Abstract
The imaging genetics approach to studying the genetic basis of disease leverages the individual strengths of both neuroimaging and genetic studies by visualizing and quantifying the brain activation patterns in the context of genetic background. Brain imaging as an intermediate phenotype can help clarify the functional link among genes, the molecular networks in which they participate, and brain circuitry and function. Integrating genetic data from a genome-wide association study (GWAS) with brain imaging as a quantitative trait (QT) phenotype can increase the statistical power to identify risk genes. A QT analysis using brain imaging (DLPFC activation during a working memory task) as a quantitative trait has identified unanticipated risk genes for schizophrenia. Several of these genes (RSRC1, ARHGAP18, ROBO1-ROBO2, GPC1, TNIK, and CTXN3-SLC12A2) have functions related to progenitor cell proliferation, migration, and differentiation, cytoskeleton reorganization, axonal connectivity, and development of forebrain structures. These genes, however, do not function in isolation but rather through gene regulatory networks. To obtain a deeper understanding how the GWAS-identified genes participate in larger gene regulatory networks, we measured correlations among transcript levels in the mouse and human postmortem tissue and performed a gene set enrichment analysis (GSEA) that identified several microRNA associated with schizophrenia (448, 218, 137). The results of such computational approaches can be further validated in animal experiments in which the networks are experimentally studied and perturbed with specific compounds. Glypican 1 and FGF17 mouse models for example, can be used to study such gene regulatory networks. The model demonstrates epistatic interactions between FGF and glypican on brain development and may be a useful model of negative symptom schizophrenia.
Introduction
Genome-wide association studies (GWAS) to date have not provided a genetic “smoking gun” for schizophrenia (Harrison and Weinberger, 2005). Given the known genetic components in the disorder, we hypothesize that it, and indeed most, complex psychiatric disorders arise through small contributions from many polymorphic loci, rather than through disruption of single genes or pathways. Certainly, GWAS results from schizophrenia, bipolar disorder, and major depression studies—in which many loci are found, each with small effect (e.g.,(Committee, 2009; Moskvina et al., 2008))--support such a view. In this sense, we see psychiatric disorders as system-level disruptions of what are large, complex, non-linear networks of gene-, protein- and cell-interactions. While GWAS results may tell us which genes in this network happen to have disease-associated polymorphisms of reasonably high frequency in the human population, it only gives us tiny glimpses of the underlying functional network itself.
Moreover, GWAS techniques create statistical challenges producing anywhere from 100,000 to more than 5,000,000 genotypes per subject (Potkin et al., 2009d). Classical statistical analytical techniques are not designed for situations where the number of variables so grossly outnumbers the number of subjects. In addition to the problem of multiple testing / multiple hypotheses, there are several other important issues that are the current focus of interest in statistical genetics. Examples of active statistical research are: 1) how to analyze “genes” (or chromosomal regions) rather than SNPs, given our primary interest in mapping putative functional elements of the genome rather than simple point variations; 2) how to address gene*gene epistatic interactions (Brzustowicz, 2008; Chapman and Clayton, 2007; Evans et al., 2006; Jiang et al., 2009; Moore, 2008), or gene*environment epigenetic (Clayton and McKeigue, 2001; Glazier et al., 2002; Hoffmann et al., 2009; Lander and Kruglyak, 1995) interactions, or 3) even how to validate a causative or regulatory network (Barabasi, 2007; Hidalgo et al., 2009). Identification and application of the networks is the focus of this paper, using schizophrenia as an example.
Using brain imaging as a quantitative trait greatly increases the statistical power of GWAS (as in (Potkin et al., 2009d)). Neuroimaging as a quantitative trait may identify dimensions of brain function that are more closely related to susceptibility genes than are more subjective assessments of clinical symptoms or features (e.g. (Gottesman and Gould, 2003)). While imaging studies by themselves reveal many aspects of function and dysfunction in neuropsychiatric disorders, their explanatory power may be limited by not considering the genetic bases of brain structure and function, as both are clearly heritable (Kennedy et al., 2003). Integrating genotypic information with brain imaging results can help identify the function of candidate genes at the level of brain function (e.g. (Meyer-Lindenberg and Weinberger, 2006)).
However, the availability of high-throughput genotyping technologies and genomic resources such as HapMap (www.hapmap.org) has made it possible to survey SNP markers throughout the entire genome, and increase the probability of discovering important unanticipated genetic influences. This allows imaging genetics to perform gene discovery - identification of new “candidate” genes related to brain function that would not be discovered by traditional candidate gene approaches (e.g. (Papassotiropoulos et al., 2006; Potkin et al., 2009b; Potkin et al., 2009c)).
Our imaging genetics GWAS approach uses brain imaging as a Quantitative Trait (QT) and determines which genes affect the QT, employing a reverse strategy compared to a candidate gene approach. We do not test a priori hypotheses regarding genetic effects on brain function based on current physiological or pathophysiological knowledge since a major limitation of the candidate gene approach is precisely that we know our current understanding of physiology or pathophysiology is woefully incomplete (Meyer-Lindenberg and Weinberger, 2006; Roffman et al., 2006). Imaging Genetics can visualize brain activation patterns in the context of a whole genome background, thereby synergizing the strengths of each individual approach (Potkin et al., 2009b; Potkin et al., 2009c) and ultimately representing a strategy for risk gene discovery.
Figure 1 depicts our approach: In the pathway from SNP to disease, the human GWAS results can reveal a link between the SNP or gene and the phenotype, but do not illuminate the causative networks of interacting mechanisms. Small but significant differences in allelic frequency in cases and controls are best not interpreted as a particular gene being the causative factor, but instead should be considered to implicate a larger network of genes. A systems biology approach can improve our understanding of the implications of these GWAS findings of relatively small effects within these interacting mechanisms.
Figure 1.
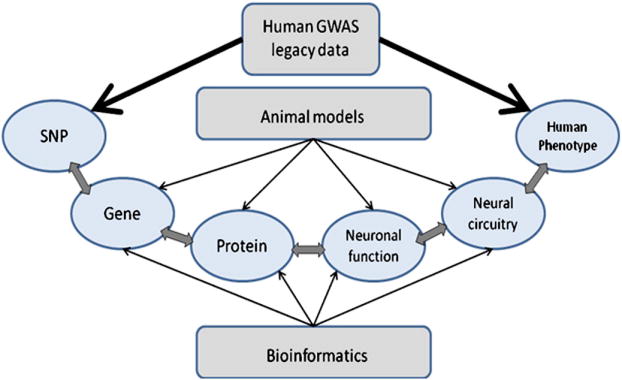
Systems biology addresses links between human phenotype and SNPs originally identified by GWAS.
As an illustration of this approach, we have used activation in the dorsolateral prefrontal cortex (DLPFC) of the middle frontal gyrus,measured using fMRI during a working memory task, as a QT phenotype to identify genes related to schizophrenia that were not anticipated a priori to this study (Potkin et al., 2009b; Potkin et al., 2009c). Employing this approach we have identified several genes related to brain development and stress that had never before been associated with schizophrenia. This is in line with the well-known Common Variants - Common Disease (CVCD) hypothesis that schizophrenia arises through small contributions from many polymorphic loci, rather than through disruption of single genes. Building on these results, we present several computational biology approaches that provide initial steps in identifying putative gene regulatory networks.
Gene regulatory networks can potentially be inferred from expression profiles, the locations of regulatory motifs, and interactions between regulatory targets and MicroRNA (miRNA). A number of methods have been proposed to infer gene regulatory networks from large-scale gene expression data (Eisen et al., 1998). A basic assumption underlying all these methods is that genes interacting together are correlated in their gene expression (positive or negative). Therefore, correlation in gene expression can be used as a measure for inferring gene interactions, using methods such as Boolean network analysis, informatics-based approaches, linear regression, or Bayesian networks (Bansal et al., 2007).
An additional approach for constructing gene interaction networks takes advantage of sequence analysis by searching for the locations of regulatory motifs in the human genome. Previously we have demonstrated the power of comparative genomics for discovering novel regulatory motifs and for identifying individual regulatory motif sites in the human genome (Xie et al., 2005; Xie et al., 2007). Recent availability of over 25 placental mammalian genomes significantly boosts our power for detecting motif sites in the human genome. While these genomes are closely related to each other, and likely share basic regulatory motifs, they are carefully chosen to represent distinct branches of the mammalian evolutionary tree. As such, they are ideal for separating regulatory sequences from neutral sequences (Margulies et al., 2005).
The indirect interaction between regulators and targets of miRNAs can also be determined. MicroRNAs (miRNAs), another important class of regulators of gene expression, are endogenous ~22-nucleotide RNAs that repress gene expression post-transcriptionally (Carthew, 2006). miRNAs are believed to regulate thousands of genes by virtue of base pairing to 3’ untranslated regions (3’UTRs). Individual miRNAs can each affect hundreds of genes. Many of the characterized miRNAs are involved in developmental regulation, including the timing and neuronal asymmetry in worm; brain morphogenesis in zebrafish; and dendritic spine development in mammals (Giraldez et al., 2005; Schratt et al., 2006). Based on a recent survey (Griffiths-Jones et al., 2008), we note that the human genome contains over 500 miRNA genes, many of which are highly or specifically expressed in neural tissues. The function of the brain-related miRNAs and the mechanisms underlying their transcriptional control are beginning to emerge and miRNA expression differences have been found in the frontal and temporal gyri of schizophrenia patients (Beveridge et al., 2008).
These statistical approaches, however, require validation by experimental models. For example, the function of RSRC1 and AHRGAP18 in schizophrenia that we identified through imaging genetics (Potkin et al., 2009b) is largely unidentified and consequently the modification of function that may be therapeutic is unknown. However, perturbation of these candidate genes with compounds that affect genetic expression in animal neurodevelopmental models can help us understand the gene regulatory networks. These in vivo models improve our understanding of the biological significance of the networks identified by the bioinformatic approaches.
We demonstrate the use of existing and novel algorithms to infer gene regulatory networks from heterogeneous datasets (e.g. literature, gene expression and genomic sequences) by employing a gene set based approach for GWAS data instead of individual SNPs or genes (Subramanian et al., 2005). We focus on the specific problem of placing loci identified through GWAS into the context of meaningful networks, the network of genes that directly or indirectly control, or are controlled by, schizophrenia risk genes, and the implication of selected genes identified from these analyses in a mouse model. Improving our understanding of the dynamic networks that underlie these disorders is key to developing interventions that restore the network to its normal regulatory state.
Materials and methods
Quantitative Trait
The full details of the neuroimaging data collection and analysis are available in (Potkin et al., 2009b; Potkin et al., 2009c). We used measures of Blood Oxygenation Level Dependent (BOLD) fMRI signal in the dorsal lateral prefrontal cortex (DLPFC) during the Sternberg Item Recognition Paradigm (SIRP), a heritable (Karlsgodt et al., 2007) working memory task, as a QT in a group of schizophrenia (n = 24 + 64) and healthy control subjects (n = 74) (Potkin et al., 2009b; Potkin et al., 2009c) in the context of GWAS to identify genes related to schizophrenia using a GWAS approach. The brain imaging phenotype was chosen based on its relevance to the neuropsychiatric disorder, e.g. DPLFC activation in the case of schizophrenia. The BOLD signal obtained during the probe condition while holding 3 items in memory contrasted with a one item memorandum was used as a quantitative phenotype in a GWAS.
GWAS analysis
Subjects were genotyped using the Illumina HumanHap370-Duo, providing 370,404 SNPs suitable for later analysis with the fMRI QT (Potkin et al., 2009b). All SNPs that passed quality control checks (Teo, 2008) were included in the GWAS analysis. The simplest model we applied was a General Linear Model (GLM) identifying the effects of SNP alleles or genotypes on the QT, thus determining how genetic variation can be related to phenotypes characterized by brain activation. With our model, we can determine the genes (SNPs) on brain activation (or structure) and then determine if these genetic effects differ by diagnosis, simply adding a term to the GLM as follows:
This model can include other variables, for example, nuisance covariates such as the site from which the subject was recruited, their age or gender, etc. To guard against false positives, loci for further consideration were identified by at least 2 independent SNPs with a P < 10-6 because the conjunction of these results is less likely than a single result alone. See (Potkin et al., 2009d) for discussion of GWAS statistical significance thresholds. This is in keeping with the WTCCC and O’Donovan et al,56 considering genome-wide thresholds of P <10_5 and 10_7 as “moderately strong” and “strong” evidence for an association.
The newly identified risk-producing genes may be involved in pathophysiological neuronal networks; their putative role within their larger genetic networks was initially determined by bioinformatics and computational biology methods.
Gene network identification
In order to obtain deeper understanding of how these genes participated into larger gene regulatory networks we applied two bioinformatics approaches: 1) correlations among transcript levels, and 2) gene set enrichment analysis.
Correlation mapping
Gene interaction networks are inferred from correlations among gene expression (mRNA) datasets. We added prefrontal cortex gene expression data derived from 42 different inbred mouse strains of the BXD recombinant inbred panel (derived from progenitor B6 and D2 strains (Wang et al., 2003)). Genes were clustered into expression networks based on the correlated variation among strains. Expression “neighbors” may represent genes that regulate one or another’s expression, or are controlled by a common regulator. Such clustering is possible because there is sufficient variation in gene expression among the tested strains. The human microarray gene expression methods are fully described in (Shao and Vawter, 2008). Briefly the gene expression values from DLPFC for the Stanley Microarray Cohort were obtained using a Codelink platform in the UCI Functional Genomics laboratory. The DLPFC was Trizol extracted for RNA at the Stanley Institute. At UCI, 105 subjects were received for analysis. High quality arrays were obtained on 27 bipolar subjects, 30 schizophrenia subjects, and 29 controls. The raw expression values were background adjusted, regressed for pH and age and used for downstream correlation analysis and over-representation analysis. The full microarray dataset is available from the Stanley Medical Research Institute upon request (www.stanleyresearch.org).
Gene set enrichment analysis (GSEA)
We adapted gene set enrichment analysis, originally developed for gene expression analysis (Subramanian et al., 2005), to discover candidate genes sets or pathways that likely contribute to schizophrenia. GSEA determines whether a group of genes is over-enriched with SNPs associated with a disease trait compared to the entire genome. It first ranks all genes in the genome according to the association with the quantitative trait or disease (in this case, the p-value of the SNP’s effect on the QT in the 24 SCZ data (Potkin et al., 2009b), or the interaction of diagnosis and SNP on the QT in the SIRP imaging genetics analysis (Potkin et al., 2009c)); it then tests whether a query gene set is enriched with low rank genes (most significant p values) using a Mann-Whitney U or a Kolmogorov-Smirnov test (KS-test). Gene sets are defined based in prior biological knowledge (e.g., canonical pathways, chemical and genetic perturbations) primarily from the MSIG dataset, plus microRNA targets, and transcriptional factors targets curated by us (referred to as C3 motif gene sets (Xie et al., 2005)) and several clusters that were generated by the correlational analysis of the BxD dataset and from the Novartis gene expression atlas (Su et al., 2004). Altogether we tested 9709 gene sets.
Animal models
Animal models can be used to both explore and validate the computational biological approaches. For example, glypican-1 (GPC1) was one of the genes identified in the gene regulatory network based on the GWAS imaging genetics analyses and the BxD data (see results section below). A useful glypican mouse model exists (Aikawa et al., 2008; Ivins et al., 1997; Lander et al., 1996; Litwack et al., 1994). GPC1 encodes a cell surface heparan sulfate proteoglycan (HSPG), a molecule that can act as a co-receptor for growth factors and other signaling molecules, including FGFs, neuregulins, Wnts, BMPs, slits and netrins (Lander et al., 1996; Selleck, 2006; Song and Filmus, 2002). In this analysis, we assessed the effects of knocking out the GPC1 gene on brain development and the epistatic interactions with FGF17. For full methods, see (Jen et al., 2009).
Results
fMRI
In our data, schizophrenics show more (BOLD) activation in the DLPFC than do healthy controls when matched for accuracy performance on the Sternberg Item Recognition Paradigm (SIRP), a working memory task, consistent with cortical inefficiency (Potkin et al., 2009a). The BOLD activation was used as the quantitative phenotype in the GWAS analyses.
Two genes, RSRC1 and ARHGAP18, were identified that had not been previously associated with cognition or schizophrenia (Potkin et al., 2009b). These two genes, based on available annotation software (Ingenuity Pathways Analysis, SWISSPROT and dbSNP), have functions related to prenatal brain development and cell migration to forebrain structures. Their role in cortical development supports the neurodevelopmental hypothesis of schizophrenia. RSRC1 is a unique marker of progenitor cells that are found in the subventricular zone (SVZ) in the developing and postnatal forebrain. These SVZ progenitor cells give rise to EGFr-responsive progenitors which in the presence of TGF-alpha bind to ERB B EGF receptors (Fallon et al., 2000; Rakic and Zecevic, 2003). ARHGAP18 is part of the family of RhoGAP proteins that participate in cell proliferation, migration, intercellular adhesion, cytokinesis, proliferation, differentiation, and apoptosis (Symons, 1996). ARHGAP18 gene products have been linked to RAS and EGFr-mediated proliferation of cells in general (Wells, 1999). Interestingly, both genes have function in prenatal brain development including neural stem cell proliferation in the SVZ and migration to forebrain structures including limbic, striatal, and amygdaloid circuitry.
In a second study using similar methodology with DLPFC activation as a quantitative trait in schizophrenia subjects and matched controls, six additional genes (or chromosomal regions) related to forebrain development and stress response, and affecting prefrontal efficiency, were also identified (ROBO1-ROBO2, TNIK, CTXN3-SLC12A2, POU3F2, TRAF, and GPC1) (Potkin et al., 2009c). Several of these genes are involved in cortical development, especially in the forebrain in midline connections. GPC1 (glypican, slit receptor) and ROBO1-ROBO2 are involved in dorsal forebrain development, specifically neural precursor migration and axonal connectivity (e.g. midline crossing and guidance of neuron axons to prefrontal cortices including DLPFC). TNIK is highly expressed in the brain (Nonaka et al., 2008) and TNIK mRNA was shown to be upregulated in the dorsolateral prefrontal cortex (DLPFC) of schizophrenia patients (Glatt et al., 2005). A SNP in TNIK was in the top 12 hits associated with schizophrenia in the African-American sample case-control analysis from the Molecular Genetics of Schizophrenia (MGS) consortium (Shi et al., 2009). SLC12A2 is involved in regulation of GABA neurotransmission and is differentially expressed in schizophrenia (Dean et al., 2007). CTXN3 (cortexin) is highly enriched in the cortex and increases postnatally. CTXN3-SLC12A2 was found linked to schizophrenia in Lewis et al (Lewis et al., 2003) meta-analysis and lies within the chromosome 5 region implicated in cognitive dysfunction found in schizophrenia (Almasy et al., 2008). These findings are consistent with the previously described abnormal callosal morphometry and cortico-subcortical connectivity (Barch et al., 2001; Brett et al., 2002; D’Esposito et al., 1998; Johnson et al., 2006; McNab and Klingberg, 2008; Tura et al., 2008). The discovery of novel associations between genes and risk for neuropsychiatric disorder offers a powerful impetus to postulate new biological mechanisms as well as support previous neurodevelopmental hypotheses.
Identification of gene networks
Correlation Mapping
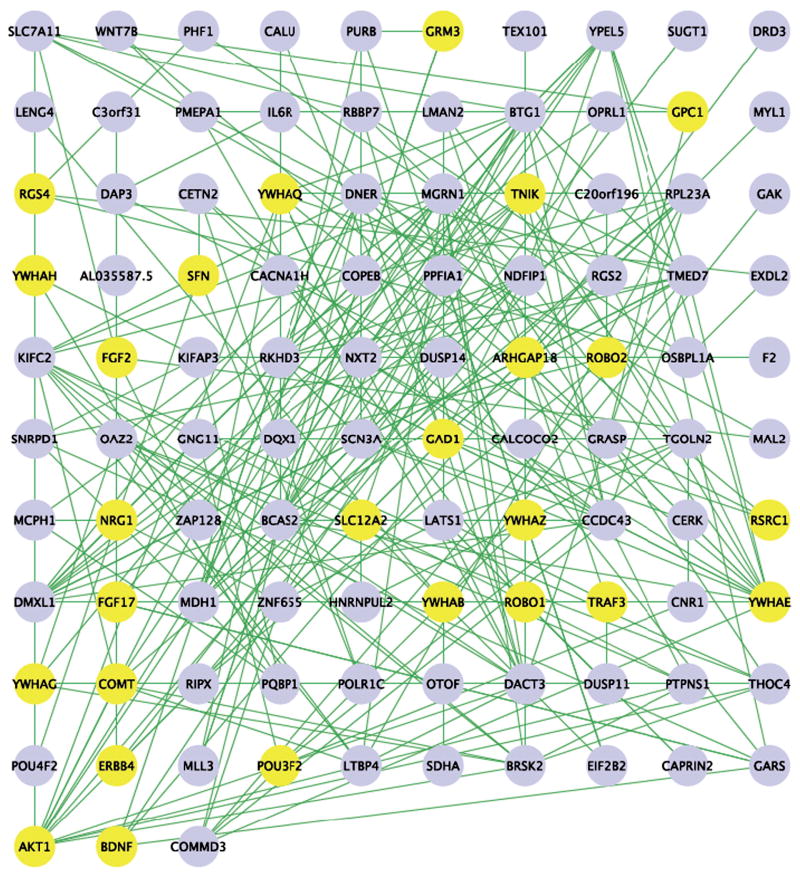
Figure 3 shows the network inferred through this approach. Variation of gene expression in this dataset can be viewed as a dynamic response to the perturbation of gene expression in a subset of genes in the network, induced by genetic variations in the coding or regulatory sequences of these genes. Thus links in this network are likely to reflect functional interactions. Figure 3 shows a large number of connections between schizophrenia candidate genes, as well as some unexpected connections: DACT3, with 3 direct and 8 indirect (one-stop) connections to our candidate genes, encodes the orthologue of an amphibian regulator of Wnt signaling, a fact that may be significant given literature linking Wnt signaling to schizophrenia (Cotter et al., 1998; Miyaoka et al., 1999; Proitsi et al., 2008).
Figure 3.
Gene interaction network inferred from prefrontal cortex gene expression in 42 different inbred mouse strains. Schizophrenia candidate genes from our GWAS and human expression data are highlighted in yellow.
NDFIP (Nedd4 family interacting protein 1) has 4 direct and 8 indirect connections. It is a Golgi protein that is ubiquitinated by the Nedd4 family of proteins (Harvey et al., 2002); the product of another gene on the list, PMEPA1, also interacts with Nedd4. DNER (delta/notch-like EGF repeat containing) with 4 direct and 8 indirect connections, regulates differentiation of glia through Notch signaling (Eiraku et al., 2005); loss of function mouse Dner gives rise to impaired cerebellar function. MCPH1 (microcephalin 1) has 2 direct and 3 indirect connections; it is expressed in fetal brain, in migrating neurons of the developing forebrain, and on the ependymal and subventricular walls of the lateral ventricles. It is related to brain size in human (mutations in it cause a form of primary microcephaly), and is positively selected for (Evans et al., 2005). GPC1 has a direct connection with Wnt and encodes a cell surface heparan sulfate proteoglycan, which acts as a co-receptor for growth factors and other signaling molecules (Lander et al., 1996; Selleck, 2006; Song and Filmus, 2002). TNIK and TRAF3 have been shown to interact with DISC1 in yeast-two hybrid experiments (Camargo et al., 2007) but did not arise in this mouse dataset. DISC1 (disrupted in schizophrenia) has been strongly implicated in schizophrenia and plays a role in brain development. Therefore we conducted human gene expression data.
Human Post-Mortem Expression Data
Gene expression data was obtained by microarray in the DLPFC of thirty patients with schizophrenia, twenty-seven with bipolar disorder, and twenty-nine healthy controls. An interesting pattern of gene co-expression is observed in the table and differs by diagnosis. The gene expression values within subjects for TNIK was negatively correlated with DISC1 (r = -0.25) in schizophrenia, but weakly correlated in bipolar disorder or healthy controls. Further support of the interaction between TNIK and DISC1 is provided by a direct binding between TNIK and DISC1 which regulates AMPA receptor activity (Wang, 2010). In rat primary hippocampal neurons, knockdown of DISC1 leads to increase in the TNIK protein level, suggesting that DISC1 negatively regulates the expression of TNIK (Wang, 2010) which is consistent with the negative gene expression correlation between DISC1 and TNIK. GPC1 was significantly correlated with FGF2 (r = 0.42) and showed a trend with FGF17 (r = 0.31) in schizophrenia but to a lesser degree in bipolar disorder at r = -0.26 and -.013 respectively, and -0.24 and 0.08 in controls. The correlations for FGF2 with GPC1 in schizophrenic patients and controls are in opposite directions.
Gene Set Enrichment Analysis
GSEA was applied to the previously described imaging genetics GWAS datasets based on differences between SZ and controls (Potkin et al., 2009c) and to an independent, smaller dataset of schizophrenic subjects only (Potkin et al., 2009b). Table 1 below shows the top 25 gene sets with Mann-Whitney test Z-score > 4.0 (p < 3×10-5) in both datasets, at which threshold the random permuted gene sets return no hits. There is significant overlap between the GSEA results from the two datasets, although individual genes identified from each dataset are different, supporting the merit of using systems and a network-based approach for testing the disease association. A number of interesting gene sets emerge from this analysis. For instance, several miRNA target gene sets (.mir 448, 218, 137) are highly enriched in both data sets, suggesting a potential role of miRNA perturbation in schizophrenia and meriting further investigation. There are nine total miRNA gene sets in the table that were over enriched in the two imaging genetics GWAS data sets.
Table 1.
Gene co-expression data in human post-mortem DLPFC in patient and control groups. Bold indicates P<0.05.
| GPC1 | FGF2 | FGF17 | TNIK | DISC1 | ||
|---|---|---|---|---|---|---|
| Group SZ (n=30) | ||||||
| GPC1 | NM_002081.1_PROBE1 | 1.000 | ||||
| FGF2 | NM_002006.2_PROBE1 | 0.421 | 1.000 | |||
| FGF17 | 334553.7_PROBE1 | 0.313 | 0.093 | 1.000 | ||
| TNIK | AB011123_PROBE1 | 0.020 | 0.205 | 0.264 | 1.000 | |
| DISC1 | NM_018662.1_PROBE1 | −0.028 | 0.047 | 0.223 | −0.246 | 1.000 |
| Group Bipolar (n=27) | ||||||
| GPC1 | NM_002081.1_PROBE1 | 1.000 | ||||
| FGF2 | NM_002006.2_PROBE1 | −0.259 | 1.000 | |||
| FGF17 | 334553.7_PROBE1 | −0.127 | 0.345 | 1.000 | ||
| TNIK | AB011123_PROBE1 | −0.139 | 0.674 | 0.439 | 1.000 | |
| DISC1 | NM_018662.1_PROBE1 | −0.147 | 0.152 | 0.635 | 0.107 | 1.000 |
| Group Control (n=29) | ||||||
| GPC1 | NM_002081.1_PROBE1 | 1.000 | ||||
| FGF2 | NM_002006.2_PROBE1 | −0.243 | 1.000 | |||
| FGF17 | 334553.7_PROBE1 | 0.075 | −0.233 | 1.000 | ||
| TNIK | AB011123_PROBE1 | 0.093 | 0.205 | 0.063 | 1.000 | |
| DISC1 | NM_018662.1_PROBE1 | −0.125 | −0.207 | 0.542 | 0.024 | 1.000 |
The traditional Gene Ontology categories were significant only once in the table (Biological Process, Nervous System Development), emphasizing the value of using GWAS with GSEA together with miRNA and gene expression datasets.
Glypican-1 and FGF17 combined mutant mouse models
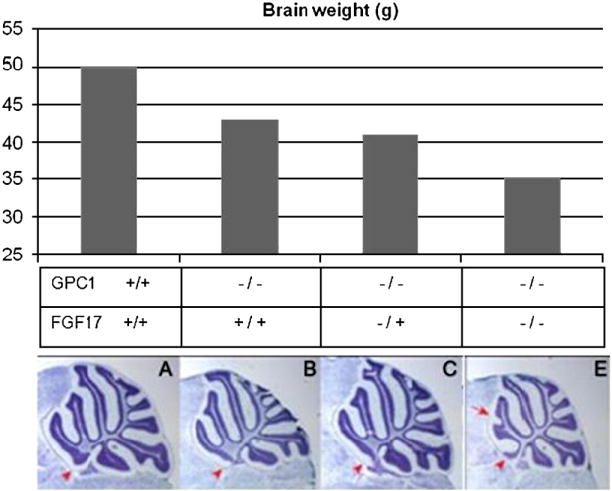
Homozygous mutant GPC1 animals are anatomically grossly normal, but possess brains that are ~15% smaller (and containing 18% fewer cells) than wildtype, and display subtle cerebellar mispatterning (Jen et al., 2009). GPC1 heterozygotes have intermediate brain size. The GPC1-related decrease in brain size is significantly affected by FGF17 status; strongest effects are observed with homozygote mutant FGF17 animals while heterozygotes have an intermediate effect, in total indicating an epistatic interaction (P < 0.005; t-test- see Figure 4). The data show the presence of either one or two copies of mutant alleles for either GPC1 or FGF17 progressively reduces brain size (P < 0.005; t-test). When animals are null for FGF17, the presence of mutant GPC1 alleles has no significant effect. Jen et al (Jen et al., 2009), assessed signaling pathways by Q-RT-PCR at embryonic day 5, the time in which brain size reduction in GPC1 mutant mice emerges. Levels of transcripts for markers of FGF signaling (Sprouty 1 and Sprouty 2) are reduced whereas markers of Hedgehog, Wnt and BMP signaling are not. Additional support for the conclusion that GPC1 regulates FGF signaling is the lower levels of endogenous MAP kinase (Erk) activity found in homozygous GPC1 mutants.
Figure 4.
Epistasis: GPC1 phenotypes require FGF17. The effects of compound GPC1/FGF17 genotypes demonstrate an epistatic effect on brain weight. Bars indicate fresh brain weights of the compound mutants. The genotypes are indicated by + for wildtype, - for knockout, -/+ for heterozygote). Corresponding Nissl-stained mid-sagittal sections the cerebella morphometry is shown. The red arrowheads mark the anterior-most lobe (lobe 1) and the fusion of lobes III and IV, a phenotype observed in FGF17-/- mice that disappears in GPC1 -/- and FGF17 -/- animals. GPC1 appears to be acting in a pathway upstream of FGF17 (Jen et al., 2009).
Discussion
Our approach to identifying gene regulatory networks that contribute to a risk of schizophrenia begins with the identification of new candidate risk genes using brain imaging as a QT in the context of a GWAS. We then applied computational biology methods to these GWAS candidates and to expression data sets in both humans and animals to more fully understand these candidates. This builds on the previous imaging genetics results to integrate across levels of inquiry in understanding genetic influences on system-level phenotypes, as denoted in Figure 1. The GSEA method identified multiple miRNAs in both imaging genetics GWAS datasets. As miRNAs affect the expression of many genes, this provides support for the idea of widespread genetic networks underlying schizophrenia. Finally, it highlights the potential of the GPC1/FGF mouse as an animal model of some characteristics of schizophrenia, in part based on the human GWAS data. Initially, we asked whether commercial software (Ingenuity Pathways Analysis v7, Santa Clara CA) developed to search certain published networks (e.g. protein-protein interaction data (Stelzl et al., 2005) for functional connections among sets of genes) might reveal any new relationships among the genes identified by imaging genetics GWAS (ARHGAP18, RSRC1, GPC1, ROBO2, ROBO1, CTXN3, SLC12A2, TRAF3, TNIK, POU3F2 (Potkin et al., 2008; Potkin et al., 2009c)). The Ingenuity annotation output called attention to a few connections—for example that the ligands for the ROBO1 and ROBO2 receptors, the SLITS (Killeen and Sybingco, 2008; Lopez-Bendito et al., 2007; Nguyen-Ba-Charvet and Chedotal, 2002), bind GPC1 (Ronca et al., 2001)—that are well established in the literature. The full implication of these results, however, required more innovative computational and physiological approaches. The correlational connections shown in Figures 2 and 3 were not available in any currently existing software, and the miRNA connection was not identified. Our expression data provided support for an interaction between GPC1 and FGF17 and FGF2 as well as between TNIK and DISC1. A recent set of studies found the kinase domain of TNIK binds to a small region on DISC1, a key gene consistently linked to schizophrenia risk (Wang, 2010). The potential importance of TNIK itself in schizophrenia has been highlighted by several independent studies, supporting its role as an emerging risk factor (Glatt et al., 2005; Shi et al., 2009). DISC1 has been shown to modulate TNIK kinase activity and together they function at the synapse to regulate synaptic composition and GLUR1 and AMPA activity (Wang 2010), both hypothetically related to schizophrenia (Harrison and Weinberger, 2005).
Figure 2.
BOLD signal change in DLPFC, by memory load, for healthy volunteer (HV) controls and schizophrenia patients (SZ) that are behaviorally matched on memory performance by diagnostic group (Potkin et al., 2009a). The same accuracy performance is associated with significantly greater DLPFC activation in schizophrenia subjects than in healthy controls beginning at a memory load of three.
Whether the GPC1 brain size phenotype noted in the mouse model has any relationship to a role for GPC1 in schizophrenia is unknown. It is interesting, however, that clinical studies support a small but significant decrease in brain size in schizophrenia (on the order of 3% (Steen et al., 2006; Ward et al., 1996)). In addition, although FGF17-deficient mice are behaviorally relatively normal, they show striking deficits in social recognition and affiliative interactions (Scearce-Levie et al., 2008), which is particularly intriguing given the social dysfunction characteristic of schizophrenia patients (Cholfin and Rubenstein, 2008; Scearce-Levie et al., 2008). Thus, even though FGF17 has never itself emerged by GWAS as an SZ candidate gene, its functional association with GPC1, and its mouse behavioral phenotype, strongly point to it being a potentially important component of a schizophrenia-related gene network. In support of this idea, we note that recent GWAS studies have identified FGFR2, a major FGF brain receptor, as strongly associated with schizophrenia (O’Donovan et al., 2009; Potkin et al., 2009c).
This work suggests that FGF17 and/or GPC1 mutant mice may develop into useful and valid models for the negative symptoms of schizophrenia (which are especially difficult to treat), and additionally provide a proof-of-concept model of therapeutic intervention. It is also of interest that the brain weights of heterozygous GPC1 mutants fall midway between wild type and homozygous mutants, as it suggests a strong quantitative dependency of FGF signaling on GPC1 expression levels. We have observed alterations in FGF2 gene expression in the DLPFC of SZ patients (Shao and Vawter, 2008). Because most genetic polymorphisms probably exert their effects by altering levels of expression genes, such a strong dependency may explain why GPC1 is more prone to detection by GWAS than other genes in the networks in which it acts.
The miRNA findings based on the imaging genetics GWAS data are intriguing, as they suggest a mechanism for regulation of a network of genes. Our third top miRNA, 137, was one of the five major findings from a 52,156 subject GWAS study reported by P. Gejman (Gejman, 2010). This is in accord with the underlying multiple genes models of schizophrenia in general, and specifically the potential importance of gene regulatory networks as contrasted with single-gene effects for complex illnesses. Understanding these networks in animal models offers potentially new opportunities for therapeutic modulation.
In summary, the integration of imaging genetics GWAS analyses in schizophrenia with computational biology methods, and animal models identified several key findings that extended our initial imaging GWAS studies in schizophrenia to improved understanding of the system-level phenotype. Finally, the GPC1/FGF mouse based in part on our imaging genetics human GWAS data and computational gene expression analyses may provide a useful animal model of key characteristics of schizophrenia.
Table 2.
Top 25 gene sets enriched with most significant P-value genes in two independent imaging genetics GWAS data sets; SCZ data set n=24 (Potkin et al., 2009b) and SIRP data set n=138 (Potkin et al., 2009c). Gene set codes: c2 refers to curated gene sets from canonical pathways and chemical and genetic perturbations. C3 refers to motif gene sets in MSigDB; c4, to computational gene sets, and c5 to GO gene sets. MicroRNA data sets are indicated by .mir. For more information see MSigDB (www.broad.mit.edu/gsea/msigdb/index.jsp).
| Gene set category | Gene set name | SIRP dataset
|
SIRP dataset
|
Number of genes | ||
|---|---|---|---|---|---|---|
| Mann–Whitney Z-score | KS −log10 (P value) | Mann–Whitney Z-score | KS −log10 (P value) | |||
| c3.mir | ATATGCA,MIR-448 | 8.8657 | 17.1214 | 6.9348 | 9.7789 | 177 |
| c2.cgp | UVC_TTD_ALL_DN | 8.6874 | 12.9778 | 7.4691 | 9.9803 | 318 |
| c2.cgp | UVC_XPCS_ALL_DN | 8.1193 | 12.5773 | 7.9788 | 12.225 | 419 |
| c2.cgp | UVC_XPCS_8HR_DN | 7.6156 | 10.3729 | 7.5681 | 10.7036 | 358 |
| c2.cgp | UVC_TTD_8HR_DN | 7.3774 | 8.0467 | 6.4858 | 6.8742 | 144 |
| c2.cgp. | UVC_TTD_4HR_DN | 7.3072 | 10.244 | 6.1085 | 7.1162 | 262 |
| c3.tft | V$OCT1_03 | 6.783 | 8.168 | 2.6201 | 1.6422 | 147 |
| c2.cgp | UVC_XPCS_4HR_DN | 6.6389 | 9.8161 | 5.7022 | 7.1296 | 208 |
| c2.cgp | BAF57_BT549_UP | 6.6375 | 9.8099 | 5.2192 | 4.502 | 203 |
| c3.mir | AAGCACA,MIR-218 | 6.5172 | 7.8408 | 6.9509 | 11.0283 | 332 |
| c3.tft | YNGTTNNNATT_UNKNOWN | 5.9645 | 6.611 | 7.3309 | 12.7903 | 235 |
| c3.mir | AAGCAAT,MIR-137 | 5.8681 | 7.2371 | 6.5203 | 7.6126 | 168 |
| c3.tft | V$NKX25_02 | 5.8142 | 7.1266 | 4.7564 | 3.9708 | 181 |
| GNF | cluster_all_GNF_105 | 5.784 | 6.5692 | 6.3786 | 7.582 | 187 |
| c3.tft | V$CART1_01 | 5.7749 | 6.3817 | 5.4727 | 7.8015 | 157 |
| c3.tft | V$OCT1_07 | 5.6725 | 9.2947 | 3.8748 | 3.503 | 107 |
| c3.mir | TTGCCAA,MIR-182 | 5.6675 | 5.2286 | 5.7457 | 8.175 | 263 |
| c3.mir | TCCAGAG,MIR-518C | 5.5932 | 7.203 | 3.1213 | 2.5984 | 124 |
| c3.mir | CAGTATT,MIR-200B,MIR-200C,MIR-429 | 5.5887 | 6.1427 | 6.0432 | 6.227 | 363 |
| c3.tft | V$FOXJ2_02 | 5.5746 | 5.385 | 5.9776 | 6.5761 | 160 |
| c3.mir | TATTATA,MIR-374 | 5.5734 | 5.1674 | 5.6163 | 6.0237 | 233 |
| c5.bp | NERVOUS_SYSTEM_DEVELOPMENT | 5.5395 | 6.0857 | 5.1778 | 6.3565 | 331 |
| c3.mir | GTATTAT,MIR-369-3P | 5.537 | 9.2865 | 3.6326 | 4.2507 | 170 |
| c2.cgp | BOQUEST_CD31PLUS_VS_CD31MINUS_DN | 5.5227 | 5.4404 | 5.6424 | 4.5051 | 217 |
| c3.mir | ACTGTGA,MIR-27A,MIR-27B | 5.4877 | 5.789 | 5.9505 | 6.8975 | 381 |
Acknowledgments
This research was supported by grants to The Transdisciplinary Imaging Genetics Center (TIGC-P20 RR020837-01) and to the Functional Imaging Biomedical Informatics Research Network (FBIRN-1 U24 RR021992) from the National Center for Research Resources (NCRR) at the National Institutes of Health (NIH) as well as R01-NS26862 and P50-GM076516 (to ADL) and NIAAA- U01AA016667 (to MFM). We thank Liv McMillan (UCI) for excellent editorial support.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aikawa T, Whipple CA, Lopez ME, Gunn J, Young A, Lander AD, Korc M. Glypican-1 modulates the angiogenic and metastatic potential of human and mouse cancer cells. Journal of Clinical Investigation. 2008;118:89–99. doi: 10.1172/JCI32412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almasy L, Gur RC, Haack K, Cole SA, Calkins ME, Peralta JM, Hare E, Prasad K, Pogue-Geile MF, Nimgaonkar V, Gur RE. A Genome Screen for Quantitative Trait Loci Influencing Schizophrenia and Neurocognitive Phenotypes. American Journal of Psychiatry. 2008 doi: 10.1176/appi.ajp.2008.07121869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bansal M, Belcastro V, Ambesi-Impiombato A, di Bernardo D. How to infer gene networks from expression profiles. Mol Syst Biol. 2007;3:78. doi: 10.1038/msb4100120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barabasi AL. Network medicine--from obesity to the “diseasome”. New England Journal of Medicine. 2007;357:404–407. doi: 10.1056/NEJMe078114. [DOI] [PubMed] [Google Scholar]
- Barch DM, Carter CS, Braver TS, Sabb FW, MacDonald A, 3rd, Noll DC, Cohen JD. Selective deficits in prefrontal cortex function in medication-naive patients with schizophrenia. Archives of General Psychiatry. 2001;58:280–288. doi: 10.1001/archpsyc.58.3.280. [DOI] [PubMed] [Google Scholar]
- Beveridge NJ, Tooney PA, Carroll AP, Gardiner E, Bowden N, Scott RJ, Tran N, Dedova I, Cairns MJ. Dysregulation of miRNA 181b in the temporal cortex in schizophrenia. Human Molecular Genetics. 2008;17:1156–1168. doi: 10.1093/hmg/ddn005. [DOI] [PubMed] [Google Scholar]
- Brett M, Johnsrude IS, Owen AM. The problem of functional localization in the human brain. Nat Rev Neurosci. 2002;3:243–249. doi: 10.1038/nrn756. [DOI] [PubMed] [Google Scholar]
- Brzustowicz LM. NOS1AP in schizophrenia. Curr Psychiatry Rep. 2008;10:158–163. doi: 10.1007/s11920-008-0027-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camargo LM, Collura V, Rain JC, Mizuguchi K, Hermjakob H, Kerrien S, Bonnert TP, Whiting PJ, Brandon NJ. Disrupted in Schizophrenia 1 Interactome: evidence for the close connectivity of risk genes and a potential synaptic basis for schizophrenia. Mol Psychiatry. 2007;12:74–86. doi: 10.1038/sj.mp.4001880. [DOI] [PubMed] [Google Scholar]
- Carthew RW. Gene regulation by microRNAs. Current Opinion in Genetics and Development. 2006;16:203–208. doi: 10.1016/j.gde.2006.02.012. [DOI] [PubMed] [Google Scholar]
- Chapman J, Clayton D. Detecting association using epistatic information. Genetic Epidemiology. 2007;31:894–909. doi: 10.1002/gepi.20250. [DOI] [PubMed] [Google Scholar]
- Cholfin JA, Rubenstein JL. Frontal cortex subdivision patterning is coordinately regulated by Fgf8, Fgf17, and Emx2. Journal of Comparative Neurology. 2008;509:144–155. doi: 10.1002/cne.21709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton D, McKeigue PM. Epidemiological methods for studying genes and environmental factors in complex diseases. Lancet. 2001;358:1356–1360. doi: 10.1016/S0140-6736(01)06418-2. [DOI] [PubMed] [Google Scholar]
- Committee, P.G.C.S. A framework for interpreting genome-wide association studies of psychiatric disorders. Mol Psychiatry. 2009;14:10–17. doi: 10.1038/mp.2008.126. [DOI] [PubMed] [Google Scholar]
- Cotter D, Kerwin R, al-Sarraji S, Brion JP, Chadwich A, Lovestone S, Anderton B, Everall I. Abnormalities of Wnt signalling in schizophrenia--evidence for neurodevelopmental abnormality. Neuroreport. 1998;9:1379–1383. doi: 10.1097/00001756-199805110-00024. [DOI] [PubMed] [Google Scholar]
- D’Esposito M, Aguirre GK, Zarahn E, Ballard D, Shin RK, Lease J. Functional MRI studies of spatial and nonspatial working memory. Brain Research Cognitive Brain Research. 1998;7:1–13. doi: 10.1016/s0926-6410(98)00004-4. [DOI] [PubMed] [Google Scholar]
- Dean B, Keriakous D, Scarr E, Thomas EA. Gene expression profiling in Brodmann’s area 46 from subjects with schizophrenia. Australian and New Zealand Journal of Psychiatry. 2007;41:308–320. doi: 10.1080/00048670701213245. [DOI] [PubMed] [Google Scholar]
- Eiraku M, Tohgo A, Ono K, Kaneko M, Fujishima K, Hirano T, Kengaku M. DNER acts as a neuron-specific Notch ligand during Bergmann glial development. Nat Neurosci. 2005;8:873–880. doi: 10.1038/nn1492. [DOI] [PubMed] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans DM, Marchini J, Morris AP, Cardon LR. Two-stage two-locus models in genome-wide association. PLoS Genet. 2006;2:e157. doi: 10.1371/journal.pgen.0020157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans PD, Gilbert SL, Mekel-Bobrov N, Vallender EJ, Anderson JR, Vaez-Azizi LM, Tishkoff SA, Hudson RR, Lahn BT. Microcephalin, a gene regulating brain size, continues to evolve adaptively in humans. Science. 2005;309:1717–1720. doi: 10.1126/science.1113722. [DOI] [PubMed] [Google Scholar]
- Fallon J, Reid S, Kinyamu R, Opole I, Opole R, Baratta J, Korc M, Endo TL, Duong A, Nguyen G, Karkehabadhi M, Twardzik D, Patel S, Loughlin S. In vivo induction of massive proliferation, directed migration, and differentiation of neural cells in the adult mammalian brain. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:14686–14691. doi: 10.1073/pnas.97.26.14686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraldez AJ, Cinalli RM, Glasner ME, Enright AJ, Thomson JM, Baskerville S, Hammond SM, Bartel DP, Schier AF. MicroRNAs regulate brain morphogenesis in zebrafish. Science. 2005;308:833–838. doi: 10.1126/science.1109020. [DOI] [PubMed] [Google Scholar]
- Glatt SJ, Everall IP, Kremen WS, Corbeil J, Sasik R, Khanlou N, Han M, Liew CC, Tsuang MT. Comparative gene expression analysis of blood and brain provides concurrent validation of SELENBP1 up-regulation in schizophrenia. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:15533–15538. doi: 10.1073/pnas.0507666102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazier AM, Nadeau JH, Aitman TJ. Finding genes that underlie complex traits. Science. 2002;298:2345–2349. doi: 10.1126/science.1076641. [DOI] [PubMed] [Google Scholar]
- Gottesman II, Gould TD. The endophenotype concept in psychiatry: etymology and strategic intentions. American Journal of Psychiatry. 2003;160:636–645. doi: 10.1176/appi.ajp.160.4.636. [DOI] [PubMed] [Google Scholar]
- Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. miRBase: tools for microRNA genomics. Nucleic Acids Res. 2008;36:D154–158. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison PJ, Weinberger DR. Schizophrenia genes, gene expression, and neuropathology: on the matter of their convergence. Molecular Psychiatry. 2005;10:40–68. doi: 10.1038/sj.mp.4001558. image 45. [DOI] [PubMed] [Google Scholar]
- Harvey KF, Shearwin-Whyatt LM, Fotia A, Parton RG, Kumar S. N4WBP5, a potential target for ubiquitination by the Nedd4 family of proteins, is a novel Golgi-associated protein. J Biol Chem. 2002;277:9307–9317. doi: 10.1074/jbc.M110443200. [DOI] [PubMed] [Google Scholar]
- Hidalgo CA, Blumm N, Barabasi AL, Christakis NA. A dynamic network approach for the study of human phenotypes. PLoS Comput Biol. 2009;5:e1000353. doi: 10.1371/journal.pcbi.1000353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann TJ, Lange C, Vansteelandt S, Laird NM. Gene-environment interaction tests for dichotomous traits in trios and sibships. Genetic Epidemiology. 2009 doi: 10.1002/gepi.20421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivins JK, Litwack ED, Kumbasar A, Stipp CS, Lander AD. Cerebroglycan, a developmentally regulated cell-surface heparan sulfate proteoglycan, is expressed on developing axons and growth cones. Dev Biol. 1997;184:320–332. doi: 10.1006/dbio.1997.8532. [DOI] [PubMed] [Google Scholar]
- Jen YH, Musacchio M, Lander AD. Glypican-1 controls brain size through regulation of fibroblast growth factor signaling in early neurogenesis. Neural Dev. 2009;4:33. doi: 10.1186/1749-8104-4-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang R, Tang W, Wu X, Fu W. A random forest approach to the detection of epistatic interactions in case-control studies. BMC Bioinformatics. 2009;10(Suppl 1):S65. doi: 10.1186/1471-2105-10-S1-S65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson MR, Morris NA, Astur RS, Calhoun VD, Mathalon DH, Kiehl KA, Pearlson GD. A functional magnetic resonance imaging study of working memory abnormalities in schizophrenia. Biological Psychiatry. 2006;60:11–21. doi: 10.1016/j.biopsych.2005.11.012. [DOI] [PubMed] [Google Scholar]
- Karlsgodt KH, Glahn DC, van Erp TG, Therman S, Huttunen M, Manninen M, Kaprio J, Cohen MS, Lonnqvist J, Cannon TD. The relationship between performance and fMRI signal during working memory in patients with schizophrenia, unaffected co-twins, and control subjects. Schizophrenia Research. 2007;89:191–197. doi: 10.1016/j.schres.2006.08.016. [DOI] [PubMed] [Google Scholar]
- Kennedy JL, Farrer LA, Andreasen NC, Mayeux R. The genetics of adult-onset neuropsychiatric disease: complexities and conundra? Science. 2003;302:822–826. doi: 10.1126/science.1092132. [DOI] [PubMed] [Google Scholar]
- Killeen MT, Sybingco SS. Netrin, Slit and Wnt receptors allow axons to choose the axis of migration. Developmental Biology. 2008;323:143–151. doi: 10.1016/j.ydbio.2008.08.027. [DOI] [PubMed] [Google Scholar]
- Lander AD, Stipp CS, Ivins JK. The glypican family of heparan sulfate proteoglycans: major cell-surface proteoglycans of the developing nervous system. Perspect Dev Neurobiol. 1996;3:347–358. [PubMed] [Google Scholar]
- Lander E, Kruglyak L. Genetic dissection of complex traits: guidelines for interpreting and reporting linkage results. Nature Genetics. 1995;11:241–247. doi: 10.1038/ng1195-241. [DOI] [PubMed] [Google Scholar]
- Lewis CM, Levinson DF, Wise LH, DeLisi LE, Straub RE, Hovatta I, Williams NM, Schwab SG, Pulver AE, Faraone SV, Brzustowicz LM, Kaufmann CA, Garver DL, Gurling HM, Lindholm E, Coon H, Moises HW, Byerley W, Shaw SH, Mesen A, Sherrington R, O’Neill FA, Walsh D, Kendler KS, Ekelund J, Paunio T, Lonnqvist J, Peltonen L, O’Donovan MC, Owen MJ, Wildenauer DB, Maier W, Nestadt G, Blouin JL, Antonarakis SE, Mowry BJ, Silverman JM, Crowe RR, Cloninger CR, Tsuang MT, Malaspina D, Harkavy-Friedman JM, Svrakic DM, Bassett AS, Holcomb J, Kalsi G, McQuillin A, Brynjolfson J, Sigmundsson T, Petursson H, Jazin E, Zoega T, Helgason T. Genome scan meta-analysis of schizophrenia and bipolar disorder, part II: Schizophrenia. American Journal of Human Genetics. 2003;73:34–48. doi: 10.1086/376549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litwack ED, Stipp CS, Kumbasar A, Lander AD. Neuronal expression of glypican, a cell-surface glycosylphosphatidylinositol-anchored heparan sulfate proteoglycan, in the adult rat nervous system. J Neurosci. 1994;14:3713–3724. doi: 10.1523/JNEUROSCI.14-06-03713.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Bendito G, Flames N, Ma L, Fouquet C, Di Meglio T, Chedotal A, Tessier-Lavigne M, Marin O. Robo1 and Robo2 cooperate to control the guidance of major axonal tracts in the mammalian forebrain. Journal of Neuroscience. 2007;27:3395–3407. doi: 10.1523/JNEUROSCI.4605-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA, Berka J, Braverman MS, Chen YJ, Chen Z, Dewell SB, Du L, Fierro JM, Gomes XV, Godwin BC, He W, Helgesen S, Ho CH, Irzyk GP, Jando SC, Alenquer ML, Jarvie TP, Jirage KB, Kim JB, Knight JR, Lanza JR, Leamon JH, Lefkowitz SM, Lei M, Li J, Lohman KL, Lu H, Makhijani VB, McDade KE, McKenna MP, Myers EW, Nickerson E, Nobile JR, Plant R, Puc BP, Ronan MT, Roth GT, Sarkis GJ, Simons JF, Simpson JW, Srinivasan M, Tartaro KR, Tomasz A, Vogt KA, Volkmer GA, Wang SH, Wang Y, Weiner MP, Yu P, Begley RF, Rothberg JM. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNab F, Klingberg T. Prefrontal cortex and basal ganglia control access to working memory. Nature Neuroscience. 2008;11:103–107. doi: 10.1038/nn2024. [DOI] [PubMed] [Google Scholar]
- Meyer-Lindenberg A, Weinberger DR. Intermediate phenotypes and genetic mechanisms of psychiatric disorders. Nat Rev Neurosci. 2006;7:818–827. doi: 10.1038/nrn1993. [DOI] [PubMed] [Google Scholar]
- Miyaoka T, Seno H, Ishino H. Increased expression of Wnt-1 in schizophrenic brains. Schizophr Res. 1999;38:1–6. doi: 10.1016/s0920-9964(98)00179-0. [DOI] [PubMed] [Google Scholar]
- Moore JH. Analysis of gene-gene interactions. Curr Protoc Hum Genet. 2008;Chapter 1(Unit 1):14. doi: 10.1002/0471142905.hg0114s59. [DOI] [PubMed] [Google Scholar]
- Moskvina V, Craddock N, Holmans P, Nikolov I, Pahwa JS, Green E, Owen MJ, O’Donovan MC. Gene-wide analyses of genome-wide association data sets: evidence for multiple common risk alleles for schizophrenia and bipolar disorder and for overlap in genetic risk. Mol Psychiatry. 2008 doi: 10.1038/mp.2008.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen-Ba-Charvet KT, Chedotal A. Role of Slit proteins in the vertebrate brain. Journal of Physiology, Paris. 2002;96:91–98. doi: 10.1016/s0928-4257(01)00084-5. [DOI] [PubMed] [Google Scholar]
- Nonaka H, Takei K, Umikawa M, Oshiro M, Kuninaka K, Bayarjargal M, Asato T, Yamashiro Y, Uechi Y, Endo S, Suzuki T, Kariya K. MINK is a Rap2 effector for phosphorylation of the postsynaptic scaffold protein TANC1. Biochemical and Biophysical Research Communications. 2008;377:573–578. doi: 10.1016/j.bbrc.2008.10.038. [DOI] [PubMed] [Google Scholar]
- O’Donovan MC, Norton N, Williams H, Peirce T, Moskvina V, Nikolov I, Hamshere M, Carroll L, Georgieva L, Dwyer S, Holmans P, Marchini JL, Spencer CC, Howie B, Leung HT, Giegling I, Hartmann AM, Moller HJ, Morris DW, Shi Y, Feng G, Hoffmann P, Propping P, Vasilescu C, Maier W, Rietschel M, Zammit S, Schumacher J, Quinn EM, Schulze TG, Iwata N, Ikeda M, Darvasi A, Shifman S, He L, Duan J, Sanders AR, Levinson DF, Adolfsson R, Osby U, Terenius L, Jonsson EG, Cichon S, Nothen MM, Gill M, Corvin AP, Rujescu D, Gejman PV, Kirov G, Craddock N, Williams NM, Owen MJ. Analysis of 10 independent samples provides evidence for association between schizophrenia and a SNP flanking fibroblast growth factor receptor 2. Molecular Psychiatry. 2009;14:30–36. doi: 10.1038/mp.2008.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papassotiropoulos A, Stephan DA, Huentelman MJ, Hoerndli FJ, Craig DW, Pearson JV, Huynh KD, Brunner F, Corneveaux J, Osborne D, Wollmer MA, Aerni A, Coluccia D, Hanggi J, Mondadori CR, Buchmann A, Reiman EM, Caselli RJ, Henke K, de Quervain DJ. Common Kibra alleles are associated with human memory performance. Science. 2006;314:475–478. doi: 10.1126/science.1129837. [DOI] [PubMed] [Google Scholar]
- Potkin SG, Turner JA, Brown GG, McCarthy G, Greve DN, Glover GH, Manoach DS, Belger A, Diaz M, Wible CG, Ford JM, Mathalon DH, Gollub R, Lauriello J, O’Leary D, van Erp TG, Toga AW, Preda A, Lim KO. Working memory and DLPFC inefficiency in schizophrenia: the FBIRN study. Schizophr Bull. 2009a;35:19–31. doi: 10.1093/schbul/sbn162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potkin SG, Turner JA, Fallon JA, Lakatos A, Keator DB, Guffanti G, Macciardi F. Gene discovery through imaging genetics: identification of two novel genes associated with schizophrenia. Mol Psychiatry. 2008 doi: 10.1038/mp.2008.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potkin SG, Turner JA, Fallon JA, Lakatos A, Keator DB, Guffanti G, Macciardi F. Gene discovery through imaging genetics: identification of two novel genes associated with schizophrenia. Molecular Psychiatry. 2009b;14:416–428. doi: 10.1038/mp.2008.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potkin SG, Turner JA, Guffanti G, Lakatos A, Fallon JH, Nguyen DD, Mathalon D, Ford J, Lauriello J, Macciardi F. A genome-wide association study of schizophrenia using brain activation as a quantitative phenotype. Schizophrenia Bulletin. 2009c;35:96–108. doi: 10.1093/schbul/sbn155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potkin SG, Turner JA, Guffanti G, Lakatos A, Torri F, Keator DB, Macciardi F. Genome-wide strategies for discovering genetic influences on cognition and cognitive disorders: methodological considerations. Cogn Neuropsychiatry. 2009d;14:391–418. doi: 10.1080/13546800903059829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proitsi P, Li T, Hamilton G, Di Forti M, Collier D, Killick R, Chen R, Sham P, Murray R, Powell J, Lovestone S. Positional pathway screen of wnt signaling genes in schizophrenia: association with DKK4. Biol Psychiatry. 2008;63:13–16. doi: 10.1016/j.biopsych.2007.03.014. [DOI] [PubMed] [Google Scholar]
- Rakic S, Zecevic N. Early oligodendrocyte progenitor cells in the human fetal telencephalon. Glia. 2003;41:117–127. doi: 10.1002/glia.10140. [DOI] [PubMed] [Google Scholar]
- Roffman JL, Weiss AP, Goff DC, Rauch SL, Weinberger DR. Neuroimaging-genetic paradigms: a new approach to investigate the pathophysiology and treatment of cognitive deficits in schizophrenia. Harvard Review of Psychiatry. 2006;14:78–91. doi: 10.1080/10673220600642945. [DOI] [PubMed] [Google Scholar]
- Ronca F, Andersen JS, Paech V, Margolis RU. Characterization of Slit protein interactions with glypican-1. Journal of Biological Chemistry. 2001;276:29141–29147. doi: 10.1074/jbc.M100240200. [DOI] [PubMed] [Google Scholar]
- Scearce-Levie K, Roberson ED, Gerstein H, Cholfin JA, Mandiyan VS, Shah NM, Rubenstein JL, Mucke L. Abnormal social behaviors in mice lacking Fgf17. Genes Brain Behav. 2008;7:344–354. doi: 10.1111/j.1601-183X.2007.00357.x. [DOI] [PubMed] [Google Scholar]
- Schratt GM, Tuebing F, Nigh EA, Kane CG, Sabatini ME, Kiebler M, Greenberg ME. A brain-specific microRNA regulates dendritic spine development. Nature. 2006;439:283–289. doi: 10.1038/nature04367. [DOI] [PubMed] [Google Scholar]
- Selleck SB. Shedding light on the distinct functions of proteoglycans. Sci STKE. 2006:pe17. doi: 10.1126/stke.3292006pe17. [DOI] [PubMed] [Google Scholar]
- Shao L, Vawter MP. Shared gene expression alterations in schizophrenia and bipolar disorder. Biological Psychiatry. 2008;64:89–97. doi: 10.1016/j.biopsych.2007.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J, Levinson DF, Duan J, Sanders AR, Zheng Y, Pe’er I, Dudbridge F, Holmans PA, Whittemore AS, Mowry BJ, Olincy A, Amin F, Cloninger CR, Silverman JM, Buccola NG, Byerley WF, Black DW, Crowe RR, Oksenberg JR, Mirel DB, Kendler KS, Freedman R, Gejman PV. Common variants on chromosome 6p22.1 are associated with schizophrenia. Nature. 2009;460:753–757. doi: 10.1038/nature08192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song HH, Filmus J. The role of glypicans in mammalian development. Biochim Biophys Acta. 2002;1573:241–246. doi: 10.1016/s0304-4165(02)00390-2. [DOI] [PubMed] [Google Scholar]
- Steen RG, Mull C, McClure R, Hamer RM, Lieberman JA. Brain volume in first-episode schizophrenia: systematic review and meta-analysis of magnetic resonance imaging studies. Br J Psychiatry. 2006;188:510–518. doi: 10.1192/bjp.188.6.510. [DOI] [PubMed] [Google Scholar]
- Stelzl U, Worm U, Lalowski M, Haenig C, Brembeck FH, Goehler H, Stroedicke M, Zenkner M, Schoenherr A, Koeppen S, Timm J, Mintzlaff S, Abraham C, Bock N, Kietzmann S, Goedde A, Toksoz E, Droege A, Krobitsch S, Korn B, Birchmeier W, Lehrach H, Wanker EE. A human protein-protein interaction network: a resource for annotating the proteome. Cell. 2005;122:957–968. doi: 10.1016/j.cell.2005.08.029. [DOI] [PubMed] [Google Scholar]
- Su AI, Wiltshire T, Batalov S, Lapp H, Ching KA, Block D, Zhang J, Soden R, Hayakawa M, Kreiman G, Cooke MP, Walker JR, Hogenesch JB. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc Natl Acad Sci U S A. 2004;101:6062–6067. doi: 10.1073/pnas.0400782101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symons M. Rho family GTPases: the cytoskeleton and beyond. Trends in Biochemical Sciences. 1996;21:178–181. [PubMed] [Google Scholar]
- Teo YY. Common statistical issues in genome-wide association studies: a review on power, data quality control, genotype calling and population structure. Current Opinion in Lipidology. 2008;19:133–143. doi: 10.1097/MOL.0b013e3282f5dd77. [DOI] [PubMed] [Google Scholar]
- Tura E, Turner JA, Fallon JH, Kennedy JL, Potkin SG. Multivariate analyses suggest genetic impacts on neurocircuitry in schizophrenia. Neuroreport. 2008;19:603–607. doi: 10.1097/WNR.0b013e3282fa6d8d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q. Understanding the function of DISC1 and its interacting partner TNIK at the synapse. Winter Conference on Brain Research; Breckenridge, Colorado. 2010. [Google Scholar]
- Ward KE, Friedman L, Wise A, Schulz SC. Meta-analysis of brain and cranial size in schizophrenia. Schizophr Res. 1996;22:197–213. doi: 10.1016/s0920-9964(96)00076-x. [DOI] [PubMed] [Google Scholar]
- Wells A. EGF receptor. International Journal of Biochemistry and Cell Biology. 1999;31:637–643. doi: 10.1016/s1357-2725(99)00015-1. [DOI] [PubMed] [Google Scholar]
- Xie X, Lu J, Kulbokas EJ, Golub TR, Mootha V, Lindblad-Toh K, Lander ES, Kellis M. Systematic discovery of regulatory motifs in human promoters and 3’ UTRs by comparison of several mammals. Nature. 2005;434:338–345. doi: 10.1038/nature03441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X, Mikkelsen TS, Gnirke A, Lindblad-Toh K, Kellis M, Lander ES. Systematic discovery of regulatory motifs in conserved regions of the human genome, including thousands of CTCF insulator sites. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:7145–7150. doi: 10.1073/pnas.0701811104. [DOI] [PMC free article] [PubMed] [Google Scholar]