Abstract
The core domain of p53 is extremely susceptible to mutations that lead to loss of function. We analysed the stability and DNA-binding activity of such mutants to understand the mechanism of second-site suppressor mutations. Double-mutant cycles show that N239Y and N268D act as ‘global stability’ suppressors by increasing the stability of the cancer mutants G245S and V143A—the free energy changes are additive. Conversely, the suppressor H168R is specific for the R249S mutation: despite destabilizing wild type, H168R has virtually no effect on the stability of R249S, but restores its binding affinity for the gadd45 promoter. NMR structural comparisons of R249S/H168R and R249S/T123A/H168R with wild type and R249S show that H168R reverts some of the structural changes induced by R249S. These results have implications for possible drug therapy to restore the function of tumorigenic mutants of p53: the function of mutants such as V143A and G245S is theoretically possible to restore by small molecules that simply bind to and hence stabilize the native structure, whereas R249S requires alteration of its mutant native structure.
Keywords: DNA/folding/NMR/protein/stability
Introduction
The p53 tumour suppressor protein is a sequence-specific transcription factor whose function is to maintain genome integrity. Inactivation of p53 by mutation is a key molecular event, which is detected in >50% of all human cancers. In response to stress insults, the p53 tumour suppressor protein activates a network of genes whose products mediate vital biological functions, the most critical of which are cell cycle arrest and apoptosis (Harris, 1996; Ko and Prives, 1996; Prives and Hall, 1999). Missense single-point mutations are the most common alterations in the p53 gene (http://www.iarc.fr/p53homepage.html; Levine, 1997) that give rise to a faulty protein. The majority of these mutations (96%) are mapped to the DNA-binding domain of p53 protein with very few mutations reported in the N- or C–terminal domain (http://www.iarc.fr/p53homepage.html). The most frequent missense mutations associated with cancer affect six amino acid residues (R175, G245, R248, R249, R273 and R282) known as ‘hot spots’. The tumour-derived mutants fall into two distinct categories, based on the crystal structure of the p53 DNA-binding domain in a complex with the DNA consensus sequence (Cho et al., 1994). The large majority of the mutations affect residues that are critical for maintaining the structural fold of this highly conserved DNA-binding (core) domain (Cho et al., 1994; Soussi and May, 1996; Walker et al., 1999). The inability of this class of ‘structural’ mutants to bind DNA has been ascribed to structural defects spanning from small partial to global destabilization and unfolding (Brachmann et al., 1998). On the other hand, a second class of ‘DNA-contact’ or ‘functional’ mutants is known to affect residues that are directly involved in protein–DNA interactions (Cho et al., 1994; Wieczorek et al., 1996; Brachmann et al., 1998). There are second-site suppressor mutants that have been selected in yeast that reversed the effect of common p53 cancer mutations (Brachmann et al., 1998). The restoration of p53 function was ascribed from molecular modelling either to introduction of additional DNA contacts or to enhancement of the stability of the folded state (Brachmann et al., 1998). Here, we investigate the molecular basis of the structural restoration of function to the common p53 cancer-associated mutations on introducing second-site suppressor mutations. We used double-mutant cycles (Carter et al., 1984; Serrano et al., 1990; Horovitz, 1996), DNA-binding and NMR spectroscopy studies.
Results
Single amino acid substitutions
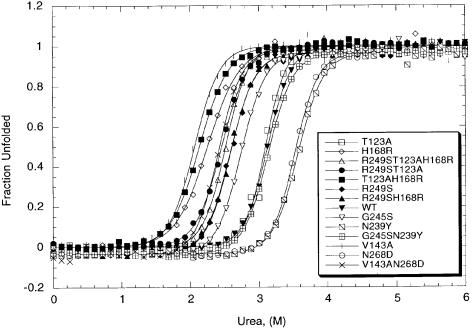
A system for analysis of the stability of human p53 DNA-binding domain (amino acids 94–312) and its mutants has been developed (Bullock et al., 1997) and used to design a p53 variant with enhanced stability and unimpaired DNA binding (Nikolova et al., 1998). The thermodynamic parameters of common missense tumorigenic and selected single suppressor mutations were determined by this procedure and compared with those of the native p53 DNA-binding domain (Table I; Figures 1 and 2). The concentration of urea at which half of the wild-type p53 core domain is unfolded (the midpoint) at 10°C was calculated to be 3.13 ± 0.01 M, the m value 3.25 ± 0.12 kcal/mol/M, and the free energy of unfolding in the absence of denaturant, ΔGH2OD–N, 9.77 kcal/mol. The stability of the single mutants ranged from 3.34 kcal/mol less stable (V143A) to –1.37 kcal/mol more stable (N239Y) than wild-type p53. The most destabilizing suppressor mutation was H168R, which lowered stability by 2.75 kcal/mol. The most stable single suppressor mutations were N239Y and N268D, which increased stability by –1.37 and –1.21 kcal/mol, respectively. The suppressor mutant T123A was nearly as stable as the wild-type p53. All m values (except in two cases) were similar to that for wild-type p53 within experimental error.
Table I. Changes in the free energies of urea-induced unfolding of p53 DNA-binding domain mutants.
| p53 variant | m (kcal/mol/M)a | [Urea]50% (M) | ΔGH2OD–N (kcal/mol) | ΔΔGH2OD–N (kcal/mol) | ΣΔΔGH2OD–N (kcal/mol)b |
|---|---|---|---|---|---|
| WT | 3.25 ± 0.12 | 3.13 ± 0.01 | 9.77 ± 0.16 | ||
| T123A | 3.76 ± 0.19 | 3.09 ± 0.01 | 9.64 ± 0.16 | 0.13 ± 0.22 | |
| V143A | 3.20 ± 0.21 | 2.06 ± 0.01 | 6.42 ± 0.13 | 3.34 ± 0.20 | |
| H168R | 2.40 ± 0.11 | 2.25 ± 0.01 | 7.02 ± 0.13 | 2.75 ± 0.21 | |
| G245S | 3.15 ± 0.12 | 2.74 ± 0.01 | 8.55 ± 0.15 | 1.22 ± 0.22 | |
| R249S | 3.13 ± 0.13 | 2.59 ± 0.01 | 8.08 ± 0.14 | 1.69 ± 0.21 | |
| N239Y | 3.01 ± 0.11 | 3.57 ± 0.01 | 11.14 ± 0.17 | –1.37 ± 0.23 | |
| N268D | 3.27 ± 0.10 | 3.52 ± 0.01 | 10.98 ± 0.17 | –1.21 ± 0.23 | |
| V143A/N268D | 3.35 ± 0.13 | 2.41 ± 0.01 | 7.52 ± 0.14 | 2.25 ± 0.21 | 2.13 |
| G245S/N239Y | 3.07 ± 0.06 | 3.13 ± 0.01 | 9.77 ± 0.16 | 0.00 ± 0.22 | –0.14 |
| R249S/T123A | 2.92 ± 0.08 | 2.46 ± 0.07 | 7.68 ± 0.19 | 2.09 ± 0.24 | 1.82 |
| R249S/H168R | 3.05 ± 0.07 | 2.58 ± 0.01 | 8.05 ± 0.14 | 1.72 ± 0.21 | 4.46 |
| T123A/H168R | 2.65 ± 0.07 | 2.13 ± 0.01 | 6.65 ± 0.13 | 3.12 ± 0.20 | 2.88 |
| R249S/T123A/H168R | 3.44 ± 0.10 | 2.52 ± 0.01 | 7.86 ± 0.14 | 1.91 ± 0.21 | 4.57 |
Data were fitted to the equation ΔGDD–N = ΔGH2OD–N – m [D], where ΔGDD–N is the free energy of unfolding in the presence of denaturant and ΔG is the free energy of unfolding in water, using the transformation in equation (1). ΔGH2OD–N = m [D]50% and ΔΔGH2OD–N = <m>Δ[D]50%. The standard errors of fitting for ΔGH2OD–N and ΔΔGH2OD–N are indicated.
aThe mean m value (<m>) of 14 denaturation curves (at 10°C in 50 mM sodium phosphate/5 mM DTT at pH 7.2) is 3.12 ± 0.088 kcal/mol and was used to calculate ΔGH2OD–N and ΔΔGH2OD–N in the table.
bSums of ΔΔGH2OD–N for individual mutations in the double and triple mutants.
Fig. 1. Urea-induced unfolding of human wild-type p53 and some mutant proteins. The experiments were performed at 10°C in 50 mM sodium phosphate and 5 mM DTT (pH 7.2). Protein concentration was 2 μM.
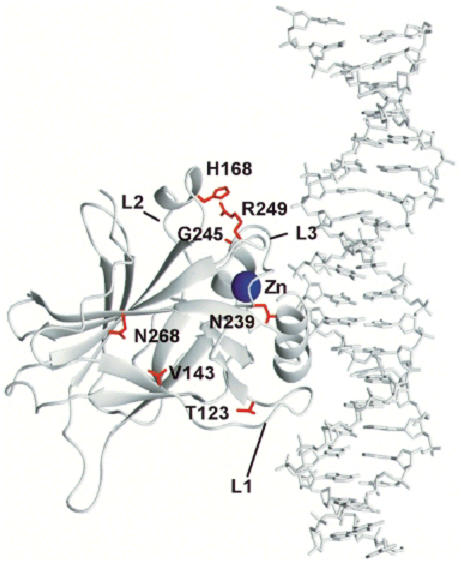
Fig. 2. Schematic model of the human p53 core domain tumour suppressor protein–DNA consensus sequence complex. The PDB (1tsr) co-ordinates were used to display only chain B of p53 (amino acids 94–312) in a complex with the DNA double helix (Cho et al., 1994). The mutated residues are shown in red and the zinc atom in blue. Figures 2 and 4 were made using the program MOLMOL (Koradi et al., 1996).
Additivity of free energy changes on mutations
Five double mutants and one triple mutant were constructed in order to evaluate quantitatively the thermodynamic stability of mutations most commonly found in cancer that also contained a second- and/or third-site suppressor mutation (Table I). All multiple mutants were destabilized relative to wild-type p53 except the double mutant G245S/N239Y. The incorporation of the secondary mutation into the cancer mutants V143A and G245S gave double mutants (V143A/N268D and G245S/N239Y) that were 1.09 and 1.22 kcal/mol, respectively, more stable than the single cancer mutants, but introduction of the suppressor mutation T123A into the cancer mutant R249S had no appreciable effect on stability. The mutants R249S/T123A and R249S/T123A/H168R were 0.22 and 0.40 kcal/mol, respectively, less stable than R249S, which was destabilized by 1.69 kcal/mol, relative to wild-type p53. The most interesting of all suppressor mutants was H168R. The stability of the double mutant R249S/H168R was nearly identical to that of the cancer mutant R249S (destabilized by 1.72 and 1.69 kcal/mol, respectively), yet the stability of the single suppressor H168R was 1.06 kcal/mol lower than that of the cancer mutant.
The sums of the individual changes in free energy on mutation are listed in Table I. The change for a multiple mutant should be equal to the sum of energy changes of the individual parental single mutations if the mutations do not interact with each other (Carter et al., 1984). This has been frequently observed for residues that are far apart in the structure and do not interact directly, or interact indirectly via structural reorganization. If, however, the residues interact directly or indirectly via structural changes, then the effects of mutations should not be additive. Of the six multiple mutants subjected to this analysis, four (R249S/T123A, V143A/N268D, T123A/H168R and G245S/N239Y) exhibited near additive effects and two (R249S/H168R and R249S/T123A/H168R) had non-additive effects. Most of the mutants are far apart, but H168R and R249S are nearly in contact (Figure 2).
DNA binding
In order to evaluate the restoration of wild-type p53 function to cancer-associated mutants, we measured the binding of a specific DNA sequence to all mutant p53 proteins. A biotinylated double-stranded gadd45 30mer oligonucleotide was immobilized into a streptavidin sensor chip. A fixed volume of p53 protein was injected at a constant flow rate and the binding was monitored by the BIAcore instrument (Table II). The concentration of wild-type p53 core domain for 50% binding was estimated to be 100 nM at 20°C in 50 mM Tris–HCl pH 7.2 and 5 mM dithiothreitol (DTT).
Table II. Binding of wild-type and mutant p53 core domain proteins to double-stranded gadd45 DNA.
| p53 core domaina | [p53]50% (nM)b |
|---|---|
| WT | 100 |
| T123A | 180 |
| V143A | 150 |
| H168Rc | 190 |
| G245S | 160 |
| R249S | >1000 |
| N239Y | 190 |
| N268D | 160 |
| V143A/N268D | 160 |
| G245S/N239Y | 130 |
| R249S/T123A | >1000 |
| R249S/H168R | 170 |
| R249S/T123A/H168R | 170 |
aMutants are indicated by the wild-type residue, followed by the sequence of the mutation. Cancer mutants are in bold.
bCalculation of p53 core domain that gives 50% binding to the DNA. The experiments were performed at 20°C in 50 mM Tris pH 7.2 in the presence of 5 mM DTT. The protein concentration ranged from 0.03 to 1.0 μM. Biotinylated double-stranded gadd 45 oligonucleotide was injected onto the streptavidin-coated chip, resulting in 400–800 response units (RU) immobilized. The data were fitted to the two-state transition curve equation using the program KALEIDAGRAPH and the best fit was used to calculate the DNA-binding activity.
cThis mutation is cancer associated according to the p53 database (http://www.iarc.fr), but in this study was used as a suppressor mutation.
The cancer mutants have reduced affinity for binding of the gadd45 promoter, with the structural mutant R249S having no detectable binding activity at 20°C (Table II). The two cancer mutants (V143A and G245S) had concentrations for 50% binding of 150 and 160 nM, respectively. The suppressor mutants as a group also exhibited reduced DNA binding to gadd45 promoter. The concentrations for 50% binding ranged from 160 (N268D) to 190 nM (H168R). It should be noted that H168R was the only mutation found in cancers in the group of four suppressors tested. The other two mutants (T123A and N239Y) had values of 180 and 190 nM, respectively, for concentrations for 50% binding. The affinities of the multiple mutations ranged from no detectable binding (R249S/T123A) to 170 nM (R249S/T123A/H168R). The most remarkable observation was that of the double mutant R249S/H168R, which manifested the most dramatic rescue effect in terms of DNA binding relative to the two single mutants R249S and H168R, both of which were found in human cancers. While R249S has no detectable binding, the introduction of suppressor mutant H168R into R249S rescued the DNA-binding affinity to a concentration of 170 nM for 50% binding of R249S/H168R. On the other hand, the suppressor mutant T123A had no apparent effect on the DNA binding of R249S (Table II). The double mutant R249S/T123A and R249S had virtually no affinity for gadd45 promoter. The triple mutant R249S/T123A/H168R and the double mutant R249S/H168R had near identical DNA binding to gadd45 promoter (Table II). The effect of the suppressor mutation N268D on the DNA binding of V143A was negligible. Incorporation of the suppressor mutant N239Y into the cancer mutation G245S had a beneficial effect and resulted in a small increase of DNA binding of the double mutant G245S/N239Y.
NMR structural studies
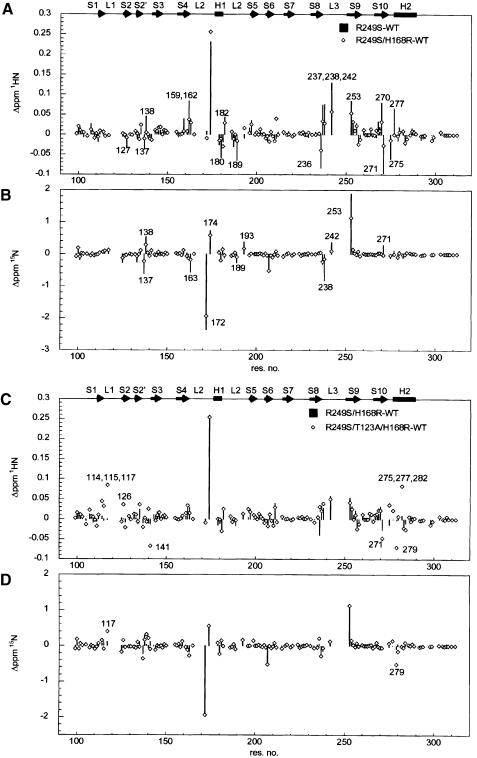
To investigate the structural basis of how the suppressor mutation H168R restores DNA-binding activity of the mutant R249S, we followed structural changes of p53 core domain mutants by NMR spectroscopy, using the deviation of backbone amide 1H and 15N chemical shifts from wild-type values on mutation. 1H–15N correlation spectra were acquired for the mutants R249S, R249S/H168R and R249S/H168R/T123A and compared with those for wild type. Based on the assignment of wild type and R249S (Wong et al., 1999; B.DeDecker, K.–B.Wong and A.R.Fersht, unpublished data), backbone resonances of the suppressor mutants were assigned by comparison of spectra of mutants with those of wild type and R249S. Deviation of amide chemical shifts for the mutants are plotted against sequence position in Figure 3.
Fig. 3. Deviation of amide 1H (A, C), 15N (B, D) chemical shifts from wild-type values. (A and B) Comparison of chemical shift changes between R249S and H168R/R249S. Residues that show recovery of wild-type chemical shifts upon the H168R mutation are indicated in (A) and (B) and colour coded red in Figure 4A. (C and D) Comparison of chemical shift changes between R249S/H168R and R249S/H168R/T123A. Residues with large chemical shift changes upon the third T123A mutation are indicated in (C) and (D) and colour coded red in Figure 4B. The chemical shifts for some residues were not determined (shown as gaps between data points) due to cross-peak overlapping or insufficient sensitivity in NMR experiments.
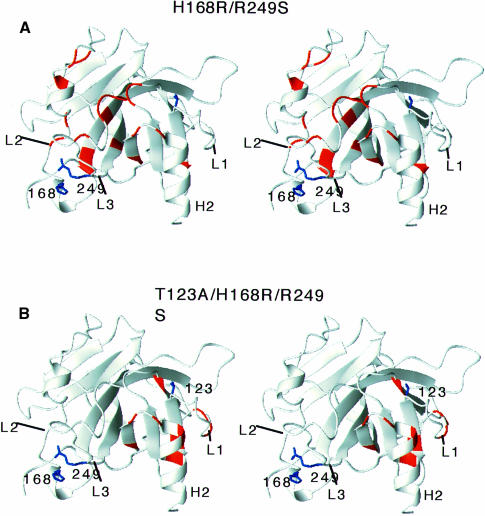
R249S/H168R. Replacement of Arg249 by Ser introduces chemical shift changes for residues in loops L2, L3 and terminal residues of strands 4, 9 and 10 (Wong et al., 1999). A number of these chemical shift changes of R249S were partially or completely reverted upon the addition of the second mutation H168R (Figure 3A and B). Recovery of the wild-type chemical shifts was found predominantly in L2 and L3 (Figure 4A). The fact that some residues of R249S/H168R still retain chemical shift deviation from wild-type values suggests that the recovery of R249S/H168R is a partial one.
Fig. 4. Structural changes introduced by (A) the second suppressor mutation H168R and (B) the third mutation T123A. Chemical shifts of R249S, R249S/H168R and R249S/H168R/T123A were compared (see Figure 3).
R249S/H168R/T123A. T123A did not introduce major chemical shift changes in H168R/R249S. Amide chemical shifts of R249S/H168R and R249S/H168R/T123A are similar (Figure 3C and D). Chemical shift changes were mainly found in the loop–sheet–helix region (Figure 4B), which is localized around the site of mutation. It is noteworthy that the mutation of T123A does not affect the structure in the L2–L3 loop.
Discussion
Malfunction of the p53 tumour-associated mutants is caused either by structural destabilization and unfolding of the native global fold of the core or by distortion of the conformation required for DNA binding, or both. In theory, improving the stability of the fold or re-establishing the proper conformation should restore biological function to p53 mutant proteins associated with human cancers. We tested the above hypothesis by dissecting the stability, DNA binding and structural changes of p53 core domain cancer-associated mutants in the presence of second-site (suppressor) mutants.
Suppressor mutant N268D rescues cancer mutant V143A by stabilizing the global fold of the p53 core
Val143 is located in strand S3 of the β–sandwich (Table III). This residue is deeply buried and participates in the packing of the hydrophobic core of the fold (Cho et al., 1994). This variant represents an example of the group of structural mutants that is temperature sensitive. DNA-binding and transcriptional activity of V143A were not detected at physiological temperature but were observed at lower temperatures (Zhang et al., 1994). Truncation of two methyl groups from Val to Ala143 dramatically destabilized the fold by 3.34 kcal/mol (Table I). The V143A mutation introduces structural changes in almost all residues in the β–sandwich and hence it is likely to affect the packing of the hydrophobic core (Wong et al., 1999). Similar substitutions in other monomeric proteins have been reported to cause destabilization by 0.3–4.7 kcal/mol, depending upon the structural contexts and the packing density in the vicinity of the mutated residue (Shortle et al., 1990; Sauer and Lim, 1992; Serrano et al., 1992; Buckle et al., 1993; Jackson et al., 1993). The decreased stability of V143A causes its temperature sensitivity. At 37°C the V143A mutant is estimated to be ∼80% denatured (A.N.Bullock and A.R.Fersht, unpublished data). At lower temperatures the V143A variant may retain a sufficient population of folded species, allowing the protein to be at least partially functional. The V143A protein gained 1.1 kcal/mol in stability and retained the DNA binding by having the second-site suppressor N268D (Tables I and II; Figure 1). The enhanced stability of V143A/N268D favourably shifts the equilibrium towards the native state relative to V143A, thus providing a mechanism for restoring the function of p53 mutant by second-site suppressor mutation.
Table III. Key characteristics of residues of human p53 protein used in the study.
| p53 core domain residues and mutationsa | Location in the structure | Solvent accessibility of side chain in wild type (%)b | Cancer occurrence of mutant p53 (%)c | Occurrence of wt residue in the p53 family (%)d |
|---|---|---|---|---|
| T123A | End of L1 loop, adjacent to S2–S2′ hairpin | 46.0 | 0 | 92 |
| V143A | S3 strand, hydrophobic core of the β-sandwich, closest to the DNA-binding interface and near the loop–sheet–helix motif | 0.2 | 0.5 | 69 |
| H168R | L2 loop between β-strands S4 and S5, near the zinc binding site and α-helix | 34.2 | 0.4 | 77 |
| G245S | L3 loop, between β-strands S8 and S9, critical for backbone conformation (allowing formation of 2 H-bonds with C247, a zinc ligand and R249) | 34.5 | 3.4 | 100 |
| R249S | L3 loop, adjacent to the minor groove contact residue Arg248, between β-strands S8 and S9 in the sandwich | 13.4 | 3.8 | 96 |
| N239Y | L3 loop between β-strands S8 and S9, adjacent to the zinc co-ordinating Cys238 and Cys 242 in the same loop | 17.7 | 0.6 | 93 |
| N268D | S10 strand, which participates in the small three-stranded sheet and in the β-sandwich | 13.8 | 0.07 | 61 |
aThe position according to the sequence of the human 53 protein and the PDB (1tsr).
bSolvent accessibility (%), defined as solvent-accessible surface area of the amino acid residue in its parent protein calculated by WHATIF, divided by the solvent accessibility of that residue in an extended Ala-X-Ala peptide.
cThe relative frequency of occurrence of the mutant residue found in cancer was calculated from the p53 database.
dThe frequency of occurrence of wt residue (%) in other p53 family proteins was calculated based on the sequence alignment of 28 different species using human p53 residue as a reference (Soussi and May, 1996). We also used the sequences of human p40, human p51, human p73, human p63 and rat Ket proteins.
The suppressor, N268D, was 1.21 kcal/mol more stable than wild-type p53 (Table I). Residue Asn268 is located in strand S10 of the β-sandwich (Figure 2; Table III) and its substitution to Asp was suggested to participate in a hydrogen-bonding network that might keep the two sheets of the β-sandwich together and, therefore, result in a more stable fold (Brachmann et al., 1998).
Suppressor N239Y restores wild-type stability to G245S
The cancer-associated hot spot mutation G245S is destabilized by 1.22 kcal/mol relative to wild-type p53, and has free energy of unfolding in the absence of denaturant of 8.55 kcal/mol (Table I). This residue is located in the L3 loop of the β-sandwich and is partially buried in the core domain (Figure 2; Table III). In the wild-type structure, it adopts an unusual backbone conformation, which facilitates the formation of two hydrogen bonds between the backbone amide of Gly245 and the backbone carbonyl of Cys242 (one of the three Cys residues involved in zinc binding), and between the backbone of Gly245 and the guanidinium group of Arg249 (Cho et al., 1994). Structural changes are found extensively in the L2 and L3 loops, suggesting distortion of the conformation required for DNA binding (Wong et al., 1999), consistent with the decreased DNA binding observed in this study (Table II). The mechanism of inactivation of p53 by G245S is more complex. It is probably caused by a combination of decreased stability of the fold and distortion of the conformation critical for DNA binding. The N239Y mutation recovered the stability of G245S back to wild-type level and improved the DNA binding, suggesting a plausible explanation for how N239Y acts as a suppressor.
Suppressor mutation H168R may recover the DNA binding of R249S by partially restoring wild-type conformation in loops L2 and L3
Both R249S and H168R are cancer-associated mutants of p53. In the wild-type structure, Arg249 is a well-buried residue (Figure 2; Table III) whose replacement with a number of other amino acids results in a complete loss of function (Cho et al., 1994). The mutation of R249S destabilized the protein by 1.69 kcal/mol and abolished the DNA binding (Tables I and II). The suppressor mutation H168R destabilized wild-type protein by 2.75 kcal/mol and decreased the DNA binding (Tables I and II). These effects are probably caused by the introduction of a guanidinium group that may affect the packing interactions between the L2 and L3 loops. However, the introduction of H168R to the cancer ‘hot spot’ R249S resulted in a double mutant with dramatically rescued DNA binding for gadd45 promoter. The insertion of H168R into R249S did not further destabilize the mutant (Table I). To understand better how H168R salvages the DNA binding of R249S cancer mutant, the structure of R249S/H168R was compared with that of R249S and wild-type p53 core by NMR spectroscopy. Structural changes in R249S involved L2–L3 loops and terminal residues of strands 4, 9 and 10 (Wong et al., 1999). Inter-proton distances and backbone dynamics analysis revealed that the L3 loop is locally unfolded (B.DeDecker and A.R.Fersht, unpublished data), suggesting that the replacement of Arg249 by Ser disrupts the conformation of L3 required for DNA binding. Here, we have shown that in the R249S/H168R mutant the second mutation H168R reverted some of the chemical shift changes in L2 and L3 introduced by R249S. Taken together, our data strongly suggest that H168R restores, at least partially, the wild-type conformation of L2 and L3 that is necessary for DNA binding. A partially recovered structure of R249S/H168R mutant may either (i) adopt a conformation that resembles a hybrid between the wild-type and R249S structure or, more likely, (ii) represent an ensemble of structures that sample active (wild-type-like) conformations and inactive (R249S-like) conformations. The guanidinium group of Arg249 in the wild-type structure plays an important role in stabilizing the structure of L2–L3 loops by salt bridging to the carboxylate group of E171 in L2 and by hydrogen bonding to backbone oxygens of G245 and M246. The side chains of 168 and 249 are in close proximity. It is likely that the mutation of H168R introduces a guanidinium group that mimics the role of Arg249 in wild type and hence restores the conformation of L2–L3 that is required for DNA binding.
T123A does not act as a suppressor for stability of R249S core domain
Mutation of T123A does not affect the stability of the core domain significantly (Table I). On the other hand, it decreases the DNA-binding affinities (Table II). T123A is located in the L1 loop of the loop–sheet–helix motif (Figure 2). In the native structure Thr123 is 46% solvent exposed (Table III). The substitution to Ala resulted in a protein that is only marginally less stable (0.13 kcal/mol) than wild-type p53. Structural changes induced by T123A are localized in the loop–sheet–helix motif, as shown by NMR spectroscopy (Figure 4B). Local structural changes in the loop–sheet–helix motif probably compromise the DNA-binding activities of p53 core domain, consistent with the impaired DNA binding (Table II). In contrast to the H168R mutation, T123A does not introduce structural changes in L2 and L3 loops. These findings, along with the observed lack of DNA binding for R249S/T123A (Table II), suggest that T123A does not restore the conformation required for DNA binding. Using a full-length p53 construct, Brachmann et al. (1998) have shown that both T123A and H168R mutations were required to suppress R249S and yield a functional p53 molecule in a yeast-based assay. From the present studies of p53 core domain, it is unclear how T123A plays a role as a suppressor for the R249S mutation. Modelling studies of two p53 core domain molecules bound to DNA suggest that Thr140, Glu198, Gly199 and Glu224 participate in the dimer interface formation (Cho et al., 1994). T123A co-localizes with these residues constituting the protein–protein interface. It might be possible that T123A mediates its ‘suppressor’ effect in full-length p53 via domain–domain interactions.
Compensatory and additivity effects of multiple mutations on the stability of p53 core domain
During molecular evolution, compensatory neutral mutations have evolved as a salvage mechanism against deleterious changes in the functional properties of proteins (Kimura, 1991; Jucovic and Poteete, 1998). These are defined as a pair of substitutions at different sites in a protein that have a detrimental effect individually but restore normal function in combination. This type of effect is reported to occur between amino acid residues that are in close proximity to each other in the native (folded) state of a protein (Kimura, 1991). Multiple mutations involving two or more substitutions may also have a cumulative effect, the net result of which equals the sum of the individual effects of the mutations, i.e. additivity effect (Wells, 1990; Serrano et al., 1993). In the former case, while two or more individual amino acid replacements may have mutually compensatory effects that revert the abnormal effect of the individual mutations, the effect on global stability or function may remain non-additive (Wells, 1990; Mateu and Fersht, 1999).
The thermodynamic analysis of multiple mutants of human p53 core domain (Table I; Figure 2) showed that these mutants fall into two different categories. The single (suppressor) mutation, H168R, with the greatest compensatory effect for functional repair (but not for stability) of R249S, falls in the group that represents the non-additivity effect. The sum of the stability of single mutants (H168R and R249S), 4.46 kcal/mol, was not equal to the stability of the double mutant (R249S/H168R), 1.72 kcal/mol. This non-additivity is in agreement with the NMR structural observations for the two residues in R249S/H168R, which are in very close proximity though not in direct contact (Figures 2 and 4A). In the wild-type structure, Nɛ of His168 is 3.16 Å away from the NH group of Arg249. Similarly, the triple mutant R249S/T123A/H168R was destabilized by 1.91 kcal/mol compared with the expected additivity value of 4.57 kcal/mol. Here T123 and R249 are ∼16 Å apart while T123 and H168 are ∼24 Å apart. The rest of the double mutants exhibited near additive effects on stability, consistent with the observed distances between the residues concerned.
Suppressor mutants and their role in restoring function to tumorigenic p53 variants: potential implications for cancer therapy
Understanding the structural mechanism of how suppressor mutations rescue the function of p53 cancer mutants could provide insights for the discovery of drugs against cancer. The present study highlights two mechanisms of restoring function to cancer-associated mutants: either by improving the stability or by restoring the conformation for DNA binding. For mutants that inactivate p53 by destabilizing the global fold, drugs improving the stability of p53 might be used as a ‘global suppressor’ to restore the function of p53 mutants. All that is required is for a small molecule to bind to the native structure of p53 and not the denatured state, and that the binding does not inhibit the activity of p53. The law of mass action then dictates that the denatured state is converted to the active native state. Other cancer-associated mutations, such as R249S, affect the required conformation for DNA binding and in some cases also destabilize the native fold. For this class of mutants, restoring the stability will not be sufficient to rescue the function of p53. Drugs targeting this class of mutants must also restore the conformation for DNA binding.
Materials and methods
Site-directed mutagenesis, protein expression and purification
The human p53 DNA-binding domain (residues 94–312) was produced from the plasmid pT7hp53 in Escherichia coli as described (Bullock et al., 1997). Single and multiple mutations were introduced by QuickChange site-directed mutagenesis kit (Stratagene, La Jolla, CA). The oligonucleotides were synthesized by Operon Technologies, Inc. (Alameda, CA). All mutants were sequenced, expressed and purified to homogeneity as described (Bullock et al., 1997; Nikolova et al., 1998). Proteins were purified using HiTrap SP and heparin columns (Pharmacia) and elution with a 0–1 M NaCl gradient in 50 mM Tris–HCl pH 7.2 and 5 mM DTT. Protein concentrations were determined spectrophotometrically using an extinction coefficient ɛ280 = 17 130, calculated by the method of Gill and von Hippel (1989). The purified proteins were analysed by electrospray mass spectroscopy and SDS Phastgel electrophoresis. All proteins were flash frozen in small aliquots in liquid nitrogen after purification and stored at –80°C.
Equilibrium urea denaturation
Urea-mediated equilibrium unfolding of p53 core domain was performed at 10°C in 50 mM sodium phosphate buffer and 5 mM DTT at pH 7.2 using an Aminco-Bowman luminescence spectrofluorimeter with excitation at 280 nm and emission scans between 300 and 370 nm as described (Bullock et al., 1997). DTT was included in all solutions in order to keep the 10 cysteine residues reduced. For each data point of the denaturation curve, 100 μl of 18 μM protein in the above buffer were added to 800 μl of the corresponding denaturant solution. Samples were incubated overnight at 10°C constant temperature and analysed at 10°C in 1 ml thermostatted cuvettes. The temperature of each sample was monitored throughout the measurements, using a thermocouple. All the proteins unfolded reversibly at 10°C and followed two-state co-operative transitions from native to denatured state. Experimental data were fitted to a two-state transition curve for which the fluorescence of the folded and unfolded states is dependent on the denaturant concentration according to equation (1):
F=(αN+βN[D]) + (αD+βD[D])exp{m([D] – [D]50%)/RT}1 + exp{m([D] – [D]50%)/RT} (1)
where F is the fluorescence at a given concentration of urea, [U], αN and αD are the intercepts, and βN and βD are the slopes of the baselines at low and high denaturant concentration, respectively (Clarke and Fersht, 1993). The concentration of denaturant at which half of the protein is denatured is denoted as [D]50%, R is the gas constant, T is the temperature in K, and m is a constant that is proportional to the increase in solvent-accessible surface area between the native and denatured state of the protein (Pace, 1986). The data were fitted to the above equation by non-linear least squares analysis using the KALEIDAGRAPH program (Abelbeck Software, Reading, PA), which gave the values of the linear slope of the transition from native to denatured state, m, and [D]50%, as well as their standard errors.
DNA-binding studies
The capacity of wild-type p53 DNA-binding domain and the mutant proteins to bind dsDNA was tested using surface plasmon resonance binding experiments at 20°C, using immobilized gadd45 promotor on the surface of a streptavidin-coated chip (SA5) and a BIAcore 2000 Pharmacia Biosensor (AB) as described (Nikolova et al., 1998). The sequence of the gadd45 promoter is:
Gadd45: 5′-biotin-GTACAGAACATGTCTAAGCATGCTGGGGAC-3′
Gadd45: 3′-CATGTCTTGTACAGATTCGTACGACCCCTG-5′
The relative change in resonance units was monitored over a period of 500 s. The changes in resonance units upon DNA binding were plotted as a function of protein concentration. The experimental data were fitted to a two-state transition equation using the KALEIDAGRAPH program. The midpoint of the transition curves was used as an estimate of the affinity of p53 core domain for gadd45 promoter.
NMR spectroscopy
15N-labelled samples of wild-type and mutant p53 core domains were prepared as described (Wong et al., 1999). The final sample concentration was 500 μM in 150 mM KCl, 5 mM DTT, 5% (v/v) D2O and 50 mM sodium phosphate buffer at pH 7.1. All 1H, 15N HSQC spectra were acquired at 20°C on a Bruker DRX600 spectrometer equipped with a 5 mm inverse detection triple resonance probe with x, y, z gradient coils. Selective flip-back pulses were applied to minimize attenuation of signals due to solvent exchange (Grzesiek and Bax, 1993). Suppression of the residual water signal was achieved by WATERGATE (Piotto et al., 1992).
Acknowledgments
Acknowledgements
We wish to acknowledge Drs M.Bycroft and A.Bullock for scientific discussions, and Mr M.Proctor for producing protein for the NMR. This work was supported by a grant from the CRC of the UK. P.V.N. is a recipient of a Post-Doctoral Research Fellowship from MRC of Canada, J.H. holds a Marie Curie Research Training Grant of the European Commission and B.D.D. holds a Burroughs-Wellcome Fund Hitchings–Elion Fellowship.
References
- Brachmann R.K., Yu, K., Eby, Y., Pavletich, N.P. and Boeke, J.D. (1998) Genetic selection of intragenic suppressor mutations that reverse the effect of common p53 cancer mutations. EMBO J., 17, 1847–1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckle A.M., Henrick, K. and Fersht, A.R. (1993) Crystal structural analysis of mutations in the hydrophobic cores of barnase. J. Mol. Biol., 234, 847–860. [DOI] [PubMed] [Google Scholar]
- Bullock A.B., Henckel, J., DeDecker, B.S., Johnson, C.M., Nikolova, P.N., Procter, M.R., Lane, D.P. and Fersht, A.R. (1997) Thermodynamic stability of wild-type and mutant p53 core domain. Proc. Natl Acad. Sci. USA, 94, 14338–14342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter P.G., Winter, G., Wilkinson, A.J. and Fersht, A.R. (1984) The use of double mutants to detect structural changes in the active site of the tyrosyl-tRNA synthetase (Bacillus stearothermophilus). Cell, 38, 835–840. [DOI] [PubMed] [Google Scholar]
- Cho Y., Gorina, S., Jeffrey, P.D. and Pavletich, N.P. (1994) Crystal structure of a p53 tumor suppressor–DNA complex: understanding tumorigenic mutations. Science, 265, 346–355. [DOI] [PubMed] [Google Scholar]
- Clarke J. and Fersht, A.R. (1993) Engineering disulfide bonds as probes of the folding pathway of barnase: increasing the stability of proteins against the rate of denaturation. Biochemistry, 32, 4322–4329. [DOI] [PubMed] [Google Scholar]
- Gill C.S. and von Hippel, P.H. (1989) Calculation of protein extinction coefficients from amino acid sequence data. Anal. Biochem., 182, 319–326. [DOI] [PubMed] [Google Scholar]
- Grzesiek S. and Bax, A. (1993) The importance of not saturating H2O in protein NMR. Application to sensitivity enhancement and NOE measurements. J. Am. Chem. Soc., 115, 12593–12594. [Google Scholar]
- Harris C.C. (1996) Structure and function of the p53 tumor suppressor gene: clues for rational cancer therapeutic strategies. J. Natl Cancer Inst., 88, 1442–1455. [DOI] [PubMed] [Google Scholar]
- Horovitz A. (1996) Double-mutant cycles: a poweful tool for analyzing protein structure and function. Fold. Des., 1, R121–R126. [DOI] [PubMed] [Google Scholar]
- Jackson S., Moracci, M., el Masry, N., Johnson, C.M. and Fersht, A.R. (1993) Effect of cavity-creating mutations in the hydrophobic core of chymotrypsin inhibitor 2. Biochemistry, 32, 11259–11269. [DOI] [PubMed] [Google Scholar]
- Jucovic M. and Poteete, A.R. (1998) Delineation of evolutionary salvage pathway by compensatory mutations of a defective lysozyme. Protein Sci., 7, 2200–2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M. (1991) Recent development of the neutral theory viewed from the Wrightian tradition of theoretical population genetics. Proc. Natl Acad. Sci. USA, 88, 5969–5973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko L.J. and Prives, C. (1996) p53: puzzle and paradigm. Genes Dev., 10, 1054–1072. [DOI] [PubMed] [Google Scholar]
- Koradi R., Billeter, M. and Wüthrich, K. (1996) MOLMOL: a program for display and analysis of macromolecular structures. J. Mol. Graph., 14, 51–55. [DOI] [PubMed] [Google Scholar]
- Levine A.J. (1997) p53, the cellular gatekeeper for growth and division. Cell, 88, 323–331. [DOI] [PubMed] [Google Scholar]
- Mateu M.G. and Fersht, A.R. (1999) Mutually compensatory mutations during evolution of the tetramerization domain of tumor suppressor p53 lead to impaired hetero-oligomerization. Proc. Natl Acad. Sci. USA, 96, 3595–3599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolova P.V., Henckel, J., Lane, D.P. and Fersht, A.R. (1998) Semirational design of active tumor suppressor p53 DNA binding domain with enhanced stability. Proc. Natl Acad. Sci. USA, 94, 14675–14680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pace C.N. (1986) Determination and analysis of urea and guanidinium denaturation curves. Methods Enzymol., 131, 266–279. [DOI] [PubMed] [Google Scholar]
- Piotto M., Saudek, V. and Sklenar, V. (1992) Gradient-tailored excitation for single-quantum NMR spectroscopy of aqueous solutions. J. Biomol. NMR, 2, 661–665. [DOI] [PubMed] [Google Scholar]
- Prives C. and Hall, P. (1999) The p53 pathway. J. Pathol., 187, 112–126. [DOI] [PubMed] [Google Scholar]
- Sauer R.T. and Lim, W.A. (1992) Mutational analysis of protein stability. Curr. Opin. Struct. Biol., 2, 46–51. [Google Scholar]
- Serrano L., Horovitz, A., Avron, B., Bycroft, M. and Fersht, A.R. (1990) Estimating the contribution of the engineered surface electrostatic interactions to protein stability by using double-mutant cycles. Biochemistry, 29, 9343–9352. [DOI] [PubMed] [Google Scholar]
- Serrano L., Kellis, J.T.,Jr, Cann, P., Matouscheck, A. and Fersht, A.R. (1992) The folding of an enzyme II. Substructure of barnase and the contribution of different interactions to protein stability. J. Mol. Biol., 224, 783–804. [DOI] [PubMed] [Google Scholar]
- Serrano L., Day, A.G. and Fersht, A.R. (1993) Step-wise mutation of barnase to binase: a procedure for engineering increased stability of proteins and an experimental analysis of the evolution of protein stability. J. Mol. Biol., 233, 305–312. [DOI] [PubMed] [Google Scholar]
- Shortle D., Stites, W.E. and Meeker, A.K. (1990) Contribution of large hydrophobic amino acids to the stability of staphylococcal nuclease. Biochemistry, 29, 8033–8041. [DOI] [PubMed] [Google Scholar]
- Soussi T. and May, P. (1996) Structural aspects of the p53 protein in relation to gene evolution: a second look. J. Mol. Biol., 260, 623–637. [DOI] [PubMed] [Google Scholar]
- Walker D.R., Bond, J.P., Tarone, R.E., Harris, C.C., Makalowski, W., Boguski, M.S. and Greenblatt, M.S. (1999) Evolutionary conservation and somatic mutation hotspot maps of p53: correlation with p53 protein structural and functional features. Oncogene, 19, 211–218. [DOI] [PubMed] [Google Scholar]
- Wells J.A. (1990) Additivity of mutational effects in proteins. Biochemistry, 29, 8509–8517. [DOI] [PubMed] [Google Scholar]
- Wieczorek A.M., Watermann, J.L.F., Watermann, M.J.F. and Halazonetis, T.D. (1996) Structure-base rescue of common tumor-derived p53 mutants. Nature Med., 2, 1143–1146. [DOI] [PubMed] [Google Scholar]
- Wong K.-B., DeDecker, B.S., Freund, S.M.V., Proctor, M.R., Bycroft, M. and Fersht, A.R. (1999) Hot-spot mutants of p53 core domain reveal characteristic local structural changes. Proc. Natl Acad. Sci. USA, 96, 8438–8442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Guo, X.-Y., Hu, G.-Y., Liu, W.-B., Shay, J.W. and Deisseroth, A.B. (1994) A temperature-sensitive mutant of human p53. EMBO J., 13, 2535–2544. [DOI] [PMC free article] [PubMed] [Google Scholar]