Abstract
DNA ligases catalyze the crucial step of joining the breaks in duplex DNA during DNA replication, repair and recombination, utilizing either ATP or NAD+ as a cofactor. Despite the difference in cofactor specificity and limited overall sequence similarity, the two classes of DNA ligase share basically the same catalytic mechanism. In this study, the crystal structure of an NAD+-dependent DNA ligase from Thermus filiformis, a 667 residue multidomain protein, has been determined by the multiwavelength anomalous diffraction (MAD) method. It reveals highly modular architecture and a unique circular arrangement of its four distinct domains. It also provides clues for protein flexibility and DNA-binding sites. A model for the multidomain ligase action involving large conformational changes is proposed.
Keywords: DNA ligase/helix–hairpin–helix motif/nucleotidyl transferase/oligomer-binding fold/zinc finger motif
Introduction
All life forms on earth depend on DNA ligases for joining the breaks in double-stranded DNA, which is a crucial step in replication and repair of DNA and in genetic recombination (Lehman, 1974; Lindahl and Barnes, 1992; Tomkinson and Levin, 1997). They catalyze the formation of phosphodiester bonds at single-stranded or double-stranded breaks between adjacent 3′–hydroxyl and 5′–phosphate termini (Lehman, 1974; Tomkinson and Levin, 1997). DNA ligases fall into two classes depending on the cofactor specificity. ATP-dependent ligases are ubiquitous in eukaryotes and they are also encoded by eukaryotic DNA viruses, bacteriophages of the T series and archaebacteria. In contrast, NAD+-dependent ligases are found exclusively in eubacteria. This makes NAD+-dependent DNA ligases a potential target for developing novel antibiotics. Thermostable bacterial DNA ligases with high fidelity have applications in detecting disease-associated mutations by ligase chain reaction (Barany, 1991). The sequence similarity between these two classes is not detected readily except for the Lys-Xaa-Asp-Gly (KXDG) sequence motif, which is one of the six conserved sequence elements (I, III, IIIa, IV, V and VI) present among members of the covalent nucleotidyl transferase superfamily including ATP-dependent DNA ligases, RNA and tRNA ligases, and the eukaryotic mRNA guanylyl transferases (capping enzymes) (Shuman and Schwer, 1995). The recently developed iterative sequence database search method, however, allowed the detection of five of the conserved sequence motifs, with the exception being VI (Aravind and Koonin, 1999).
Apart from the difference in cofactor requirement, the reactions catalyzed by the two classes of DNA ligase are identical. In the first step, an AMP group derived from either cofactor is covalently attached to the conserved lysine residue within the KXDG motif. The AMP moiety is then transferred from the adenylated enzyme intermediate to the free 5′–phosphoryl group at a nicked site of duplex DNA. Finally, the AMP group is released from the adenylated DNA intermediate as the phosphodiester bond is formed (Lehman, 1974; Tomkinson and Levin, 1997). Several structures of nucleotidyl transferases including the 41 kDa ATP-dependent DNA ligase from bacteriophage T7 (Subramanya et al., 1996), a GTP-dependent mRNA guanylyl transferase (Håkansson et al., 1997; Håkansson and Wigley, 1998) and the N–terminal fragment (‘adenylation’ domain) of an NAD+-dependent DNA ligase from Bacillus stearothermophilus (Bst ligase) (Singleton et al., 1999) provided the details on the cofactor binding site and adenylation/guanylation step of the reaction. However, our understanding of the architecture of multidomain DNA ligases of more typical size, the DNA-binding mode and possible domain rearrangements during catalytic steps is very limited. To gain insight into such questions, we solved the crystal structure of the entire NAD+-dependent DNA ligase from Thermus filiformis (Tfi ligase), a monomeric protein of 667 amino acid residues (Mr 75 936 Da). This study provides the first view of the full-length NAD+-dependent DNA ligase, including the C–terminal half, which was reported to play an important role in DNA binding (Timson and Wigley, 1999). It reveals highly modular architecture suited for the possible large conformational changes that seem to be necessary for its action.
Results and discussion
Structure determination
Recombinant Tfi DNA ligase, in both native and selenomethionine (SeMet)-substituted forms, was expressed, purified and crystallized as described in Materials and methods. Due to the severe non-isomorphism of heavy atom derivative crystals, initial trials with the multiple isomorphous replacement method were not successful and the structure had to be determined using three-wavelength multiwavelength anomalous diffraction (MAD) data collected from a crystal of the SeMet-substituted enzyme (see Table I). Subsequently, the structure of the native enzyme was also refined. Two molecules of Tfi ligase in the asymmetric unit of the crystal take somewhat different conformations. The conformational difference between the two copies is more pronounced in the crystal of the native enzyme than in the crystal of the SeMet-substituted enzyme. Root-mean-square (r.m.s.) deviations are 2.1 and 1.1 Å for 581 Cα atom pairs, respectively. The electron density for residues beyond 581 is missing for all but one molecule of the native enzyme in the most closed conformation. Since the electron density for this part was rather poor, only a polyalanine model was built for most of the residues 582–660 in this molecule.
Table I. Crystallographic data, phasing and refinement statistics.
| Data set | Radiation | Resolution (Å) | Total/unique reflections | Completeness (%)a | I/∣σ (I) | Rsym (%)b | Phasing powerc acentric/centric |
|---|---|---|---|---|---|---|---|
| Native | 1.009 Å, X8-C | 30–2.8 (2.9–2.8) | 142 175/43 634 | 97.9 (95.3) | 11.5 (5.7) | 6.4 (45.6) | |
| SeMet-1 (remote) | 0.9400 Å, X12-C | 30–2.9 (3.0–2.9) | 215 788/40 641 | 97.9 (79.2) | 9.8 (4.0) | 7.0 (46.3) | |
| SeMet-2 (inflection) | 0.9788 Å, X12-C | 30–2.9 (3.0–2.9) | 311 355/40 546 | 98.1 (80.8) | 11.8 (5.4) | 5.9 (34.1) | 1.2/0.8 |
| SeMet-3 (peak) | 0.9784 Å, X12-C | 30–2.9 (3.0–2.9) | 315 581/40 572 | 98.2 (81.9) | 12.0 (5.5) | 6.4 (32.1) | 1.0/0.7 |
| Figure of meritd for MAD phasing using SeMet-1–3 data (calculated for 30–2.9 Å) | 0.52 (before DM)/0.80 (after DM) | ||||||
| Data set | Resolution (Å) | No. of reflectionse | Total no. of atomsf | R-factor/Rfree (%)g | R.m.s. deviationsh |
|
|---|---|---|---|---|---|---|
| bond lengths (Å) | bond angles (°) | |||||
| Native | 8.0–2.8 | 38 138 (90.7%) | 10 080 (264) | 25.1/31.6 | 0.012 | 1.91 |
| SeMet-1 | 20–2.9 | 36 053 (89.5%) | 9674 (242) | 22.8/29.8 | 0.007 | 1.52 |
aCompleteness for I/σ (I) >1.0, high-resolution shell in parentheses.
bRsym = ΣhΣi∣I(h)i – <I(h)>∣/ΣhΣiI(h)i, where I(h) is the intensity of reflection h, Σh is the sum over all reflections and Σi is the sum over i measurements of reflection h. Numbers in parentheses reflect statistics for the last shell.
cPhasing power is the mean value of the heavy atom structure factor amplitude divided by the residual lack of closure error for both acentric and centric reflections.
dFigure of merit = <∣ΣP(α)eiα/ΣP(α)∣>, where α is the phase and P(α) is the phase probability distribution.
eNumbers reflect the reflections with F/σ (F) >2.0, overall completeness in parentheses.
fNumbers in parentheses are water molecules.
gR = Σ∣∣Fobs∣ – ∣Fcalc∣∣/Σ∣Fobs∣, where Rfree (Brünger, 1992b) is calculated for a randomly chosen 10% of reflections [F/σ (F) >2.0] that were not used for structure refinement, and the R-factor is calculated for the remaining reflections.
hDeviations from ideal bond lengths and angles.
Modular architecture of Tfi ligase
Tfi ligase is toroidal with approximate dimensions of 95 × 75 × 55 Å, displaying highly modular architecture (Figure 1A and B). The polypeptide chain is folded into four discrete domains. Domain 1 (residues 1–317) consists of two subdomains. Subdomain 1a (residues 1–73) is mainly α–helical. The ‘adenylation’ subdomain 1b (residues 74–317) comprises two mainly antiparallel β–sheets flanked by α–helices and contains the adenylation site (Lys116) within the KXDG motif. Domain 2 (residues 318–403) contains a five-stranded antiparallel β–barrel of the oligomer-binding (OB) fold (Murzin, 1993), which is formed by β–strands 12–16 and α–helix M. The latter helix, located between strands 14 and 15, covers one end of the β–barrel. Domain 3 (residues 404–581) consists of two subdomains. Subdomain 3a (residues 404–429) is a Cys4-type zinc finger (Schmiedeskamp and Klevit, 1994; Klug and Schwabe, 1995; Mackay and Crossley, 1998), which includes a β–hairpin formed by strands 17 and 18. Subdomain 3b (residues 430–581) comprises four helix–hairpin–helix (HhH) motifs (helix pairs O–P, R–S, U–V and X–Y) (Thayer et al., 1995; Doherty et al., 1996). Domain 4 (residues 582–660) is a distinct member of the BRCT (BRCA1 C–terminus) domain superfamily (Bork et al., 1997; Callebaut and Mornon, 1997). All adjacent (sub-) domains are linked by a single chain. A circular arrangement of the four domains leads to a hole large enough to hold a double-stranded DNA with average side chain–side chain distances of 15 Å (height) × 25 Å (width) × 25–50 Å (depth).
Fig. 1. The structure of Tfi DNA ligase. (A) Domains and conserved sequence motifs of Tfi DNA ligase. Domains are in different colors: subdomain 1a, blue; subdomain 1b, cyan; domain 2, green; subdomain 3a, yellow; subdomain 3b, orange; domain 4, light pink. Residues in red are more strongly conserved than others. (B) Stereo ribbon diagram of Tfi DNA ligase. Drawn with MOLSCRIPT (Kraulis, 1991) and RASTER3D (Merritt and Bacon, 1997). Domain colors are the same as in (A). The covalently bound AMP moiety (stick model in purple) and a zinc ion (a cyan ball) are shown. Secondary structures were defined by PROCHECK (Laskowski et al., 1993). (C) Electron density map calculated using MAD solvent-flattened phases around the covalently bound AMP group. Atoms are yellow for carbon, red for oxygen, blue for nitrogen and cyan for phosphorus. (D) Residues around the AMP moiety. Atom colors are the same as above except black for carbon. Hydrogen bonds between the AMP and protein residues are represented by green dotted lines. Drawn with LIGPLOT (Wallace et al., 1995).
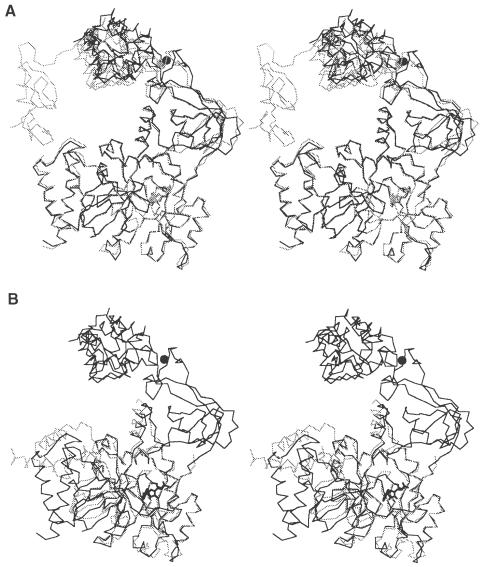
The C–terminal BRCT domain was invisible in the MAD-phased electron density map of the SeMet-substituted enzyme, probably because it is disordered in the crystal. However, one of the two ligase molecules in the asymmetric unit of the native enzyme crystal, which adopted the most closed conformation, showed a weak and discontinuous electron density for the BRCT domain. This indicates that the BRCT domain of Tfi ligase is rather mobile as a whole in the open conformation but its mobility is somewhat restricted in the closed conformation. Compared with the most open conformation, domain 3 in the most closed conformation is shifted by ∼6.4 Å toward domain 1, by a rigid-body rotation around the hinge in the loop between domains 2 and 3 (Figure 2A). As a consequence, the BRCT domain nearly contacts the conserved segment around the N–terminus of helix B. The orientation of subdomain 1a relative to subdomain 1b in the N–terminal fragment of Bst ligase is significantly different from that of Tfi ligase (Singleton et al., 1999). Subdomain 1a of Bst ligase is rotated by ∼90° around Pro68, which is also conserved in Tfi ligase (Figure 2B). If subdomain 1a of Tfi ligase takes the orientation of Bst ligase, its interaction with the BRCT domain is more extensive. It is not clear whether this discrepancy is related to the adenylation state of domain 1b. However, it suggests that subdomain 1a can make a large domain movement relative to subdomain 1b.
Fig. 2. Stereo Cα superposition of Tfi DNA ligase. (A) One of the two crystallographically independent ligase molecules in the native structure takes a more closed conformation (gray) than the other (black), and its BRCT domain is visible in the electron density map. Superposition is made for domain 1. (B) Subdomain 1a of Bst ligase (gray) takes a very different orientation from that of Tfi ligase (black). Superposition is made for subdomain 1b.
Adenylation domain
Although the cofactor NAD+ was not added deliberately during protein purification and crystallization, an AMP moiety was covalently attached to Lys116 within the KXDG motif, as indicated by the initial MAD-phased, solvent-flattened electron density map (Figure 1C). Thus, the present structure provides a direct view of the covalent enzyme–adenylate intermediate of the proposed catalytic mechanism. The AMP-binding pocket is located between the two β–sheets of subdomain 1b (Figure 1B). The interactions between the covalently bound AMP and the enzyme are depicted in Figure 1D. The negative charge of α–phosphate is compensated for by the positive charge on Lys312 (distance 3.1 Å). The adenine ring is stacked against the side chain of Tyr221 in a deep pocket, which is lined with Leu85, Leu120, Val286 and the aliphatic portions of Glu169, Tyr221, His253 and Lys288. The 6–amino group of the adenine ring makes hydrogen bonds with the main chain carbonyl of His115 and the side chain of Glu114. Lys288 and Glu114 form an ion pair at the base of the AMP-binding pocket. A similar ion pair is also present in the ATP-dependent DNA ligase from bacteriphage T7 (Lys222 and Glu32) (Subramanya et al., 1996). Many of the residues that line the AMP-binding pocket of Tfi ligase belong to five (motifs I, III, IIIa, IV and V) of the six sequence elements conserved among covalent nucleotidyl transferases (Shuman and Schwer, 1995). The observed binding mode is conserved in other non-covalent or covalent complexes of T7 DNA ligase (Subramanya et al., 1996) and Chlorella virus mRNA guanylyl transferase (Håkansson et al., 1997; Håkansson and Wigley, 1998). Motif VI encompasses the last strand of the ‘OB-fold’ domain 2 (strand 16). It was proposed that the corresponding motif in the mRNA guanylyl transferase binds and positions the triphosphate tail of GTP (Håkansson et al., 1997). Site-directed mutagenesis studies on human DNA ligase III also implicated this motif in the interaction with nicked DNA (Mackey et al., 1999). The location of this motif in the Tfi ligase structure does not seem to rule out the possibility of its involvement in interactions with both the cofactor NAD+ and DNA.
OB-fold domain
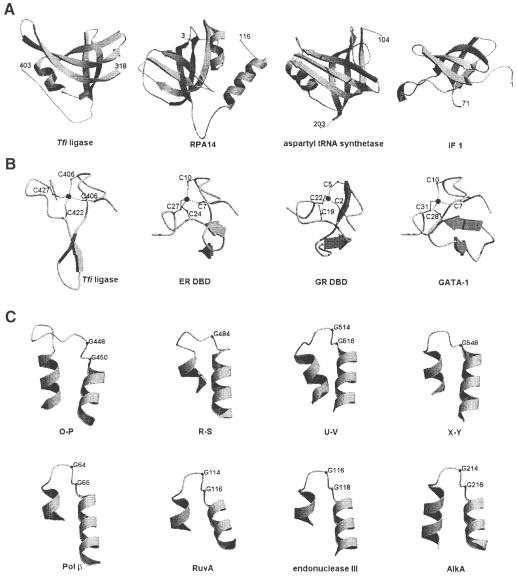
Domain 2 of Tfi ligase contains all of the three structural determinants of the OB-fold as defined previously (Bycroft et al., 1997). This fold is common among RNA or single-stranded DNA-binding (SSB) proteins (Murzin, 1993; Draper and Reynaldo, 1999), including the bacterial ribosomal proteins S1 (Bycroft et al., 1997) and S17 (Jaishree et al., 1996), the subunits of replication protein A (the eukaryotic SSB protein) (Bochkarev et al., 1997, 1999), the Oxytricha nova telomere end-binding protein (Horvath et al., 1998), bacterial cold-shock proteins CspA and CspB (Schindler et al., 1998), Pyrobaculum aerophilum translation initiation factor (IF) 5A (Peat et al., 1998), Escherichia coli translation IF 1 (Sette et al., 1997), E.coli SSB protein (Raghunathan et al., 1997), E.coli RuvA DNA recombination protein (Roe et al., 1998), staphylococcal nuclease (Murzin, 1993) and several tRNA synthetases (Murzin, 1993). A structural comparison is shown in Figure 3A. Many of the OB-fold proteins bind their ligands on the side surface of the β–barrel distant from the middle of the first strand (strand 12 in Tfi ligase). The equivalent domain in T7 DNA ligase is a much shortened version of the OB-fold. Also, compared with Tfi ligase, its orientation in the non-covalent ATP complex is rotated around the loop just before the first strand of the OB-fold domain so that its expected DNA-binding surface is not exactly facing the active site (Subramanya et al., 1996). This may be understandable, because the adenylation site should not be blocked by binding the DNA until the conserved lysine at the active site is adenylated. This orientation may change upon self-adenylation so that the putative DNA-binding groove will be completed. Since T7 DNA ligase with a more compact OB-fold domain is fully functional, we suggest that domain 1 together with the OB-fold domain is the minimal unit for the bacterial DNA ligases and that this minimal ligase should have the nick sensing as well as the ligation activities. Our suggestion is supported further by the recent finding that the 298 residue ATP-dependent DNA ligase of Chlorella virus, the smallest eukaryotic DNA ligase known, has intrinsic specificity for binding to nicked duplex DNA (Odell and Shuman, 1999).
Fig. 3. Comparison of OB-fold domains, zinc fingers and HhH motifs. (A) OB-fold domains of Tfi ligase, human replication protein A subunit (RPA14), yeast aspartyl tRNA synthetase and E.coli translation initiation factor 1 (IF 1) are shown in similar orientations. (B) Cys4-type zinc fingers of Tfi ligase, human estrogen receptor DNA-binding domain (ER DBD), rat glucocorticoid receptor DBD (GR DBD) and chicken erythroid transcription factor (GATA–1) are shown in similar orientations. Cysteines that coordinate the zinc ion are labeled. (C) HhH motifs of Tfi ligase (top row), human DNA polymerase β, Mycobacterium leprae RuvA, E.coli endonuclease III and E.coli AlkA (bottom row) are shown in similar orientations. Conserved glycine residues are indicated.
Zinc finger motif
A very strong peak of electron density tetrahedrally liganded by the four conserved cysteine residues (Cys406, Cys409, Cys422 and Cys427) was interpreted as a zinc ion. The presence of zinc ions in stoichiometric amounts (one Zn2+ ion per Tfi ligase molecule) was confirmed by inductively coupled plasma atomic emission spectrometry. Since the four cysteine residues that coordinate the zinc ion are strictly conserved among bacterial DNA ligases, it is expected that other ligases have a similar zinc finger. The overall fold of this zinc finger is similar to other Cys4-type zinc fingers (Figure 3B), including the first of the two zinc fingers in the DNA-binding domain (DBD) of steroid/nuclear hormone receptors such as estrogen receptor (ER) (Schmiedeskamp and Klevit, 1994; Klug and Schwabe, 1995; Mackay and Crossley, 1998). The two zinc-binding motifs of the ER DBD are folded to form a single structural unit in each monomer, and two such monomers form a symmetrical dimer when bound to the cognate DNA. Similar DBDs of receptor proteins can also bind to cognate or non-cognate DNA targets as a monomer (Gewirth and Sigler, 1995; Meinke and Sigler, 1999). In all these cases, the phosphate backbone of DNA interacts with the residues on the β–hairpin and the α–helix of the first zinc finger, with the latter helix sitting in the major groove of double-stranded DNA. Conceivable roles for the Tfi ligase zinc finger motif (subdomain 3a) may include a direct interaction with the nicked DNA as well as a structural support for subdomain 3b and domain 4. This suggestion is largely consistent with the results of mutagenesis of the zinc-coordinating cysteines, which abolished the DNA-binding activity of Thermus thermophilus DNA ligase (Tth ligase) (Luo and Barany, 1996). It is also interesting to note that human DNA ligase III possesses a Cys-Cys/His-Cys-type zinc finger motif that is homologous to the two zinc fingers present in human poly(ADP-ribose) polymerase (PARP) (Taylor et al., 1998; Mackey et al., 1999), the second of which is involved in the specific recognition of a DNA strand break (Gradwohl et al., 1990). It has been demonstrated that the human DNA ligase III zinc finger forms a specific complex with a nick in duplex DNA (Mackey et al., 1999). The possibility that the zinc finger in Tfi ligase may be involved in recognizing the nick in duplex DNA deserves further study.
HhH motif domain
Tfi ligase provides a unique example in which the four clustered HhH motifs form a single compact structure (subdomain 3b). They are helix pairs O–P, R–S, U–V and X–Y with the intervening hairpins (residues 430–460, 474–498, 502–528 and 537–560, respectively) (Figure 1B). Interestingly, all the hairpins are located in a linear chain at the bottom of this subdomain. This surface is also rich in positively charged residues. Similar HhH motifs are present in a number of DNA repair enzymes (Doherty et al., 1996; Aravind et al., 1999), including E.coli endonuclease III (Thayer et al., 1995), E.coli AlkA (Labahn et al., 1996), E.coli MutY (Guan et al., 1998) and human DNA polymerase β (Pol β) (Mullen and Wilson, 1997). A structural comparison is shown in Figure 3C. Compared with the two HhH motifs of Pol β, the second and third HhH motifs of Tfi ligase show r.m.s. deviations of 0.8–1.5 Å for 18 Cα atoms. The first and last HhH motifs are more divergent (1.9–2.7 Å r.m.s. deviations for 18 Cα atoms). The sequence of the third HhH motif of Tfi ligase, Leu-Pro-Gly-Val-Gly-(Xaa)3-Ala, is conserved in endonuclease III, MutY and Pol β. The HhH motif has been implicated in non-sequence-specific DNA binding (Thayer et al., 1995; Doherty et al., 1996). This HhH motif subdomain is suggested to provide one of the two DNA-binding sites in Tfi ligase, as discussed below.
BRCT domain
Our structure of Tfi ligase is the first case in which the BRCT domain is seen as part of a multidomain protein. It consists of a four-stranded parallel β–sheet flanked by three α–helices. Its overall fold is grossly similar to that of the C–terminal BRCT domain of the human DNA repair protein, X–ray cross-complementing group I (XRCC1) (Zhang et al., 1998). At present, a quantitative comparison is not possible, because the coordinates of the XRCC1 BRCT domain are not yet available. The most significant characteristic of the Tfi ligase BRCT domain is its high mobility as a whole. As a consequence, it is completely disordered in the open conformation and exhibits a high average B–factor in the closed conformation. For the ligase molecule in the most closed conformation, the average B–factors and the real space correlation coefficients of domains 1–4 are 56/72/55/84 Å2 and 0.69/0.62/0.67/0.53, respectively, for the main chain atoms only.
The BRCT domain present in NAD+-dependent DNA ligases is a distinct version of its kind and is shared by the large subunits of eukaryotic replication factor C and PARP (Bork et al., 1997). Evolutionarily, it must be the ancestor of eukaryotic BRCT domains. It was suggested that BRCT domains are likely to perform critical functions in the cell cycle control of organisms from bacteria to humans (Bork et al., 1997; Callebaut and Mornon, 1997). They may act as a signal transducer that transmits the signal from DNA damage sensors such as, for example, the central region (301–402) of PARP, to other components of the DNA damage-responsive checkpoint machinery via specific protein–protein interactions (Bork et al., 1997). Mammalian XRCC1, a multidomain protein functioning in the repair of single strand breaks in DNA, forms repair complexes with DNA ligase III, PARP and Pol β. The two BRCT domains of XRCC1 interact with PARP and DNA ligase III, while the N–terminal domain (NTD) of XRCC1 interacts with Pol β (Marintchev et al., 1999). The XRCC1 C–terminal BRCT domain forms a specific heterodimer in vitro with the BRCT domain of mammalian DNA ligase IIIα (Nash et al., 1997). A recent solution structure of XRCC1 NTD showed that it binds a gapped DNA–Pol β complex (Marintchev et al., 1999). All these available data suggest a plausible scenario for Tfi ligase function: after other DNA repair proteins/enzymes recognize and repair the damaged DNA, it is recruited to the nick site for ligation through protein–protein interactions with its BRCT domain. However, the possibility of the BRCT domain being involved in other uncharacterized functions should not be ruled out.
Two distinct DNA-binding sites
The present structure of Tfi ligase in the covalently adenylated state is likely to represent the enzyme conformation that is ready to bind the nicked duplex DNA so that the ligation reaction continues after self-adenylation. The electrostatic potential at the molecular surface and shape complementarity (Figure 4), together with the nucleic acid-binding properties of protein modules related to domains 2 and 3, suggest that two distinct putative DNA-binding sites exist in Tfi ligase. One is a positively charged groove at the interface between domains 1 and 2. This is called the ‘catalytic’ DNA-binding site, because it encompasses the active site. One side of this site is lined by residues from the conserved sequence motif VI (a blue surface around the arrow in Figure 4A). The ‘catalytic’ DNA-binding site of Tfi ligase is not symmetrical about Lys116, with the lower half a little shorter than the upper half (Figure 4A). Therefore, the nicked duplex DNA protected by binding to the ‘catalytic’ DNA-binding site is expected to be asymmetrical in length about the nick. This is supported by the footprinting study on 298 residue Chlorella virus DNA ligase bound at a nick in duplex DNA, which showed that the footprint is asymmetric, extending 8 or 9 nucleotides on the 3′–hydroxyl side of the nick and 11 or 12 nucleotides on the 5′–phosphate side (Odell and Shuman, 1999). This small ligase is roughly equivalent to a more compact version of subdomain 1b and domain 2 of Tfi ligase. The ‘catalytic’ DNA-binding site is connected to the positively charged surface at the tip between strands 12 and 13 of the OB-fold domain (around the conserved Arg331) and continues to the bottom of the HhH motif subdomain 3b (Figure 1B). All four HhH motifs seem to be involved in DNA binding, as suggested by the observation that their hairpins are all located at the bottom of subdomain 3b. We propose that this is the second DNA-binding site; it is called the ‘non-catalytic’ DNA-binding site because it is well separated from the adenylation site.
Fig. 4. Possible interactions between a duplex DNA and Tfi ligase in the observed adenylated structure. (A) Stereo view of the electrostatic potential surface (Nicholls et al., 1991) of Tfi ligase with a duplex DNA interacting with the two putative binding sites. The surface is color-coded according to the potential: red, –15 kT; white, 0 kT; blue, +10 kT. The covalently bound AMP is indicated in a ball-and-stick model. The BRCT domain is in gray because its model lacks side chains. The arrow indicates the highly negatively charged site near Lys116 that is formed by Asp118, Glu281 and Asp283. This view shows the ‘catalytic’ DNA-binding site between domains 1 and 2. (B) This was obtained by rotating the view in (A) by 90° around the vertical axis. In order to show the ‘non-catalytic’ DNA-binding site, the BRCT domain has been omitted from this figure.
Our proposal is largely consistent with the result of a limited proteolysis study on homologous Bst ligase (Timson and Wigley, 1999). It was shown that the DNA-binding activity of the C–terminal fragment of Bst ligase (residues 397–670), corresponding to domains 3 and 4 of Tfi ligase, is comparable to the full-length protein, and the activity of this fragment is independent of the N–terminal fragment (residues 1–318), which is responsible for self-adenylation (Timson and Wigley, 1999). The N–terminal fragment itself, corresponding to domain 1 of Tfi ligase, showed minimal DNA-binding activity. This suggests that the C–terminal domains 3 and 4 play an important role in DNA binding and that domain 1 alone is not sufficient for high-affinity DNA binding. Our structure of Tfi ligase suggests that the inability of the N–terminal fragment of Bst ligase to bind strongly to duplex DNA may be due to the loss of the OB-fold domain 2 that occurred during limited proteolysis, because this domain provides one side of the ‘catalytic’ DNA-binding site. In the case of human DNA ligase III, two functionally distinct DNA-binding regions have been identified (Mackey et al., 1999).
Figure 4 shows one possible mode of DNA binding to Tfi ligase, in which the duplex DNA is bound to both the ‘catalytic’ and ‘non-catalytic’ DNA-binding sites without causing any change in the ligase structure, as modeled using the graphics program O (Jones et al., 1991). In this hypothetical model, DNA is kinked by ∼110° around the β–hairpin of the zinc finger motif, with the kink occurring at the nick site of duplex DNA; the active site lies ∼11 bp away from the kinked site. This implies that a major rearrangement of domains in Tfi ligase is very likely to be required for the nick to be placed near the active site.
A model for the active site
The final step of ligation reaction, deadenylation of the adenylated DNA intermediate and phosphodiester bond formation, is analogous to the polymerizing step by DNA polymerases, for which a divalent metal ion mechanism was proposed (Steitz et al., 1994). The adenylated DNA intermediate in ligation corresponds to deoxyribonucleoside 5′–triphosphate in polymerization. Therefore, it is reasonable to expect a fundamentally related active site configuration of divalent metal ions, their ligands and the adenylated DNA intermediate in DNA ligases. A homologous Thermus DNA ligase demonstrated a divalent metal ion-dependent activity, showing the maximum activity with magnesium or manganese ions and approximately half the maximum activity with calcium ions (Tong et al., 1999). The last step of the ligation reaction by human DNA ligase I was shown to require magnesium ions and it was inhibited by ATP and pyridoxal phosphate, indicating that the availability of the AMP-binding pocket in the enzyme is essential for completion of the reaction (Yang and Chan, 1992).
Some of the highly conserved residues Asp118, Glu169, Glu281, Asp283 and Glu317 are likely to participate in magnesium ion binding. This is supported by the identification of a putative metal ion-binding site by the program package SPASM (Kleywegt, 1999). It was found that the side chain configuration of Asp118, Glu281 and Asp283 in Tfi ligase matches well with that of Asp97, Glu235 and Asp108 in E.coli methionine aminopeptidase, which form the binding site for two cobalt ions. The former three residues form a highly negatively charged pocket near Lys116 (Figure 4A). The strictly conserved Arg196 is likely to interact with the 5′–phosphate end of the nicked strand, while the strictly conserved Phe192, exposed to the solvent, seems to be involved in stacking with DNA bases. A highly schematic model proposed for the Tfi ligase active site is shown in Figure 5. The crystal structure of Chlorella virus mRNA guanylyl transferase in complex with a cap analog revealed that the 5′ RNA base is stacked against the hydrophobic Ile86, the fourth residue from the adenylated Lys82 toward the C–terminus (Håkansson and Wigley, 1998). By analogy, the base at the 5′ end is likely to be in contact with Leu120 in Tfi ligase. High fidelity of homologous Tth ligase against the mismatch at the 3′ side of the nick was found to be influenced by mutation of Lys294 (Luo et al., 1996). This residue corresponds to Lys288 of Tfi ligase, which forms a conserved ion pair with Glu114. These findings are all consistent with the 5′ side of the nick lying above the AMP-linked Lys116 in Figure 1B.
Fig. 5. Schematic model proposed for the Tfi ligase active site. Residues that are likely to participate in binding metal ions and the 5′–phosphate end of the nick are indicated.
Possible conformational changes: a model for the multidomain DNA ligase action
A model for Tfi ligase action involving conformational changes is described in Figure 6. The proposed DNA-binding sites and the toroidal architecture of Tfi ligase suggest that its BRCT domain may act as a gate for DNA binding and release. Analogous opening and closing of the DNA-binding hole was suggested for E.coli DNA topoisomerase I (Lima et al., 1994). This suggests that Tfi ligase may also clamp around and slide along the DNA until it encounters the nick site or interacting partners bound to the damaged site. A precedent for this type of mechanism is provided by mammalian DNA ligase I, which interacts with the sliding clamp, proliferating cell nuclear antigen, through its N–terminal domain (Levin et al., 1997). When the nick is recognized, perhaps by the zinc finger motif, the duplex DNA may be kinked at the nick so that it contacts the ‘catalytic’ DNA-binding site. This may trigger a major rearrangement of domains and in this process the nick would get close enough to the active site for ligation to take place. Modeling studies indicate that such a domain rearrangement is possible by simple concerted hinge motions of domain 2 around Pro314 and of domain 3 around Pro403. This domain movement would allow a tighter binding of duplex DNA to Tfi ligase without imposing substantial bending or kinking on DNA. An alternative model of the duplex DNA remaining unchanged during the domain rearrangement is also feasible. It is interesting to note that the two prolines are located at the interdomain regions and are strongly conserved. It is also worth mentioning that Pro314 is strategically located next to the strictly conserved Lys312, which interacts with the α–phosphate group of the covalently bound AMP (Figure 1C and D). Although no suitable methods are available to observe the proposed conformational changes directly, it may be possible to obtain evidence for this speculative model through structure determination of the DNA complex of Tfi ligase. However, we have not yet been able to grow crystals of the enzyme in complex with DNA.
Fig. 6. Model for Tfi ligase action. Domains are color-coded: domain 1, blue; domain 2, green; domain 3, orange; domain 4, gray. DNA is in red and the bound AMP is in cyan. Apo enzyme (A) is self-adenylated (B) and a duplex DNA is bound to the ‘non-catalytic’ DNA-binding site (C). Tfi ligase slides along DNA and recognizes a nick (D). The duplex DNA is kinked at the nick (D) and the kinked DNA is bound to both the ‘catalytic’ and ‘non-catalytic’ DNA-binding sites, triggering a major domain rearrangement (E). The AMP is de-adenylated from Lys116 and is transferred to the 5′–phosphate of the nicked site, and magnesium ions are bound (E). Nick closure occurs and the ligated duplex DNA is now detached from the ‘catalytic’ DNA-binding site, and the second domain movement restores the ligase conformation (F). The duplex DNA is released to continue another reaction cycle.
The modular architecture of Tfi ligase seems to be highly suited for the proposed large-scale domain movements. The presence of two conformers within the same crystal lattice and the positional variability of the BRCT domain provide additional support for domain motions. Proposed domain rearrangements are also hinted at by the significantly different orientations of the OB-fold domain in Tfi ligase and T7 ligase and the very different orientations of the N–terminal subdomain 1a in Tfi ligase and Bst ligase. There are numerous examples in which a large movement of domains was actually observed (Gerstein et al., 1994). For example, GTP binding by Thermus aquaticus elongation factor EF–Tu leads to dramatic conformational changes that expose the tRNA-binding site (Kjeldgaard et al., 1993). In the case of PcrA DNA helicase from E.coli, large and distinct conformational changes occur on binding DNA and the nucleotide cofactor (Velankar et al., 1999). The E.coli Rep helicase bound to single-stranded DNA undergoes a large reorientation of one of the domains upon binding ADP (Korolev et al., 1997).
Conclusions
The present structure of Tfi ligase reveals an interesting organization of the OB-fold domain (domain 2), the zinc finger motif (subdomain 3a) and the HhH motif domain (subdomain 3b), representing a unique combination and spatial arrangement of these protein modules, which are all known to bind to nucleic acids. These modules, along with the BRCT domain, may have fused with domain 1 during evolution to yield a multidomain protein of highly specific function. As a member of a large superfamily of covalent nucleotidyl transferases that includes mammalian DNA ligases, the structure of Tfi ligase should provide a framework for understanding the domain organization, catalytic function and evolution of this important class of enzymes. It should also serve as a useful model for other enzymes that share the protein modules such as the OB-fold, the zinc finger, the HhH motif and the BRCT domain. For example, human DNA ligase IIIα possesses one BRCT domain at the C–terminus and a zinc finger at the N–terminus, while human DNA ligase IV possesses two BRCT domains at the C–terminus (Chen et al., 1995; Wei et al., 1995; Tomkinson and Levin, 1997; Tomkinson and Mackey, 1998). Terminal nucleotidyl transferase, involved in immunoglobulin gene somatic recombination, also has both a BRCT domain and two HhH motifs at the N–terminus (Bork et al., 1997). Tfi ligase now joins a large group of structures available for DNA polymerases, an ATP-dependent DNA ligase and DNA repair enzymes. This study helps to secure a more comprehensive picture of the machinery for preserving the intact structure of DNA at the atomic level and enhances our understanding of one of the most fundamental processes of biological systems. The proposed model for Tfi ligase action may be generally applicable to eukaryotic multidomain DNA ligases. It would not be surprising if the structural principles related to that observed in this study are found to work in other functionally related enzymes such as the eukaryotic multidomain DNA ligases.
Materials and methods
Purification, crystallization and data collection
Recombinant Tfi ligase was expressed and purified as described previously (Kim and Kwon, 1998). Crystallization of the native enzyme will be described elsewhere (Lee et al., 2000). Neither NAD+ nor zinc ions were added deliberately during purification and crystallization. The reservoir solution for the vapor diffusion crystallization consisted of 100 mM sodium citrate pH 5.6, 5% methoxyPEG 5000 and 5 mM calcium chloride. The native enzyme crystallized into the P21 space group with unit cell parameters of a = 89.51, b = 115.63, c = 97.17 Å and β = 115.73°. The asymmetric unit contains two molecules of monomeric enzyme. SeMet-substituted Tfi ligase was produced in E.coli B834(DE3) cells by growth in minimal media supplemented with SeMet. Its purification procedure was identical to that of the native protein except that buffers contained 5 mM dithiothreitol to prevent selenium oxidation. The crystals of SeMet-substituted enzyme were isomorphous to the native crystals, with slightly different unit cell parameters of a = 89.21, b = 117.33, c = 97.48 Å and β = 115.09°. For the cryo-cooling, a crystal of Tfi DNA ligase was transferred initially to a solution of 100 mM sodium citrate pH 5.6 containing 10% methoxyPEG 5000 and 5 mM calcium chloride. The glycerol concentration was increased stepwise from 0 to 25% (v/v) over a period of 2 h before the crystal was flash-frozen in a cold nitrogen stream at 100 K. Data were collected on beamlines X12–C and X8–C at the National Synchrotron Light Source, Brookhaven National Laboratory. An ADSC Quantum 4R charge-coupled device (CCD) detector or a Brandeis CCD detector was used. Diffraction data were processed and scaled using the HKL software package (Otwinowski and Minor, 1997).
Structure determination and refinement
Twelve of 14 possible selenium sites in the asymmetric unit were located with SOLVE (Terwilliger and Berendzen, 1999). The phases were calculated with SHARP (de la Fortelle and Bricogne, 1997) and were improved by the 2–fold non-crystallographic symmetry (NCS) averaging, solvent flattening and histogram matching with DM (CCP4, 1994). Two zinc sites were also located by SOLVE but were omitted from the phase calculation. The model was built with O (Jones et al., 1991). The protein model was refined with X–PLOR (Brünger, 1992a) and CNS (Brünger et al., 1998), including the bulk solvent correction. The 2–fold NCS was tightly maintained for the SeMet-substituted model, whereas it had to be relaxed for the native model. The model of the SeMet-substituted enzyme accounts for 1162 residues in two molecules of Tfi ligase (residues 1–581) in the crystallographic asymmetric unit, two AMP moieties covalently bound to Lys116, two zinc ions and 242 water molecules. No electron density was observed for the C–terminal residues 582–667. Subsequently, the crystal structure of the native enzyme was also refined. The model of the native enzyme accounts for 1162 residues in two molecules of Tfi ligase (residues 1–581) in the asymmetric unit, two AMP moieties, two zinc ions and 264 water molecules. In addition, it includes a polyalanine model of the C–terminal residues 582–660 in one of the two ligase molecules. The occupancy of all atoms including those in the AMP moieties was assumed to be 1.0. An average B–factor for the AMP moiety in the closed conformation is higher than that for the other AMP in the open conformation (76 versus 45 Å2 for the SeMet-substituted enzyme and 72 versus 41 Å2 for the native enzyme). Coordinates for the SeMet-substituted enzyme and the native enzyme have been deposited in the Protein Data Bank under ID codes 1DGS and 1DGT, respectively.
Acknowledgments
Acknowledgements
We thank the staff at beamlines X12-C and X8-C of NSLS, BNL, USA and at beamlines BL-6A and BL-6B of Photon Factory, Japan. We also thank Drs S.-H.Kim, Y.Cho, S.E.Ryu and S.H.Eom for sharing the X-ray facilities. This work was supported by Korea Research Foundation (Non-directed Research Fund, 1996). S.W.S. is a member of the Center for Molecular Catalysis at Seoul National University.
References
- Aravind L. and Koonin, E.V. (1999) Gleaning non-trivial structural, functional and evolutionary information about proteins by iterative database searches. J. Mol. Biol., 287, 1023–1040. [DOI] [PubMed] [Google Scholar]
- Aravind L., Walker, D.R. and Koonin, E.V. (1999) Conserved domains in DNA repair proteins and evolution of repair systems. Nucleic Acids Res., 27, 1223–1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barany F. (1991) Genetic disease detection and DNA amplification using cloned thermostable ligase. Proc. Natl Acad. Sci. USA, 88, 189–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bochkarev A., Pfuetzner, R.A., Edwards, A.M. and Frappier, L. (1997) Structure of the single-stranded-DNA-binding domain of replication protein A bound to DNA. Nature, 385, 176–181. [DOI] [PubMed] [Google Scholar]
- Bochkarev A., Bochkareva, E., Frappier, L. and Edwards, A.M. (1999) The crystal structure of the complex of replication protein A subunits RPA32 and RPA14 reveals a mechanism for single-stranded DNA binding. EMBO J., 18, 4498–4504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bork P., Hofmann, K., Bucher, P., Neuwald, A.F., Altschul, S.F. and Koonin, E.V. (1997) A superfamily of conserved domains in DNA damage-responsive cell cycle checkpoint proteins. FASEB J., 11, 68–76. [PubMed] [Google Scholar]
- Brünger A.T. (1992a) X-PLOR Version 3.1: A System for X-ray Crystallography & NMR. Yale University Press, New Haven, CT. [Google Scholar]
- Brünger A.T. (1992b) The free R-value: a novel statistical quantity for assessing the accuracy of crystal structures. Nature, 355, 472–474. [DOI] [PubMed] [Google Scholar]
- Brünger A.T., et al. (1998) Crystallography and NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D, 54, 905–921. [DOI] [PubMed] [Google Scholar]
- Bycroft M., Hubbard, T.J.P., Proctor, M., Freund, S.M.V. and Murzin, A.G. (1997) The solution structure of the S1 RNA binding domain: a member of an ancient nucleic acid-binding fold. Cell, 88, 235–242. [DOI] [PubMed] [Google Scholar]
- Callebaut I. and Mornon, J.P. (1997) From BRCA1 to RAP1: a widespread BRCT module closely associated with DNA repair. FEBS Lett., 400, 25–30. [DOI] [PubMed] [Google Scholar]
- CCP4 (1994) Collaborative Computational Project Number 4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D, 50, 760–763. [DOI] [PubMed] [Google Scholar]
- Chen J., Tomkinson, A.E., Ramos, W., Mackey, Z.B., Danehower, S., Walter, C.A., Schultz, R.A., Besterman, J.M. and Husain, I. (1995) Mammalian DNA ligase III: molecular cloning, chromosomal localization, and expression in spermatocytes undergoing meiotic recombination. Mol. Cell. Biol., 15, 5412–5422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Fortelle E. and Bricogne, G. (1997) Maximum-likelihood heavy-atom parameter refinement for multiple isomorphous replacement and multiwavelength anomalous diffraction methods. Methods Enzymol., 276, 472–494. [DOI] [PubMed] [Google Scholar]
- Doherty A.J., Serpell, L.C. and Ponting, C.P. (1996) The helix–hairpin–helix DNA-binding motif: a structural basis for non-sequence-specific recognition of DNA. Nucleic Acids Res., 24, 2488–2497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draper D.E. and Reynaldo, L.P. (1999) RNA binding strategies of ribosomal proteins. Nucleic Acids Res., 27, 381–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerstein M., Lesk, A.M. and Chothia, C. (1994) Structural mechanisms for domain movements in proteins. Biochemistry, 33, 6739–6749. [DOI] [PubMed] [Google Scholar]
- Gewirth D.T. and Sigler, P.B. (1995) The basis for half-site specificity explored through a non-cognate steroid receptor–DNA complex. Nature Struct. Biol., 2, 386–394. [DOI] [PubMed] [Google Scholar]
- Gradwohl G., Menissier de Murcia, J.M., Molinete, M., Simonin, F., Koken, M., Hoeijmakers, J.H. and de Murcia, G. (1990) The second zinc-finger domain of poly(ADP-ribose) polymerase determines specificity for single-stranded breaks in DNA. Proc. Natl Acad. Sci. USA, 87, 2990–2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan Y., Manuel, R.C., Arvai, A.S., Parikh, S.S., Mol, C.D., Miller, J.H., Lloyd, S. and Tainer, J.A. (1998) MutY catalytic core, mutant and bound adenine structures define specificity for DNA repair enzyme superfamily. Nature Struct. Biol., 5, 1058–1064. [DOI] [PubMed] [Google Scholar]
- Håkansson K. and Wigley, D.B. (1998) Structure of a complex between a cap analogue and mRNA guanylyl transferase demonstrates the structural chemistry of RNA capping. Proc. Natl Acad. Sci. USA, 95, 1505–1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Håkansson K., Doherty, A.J., Shuman, S. and Wigley, D.B. (1997) X-ray crystallography reveals a large conformational change during guanyl transfer by mRNA capping enzymes. Cell, 89, 545–553. [DOI] [PubMed] [Google Scholar]
- Horvath M.P., Schweiker, V.L., Bevilacqua, J.M., Ruggles, J.A. and Schultz, S.C. (1998) Crystal structure of the Oxytricha nova telomere end binding protein complexed with single strand DNA. Cell, 95, 963–974. [DOI] [PubMed] [Google Scholar]
- Jaishree T.N., Ramakrishnan, V. and White, S.W. (1996) Solution structure of prokaryotic ribosomal protein S17 by high-resolution NMR spectroscopy. Biochemistry, 35, 2845–2853. [DOI] [PubMed] [Google Scholar]
- Jones T.A., Zou, J.-Y., Cowan, S.W. and Kjeldgaard, M. (1991) Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A, 47, 110–119. [DOI] [PubMed] [Google Scholar]
- Kim H.-K. and Kwon, S.-T. (1998) Cloning, nucleotide sequence, and expression of the DNA ligase-encoding gene from Thermus filiformis. Mol. Cell, 8, 438–443. [PubMed] [Google Scholar]
- Kjeldgaard M., Nissen, P., Thirup, S. and Nyborg, J. (1993) The crystal structure of elongation factor EF-Tu from Thermus aquaticus in the GTP conformation. Structure, 1, 35–50. [DOI] [PubMed] [Google Scholar]
- Kleywegt G.J. (1999) Recognition of spatial motifs in protein structures. J. Mol. Biol., 285, 1887–1897. [DOI] [PubMed] [Google Scholar]
- Klug A. and Schwabe, J.W.R. (1995) Zinc fingers. FASEB J., 9, 597–604. [PubMed] [Google Scholar]
- Korolev S., Hsieh, J., Gauss, G.H., Lohman, T.M. and Waksman, G. (1997) Major domain swiveling revealed by the crystal structures of complexes of E.coli Rep helicase bound to single-stranded DNA and ADP. Cell, 90, 635–647. [DOI] [PubMed] [Google Scholar]
- Kraulis P.J. (1991) MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr., 24, 946–950. [Google Scholar]
- Labahn J., Scharer, O.D., Long, A., Ezaz-Nikpay, K., Verdine, G.L. and Ellenberger, T.E. (1996) Structural basis for the excision repair of alkylation-damaged DNA. Cell, 86, 321–329. [DOI] [PubMed] [Google Scholar]
- Laskowski R.A., MacArthur, M.W., Moss, D.S. and Thornton, J.M. (1993) PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr., 26, 283–291. [Google Scholar]
- Lee J.Y., Kim,H.-K., Chang,C., Eom,S.H., Hwang,K.Y., Cho,Y., Yu,Y.G., Ryu,S.E., Kwon,S.-T. and Suh,S.W. (2000) Crystallization and preliminary X-ray crystallographic analysis of NAD+-dependent DNA ligase from Thermus filiformis. Acta Crystallogr. D, 56, in press. [DOI] [PubMed] [Google Scholar]
- Lehman I.R. (1974) DNA ligase: structure, mechanism, and function. Science, 186, 790–797. [DOI] [PubMed] [Google Scholar]
- Levin D.S., Bai, W., Yao, N., O'Donnell, M. and Tomkinson, A.E. (1997) An interaction between DNA ligase I and proliferating cell nuclear antigen: implications for Okazaki fragment synthesis and joining. Proc. Natl Acad. Sci. USA, 94, 12863–12868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lima C.D., Wang, J.C. and Mondragón, A. (1994) Three-dimensional structure of the 67K N–terminal fragment of E.coli DNA topoisomerase I. Nature, 367, 138–146. [DOI] [PubMed] [Google Scholar]
- Lindahl T. and Barnes, D.E. (1992) Mammalian DNA ligases. Annu. Rev. Biochem., 61, 251–281. [DOI] [PubMed] [Google Scholar]
- Luo J. and Barany, F. (1996) Identification of essential residues in Thermus thermophilus DNA ligase. Nucleic Acids Res., 24, 3079–3085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo J., Bergstrom, D.E. and Barany, F. (1996) Improving the fidelity of Thermus thermophilus DNA ligase. Nucleic Acids Res., 24, 3071–3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay J.P. and Crossley, M. (1998) Zinc fingers are sticking together. Trends Biochem. Sci., 23, 1–4. [DOI] [PubMed] [Google Scholar]
- Mackey Z.B., Niedergang, C., Murcia, J.M., Leppard, J., Au, K., Chen, J., de Murcia, G. and Tomkinson, A.E. (1999) DNA ligase III is recruited to DNA strand breaks by a zinc finger motif homologous to that of poly(ADP-ribose) polymerase. Identification of two functionally distinct DNA binding regions within DNA ligase III. J. Biol. Chem., 274, 21679–21687. [DOI] [PubMed] [Google Scholar]
- Marintchev A., Mullen, M.A., Maciejewski, M.W., Pan, B., Gryk, M.R. and Mullen, G.P. (1999) Solution structure of the single-strand break repair protein XRCC1 N–terminal domain. Nature Struct. Biol., 6, 884–893. [DOI] [PubMed] [Google Scholar]
- Meinke G. and Sigler, P.B. (1999) DNA-binding mechanism of the monomeric orphan nuclear receptor NGFI-B. Nature Struct. Biol., 6, 471–477. [DOI] [PubMed] [Google Scholar]
- Merritt E.A. and Bacon, D.J. (1997) RASTER3D: photorealistic molecular graphics. Methods Enzymol., 277, 505–524. [DOI] [PubMed] [Google Scholar]
- Mullen G.P. and Wilson, S.H. (1997) DNA polymerase β in abasic site repair: a structurally conserved helix–hairpin–helix motif in lesion detection by base excision repair enzymes. Biochemistry, 36, 4713–4717. [DOI] [PubMed] [Google Scholar]
- Murzin A.G. (1993) OB (oligonucleotide/oligosaccharide binding)-fold: common structural and functional solution for non-homologous sequences. EMBO J., 12, 861–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nash R.A., Caldecott, K.W., Barnes, D.E. and Lindahl, T. (1997) XRCC1 protein interacts with one of two distinct forms of DNA ligase III. Biochemistry, 36, 5207–5211. [DOI] [PubMed] [Google Scholar]
- Nicholls A., Sharp, K.A. and Honig, B. (1991) Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins, 11, 281–296. [DOI] [PubMed] [Google Scholar]
- Odell M. and Shuman, S. (1999) Footprinting of Chlorella virus DNA ligase bound at a nick in duplex DNA. J. Biol. Chem., 274, 14032–14039. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z. and Minor, W. (1997) Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol., 276, 307–326. [DOI] [PubMed] [Google Scholar]
- Peat T.S., Newman, J., Waldo, G.S., Berendzen, J. and Terwilliger, T.C. (1998) Structure of translation initiation factor 5A from Pyrobaculum aerophilum at 1.75 Å resolution. Structure, 6, 1207–1214. [DOI] [PubMed] [Google Scholar]
- Raghunathan S., Ricard, C.S., Lohman, T.M. and Waksman, G. (1997) Crystal structure of the homo-tetrameric DNA binding domain of Escherichia coli single-stranded DNA-binding protein determined by multiwavelength X-ray diffraction on the selenomethioninyl protein at 2.9-Å resolution. Proc. Natl Acad. Sci. USA, 94, 6652–6657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roe S.M., Barlow, T., Brown, T., Oram, M., Keeley, A., Tsaneva, I.R. and Pearl, L.H. (1998) Crystal structure of an octameric RuvA–Holliday junction complex. Mol. Cell, 2, 361–372. [DOI] [PubMed] [Google Scholar]
- Schindler T., Perl, D., Graumann, P., Sieber, V., Marahiel, M.A. and Schmid, F.X. (1998) Surface-exposed phenylalanines in the RNP1/RNP2 motif stabilize the cold-shock protein CspB from Bacillus subtilis. Proteins, 30, 401–406. [DOI] [PubMed] [Google Scholar]
- Schmiedeskamp M. and Klevit, R.E. (1994) Zinc finger diversity. Curr. Opin. Struct. Biol., 4, 28–35. [Google Scholar]
- Sette M., van Tilborg, P., Spurio, R., Kaptein, R., Paci, M., Gualerzi, C.O. and Boelens, R. (1997) The structure of the translational initiation factor IF1 from E.coli contains an oligomer-binding motif. EMBO J., 16, 1436–1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuman S. and Schwer, B. (1995) RNA capping enzyme and DNA ligase: a superfamily of covalent nucleotidyl transferases. Mol. Microbiol., 17, 405–410. [DOI] [PubMed] [Google Scholar]
- Singleton M.R., Håkansson, K., Timson, D.J. and Wigley, D.B. (1999) Structure of the adenylation domain of an NAD+-dependent DNA ligase. Structure, 7, 35–42. [DOI] [PubMed] [Google Scholar]
- Steitz T.A., Smerdon, S.J., Jäger, J. and Joyce, C.M. (1994) A unified polymerase mechanism for nonhomologous DNA and RNA polymerases. Science, 266, 2022–2025. [DOI] [PubMed] [Google Scholar]
- Subramanya H.S., Doherty, A.J., Ashford, S.R. and Wigley, D.B. (1996) Crystal structure of an ATP-dependent DNA ligase from bacteriophage T7. Cell, 85, 607–615. [DOI] [PubMed] [Google Scholar]
- Taylor R.M., Whitehouse, J., Cappelli, E., Frosina, G. and Caldecott, K.W. (1998) Role of the DNA ligase III zinc finger in polynucleotide binding and ligation. Nucleic Acids Res., 26, 4804–4810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terwilliger T.C. and Berendzen, J. (1999) Automated MAD and MIR structure solution. Acta Crystallogr. D, 55, 849–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thayer M.M., Ahern, H., Xing, D., Cunningham, R.P. and Tainer, J.A. (1995) Novel DNA binding motifs in the DNA repair enzyme endonuclease III crystal structure. EMBO J., 14, 4108–4120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timson D.J. and Wigley, D.B. (1999) Functional domains of an NAD+-dependent DNA ligase. J. Mol. Biol., 285, 73–83. [DOI] [PubMed] [Google Scholar]
- Tomkinson A.E. and Levin, D.S. (1997) Mammalian DNA ligases. BioEssays, 19, 893–901. [DOI] [PubMed] [Google Scholar]
- Tomkinson A.E. and Mackey, Z.B. (1998) Structure and function of mammalian DNA ligases. Mutat. Res., 407, 1–9. [DOI] [PubMed] [Google Scholar]
- Tong J., Cao, W. and Barany, F. (1999) Biochemical properties of a high fidelity DNA ligase from Thermus species AK16D. Nucleic Acids Res., 27, 788–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velankar S.S., Soultanas, P., Dillingham, M.S., Subramanya, H.S. and Wigley, D.B. (1999) Crystal structures of complexes of PcrA DNA helicase with a DNA substrate indicate an inchworm mechanism. Cell, 97, 75–84. [DOI] [PubMed] [Google Scholar]
- Wallace A.C., Laskwoski, R.A. and Thornton, J.M. (1995) LIGPLOT: a program to generate schematic diagrams of protein–ligand interactions. Protein Eng., 8, 127–134. [DOI] [PubMed] [Google Scholar]
- Wei Y.F., et al. (1995) Molecular cloning and expression of human cDNAs encoding a novel DNA ligase IV and DNA ligase III, an enzyme active in DNA repair and recombination. Mol. Cell. Biol., 15, 3206–3216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S.-W. and Chan, Y.H. (1992) Analysis of the formation of AMP–DNA intermediate and the successive reaction by human DNA ligases I and II. J. Biol. Chem., 267, 8117–8122. [PubMed] [Google Scholar]
- Zhang X., et al. (1998) Structure of an XRCC1 BRCT domain: a new protein–protein interaction module. EMBO J., 17, 6404–6411. [DOI] [PMC free article] [PubMed] [Google Scholar]