Abstract
Membrane fusion is required for diverse biological functions ranging from viral infection to neurotransmitter release. Fusogenic proteins increase the intrinsically slow rate of fusion by coupling energetically downhill conformational changes of the protein to kinetically unfavorable fusion of the membrane–phospholipid bilayers. Class I viral fusogenic proteins have an N-terminal hydrophobic fusion peptide (FP) domain, important for interaction with the target membrane, plus a C-terminal transmembrane (C-term-TM) helical membrane anchor. The role of the water-soluble regions of fusogenic proteins has been extensively studied, but the contributions of the membrane-interacting FP and C-term-TM peptides are less well characterized. Typically, FPs are thought to bind to membranes at an angle that allows helix penetration but not traversal of the lipid bilayer. Here, we show that the FP from the paramyxovirus parainfluenza virus 5 fusogenic protein, F, forms an N-terminal TM helix, which self-associates into a hexameric bundle. This FP also interacts strongly with the C-term-TM helix. Thus, the fusogenic F protein resembles SNARE proteins involved in vesicle fusion by having water-soluble coiled coils that zipper during fusion and TM helices in both membranes. By analogy to mechanosensitive channels, the force associated with zippering of the water-soluble coiled-coil domain is expected to lead to tilting of the FP helices, promoting interaction with the C-term-TM helices. The energetically unfavorable dehydration of lipid headgroups of opposing bilayers is compensated by thermodynamically favorable interactions between the FP and C-term-TM helices as the coiled coils zipper into the membrane phase, leading to a pore lined by both lipid and protein.
The basic mechanisms of viral membrane fusion have been studied extensively, but major gaps remain in our understanding of the relative roles of lipidic intermediates and viral fusogenic proteins in lowering the energy barrier for the overall process (1–4). The most common mechanistic hypothesis concerning enveloped viral fusion is that fusogenic proteins primarily serve to bring the target cell and viral membranes into proximity. Fusion occurs in a multistep process, in which the virus first binds to a specific receptor; this event and/or other environmental cues then cause a conformational change in the protein, leading to a metastable state with an exposed hydrophobic fusion peptide (FP) that binds to the target membrane. Once engaged with the bilayer, a second energetically favorable conformational change in the fusogenic protein then exerts a force pulling the FP toward the viral membrane, in effect reeling the host and viral membranes together.
The conformational changes involved in the water-soluble portions of viral fusogenic proteins have been largely elucidated, but the roles of the membrane-binding FP and the C-terminal transmembrane (C-term-TM) anchor are less clear. After the crystal structure of the prefusogenic form of influenza hemagglutinin (HA) was solved (5), experimental studies suggested that its FP inserted into the bilayer (6). The FP helix was thought to bind sufficiently deeply to act as a hydrophobic wedge that not only served as an anchor but also destabilized the bilayer and facilitated fusion. Many biochemical, biophysical, and mutagenesis studies on the fusion proteins of the influenza virus, HIV, and other viruses are consistent with, and have added considerable detail to, this initial suggestion (7–34). However, a number of intriguing findings suggest that the FPs and C-term-TM helices might play additional, more specific, roles in bilayer fusion than mere membrane binding and disruption.
Surprisingly subtle mutations in the C-term-TM sequence of fusogenic proteins can be quite deleterious to their ability to induce fusion, while retaining normal processing and the ability to change conformations (35–38). Also, replacing C-term-TM helices of fusogenic proteins with lipid anchors results in a loss of fusion (39–42) and, surprisingly, FP sequences often show greater conservation than might be expected from the functional requirements for membrane binding (43). Moreover, the very strong conservation of polar and small residues at regularly spaced intervals as found in GXXXG (44–46), glycine zippers (47), and GAS (glycine-alanine-serine) motifs (48) (Table S1), is intriguing. These patterns are known to stabilize TM helix association and also figure in the helix–helix packing of proteins that undergo large-scale conformational changes in response to lipidic environment, such as in mechanosensitive (MS) channels (49).
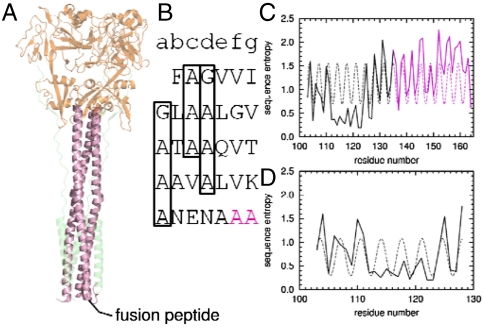
Thus, we investigated whether FPs might adopt TM helical rather than surface orientations, and how this might relate to their mechanisms of action. If indeed FPs adopt a TM orientation, then one might envision a mechanism akin to SNARE proteins (50–53), in which both the target and vesicular proteins have TM helices that associate as membrane fusion progresses. Accordingly, we investigate herein a class I fusogenic protein, F, from parainfluenza virus 5 (PIV5), which, like influenza virus HA and HIV gp41, has an N-terminal FP in the mature, cleaved protein (3). High-resolution structures are available for both the pre- and postfusion forms of the ectodomain of the F protein (54–56), and the structure and function of its C-term-TM domain has been extensively investigated by scanning mutagenesis and Cys crosslinking (57). The sequence of the FP suggests that it also forms a TM helix (58). Moreover, it has a single polar Gln residue, a residue known to promote helix–helix interactions in membranes (59), plus glycine and alanine residues in a heptad repeat pattern (Fig. 1B), known to stabilize TM helix–helix assembly (60–62) and pore formation (47). The sequence variability of the FP across homologous viruses shows a heptad repeat in phase with the heptad repeat of the long water-soluble coiled coil, which directly follows it (Fig. 1 C and D), interrupted only by a highly conserved region (residues 112–117) that is constrained by packing in the prefusion trimer (55, 63). Consistent with this, the postfusion structure of PIV5 (54) showed that the C-terminus of the FP is helical. Here, we show that this FP adopts a TM orientation in phospholipid membranes, specifically oligomerizing into a homohexameric bundle (6HB), and it also associates with the C-term-TM domain in micelles. Computational studies suggest that conformational changes involving zippering of the water-soluble coiled coil in the ectodomain drive changes in helix-crossing angles that may lead to an initial heteromeric contact or “pinprick” between the FP and the C-term-TM leading to a fusion pore possibly lined by both protein and lipid.
Fig. 1.
Sequence conservation suggests a continuous helix including HRA and the FP. (A) Postfusion crystal structure of the soluble domain of closely related hPIV3 virus F protein (54). Shown in magenta is HRA. Below HRA, in the postfusion membrane, is the predicted location of the FP. (B) Heptad repeat of the FP and HRA. The beginning of the crystallographic resolved region of HRA is shown in magenta. Heptad repeats of small residues in the FP are boxed. (C) Sequence entropy of the FP and HRA can be fit to a single sinusoidal function with period of 3.47 ± 0.02 residues/turn (r = 0.51). (D) Sequence entropy of the FP alone can be fit to a single sinusoidal function with a period of 3.51 ± 0.08 residues/turn (r = 0.59).
Results
Association of Fusion and C-term-TM Peptides in Detergent Micelles.
Analytical ultracentrifugation (AUC) of the FP from PIV5 F protein in phospholipid micelles reveals cooperative assembly into hexamers (Fig. S1A). The FP was dissolved in dodecylphosphocholine (DPC) micelles, and the density of the solution was adjusted to precisely match that of the DPC detergent (64) so that only the protein component contributes to the sedimentation equilibrium. Three samples prepared at differing peptide-to-detergent ratios were each centrifuged at four to five rotor speeds, respectively, for the wild-type and mutant Q120A. The data were then globally analyzed to extract the number of peptides per oligomer as well as the free energy of association (62, 64, 65). The data conform very well to tightly associating and fully cooperative monomer-hexamer equilibrium (Fig. S2). The addition of lower-order intermediate states failed to improve the quality of the fit, indicating that the association was highly cooperative and specific for the formation of hexamers relative to other possible association states.
The mutant Q120A also forms hexamers (Figs. S1 C and D and S2B), but its association is weaker than that of the wild-type peptide by 13.4 kcal/mol of hexamer, or 2.2 kcal/mol of monomer. Glutamine (Gln) is well known to stabilize the association of TM helices (59), and the magnitude of the effect is similar. Thus, it is likely that the Gln helps stabilize TM helix association within the structure, although this residue is not absolutely essential for forming the 6HB. Q120 is strongly conserved in related viruses (Table S1) and is a promising target for future studies using reverse-engineered viruses.
Although the C-term-TM domain has been shown to associate in the full-length protein (57), the C-term-TM peptide alone does not associate in DPC micelles (Fig. S3A). However, when unlabeled wild-type FP is introduced at a 1∶1 ratio, the C-term-TM-peptide strongly associates (Fig. S3B), perhaps adopting a structure relevant for the postfusion state. Analysis of the sedimentation curves indicates that the TM peptide self-associates with the FP at least 20-fold less tightly than the corresponding heteromeric interaction with the C-term-TM peptide.
FPs Adopt a TM Orientation in Lipid Bilayers.
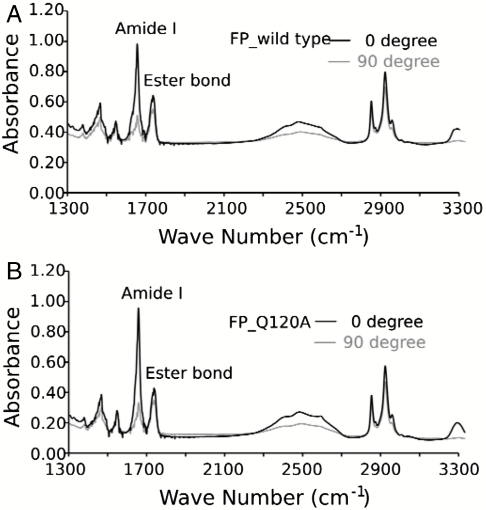
The secondary structure and orientation of the wild-type and mutant FP in micelles and deuterium oxide (D2O) hydrated bilayers were evaluated using circular dichroism (CD) and attenuated total reflection IR spectroscopy (ATR-IR), respectively. The CD spectra of both peptides in DPC micelles are typical of an α-helix (Fig. S4) indicating that the association observed by AUC corresponded to the formation of helical bundles. The IR spectra in the amide I region of the FPs shows a single, sharp peak at 1656 cm-1, indicative of a dehydrated helical conformation (66) in bilayers (Fig. 2 A and B). The dichroic ratio for parallel versus perpendicularly polarized light was 3.2 and 3.6 for the wild type and mutant, respectively. These values correspond to an orientation of approximately 29° and 22° relative to the membrane normal (67, 68), assuming the entire peptide is fully helical and the bilayers are well ordered. Deviation from helical geometry or disorder of the bilayer would result in somewhat lower dichroic ratios. In this case, the true angles would be even closer to parallel to the bilayer normal. Thus, both peptides have a strong preference to adopt a TM orientation relative to other possibilities in which the helix was either randomly oriented or oriented parallel to the membrane surface.
Fig. 2.
ATR-IR of FP wild-type (A) and mutant Q120A (B) in phopholipid (POPC) bilayers. The sharp peak at 1656 cm-1 is indicative of alpha helical secondary structure. The TM orientation is demonstrated by the much greater intensity of the 1656 cm-1 amide I bond for parallel (0°) versus perpendicular (90°) polarized incident light (relative to the membrane normal).
Computational Modeling of the 6HB.
To model the FP 6HB, the possible structural space was systematically sampled and scored using a protocol akin to the conformational search of Brunger, Arkin, and coworkers (69). The strong heptad repeat (Fig. 1 C and D) is indicative of a left-handed helical bundle. A left-handed bundle also would be consistent with a continuous helical structure beginning in the soluble heptad repeat A (HRA) and continuing directly to FP, as the conservation pattern suggests (Fig. 1C). Moreover, the nature of viral fusion, with asymmetric insertion of peptides into the target membrane, suggests that the FPs comprising the 6HB should adopt a parallel orientation. Symmetric, parallel coiled coils can be described by a limited number of variables (70). Three of these—α-helical phase (ϕ), pitch angle (α), and superhelical radius (R)—were allowed to vary and were sampled systematically in search of optimal coiled-coil structures. For each structure, optimal rotamers were selected, and the structure was then minimized. Each structure was scored using the CHARMM energy function in an implicit membrane environment (71, 72) to select candidate models.
The energy of a particular 6HB conformation depends primarily on the phase, ϕ. Multiple energy minima are observed as the helices are rotated (Fig. S5A). Five left-handed structures were selected, corresponding to the lowest energy basins, for further refinement using molecular dynamics (MD) simulations in an explicit fully hydrated lipid bilayer. The lowest predicted energy for an antiparallel orientation 6HB was selected as a negative control. It was less stable in MD simulations than the low-energy parallel models and was not further pursued (Figs. S5 and S6).
MD simulations on the five parallel 6HB structures (labeled according to their phases, ϕ = 40°, 43°, 88°, 196°, and 300°, respectively) show that the orientation of the Gln side chain is crucial for 6HB stability. Two closely related structures, ϕ = 40° and 43° (Cα rmsd = 1.6 Å), place the Gln in a “d” position within the coiled coil, whereas the phase 88° structure places the Gln in an “a” position. The remaining two structures have Gln facing the lipid (phases 196° and 300°), are much less stable than the interior-facing ones (Figs. S5B and S6), and rapidly depart from their initial structures (as measured by Cα rmsd), whereas structures with an interior Gln are stable near the initial structure for 50 ns of MD simulation.
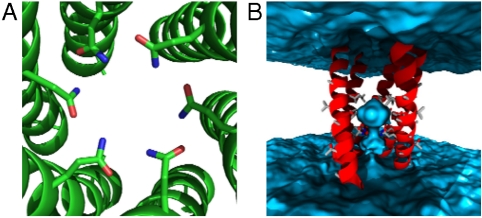
Of the interior-facing Gln structures, the phase 40° and 43° models are most stable during the MD simulation and best maintain a symmetric coiled-coil structure (Fig. S6). These models form a highly stable hydrogen bond network in the interior of the 6HB coiled coil (Fig. 3A), consistent with the important role Gln plays in oligomerization (Fig. S1). The periodically conserved small residues of the FP are found at the helix interface in this model. Of note is the penetration of water into the core of the 6HB from the viral side of the membrane (Fig. 3B). It is possible that the formation of the FP 6HB structure may reduce the barrier to fusion by initiating formation of a nucleus for expansion into the later, much larger fusion pore. In the less stable models (phases 88°, 196°, and 300°) the water distribution is not stable because of either a less favorable arrangement of the Gln side chains (88°) or their location outside the pore (196° and 300°) (Fig. S7).
Fig. 3.
Computational model of the PIV5 F FP hexameric bundle. (A) The Q120 residues form hydrogen bonds with one another as well as waters on the interior. (B) Side view shows the bundle oriented with the N-terminal end (which presumably faces the cellular interior) up. Water is shown in blue. Not shown for clarity are the phospholipids as well as one helix closest to the viewer.
The most stable 6HB (ϕ = 40°) structure was then used to compute an FTIR dichroism ratio following the method of Arkin and coworkers (73, 74) where the individual residue dipoles are combined. The computed dichroism ratio is 2.95 ± 0.07 (mean and standard deviation over the MD simulation), in good agreement with the experimental value (Fig. 2).
Discussion
Comparison of the Properties of the FP from PIV5 with Other Systems.
Here, we provide experimental evidence that the FP from the PIV5 F protein is able to adopt a TM helical conformation when incorporated into lipid bilayers, and that it associates with the C-term-TM helix. Similarly, a FRET assay (75) suggested the C-term-TM domain of influenza HA interacts with its FP, although the orientation of the peptide in the complex was not determined. These findings extend the structural and mechanistic similarity between the PIV5 fusogenic F protein and SNARE proteins to include not only their water-soluble coiled-coil domains, but also their membrane-interactive domains. Recent biochemical and structural studies on SNARE proteins (50) suggest a zippering motion of the water-soluble coiled coils that continues into the TM domains promoting a heteromeric interaction between the two TM helices to provide part of the driving force for bilayer fusion.
The conformation and TM orientation of the FP from the PIV5 F protein is clearly defined by IR dichroism (Fig. 2), which showed an average helical tilt of 20° to 30° relative to the membrane normal, and also ruled out the possibility of significant amounts of β-structure. The situation is less clear for other FPs, which often are found to adopt more “oblique-oriented” or “tilted helical conformation” (76), in which the helix is oriented at 30° to 70° relative to the bilayer normal, either spanning the bilayer or penetrating a single leaflet, depending on the length of the synthetic peptide investigated (12–14, 25, 32–34). For example, the N-terminal peptide of gp41 has been reported to adopt a TM (15), tilted (28–30), and beta (19, 26–28, 31) conformation in various membrane mimetics. Synthetic versions of the FP from influenza virus HA2 span approximately half of the bilayer width, but as a bent helix (12) or helical hairpin (18) in micelles. However, the hydrophobic region of the FP in the intact virus spans residues up to Arg25, and NMR studies have been conducted with peptides spanning between 20 to 23 residues, with an artificial oligo-Lys tail added to enhance water solubility. The dynamics and conformational properties of the 20- versus 23-residue peptide differ significantly (18), as expected for a finely tuned system with multiple low-lying energy wells that are progressively populated during fusion. These distinct structural states, and their sensitivity to small changes in sequence and environment, may be both functionally relevant and reflect the energetic fine-tuning of the landscape and the dynamic nature of fusion.
Within a family, the FPs of viral fusion proteins have highly conserved sequence motifs, such as heptad repeats of small residues, that are similar to those important for association of other oligomeric TM helical bundles (45, 47, 48) (Table S1), suggesting that TM helix–helix association might be relevant to fusion. In this regard, it is interesting to compare the avidity of homo- and heterooligomer formation for the C-term-TM and FP of the PIV5 F protein. Isolated FPs homooligomerize strongly and specifically to a 6HB. The C-term-TM peptide also engages in helix–helix interactions, which have been experimentally demonstrated using disulfide crosslinking of the full-length protein (57). However, the present study shows that the C-term-TM helix homooligomerizes more weakly than the FP in the absence of the trimeric ectodomain, but associates tightly with the FP (Fig. S3). The hierarchy of association strengths mimics the assembly process of the ectodomains, in which the weakly associated parallel C-terminal coiled-coil trimer (contiguous with the C-term-TM helix) dissociates and zippers up along the N-terminal coiled coil (contiguous with the FP) to form the final antiparallel bundle (1–4). The C-term-TM and FP may likewise zipper as an antiparallel bundle in forming the postfusion state.
A Provisional Model for Membrane Fusion by Class I Proteins: Lipid-Centric and Pinprick Mechanisms.
In the absence of fusion proteins, the process of bilayer fusion is a physical process with multiple high-energy intermediates (77, 78) corresponding to: (i) diffusion of the membranes together, (ii) dehydration of the bilayers as the headgroups of opposing bilayers come into still closer proximity, (iii) formation of a lipidic stalk, (iv) hemifusion, (v) pore formation and expansion. Viral fusion proteins and SNARE proteins utilize essentially irreversible, energetically favorable conformational transitions to lower the activation energy for membrane fusion (50, 51, 77, 78). Thus, they are active participants that shape the energy landscape. There are multiple classes of fusogenic proteins, and there is significant variation in the number of fusion proteins per particle, suggesting additional biological requirements, presence of accessory proteins, or lipid compositions (2). Here, we consider how the class I fusogenic proteins might orchestrate energetic landscape-shaping mechanisms. The present observations provide molecular detail to two limiting hypothetical models, representing extremes in a continuum of kinetic pathways that depend on the protein and experimental variables.
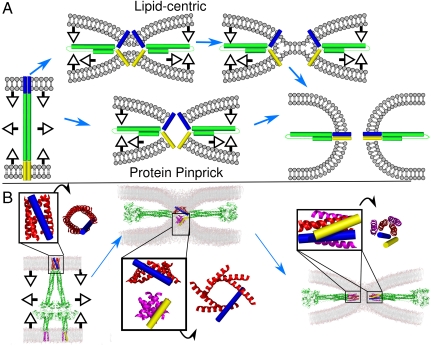
In a lipid-centric model of viral fusion, the proteins hold the bilayers in close proximity to promote the progression through lipidic intermediates of fusion (Fig. 4A). The FP and C-term-TM domains are hypothesized to remain outside of the point of membrane apposition, which is instead made up exclusively of lipids. The ability of the FP to embed deeply into the membrane and engage in favorable C-term-TM to FP interactions provides a mechanism for forcing the two bilayers into close proximity within a very small area, as the coiled-coil domains of preassociated proteins zipper through the water-soluble regions and extend into the membrane. Favorable FP to C-term-TM interaction provides a continuously downhill process for the protein component, facilitating bilayer–bilayer apposition. Moreover, for systems in which many fusion proteins are required for fusion, the association of the FP in target membranes might bring sufficient fusion proteins near the protein-free zone.
Fig. 4.
Provisional model of PIV5 fusion. (A) Schematic diagrams of the limiting extremes of lipid-centric and pinprick fusion. (B) Shown is a model of the conformational change of the F protein (FP) hexamer (6HB) from a prehairpin, extended intermediate (Left) to a point of membrane apposition (Center) and finally to the postfusion state (Right). Proposed conformations of the FP 6HB are shown in the Insets along with 90° rotations. Note the increased tilt of the FP moving from the extended intermediate to the point of membrane apposition as well as the recruitment of lipid headgroups to the nascent pore. FPs are shown in red and blue, C-term-TMs are shown in magenta and yellow. The C-term-TM in the middle image contains two trimeric structures (57).
A second model of fusion envisions that fusion proceeds via an initial contact between the TM domains in the two bilayers. The central point of protein contact can be thought of as a pinprick that expands into a fusion pore. This model is in contrast to those that propose a gap junction-like pore (23) as only the initial contact is mediated solely by protein domains. The 6HB is hypothesized to be at the center of the contact region between the two membranes. Subsequent pore formation involves the initial protein contact expanding with recruitment of additional lipids with their headgroups facing the growing fusion pore (note the incursion of phospholipids in Fig. 4B).
To probe the hypothesized mechanism further, we built models of two bilayers in the process of fusion and asked how the previously defined structural intermediates of the water-soluble and membrane domains of the protein might map onto likely lipid intermediates, lowering the activation energy of the process. Fig. 4 (and Movie S1) shows how a hexamer of the PIV5 FPs might serve as a pinprick to nucleate a pore at three critical points: the extended prehairpin intermediate (Left), membrane apposition (Center), and postfusion (Right). As the conformational change progresses, the TM bundles formed by the C-term-TM and FP helices first dock, then coalesce into heteromeric bundles. The initial zone of intermembrane contact involves favorable protein–protein interactions rather than energetically unfavorable dehydration of the bilayer headgroups, and the fusion of these two helical bundles provides a low-energy pathway to direct fusion of the bilayers, which remain associated with the TM bundles throughout the process.
The latter mechanism provides a rationale for the multiple conformational forms and strong intrafamily conservation in the FP sequences, which must associate with graded affinities in a homomeric as well as heteromeric fashion. It also explains how the addition of various shaped lipids can either promote or inhibit fusion. As the protein conformational change proceeds, the C-term-TM and FP become more tilted (relative to the normal of the initial bilayer). The driving force for tilting includes the zippering of the coiled coil and favorable heteromeric TM interactions. The recent structure by Rees and coworkers of the MscL MS channel (49) illustrates how mechanical forces from external domains and lipid-specific effects result in changes in helical tilt and channel radius. Changes in the membrane lateral surface pressure profile cause helices comprising the MscL channel to slide relative to each other, increasing their tilt and opening the channel like a diaphragm. In a similar manner, mechanical forces from conformational changes as well as lateral surface pressure effects associated with the lipid composition would couple to the energetics of protein-mediated bilayer fusion.
The highly conserved small glycine and alanine residues, which are found in both MS channels and class I viral FP (Fig. 1 and Table S1), are ideally suited for helix sliding because they present relatively smooth interfaces (45, 46). The channel formed by MscL also expands with these conformational changes, both opening the channel and increasing the surface area available for protein–protein interaction. The hypothetical tilting of the FP and C-term-TM domains would increase the number of residues in contact with the hydrophobic region of the bilayer beyond the length of 20 residues typically seen for TM helices. This longer membrane-suitable region is observed for the PIV5 C-term-TM and contributes to fusion (57). It is also observed in other viruses such as influenza (51) and HIV, where shortening the length of C-term-TM helices can halt the fusion process (79, 80).
Fig. 4A compares the lipid-only and pinprick mechanisms; in both cases, protein–protein interactions between membrane-embedded helices bring the two bilayers into intimate contact. After the bilayers are brought close together, different proteins might take different pathways to achieve fusion. The zone of adhesion can widen to create a hemifusion intermediate, particularly for situations in which one of the two helices does not fully span the bilayer (Fig. 4A). Alternatively, the protein might act as a pinprick to nucleate the fusion pore (Fig. 4B). The requirements for tight and specific interactions between the membrane-embedded helices will also vary depending on the specifics of the mechanism.
Overall, it seems likely that a continuum of mechanisms is needed, with protein-rich and lipid-rich patches in the fusion pore for many proteins. This in turn will allow for failures leading to lipid mixing-arrested hemifusion when the fusogenic peptides are mutated. The present work favors a protein-centric but not a protein-only fusion mechanism. The pinprick mechanism should face a less difficult pathway for interbilayer interaction to initiate the pore. In addition, it bridges SNARE-like and virus-like mechanisms while explaining why different angles of insertion have been observed for various FPs. Thus, this mechanism provides a general framework for understanding protein-mediated membrane fusion.
Methods
Wild-type and mutant PIV5 FPs and the C-term-TM were chemically synthesized. AUC experiments in DPC used D2O to match the density of the detergent. ATR-IR experiments used 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) lipids in a peptide to POPC ratio of 1∶20. Detailed experimental and computational methods are available in SI Text.
Supplementary Material
Acknowledgments.
We thank James D. Lear, Paul C. Billings, Kathryn B. Smith-Dupont, Gregory A. Caputo, and Ivan V. Korendovych for help with data acquisition and useful discussions. This work was supported by the National Institutes of Health (GM040712). MD studies were made possible by the National Science Foundation through TeraGrid resources provided by the Pittsburgh Supercomputing Center and the National Institute for Computational Sciences (allocation number TG-MCA93S020).
Footnotes
The authors declare no conflict of interest.
See Commentary on page 3827.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1019668108/-/DCSupplemental.
References
- 1.Harrison SC. Viral membrane fusion. Nat Struct Mol Biol. 2008;15:690–698. doi: 10.1038/nsmb.1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.White JM, Delos SE, Brecher M, Schornberg K. Structures and mechanisms of viral membrane fusion proteins: Multiple variations on a common theme. Crit Rev Biochem Mol Biol. 2008;43:189–219. doi: 10.1080/10409230802058320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lamb RA, Jardetzky TS. Structural basis of viral invasion: Lessons from paramyxovirus F. Curr Opin Struct Biol. 2007;17:427–436. doi: 10.1016/j.sbi.2007.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Colman PM, Lawrence MC. The structural biology of type I viral membrane fusion. Nat Rev Mol Cell Biol. 2003;4:309–319. doi: 10.1038/nrm1076. [DOI] [PubMed] [Google Scholar]
- 5.Wilson IA, Skehel JJ, Wiley DC. Structure of the haemagglutinin membrane glycoprotein of influenza virus at 3 A resolution. Nature. 1981;289:366–373. doi: 10.1038/289366a0. [DOI] [PubMed] [Google Scholar]
- 6.Stegmann T, Delfino JM, Richards FM, Helenius A. The HA2 subunit of influenza hemagglutinin inserts into the target membrane prior to fusion. J Biol Chem. 1991;266:18404–18410. [PubMed] [Google Scholar]
- 7.Blumenthal R, Sarkar DP, Durell S, Howard DE, Morris SJ. Dilation of the influenza hemagglutinin fusion pore revealed by the kinetics of individual cell-cell fusion events. J Cell Biol. 1996;135:63–71. doi: 10.1083/jcb.135.1.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cross KJ, Wharton SA, Skehel JJ, Wiley DC, Steinhauer DA. Studies on influenza haemagglutinin fusion peptide mutants generated by reverse genetics. EMBO J. 2001;20:4432–4442. doi: 10.1093/emboj/20.16.4432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Durrer P, et al. H+-induced membrane insertion of influenza virus hemagglutinin involves the HA2 amino-terminal fusion peptide but not the coiled coil region. J Biol Chem. 1996;271:13417–13421. doi: 10.1074/jbc.271.23.13417. [DOI] [PubMed] [Google Scholar]
- 10.Freitas MS, et al. Structure of the Ebola fusion peptide in a membrane-mimetic environment and the interaction with lipid rafts. J Biol Chem. 2007;282:27306–27314. doi: 10.1074/jbc.M611864200. [DOI] [PubMed] [Google Scholar]
- 11.Ge M, Freed JH. Fusion peptide from influenza hemagglutinin increases membrane surface order: An electron-spin resonance study. Biophys J. 2009;96:4925–4934. doi: 10.1016/j.bpj.2009.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Han X, Bushweller JH, Cafiso DS, Tamm LK. Membrane structure and fusion-triggering conformational change of the fusion domain from influenza hemagglutinin. Nat Struct Biol. 2001;8:715–720. doi: 10.1038/90434. [DOI] [PubMed] [Google Scholar]
- 13.Han X, Steinhauer DA, Wharton SA, Tamm LK. Interaction of mutant influenza virus hemagglutinin fusion peptides with lipid bilayers: Probing the role of hydrophobic residue size in the central region of the fusion peptide. Biochemistry. 1999;38:15052–15059. doi: 10.1021/bi991232h. [DOI] [PubMed] [Google Scholar]
- 14.Macosko JC, Kim CH, Shin YK. The membrane topology of the fusion peptide region of influenza hemagglutinin determined by spin-labeling EPR. J Mol Biol. 1997;267:1139–1148. doi: 10.1006/jmbi.1997.0931. [DOI] [PubMed] [Google Scholar]
- 15.Jaroniec CP, et al. Structure and dynamics of micelle-associated human immunodeficiency virus gp41 fusion domain. Biochemistry. 2005;44:16167–16180. doi: 10.1021/bi051672a. [DOI] [PubMed] [Google Scholar]
- 16.Lai AL, Park H, White JM, Tamm LK. Fusion peptide of influenza hemagglutinin requires a fixed angle boomerang structure for activity. J Biol Chem. 2006;281:5760–5770. doi: 10.1074/jbc.M512280200. [DOI] [PubMed] [Google Scholar]
- 17.Li Y, et al. Membrane structures of the hemifusion-inducing fusion peptide mutant G1S and the fusion-blocking mutant G1V of influenza virus hemagglutinin suggest a mechanism for pore opening in membrane fusion. J Virol. 2005;79:12065–12076. doi: 10.1128/JVI.79.18.12065-12076.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lorieau JL, Louis JM, Bax A. The complete influenza hemagglutinin fusion domain adopts a tight helical hairpin arrangement at the lipid:water interface. Proc Natl Acad Sci USA. 2010;107:11341–11346. doi: 10.1073/pnas.1006142107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sackett K, et al. Comparative analysis of membrane-associated fusion peptide secondary structure and lipid mixing function of HIV gp41 constructs that model the early pre-hairpin intermediate and final hairpin conformations. J Mol Biol. 2010;397:301–315. doi: 10.1016/j.jmb.2010.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schoch C, Blumenthal R. Role of the fusion peptide sequence in initial stages of influenza hemagglutinin-induced cell fusion. J Biol Chem. 1993;268:9267–9274. [PubMed] [Google Scholar]
- 21.Spruce AE, Iwata A, Almers W. The first milliseconds of the pore formed by a fusogenic viral envelope protein during membrane fusion. Proc Natl Acad Sci USA. 1991;88:3623–3627. doi: 10.1073/pnas.88.9.3623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Steinhauer DA, Wharton SA, Skehel JJ, Wiley DC. Studies of the membrane fusion activities of fusion peptide mutants of influenza virus hemagglutinin. J Virol. 1995;69:6643–6651. doi: 10.1128/jvi.69.11.6643-6651.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tse FW, Iwata A, Almers W. Membrane flux through the pore formed by a fusogenic viral envelope protein during cell fusion. J Cell Biol. 1993;121:543–552. doi: 10.1083/jcb.121.3.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tsurudome M, et al. Lipid interactions of the hemagglutinin HA2 NH2-terminal segment during influenza virus-induced membrane fusion. J Biol Chem. 1992;267:20225–20232. [PubMed] [Google Scholar]
- 25.Han X, Tamm LK. A host–guest system to study structure–function relationships of membrane fusion peptides. Proc Natl Acad Sci USA. 2000;97:13097–13102. doi: 10.1073/pnas.230212097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Qiang W, Sun Y, Weliky DP. A strong correlation between fusogenicity and membrane insertion depth of the HIV fusion peptide. Proc Natl Acad Sci USA. 2009;106:15314–15319. doi: 10.1073/pnas.0907360106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li Y, Tamm LK. Structure and plasticity of the human immunodeficiency virus gp41 fusion domain in lipid micelles and bilayers. Biophys J. 2007;93:876–885. doi: 10.1529/biophysj.106.102335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rafalski M, Lear JD, DeGrado WF. Phospholipid interactions of synthetic peptides representing the N-terminus of HIV gp41. Biochemistry. 1990;29:7917–7922. doi: 10.1021/bi00486a020. [DOI] [PubMed] [Google Scholar]
- 29.Martin I, Schaal H, Scheid A, Ruysschaert JM. Lipid membrane fusion induced by the human immunodeficiency virus type 1 gp41 N-terminal extremity is determined by its orientation in the lipid bilayer. J Virol. 1996;70:298–304. doi: 10.1128/jvi.70.1.298-304.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martin I, et al. Orientation and structure of the NH2-terminal HIV-1 gp41 peptide in fused and aggregated liposomes. Biochim Biophys Acta. 1993;1145:124–133. doi: 10.1016/0005-2736(93)90389-h. [DOI] [PubMed] [Google Scholar]
- 31.Nieva JL, Nir S, Muga A, Goni FM, Wilschut J. Interaction of the HIV-1 fusion peptide with phospholipid vesicles: Different structural requirements for fusion and leakage. Biochemistry. 1994;33:3201–3209. doi: 10.1021/bi00177a009. [DOI] [PubMed] [Google Scholar]
- 32.Ishiguro R, Kimura N, Takahashi S. Orientation of fusion-active synthetic peptides in phospholipid bilayers: Determination by Fourier transform infrared spectroscopy. Biochemistry. 1993;32:9792–9797. doi: 10.1021/bi00088a034. [DOI] [PubMed] [Google Scholar]
- 33.Luneberg J, Martin I, Nussler F, Ruysschaert JM, Herrmann A. Structure and topology of the influenza virus fusion peptide in lipid bilayers. J Biol Chem. 1995;270:27606–27614. doi: 10.1074/jbc.270.46.27606. [DOI] [PubMed] [Google Scholar]
- 34.Gray C, Tatulian SA, Wharton SA, Tamm LK. Effect of the N-terminal glycine on the secondary structure, orientation, and interaction of the influenza hemagglutinin fusion peptide with lipid bilayers. Biophys J. 1996;70:2275–2286. doi: 10.1016/S0006-3495(96)79793-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Miyauchi K, et al. Mutations of conserved glycine residues within the membrane-spanning domain of human immunodeficiency virus type 1 gp41 can inhibit membrane fusion and incorporation of Env onto virions. Jpn J Infect Dis. 2006;59:77–84. [PubMed] [Google Scholar]
- 36.Miyauchi K, et al. Role of the specific amino acid sequence of the membrane-spanning domain of human immunodeficiency virus type 1 in membrane fusion. J Virol. 2005;79:4720–4729. doi: 10.1128/JVI.79.8.4720-4729.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Owens RJ, Burke C, Rose JK. Mutations in the membrane-spanning domain of the human immunodeficiency virus envelope glycoprotein that affect fusion activity. J Virol. 1994;68:570–574. doi: 10.1128/jvi.68.1.570-574.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Taylor GM, Sanders DA. The role of the membrane-spanning domain sequence in glycoprotein-mediated membrane fusion. Mol Biol Cell. 1999;10:2803–2815. doi: 10.1091/mbc.10.9.2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kemble GW, Danieli T, White JM. Lipid-anchored influenza hemagglutinin promotes hemifusion, not complete fusion. Cell. 1994;76:383–391. doi: 10.1016/0092-8674(94)90344-1. [DOI] [PubMed] [Google Scholar]
- 40.Markosyan RM, Cohen FS, Melikyan GB. The lipid-anchored ectodomain of influenza virus hemagglutinin (GPI-HA) is capable of inducing nonenlarging fusion pores. Mol Biol Cell. 2000;11:1143–1152. doi: 10.1091/mbc.11.4.1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Melikyan GB, White JM, Cohen FS. GPI-anchored influenza hemagglutinin induces hemifusion to both red blood cell and planar bilayer membranes. J Cell Biol. 1995;131:679–691. doi: 10.1083/jcb.131.3.679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tong S, Compans RW. Oligomerization, secretion, and biological function of an anchor-free parainfluenza virus type 2 (PI2) fusion protein. Virology. 2000;270:368–376. doi: 10.1006/viro.2000.0286. [DOI] [PubMed] [Google Scholar]
- 43.Nobusawa E, et al. Comparison of complete amino acid sequences and receptor-binding properties among 13 serotypes of hemagglutinins of influenza A viruses. Virology. 1991;182:475–485. doi: 10.1016/0042-6822(91)90588-3. [DOI] [PubMed] [Google Scholar]
- 44.Lemmon MA, Flanagan JM, Treutlein HR, Zhang J, Engelman DM. Sequence specificity in the dimerization of transmembrane alpha-helices. Biochemistry. 1992;31:12719–12725. doi: 10.1021/bi00166a002. [DOI] [PubMed] [Google Scholar]
- 45.Senes A, Engel DE, DeGrado WF. Folding of helical membrane proteins: The role of polar, GxxxG-like and proline motifs. Curr Opin Struct Biol. 2004;14:465–479. doi: 10.1016/j.sbi.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 46.Senes A, Gerstein M, Engelman DM. Statistical analysis of amino acid patterns in transmembrane helices: The GxxxG motif occurs frequently and in association with beta-branched residues at neighboring positions. J Mol Biol. 2000;296:921–936. doi: 10.1006/jmbi.1999.3488. [DOI] [PubMed] [Google Scholar]
- 47.Kim S, et al. Transmembrane glycine zippers: Physiological and pathological roles in membrane proteins. Proc Natl Acad Sci USA. 2005;102:14278–14283. doi: 10.1073/pnas.0501234102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Walters RF, DeGrado WF. Helix-packing motifs in membrane proteins. Proc Natl Acad Sci USA. 2006;103:13658–13663. doi: 10.1073/pnas.0605878103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu Z, Gandhi CS, Rees DC. Structure of a tetrameric MscL in an expanded intermediate state. Nature. 2009;461:120–124. doi: 10.1038/nature08277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stein A, Weber G, Wahl MC, Jahn R. Helical extension of the neuronal SNARE complex into the membrane. Nature. 2009;460:525–528. doi: 10.1038/nature08156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Armstrong RT, Kushnir AS, White JM. The transmembrane domain of influenza hemagglutinin exhibits a stringent length requirement to support the hemifusion to fusion transition. J Cell Biol. 2000;151:425–437. doi: 10.1083/jcb.151.2.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tamm LK, Crane J, Kiessling V. Membrane fusion: A structural perspective on the interplay of lipids and proteins. Curr Opin Struct Biol. 2003;13:453–466. doi: 10.1016/s0959-440x(03)00107-6. [DOI] [PubMed] [Google Scholar]
- 53.Tamm LK. Hypothesis: Spring-loaded boomerang mechanism of influenza hemagglutinin-mediated membrane fusion. Biochim Biophys Acta. 2003;1614:14–23. doi: 10.1016/s0005-2736(03)00159-7. [DOI] [PubMed] [Google Scholar]
- 54.Yin HS, Paterson RG, Wen X, Lamb RA, Jardetzky TS. Structure of the uncleaved ectodomain of the paramyxovirus (hPIV3) fusion protein. Proc Natl Acad Sci USA. 2005;102:9288–9293. doi: 10.1073/pnas.0503989102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yin HS, Wen X, Paterson RG, Lamb RA, Jardetzky TS. Structure of the parainfluenza virus 5 F protein in its metastable, prefusion conformation. Nature. 2006;439:38–44. doi: 10.1038/nature04322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Baker KA, Dutch RE, Lamb RA, Jardetzky TS. Structural basis for paramyxovirus-mediated membrane fusion. Mol Cell. 1999;3:309–319. doi: 10.1016/s1097-2765(00)80458-x. [DOI] [PubMed] [Google Scholar]
- 57.Bissonnette ML, Donald JE, DeGrado WF, Jardetzky TS, Lamb RA. Functional analysis of the transmembrane domain in paramyxovirus F protein-mediated membrane fusion. J Mol Biol. 2009;386:14–36. doi: 10.1016/j.jmb.2008.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Krogh A, Larsson B, von Heijne G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. J Mol Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 59.Gratkowski H, Lear JD, DeGrado WF. Polar side chains drive the association of model transmembrane peptides. Proc Natl Acad Sci USA. 2001;98:880–885. doi: 10.1073/pnas.98.3.880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.DeGrado WF, Gratkowski H, Lear JD. How do helix-helix interactions help determine the folds of membrane proteins? Perspectives from the study of homo-oligomeric helical bundles. Protein Sci. 2003;12:647–665. doi: 10.1110/ps.0236503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Moore DT, Berger BW, DeGrado WF. Protein–protein interactions in the membrane: Sequence, structural, and biological motifs. Structure. 2008;16:991–1001. doi: 10.1016/j.str.2008.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang Y, Kulp DW, Lear JD, DeGrado WF. Experimental and computational evaluation of forces directing the association of transmembrane helices. J Am Chem Soc. 2009;131:11341–11343. doi: 10.1021/ja904625b. [DOI] [PubMed] [Google Scholar]
- 63.Horvath CM, Lamb RA. Studies on the fusion peptide of a paramyxovirus fusion glycoprotein: Roles of conserved residues in cell fusion. J Virol. 1992;66:2443–2455. doi: 10.1128/jvi.66.4.2443-2455.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yin H, et al. Computational design of peptides that target transmembrane helices. Science. 2007;315:1817–1822. doi: 10.1126/science.1136782. [DOI] [PubMed] [Google Scholar]
- 65.Cristian L, Lear JD, DeGrado WF. Determination of membrane protein stability via thermodynamic coupling of folding to thiol-disulfide interchange. Protein Sci. 2003;12:1732–1740. doi: 10.1110/ps.0378603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mukherjee S, Chowdhury P, Gai F. Infrared study of the effect of hydration on the amide I band and aggregation properties of helical peptides. J Phys Chem B. 2007;111:4596–4602. doi: 10.1021/jp0689060. [DOI] [PubMed] [Google Scholar]
- 67.Menikh A, Saleh MT, Gariepy J, Boggs JM. Orientation in lipid bilayers of a synthetic peptide representing the C-terminus of the A1 domain of shiga toxin. A polarized ATR-FTIR study. Biochemistry. 1997;36:15865–15872. doi: 10.1021/bi970944+. [DOI] [PubMed] [Google Scholar]
- 68.Tucker MJ, Getahun Z, Nanda V, DeGrado WF, Gai F. A new method for determining the local environment and orientation of individual side chains of membrane-binding peptides. J Am Chem Soc. 2004;126:5078–5079. doi: 10.1021/ja032015d. [DOI] [PubMed] [Google Scholar]
- 69.Kukol A, Adams PD, Rice LM, Brunger AT, Arkin TI. Experimentally based orientational refinement of membrane protein models: A structure for the influenza A M2 H+ channel. J Mol Biol. 1999;286:951–962. doi: 10.1006/jmbi.1998.2512. [DOI] [PubMed] [Google Scholar]
- 70.Crick FHC. The Fourier transform of a coiled-coil. Acta Crystallogr. 1953;6:685–689. [Google Scholar]
- 71.MacKerell AD, Jr, et al. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J Phys Chem B. 1998;102:3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 72.Lazaridis T. Effective energy function for proteins in lipid membranes. Proteins. 2003;52:176–192. doi: 10.1002/prot.10410. [DOI] [PubMed] [Google Scholar]
- 73.Torii H, Tasumi M. Model calculations on the amide-I infrared bands of globular proteins. J Chem Phys. 1992;96:3379–3387. [Google Scholar]
- 74.Manor J, et al. Gating mechanism of the influenza A M2 channel revealed by 1D and 2D IR spectroscopies. Structure. 2009;17:247–254. doi: 10.1016/j.str.2008.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chang DK, Cheng SF, Kantchev EA, Lin CH, Liu YT. Membrane interaction and structure of the transmembrane domain of influenza hemagglutinin and its fusion peptide complex. BMC Biol. 2008;6:2. doi: 10.1186/1741-7007-6-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Brasseur R, Pillot T, Lins L, Vandekerckhove J, Rosseneu M. Peptides in membranes: Tipping the balance of membrane stability. Trends Biochem Sci. 1997;22:167–171. doi: 10.1016/s0968-0004(97)01047-5. [DOI] [PubMed] [Google Scholar]
- 77.Chernomordik LV, Kozlov MM. Mechanics of membrane fusion. Nat Struct Mol Biol. 2008;15:675–683. doi: 10.1038/nsmb.1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kasson PM, Lindahl E, Pande VS. Atomic-resolution simulations predict a transition state for vesicle fusion defined by contact of a few lipid tails. PLoS Comput Biol. 2010;6:e1000829. doi: 10.1371/journal.pcbi.1000829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lorizate M, Huarte N, Saez-Cirion A, Nieva JL. Interfacial pre-transmembrane domains in viral proteins promoting membrane fusion and fission. Biochim Biophys Acta. 2008;1778:1624–1639. doi: 10.1016/j.bbamem.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yue L, Shang L, Hunter E. Truncation of the membrane-spanning domain of human immunodeficiency virus type 1 envelope glycoprotein defines elements required for fusion, incorporation, and infectivity. J Virol. 2009;83:11588–11598. doi: 10.1128/JVI.00914-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.