Abstract
3-hydroxy-3-methylglutaryl-CoA reductase (HMGR) catalyzes the formation of mevalonate, the committed step in the biosynthesis of sterols and isoprenoids. The activity of HMGR is controlled through synthesis, degradation and phosphorylation to maintain the concentration of mevalonate-derived products. In addition to the physiological regulation of HMGR, the human enzyme has been targeted successfully by drugs in the clinical treatment of high serum cholesterol levels. Three crystal structures of the catalytic portion of human HMGR in complexes with HMG-CoA, with HMG and CoA, and with HMG, CoA and NADP+, provide a detailed view of the enzyme active site. Catalytic portions of human HMGR form tight tetramers. The crystal structure explains the influence of the enzyme's oligomeric state on the activity and suggests a mechanism for cholesterol sensing. The active site architecture of human HMGR is different from that of bacterial HMGR; this may explain why binding of HMGR inhibitors to bacterial HMGRs has not been reported.
Keywords: catalysis/cholesterol biosynthesis/HMG-CoA binding/NADPH binding/statins
Introduction
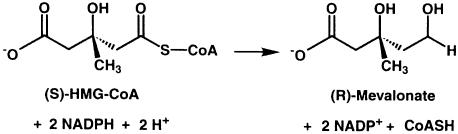
Mevalonic acid is the precursor of isoprenoids, a class of compounds involved in diverse cellular functions such as sterol synthesis and growth control. Products of the mevalonate pathway include cholesterol, heme and farnesyl-pyrophosphate. Within cells, the concentration of mevalonate is tightly controlled through the activity of 3-hydroxy-3-methylglutaryl-CoA reductase (HMGR), the enzyme that catalyzes the four-electron reduction of HMG-CoA to mevalonate (Scheme 1).
Scheme 1.
HMGR is among the most highly regulated enzymes known (Goldstein and Brown, 1990). Transcription and translation of HMGR increase when the concentrations of products of the mevalonate pathway are low. Conversely, when sterol concentrations are high, the intracellular HMGR concentration decreases rapidly (Nakanishi et al., 1988). A third level of regulation is achieved by phos– phorylation of S872 (human enzyme) by AMP-activated protein kinase, which decreases the enzyme's activity (Omkumar et al., 1994).
Elevated cholesterol levels have been identified as a primary risk factor of coronary artery disease. In the USA, coronary artery disease affects 13–14 million adults and is a major cause of disability and death (Eisenberg, 1998). As shown in large-scale clinical trials, inhibition of HMGR significantly decreases cholesterol levels and reduces the risks of stroke by 29% and the overall mortality by 22% (Hebert et al., 1997). Potent HMGR inhibitors with Ki values in the nanomolar range are available (Endo, 1985). These compounds, commonly referred to as statins, are very effective in lowering serum cholesterol levels and are prescribed widely in treatment of hypercholesterolemia (Gotto, 1997). All statins share an HMG moiety and are thought to bind competitively in the active site of HMGR. Inhibition of HMGR also induces growth arrest and cell death in several cancer cell types, presumably through the reduction of non-sterol, mevalonate-derived products (Bennis et al., 1993; Hawk et al., 1996; Lee et al., 1998; Caruso et al., 1999). A new and unexpected role for HMGR was demonstrated recently in studies on Drosophila melanogaster development, where it was shown that the columbus gene, expression of which is required to guide primordial germ cells, encodes HMGR (Van Doren et al., 1998).
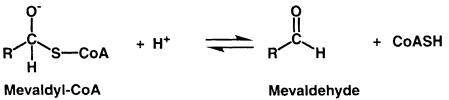
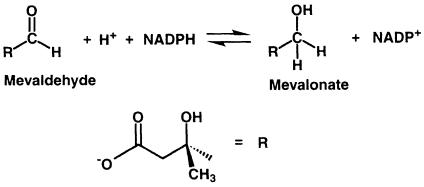
Sequence comparison among HMGRs reveals two distinct classes: eukaryotic HMGRs (class I) and prokaryotic HMGRs (class II) (Bochar et al., 1999a). The majority of archeal HMGRs are assigned to class I; however, HMGR from the archaeon Archaeoglobus fulgidus appears to be more closely related to class II HMGRs. All class I HMGRs, with the exception of the archeal members of the family, contain N–terminal membrane domains that are involved in the sterol-regulated degradation of the HMGR protein, while class II HMGRs lack a membrane domain and are soluble. The membrane domains of class I HMGRs are diverse, containing two (plants), seven (yeast) or eight (mammals) membrane-spanning helices. The amino acid sequences of catalytic portions are well conserved within each class (Hampton et al., 1996); they are quite different, with sequence identities of only 14–20%, between class I and II. Detailed studies on the reaction mechanism in Saccharomyces cerevisiae (Veloso et al., 1981), Pseudomonas mevalonii (Wang et al., 1990; Darnay et al., 1992; Bochar et al., 1999b) and hamster HMGR (Frimpong and Rodwell, 1994a,b) have shown that the reductive cleavage of HMG-CoA to mevalonate uses two molecules of NADPH and proceeds via two successive hydride transfers. The overall reaction can be divided into three steps. The first reduction results in the formation of a mevaldyl-CoA hemi-thioacetal intermediate (Scheme 2). This intermediate decomposes to mevaldehyde and CoAS– and is subsequently protonated (Scheme 3). A second NADPH molecule then replaces NADP+ and reduces mevaldehyde to mevalonate (Scheme 4).
Scheme 2.
Scheme 3.
Scheme 4.
The crystal structure of HMGR from the eubacterium P.mevalonii revealed a novel protein fold and showed that the enzyme forms a dimer (Lawrence et al., 1995). Pseudomonas mevalonii HMGR normally catalyzes the reverse reaction of human HMGR, namely the synthesis of HMG-CoA from mevalonate and CoA, and the bacterium can utilize mevalonate as a sole carbon source (Gill et al., 1985; Beach and Rodwell, 1989). A catalytic mechanism for the reduction of HMG-CoA to mevalonate in prokaryotic HMGRs was proposed recently (Tabernero et al., 1999). Mutagenesis studies (Darnay et al., 1992; Darnay and Rodwell, 1993; Frimpong and Rodwell, 1994a,b) and the crystal structure of P.mevalonii HMGR (Lawrence et al., 1995) have identified a number of residues that are thought to participate in substrate binding or catalysis.
Here we report the three-dimensional structure of the catalytic portion of human HMGR (residues 426–888), providing the first structural insights into class I reductases. The structure of human HMGR in the complex with substrates allows for a detailed characterization of the active site and shows that it differs in architecture from the bacterial HMGR active site. The crystallographic analysis, as well as solution studies, reveals that the catalytic portion of human HMGR is a tetramer, suggesting a revised mechanism of sterol sensing by HMGR.
Results and discussion
Structure determination
The catalytic portion (residues 426–888) of recombinant human HMGR was expressed and purified as described in Materials and methods. Three crystal structures were determined with different substrates bound to the protein: form A contains HMG and CoA (refined to 2.1 Å resolution); form B (refined to 2.8 Å resolution) contains HMG-CoA; and form C (refined to 2.0 Å resolution) contains HMG, CoA and NADP+. The structure of form A crystals was determined by multi-wavelength anomalous dispersion (MAD) from 58 selenomethionine-substituted positions (Table I). The structures of crystal forms B and C were determined by molecular replacement using the structure of form A as the search model. Each of the three structures contains four monomers in the asymmetric unit. The structures of the 12 crystallographically independent monomers are very similar but not identical. All 12 models contain residues 477–863, while additional N- (439–476) and C–terminal residues (863–872) are present in only some of the monomers (Table II). Residues 426–438 and 873–888 are not visible in any of the structures. With the exception of residues 439–476, which are poorly ordered in all structures, the electron density maps are of excellent quality and the models are well refined, as indicated by low R–factors and small deviations from ideal geometry (Table II).
Table I. MAD data collection and phasing statistics.
| λ1 (peak) | λ2 (inflection) | λ3 (high energy remote) | |
|---|---|---|---|
| Wavelength (Å) | 0.979471 | 0.979595 | 0.964237 |
| f′/f′ | –7.65/4.74 | –9.30/2.98 | –5.20/3.70 |
| No. of unique reflections | 105 550 | 105 651 | 105 088 |
| Average multiplicity | 2.1 | 2.1 | 2.2 |
| Completeness (%) | 99.4 (99.5) | 99.4 (99.5) | 99.4 (99.5) |
| <I>/σI | 18.0 (2.9) | 17.8 (2.6) | 20.3 (2.7) |
| Rmergea | 5.6 (22.2) | 6.1 (33.4) | 4.3 (29.8) |
| Phasing powerb | 2.95 | 2.19 | 2.18 |
| Space group | P21 | ||
| Resolution range (Å) | 51.0–2.6 | ||
| Unit cell dimensions | a = 75.25, b = 128.26, c = 94.91 Å, β = 105.01° |
aRmerge = Σ∣(Ihkl) – <I>∣/Σ(Ihkl), where Ihkl is the integrated intensity of a given reflection. For Rmerge, completeness and I/σI, the numbers in parentheses refer to data in the highest resolution shell.
bPhasing power is defined as <fH>/residual, where <fH> is the r.m.s. of the heavy atom structure factor amplitudes in the derivative, and the residual is the r.m.s. of the lack of closure error.
Table II. Native data collection and refinement statistics.
| Form A | Form B | Form C | |
|---|---|---|---|
| Space group | P21 | P21 | P21 |
| Unit cell dimensions | a = 75.30, b = 130.18, | a = 74.58, b = 171.20, | a = 73.92, b = 172.63, |
| c = 92.55 Å, β = 106.48° | c = 80.37 Å, β = 116.49° | c = 73.99 Å, β = 117.50° | |
| Resolution range (Å) | 38.0–2.1 | 25.0–2.8 | 44.0–2.0 |
| Wavelength (Å) | 1.012 | 1.5418 | 1.083 |
| No. of unique reflections | 101 445 | 37 393 | 109 778 |
| Average multiplicity | 3.1 | 2.5 | 3.9 |
| Completeness (%) | 99.1 (97.5) | 85.6 (41.0) | 100.0 (100.0) |
| <I>/σI | 16.0 (2.5) | 13.3 (1.9) | 27.4 (7.2) |
| Rmerge (I> –3σI) | 5.9 (38.5) | 7.3 (39.8) | 5.0 (19.3) |
| R/Rfreea | 0.199/0.239 | 0.209/0.253 | 0.168/0.197 |
| Residues | |||
| monomer 1α | 439–451, 460–863 | 461–865 | 462–870 |
| monomer 1β | 461–865 | 460–864 | 462–866 |
| monomer 2α | 464–865 | 460–866 | 468–871 |
| monomer 2β | 458–864 | 453–865 | 477–872 |
| Ligands | 4 HMG, 4 CoA | 4 HMG-CoA | 4 HMG, 4 CoA, 4 NADP+ |
| Waters | 282 | 472 | |
| Other molecules | 2 DTT | ||
| No. of atoms in refinement | 12 669 | 12 351 | 12 911 |
| Deviation from ideality | 0.0105/1.51 | 0.0157/1.62 | 0.0120/1.58 |
| R.m.s.d. bonds (Å)/angles (°) | |||
| average protein B–factors | 56.3/30.3/36.0 | 71.0/46.6/52.44 | 34.8/16.3/19.3 |
| N/L/S domain (Å2)b | |||
| average ligand B–factors (Å2) | 41.6 | 58.6 | 23.0 |
| B–factor r.m.s.d. of bonded atoms main/side chain | 1.190/2.035 | N/Ac | 1.39/2.54 |
aR = (Σ∣Fobs – Fcalc∣)/(ΣFobs), where Fobs and Fcalc are observed and calculated structure factors, respectively.
bThe three domains are indicated in Figure 1.
cB–factors were grouped for residues.
Molecular architecture
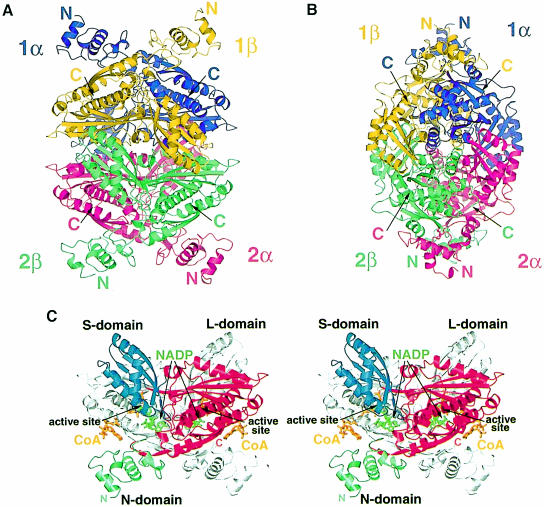
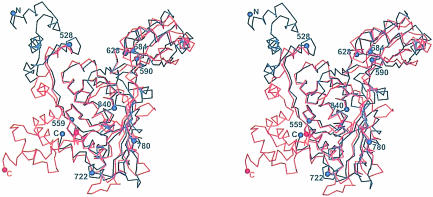
Catalytic portions of human HMGR form tetramers with approximate D2 symmetry and overall dimensions of ∼110 × 80 × 70 Å. The individual monomers wind around each other in an intricate fashion (Figure 1A and 1B). In the tetramer, the monomers are arranged in two dimers (called ‘1’ and ‘2’), each of which has two active sites. The active sites are formed by residues from both monomers (called ‘α’ and ‘β’) (Figure 1C).
Fig. 1. Ribbon diagrams of human HMGR. (A and B) The structure of the HMGR homotetramer. Dimer 1 consists of the monomers called ‘1α’ (purple) and ‘1β’ (yellow), while dimer 2 consists of monomers ‘2α’ (pink) and ‘2β’ (green). (C) Stereo diagram of the HMGR dimer structure. For clarity, only one monomer is colored. The N–terminal N–domain is in green, the large L–domain is in red and the small S–domain is in blue. The N–terminus (residue 462) and the C–terminus (residue 870) for the colored monomer are indicated. CoA (orange) and NADP(H) (green) are labeled and the two active sites are marked. This figure and Figures 5–8 were prepared with Bobscript (Esnouf, 1999), gl_render (L.Esser, personal communication) and POV-Ray (Persistence of Vision Ray Tracer v3.02, Copyright 1997 POV-Team, http://www.povray.org).
Structure of the monomer
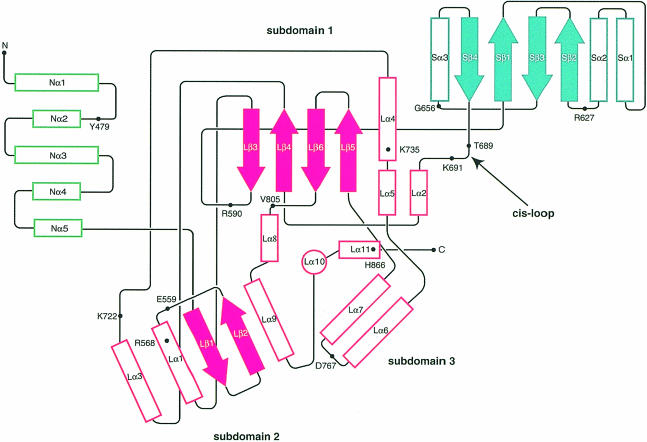
Monomers of the catalytic portion of human HMGR consist of three domains: an N–terminal ‘N–domain’, a large ‘L–domain’ and a small ‘S–domain’ (Figures 1C and 2). The N–domain (residues 460–527) is the smallest of the three domains and is α-helical. Its high B–factors (Table II) and weak electron density for residues 460–477 indicate mobility. In the full-length protein, the N–domain connects the catalytic portion of HMGR to the membrane domain. The fold of the L–domain (residues 528–590 and 694–872) is unique to HMGRs. Its architecture resembles a prism, with a 27–residue α-helix (Lα10) forming the central structural element. This helix is surrounded by three subdomains (Figure 2). The S–domain (residues 592–682) is inserted into the L–domain between Lβ3 and Lα2. It forms the binding site for NADP(H) and folds as an α/β sandwich that resembles ferredoxin and not the Rossmann fold usually observed in NAD(P)-dependent enzymes (Rossmann et al., 1975).
Fig. 2. Topology diagram of the human HMGR monomer. Colors for the three domains are as in Figure 1C. Helices are shown as outlined rectangles and strands are shown as solid arrows. The central helix Lα10 in the L–domain is indicated by the outlined circle. Residues that participate in substrate binding as well as the N- and C–termini are indicated.
The S- and L–domains are connected by an almost continuous β–strand (Lβ3 and Sβ1) and a loop (residues 682–694). This loop, which we call the ‘cis-loop’ because it contains a cis-peptide between residues C688 and T689, is essential in the formation of the HMG-binding site.
Oligomerization
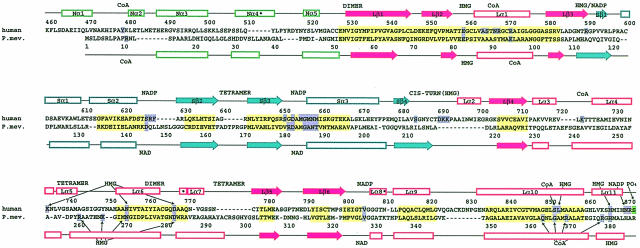
All three domains of a monomer participate in the formation of a dimer (Figure 1C). The N–terminal residues of the L–domain wrap around the equivalent amino acids of the neighboring monomer to establish a β–sheet with nine main chain hydrogen bonds between two monomers (Figure 1C). A prominent feature of this region is the sequence element ENVIGX3I/LP, which is conserved in both classes of HMGRs (Figure 3), supporting a role as a key dimerization element for the enzyme. A second dimerization element is located in the L–domain's helical subdomain: helices Lα6 and Lα7 fold antiparallel against the equivalent helices of the neighboring monomer to establish a buried four-helix bundle. Additional interactions between L- and S–domains of neighboring monomers involve the S–domain's four-stranded antiparallel β-sheet of monomer α, which is packed next to helices Lα4 and Lα5 of monomer β. A salt bridge between residues R595 and E730, and numerous hydrogen bonds stabilize the interactions between the domains. In the N–domain of human HMGR, monomer–monomer interactions are formed by the 310–helix Nα4 that packs against helix Lα9 of the L–domain.
Fig. 3. Structure-based sequence alignment of human and P.mevalonii HMGR (PDB code 1qax; Tabernero et al., 1999). Colors of the secondary structure elements are as in Figure 1C; solid arrows indicate β–strands, outlined rectangles indicate α–helices and an asterisk indicates a 310-helix. Regions of high structural similarity are in yellow, residues involved directly in substrate binding are in purple and regions involved in dimerization or tetramerization are marked by the words ‘dimer’ and ‘tetramer’. The phosphorylation site (S872) of human HMGR is marked in green.
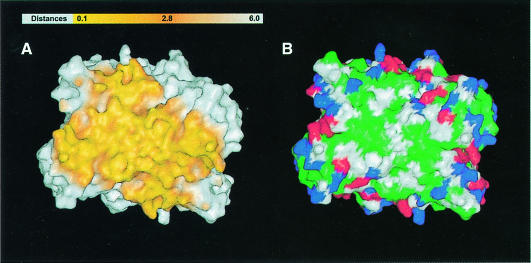
The formation of the human HMGR tetramer buries a total solvent-accessible area of 24 260 Å2 or 46% of the tetramer surface. Of this area, each monomer contributes 4480 Å2 in the formation of the dimer, while an additional 6340 Å2 (3170 Å2 per dimer) are buried upon formation of the tetramer. The surface of the dimer–dimer interface has the shape of a saddle (Figure 4A and B). Residues at the interface are typical of protein–protein interactions (Figure 4B) (Jones and Thornton, 1995). Carbon atoms contribute 64% and polar atoms (N, O and S) contribute 36% of the contact surface. Entropic considerations suggest that few surface water molecules become buried in the formation of protein–protein interfaces (Horton and Lewis, 1992). In the form C structure, 32 out of 472 water molecules are located at the tetramer contact surface and only 11 of these 32 water molecules are solvent inaccessible.
Fig. 4. Molecular surface of the dimer–dimer interface. (A) Atoms between 0.1 and 6 Å of the dimer–dimer interface are indicated by a color gradient of yellow to orange. (B) Colored according to residue type with hydrophobic residues (A, V, F, P, M, I, L, Y, W and G) in green, polar residues (S, T, H, C, N and Q) in white, K and R in blue, and D and E in red. This figure was prepared with GRASP (Nicholls et al., 1991), gl_render and POV-Ray.
The D2 symmetry of the tetramer results in unique interactions between each monomer of one dimer with the two monomers of the neighboring dimer. Although the interface is predominantly hydrophobic (Figure 4B), buried salt bridges between residues R641 and E782 and hydrogen bonds between residues E700 and E700 from neighboring monomers establish four anchor points of the saddle. The majority of monomer α–monomer β interactions of neighboring dimers are formed by the slightly twisted antiparallel β-sheet of the L–domain, which packs against the S–domain (Figure 1A and B). Contacts between monomer 1α and monomer 2α are less extensive and limited to helix Lα5, the loop connecting Lα5 and Lα6 and the 310–helix Lα8.
The large contact areas between the dimers observed in all our crystal forms suggest that the catalytic portion of human HMGR exists as a tetramer in solution; this is supported by the results of analytical ultracentrifugation experiments. Both sedimentation velocity and sedimentation equilibrium studies of the catalytic portion are best interpreted in terms of a population of particles of ∼200 000 Da, which corresponds well to the molecular weight of four 49 953 Da monomers. On the basis of these results, we conclude that full-length human HMGR is likely to form a tetramer as well. Human HMGR contains an N–terminal transmembrane domain (residues 1–340) that resides in the endoplasmic reticulum (ER) membrane (Roitelman et al., 1992) and that is missing in our construct. A 120–residue linker connects the membrane domain to the catalytic portion. This linker would allow the HMGR tetramer to position itself with its long dimension parallel to the ER membrane such that the four membrane domains are close to one another.
Mammalian HMGR was first solubilized and partially purified by Brown and co-workers (Brown et al., 1973). Molecular cloning revealed that full-length HMGR is easily proteolyzed and that the solubilized protein was a fragment containing the complete catalytic portion (Chin et al., 1982b). This catalytic fragment, which is similar to the one studied here, migrated on denaturing gels with an apparent mol. wt of 50 kDa and on gel filtration as a 200 kDa species (Brown et al., 1973; Chin et al., 1982a). The enzyme had the unusual property that it was inactivated by exposure to low temperatures and protected against cold inactivation by high concentrations of salt. Brown and co-workers (Brown et al., 1973) speculated that the purified enzyme was a multimer and was held together by hydrophobic forces that were disrupted at low temperature and stabilized at high ionic strength. The functional size of full-length mammalian HMGR was studied further by radiation inactivation of microsomes, and was suggested to be dimeric (Edwards et al., 1985) or to change between dimeric and monomeric forms (Ness et al., 1988). The molecular mass of a tetramer of catalytic portions is similar to that of a dimer of full-length proteins (∼200 kDa), and it is possible that in some cases a tetramer of soluble catalytic subunit rather than a dimer of the full-length enzyme was studied. If HMGR indeed exists in the ER membrane as a dimeric enzyme, it may be catalytically active, since the dimer–dimer interface is distant from the active site. However, the hydrophobic nature of the surfaces involved in the formation of a tetramer suggests that dimeric HMGR may be less stable than tetrameric HMGR.
The membrane domains of mammalian HMGRs contain a 167–residue segment, termed the sterol-sensing domain, which shares ∼25% sequence identity with membrane regions of other proteins that are influenced by cholesterol (Brown and Goldstein, 1999). This domain is responsible for the enhanced degradation of HMGR in response to increased concentrations of oxysterols (Kumagai et al., 1995; McGee et al., 1996). Degradation of HMGR appears to be initiated when a membrane-bound cysteine protease cleaves within the transmembrane region of HMGR (Moriyama et al., 1998). The rate of protein degradation is influenced not only by the concentration of sterols, but also by the oligomeric state of the enzyme. When monomeric soluble proteins are fused to the HMGR membrane domain, the rate of degradation of the fusion protein is increased even in the absence of sterols (Cheng et al., 1999). These experiments, together with the crystallographic data, suggest that the soluble domains may initiate the tetramerization of the membrane domains, and that dissociation of the membrane domains increases the accessibility of HMGR for the protease cleavage, resulting in the inactivation of the enzyme.
This mechanism of control may be general to proteins containing sterol-sensing domains. For example, the protein SCAP activates membrane-bound transcription factors that modulate the expression of enzymes involved in cholesterol biosynthesis and uptake (SREBPs) and contains a sterol-sensing domain (Brown and Goldstein, 1999). Similarly to HMGR, the activity of SCAP is decreased when sterol concentrations are high. Although the functional size of SCAP is unknown, the protein interacts with the membrane-bound site–1 protease (S1P) and SREBPs, suggesting that the membrane domain of SCAP forms hetero-oligomers (Brown and Goldstein, 1999). It may be that the disruption of these oligomers is at least partially responsible for the decrease in SCAP activity. Sterols also affect the ability of SCAP to cycle between the ER and the Golgi. This cycling of SCAP may influence the transport of SREBPs to a post-ER compartment, where the transcription factors become activated (Nohturfft et al., 1999).
Substrate binding
The extended HMG-CoA and NADPH molecules make numerous contacts with the L- and S–domains of the protein to form the four active sites in the tetramer. At each active site, the HMG moiety of one HMG-CoA molecule, which is bound predominantly to a single monomer, comes into the proximity of the nicotinamide ring of an NADPH molecule, whose binding pocket is located in the neighboring monomer (Figure 1C). Thus, the active sites are positioned at the interface of the two monomers of a dimer. The formation of the tetramer does not appear to be involved in substrate binding.
CoA binding
CoA binds in an extended conformation (Figure 1C): the ADP moiety of CoA is located in a positively charged pocket near the enzyme surface, with the pantothenic acid part extending deep into the interior of the protein. All but one of the interactions between HMGR and CoA are formed by the L–domain of a single monomer (Table IIIA). The neighboring monomer contributes a single tyrosine residue (Y479) to the binding of CoA. This tyrosine residue, which is located in the N–domain, forms a hydrophobic shield over the CoA adenine base and appears to close the extended binding pocket. In our crystals, CoA is tightly bound and is not removed when crystals are soaked in CoA-free solutions for several days. It is likely that in order for CoA to be released, Y479 must move. In solution, conformational changes of the N–domain could assist in the liberation of the reaction product from the binding pocket.
Table III. HMGR interactions with substrate.
|
A. Interactions with CoA | |||
|---|---|---|---|
| Group | CoA atom | HMGR atom | Distancea |
| Adenine | N3A | N567 ND2 | 3.0–3.2 |
| ring | Y479 side chain | ∼3.0–4.0 | |
| C5A or C6A | A564 CB | 3.6–3.7 | |
| Ribose-3′P | O7A | Y479 OH | 2.6 |
| O9A | R571 NH2 | 4.0–5.0 | |
| Ribose | O2*A (2′OH) | N567 OD1 | 3.0–3.2 |
| diphosphate | O1A (Pα) | K722 NZ | 2.6–2.7 |
| O4A (Pβ) | K722 NZ | 2.6–2.7 | |
| R568 NH1 | 3.0–3.1 | ||
| Pantothenate | O9P | S852 OG | 2.9–3.0 |
| O5P | S565 OG | 2.7–2.8 | |
| N4P | H866 NE2 | 2.9–3.8 | |
| |
N8P |
S865 OG |
3.1–3.4 |
|
B. Interactions with NADP+ | |||
| Groupb |
NADP atom |
HMGR atom |
Distancea |
| Adenine | AN6 | D653 OD2 | 3.1–3.4 |
| AN1 | D653 N | 2.8–2.9 | |
| ring | F628 side chain | ∼3.5 | |
| ring | V805 CG1 and CG2 | ∼3.8–4.5 | |
| Ribose-A-2′P | AOP1 | S626 OG | 2.5–2.6 |
| Water | 2.6–2.9 | ||
| AOP3 | R627 NH1 | 3.1–3.8 | |
| R627 N | 2.8–3.1 | ||
| Ribose-A | AO5* | R871 NH1 | 3.1–3.2 |
| diphosphate | AO2 (Pα) | N658 ND2 | 2.9–3.0 |
| R871 NH1 | 3.9–3.1 | ||
| NO1 (Pβ) | G656 N | 2.6–2.8 | |
| M655 N | 3.1–3.4 | ||
| Water | 2.7–2.9 | ||
| NO2 (Pβ) | M657 N | 3.6–3.7 | |
| M659 N | 2.9–3.1 | ||
| Ribose-N | NO4* | N658 ND2 | 3.4–3.5 |
| NO2* | D767 OD2 | 2.6–2.7 | |
| NO3* | N870 ND2 | 3.0–3.6 | |
| Water | 2.8–3.0 | ||
| Nicotinamide | NN1 | E559 OE1 | 3.1–3.3 |
| NN7 | N658 OD1 | 3.2–3.6 | |
| NC4 | M657 CG | 2.9–3.6 | |
| NO7 | R590 NH2 | 2.4–2.7 | |
|
C. Interactions of HMG with hHMGRc or pmHMGRd |
|||||
|---|---|---|---|---|---|
| Group | Atom 1 | hHMGR | Distancea | pmHMGR | Distancea |
| Carboxylate | O3 | K735 NZ | 2.6–2.7 | ||
| O4 | S684 OG | 2.5–2.6 | R261 NH2 | 2.6 | |
| K692 NZ | 3.1–3.3 | ||||
| 3-methyl | C6 | L853 CD2 | 3.9 | A368 CB | 3.7 |
| 3-hydroxyl | O7 | D690 OD2 | 2.6–2.8 | ||
| R590 NH2 | 3.0–3.3 | ||||
| Carbonyl | O2 | K691 NZ | 2.7–2.9 | K267 NZ | 2.8 |
| E559 OE2 | 2.5–2.6 | E83 OE2 | 2.9 | ||
| N755 ND2 | 2.7–2.8 | N271 OD1 | 3.2 | ||
| C1 | NADP NC4 | 2.9–3.1 | NAD NC4 | 4.0 | |
| hHMGR-D767 | OD2 | E559 OE1 | 3.6–3.7 | E83 OE1 | 3.0 |
| (pmHMGR-D283) | OD2 | K691 NZ | 2.6–2.7 | K267 NZ | 3.3 |
aAll distances are in Å. For human HMGR, distances are taken from the form C structure, and the distance range for the four crystallographically independent molecules is reported.
bRibose-A corresponds to the ADP ribose, ribose-N to the nicotinamide ribose of NADP.
chHMGR, human HMGR.
dpmHMGR, P.mevalonii HMGR (PDB code 1qax).
NADP(H) binding
The NADP(H)-binding site is formed primarily by the S–domain (Figure 1C and Table IIIB). Among the residues with specific interactions with NADP(H), the loop region between helix Sα2 and strand Sβ2 (residues 626–628) participates in binding of the NADP(H) ADP moiety (Table IIIB). The diphosphate is stabilized primarily by main chain interactions with residues located in the highly conserved sequence element DAMGXN and by the dipole of helix Sα3. Residues N870 and R871 and the C–terminal residues of helix Lα11 are only ordered in the NADP+-containing structure (Table II), indicating that it is the binding of NADP(H) that causes a conformational change in the C–terminus of the enzyme, resulting in the complete closure of the active site. A substrate-free structure of human HMGR does not exist. However, such a structure has been described for bacterial HMGR (Lawrence et al., 1995). In this enzyme, the active site is partially formed and the only major conformational changes upon substrate binding result in the ordering of a C–terminal domain (Tabernero et al., 1999). These changes are similar to the ordering of C–terminal residues in human HMGR when NADP+ is bound in addition to HMG-CoA, as described here.
HMG binding
The HMG-binding pocket is located between the L- and S–domains, and residues from neighboring monomers contribute to the binding (Figure 5). It is the site of catalysis and the active site in HMGR. Although we used HMG-CoA as ligand in all our co-crystallization trials, the electron density unambiguously shows that the substrate is cleaved to HMG and CoA in form A and form C structures (Figure 6). The lower resolution form B structure, on the other hand, is best interpreted as containing intact HMG-CoA. The presence of cleaved HMG-CoA in our crystals is possible due to the fact that HMG-CoA is hydrolyzed to HMG and CoASH in buffer over time, as confirmed by mass spectrometric analysis. In our experiments, HMG-CoA is added to the crystallization drop and crystals grow over several weeks (see Materials and methods). It is unclear why HMG-CoA does not appear to be cleaved in the crystal used to solve the form B structure. All structural differences between the core region (residues 500–860) of the monomers are very small, with Cα r.m.s. deviations of ∼0.2 Å between monomers of different crystal forms and deviations of 0.13–0.23 Å between monomers of the same crystal form. Furthermore, atoms from residues within 5 Å of the HMG-binding site also superimpose very well, with r.m.s. deviations of 0.2 Å. Only the CoA thiol group, which rotates away and towards the back of the active site in the cleaved HMG-CoA structures, is noticeably different in the three crystal forms. This rotation of the CoA thiol may simply be a consequence of limiting steric overlap between the thiol and the oxygen atom of HMG introduced in the cleavage of HMG-CoA.
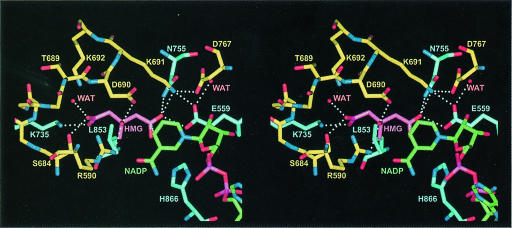
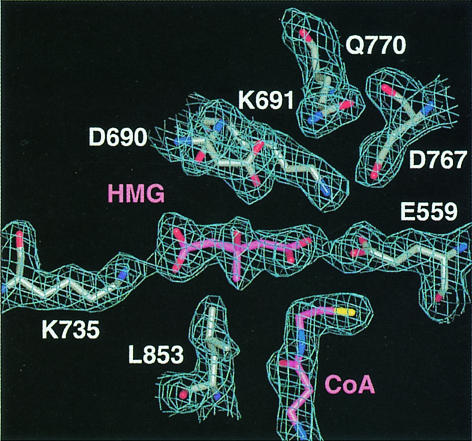
Fig. 5. Stereo diagram showing residues involved in HMG binding based on the form C structure at a resolution of 2.0 Å. Residues from monomer α are in yellow and residues from monomer β are in blue. HMG is magenta and NADP+ is in green. All distances within 3.0 Å or closer are indicated by dotted lines.
Fig. 6. Simulated annealing omit 2Fo – Fc map at 2.1 Å and contoured at 1σ showing the cleaved HMG and CoA of the form A structure. HMG and CoA are colored in magenta.
Many specific interactions between human HMGR side chains and HMG are formed (Figure 5 and Table IIIC). The ‘cis-loop’ (residues 684–692) that bends over the top of HMG is the most important structural element in the binding of HMG. Its cis-peptide bond between residues C688 and T689 is critical in positioning residues D690, K691 and K692. Residues E559 and D767 are located in the front of the active site (Figure 5). K691 is also positioned in the active site with its NZ atom 2.7–2.9 Å from the HMG O2 carbonyl oxygen (Table IIIC).
Catalysis
Analysis of the three structures of human HMGR presented here provides new insights into the mechanism by which the reductive cleavage is catalyzed. The product of the first reduction reaction, mevaldyl-CoA (Scheme 2), has a negatively charged oxygen and must be stabilized in the enzyme. This is accomplished by K691, whose side chain is ideally positioned in the middle of the active site. The ordering of C–terminal residues, including H866, upon NADP+ binding results in the completion and closure of the active site. When HMG-CoA, as is found in the form B structure, is modeled in the form C structure, H866 is within hydrogen bonding distance of the thiol. This is consistent with its proposed role as a proton donor to the thioanion (Frimpong and Rodwell, 1994b). Several residues participate in the reduction of mevaldehyde (Scheme 4). The OE1 atom of E559 is located 2.5–2.6 Å from the HMG carbonyl oxygen and 3.6–3.7 Å from D767 (atom OD2) (Table IIIC). The proximity of E559 to D767 potentially could raise the pKa of the glutamic acid side chain such that it may be protonated. Consequently, we propose that E559 is the proton donor for mevaldehyde. D767 is critical in the catalysis, because its side chain is positioned near D559 and it also forms ionic interactions with K691, stabilizing the lysine side chain in the active site. A possible mechanism for bacterial HMGR has been proposed recently (Tabernero et al., 1999). In this model, K267 is the proton donor in the reduction of mevaldehyde to mevalonate, whereas in human HMGR the analogous K691 most probably stabilizes the mevaldyl-CoA intermediate and E559 donates the proton to mevaldehyde. Given the short distance between E559 and the HMG carbonyl oxygen (Table IIIC), it is likely that E559 and the analogous E83 in P.mevalonii participate directly in the reduction of mevalonate. However, differences in the two catalytic models could also be due to different mechanisms between class I and class II HMGRs.
Phosphorylation
HMGR activity is modulated by phosphorylation. The level of phosphorylation is controlled by a pair of enzymes: AMP-activated protein kinase and HMG-CoA phosphorylase (Omkumar et al., 1994). Phosphorylation at S872 reduces the activity of the protein. A similar effect can be observed when S872 is mutated to an aspartate (Omkumar et al., 1994). Because S872 is located close to the catalytically important residue H866 in the primary structure, it was proposed that the phosphoserine interacts directly with H866 and abstracts its imidazolium proton (Omkumar and Rodwell, 1994). In our structure, S872 is not close to H866, but in the vicinity of the α–phosphate of NADP+ and the side chain of R871. This suggests that phosphorylation is likely to result in a decrease in affinity for NADPH. However, replacement of S872 by aspartate does not significantly change the apparent Km for NADPH (Omkumar et al., 1994). A possible explanation for this observation is that phosphoserine is significantly larger than aspartate and has two negative charges rather than one. These differences may contribute to a reduction in NADP(H) affinity in the phosphorylated enzyme that was not observed in the mutant protein.
Comparison of human and bacterial HMGR
A superposition of the Cα atoms of the human with the P.mevalonii monomer structures (Figure 7) shows large differences in the N- and C–terminal domains, while the cores adopt similar folds. A closer inspection of the structures and sequence alignments (Figure 3) indicates that elements important for the formation of the tetramer in the human enzyme are not present in the bacterial structure: insertions at residues 640 and 776 as well as large conformational differences at residue 740 (residues 168, 293 and 258, respectively, in the P.mevalonii HMGR) prevent a close packing of neighboring dimers as seen in the human structure. Sequence elements important for dimerization, on the other hand, are conserved (Figure 3), and the HMGR class I and class II dimers are similar.
Fig. 7. Stereo drawing of the Cα superposition of human (in blue) and P.mevalonii (in red) (PDB code 1qax) HMGR monomer. The superposition was done with the program LSQMAN (Kleywegt, 1997). Selected residues in human HMGR are indicated by blue spheres and numbered. The N- and C–termini are indicated.
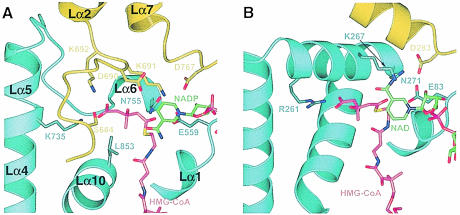
The most striking difference between the bacterial and the human active sites is the ‘cis-loop’ in human HMGR. This key HMG-binding module in the human enzyme is not present in P.mevalonii HMGR (Figures 3 and 8). The catalytic lysine residue (K691), which resides in the ‘cis-loop’ of one monomer α in human HMGR, is replaced by a lysine from the neighboring monomer β (K267) in P.mevalonii HMGR (Figure 8 and Table IIIC). As a result, the position and orientation of key catalytic residues are conserved in the class I compared with class II HMGRs despite the large differences in the amino acid sequence and architecture.
Fig. 8. Ribbon diagram of the human HMGR (A) and P.mevalonii HMGR (B) active site. Residues from monomer α are in yellow, residues from monomer β are in blue, HMG-CoA is in magenta and NAD(P) is in green. For human HMGR, secondary structure elements are labeled. For clarity, not all CoA atoms are shown. Side chains of selected residues are indicated.
The orientation of the NAD(P) dihydronicotinamide ring is not conserved in the two HMGR classes. NAD(P)-dependent oxidation and reductions are classified according to which of the two C-4 protons of the dihydronicotinamide ring is transferred. The position of the nicotinamide in the structure of human HMGR suggests that the human enzyme transfers the pro–(R) hydrogen and belongs to class A NADP-dependent enzymes. For P.mevalonii HMGR, on the other hand, the nicotinamide ring is rotated by ∼180° compared with human HMGR (Figure 8). It appears that the bacterial enzyme transfers the pro–(S) hydrogen atom and therefore belongs to class B NAD(P)-dependent enzymes.
Differences in the mode of substrate binding in the two enzymes are not limited to the active site. The P.mevalonii HMGR utilizes NAD(H) rather than NADP(H) for its cofactor and consequently lacks structural elements that are involved in the binding of the NADP(H) 2′–phosphate in the human enzyme (Figure 3). Furthermore, a number of residues that participate in CoA binding in human HMGR are replaced by functionally similar residues in P.mevalonii HMGR that are located in different secondary structural elements (Figure 3).
Statin binding and inhibitor design
Statins are potent competitive HMGR inhibitors (Brown et al., 1978). All statins share an HMG moiety and rigid hydrophobic groups linked to it (Endo and Hasumi, 1989; Corsini et al., 1995). Inspection of the active site suggests possible binding modes of these inhibitors. Because the CoA-binding pocket is elongated and narrow, it is unlikely that the statins' cyclic groups are located in this pocket. The nicotinamide of NADP(H), however, is 2.9–3.1 Å from the HMG carbonyl carbon C1. It is possible that the statins' cyclic groups mimic the nicotinamide ring of NADP(H) and that the inhibitors exploit binding pockets originating from both substrates, namely the HMG-binding site and the nicotinamide-binding site. Clearly, a high resolution crystal structure of human HMGR with statin is required to confirm that statins indeed bind in the active site of the enzyme and in order to visualize specific interactions.
The large differences in architecture between the active sites of human HMGR and P.mevalonii HMGR suggest that statins are specific to class I reductases and that the P.mevalonii HMGR structure is a poor model for the design of inhibitors for human HMGR. On the other hand, it may be feasible to develop novel bacterial HMGR inhibitors that do not bind strongly to human HMGR. In the era of rapidly developing antibiotic-resistant pathogens, it is essential that new antimicrobial agents become available and new metabolic pathways need to be exploited.
Materials and methods
Protein expression and purification
A plasmid containing the gene for human HMGR was purchased from the American Type Culture Collection. The segment encoding the catalytic portion of the protein (residues 426–888) was amplified by PCR and inserted into the pGEX-cs expression plasmid (Parks et al., 1994). The recombinant fusion protein containing an N–terminal glutathione S–transferase (GST) protein was expressed in Escherichia coli DH5α cells. Selenomethionine variant protein was produced in the E.coli methionine auxotroph B834. Cells were grown at 24°C, and the medium was supplemented with 0.5 M sorbitol and 2.5 mM betaine (Blackwell and Horgan, 1991). We observed the expression of two protein products: a 50 kDa protein, which corresponds to the full-length product, and a 44 kDa protein. Western blot analysis and protein sequencing confirmed that the smaller protein was a fragment of HMG-CoA reductase; its expression was caused by an internal translation initiation site consisting of an ATG preceded by a ribosome-binding site at residue 485 (Mayer et al., 1988). To facilitate purification, we mutated M485 to isoleucine using the pALTER mutagenesis system (Novagen). The enzymatic activity of the M485I mutant protein is unchanged compared with the wild-type protein (data not shown). The recombinant protein was purified using a GST affinity matrix (Sigma) and released from the GST fusion protein by cleaving with a recombinant tobacco etch virus protease–GST fusion protein (Parks et al., 1994). Additional purification was achieved using anion exchange chromatography on a Mono Q column (Pharmacia). At this point, the protein appeared homogenous in size as shown on SDS–PAGE and by mass spectrometry, but purification on phenyl–Superose (Pharmacia) separated the protein further into a hydrophilic and a hydrophobic fraction, of which only the hydrophobic species produced well diffracting crystals. The last purification step was size exclusion chromatography with a Superose S–200 column (Pharmacia) equilibrated with buffer A [10 mM HEPES–NaOH pH 7.5, 1 mM EDTA, 50 mM NaCl, 10% glycerol and 30–50 mM dithiothreitol (DTT)]. Typical yields were 12–20 mg from a 1 l culture. Mass spectrometric analysis confirmed that incorporation of selenomethione in the variant protein was <95%.
Analytical ultracentrifugation
Sedimentation velocity and sedimentation equilibrium experiments were performed with a Beckman XL-A analytical ultracentrifuge and scanning absorption optics at a wavelength of 280 nm. We used an An–60Ti rotor with double sector cells of 1.2 cm path length and buffer A at 20°C for all experiments. The sedimentation velocity experiment was carried out at 40 000 r.p.m. and a protein concentration of 7.6 μM; the sedimentation equilibrium experiment was carried out at 9000 r.p.m. and protein concentrations between 3.6 and 27 μM. The partial specific volumes of the sample were calculated on the basis of the amino acid composition to be 0.734 cm3/g. The protein concentration was calculated from UV absorbance based on an extinction coefficient of 29 600 M–1cm–1 at 280 nm, which was determined independently using the Edelhoch method (Pace et al., 1995). All centrifugation data were analyzed with the Optima™ data analysis software.
Crystallization and data collection
Crystals of purified HMG-CoA reductase (protein concentration 24–30 mg/ml) were grown by the batch method with microseeding in 12–15% (w/v) PEG 4000, 30–50 mM DTT, 10% glycerol, 0.1–0.2 M ammonium acetate and 30 mM Na–HEPES pH 7.5 at 21°C. (R,S)–HMG-CoA (purchased from Sigma) was added to all crystallization trials at a molar ratio of 1:1. Crystal form C grew in the presence of 1 mM NADP+ as well as HMG-CoA. Crystals appeared after 1–2 days and grew within 10 days to a final size up to ∼0.4 × ∼0.2 × 0.03 mm3. The crystals were harvested after 20–30 days. For cryoprotection, crystals were soaked in solutions with increasing glycerol concentrations up to 25% (v/v) and flash-cooled in liquid propane. The native form A crystal contained 15% saturated glucose in addition to 25% (v/v) glycerol as cryoprotectant. All diffraction data were collected at 100 K. MAD data for form A crystals were collected at the APS beamline 19ID using the SBC CCD detector. Native form A data were collected at beamline X12B at NSLS on an ADSC Quantum 4 CCD detector. Data for native form B were collected on a Rigaku R–axis II detector using focused CuKα radiation from a Rigaku RU–300 anode X–ray generator equipped with mirror optics (Molecular Structure Corporation). Data for the high resolution form C were collected at the SSRL Beamline 7–1 on an MAR detector. All data were processed with DENZO and merged with SCALEPACK (Otwinowski, 1997).
Structure determination and refinement
The structure of the catalytic portion of human HMGR was determined by MAD. Data measured at λ2 (inflection) were used as the native reference set. Of 68 selenium sites, 45 were identified with the program Shake-n-Bake v2.0 (Smith et al., 1998). An additional 13 sites were located with difference Fourier methods. The remaining 10 selenium atoms were disordered in the crystal structure. The positions, occupancies and B–factors of the sites were refined with CNS v0.5 (Brünger et al., 1998), and phases were calculated resulting in a figure of merit of 0.65 at 2.6 Å resolution. Density modification, consisting of 4–fold averaging, solvent flipping and histogram matching, resulted in improved phases and a figure of merit of 0.78. The initial model was built into the resulting electron density at 2.6 Å resolution using the program O (Jones et al., 1991) and refined making use of the prior phase information as a restraint in energy minimization. Once no new density was observed in maps calculated with the MAD data, the model was refined against the 2.1 Å resolution form A data. This was done by rigid body refinement, followed by iterative cycles of simulated annealing, conjugate gradient minimization and individual B–factor refinement. Form B was solved by molecular replacement using the structure of the form A dimer as the search model. The rotation and translation functions were calculated with the program AMoRe (Navaza and Saludjian, 1997). Form C was solved by molecular replacement using the structure of the form A tetramer as the search model and with the program EPMR (Kissinger et al., 1999). Structure refinement for form B and form C structures was carried out as above with CNS, except that B–factors were grouped for residues in the refinement of the form B structure rather than refined individually. For the calculation of the Rfree value, 2275 reflections were used for form A, 2241 for form B and 2186 for form C. With the exception of native data from form A, all reflections with F >0 were included in the refinement. A weak ice ring between 2.24 and 2.26 Å resolution in the diffraction images for form A native data resulted in high R–factors, and we removed all 2129 reflections within this resolution shell. The structures of the individual monomers in the three crystal forms are very similar but not identical. Specific residues included in the each of the 12 models are listed in Table II. Substrate atoms were not included in the refinement until the last stages. Electron density for the substrates was excellent for HMG, CoA and NADP+ in all structures, while the electron density for the pantothenic acid of HMG-CoA in the form B structure was continuous only for two of the four monomers in the asymmetric unit. HMG-CoA appeared to be uncleaved in the form B structure; however, the data were only complete to 3.0 Å resolution and the electron density could not be interpreted unambiguously. Water molecules were included only when density was observed in both 2Fo – Fc and Fo – Fc maps and where they are chemically plausible.
Coordinates
The Protein Data Bank accession numbers are 1DQ8 for the form A structure, 1DQ9 for the form B structure and 1DQA for the form C structure of human HMGR.
Acknowledgments
Acknowledgements
We thank all members of the Deisenhofer laboratory for invaluable discussions, staff at the synchrotron beamlines for assistance, Drs L.Esser and Z.Otwinowski for help with phasing, and Drs M.Machius, K.Fischer Lindahl and M.S.Brown for critical reading of the manuscript.
References
- Beach M.J. and Rodwell, V.W. (1989) Cloning, sequencing and overexpression of mvaA, which encodes Pseudomonas mevalonii 3-hydroxy-3-methylglutaryl coenzyme A reductase. J. Bacteriol., 171, 2994–3001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennis F., Favre, G., Le Gaillard, F. and Soula, G. (1993) Importance of mevalonate-derived products in the control of HMG-CoA reductase activity and growth of human lung adenocarcinoma cell line A549. Int. J. Cancer, 55, 640–645. [DOI] [PubMed] [Google Scholar]
- Blackwell J.R. and Horgan, R. (1991) A novel strategy for production of a highly expressed recombinant protein in an active form. FEBS Lett., 295, 10–12. [DOI] [PubMed] [Google Scholar]
- Bochar D.A., Stauffacher, C.V. and Rodwell, V.W. (1999a) Sequence comparisons reveal two classes of 3-hydroxy-3-methylglutaryl coenzyme A reductase. Mol. Genet. Metab., 66, 122–127. [DOI] [PubMed] [Google Scholar]
- Bochar D.A., Tabernero, L., Stauffacher, C.V. and Rodwell, V.W. (1999b) Aminoethylcysteine can replace the function of the essential active site lysine of Pseudomonas mevalonii 3-hydroxy-3-methylglutaryl coenzyme A reductase. Biochemistry, 38, 8879–8883. [DOI] [PubMed] [Google Scholar]
- Brown M.S. and Goldstein, J.L. (1999) A proteolytic pathway that controls the cholesterol content of membranes, cells and blood. Proc. Natl Acad. Sci. USA, 96, 11041–11048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown M.S., Dana, S.E., Dietschy, J.M. and Siperstein, M.D. (1973) 3-hydroxy-3-methylglutaryl coenzyme A reductase. Solubilization and purification of a cold-sensitive microsomal enzyme. J. Biol. Chem., 248, 4731–4738. [PubMed] [Google Scholar]
- Brown M.S., Faust, J.R., Goldstein, J.L., Kaneko, I. and Endo, A. (1978) Induction of 3-hydroxy-3-methylglutaryl coenzyme A reductase activity in human fibroblasts incubated with compactin (ML-236B), a competitive inhibitor of the reductase. J. Biol. Chem., 253, 1121–1128. [PubMed] [Google Scholar]
- Brünger A.T., et al. (1998) Crystallography and NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D, 54, 905–921. [DOI] [PubMed] [Google Scholar]
- Caruso M.G., Notarnicola, M., Santillo, M., Cavallini, A. and Di Leo, A. (1999) Enhanced 3-hydroxy-3-methyl-glutaryl coenzyme A reductase activity in human colorectal cancer not expressing low density lipoprotein receptor. Anticancer Res., 19, 451–454. [PubMed] [Google Scholar]
- Cheng H.H., Xu, L., Kumagai, H. and Simoni, R.D. (1999) Oligomerization state influences the degradation rate of 3-hydroxy-3-methylglutaryl-CoA reductase. J. Biol. Chem., 274, 17171–17178. [DOI] [PubMed] [Google Scholar]
- Chin D.J., Luskey, K.L., Anderson, R.G.W., Faust, J.R., Goldstein, J.L. and Brown, M.S. (1982a) Appearance of crystalloid endoplasmic reticulum in compactin-resistant Chinese hamster cells with a 500-fold increase in 3-hydroxy-3-methylglutaryl-coenzyme A reductase. Proc. Natl Acad. Sci. USA, 79, 1185–1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin D.J., Luskey, K.L., Faust, J.R., MacDonald, R.J., Brown, M.S. and Goldstein, J.L. (1982b) Molecular cloning of 3-hydroxy-3-methylglutaryl coenzyme A reductase and evidence for regulation of its mRNA. Proc. Natl Acad. Sci. USA, 79, 7704–7708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corsini A., Maggi, F.M. and Catapano, A.L. (1995) Pharmacology of competitive inhibitors of HMG-CoA reductase. Pharmacol. Res., 31, 9–27. [DOI] [PubMed] [Google Scholar]
- Darnay B.G. and Rodwell, V.W. (1993) His865 is the catalytically important histidyl residue of Syrian hamster 3-hydroxy-3-methylglutaryl-coenzyme A reductase. J. Biol. Chem., 268, 8429–8435. [PubMed] [Google Scholar]
- Darnay B.G., Wang, Y. and Rodwell, V.W. (1992) Identification of the catalytically important histidine of 3-hydroxy-3-methylglutaryl-coenzyme A reductase. J. Biol. Chem., 267, 15064–15070. [PubMed] [Google Scholar]
- Edwards P.A., Kempner, E.S., Lan, S.F. and Erickson, S.K. (1985) Functional size of rat hepatic 3-hydroxy-3-methylglutaryl coenzyme A reductase as determined by radiation inactivation. J. Biol. Chem., 260, 10278–10282. [PubMed] [Google Scholar]
- Eisenberg D.A. (1998) Cholesterol lowering in the management of coronary artery disease: the clinical implications of recent trials. Am. J. Med., 104, 2S–5S. [DOI] [PubMed] [Google Scholar]
- Endo A. (1985) Compactin (ML-236B) and related compounds as potential cholesterol-lowering agents that inhibit HMG-CoA reductase. J. Med. Chem., 28, 401–405. [DOI] [PubMed] [Google Scholar]
- Endo A. and Hasumi, K. (1989) Biochemical aspect of HMG-CoA reductase inhibitors. Adv. Enzyme Regul., 28, 53–64. [DOI] [PubMed] [Google Scholar]
- Esnouf R.M. (1999) Further additions to MolScript version 1.4, including reading and contouring of electron-density maps. Acta Crystallogr. D, 55, 938–940. [DOI] [PubMed] [Google Scholar]
- Frimpong K. and Rodwell, V.W. (1994a) The active site of hamster 3-hydroxy-3-methylglutaryl-CoA reductase resides at the subunit interface and incorporates catalytically essential acidic residues from separate polypeptides. J. Biol. Chem., 269, 1217–1221. [PubMed] [Google Scholar]
- Frimpong K. and Rodwell, V.W. (1994b) Catalysis by Syrian hamster 3-hydroxy-3-methylglutaryl-coenzyme A reductase. Proposed roles of histidine 865, glutamate 558, and aspartate 766. J. Biol. Chem., 269, 11478–11483. [PubMed] [Google Scholar]
- Gill J.F. Jr, Beach, M.J. and Rodwell, V.W. (1985) Mevalonate utilization in Pseudomonas sp. M. Purification and characterization of an inducible 3-hydroxy-3-methylglutaryl coenzyme A reductase. J. Biol. Chem., 260, 9393–9398. [PubMed] [Google Scholar]
- Goldstein J.L. and Brown, M.S. (1990) Regulation of the mevalonate pathway. Nature, 343, 425–430. [DOI] [PubMed] [Google Scholar]
- Gotto A.M. Jr (1997) Results of recent large cholesterol-lowering trials and implications for clinical management. Am. J. Cardiol., 79, 1663–1666. [DOI] [PubMed] [Google Scholar]
- Hampton R., Dimster-Denk, D. and Rine, J. (1996) The biology of HMG-CoA reductase: the pros of contra-regulation. Trends Biochem. Sci., 21, 140–145. [PubMed] [Google Scholar]
- Hawk M.A., Cesen, K.T., Siglin, J.C., Stoner, G.D. and Ruch, R.J. (1996) Inhibition of lung tumor cell growth in vitro and mouse lung tumor formation by lovastatin. Cancer Lett., 109, 217–222. [DOI] [PubMed] [Google Scholar]
- Hebert P.R., Gaziano, J.M., Chan, K.S. and Hennekens, C.H. (1997) Cholesterol lowering with statin drugs, risk of stroke and total mortality. An overview of randomized trials. J. Am. Med. Assoc., 278, 313–321. [PubMed] [Google Scholar]
- Horton N. and Lewis, M. (1992) Calculation of the free energy of association for protein complexes. Protein Sci., 1, 169–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones S. and Thornton, J.M. (1995) Protein–protein interactions: a review of protein dimer structures. Prog. Biophys. Mol. Biol., 63, 31–65. [DOI] [PubMed] [Google Scholar]
- Jones T.A., Zou, J.Y., Cowan, S.W. and Kjeldgaard, M. (1991) Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A, 47, 110–119. [DOI] [PubMed] [Google Scholar]
- Kissinger C.R., Gehlhaar, D.K. and Fogel, D.B. (1999) Rapid automated molecular replacement by evolutionary search. Acta Crystallogr. D, 55, 484–491. [DOI] [PubMed] [Google Scholar]
- Kleywegt G.J. and Jones, T.A. (1997) Detecting folding motifs and similarities in protein structures. Methods Enzymol., 277, 525–545. [DOI] [PubMed] [Google Scholar]
- Kumagai H., Chun, K.T. and Simoni, R.D. (1995) Molecular dissection of the role of the membrane domain in the regulated degradation of 3-hydroxy-3-methylglutaryl coenzyme A reductase. J. Biol. Chem., 270, 19107–19113. [DOI] [PubMed] [Google Scholar]
- Lawrence C.M., Rodwell, V.W. and Stauffacher, C.V. (1995) Crystal structure of Pseudomonas mevalonii HMG-CoA reductase at 3.0 Å resolution. Science, 268, 1758–1762. [DOI] [PubMed] [Google Scholar]
- Lee S.J., et al. (1998) Inhibition of the 3-hydroxy-3-methylglutaryl-coenzyme A reductase pathway induces p53-independent transcriptional regulation of p21(WAF1/CIP1) in human prostate carcinoma cells. J. Biol. Chem., 273, 10618–10623. [DOI] [PubMed] [Google Scholar]
- Mayer R.J., Debouck, C. and Metcalf, B.W. (1988) Purification and properties of the catalytic domain of human 3-hydroxy-3-methylglutaryl-CoA reductase expressed in Escherichia coli. Arch. Biochem. Biophys., 267, 110–118. [DOI] [PubMed] [Google Scholar]
- McGee T.P., Cheng, H.H., Kumagai, H., Omura, S. and Simoni, R.D. (1996) Degradation of 3-hydroxy-3-methylglutaryl-CoA reductase in endoplasmic reticulum membranes is accelerated as a result of increased susceptibility to proteolysis. J. Biol. Chem., 271, 25630–25638. [DOI] [PubMed] [Google Scholar]
- Moriyama T., Sather, S.K., McGee, T.P. and Simoni, R.D. (1998) Degradation of HMG-CoA reductase in vitro. Cleavage in the membrane domain by a membrane-bound cysteine protease. J. Biol. Chem., 273, 22037–22043. [DOI] [PubMed] [Google Scholar]
- Nakanishi M., Goldstein, J.L. and Brown, M.S. (1988) Multivalent control of 3-hydroxy-3-methylglutaryl coenzyme A reductase. Mevalonate-derived product inhibits translation of mRNA and accelerates degradation of enzyme. J. Biol. Chem., 263, 8929–8937. [PubMed] [Google Scholar]
- Navaza J. and Saludjian, P. (1997) AMoRe: an automated molecular replacement program package. Methods Enzymol., 276, 581–594. [DOI] [PubMed] [Google Scholar]
- Ness G.C., Pendleton,L.C. and McCreery,M.J. (1988) In situ determination of the functional size of hepatic 3-hydroxy-3-methylglutaryl-CoA reductase by radiation inactivation analysis. Biochim. Biophys. Acta, 953, 361–364. [DOI] [PubMed] [Google Scholar]
- Nicholls A., Sharp, K.A. and Honig, B. (1991) Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins, 11, 281–296. [DOI] [PubMed] [Google Scholar]
- Nohturfft A., DeBose-Boyd, R.A., Scheek, S., Goldstein, J.L. and Brown, M.S. (1999) Sterols regulate cycling of SREBP cleavage-activating protein (SCAP) between endoplasmic reticulum and Golgi. Proc. Natl Acad. Sci. USA, 96, 11235–11240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omkumar R.V. and Rodwell, V.W. (1994) Phosphorylation of Ser871 impairs the function of His865 of Syrian hamster 3-hydroxy-3-methylglutaryl-CoA reductase. J. Biol. Chem., 269, 16862–16866. [PubMed] [Google Scholar]
- Omkumar R.V., Darnay, B.G. and Rodwell, V.W. (1994) Modulation of Syrian hamster 3-hydroxy-3-methylglutaryl-CoA reductase activity by phosphorylation. Role of serine 871 [published erratum appears in J. Biol. Chem., 1994, 269, 16518]. J. Biol. Chem., 269, 6810–6814. [PubMed] [Google Scholar]
- Otwinowski Z. and Minor, W. (1997) Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol., 276, 307–326. [DOI] [PubMed] [Google Scholar]
- Pace C.N., Vajdos, F., Fee, L., Grimsley, G. and Gray, T. (1995) How to measure and predict the molar absorption coefficient of a protein. Protein Sci., 4, 2411–2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks T.D., Leuther, K.K., Howard, E.D., Johnston, S.A. and Dougherty, W.G. (1994) Release of proteins and peptides from fusion proteins using a recombinant plant virus proteinase. Anal. Biochem., 216, 413–417. [DOI] [PubMed] [Google Scholar]
- Roitelman J., Olender, E.H., Bar-Nun, S., Dunn, W.A., Jr and Simoni, R.D. (1992) Immunological evidence for eight spans in the membrane domain of 3-hydroxy-3-methylglutaryl coenzyme A reductase: implications for enzyme degradation in the endoplasmic reticulum. J. Cell Biol., 117, 959–973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossmann M.G., Liljas,A., Brändén,C.-I. and Banaszak,L.J. (1975) Evolutionary and structural relationships among dehydrogenases. In Boyer,P.D. (ed.), The Enzymes. Academic Press, New York, NY, Vol. 11, pp. 61–102. [Google Scholar]
- Smith G.D., Nagar, B., Rini, J.M., Hauptman, H.A. and Blessing, R.H. (1998) The use of SNB to determine an anomalous scattering substructure. Acta Crystallogr. D, 54, 799–804. [DOI] [PubMed] [Google Scholar]
- Tabernero L., Bochar, D.A., Rodwell, V.W. and Stauffacher, C.V. (1999) Substrate-induced closure of the flap domain in the ternary complex structures provides insights into the mechanism of catalysis by 3-hydroxy-3-methylglutaryl-CoA reductase. Proc. Natl Acad. Sci. USA, 96, 7167–7171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Doren M., Broihier, H.T., Moore, L.A. and Lehmann, R. (1998) HMG-CoA reductase guides migrating primordial germ cells. Nature, 396, 466–469. [DOI] [PubMed] [Google Scholar]
- Veloso D., Cleland, W.W. and Porter, J.W. (1981) pH properties and chemical mechanism of action of 3-hydroxy-3-methylglutaryl coenzyme A reductase. Biochemistry, 20, 887–894. [DOI] [PubMed] [Google Scholar]
- Wang Y., Darnay, B.G. and Rodwell, V.W. (1990) Identification of the principal catalytically important acidic residue of 3-hydroxy-3-methylglutaryl coenzyme A reductase. J. Biol. Chem., 265, 21634–21641. [PubMed] [Google Scholar]