Abstract
Hormones and growth factors induce protein translation in part by phosphorylation of the eukaryotic initiation factor 4E (eIF4E) binding protein 1 (4E-BP1). The rapamycin and FK506-binding protein (FKBP)-target 1 (RAFT1, also known as FRAP) is a mammalian homolog of the Saccharomyces cerevisiae target of rapamycin proteins (mTOR) that regulates 4E-BP1. However, the molecular mechanisms involved in growth factor-initiated phosphorylation of 4E-BP1 are not well understood. Here we demonstrate that protein kinase Cδ (PKCδ) associates with RAFT1 and that PKCδ is required for the phosphorylation and inactivation of 4E-BP1. PKCδ-mediated phosphorylation of 4E-BP1 is wortmannin resistant but rapamycin sensitive. As shown for serum, phosphorylation of 4E-BP1 by PKCδ inhibits the interaction between 4E-BP1 and eIF4E and stimulates cap-dependent translation. Moreover, a dominant-negative mutant of PKCδ inhibits serum-induced phosphorylation of 4E-BP1. These findings demonstrate that PKCδ associates with RAFT1 and thereby regulates phosphorylation of 4E–BP1 and cap-dependent initiation of protein translation.
Keywords: cap-dependent translation/eukaryotic initiation factor 4E/PKCδ/mTOR/4E-BP1 phosphorylation
Introduction
Growth factors and hormones induce translation of certain proteins that are essential for the proliferation and survival responses of cells (Sonenberg, 1994, 1996; Brown et al., 1995; Nielsen et al., 1995; Brown and Schreiber, 1996). However, little is known about signaling events that connect growth stimuli to activation of the protein synthesis machinery. Studies with rapamycin, an immunosuppressive macrolide that interacts with the cytosolic 12 kDa FK506-binding protein (FKBP) (Kunz and Hall, 1993), have elucidated a signaling pathway that regulates protein synthesis in both animals and yeast (Barbet et al., 1996; Beretta et al., 1996; von Manteuffel et al., 1996). Pretreatment of cells with rapamycin affects mitogen-stimulated phosphorylation of certain modulators of protein translation that include the ribosomal S6 protein, p70s6k (Chung et al., 1992; Kuo et al., 1992; Price et al., 1992; Lane et al., 1993; Reinhard et al., 1994; Pearson et al., 1995; Lin and Lawrence, 1996), 4E-binding protein 1 (4E-BP1)/PHAS-I (Hu et al., 1994; Beretta et al., 1996; von Manteuffel et al., 1996), 4E-BP2 (Lin and Lawrence, 1996) and elongation factor 2 (Redpath et al., 1996). In yeast, rapamycin is a potent inhibitor of translation (>90%) (Barbet et al., 1996), but causes only partial inhibition of translation in various mammalian cells (Jefferies et al., 1994, 1997; Terada et al., 1994; Beretta et al., 1996; Jefferies and Thomas, 1996). Rapamycin inhibits G1 cell cycle progression as a consequence of decreases in mRNA translation (Brown et al., 1994; Brown and Schreiber, 1996). In concert with these findings, the yeast target of rapamycin (TOR) regulates G1 progression through a translational mechanism (Barbet et al., 1996).
The rapamycin–FKBP complex binds to FKBP– rapamycin-associated protein (FRAP) in humans, rapamycin and FKBP12 target (RAFT1) in rats and TOR in yeast (Heitman et al., 1991; Kunz et al., 1993; Brown et al., 1994; Sabatini et al., 1994; Stan et al., 1994; Sabers et al., 1995). RAFT1 is a 220 kDa polypeptide that contains at the C-terminus a protein and/or lipid kinase catalytic domain, most closely related to those of the DNA-dependent protein kinase (DNA-PK) and the ataxia-telangectasia mutated (ATM), MEC1 and Tel1 checkpoint gene products, and somewhat distantly related to that of the phosphoinositide 3-kinase (PI3-K) (Keith and Schreiber, 1995). Mammalian TOR (mTOR) is an upstream regulator of 4E-BP1 phosphorylation (Brunn et al., 1997; Hara et al., 1997; Burnett et al., 1998; Gingras et al., 1999). Moreover, recent studies have shown that the PI3-K–Akt(PKB) signaling pathway induces phosphorylation of 4E-BP1 (Gingras et al., 1998). PI3-K-mediated phosphorylation of 4E-BP1 is wortmannin- and rapamycin-sensitive, but Akt-mediated phosphorylation is wortmannin insensitive (Gingras et al., 1998). These findings demonstrate that PI3-K, Akt and FRAP/mTOR are key regulators in the signaling pathway that confers phosphorylation of 4E-BP1.
The protein kinase C (PKC) family, a group of 11 known members that contain phospholipid-dependent serine/threonine kinase activity, plays a key role in cellular signal transduction and is involved in the regulation of various cellular processes (Nishizuka, 1988, 1995; Dekker and Parker, 1994). PKC isoenzymes are differentially expressed and respond differently to physiological inducers in diverse tissues and cell types (Dekker et al., 1995). PKCδ is a novel PKC (nPKC) that is activated by diacylglycerol (DAG) or 12-O-tetradecanoylphorbol 13-acetate (TPA), but is unresponsive to Ca2+ (Osada et al., 1992). Recent work has demonstrated that insulin induces its proliferative effects in part through a PKCδ-dependent pathway (Reks et al., 1998). Treatment of H4 hepatoma cells with insulin is associated with translocation of PKCδ from cytosol to membrane and thereby activation of PKCδ (Reks et al., 1998).
Recent studies have shown that phosphoinositide-dependent kinase 1 (PDK1) is at the hub of many signaling pathways, activating Akt and PKC isoenzymes (Belham et al., 1999). The activity of PDK1 expressed in mammalian cells is unaffected by stimuli that strongly activate Akt through PI3-K (Alessi et al., 1997). Furthermore, recent studies have shown that PDK1 associates with PKCδ in vivo and directly phosphorylates the activation loop of PKCδ in vitro (Le Good et al., 1998). The findings that serum-stimulated phosphorylation of PKCδ is enhanced by coexpression of PDK1 and that this response is also sensitive to wortmannin indicate that PKCδ plays a key role in regulating the serum-induced signaling pathway and acts downstream to PI3-K. Moreover, a recent study has shown that, like PDK1, phosphorylation of PKCδ is independently regulated by a pathway involving mTOR/RAFT1 (Parekh et al., 1999). Taken together, these findings demonstrate that serum-induced phosphorylation and activation of PKCδ is regulated by multiple upstream effectors.
Since phosphorylation and activation of PKCδ by serum and insulin is mediated by PI3-K→PDK1 signaling, we sought to determine whether PKCδ contributes to the regulation of the FRAP/mTOR→→4E-BP1 phosphoryl– ation pathway and thereby mediates cap-dependent translation. The results demonstrate that PKCδ interacts with FRAP/mTOR and is required for phosphorylation of 4E-BP1 in vivo. The functional significance of the PKCδ–mTOR interaction is supported by the finding that PKCδ stimulates eukaryotic initiation factor 4E (eIF4E)dependent protein translation.
Results
PKCδ constitutively associates with RAFT1
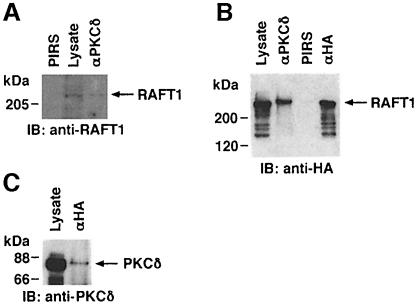
To determine whether PKCδ associates with RAFT1, total cell lysates from 293T cells were subjected to immunoprecipitation with anti-PKCδ and the precipitates were analyzed by immunoblotting with anti-RAFT1. The results demonstrate that PKCδ interacts constitutively with RAFT1 (Figure 1A). The finding that RAFT1 does not interact with DNA-PK supports a specific interaction between PKCδ and RAFT1 (data not shown). To confirm the interaction of PKCδ with RAFT1, 293T cells were transiently transfected with hemagglutanin (HA)-RAFT1. Anti-PKCδ immunoprecipitates from cytoplasmic extracts were analyzed by immunoblotting with anti-HA. Extracts were also subjected to incubation with pre-immune rabbit serum (PIRS) or anti-HA as controls. Reactivity of anti-PKCδ with a >200 kDa protein supported the coprecipitation of RAFT1 with PKCδ (Figure 1B). In a reciprocal experiment, anti-HA immunoprecipitates were analyzed by immunoblotting with anti-PKCδ. The results confirmed the presence of RAFT1 in a cytoplasmic complex of PKCδ (Figure 1C).
Fig. 1. Association of PKCδ with RAFT1. (A) Total lysates from 293T cells were subjected to immunoprecipitation with anti-PKCδ or pre-immune rabbit serum (PIRS). The precipitates and total lysate were separated by SDS–PAGE and analyzed by immunoblotting with anti-RAFT1. (B) 293T cells were transiently transfected with HA-RAFT1. Total lysates were subjected to immunoprecipitation with anti-PKCδ, anti-HA or PIRS. The precipitates and the lysates were analyzed by immunoblotting with anti-HA. (C) 293T cells were transiently transfected with HA-RAFT1. Lysates were subjected to immunoprecipitation with anti-HA. The precipitates and lysates were analyzed by immunoblotting with anti-PKCδ.
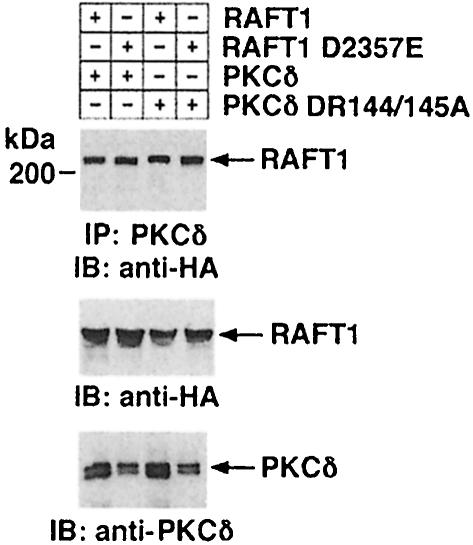
To determine whether the kinase function of RAFT1 is necessary for the interaction with PKCδ, we transiently transfected HA-RAFT1 or a kinase-inactive HA-RAFT1 D2357E (RAFT1 D–E) mutant (Burnett et al., 1998) with PKCδ in 293T cells. Anti-PKCδ immunoprecipitates were analyzed by immunoblotting with anti-HA. As controls, anti-HA and anti-PKCδ immunoprecipitates were analyzed by immunoblotting with anti-HA and anti-PKCδ, respectively. The association of RAFT1 and PKCδ was detected in cells overexpressing HA-RAFT1 or HA-RAFT1 D–E (Figure 2). To determine whether the association of PKCδ with RAFT1 is dependent on the kinase function of PKCδ, 293T cells were transfected with PKCδ DR144/145A mutant and HA-RAFT1 or HA-RAFT1 D2357E. PKCδ DR144/145A harbors two amino acid substitutions in its pseudosubstrate region (Hirai et al., 1994), and thereby functions as a highly active kinase compared with full-length PKCδ (PKCδ FL) (data not shown). Total cell lysates were subjected to immunoprecipitation with anti-PKCδ and analyzed by immunoblotting with anti-HA. The results demonstrate that PKCδ DR144/145A associates both with wild-type as well as the D2357E mutant of RAFT1 (Figure 2). Taken together, these findings indicate that the interaction of PKCδ and RAFT1 is independent of the kinase functions of RAFT1 and PKCδ.
Fig. 2. Kinase activities of PKCδ and RAFT1 are not required for their association. 293T cells were transiently transfected with PKCδ FL or PKCδ DR144/145A with HA-RAFT1 or HA-RAFT1 D2357E. Lysates were subjected to immunoprecipitation with anti-PKCδ and the precipitates were analyzed by immunoblotting with anti-HA (top panel). Anti-HA immunoprecipitates were analyzed by immunoblotting with anti-HA (middle panel). Lysates were also analyzed by immunoblotting with anti-PKCδ (bottom panel).
PKCδ mediates the phosphorylation of 4E-BP1
To assess in part the functional significance of the interaction of PKCδ and RAFT1, we investigated whether RAFT1 is a substrate for PKCδ. To address this issue, 293T cells were transfected with vectors expressing HA-RAFT1 or the HA-RAFT1 D2357E mutant. Cell lysates were subjected to immunoprecipitation with anti-HA and half of the protein precipitates were incubated with the constitutively active glutathione S-transferase (GST)–PKCδ catalytic fragment (PKCδ CF) fusion protein (Emoto et al., 1995) in the presence of [γ-32P]ATP. As a control, the precipitates were also incubated with a kinase-inactive GST–PKCδ CF(K–R) mutant. Analysis of the reaction products demonstrated that, in contrast to myelin basic protein (MBP), RAFT1 (wild type or D2357E) is not phosphorylated by PKCδ (data not shown). To assess the amount of RAFT1 protein, the precipitates were also analyzed by immunoblotting with anti-HA. Detection of RAFT1 protein in the anti-HA immunoprecipitates ruled out the possibility of substrate limitation (data not shown). These findings indicate that RAFT1 is not a substrate of PKCδ in vitro.
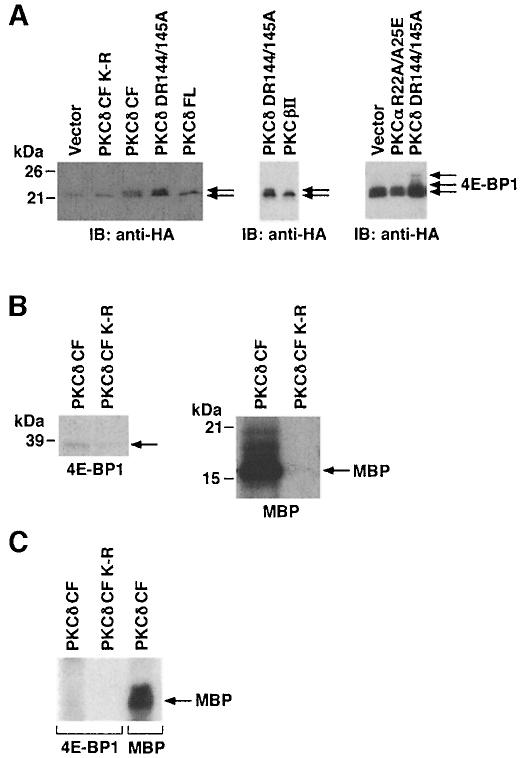
Previous studies have shown that RAFT1 immunoprecipitates contain a kinase activity that phosphorylates 4E-BP1 (Brunn et al., 1997) on two sites (Thr37 and Thr46) (Burnett et al., 1998; Gingras et al., 1998; Heesom and Denton, 1999). These sites are not those phosphorylated in the response of cells to serum and it is not clear whether FRAP/mTOR itself or an associated kinase is responsible for 4E-BP1 phosphorylation. To study the role of PKCδ in the phosphorylation of 4E-BP1, HA-4E-BP1 (Gingras et al., 1998) was transiently cotransfected into 293T cells with empty vector, PKCδ FL or PKCδ DR144/145A. As a control, PKCβII or an active mutant of PKCα (PKCα R22A/A25E) (Ueda et al., 1996) was cotransfected with HA-4E-BP1. After serum starvation for 48 h, the cell extracts were analyzed by immunoblotting with anti-HA. The results demonstrate that cotransfection of HA-4E-BP1 with the active PKCδ DR144/145A, but not PKCβII or PKCα R22A/A25E, induced phosphorylation of 4E-BP1 and thereby a shift in its mobility (Figure 3A). Since PKCδ FL, but not PKCδ DR144/145A, failed to induce phosphorylation of 4E-BP1, we assessed the activity of PKCδ after transfecting FL or DR144/145A. In contrast to PKCδ DR144/145A, overexpression of PKCδ FL followed by serum starvation was associated with little, if any, increase in PKCδ activity (data not shown). However, treatment of starved cells with serum + TPA was associated with an increase in the kinase activity of transiently overexpressed PKCδ and an increase in phosphorylation of 4E-BP1 (data not shown). In this context, a recent study has shown that treatment of serum-starved cells with serum + TPA is associated with mTOR-mediated increases in PKCδ activity (Parekh et al., 1999). To confirm a role for PKCδ in phosphorylation of 4E-BP1, we transiently cotransfected HA-4E-BP1 with the kinase-active PKCδ CF into 293T cells. As a control, an empty vector or a kinase-dead catalytic fragment of PKCδ (PKCδ CF K–R) were separately overexpressed with HA-4E-BP1. Cells were serum starved for 48 h and then lysates were analyzed by immunoblotting with anti-HA. The results demonstrate that overexpression of PKCδ CF, but not empty vector or PKCδ CF(K–R), is associated with phosphorylation of 4E-BP1 (Figure 3A). These findings demonstrate that, in contrast to inactive forms of PKCδ or other isoforms of PKC (PKCβII and PKCα R22A/A25E), overexpression of an active form of PKCδ (PKCδ DR144/145A or PKCδ CF) is sufficient to induce a mobility shift of 4E-BP1. Analysis of the results from replicative experiments indicate that there is a 4- to 5-fold increase in phosphorylation of 4E-BP1 by over– expressing active mutants of PKCδ. To assess whether PKCδ directly phosphorylates 4E-BP1, in vitro kinase assays were performed in which a GST–4E-BP1 fusion protein was incubated with active or inactive PKCδ in the presence of [γ-32P]ATP. The phosphorylated reaction products were analyzed by SDS–PAGE and autoradiography. The results demonstrate that, in contrast to MBP (Figure 3B, right panel), there is little, if any, phosphoryl– ation of GST–4E-BP1 by PKCδ (Figure 3B, left panel). Previous studies have shown that anti-RAFT1/mTOR immunoprecipitates contain kinase activities that are specifically targeted to the eIF4E–4E-BP1 complex and not free 4E-BP1 (Gingras et al., 1998; Heesom and Denton, 1999). To determine whether PKCδ phosphoryl– ates 4E-BP1 complexed with eIF4E, 293 cells were serum starved for 48 h and total cell lysates were subjected to precipitation with 7-methyl-GTP (m7GTP). eIF4E–4E-BP1 complexes were then incubated with PKCδ CF in the presence of [γ-32P]ATP and analyzed by SDS–PAGE and autoradiography. The results demonstrate that PKCδ has no detectable effect on phosphorylation of 4E-BP1 in a complex with eIF4E (Figure 3C). Thus, PKCδ is not likely to be a direct 4E-BP1 kinase.
Fig. 3. PKCδ induces phosphorylation of 4E-BP1. (A) 293T cells were transiently transfected with HA-4E-BP1 and vector, PKCδ FL or PKCδ DR144/145A. Cells were serum starved for 48 h and lysates were resolved by SDS–PAGE and analyzed by immunoblotting with anti-HA. 293T cells were transiently cotransfected with HA-4E-BP1 and vector, PKCδ CF or PKCδ CF(K–R). Cells were serum starved for 48 h. Lysates were resolved by SDS–PAGE and analyzed by immunoblotting with anti-HA (left panel). 293T cells were transiently transfected with HA-4E-BP1 and PKCβII or PKCδ DR144/145A (middle panel). 293T cells were transfected with HA-4E-BP1 and vector, PKCα R22A/A25E or PKCδ DR144/145A (right panel). Cells were serum starved for 48 h. Lysates were resolved by SDS–PAGE and analyzed by immunoblotting with anti-HA. (B) GST–PKCδ CF or GST–PKCδ CF(K–R) fusion proteins were incubated with purified GST–4E-BP1 in the presence of [γ-32P]ATP for 15 min at 30°C (left panel). GST–PKCδ CF or GST–PKCδ CF(K–R) were also separately incubated with MBP (right panel). The reaction products were resolved by SDS–PAGE and analyzed by autoradiography. (C) 293 cells were serum starved for 36 h. Lysates were incubated with m7GTP resin and the precipitates were incubated with PKCδ CF in the presence of [γ-32P]ATP and MBP and analyzed by SDS–PAGE and autoradiography.
PKCδ inhibits the ability of 4E-BP1 to interact with eIF4E and stimulates cap-dependent initiation of protein translation
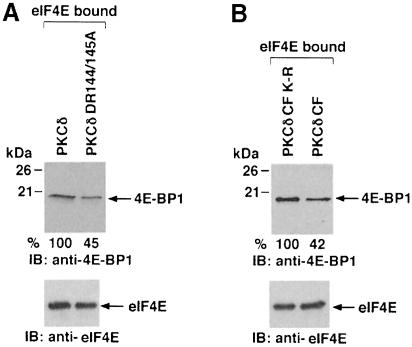
Since PKCδ stimulates phosphorylation of 4E-BP1, we asked whether PKCδ regulates the interaction of 4E-BP1 with eIF4E. To address this issue, 293 cells were transiently transfected with PKCδ FL or PKCδ DR144/145A. After serum starvation for 48 h, the cells were lysed and the lysates were incubated with an m7GTP-coupled agarose resin. Bound proteins were eluted and analyzed by immunoblotting with anti-4E-BP1. As expected, eIF4E, which recognizes the cap structure, bound to the resin in equal amounts (Figure 4A, bottom panel). The results also demonstrate that the 4E-BP1 in cells expressing PKCδ is retained by the eIF4E-bound resin to a greater extent than that in cells expressing the PKCδ DR144/145A mutant (Figure 4A, upper panel). To confirm a role for PKCδ in regulating the interaction of 4E-BP1 and eIF4E, we transiently overexpressed PKCδ CF or PKCδ CF(K–R). Cells were serum starved for 48 h and then lysates were incubated with an m7GTP-coupled agarose resin. Bound proteins were eluted and analyzed by immunoblotting with anti-4E-BP1. The results demonstrate that overexpression of PKCδ CF, but not PKCδ CF(K–R) is associated with significant inhibition (50–55%) of the interaction between 4E-BP1 and eIF4E (Figure 4B). Taken together, these findings indicate that catalytic activity of PKCδ is required to inhibit the formation of 4E-BP1–eIF4E complexes.
Fig. 4. PKCδ inhibits the interaction of eIF4E with 4E-BP1. (A) 293T cells were transiently cotransfected with PKCδ FL or PKCδ DR144/145A and serum starved for 48 h. Lysates were incubated with m7GTP–agarose beads for 45 min at 4°C. After incubation, the proteins were resolved by 15% SDS–PAGE and analyzed by immunoblotting with anti-4E-BP1 (upper panel) or anti-eIF4E (bottom panel). (B) 293T cells were transiently transfected with PKCδ CF or PKCδ CF(K–R) and serum starved for 48 h. Lysates were incubated with m7GTP–agarose beads for 45 min at 4°C. After incubation, the proteins were resolved by 15% SDS–PAGE and analyzed by immuno- blotting with anti-4E-BP1 (upper panel) or anti-eIF4E (bottom panel).
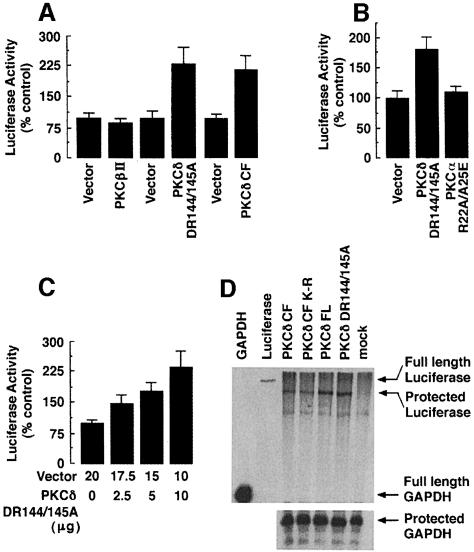
Translation of cap-dependent mRNAs is inhibited by binding of 4E-BP1 to eIF4E (Pause et al., 1994; Sonenberg, 1996) and thereby abrogation of the interaction of eIF4E with the N7-methylguanosine cap structure. To determine whether PKCδ regulates cap-dependent translation, we transfected 293T cells with the pcDNA3-LUC-pol-CAT vector that exhibits cap-dependent translation of luciferase (LUC) (Craig et al., 1998). Cotransfection of pcDNA3-LUC-pol-CAT with PKCβII had no effect on luciferase activity. In contrast, PKCδ DR144/145A induced the expression of luciferase by >2-fold (Figure 5A). To assess further the role of PKCδ in stimulation of cap-dependent translation, 293 cells were cotransfected with pcDNA3-LUC-pol-CAT and empty vector or PKCδ CF. Cells were serum starved for 48 h and the luciferase activity was measured in total cell lysates. The results demonstrate that transfection of PKCδ CF, but not empty vector, is associated with induction of luciferase activity (Figure 5A). To determine the specificity for PKCδ, 293 cells were cotransfected with pcDNA3-LUC-pol-CAT and PKCδ DR144/145A or PKCα R22A/A25E. Cells were serum starved for 48 h and the luciferase activity was measured in total cell lysates. The results demonstrate that transfection of PKCδ DR144/145A, but not PKCα R22A/A25E, is associated with induction of luciferase activity (Figure 5B). To exclude the possibility that the effects observed reflect differences in mRNA concentration, RNase protection assays were performed on RNAs extracted from parallel transfections using antisense GAPDH and luciferase probes (Figure 5C). There were no protected fragments for the transfer RNA control (data not shown), and only GAPDH mRNA was detected in mock-transfected cells (Figure 5C). The luciferase: GAPDH mRNA ratio for PKCδ DR144/145A was 1.09 ± 0.17 relative to PKCδ FL (normalized at 1). The luciferase:GAPDH mRNA ratio for PKCδ CF was 1.16 ± 0.22 relative to PKCδ CF(K–R) (normalized at 1) (Figure 5C). To confirm the effects of PKCδ on cap-dependent initiation of protein translation, we cotransfected cells with pcDNA3-LUC-pol-CAT and increasing amounts of PKCδ DR144/145A. Cells were serum starved for 48 h before assessing luciferase activity. Transfection of PKCδ DR144/145A was associated with a dose-dependent increase in luciferase activity compared with that obtained for the vector alone (Figure 5D). Collectively, these findings indicate that overexpression of active forms of PKCδ (i) stimulates the phosphorylation of 4E-BP1; (ii) inhibits the interaction of 4E-BP1 with eIF4E; and (iii) stimulates cap-dependent initiation of protein translation.
Fig. 5. Effect of PKCδ on cap-dependent protein translation. (A) 293T cells were transiently cotransfected with pcDNA3-LUC-pol-CAT (2 μg) and empty vector, PKCβII or PKCδ DR144/145A. Cells were serum starved for 48 h and lysates were analyzed for luciferase activity. Activity is expressed as percentage control (mean ± SD of three independent transfections). 293T cells were cotransfected with pcDNA3-LUC-pol-CAT (2 μg) and vector or PKCδ CF. Following serum starvation for 48 h, total cell lysates were analyzed for luciferase activity. (B) 293T cells were transiently cotransfected with pcDNA3-LUC-pol-CAT (2 μg) and vector (10 μg), PKCα R22A/A25E (10 μg) or PKCδ DR144/145A (10 μg). Following serum starvation for 48 h, lysates were assayed for luciferase activity. (C) 293T cells were transiently transfected with vector, pcDNA3-LUC-pol-CAT and the amounts of PKCδ DR144/145A indicated. Lysates were assayed for luciferase activity. Luciferase activity is expressed as percentage control (mean ± SD of four independent transfections). (D) Antisense GAPDH and luciferase probes (lanes 1 and 2) were tested against RNA from PKCδ CF (lane 3), PKCδ CF(K–R) (lane 4), PKCδ FL (lane 5), PKCδ DR144/145A (lane 6) or mock (lane 7) transfected cells. Positions of FL probes and protected fragments are indicated on the right.
PKCδ-mediated, cap-dependent translation requires RAFT1 activity and is resistant to wortmannin and sensitive to rapamycin
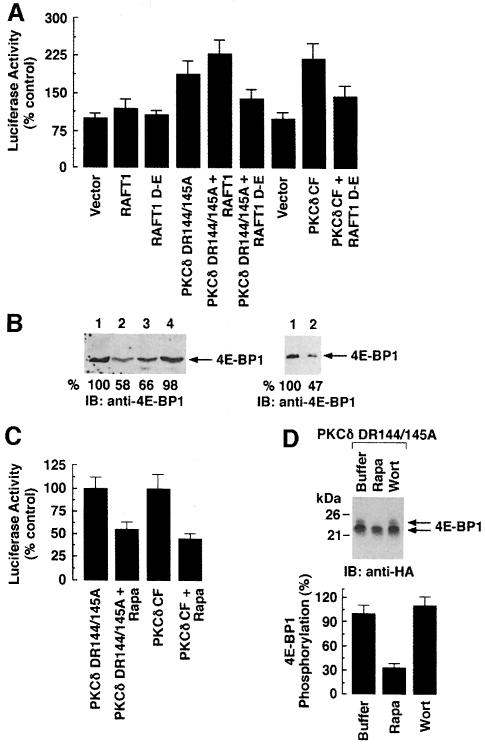
4E-BP1 phosphorylation and cap-dependent translation are mediated by the RAFT1 signaling pathway both in yeast and mammalian cells (Barbet et al., 1996; Beretta et al., 1996; Burnet et al., 1998; Gingras et al., 1998; Hara et al., 1997). To determine whether a dominant-negative mutant of RAFT1 (RAFT1 D2357E) inhibits PKCδ DR144/145A-mediated increases in cap-dependent translation, cells were transfected with vector, RAFT1 wild type, RAFT1 D2357E, PKCδ DR144/145A, PKCδ DR144/145A + RAFT1 D2357E or PKCδ DR144/145A + RAFT1. Cells were cotransfected with pcDNA3-LUC-pol-CAT. Following transfection, cells were serum starved for 48 h and total lysates were analyzed by luciferase activity assays. The results demonstrate that overexpression of PKCδ DR144/145A with RAFT1 increases cap-dependent translation by ∼2.5-fold compared with vector (Figure 6A). Importantly, this increase in protein translation was inhibited by >50% in cells coexpressing PKCδ DR144/145A and RAFT1 D2357E (Figure 6A). Similar experiments were performed in which cells were transfected with PKCδ CF with or without RAFT1 D2357E. Cells were cotransfected with pcDNA3-LUC-pol-CAT, serum starved for 48 h and luciferase activity was measured in total lysates. The results demonstrate that the PKCδ CF-mediated increase in luciferase activity was significantly inhibited by overexpression of RAFT1 D2357E (Figure 6A). To confirm a role for PKCδ–RAFT1 complexes in regulating the interaction of 4E-BP1 and eIF4E, we transiently cotransfected PKCδ DR144/145A, PKCδ CF or PKCδ CF(K–R) with RAFT1 or RAFT1 D2357E. Cells were serum starved for 48 h and lysates were incubated with m7GTP-coupled agarose resin. Bound proteins were eluted and analyzed by immunoblotting with anti-4E-BP1. The results demonstrate that, in contrast to RAFT1 D2357E, overexpression of PKCδ DR144/145A or PKCδ CF with RAFT1 wild type is associated with significant inhibition (45–55%) of the interaction between 4E-BP1 and eIF4E (Figure 6B). Taken together, these findings demonstrate that the PKCδ-dependent increase in protein translation also requires RAFT1 activity.
Fig. 6. PKCδ-mediated increase in cap-dependent protein translation requires activity of RAFT1 and is resistant to wortmannin and sensitive to rapamycin. (A) 293 cells were cotransfected with pcDNA3-LUC-pol-CAT (2 μg) and vector (10 μg) (bar 1), RAFT1 (10 μg) (bar 2), RAFT1 D2357E (10 μg) (bar 3), PKCδ DR144/145A (10 μg) (bar 4), PKCδ DR144/145A + RAFT1 (bar 5) or PKCδ DR144/145A + RAFT1 D2357E (bar 6). Cells were serum starved for 48 h and lysates were assayed for luciferase activity. Luciferase activity is expressed as percentage control (mean ± SD of three independent transfections). 293 cells were cotransfected with pcDNA3-LUC-pol-CAT (2 μg) and vector or PKCδ CF in the presence or absence of HA-RAFT1 D2357E. Cells were serum starved for 48 h and lysates were assayed for luciferase activity. (B) Left panel: 293 cells were transfected with HA-4E-BP1 with vector (lane 1); PKCδ DR144/145A + RAFT1 wild type (lane 2); PKCδ DR144/145A (lane 3) and PKCδ DR144/145A + RAFT1 D2357E (lane 4). Lysates were incubated with m7GTP resin and the precipitates were analyzed by immunoblotting with anti-HA (left panel). Right panel: 293 cells were also separately cotransfected with HA-4E-BP1 and PKCδ CF + RAFT1 D–E (lane 1) or PKCδ CF + RAFT1 wild type (lane 2). Lysates were incubated with m7GTP resin and the precipitates were analyzed by immunoblotting with anti-HA. (C) 293 cells were cotransfected with pcDNA3-LUC-pol-CAT (2 μg) and PKCδ DR144/145A mutant or PKCδ CF. After transfection, cells were serum starved for 48 h and then treated with 20 ng/ml rapamycin. Lysates were assayed for luciferase activity. (D) 293 cells were cotransfected with HA-4E-BP1 and PKCδ DR144/145A. Cells were also separately transfected with vector and HA-4E-BP1. Cells were serum starved for 48 h after transfection and then treated with buffer (lane 1), 20 ng/ml rapamycin (lane 2) or 200 nM wortmannin (lane 3). Lysates were analyzed by immunoblotting with anti-HA (top panel). The results are expressed as percentage inhibition (mean ± SD) of three independent experiments (bottom panel).
Since the rapamycin–FKBP12 complex inactivates RAFT1 kinase function, we overexpressed PKCδ DR144/145A or PKCδ CF, treated the transfectants with rapamycin and then assessed luciferase activity. The results demonstrate that, as shown with expression of RAFT1 D2357E, the PKCδ-induced increase in luciferase activity was significantly inhibited by treatment with rapamycin (Figure 6C). Previous studies have supported a role for PI3-K in the phosphorylation of 4E-BP1 (von Manteuffel et al., 1996; Gingras et al., 1998). To determine whether PKCδ acts downstream to PI3-K in mediating 4E-BP1 phosphorylation, we overexpressed PKCδ DR144/145A and HA-4E-BP1. After transfection, cells were deprived of serum for 48 h and then treated with buffer, wortmannin or rapamycin. Lysates were analyzed by immunoblotting with anti-HA antibody. The results demonstrate that the mobility shift in 4E-BP1 elicited by PKCδ DR144/145A is resistant to wortmannin and sensitive to rapamycin (Figure 6D). There was 67 ± 6.8% (mean ± SD of three independent experiments) inhibition of the PKCδ DR144/145A-mediated increase in phosphorylation of 4E-BP1 by rapamycin (Figure 6D, bottom panel). This effect is similar to that observed previously for Akt (Gingras et al., 1998). Taken together, these findings support a pathway that (i) involves activation of PI3-K and thereby stimulation of PKCδ and Akt through PDK1 (Le Good et al., 1998); and (ii) confers phosphorylation of 4E-BP1. The findings also indicate that RAFT1 activity is required, at least in part, for PKCδ-dependent initiation of protein translation.
Role of PKCδ in phosphorylation of 4E-BP1 and regulation of cap-dependent translation in vivo
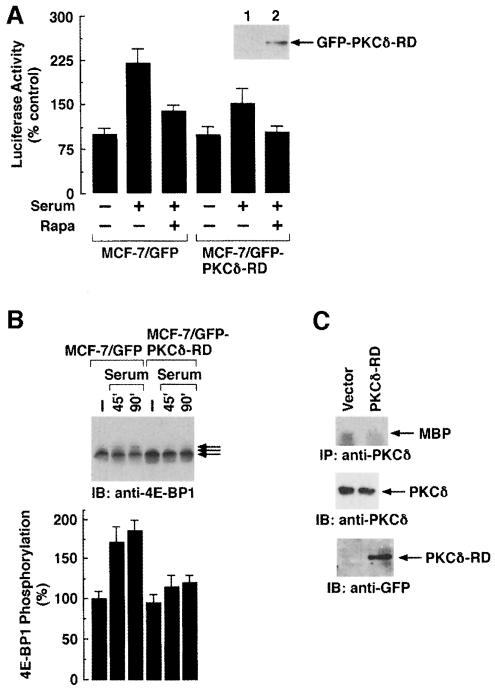
Previous studies have indicated a role for PI3-K and Akt(PKB) kinase in the phosphorylation of 4E-BP1 by serum and growth factors (von Manteuffel et al., 1996; Gingras et al., 1998). Moreover, the effects of serum or insulin on 4E-BP1 phosphorylation in vivo are mediated by RAFT1 (Beretta et al., 1996; Brunn et al., 1997; Gingras et al., 1998). To examine whether PKCδ regulates cap-dependent translation in vivo, we generated an MCF-7 cell line that stably expresses a green fluorescence protein (GFP) fused to the N-terminal regulatory domain (RD) of PKCδ (MCF-7/GFP–PKCδ-RD). Overexpression of PKCδ-RD competes with endogenous PKCδ for binding to its targets (our unpublished data). Detection of the GFP–PKCδ-RD protein with anti-GFP demonstrated that MCF-7/GFP–PKCδ-RD cells overexpress this protein (Figure 7A, inset). MCF-7/GFP and MCF-7/GFP–PKCδ-RD cells were transiently transfected with pcDNA3-LUC-pol-CAT. After transfection, the cells were deprived of serum for 36 h and then stimulated with serum for 12 h. The results demonstrate that the serum-induced increase in luciferase activity is partially inhibited in cells overexpressing GFP–PKCδ-RD (Figure 7A). Previous studies have shown that Akt inhibits the binding of 4E-BP1 to eIF4E and thereby stimulates cap-dependent translation (Gingras et al., 1998). The partial inhibition in serum-induced, cap-dependent translation by PKCδ-RD observed in the present study therefore supports the role of Akt in contributing to this effect. Serum-starved MCF-7/GFP or MCF-7/GFP–PKCδ-RD cells were also stimulated with serum in the presence and absence of rapamycin. As expected, the results demonstrate that treatment of cells with rapamycin inhibited serum-induced, cap-dependent translation in vivo (Figure 7A). To extend the analysis of PKCδ in cap-dependent translation in vivo, MCF-7/GFP and MCF-7/GFP–PKCδ-RD cells were serum starved for 36 h. Serum was added for 45 or 90 min and lysates were analyzed by immunoblotting with anti-4E-BP1. The results demonstrate that serum-induced phosphorylation of endogenous 4E-BP1 is partially inhibited in cells expressing GFP–PKCδ-RD (Figure 7B). To demonstrate whether PKCδ-RD specifically inhibits endogenous PKCδ activity, 293 cells were transiently transfected with different amounts of PKCδ-RD. Total cell lysates were subjected to immunoprecipitation with anti-PKCδ and the precipitates were assayed for phosphorylation of MBP. The results demonstrate that overexpression of PKCδ-RD significantly inhibits endogenous PKCδ activity (Figure 7C).
Fig. 7. Role of PKCδ in cap-dependent protein translation in vivo. (A) MCF-7/GFP and MCF-7/GFP–PKCδ-RD cells were transfected with pcDNA3-LUC-pol-CAT. Cells were serum starved for 36 h and stimulated with serum in the presence or absence of 50 ng/ml rapamycin for an additional 12 h. Lysates were assayed for luciferase activity. Luciferase activity is expressed as percentage control (mean ± SD of three independent experiments). Total cell lysates from MCF-7/GFP and MCF-7/GFP–PKCδ-RD cells were also analyzed by immunoblotting with anti-GFP (inset). (B) MCF-7/GFP and MCF-7/GFP–PKCδ-RD cells were transfected with HA-4E-BP1. Cells were serum starved for 36 h and stimulated with serum for 45 and 90 min. Lysates were analyzed by immunoblotting with anti-HA (upper panel). The results are expressed as percentage 4E-BP1 phosphorylation (mean ± SD) of two independent experiments (lower panel). (C) 293 cells were transfected with vector or GFP–PKCδ-RD. Total cell lysates were subjected to immunoprecipitation with anti-PKCδ and the precipitates were incubated with MBP in the presence of [γ-32P]ATP. The reaction products were analyzed by SDS–PAGE and auto- radiography (top panel). Anti-PKCδ immunoprecipitates were analyzed by immunoblotting with anti-PKCδ (middle panel). Lysates were also analyzed by immunoblotting with anti-GFP (bottom panel).
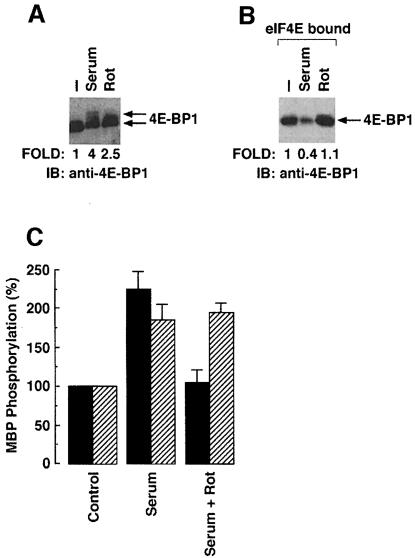
To confirm these findings, further studies have been performed by utilizing rottlerin, an inhibitor of PKCδ (Gschwendt et al., 1994). Cells were serum starved for 36 h and stimulated with serum in the presence or absence of rottlerin. Lysates were analyzed by immunoblotting with anti-4E-BP1. Pretreatment of cells with rottlerin inhibited serum-induced phosphorylation of 4E-BP1 (Figure 8A). Lysates were also incubated with an m7GTP-coupled agarose resin. Bound proteins were eluted and analyzed by immunoblotting with anti-4E-BP1. 4E-BP1 was retained by the eIF4E-bound resin in cells pretreated with rottlerin, but not in the absence of the PKCδ inhibitor (Figure 8B). Taken together, these findings support a model in which the kinase function of PKCδ is required to stimulate cap-dependent translation in vivo. To determine whether rottlerin specifically inhibits PKCδ, lysates were subjected to immunoprecipitation with anti-PKCδ or anti-Akt. In vitro immune complex kinase assays were performed using MBP as substrate. The results demonstrate that pretreatment of 293T cells with rottlerin significantly inhibits serum-induced activation of PKCδ, and not Akt (Figure 8C).
Fig. 8. PKCδ inhibitor, rottlerin, inhibits serum-induced phosphoryl- ation of 4E-BP1. (A) 293 cells were serum starved for 36 h and stimulated with serum in the presence or absence of PKCδ inhibitor, rottlerin (Rot). Lysates were analyzed by immunoblotting with anti-4E-BP1. (B) 293 cells were serum starved for 36 h and then stimulated with serum in the presence or absence of rottlerin. Lysates were incubated with m7GTP–agarose beads for 45 min at 4°C. After incubation, the bound proteins were resolved by 15% SDS–PAGE and analyzed by immunoblotting with anti-4E-BP1. (C) 293 cells were serum starved for 36 h and stimulated with serum in the presence or absence of rottlerin (Rot). Total cell lysates were subjected to incubation with anti-PKCδ (filled bars) or anti-Akt (hatched bars). Precipitates were incubated in kinase buffer containing [γ-32P]ATP and MBP. The reaction products were analyzed by SDS–PAGE and autoradiography. The results are expressed as PKCδ activity (mean ± SD) of two independent experiments.
Discussion
PKCδ interacts with multiple members of the PIK family
RAFT1/mTOR/FRAP is related to members of the PI3-kinase (PIK) family that include Tor1p, Tor2p, DNA-PK catalytic subunit (DNA-PKcs), ATR (ataxia-telangiectasia related) and ATM, which are involved in cell cycle control and DNA repair (Kunz et al., 1993; Keith and Schreiber, 1995; Abraham, 1996; Thomas and Hall, 1997). Few insights, however, are available regarding regulation of the catalytic function of PIK-related kinases. Recent studies have demonstrated that PKCδ constitutively interacts with DNA-PKcs. The functional importance of this interaction is supported by the finding that PKCδ phosphorylates and inhibits DNA-PKcs in response to DNA damage (Bharti et al., 1998). Other studies have shown that treatment of cells with cytokines increases association of PKCδ with PI3-K (Ettinger et al., 1996). Activation loop phosphorylation of PKCδ in response to serum stimulation of cells is PI3-K dependent. These findings have indicated that there are separate pools of PI3-K and PKCδ (Le Good et al., 1998). The present work extends the relationship between PKCδ and PIK family members with the demonstration that PKCδ binds constitutively and directly to RAFT1. The functional importance of the PKCδ–RAFT1 interaction is supported by the finding that this complex plays a role in the regulation of cap-dependent initiation of protein translation.
Role for PKCδ in phosphorylation of 4E-BP1 and cap-dependent initiation of protein translation
Growth factors induce rapid increases in protein synthesis by stimulating mRNA translation (Sonenberg and Gingras, 1998). The translation of mRNA into protein is controlled at several levels by hormones, growth factors and other stimuli. A major target of extracellular stimuli is the translation initiation factor eIF4E (Sonenberg and Gingras, 1998). The activity of eIF4E is controlled in part by 4E-BPs that interact with eIF4E in their hypophosphorylated, but not hyperphosphorylated, form (Lin et al., 1994; Pause et al., 1994). Recent studies have demonstrated that PI3-K is required for the phosphorylation and inactivation of 4E-BP1 (Beretta et al., 1996; von Manteuffel et al., 1996). However, other studies have shown that PI3-K is not the direct kinase of 4E-BP1 (Scott et al., 1998) and that it controls the activity of RAFT1, which is required for 4E-BP1 phosphorylation (Brunn et al., 1996). Furthermore, previous studies have shown that Akt, which also regulates 4E-BP1 phosphorylation, functions downstream of PI3-K (Gingras et al., 1998). The present work demonstrates that PKCδ is required for the phosphorylation and inactivation of 4E-BP1. The catalytic subunit of PKCδ promotes the phosphorylation of 4E-BP1 when over– expressed transiently in 293T cells. Moreover, we show that a constitutively active form of PKCδ induces phos– phorylation of 4E-BP1 in the absence of growth factors and that this effect is resistant to wortmannin. Taken together, these findings indicate that PKCδ acts downstream of PI3-K in mediating phosphorylation of 4E-BP1. In this context, recent studies have shown that addition of serum to serum-starved cells is associated with activation of PKCδ by increasing its phosphorylation on the activation loop (Le Good et al., 1998). Moreover, phosphorylation of the PKCδ activation loop by its upstream modulator is sensitive to PI3-K inhibitors (Le Good et al., 1998). To assess the role of PKCδ in vivo, we generated a cell line that stably expresses the N-terminal regulatory domain of PKCδ (MCF-7/PKCδ-RD). Overexpression of PKCδ-RD competes with the endogenous PKCδ for binding to its targets. The finding that serum-induced phosphorylation of 4E-BP1 is significantly inhibited in cells expressing PKCδ-RD indicates that PKCδ mediates phosphorylation of 4E-BP1 in vivo.
The translation of most eukaryotic mRNAs is mediated by cap-dependent mechanisms (Jackson, 1993). Unphosphorylated 4E-BP1 interacts with eIF4E and thereby inhibits cap-dependent translation (Pause et al., 1994). 4E-BP1 phosphorylation and cap-dependent translation are mediated by RAFT1 signaling in yeast and mammalian cells (Barbet et al., 1996; Beretta et al., 1996; Burnett et al., 1998; Gingras et al., 1998; Hara et al., 1997). Moreover, inhibition of RAFT1 activity by the rapamycin–FKBP12 complex contributes to translational arrest by increasing the affinity of 4E-BP1 for eIF4E (Brown et al., 1995; Hara et al., 1997). Other than the rapamycin–FKBP12 complex, there are no known inhibitors of RAFT1 activity. The results of the present study demonstrate that overexpression of activated forms of PKCδ (PKCδ DR144/145A mutant and PKCδ CF) inhibits the interaction of 4E-BP1 with eIF4E. Other studies have shown that an activated form of Akt (Myr-Akt) also inhibits the binding of 4E-BP1 to eIF4E (Gingras et al., 1998).
The present results demonstrate that PKCδ coimmunoprecipitates with RAFT1 and thereby potentiates the phosphorylation of 4E-BP1. Since PKCδ does not directly phosphorylate RAFT1, and PKCδ-mediated phosphoryl– ation of 4E-BP1 is inhibited by pretreatment with rapamycin, the rapamycin-sensitive component resides parallel to PKCδ in this signaling cascade. The finding that PKCδ-mediated phosphorylation of 4E-BP1 is also inhibited by a kinase-dead mutant of RAFT1 (RAFT1 D2357E) further indicates that PKCδ and RAFT1 activities are both required to stimulate phosphorylation of 4E-BP1 maximally.
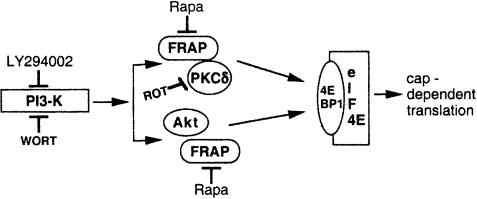
Recent work has shown that RAFT1 immunoprecipitated from cells is able to phosphorylate 4E-BP1 in vitro (Brunn et al., 1997). However, this observation does not preclude the possibility that a downstream effector of RAFT1 that coimmunoprecipitates with RAFT1 is responsible for 4E-BP1 phosphorylation. Taken together with the results of the present study, the findings suggest that PKCδ may not be a direct 4E-BP1-kinase in vivo, although kinase activities of RAFT1 and PKCδ are required to stimulate 4E-BP1 phosphorylation. Moreover, the large size of RAFT1 raises the possibility that it may act as a scaffolding protein that can interact with multiple kinases. Thus, one of these kinases could be PKCδ. Therefore, the available evidence supports a model in which the RAFT1–PKCδ complex plays an active role in 4E-BP1 phosphorylation and 4E-BP1-mediated, cap-dependent initiation of protein translation (Figure 9).
Fig. 9. Schematic model depicting the role of the mTOR/FRAP/RAFT1–PKCδ complex in 4E-BP1-mediated, cap-dependent translation. PI3-K acts upstream to the mTOR/FRAP–Akt pathway (Gingras et al., 1998). The present results demonstrate that overexpression of active PKCδ DR144/145A is associated with induction in phosphorylation of 4E-BP1 and that this effect is insensitive to wortmannin. These findings indicate that the mTOR/FRAP–PKCδ complex lies downstream to PI3-K. Moreover, there is no detectable interaction between Akt and PKCδ (data not shown). Our results and those from others (Gingras et al., 1998) indicate that FRAP/mTOR functions as a scaffolding protein through which multiple upstream effectors converge and thereby initiate cap-dependent translation. [PKCδ–mTOR/FRAP; Akt–mTOR/FRAP]→→4E-BP1.
Function for PKCδ in cell proliferation and apoptosis
PKCδ belongs to the nPKC group and is activated by DAG by a calcium-independent mechanism. Activation of the platelet-derived growth factor (PDGF) and epidermal growth factor (EGF) receptors is associated with induction of PKCδ (Li et al., 1994; Denning et al., 1996). Moreover, transformation by Ras or v-Src results in activation of PKCδ (Denning et al., 1993; Zang et al., 1997). In contrast, other studies have shown that in response to stress, PKCδ is involved in the induction of growth arrest and apoptosis (Watanabe et al., 1992; Emoto et al., 1995; Ghayur et al., 1996; Lu et al., 1997; Bharti et al., 1998). An apparent paradox with regard to the role of PKCδ in apoptosis in response to stress, however, is raised by the recent finding that activation loop phosphorylation of PKCδ in response to serum stimulation of cells is PI3-K dependent and mediated by PDK1 (Le Good et al., 1998). Thus, PKCδ may be functional in both apoptotic and proliferative pathways. In this setting, PKCδ could represent a switch that determines cell fate, such that PKCδ effects proliferative signals following growth-mediated phosphorylation and apoptotic signals as a consequence of stress-mediated phosphorylation. Since 4E-BP1 is a negative regulator of cell growth (Rousseau et al., 1996), it is possible that the growth-mediated effects of PKCδ are governed, at least in part, by phosphorylation of 4E-BP1 and thereby activation of eIF4E. Because of potential roles for PKCδ in both cell proliferation and survival, its downstream targets in the proliferative response might be different from its targets in response to stress.
Materials and methods
Cell culture and reagents
293, 293T, MCF-7, MCF-7/GFP and MCF-7/PKCδ-RD cells were grown as described (Kharbanda et al., 1995b). Cells were treated with 20–50 ng/ml rapamycin (Sigma), 200 nM wortmannin (Sigma) or 10 μM rottlerin (Calbiochem).
Antibodies and other reagents
The antibodies used were from the following sources: anti-HA, Boehringer Mannheim; anti-PKCδ and anti-PKCβ, Santa Cruz Biotechnology (Santa Cruz, CA); anti-P-Tyr, Upstate Biotechnology Inc., (UBI, Upstate, NY); anti-4E-BP1, clone 11208 (Gingras et al., 1996). m7GTP coupled to agarose resin was purchased from Pharmacia Biotech (Uppsala, Sweden).
Plasmids
FL, CF and CF(K–R) of PKCδ were as described (Bharti et al., 1998). pGEX-PKCδ FL, pGEX-PKCδ CF, pGEX-PKCδ CF(K–R), pEGFP-PKCδ CF and pEGFP-PKCδ CF(K–R) were as described (Bharti et al., 1998). HA-RAFT1 and HA-RAFT1 D2357E (Sabatini et al., 1994; Burnett et al., 1998), 4E-BP1 cDNA (Gingras et al., 1998) and pCDNA3-Luc-Pol-CAT were as described (Craig et al., 1998). PKCδ DR144/145A and PKCα R22A/A25E expression vectors were provided by S.Ohno (Ueda et al., 1996). The pE1-PKCβII expression plasmid was constructed as described (Kaneki et al., 1999).
Transient transfections
Cells were grown in 100-mm cell culture dishes and were transiently transfected by SuperFect™ or calcium phosphate as described (Kharbanda et al., 1997; Kumar et al., 1998). The transfection efficiency, as determined by analysis of β-galactosidase activity and by GFP immunofluorescence, was 60–70%. After 12 h of incubation at 37°C, the medium was replaced and the cells were incubated for another 24–36 h.
Immunoprecipitation and immunoblot analysis
Preparation of cell lysates and immunoprecipitations was performed as described (Kharbanda et al., 1995a,b). Soluble proteins (150 μg) were incubated with anti-HA, anti-PKCδ or anti-PKCβ as indicated for 2–3h and precipitated with protein A–Sepharose for an additional 1h. The resulting immune complexes were analyzed by SDS–PAGE and immunoblotting. Signal intensities were determined by densitometric analysis.
PKCδ activity assays
293T cells were transiently transfected with HA-RAFT1. Total cell lysates were subjected to immunoprecipitation with anti-HA. The immune complex kinase assays were performed using MBP as a substrate as described (Bharti et al., 1998).
Kinase assays
293 cells were serum starved for 36 h. Following serum starvation, media containing 15% fetal bovine serum was added in the presence or absence of 10 μM rottlerin for different time intervals. Total cell lysates were subjected to immunoprecipitation with anti-PKCδ or anti-Akt (Santa Cruz) antibodies. The protein precipitates were assayed for kinase activity as described (Kharbanda et al., 1996).
Phosphorylation of 4E-BP1
293T cells were transiently cotransfected with HA-4E-BP1 with pCDNA3 PKCδ FL, pCDNA3 PKCδ DR144/145A, pCDNA3 PKCδ CF or pCDNA3 PKCδ CF(K–R). Cells were also separately cotransfected with HA-4E-BP1, RAFT1 and different amounts of pCDNA3 PKCδ DR144/145A. Total cell lysates were analyzed by immunoblotting with anti-HA. 293 cells were serum starved for 36 h. Lysates were subjected to protein precipitation with m7GTP resin. The precipitates were then incubated with active PKCδ CF in the presence of [γ-32P]ATP and kinase buffer at 30°C for 20 min. The reaction products were analyzed by SDS–PAGE and autoradiography.
m7GTP and eIF4E affinity chromatography
To purify endogenous eIF4E, 50 μl of a 50% slurry of m7GTP Sepharose were added to the lysate and the mixture was incubated for 45 min at 4°C. After washing the resin twice with 50 mM HEPES pH 7.4, 50 mM NaCl, 2 mM EDTA, 0.1% Triton X-100, bound proteins were eluted and analyzed by SDS–PAGE and immunoblotting with anti-HA or anti–4E-BP1.
Luciferase activity assays
293T cells were transiently transfected with the plasmid pcDNA3-LUC-pol-CAT (Beretta et al., 1996, Craig et al., 1998) using the calcium phosphate method as described (Bharti et al., 1998). After transfection, cells were serum starved for 24 h. Serum was added for 12 h with or without 20 ng/ml rapamycin. Cell extracts were prepared and assayed for luciferase activity in a luminometer (Turner) using an Enhanced Luciferase Assay Kit (Analytical Luminescence Laboratories, Ann Arbor, MI).
Isolation of RNA and RNase protection assays
Forty-eight hours after cell transfection, total RNA was isolated using the single-step guanidinium isothiocyanate method as described (Kumar and Carmichael, 1997). Internally labeled RNA probes were made by in vitro transcription by T3 or T7 RNA polymerase in the presence of [α-32P]UTP as described (Kumar and Carmichael, 1997). In brief, DNA templates were removed by RQ1 DNase digestion followed by phenol/chloroform extraction. RNA was hybridized overnight to 32P-labeled antisense RNA probes specific for GAPDH (Pharmingen, San Diego, CA) and luciferase as described (Donze et al., 1995; Kumar and Carmichael, 1997).
Acknowledgments
Acknowledgements
We thank Drs Joseph Avruch and Robert T.Abraham for anti-mTOR antibody and S.Ohno for the PKCδ DR144/145A and PKCα R22A/A25E. We also thank Drs Nahum Sonenberg and Ann-Claude Gingras for critical reading of the manuscript and excellent suggestions. This investigation was supported by PHS grant CA75216 (S.K.) awarded by the National Cancer Institute, DHHS.
References
- Abraham R.T. (1996) Phosphoinositol 3-kinase related kinases. Curr. Opin. Immunol., 8, 412–418. [DOI] [PubMed] [Google Scholar]
- Alessi D.R., et al. (1997) Phosphoinositide-dependent protein kinase-1 (PDK1): structural and functional homology with the Drosophila DSTPK61 kinase. Curr. Biol., 7, 776–789. [DOI] [PubMed] [Google Scholar]
- Barbet N., Schneider, U., Helliwell, S.B., Stansfield, I., Tuite, M.F. and Hall, M.N. (1996) TOR controls translation initiation and early G1 progression in yeast. Mol. Cell. Biol., 7, 25–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belham C., Wu, S. and Avruch, J. (1999) Intracellular signaling: PDK1–a kinase at the hub of things. Curr. Biol., 9, R93–R96. [DOI] [PubMed] [Google Scholar]
- Beretta L., Gingras, A.C., Svitkin, Y.V., Hall, A.N. and Sonenberg, N. (1996) Rapamycin blocks the phosphorylation of 4E-BP1 and inhibits cap-dependent initiation of translation. EMBO J., 15, 658–664. [PMC free article] [PubMed] [Google Scholar]
- Bharti A., et al. (1998) Inactivation of DNA-dependent protein kinase by protein kinase Cδ: implications for apoptosis. Mol. Cell. Biol., 18, 6719–6728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown E.J. and Schreiber, S.L. (1996) A signaling pathway to translational control. Cell, 86, 517–520. [DOI] [PubMed] [Google Scholar]
- Brown E.J., Albers, M.W., Shin, T.B., Ichikawa, K., Keith, C.T., Lane, W.S. and Schreiber, S.L. (1994) A mammalian protein targeted by G1-arresting rapamycin–receptor complex. Nature, 369, 756–758. [DOI] [PubMed] [Google Scholar]
- Brown E.J., Beal, P.A., Keith, C.T., Chen, J., Shin, T.B. and Schreiber, S.L. (1995) Regulation of p70 S6 kinase activity of FRAP in vivo. Nature, 377, 441–446. [DOI] [PubMed] [Google Scholar]
- Brunn G.J., Hudson, C.C., Sekulic, A., Williams, J.M., Hosoi, H., Houghton, P.J., Lawrence, J.C., Jr and Abraham, R.T. (1997) Phosphorylation of the translational repressor PHAS-I by the mammalian target of rapamycin. Science, 277, 99–101. [DOI] [PubMed] [Google Scholar]
- Burnett P.E., Barrow, R.K., Cohen, N.A., Snyder, S.H. and Sabatini, D.M. (1998) RAFT1 phosphorylation of the translational regulators p70S6 kinase and 4E-BP1. Proc. Natl Acad. Sci. USA, 95, 1432–1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung J., Kuo, C.J., Crabtree, G.R. and Blenis, J. (1992) Rapamycin–FKBP specifically blocks growth-dependent activation of and signaling by the 70kd S6 protein kinases. Cell, 69, 1–20. [DOI] [PubMed] [Google Scholar]
- Craig A.W.B., Haghighat, A., Yu, A.T.K. and Sonenberg, N. (1998) Interaction of polyadenylate-binding protein with the eIF4G homologue PAIP enhances translation. Nature, 392, 520–523. [DOI] [PubMed] [Google Scholar]
- Dekker L.V. and Parker, P.J. (1994) Protein kinase C–a question of specificity. Trends Biochem. Sci., 19, 73–77. [DOI] [PubMed] [Google Scholar]
- Dekker L.V., Palmer, R.H. and Parker, P.J. (1995) The protein kinase C and protein kinase C related families. Curr. Opin. Struct. Biol., 5, 396–402. [DOI] [PubMed] [Google Scholar]
- Denning M.F., Dlugosz, A.A., Howett, M.K. and Yushpa, S.H. (1993) Expression of an oncogenic rasHa gene in murine keratinocytes induces tyrosine phosphorylation and reduced activity of protein kinase C δ. J. Biol. Chem., 268, 26069–26081. [PubMed] [Google Scholar]
- Denning M.F., Dlugosz, A.A., Threadgill, T., Magnuson, T. and Yushpa, S.H. (1996) Activation of the epidermal growth factor receptor signal transduction pathways stimulates tyrosine phosphorylation of protein kinase C δ. J. Biol. Chem., 271, 5325–5331. [DOI] [PubMed] [Google Scholar]
- Donze O., Damy, P. and Spahr, P.F. (1995) The first and third uORFs in RSV leader RNA are efficiently translated: implications for translational regulation and viral RNA packaging. Nucleic Acids Res., 23, 861–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emoto Y., et al. (1995) Proteolytic activation of protein kinase C δ by an ice-like protease in apoptotic cells. EMBO J., 14, 6148–6156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ettinger S.L., Lauener, R.W. and Duronio, V. (1996) Protein kinase C δ specifically associates with phosphatidyl 3-kinase following cytokine stimulation. J. Biol. Chem., 271, 14514–14518. [DOI] [PubMed] [Google Scholar]
- Ghayur T., et al. (1996) Proteolytic activation of protein kinase C δ by an ICE/CED3-like protease induces characteristics of apoptosis. J. Exp. Med., 184, 2399–2404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gingras A., Svitkin, Y., Belsham, G., Pause, A. and Sonenberg, N. (1996) Activation of the translational suppressor 4E–BP1 following infection with encephalomyocarditis virus and poliovirus. Proc. Natl Acad. Sci. USA, 93, 5578–5583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gingras A.-C., Gygi, S.P., Raught, B., Polakiewicz, R.D., Abraham, R.T., Hoekstra, M.F., Aebersold, R. and Sonenberg, N. (1999) Regulation of 4E-BP1 phosphorylation: a novel two-step mechanism. Genes Dev., 13, 1422–1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gschwendt M., Muller, H.J., Kielbassa, K., Zang, R., Kittstein, W., Rincke, G. and Marks, F. (1994) Rottlerin, a novel protein kinase inhibitor. Biochem. Biophys. Res. Comm., 199, 93–98. [DOI] [PubMed] [Google Scholar]
- Hara K., Yonezawa, K., Kozlowski, M.T., Sugimoto, T., Andrabi, T., Weng, Q.P., Kauga, M., Nishimoto, I. and Avruch, J. (1997) Regulation of eIF-4E BP1 phosphorylation by mTOR. J. Biol. Chem., 272, 26457–26463. [DOI] [PubMed] [Google Scholar]
- Heesom K.J. and Denton, R.M. (1999) Dissociation of the eukaryotic initiation factor-4E/4E-BP1 complex involves phosphorylation of 4E–BP1 by an mTOR-associated kinase. FEBS Lett., 457, 489–493. [DOI] [PubMed] [Google Scholar]
- Heitman J., Movva, N.R. and Hall, M.N. (1991) Targets for cell cycle arrest by the immunosuppressant rapamycin in yeast. Science, 253, 905–909. [DOI] [PubMed] [Google Scholar]
- Hirai S., Izumi, Y., Higa, K., Kaibuchi, K., Mizuno, K., Osada, S., Suzuki, K. and Ohno, S. (1994) Ras-dependent signal transduction is indispensable but not sufficient for the activation of AP1/JUN by PKCδ. EMBO J., 13, 2331–2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu C., Pang, S., Kong, X., Velleca, M. and Lawrence, J.C.,Jr (1994) Molecular cloning and tissue distribution of PHAS-I, an intracellular target for insulin and growth factors. Proc. Natl Acad. Sci. USA, 91, 3730–3734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson R.J. (1993) Cytoplasmic regulation of mRNA function: the importance of the 3′ untranslated region. Cell, 74, 9–14. [DOI] [PubMed] [Google Scholar]
- Jefferies H.B.J. and Thomas,G. (1996) Ribosomal protein S6 phosphorylation and signal transduction. In Hershey,J.W.B., Mathews,M.B. and Sonenberg,N. (eds), Translational Control. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 389–409. [Google Scholar]
- Jefferies H.B.J., Reinhard, C., Kozma, S.C. and Thomas, G. (1994) Rapamycin selectively represses translation of the ‘polypyrimidine tract’ mRNA family. Proc. Natl Acad. Sci. USA, 91, 4441–4445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferies H.B.J., Fumagalli, S., Dennis, P.B., Reinhard, C., Pearson, R.B. and Thomas, G. (1997) Rapamycin suppresses 5′ TOP mRNA translation through inhibition of p70s6k. EMBO J., 15, 3693–3704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneki M., Kharbanda, S., Pandey, P., Yoshida, K., Takekawa, M., Liou, J.R., Stone, R. and Kufe, D. (1999) Functional role for protein kinase Cβ as a regulator of stress-activated protein kinase activation and monocytic differentiation of myeloid leukemia cells. Mol. Cell. Biol., 19, 461–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keith C.T. and Schreiber, S.L. (1995) PIK-related kinases: DNA repair, recombination and cell cycle checkpoints. Science, 270, 50–51. [DOI] [PubMed] [Google Scholar]
- Kharbanda S., Ren, R., Pandey, P., Shafman, T.D., Feller, S.M., Weischlbaum, R. and Kufe, D. (1995a) Activation of the c-Abl tyrosine kinase in the stress response to DNA-damaging agents. Nature, 376, 785–788. [DOI] [PubMed] [Google Scholar]
- Kharbanda S., Pandey, P., Ren, R., Meyer, B., Zon, L. and Kufe, D. (1995b) The c-Abl tyrosine kinase is activated by ara-C. J. Biol. Chem., 270, 30278–30281. [DOI] [PubMed] [Google Scholar]
- Kharbanda S., Bharti, A., Wang, J., Pandey, P., Pei, D., Ren, R., Walsh, C. and Kufe, D. (1996) The stress response to ionizing radiation involves c-Abl-dependent phosphorylation of SHPTP1. Proc. Natl Acad. Sci. USA, 93, 6898–6901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kharbanda S., Pandey, P., Jin, S., Inoue, S., Bharti, A., Yuan, Y.M., Weichselbaum, R., Weaver, D. and Kufe, D. (1997) Functional interaction between DNA-PK and c-Abl in response to DNA damage. Nature, 386, 732–735. [DOI] [PubMed] [Google Scholar]
- Kumar M. and Carmichael, G.G. (1997) Nuclear antisense RNA induces extensive adenosine modifications and nuclear retention of target transcripts. Proc. Natl Acad. Sci. USA, 94, 3542–3547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Pandey, P., Bharti, A., Jin, S., Weichselbaum, R., Weaver, D., Kufe, D. and Kharbanda, S. (1998) Regulation of DNA-dependent protein kinase by the Lyn tyrosine kinase. J. Biol. Chem., 273, 25654–25658. [DOI] [PubMed] [Google Scholar]
- Kunz J. and Hall, M.N. (1993) Cyclosporin A, FK506 and rapamycin: more than just immunosuppression. Trends Biol. Sci., 18, 334–338. [DOI] [PubMed] [Google Scholar]
- Kunz J., Henriquez, R., Schneider, U., Deuter-Reinhard, M., Movva, M.R. and Hall, M.N. (1993) Target of rapamycin in yeast, TOR2, is an essential phosphatidylinositol kinase homolog required for G1 progression. Cell, 73, 585–596. [DOI] [PubMed] [Google Scholar]
- Kuo C.J., Chung, J., Fiorntino, D.F., Flanagan, W.M., Blenis, J. and Crabtree, G.R. (1992) Rapamycin selectively inhibits IL-2 activation of p70 S6 kinase. Nature, 358, 70–73. [DOI] [PubMed] [Google Scholar]
- Lane H.A., Fernandez, A., Lamb, N.J.C. and Thomas, G. (1993) p70s6k function is essential for G1 progression. Nature, 363, 170–172. [DOI] [PubMed] [Google Scholar]
- Le Good J.A., Ziegler, W.H., Parekh, D.B., Alessi, D.R, Cohen, D.P. and Parker, P.J. (1998) Protein kinase C isotypes controlled by phosphoinositide 3-kinase through the protein kinase PDK1. Science, 281, 2042–2045. [DOI] [PubMed] [Google Scholar]
- Li W., Yu, J.C., Michieli, P., Beeler, J.F., Ellmore, N., Heidaran, M.A. and Pierce, J.H. (1994) Stimulation of the PDGF β receptor signaling pathway activates protein kinase-C δ. Mol. Cell. Biol., 14, 6727–6735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin T.A. and Lawrence, J.C.,Jr (1996) Control of the translation regulators PHAS-I and PHAS-II by insulin and cAMP in 3T3-L1 adipocytes. J. Biol. Chem., 271, 30199–30204. [DOI] [PubMed] [Google Scholar]
- Lin T.A., Kong, X., Haystead, T.A.J., Pause, A., Belsham, G., Sonenberg, N. and Lawrence, J.C.,Jr (1994) PHAS-I as a link between mitogen-activated protein kinase and translation initiation. Science, 266, 653–656. [DOI] [PubMed] [Google Scholar]
- Lu Z., Hornia, H., Jiang, Y-W., Zang, Q., Ohno, S. and Foster, D.A. (1997) Activation of protein kinase C triggers its ubiquitination and degradation. Mol. Cell. Biol., 17, 3418–3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen F.C., Ostergaard, L., Nielsen, J. and Christiansen, J. (1995) Growth-independent translation of IGF-II mRNA by a rapamycin-sensitive pathway. Nature, 377, 358–362. [DOI] [PubMed] [Google Scholar]
- Nishizuka Y. (1988) The heterogeneity and differential expression of multiple species of the protein kinase family. Biofactors, 1, 17–20. [PubMed] [Google Scholar]
- Nishizuka Y. (1995) Protein kinase C and lipid signaling for sustained cellular responses. FASEB J., 9, 484–496. [PubMed] [Google Scholar]
- Osada S., Mizuno, K., Saido, T.C., Suzuki, K., Kuroki, T. and Ohno, S. (1992) A new member of the protein kinase C family, nPKC θ predominantly expressed in skeletal muscle. Mol. Cell. Biol., 12, 3930–3938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parekh D., Ziegler, W., Yonezawa, K., Hara, K. and Parker, P.J. (1999) Mammalian TOR controls one of two kinase pathways acting upon nPKCδ and nPKCɛ. J. Biol. Chem., 274, 34758–34764. [DOI] [PubMed] [Google Scholar]
- Pause A., Belsham, G.J., Gingras, A.-C., Dounze, O., Lin, T.A., Lawrence, J.C., Jr and Sonenberg, N. (1994) Insulin-dependent stimulation of protein synthesis by phosphorylation of a regulator of 5′-cap function. Nature, 371, 762–767. [DOI] [PubMed] [Google Scholar]
- Pearson R.B., Dennis, P.B., Han, J.W., Williamson, N.A., Kozma, S.C., Wettenhall, R.E.H. and Thomas, G. (1995) The principal target of rapamycin-induced p70s6k inactivation is a novel phosphorylation site within a conserved hydrophobic domain. EMBO J., 21, 5279–5287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price D.J., Grove, J.R., Calvo, V., Avruch, J. and Bierer, B.E. (1992) Rapamycin-induced inhibition of the 70 kd S6 protein kinase. Science, 257, 973–977. [DOI] [PubMed] [Google Scholar]
- Redpath N.T., Price, N.T. and Proud, C.G. (1996) Cloning and expression of cDNA encoding protein synthesis elongation factor-2 kinase. J. Biol. Chem., 271, 17547–17554. [PubMed] [Google Scholar]
- Reinhard G., Fernandez, A., Lamb, N.J.C. and Thomas, G. (1994) Nuclear localization of p85s6k: functional requirement for entry into S phase. EMBO J., 1, 1557–1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reks S.E., Smith, P.H., Messina, J.L. and Weinstock, R.S. (1998) Translocation PKCδ by insulin in a rat hepatoma cell line. Endocrine, 8, 161–167. [DOI] [PubMed] [Google Scholar]
- Rousseau D., Kasper, R., Rosenwald, I., Gehrke, L. and Sonenberg, N. (1996) Translation initiation of ornithine decarboxylase and nucleocytoplasmic transport of cyclin D1 mRNA are increased in cells overexpressing eukaryotic initiation factor 4E. Proc. Natl Acad. Sci. USA, 93, 1065–1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabatini D.M., Erdjument-Bromage, H., Lui, M., Tempst, P. and Snyder, S.H. (1994) RAFT1: A mammalian protein that binds to FKBP12 in a rapamycin-dependent fashion and is homologous to yeast TORs. Cell, 78, 35–43. [DOI] [PubMed] [Google Scholar]
- Sabers C.J., Wiederrecht, G., Williams, J.M., Martin, M.M., Dumont, F.J. and Abraham, R.T. (1995) Isolation of a protein target of the FKBP12–rapamycin complex in mammalian cells. J. Biol. Chem., 270, 815–822. [DOI] [PubMed] [Google Scholar]
- Scott P.H., Brunn, G.J., Kohn, A.D., Roth, R.A. and Lawrence, J.C. (1998) Evidence of insulin-stimulated phosphorylation and activation of the mammalian target of rapamycin mediated by a protein kinase B signaling pathway. Proc. Natl Acad. Sci. USA, 95, 7772–7777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonenberg N. (1994) mRNA translation: influence of the 5′ and 3′ untranslated regions. Curr. Opin. Genet. Dev., 4, 310–315. [DOI] [PubMed] [Google Scholar]
- Sonenberg N. (1996) mRNA 5′ cap-binding protein eIF-4E and control of cell growth. In Hershey,J.W., Mathews,M.B. and Sonenberg,N. (eds), Translational Control. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 245–269. [Google Scholar]
- Sonenberg N. and Gingras, A.C. (1998) The mRNA 5′ cap-binding eIF4E and control of cell growth. Curr. Opin. Cell Biol., 10, 268–275. [DOI] [PubMed] [Google Scholar]
- Stan R., McLaughlin, M.M., Cafferkey, R., Johnson, R.K., Rosenberg, M. and Livi, G.P. (1994) Interaction between FKBP12–rapamycin and mTOR involves a conserved serine residue. J. Biol. Chem., 269, 32027–32030. [PubMed] [Google Scholar]
- Terada N., Patel, H.R., Takase, K., Kohno, K., Nairns, A.C. and Gelfand, E.W. (1994) Rapamycin selectively inhibits translation of mRNAs encoding elongation factors and ribosomal proteins. Proc. Natl Acad. Sci. USA, 91, 11477–11481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas G. and Hall, M.N. (1997) TOR signalling and control of cell growth. Curr. Opin. Cell Biol., 9, 782–787. [DOI] [PubMed] [Google Scholar]
- Ueda Y., Hirai, S., Osada, S., Suzuki, A., Mizuno, K. and Ohno, S. (1996) Protein kinase C δ activates the MEK–ERK pathway in a manner independent of Ras and dependent on Raf. J. Biol. Chem., 271, 23512–23519. [DOI] [PubMed] [Google Scholar]
- von Manteuffel S.R., Gingras, A.-C., Ming, X.F., Sonenberg, N. and Thomas, G. (1996) 4E-BP1 phosphorylation is mediated by the FRAP–p70s6k pathway and is independent of mitogen-activated protein kinase. Proc. Natl Acad. Sci. USA, 93, 4070–4080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T., Ono, Y., Taniyama, Y., Hazama, K., Igarashi, K., Ogita, K., Kikkawa, U. and Nishizuka, Y. (1992) Cell division arrest induced by phorbol ester in CHO cells overexpressing PKCδ subspecies. Proc. Natl Acad. Sci. USA, 89, 10159–10163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zang Q., Lu, Z., Curto, M., Barile, N., Shalloway, D. and Foster, D.A. (1997) Association between v-Src and protein kinase C δ in v-Src-transformed fibroblasts. J. Biol. Chem., 272, 13275–13280. [DOI] [PubMed] [Google Scholar]