of 28 different combinatorially labeled E. coli. (A and C) Raw spectral images in which the color at each pixel corresponds to a merge of all eight fluorophore channels after unmixing. (B and D) Segmented images of the fields of view in A and C, respectively. The color of each segmented cell corresponds to 1 of 28 different label types; each label type is assigned an arbitrary color. (Scale bars: 50 μm in A and B, 15 μm in C and D.) (E) Quantification of input into the mixture of E. coli with output from the CLASI analysis. White bars represent input into the mixture, and gray bars represent output. Error bars in the input represent SD in the counts of cells in six different 40-μm2 squares of a hemocytometer, and error bars in the output represent SD in the calculated percent of cells of each label type from four fields of view of CLASI-FISH–labeled E. coli from two separate experiments.
Linear unmixing was applied to the spectral images to generate multichannel images in which each channel consisted of measured intensities assigned to one of the eight fluorophores used in the experiment. To facilitate display of the raw data, we show the raw spectral image merge (Fig. 2 A and C) in which the pixel-based intensity data from each of the fluorophore channels were pseudocolored in one of eight colors and combined. The data set was converted to a particle basis by segmentation from background in each channel using an intensity threshold and a size discriminator to exclude spurious fluorescence and aggregates of cells from the analysis. For each particle, the mean intensities of all eight fluorophores were tabulated to determine the label type, defined simply as the combination of the two fluorophores with the highest mean intensities in each particle. The taxon-assigned segmented image (Fig. 2 B and D) discards the intensity information and assigns 1 of 28 different colors to each label type. For each field of view, the number of each of the 28 different label types was determined (Fig. 2E).
To assess the accuracy of the assay, the same acquisition and image-processing protocols used to characterize the microbial mixture were applied to preparations of the 28 pure populations of labeled E. coli (Fig. S1). In each case, more than 98% of cells were correctly identified as to their label type (Fig. S2). To confirm the cell density in each of the 28 E. coli tubes, a hemocytometer was used with semiautomatic cell counting. This input is plotted alongside the output as measured in the mixture with the spectral imaging assay (Fig. 2E). We conclude that input into the mixture correlates with output. Together, these results confirm the accuracy of CLASI-FISH and establish as proof of principle that a mixture of microbes labeled with binary combinations of eight different fluorophores can be distinguished correctly in a single spectrally acquired image after linear unmixing.
Biological Proof of Principle.
In a natural community we expect to find tens to hundreds, if not more, of different species simultaneously present. In establishing biological proof of principle, we sought to demonstrate that multiple different species of microbes, mixed before labeling, could be labeled and distinguished with taxon-specific probes in a single FISH reaction. We identified previously published oligonucleotide probes or de novo designed genus- or family-level probes that targeted the 16S small subunit (SSU) rRNA of 15 of the most abundant taxa of microbes resident in the human oral cavity and tailored the probes to be specific for their targets within the context of all microbes included in the Human Oral Microbiome Database (31). This curated database consists of the nearly full-length SSU rRNA sequences of 619 microbial taxa previously identified as resident in the human oral cavity and is assumed to include the majority of human oral microbial phylotypes identified by traditional sequencing methods. Specific details of probe design are given in Materials and Methods and Table S1. Two versions of each probe were synthesized, each conjugated to one of six fluorophores to give 15 unique binary fluorophore combinations.
Laboratory stocks of representative species of these 15 taxa were procured from the American Type Culture Collection (ATCC), were grown as monocultures in media and under atmosphere ideal for each species’ growth, and then were fixed in 2% paraformaldehyde. We prepared a mixture of the 15 stock oral microbial cultures. To this cell mixture, all 30 of our probes (15 different taxon-specific oligonucleotide probes, each in two fluorophore versions) were added in a CLASI-FISH experiment. Spectral images were acquired sequentially with separate wavelength excitation; linear unmixing was applied to the spectral images; then the images were processed to segment cells from background and from each other. Particles were analyzed for their mean intensity over all the pixels within each segmented object in each fluorophore channel, giving each particle six fluorophore intensity values. As previously described for E. coli, the two highest-intensity fluorophores were identified in each particle, revealing the binary fluorophore label of each particle (Fig. 3 A and B). For all oral microbe experiments, an additional level of analysis was applied before taxon identity was declared. In addition to the two highest-intensity fluorophores in each particle, the third highest fluorophore was inspected. If the intensity of this fluorophore was ≥60% of the second-highest fluorophore, the particle was declared ambiguous in its binary label composition and was assumed to result from inadequate image segmentation so that two or more microbes of different taxa were identified incorrectly as a single object. Ambiguous cells were declared unknown in their taxon identity, were removed from further analysis, and are colored gray in Fig. 3 A and B. In this experiment, we successfully distinguished 15 different taxon-specific binary spectral markers, thereby identifying all 15 different species simultaneously in one FISH experiment. The fraction of each taxon input into the artificial mixture correlated (r = 0.91) with the output result from our CLASI-FISH assay (Fig. 3C).
Fig. 3.
CLASI-FISH with human oral microbes. Color in raw spectral images represents the merge of six different fluorophore channels after unmixing. Color in the segmented image represents 1 of 15 taxa; each taxon is assigned its own arbitrary color. (A) Field of view of a mixture of 15 different species of laboratory-grown microbes that are resident members of the human oral microbiome. (Scale bar: 20 μm.) (B) Detail images taken from A. (Scale bar: 10 μm.) (C) Quantification of input into the mixture of oral microbes with output from the CLASI analysis. Error bars in input represent SD in the count of cells from five different fields of view of single-species preparations; error bars in the output measurements represent SD in the percent of each species counted in 15 different fields of view of CLASI-FISH–labeled microbes.
We conclude that input into the mixture of laboratory-grown microbes correlated with output after image analysis, thereby establishing biological proof of principle of CLASI-FISH.
Application of CLASI-FISH to a Natural Microbial Community.
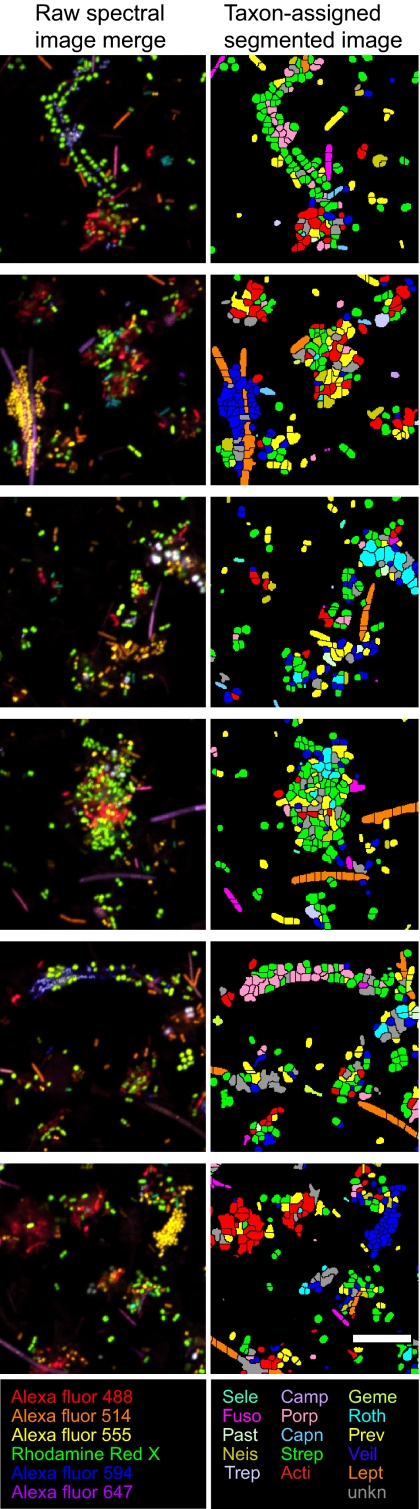
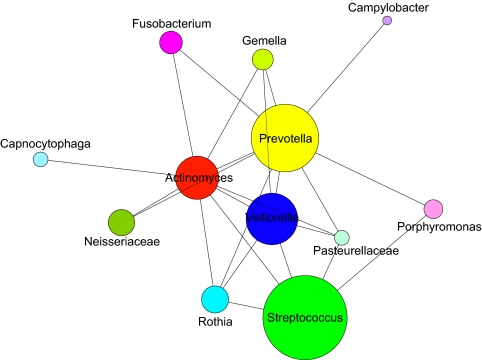
We next tested CLASI-FISH as a tool to characterize and quantify the spatial relationships of 15 different taxa of microbes in semidispersed human dental plaque. Volunteers provided samples of dental plaque removed using dental floss. These samples are assumed to consist mostly of supragingival plaque from between teeth as well as microbes from saliva, subgingival plaque, and plaque from soft tissue surfaces of the mouth. The plaque biofilm was dispersed partially in PBS with vortex mixing to break up large aggregates of cells while preserving smaller aggregates composed of a few to less than a hundred cells. We applied the same mixture of 15 human oral taxon-specific probes to the fixed plaque samples; then the FISH-labeled plaque was settled onto a glass slide and mounted for imaging (Fig. 4). For illustrative purposes, we present data from a single donor taken at a single time point. All 15 taxa were identified in the plaque sample, and the relative abundance of each taxon was calculated (Fig. S3). Relative abundance of each genus or family agreed with previous studies using molecular techniques to quantify the microbial taxa composition of plaque and saliva (12). We next performed a quantitative analysis of the intertaxon spatial relationships of cells within multicell aggregates. We developed an image analysis program that simply calculated the number of cells of each taxon that touched other cells of either the same or a different taxon. To assess the statistical significance of the observed associations, we constructed model images of randomly placed cells (Fig. S4) and compared the observed touching frequencies of taxa in the random model with the frequencies in natural plaque. This analysis identified 36 statistically significant (P < 0.05) associations in dental plaque cell aggregates (Fig. 5 and Tables S2–S5). Two genera, Leptotrichia and Treponema, were shown to have no significant associations. Cells of the genera Streptococcus and Prevotella were the most abundant, whereas cells of the genera Veillonella, Prevotella, and Actinomyces showed the most interspecies associations including those with Streptococcus, Rothia, Gemella, Pasteurellaceae, Porphyromonas and Neisseriaceae. In addition, Actinomyces and Prevotella were found to have significant association frequencies with Fusobacterium.
Fig. 4.
Representative detail images of CLASI-FISH–labeled semidispersed human dental plaque. Color in raw spectral images represents merge of six different fluorophore channels. Color in the segmented image represents 1 of 15 taxa. (Scale bar: 10 μm.)
Fig. 5.
Spatial analysis of human dental plaque. All intertaxon associations observed in 15-taxon labeled dental plaque are represented in this 2D plot. The area of the circle represents the relative abundance of each taxon. A line connecting two taxa indicates that cells of the lower-abundance taxon of any pair were observed to associate with cells of the higher-abundance taxon with >3% frequency and more frequently than would be expected from random associations.
Discussion
We have demonstrated a fluorescence imaging assay capable of distinguishing 28 differently labeled microbes in a single field of view, significantly improving the number of fluorescent labels previously distinguished in microbial FISH (32, 33). Our 15-taxon mixture of laboratory-grown oral microbes consisted of cells of varying complexity in their cell wall composition and represented both Gram-positive and Gram-negative organisms. Thus, the labeling strategy reported here is adequate to achieve specific labeling of multiple taxa in a single FISH reaction. Application of CLASI-FISH to human dental plaque, a microbial biofilm, enabled a quantitative analysis of the microscopic spatial relationships of microbes of 15 different taxa in a complex microbial community. We expect this method to provide a rapid, cost-effective characterization of microbial diversity and community composition. More importantly, we have demonstrated that this potentially high-throughput imaging assay is well suited to test hypotheses regarding the spatial distribution of microbes in complex multispecies communities.
Proximity analysis of a partially disrupted human oral biofilm revealed 26 statistically significant intertaxon cell-to-cell associations. Cells of two genera, Actinomyces and Prevotella, each showed nine intertaxon associations, appreciably more than cells of any other taxon. This result suggests an important role for cells of these two genera in maintaining or establishing biofilm complexity. In addition, the relative abundance of all 15 probed taxa, and in particular that of Prevotella, is consistent with this sample being a mixture of plaque, predominantly supragingival plaque, and saliva, a result that is consistent with the sample extraction protocol used (Materials and Methods).
Successful spectral imaging requires effective handling of potential sources of error. For this initial test of the CLASI method, we restricted our approach to binary labeling, meaning that all cells of interest were labeled with exactly two different fluorophores. This restriction provided an a priori constraint that greatly increased the accuracy and robustness of our results. In our experiments, the linear unmixing of fluorophores in cell images typically resulted in high intensity assigned to two fluorophores, whereas a smaller amount of intensity was assigned to the remaining fluorophores. The binary constraint permitted us to reject the residual off-channel intensities as artifacts resulting from limitations of the unmixing algorithm, photon shot noise, and detector dark noise (Fig. S2). Another consideration is the accuracy of image segmentation. Cells of different taxa that overlap within the plane of focus or cells that otherwise touch and are not identified as separate objects with our segmentation procedure result in ambiguous binary label assignment, because more than two fluorophores contribute to the total fluorescence intensity within that segmented feature. In the future, we expect more fluorophores and more fluorophore combinations to be unmixed successfully. This advance will be made possible by more accurate fluorophore assignment using improved unmixing algorithms with more appropriate noise consideration, better treatment of uncertainty in the unmixing solution (34), and better image segmentation. For example, with 15 fluorophores, there exist 105 binary combinations and 455 ternary combinations. We believe this number of identifiable fluorophores and combinations is well within reach.
It has been reported that some species of oral microbes (e.g., species of the genera Prevotella and Porphyromonas, among others) exhibit endogenous autofluorescence when excited with short-wavelength light (35). Indeed, we observed long-wavelength autofluorescence from some bacteria in dental plaque when illuminated with 405-nm laser excitation but not with longer-wavelength excitation. To avoid the problems of autofluorescence, we eschewed coupling short-wavelength fluorophores with taxon-specific probes and only used the short-wavelength fluorophore, Pacific Blue, with the general bacterial probe Eub338, which labels most bacteria and was not used to discriminate taxa. In the future, to increase the number of distinguishable targets, the use of short-wavelength excitation probes may be necessary. In this case, to reduce the influence of systematic errors from autofluorescence, one can measure the autofluorescence using an unlabeled sample and include these spectra in the library of reference spectra used for unmixing. Other unknown systematic biases can be detected through the analysis of the goodness of fit of unmixed fluorophores to recorded image data, for identifying particles or pixels with low confidence in fluorophore assignment.
It has been estimated that upwards of six to seven hundred species of microbes make up the human oral microbial community. In our analyses of human dental plaque, the nonlabeled cells outnumbered the taxon-labeled cells; therefore, we expect that further development of taxon-specific probes will allow a more complete analysis of this community. In addition, we expect the CLASI approach to be compatible with other forms of fluorescence labeling including immunohistochemistry with primary labeled antibodies, live-cell imaging with combinations of fluorescent proteins, and other molecular probes, expanding further the number and kinds of targets suitable for CLASI analysis. We expect future application of CLASI-FISH to intact oral biofilms or to model biofilms, as well as spatial analysis in three dimensions, to provide more detailed information on the spatial structure of these complex communities.
Finally, the CLASI approach is not limited to the analysis of the species composition of microbial aggregates. Many subdisciplines of biology could benefit from this method to expand the number of distinguishable fluorescent reporters within a single image. For example, the approach could be useful for the analysis of the spatiotemporal arrangements of complex biological structures such as cellular organelles and macromolecular assemblies within cells, allowing the precise microscopic analysis of many dynamic biological complexes. In general, to the extent that function is implicit in structure, the systems-level analysis of complex biological structures afforded by our combinatorial labeling and spectral imaging strategy offers an approach for deconstructing biological organization.
Materials and Methods
Microbial Strains and Culture Conditions.
E. coli K12 (ATCC 10798) were grown in LB broth (Beckton Dickinson) at 37 °C under aerobic conditions and were harvested at an OD600 of 0.5. A full list of oral microbial strains and culture conditions is provided in SI Materials and Methods.
Oligonucleotide Probe Design.
For E. coli experiments, eight versions of the Eub338 probe were custom synthesized. The Eub338 probe targets a conserved region of the bacterial 16S rRNA which is predicted to be present in most bacteria (http://www.microbial-ecology.net/probebase) and was verified to be present in E. coli. Each version of the probe had a different fluorophore conjugated to its 5′ end.
Oligonucleotide probes used to label 15 of the most abundant genera or families of oral microbes were designed to target the 16S rRNA and are listed in Table S1. The probes used were identified as specific genus-level probes for target organisms in the literature or were designed de novo using the probe design function of the ARB program (www.arb-home.de) with a database of 16S sequences from the Human Oral Microbiome Database (www.homd.org). Two versions of each of the 15 taxon-specific probes were synthesized. Both versions of each oligonucleotide probe were conjugated to one of two different fluorophores. Additional details on probe design are given is SI Materials and Methods.
Dental Plaque Sample Collection.
The use of human subjects in this study was approved by the New England Institutional Review Board. Plaque was provided by donors who had given informed consent. Donors refrained from normal oral hygiene (tooth brushing, flossing, and mouthwash) for 24 h before plaque samples were taken. Donors used acetate dental floss (Proctor & Gamble) to collect plaque from above and below the gum line, mostly between teeth from all parts of the mouth. Additional details on plaque sample collection are given in SI Materials and Methods.
FISH.
FISH was performed in Eppendorf tubes using a protocol modified from Pernthaler et al. (36). Details of the modified FISH protocols used in this study are given in SI Materials and Methods.
Image Acquisition.
Spectral images were acquired with a laser scanning confocal microscope equipped with a 32-channel multianode photomultiplier tube. Multiple acquisitions were made of each field of view, in series, using laser excitation appropriate for the fluorophores used in any experiment. Details on image acquisition and instrumentation are given in SI Materials and Methods.
Image Analysis.
Spectrally acquired images were subjected to linear unmixing using the microscope manufacturer's image acquisition and processing software. Unmixed images were imported into Image J (37) for image segmentation with a global intensity threshold. Foreground objects were segmented further with a watershed operation. Computed fluorophore intensities were averaged over all the pixels within a segmented object, and the two highest-intensity fluorophores were identified to assign a binary fluorophore label to each cell. Specific details on image analysis are given in SI Materials and Methods.
Spatial Analysis.
A computer program to analyze the spatial distribution of microbes in semidispersed dental plaque was developed on the Mathematica platform. The number of cells of each label type that touch each other or touch cells of a different label type was estimated as the difference between the number of objects in segmented images before and after two rounds of image dilation. Details on spatial analysis are given in SI Materials and Methods.
Image Modeling.
Model images of randomly distributed dental plaque microbes were created in Mathematica with parameters of cell size, cell shape, and cell abundances derived from measurements made on images of plaque microbes. Details on model image construction are given in SI Materials and Methods.
Spatial Plot.
A network diagram that summarizes all intertaxon cell-to-cell associations observed in dental plaque was constructed in Mathematica. Details on construction of this plot are given in SI Materials and Methods.
Supplementary Material
Acknowledgments
We are grateful to Louis M. Kerr for invaluable assistance with microscopy. We thank Nikon Instruments and Carl Zeiss for gracious instrumentation support. This work was supported by Grant 2007-3-13 from the Alfred P. Sloan Foundation (to G.G.B.), National Institutes of Health Grant 1RC1-DE020630 from the National Institute of Dental and Craniofacial Research (NIDCR) (to G.G.B. and F.E.D.) and by National Institutes of Health Fellowship 1F31-DE019576 from NIDCR (to A.M.V.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1101134108/-/DCSupplemental.
References
- 1.van Leeuwenhoek A. Letter to the Royal Society. In: Dobell C, editor. Antony van Leeuwenhoek and his “Little Animals,” Being Some Account of the Father of Protozoology and Bacteriology and his Multifarious Discoveries in These Disciplines. New York: Harcourt, Brace, and Co; 1960. [Google Scholar]
- 2.DeLong EF, Wickham GS, Pace NR. Phylogenetic stains: Ribosomal RNA-based probes for the identification of single cells. Science. 1989;243:1360–1363. doi: 10.1126/science.2466341. [DOI] [PubMed] [Google Scholar]
- 3.Amann R, Fuchs BM. Single-cell identification in microbial communities by improved fluorescence in situ hybridization techniques. Nat Rev Microbiol. 2008;6:339–348. doi: 10.1038/nrmicro1888. [DOI] [PubMed] [Google Scholar]
- 4.Waters JC. Accuracy and precision in quantitative fluorescence microscopy. J Cell Biol. 2009;185:1135–1148. doi: 10.1083/jcb.200903097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Costerton JW, Lewandowski Z, Caldwell DE, Korber DR, Lappin-Scott HM. Microbial biofilms. Annu Rev Microbiol. 1995;49:711–745. doi: 10.1146/annurev.mi.49.100195.003431. [DOI] [PubMed] [Google Scholar]
- 6.Daniel R. The metagenomics of soil. Nat Rev Microbiol. 2005;3:470–478. doi: 10.1038/nrmicro1160. [DOI] [PubMed] [Google Scholar]
- 7.Turnbaugh PJ, et al. A core gut microbiome in obese and lean twins. Nature. 2009;457:480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Woyke T, et al. Assembling the marine metagenome, one cell at a time. PLoS ONE. 2009;4:e5299. doi: 10.1371/journal.pone.0005299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Roeder J, Schink B. Syntrophic degradation of cadaverine by a defined methanogenic coculture. Appl Environ Microbiol. 2009;75:4821–4828. doi: 10.1128/AEM.00342-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Taga ME, Bassler BL. Chemical communication among bacteria. Proc Natl Acad Sci USA. 2003;100(Suppl 2):14549–14554. doi: 10.1073/pnas.1934514100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hibbing ME, Fuqua C, Parsek MR, Peterson SB. Bacterial competition: Surviving and thriving in the microbial jungle. Nat Rev Microbiol. 2010;8:15–25. doi: 10.1038/nrmicro2259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rickard AH, Gilbert P, High NJ, Kolenbrander PE, Handley PS. Bacterial coaggregation: An integral process in the development of multi-species biofilms. Trends Microbiol. 2003;11:94–100. doi: 10.1016/s0966-842x(02)00034-3. [DOI] [PubMed] [Google Scholar]
- 13.Raes J, Bork P. Molecular eco-systems biology: Towards an understanding of community function. Nat Rev Microbiol. 2008;6:693–699. doi: 10.1038/nrmicro1935. [DOI] [PubMed] [Google Scholar]
- 14.Foster JS, Palmer RJ, Jr., Kolenbrander PE. Human oral cavity as a model for the study of genome-genome interactions. Biol Bull. 2003;204:200–204. doi: 10.2307/1543559. [DOI] [PubMed] [Google Scholar]
- 15.Dewhirst FE, et al. The human oral microbiome. J Bacteriol. 2010;192:5002–5017. doi: 10.1128/JB.00542-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zaura E, Keijser BJ, Huse SM, Crielaard W. Defining the healthy “core microbiome” of oral microbial communities. BMC Microbiol. 2009;9:259. doi: 10.1186/1471-2180-9-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ding AM, Palmer RJ, Jr., Cisar JO, Kolenbrander PE. Shear-enhanced oral microbial adhesion. Appl Environ Microbiol. 2010;76:1294–1297. doi: 10.1128/AEM.02083-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kolenbrander PE. Intergeneric coaggregation among human oral bacteria and ecology of dental plaque. Annu Rev Microbiol. 1988;42:627–656. doi: 10.1146/annurev.mi.42.100188.003211. [DOI] [PubMed] [Google Scholar]
- 19.Yoshida Y, Palmer RJ, Yang J, Kolenbrander PE, Cisar JO. Streptococcal receptor polysaccharides: Recognition molecules for oral biofilm formation. BMC Oral Health. 2006;6(Suppl 1):S12. doi: 10.1186/1472-6831-6-S1-S12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Park Y, et al. Short fimbriae of Porphyromonas gingivalis and their role in coadhesion with Streptococcus gordonii. Infect Immun. 2005;73:3983–3989. doi: 10.1128/IAI.73.7.3983-3989.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nagaoka S, et al. Interactions between salivary Bifidobacterium adolescentis and other oral bacteria: In vitro coaggregation and coadhesion assays. FEMS Microbiol Lett. 2008;281:183–189. doi: 10.1111/j.1574-6968.2008.01092.x. [DOI] [PubMed] [Google Scholar]
- 22.Kolenbrander PE, et al. Bacterial interactions and successions during plaque development. Periodontol 2000. 2006;42:47–79. doi: 10.1111/j.1600-0757.2006.00187.x. [DOI] [PubMed] [Google Scholar]
- 23.Periasamy S, Kolenbrander PE. Central role of the early colonizer Veillonella sp. in establishing multispecies biofilm communities with initial, middle, and late colonizers of enamel. J Bacteriol. 2010;192:2965–2972. doi: 10.1128/JB.01631-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chalmers NI, et al. Use of quantum dot luminescent probes to achieve single-cell resolution of human oral bacteria in biofilms. Appl Environ Microbiol. 2007;73:630–636. doi: 10.1128/AEM.02164-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chalmers NI, Palmer RJ, Jr., Cisar JO, Kolenbrander PE. Characterization of a Streptococcus sp.-Veillonella sp. community micromanipulated from dental plaque. J Bacteriol. 2008;190:8145–8154. doi: 10.1128/JB.00983-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dige I, Raarup MK, Nyengaard JR, Kilian M, Nyvad B. Actinomyces naeslundii in initial dental biofilm formation. Microbiology. 2009;155:2116–2126. doi: 10.1099/mic.0.027706-0. [DOI] [PubMed] [Google Scholar]
- 27.Zijnge V, et al. Oral biofilm architecture on natural teeth. PLoS ONE. 2010;5:e9321. doi: 10.1371/journal.pone.0009321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Garini Y, Young IT, McNamara G. Spectral imaging: Principles and applications. Cytometry A. 2006;69:735–747. doi: 10.1002/cyto.a.20311. [DOI] [PubMed] [Google Scholar]
- 29.Dickinson ME, Bearman G, Tille S, Lansford R, Fraser SE. Multi-spectral imaging and linear unmixing add a whole new dimension to laser scanning fluorescence microscopy. Biotechniques. 2001;31:1272–, 1274–1276, 1278. doi: 10.2144/01316bt01. [DOI] [PubMed] [Google Scholar]
- 30.Amann RI, et al. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl Environ Microbiol. 1990;56:1919–1925. doi: 10.1128/aem.56.6.1919-1925.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen T, et al. The Human Oral Microbiome Database: A web accessible resource for investigating oral microbe taxonomic and genomic information. Database (Oxford) 2010 doi: 10.1093/database/baq013. 10.1093/database/baq013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thurnheer T, Gmür R, Guggenheim B. Multiplex FISH analysis of a six-species bacterial biofilm. J Microbiol Methods. 2004;56:37–47. doi: 10.1016/j.mimet.2003.09.003. [DOI] [PubMed] [Google Scholar]
- 33.Amann R, Snaidr J, Wagner M, Ludwig W, Schleifer KH. In situ visualization of high genetic diversity in a natural microbial community. J Bacteriol. 1996;178:3496–3500. doi: 10.1128/jb.178.12.3496-3500.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Neher R, Neher E. Optimizing imaging parameters for the separation of multiple labels in a fluorescence image. J Microsc. 2004;213:46–62. doi: 10.1111/j.1365-2818.2004.01262.x. [DOI] [PubMed] [Google Scholar]
- 35.van der Veen MH, Thomas RZ, Huysmans MC, de Soet JJ. Red autofluorescence of dental plaque bacteria. Caries Res. 2006;40:542–545. doi: 10.1159/000095655. [DOI] [PubMed] [Google Scholar]
- 36.Pernthaler J, Glöckner F-O, Schönhuber W, Amann R. Fluorescence in situ hybridization (FISH) with rRNA-targeted oligonucleotide probes. Methods Microbiol. 2001;30:207–226. [Google Scholar]
- 37.Abramoff MD, Magelhaes PJ, Ram SJ. Image processing with ImageJ. Biophotonics International. 2004;11:36–42. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.