Abstract
E26 Transformation specific (Ets) family transcription factors control the expression of a large number of genes regulating hematopoietic cell development and function. Two such transcription factors, Ets-1 and myeloid Elf-1–like factor (MEF), have been shown to play critical roles in both natural killer (NK)– and NKT-cell development, but not in the development of conventional T cells. In this study, we address the role of E74-like factor 1 (Elf-1), another Ets family transcription factor that is closely related to MEF but divergent from Ets-1, in NK- and NKT-cell development using Elf-1–deficient (Elf-1−/−) mice. Whereas the proportion of NK cells in Elf-1−/− mice was normal, the proportion of NKT cells was significantly reduced in the thymus and periphery of Elf-1−/− mice compared with wild-type (WT) mice. Although Ets-1–deficient mice lack NKT cells altogether, Elf-1−/− mice exhibited only a partial block in NKT-cell development caused by a cell-intrinsic defect in the selection, survival, and maturation of NKT cells. In addition, residual NKT cells found in Elf-1−/− mice produced less cytokine upon antigen stimulation compared with WT NKT cells. Our data demonstrate that Elf-1 plays an important and nonredundant role in the development and function of NKT cells, but is not involved in NK-cell development.
Introduction
CD1d-restricted natural killer T (NKT) cells represent a unique lineage of T cells that shares properties with both natural killer (NK) cells and memory T cells. NKT cells rapidly produce an array of cytokines on activation and play critical roles in the regulation of a variety of immune responses, including control of autoimmune diabetes, antitumor immunity, and protection from infectious diseases.1 To date, 2 NKT-cell subsets have been defined. Type I NKT cells, also referred to as invariant NKT (iNKT) cells, express an invariant T-cell receptor α (TCRα) chain (Vα14Jα18 in mice and Vα24-Jα18 in humans) that pairs with a limited repertoire of TCRβ chains (Vβ8, Vβ7, or Vβ2 in mice, and Vβ11 in humans).2 iNKT cells can be identified using CD1d tetramer loaded with the glycosphingolipid antigen α-galactosylceramide (αGalCer).3 Type II NKT cells represent the second subset of NKT cells; they exhibit diverse TCRα and TCRβ chain usage and do not bind to CD1d/αGalCer tetramers.4 This study focuses on iNKT cells, because the various stages of iNKT-cell maturation and differentiation have been clearly defined.
Like conventional T cells, iNKT cells originate from thymic CD4+CD8+ double-positive (DP) progenitors.5 However, the iNKT-cell lineage deviates from conventional T cells at the DP stage, and their positive selection is distinct from that of conventional T cells.6,7 Rare DP-precursor cells that express a rearranged Vα14Jα18 TCRα chain are positively selected by CD1d-expressing DP thymocytes that provide unique costimulatory signals to iNKT-cell precursors through homotypic interactions with signaling lymphocytic activation molecules (SLAM) family receptors. These interactions led to the recruitment of SLAM-associated protein (SAP) and the Src kinase Fyn, as well as downstream activation of nuclear factor-κB (NF-κB).8–12 After positive selection, iNKT-cell precursors down-regulate their expression of CD24 and transition through several maturation stages that can be defined based on the cell-surface expression of CD44 and NK1.1.13 Stage I iNKT cells display an NK1.1−CD44low phenotype and undergo several rounds of cell division. This expansion is accompanied by the up-regulation of CD44 (NK1.1−CD44high, stage II iNKT cells). Some of these NK1.1−CD44high iNKT cells continue to differentiate into mature NK1.1+CD44high (stage III) iNKT cells in the thymus, while others exit the thymus and mature into NK1.1+ iNKT cells in the periphery.1,13,14 iNKT cells can also be subdivided into CD4+ and CD4−CD8− (double-negative [DN]) subsets. The earliest iNKT cells are CD4+, with the DN subset diverging at the immature NK1.1− stage in the thymus.13,14 Recent studies have shown that the transcription factor Th-Pok is required for the repression of CD8 expression and the functional maturation of iNKT cells.15,16
The unique developmental program of iNKT cells is controlled by several transcription factors/molecules that are distinct from those required for the development of conventional T cells.17 For example, the transcription factor PLZF (promyelocytic leukemia zinc finger) has been shown to specifically control the development and function of iNKT cells.18,19 c-Myc also plays a critical role at an early stage of iNKT-cell development, whereas it has little effect on conventional T-cell development.20,21 Furthermore, genes encoding factors that promote the survival of DP thymocytes, such as RORγt, Bcl-xL, and c-Myb, also contribute to iNKT-cell development by extending the DP-thymocyte lifespan to allow for the rearrangement of distal Vα and Jα gene segments, including Vα14-Jα18.22–24 Similar to NK cells, interleukin-15 (IL-15) is required for the maturation and homeostasis of iNKT cells.25 Deficiency in the transcription factor T-bet results in a severe block in iNKT-cell maturation, possibly because of a lack of CD122 expression, which is important for IL-15 signaling.26 In addition to the IL-15–signaling pathway, several other factors affect both NK- and NKT-cell development, highlighting the close developmental relationship between these 2 lineages.27
The Ets family of transcription factors is composed of more than 20 members. Each shares a conserved ETS domain that specifically recognizes DNA sequences containing a GGAA/T core element.28 Several Ets transcription factors are expressed within the hematopoietic system, including PU.1, Spi-B, Ets-1, MEF, and Elf-1.29,30 Gene-targeting experiments have revealed an important role for these transcription factors in the development and maintenance of various hematopoietic lineages.28 Two such Ets family transcription factors, Ets-1 and MEF, have been shown to play a critical role in the development of both NK and NKT cells.31–33 Elf-1, an Ets transcription factor family member that is closely related to MEF, is highly expressed in both embryonic and adult lymphoid tissues, as well as in a variety of endothelial cells.34 Elf-1 has been shown to regulate the function of endothelial cells during the development of tumor angiogenesis.35 In addition, Elf-1 has been implicated in the transcriptional regulation of several T cell–specific genes, including CD4,36 CD3ζ,37 IL-2,38 and LAT.39 However, no major defects in conventional T-cell development have been observed in Elf-1–deficient (Elf-1−/−) mice.40
In this study, we examined the iNKT cells found in Elf-1−/− mice and compared their development with that in Ets-1−/− mice. We found that Elf-1−/− mice exhibited significantly reduced numbers of iNKT cells in the thymus and in the periphery, although this decrease was not as severe as that seen in Ets-1−/− mice. Unlike Ets-1−/− mice, the impairment in iNKT-cell development observed in Elf-1−/− mice was not due to a defective rearrangement of the Vα14-Jα18 TCRα chain preceding positive selection. Instead, Elf-1 deficiency affected the survival and maturation of iNKT cells. Our study highlights the nonredundant roles and distinct molecular mechanisms involved in the regulation of iNKT-cell development by the Ets family of transcription factors.
Methods
Mice
C57BL/6 (B6) and CD45.1 congenic B6 mice were purchased from The Jackson Laboratory. Elf-1−/− and Ets-1−/− mice40 on a mixed B6.129 background were provided by Drs Kevin Barton (Loyola University Medical Center) and Eric Svensson (University of Chicago), respectively, and were backcrossed onto a B6 background (11 generations for Elf-1−/− and 6 generations for Ets-1−/− mice). Vα14Tg and Jα18−/− mice on a B6 background were provided by Drs Albert Bendelac (University of Chicago) and Luc Van Kaer (Vanderbilt University), respectively. All experiments involving animals were performed in a specific pathogen-free facility at Northwestern University in compliance with institutional guidelines, and were approved by the institutional animal care and use committee.
Primary cell preparation, antibodies, and flow cytometry
Fluorescein isothiocyanate (FITC)–conjugated CD1d (5C6) and allophycocyanin (APC)–conjugated CD1d/αGalCer tetramer were generated in-house.3,41 The FITC–annexin V apoptosis detection kit, FITC bromodeoxyuridine (BrdU) flow kit; FITC-conjugated monoclonal antibody (mAb) against CD4, CD24, CD44, CD122, interferon-γ (IFN-γ), and TCRβ; biotin-conjugated anti–CD45.1, phycoerythrin (PE)–conjugated anti–CD8β, CD25, IL-4, and NK1.1; and PerCP-conjugated anti–CD4, NK1.1, and TCRβ were purchased from BD Pharmingen. Thymocytes, splenocytes, and hepatic lymphocytes were isolated and stained as described previously.42,43 Flow cytometry was performed using a FACSCanto II and data analysis was performed using FlowJo Version 8.7 software.
Antigen presentation and cytokine release assay
For activation of iNKT-cell hybridomas, 5 × 104 hybridoma cells were cultured together with 5 × 105 thymocytes in the absence or presence of αGalCer (100 ng/mL). After 24 hours, IL-2 production was quantitated by enzyme-linked immunosorbent assay (ELISA). For activation of primary iNKT cells, splenic dendritic cells (DCs) were purified using CD11c-microbeads (Miltenyi Biotec; > 85% purity). iNKT cells were enriched from the spleens of Vα14Tg mice through depletion of B220+ and MHC class II+ cells using magnetic beads; 1 × 105 enriched iNKT cells were cultured with purified DCs at a 1:1 ratio in the absence or presence of αGalCer for 48 hours. For the activation of whole splenocytes and liver leukocytes, cells were stimulated with αGalCer for 48 hours. The levels of IFN-γ, IL-4, and granulocyte-macrophage colony-stimulating factor (GM-CSF) in supernatants were quantitated by ELISA. ELISA mAb pairs were purchased from BD Pharmingen.
Adaptive transfer and generation of BM chimeras
Donor bone marrow (BM) was depleted of T cells using anti–Thy-1.2 (J1j.10) coupled with rabbit complement (Cedarlane Laboratories). Recipient mice received 980 rad of radiation 4 hours before intravenous injection with 1 × 107 total donor BM cells. Six weeks after adoptive transfer, splenocytes were isolated from recipient mice and analyzed using flow cytometry.
RNA extraction and quantitative real-time PCR
Thymocytes were stained with anti–CD4 and anti–CD8β, and DP thymocytes were purified using a MoFlo cell sorter. Total RNA was extracted from cells using an RNeasy kit (QIAGEN). cDNA was generated using Superscript II reverse transcriptase (Invitrogen), and real-time polymerase chain reaction (PCR) was performed using a MyiQ real-time detection system (BioRad). Each PCR was run in duplicate, and the level of Vα14Jα18 message was normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) using iCycler MyiQ Version 2.0 software. PCR of cDNA was conducted using SYBR Green PCR master mix along with the following primers: Vα14Jα18 forward, 5′-TCTAGAATTCTAAGCACAGCAC GCTG-3′; Vα14Jα18 reverse, 5′-CAATCAGCTGAGTCCCAGCT-3′; GAPDH forward primer, 5′-TTCACCACCATGGAGAAGGC-3′; and GAPDH reverse primer, 5′-GGCATGGACTGTGG TCATGA-3′.
Measuring BrdU incorporation and apoptosis in iNKT cells
WT and Elf-1−/− mice were injected intraperitoneally with 100 μL of BrdU (10 mg/mL) and analyzed 3 days later. Thymocytes were isolated and enriched for iNKT cells using PE-conjugated CD1d/αGalCer tetramers and anti–PE magnetic beads. Enriched tetramer+ cells were stained with APC-CD1d/αGalCer tetramer and mAb against TCRβ, CD44, and NK1.1. After cell-surface staining, cells were analyzed for BrdU incorporation using a BrdU flow kit according to the manufacturer's instructions. To detect apoptotic cells, an annexin V–FITC labeling kit was used according to the manufacturer's instructions.
Intracellular cytokine staining
To determine the capacity of iNKT cells to secrete cytokine, mice were injected with 2 μg of αGalCer. After 1 hour, liver leukocytes were isolated and stained with CD1d/αGalCer tetramer and anti–TCRβ antibody. After fixation with 4% paraformaldehyde and permeabilization with 0.15% saponin, cells were stained with FITC-anti–IFN-γ and PE-anti–IL-4.
Statistical analysis
All mice were examined individually for each experiment, and statistical analyses were performed using a Student t test. All statistical analyses were performed using the Prism Version 4.0 program (GraphPad). A P value less than .05 was considered statistically significant.
Results
The development of NKT cells, but not NK cells, is impaired in Elf-1−/− mice
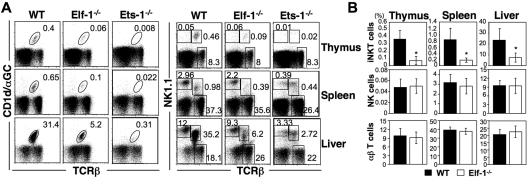
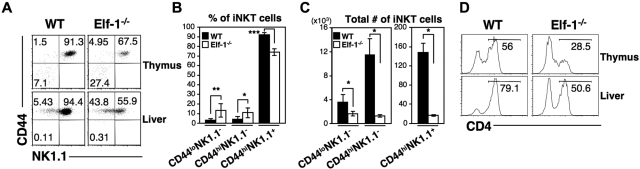
Previous studies have shown that deficiencies in the Ets family transcription factors Ets-1 and MEF result in severe defects in both NKT- and NK-cell development.31–33 Because the DNA-binding domain of Elf-1 shares a high degree of sequence similarity with that of MEF, we hypothesized that Elf-1 and MEF may overlap in function and that Elf-1 could also play a critical role in the development of the hematopoietic lineages. To investigate the role of Elf-1 in the development of iNKT cells, we isolated thymocytes, splenocytes, and liver leukocytes from Elf-1−/− and WT mice and stained for iNKT cells using αGalCer-loaded CD1d tetramer. Because CD1d/αGalCer tetramer was not yet available during our previous investigation of the role of Ets-1 in NKT-cell development,33 we also examined the iNKT-cell population in Ets-1−/− mice for comparison. The frequencies and total numbers of iNKT cells in the thymus, spleen, and liver of Elf-1−/− mice were reduced 5- to 6-fold compared with those of WT controls, although these reductions were not as profound as those observed in Ets-1−/− mice (Figure 1A left panel and 1B). To determine whether Elf-1 is also involved in the development of NK cells, we compared the frequencies of the NK1.1+TCRβ− populations in Elf-1−/− and WT mice. Whereas the proportion of NK1.1+TCRβ− NK cells was significantly reduced in the thymus, spleen, and liver of Ets-1−/− mice, the proportion of NK cells in corresponding tissues of Elf-1−/− mice was comparable with that of WT mice (Figure 1A right panel and B). In addition, the proportion of NK1.1− conventional αβ T cells in Elf-1−/− mice was comparable with that observed in WT mice (Figure 1A right panel and B). These data indicate that Elf-1 is selectively required for the normal development of iNKT cells, but not for that of NK or conventional T cells.
Figure 1.
Elf-1−/− mice display a severe defect in iNKT-cell development. Single-cell suspensions from the thymus, spleen, and liver of WT, Elf-1−/−, and Ets-1−/− mice were stained with CD1d/αGalCer tetramer, anti–TCRβ, and anti–NK1.1, and then analyzed by flow cytometry. (A) Left, numbers represent the percentage of iNKT cells in the indicated organs. Right, numbers represent the percentages of NK, conventional T cells, and total NKT cells in the indicated organs. Results are representative of 3 experiments. (B) Bar graphs depict the means ± SD for the proportion of iNKT, NK, and NK1.1−αβ T cells in the indicated organs of WT and Elf-1−/− mice (n = 15 for each group; *P < .05).
Elf-1 deficiency does not affect CD1d expression or antigen presentation
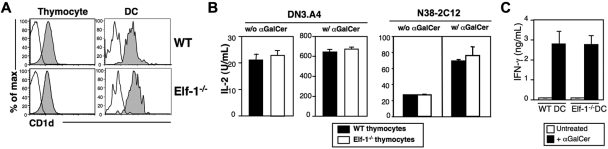
We have previously found that Elf-1 is involved in the transcriptional regulation of CD1d expression on B cells.44 Our finding that iNKT-cell development is severely impaired in Elf-1−/− mice led us to examine whether Elf-1 deficiency also affects CD1d expression on thymocytes and DCs, the 2 respective cell types required for positive and negative selection of iNKT cells. We found that CD1d expression levels on thymocytes and DCs of Elf-1−/− mice were comparable with those observed in WT controls (Figure 2A). Therefore, the effect of Elf-1 on iNKT-cell development is likely independent of its role in the regulation of CD1d transcription. We next investigated whether Elf-1 affects CD1d-mediated lipid antigen presentation by examining the ability of Elf-1−/− thymocytes to stimulate cytokine release from 2 iNKT-cell hybridomas, DN3.A4 and N38-2C12. We found that Elf-1−/− thymocytes were as efficient as WT thymocytes at stimulating IL-2 production by DN3.A4 and N38-2C12 cells in the absence or presence of αGalCer (Figure 2B), indicating that the presentation of both endogenous and exogenous glycolipid antigens is unaffected in Elf-1−/− mice. Moreover, primary iNKT cells isolated from mice transgenic for a functionally rearranged Vα14-Jα18 invariant TCRα chain (Vα14Tg) mice produced similar amounts of IFN-γ in response to αGalCer-pulsed DCs isolated from either Elf-1−/− or WT mice, further supporting the notion that glycolipid antigen presentation is fully functional in the absence of Elf-1 (Figure 2C). These results indicate that the defect in iNKT-cell development observed in Elf-1−/− mice was not due to changes in CD1d expression or antigen presentation.
Figure 2.
CD1d expression and glycolipid antigen presentation in Elf-1−/− mice are normal. (A) Representative histograms of CD1d expression on thymocytes and thymic DCs isolated from WT and Elf-1−/− mice. Cells were stained with either a control mAb (open histogram) or anti–CD1d (filled histogram) and analyzed by flow cytometry. (B) The iNKT-cell hybridomas DN3.A4 and N38-2C12 were cocultured with either WT or Elf-1−/− thymocytes in the absence (left) or presence (right) of αGalCer. After 24 hours, IL-2 levels in the supernatant were detected by ELISA. Error bars represent the SD of triplicate wells. Results are representative of 2 experiments. (C) iNKT cells were enriched from Vα14Tg mice and cocultured with either WT or Elf-1−/− DCs in the absence or presence of αGalCer. IFN-γ levels in the supernatant were detected by ELISA. Error bars represent the SD of triplicate wells. Results are representative of 2 experiments.
Elf-1 deficiency affects iNKT-cell development via a cell-intrinsic mechanism
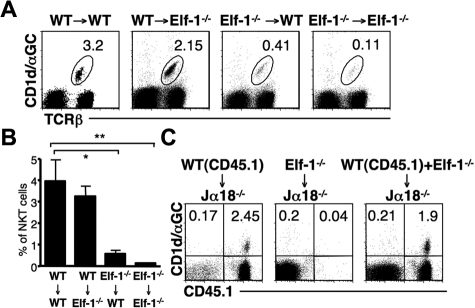
Elf-1 is expressed at high levels in lymphoid tissues, but is also expressed in nonhematopoietic tissues.34 To determine whether the defect in iNKT-cell development observed in Elf-1−/− mice resulted from abnormalities within hematopoietic or nonhematopoietic cells, we generated BM chimeras by transferring either Elf-1−/−-derived or WT-derived BM into lethally irradiated Elf-1−/− or WT recipient mice. The percentage of iNKT cells in Elf-1−/− mice that had received a transfer of WT-derived BM (WT → Elf-1−/−) was comparable with that in control mice (WT → WT), indicating that WT hematopoietic cells alone can restore proper iNKT-cell development in Elf-1−/− mice (Figure 3A-B). In contrast, Elf-1−/− BM cells failed to reconstitute an appreciable proportion of iNKT cells in Elf-1−/− → WT mice, similar to what was observed in control Elf-1−/− → Elf-1−/− mice (Figure 3A-B). These results demonstrate that the expression of Elf-1 in hematopoietic cells, but not in nonhematopoietic cells, is essential for iNKT-cell development.
Figure 3.
Defective development of iNKT cells in Elf-1−/− mice is cell intrinsic. (A-B) BM cells from WT and Elf-1−/− mice were transferred into irradiated WT or Elf-1−/− recipients. After 6 weeks, cells were harvested from recipient mice, stained with CD1d/αGalCer tetramer and anti–TCRβ, and analyzed by flow cytometry. (A) Numbers indicate the percentage of tetramer+TCRβ+ in the spleen of indicated recipients. Data shown are representative of 3 independent experiments using a minimum of 2 recipients per donor genotype. (B) Bar graphs depict the means ± SD for the proportion of iNKT cells in the spleen of indicated recipients (n = 9 for each group; *P < .05; **P < .01). (C) BM cells from Elf-1−/− or CD45.1+ WT congenic mice or mixed BM (1:1) from Elf-1−/− and WT congenic mice were transferred into irradiated Jα18−/− recipients. After 6 weeks, cells were harvested from recipient mice, stained with CD1d/αGalCer tetramer and anti–CD45.1, and analyzed by flow cytometry. Numbers in each quadrant indicate the percentage of tetramer+CD45.1+ cells in the spleen of indicated recipients. Data are representative of 3 independent experiments using a minimum of 2 recipients per donor genotype.
To further evaluate whether Elf-1 deficiency affects iNKT-cell development via a stem cell–intrinsic mechanism or via defective environmental cues, we adoptively transferred BM mixtures into irradiated Jα18−/− recipients that lack the TCRα chain required for iNKT-cell development. Jα18−/− mice received BM from Elf-1−/− mice (CD45.2), BM from CD45.1 congenic B6 (congenic WT) mice, or a mixture of BM from Elf-1−/− and congenic WT mice. As expected, the use of BM from congenic WT mice as donor cells was sufficient to reconstitute the iNKT population in Jα18−/− mice, but the use of Elf-1−/− BM cells alone was not (Figure 3C). Interestingly, the iNKT cells present in the Jα18−/− mice that received a mixture of Elf-1−/− and congenic WT mice BM cells all expressed CD45.1, indicating that they were derived solely from WT BM donor cells (Figure 3C). These data demonstrate that WT BM–derived cells are unable to rescue the development of iNKT cells that derive from Elf-1−/− BM. Thus, it is likely that the defective development observed in Elf-1−/− mice stems from a cell-autonomous defect in iNKT cells or iNKT-cell precursors.
Elf-1 is required for the proper positive selection of NKT cells
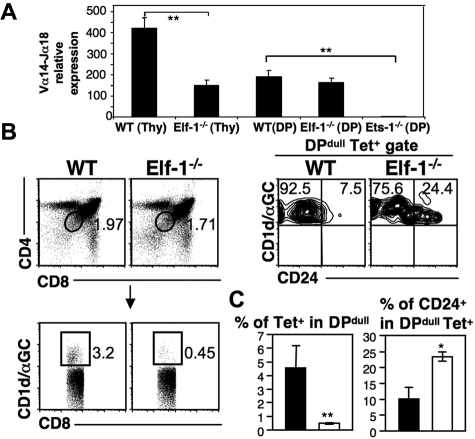
Because conventional T cells undergo normal development in Elf-1−/− mice, Elf-1 likely affects iNKT-cell development during or after the point at which DP thymocytes commit to the iNKT lineage. To investigate whether Elf-1 deficiency affects the rearrangement of the Vα14 gene segment to the Jα18 gene segment before the occurrence of positive selection, we compared the expression of Vα14Jα18 transcript in whole thymocytes and in sorted DP thymocytes isolated from WT and Elf-1−/− mice. Levels of Vα14Jα18 transcript were reduced in whole thymocytes isolated from Elf-1−/− mice compared with WT mice, which is reflective of the reduced proportion of iNKT cells found in Elf-1−/− mice. However, the levels of Vα14Jα18 transcript found in sorted DP thymocytes isolated from WT and Elf-1−/− mice were similar (Figure 4A). Despite normal levels of Vα14Jα18 rearrangement in DP thymocytes, the proportion of iNKT cells was significantly reduced in Elf-1−/− mice, suggesting that Elf-1 is likely involved in the selection and/or continued maturation of iNKT cells. In contrast, DP thymocytes isolated from Ets-1−/− mice expressed substantially less Vα14Jα18 transcript compared with WT mice (Figure 4A), indicating that Ets-1 is required for the efficient rearrangement of Vα14 with Jα18 and can therefore affect iNKT-cell development before lineage specification.
Figure 4.
Elf-1 affects the thymic selection of iNKT cells. (A) Bar graphs depict the relative expression levels of Vα14Jα18 transcripts within whole thymocytes or within sorted DP thymocytes isolated from WT, Elf-1−/−, and Ets-1−/− mice as determined by real-time PCR. Results are representative of 3 individual experiments (**P < .01). (B) Left, dot plots exhibit the gating strategy for tetramer+DPdull NKT cells. Cells were first gated based on CD4 and CD8 expression (top left), and next gated based on tetramer binding (bottom left). Right, contour plots show the proportion of CD24+ cells within the tetramer+ DPdull population. Data shown are representative of 2 independent experiments. (C) Bar graphs depict the means ± SD for the proportions of tetramer+ cells within the DPdull population (left) and of CD24+ cells within the tetramer+DPdull population (right; n = 4 for each group; *P < .05; **P < .01).
Once rearrangement of the invariant Vα14-Jα18 TCRα chain is complete, iNKT-cell precursors undergo positive selection and acquire the unique features of iNKT cells. To investigate whether Elf-1 affects the thymic selection of iNKT cells, we compared the CD1d/αGalCer tetramer+ CD4dullCD8dull (DPdull) population between Elf-1−/− and WT mice. This population represents the earliest iNKT precursors to survive positive selection.45 We found that the percentage of tetramer+ cells within the DPdull population was significantly reduced in the thymus of Elf-1−/− mice compared with WT mice (Figure 4B-C left panels). In addition, we detected a significantly higher frequency of CD24hitetramer+DPdull cells in Elf-1−/− thymus compared with WT thymus (Figure 4B-C right panels). Because CD24 is normally down-regulated after positive selection, these results suggest that Elf-1 plays an important role as a positive regulator during the positive selection of iNKT cells.
Elf-1 affects the maturation of iNKT cells and the differentiation of iNKT-cell subsets
Recent studies have suggested that iNKT cells undergo a series of maturation stages in the thymus immediately after positive selection.13,14 These stages can be identified as CD44loNK1.1− (stage I), CD44hiNK1.1− (stage II), and the terminally differentiated CD44hiNK1.1+ (stage III) iNKT cells. To determine whether Elf-1 is required for the progress of iNKT cells through this maturation process, we determined the surface expression of CD44 and NK1.1 on the iNKT cells found in Elf-1−/− mice. We found that the proportions of stage I CD44lowNK1.1− and stage II CD44highNK1.1− iNKT cells were significantly increased in the thymus and liver of Elf-1−/− mice compared with WT mice, whereas the proportions of mature stage III CD44highNK1.1+ iNKT cells were reduced (Figure 5A-B). However, because Elf-1−/− mice have fewer total numbers of iNKT cells than WT mice, the absolute number of iNKT cells at each of these 3 maturation stages was significantly lower in the thymus of Elf-1−/− mice (Figure 5C). These data suggest that Elf-1 deficiency results in a partial developmental block of iNKT cells.
Figure 5.
Elf-1 deficiency impairs proper iNKT-cell maturation and differentiation. (A-C) Thymocytes and hepatic leukocytes from WT and Elf-1−/− mice were stained with CD1d/αGalCer tetramer and mAb against TCRβ, CD44, and NK1.1, and then analyzed using flow cytometry. (A) Fluorescence-activated cell sorting plots show the CD44 and NK1.1 expression profile within the tetramer+TCRβ+ population. Data are representative of 3 independent experiments. Bar graphs depict means ± SD for the proportions (B) and absolute numbers (C) of tetramer+TCRβ+ iNKT-cell precursors in the thymus based on CD44 and NK1.1 expression (n = 6 for each group; *P < .05; ***P < .001). (D) Thymocytes and liver leukocytes from WT and Elf-1−/− mice were stained with CD1d/αGalCer tetramer and anti–CD4, and then analyzed by flow cytometry. Histograms show the proportion of CD4+ NKT cells within the tetramer+TCRβ+ population for the indicated organs. Data are representative of 3 independent experiments.
The murine iNKT-cell population consists of both CD4+ and CD4− cells. To investigate whether Elf-1 deficiency affects the development of any particular iNKT-cell subset, we determined the proportions of CD4+ and CD4− iNKT cells in the thymus and liver of Elf-1−/− mice. We found that the proportion of CD4+ iNKT cells in both the thymus and the liver of Elf-1−/− mice was reduced compared with that of WT mice (Figure 5D), suggesting that Elf-1 may either contribute to the regulation of CD4 expression during iNKT-cell development or preferentially affect the development/maintenance of the CD4+ iNKT-cell subset.
Elf-1 contributes to NKT-cell survival during early development
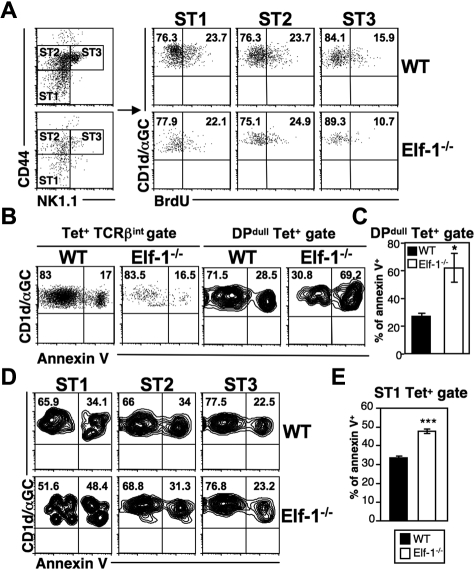
The decreases in iNKT-cell numbers observed in Elf-1−/− mice may be due to a reduction in proliferation or to an increased rate of apoptosis. To examine the former possibility, we injected Elf-1−/− mice and WT controls with BrdU and determined the rate of iNKT-cell proliferation in the thymus by analyzing BrdU incorporation at different maturation stages. In WT mice, early immature NK1.1− iNKT cells expanded more extensively than mature NK1.1+ iNKT cells. We found that stage I (CD44lowNK1.1−) and stage II (CD44highNK1.1−) iNKT cells in the thymus of Elf-1−/− mice also exhibited more proliferation than stage III (CD44highNK1.1+) iNKT cells, and that the percentages of BrdU+ iNKT cells at all of these stages were similar to those observed in WT counterparts (Figure 6A). These data suggest that the reduced proportion of iNKT cells seen in Elf-1−/− mice is not due to inefficient proliferation. To investigate whether increased cell death contributes to this reduction, we examined the level of apoptotic Elf-1−/− iNKT cells at different maturation stages using annexin V staining. The overall proportion of annexin V+ iNKT cells in the thymus of Elf-1−/− mice based on the tetramer+TCRβ+ population was shown to be similar to that seen in WT mice (Figure 6B left panel). However, the fraction of annexin V+ iNKT cells within the early-stage tetramer+DPdull population was significantly greater in the thymus of Elf-1−/− mice compared with WT mice (Figure 6B right panel and 6C). Furthermore, the increased frequency of cell death in early-stage iNKT cells of Elf-1−/− mice was observed through the initial maturation stage I (CD44−NK1.1−), whereas the intermediate stage II and mature stage III iNKT cells in Elf-1−/− mice displayed a similar proportion of annexin V staining to those observed in WT mice (Figure 6D-E). Our results suggest that Elf-1 contributes to iNKT-precursor–cell survival at the earliest stage of maturation, but is not required for the expansion of this population.
Figure 6.
NKT cells in Elf-1−/− mice display normal proliferation but increased cell death during early development. (A) WT and Elf-1−/− mice were injected with BrdU and analyzed 3 days later. iNKT cell–enriched thymocytes were stained with CD1d/αGalCer tetramer and mAb against TCRβ, CD44, and NK1.1, and then analyzed by flow cytometry. Tetramer+TCRβ+ cells were further gated to distinguish stage I (CD44lowNK1.1−, ST1), stage II (CD44highNK1.1−, ST2), and stage III (CD44highNK1.1+, ST3) NKT cells (left). BrdU incorporation by iNKT cells at each of these stages is shown in the right panel. Data shown are representative of 2 independent experiments. (B-E) Thymocytes were isolated from WT and Elf-1−/− mice, stained with CD1d-αGalCer tetramer and mAb against various cell surface markers including annexin V, and then analyzed by flow cytometry. (B) Percentages of annexin V+ cells within the tetramer+TCRβ+ (left) and tetramer+DPdull (right) gates. Data shown are representative of 3 independent experiments. (C) Bar graphs depict means ± SD for the proportion of annexin V+ cells within the tetramer+DPdull population (n = 4 for each group; *P < .05). (D) The proportions of annexin V+ cells within each iNKT-cell stage. Data shown are representative of 3 independent experiments. (E) Bar graphs depict the means ± SD for the proportion of annexin V+ cells within the stage I gate (n = 4 for each group; ***P < .001).
Residual NKT cells found in Elf-1−/− mice are deficient in cytokine production
Although Elf-1−/− mice exhibit reduced proportions of iNKT cells, a significant iNKT-cell population remains. To determine whether the function of these residual iNKT cells is affected by Elf-1 deficiency, we stimulated splenocytes and liver leukocytes isolated from Elf-1−/− and WT mice with αGalCer and measured the amount of cytokines in the culture supernatant. We found that the splenocytes and liver leukocytes from Elf-1−/− mice produced substantially reduced amounts of IFN-γ, IL-4, and GM-CSF compared with those cells isolated from WT controls (Figure 7A and supplemental Figure 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). Whereas this reduction in cytokine production could be explained by the reduced proportion of iNKT cells observed in Elf-1−/− mice, it could also be due to an impaired function of the residual iNKT cells. To distinguish between these 2 possibilities, we injected WT and Elf-1−/− mice with αGalCer and evaluated IFN-γ and IL-4 production on a per-cell basis using intracellular cytokine staining. After 1 hour of in vivo αGalCer stimulation, a substantial proportion of the iNKT cells from WT mice produced IFN-γ and IL-4 ex vivo, whereas the iNKT cells from Elf-1−/− mice exhibited a profound reduction in the production of both cytokines (Figure 7B). A reduction in IFN-γ production was also observed for iNKT cells found in Elf-1−/− mice after in vitro phorbol 12-myristate 13-acetate/ionomycin stimulation, whereas no such reduction was observed for NK cells or conventional T cells found in Elf-1−/− mice (supplemental Figure 2A), nor did the NK cells found in Elf-1−/− mice exhibit any cytolytic defect (supplemental Figure 2B). These results indicate that the function of residual NKT cells, but not that of NK cells or conventional T cells, is impaired in Elf-1−/− mice.
Figure 7.
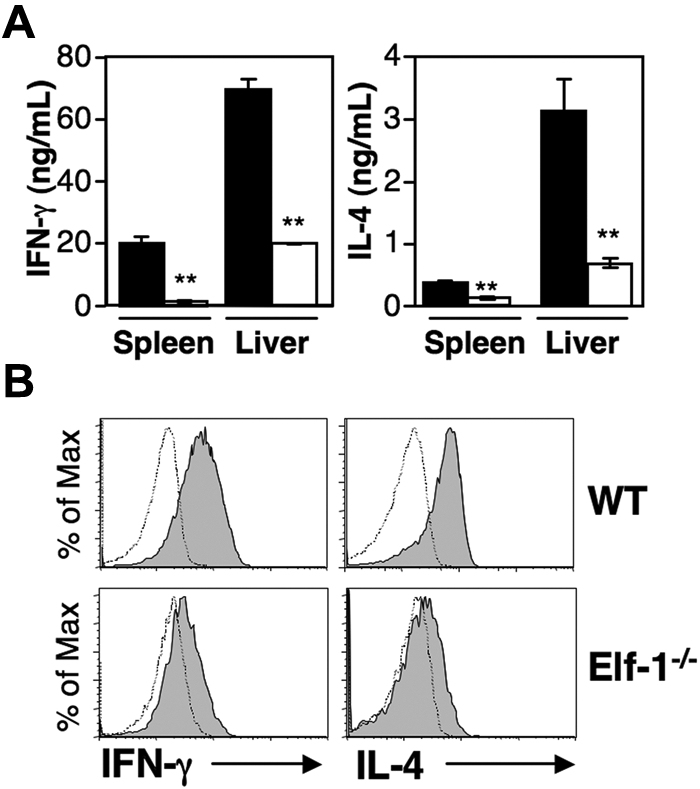
Residual NKT cells in Elf-1−/− mice are dysfunctional. (A) Splenocytes and liver leukocytes from WT and Elf-1−/− mice were stimulated with αGalCer. After 48 hours, cytokine levels in the supernatant were detected by ELISA. Error bars represent the SD of triplicate wells. Data shown are representative of 4 independent experiments (**P < .01). (B) WT and Elf-1−/− mice were injected with αGalCer. After 1 hour, liver leukocytes were harvested, stained with CD1d/αGalCer tetramer and anti–TCRβ, stained intracellularly with mAb against IFN-γ and IL-4, and then analyzed by flow cytometry. Filled histograms represent cytokine-producing iNKT cells based on tetramer+TCRβ+ gating. Control staining is represented as open histograms. Data shown are representative of 3 independent experiments.
Discussion
In this report, we demonstrate that the Ets transcription factor Elf-1 is required for the proper development and function of iNKT cells, but is not required for the development of NK cells or conventional T cells in mice. The differential requirement for Elf-1 in the development of iNKT and NK cells is distinct from that of 2 previously described Ets transcription factors, Ets-1 and MEF.31–33 Moreover, the defect in iNKT-cell development observed in Elf-1−/− mice is different from that observed in Ets-1−/− mice, because a small population of residual iNKT cells that are abnormal in phenotype and function is present in Elf-1−/− mice, while iNKT cells are almost undetectable in Ets-1−/− mice. These findings suggest that different Ets transcription factors may affect iNKT cells at different stages of development. Indeed, we found that the rearrangement of the Vα14 gene segment with Jα18 in DP thymocytes is impaired in Ets-1−/− mice, but is not affected in Elf-1−/− mice (Figure 4A), indicating that Ets-1 is required at an earlier stage of iNKT-cell development than Elf-1. It is noteworthy that the decreased proportion of tetramer+TCRβ+ iNKT cells observed in Elf-1−/− mice could account for most of the decrease in the proportion of total NK1.1+TCRβ+ cells (Figure 1A right panel). This suggests that Elf-1-deficiency has a greater effect on the development of iNKT cells than that of other NKT-cell subsets. This preferential effect on iNKT-cell development is even more profound in the absence of Ets-1, because the proportion of residual NK1.1+TCRβ+ NKT cells found in Ets-1−/− mice is far greater than the proportion of iNKT cells.
We have previously shown that Elf-1 binds to the CD1d promoter and that Elf-1 deficiency results in decreased CD1d expression on B cells.44 However, we found that the levels of CD1d expression on thymocytes and DCs were normal in Elf-1−/− mice. This phenomenon could be explained by the presence of other Ets transcription factors that may compensate for the absence of Elf-1 and regulate CD1d expression specifically in these cell types. Elf-1–deficient thymocytes not only express levels of CD1d similar to that of WT controls, they also efficiently present endogenous and exogenous glycolipid antigens to iNKT cells, suggesting that the impaired iNKT-cell development observed in Elf-1−/− mice is not due to altered antigen presentation. Consistent with this notion, we found the defect in iNKT development seen in Elf-1−/− mice to be cell autonomous, suggesting that Elf-1 signaling within iNKT cells or iNKT-cell precursors is required for their proper selection and maturation.
Our analysis of various developmental stages of thymic iNKT cells revealed that Elf-1−/− mice have fewer CD1d tetramer+ DPdull iNKT-cell precursors, and that a significant fraction of these cells remain CD24hi. Because CD24 is normally down-regulated on positive selection, these results indicate a defect in the positive selection of iNKT cells in Elf-1−/− mice. In addition, the percentages of immature iNKT cells (stage I CD44lowNK1.1− and stage II CD44hiNK1.1−) were significantly increased in Elf-1−/− mice compared with those of WT controls, suggesting that Elf-1 deficiency affects iNKT-cell maturation. Although Elf-1 deficiency does not affect the proliferative capacity of iNKT cells at any maturation stage, it results in an increased susceptibility of immature iNKT-cell populations in the thymus to apoptosis, including tetramer+DPdull cells and stage I CD44lowNK1.1− iNKT cells. Our data suggest that Elf-1 contributes to iNKT-cell development by promoting positive selection and by regulating the survival and further differentiation of immature iNKT cells.
Elf-1 has been shown to regulate the expression of many genes expressed in hematopoietic cells.36–39 One potential target of Elf-1 identified in vitro is CD4.36 Elf-1 specifically binds to the Ets-binding site found in the CD4 promoter and activates its expression. Indeed, the markedly reduced proportion of CD4+ iNKT cells observed in Elf-1−/− mice suggests that Elf-1 may directly influence the proportions of the CD4-expressing NKT-cell subsets. Nevertheless, Elf-1 deficiency has no significant impact on CD4 expression on conventional T cells, suggesting that other Ets transcription factors play a redundant role in the promotion of CD4 expression on conventional T cells. Although Elf-1 has been associated with IL-2 responsiveness,38 the levels of IL-2Rα (CD25) and IL-2Rβ (CD122) expression on the residual iNKT cells found in Elf-1−/− mice were comparable with those observed in WT mice (supplemental Figure 3). Multiple Ets transcription factors are reported to be expressed in DP and SP thymocytes, including Ets1 and Elf-1, which are both present throughout T-cell development without marked preference for any specific T-cell subset.46 Despite this potential for functional redundancy, Elf-1 deficiency selectively affects iNKT cells but not other hematopoietic cell types, suggesting that Elf-1 may have unique transcriptional targets within the iNKT-cell lineage. Elf-1, like all other Ets family members, contains domains for DNA binding, transcriptional activation, and, importantly, domains that mediate protein-protein interactions. It is possible that Elf-1 exhibits a specific effect on iNKT-cell development through preferential interaction with a molecular partner(s) required to regulate unique targets essential for iNKT-cell development and function.
Several transcription factors and signaling molecules known to regulate the development of iNKT cells in a cell-intrinsic manner have been described.17 To determine whether any of these genes could be downstream targets of Elf-1, we performed semiquantitative reverse-transcriptase PCR to compare their expression in Elf-1−/− and WT mice. However, we did not detect significant differences in the expression of any of the genes examined thus far, including those for transcription factors (Ets-1, Gata3, Id-2, MEF, NF-κB, PLZF, ROR-γt, RunX1, and T-bet) and signaling molecules (Fyn, Itk, Lck, and SAP) (data not shown). Elf-1 therefore appears to contribute to the iNKT-cell developmental pathway independently of these molecules. However, we cannot eliminate the possibility that Elf-1 may work in concert with some of these transcription factors as part of a multimolecule complex that can regulate the expression of other genes involved in iNKT-cell development.
In conclusion, the data presented here provide decisive evidence that Elf-1 acts as an important regulator in the ontogeny and function of iNKT cells. This regulation appears to be distinct from that of other transcription factors known to be involved in iNKT-cell development. Future studies focusing on the signaling events downstream of Elf-1 would provide new insight into the molecular pathways that regulate the development of this unique T-lymphocyte lineage. Several Ets transcription factors, including Ets-1 and Elf-1, have been shown to be involved in tumor angiogenesis and to act as regulators of pro-angiogenic gene expression and of the expression of genes involved in vascular development.47 Targeting Ets transcription factors in mouse tumor models through the use of a dominant-negative form of Ets-1 or membrane-permeable Elf-1–blocking peptides has now demonstrated the therapeutic potential of inhibiting selected Ets transcription factors to limit tumor growth and tumor angiogenesis.35,48 However, the results of our study suggest exercising caution regarding the potential negative impact of Ets transcription factor manipulation on antitumor immunity.
Supplementary Material
Acknowledgments
We thank Dr Kevin Barton for providing Elf-1−/− mice; Dr Eric Svensson for Ets-1−/− mice; Dr Albert Bendelac for Vα14Tg mice; Dr Luc Van Kaer for Jα18−/− mice; Kirin Brewery for α-GalCer; the National Institutes of Health Tetramer Core Facility for CD1d tetramers; the Northwestern University Flow Cytometry Core Facility for cell-sorting services; Jessica Rojas, Sharmila Shanmuganad, Stephen Wood, and Chunting Yang for technical assistance; and Yao Bian for critical review of the manuscript.
This work is supported by National Institutes of Health grant R01 AI43407 to C.-R.W.
Footnotes
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contributions: H.-J.C., Y.G., H.C., S.L., and P.K.G. performed the experiments; C.-R.W. and H.-J.C. designed the experiments and analyzed the data; and H.-J.C., C.-R.W., and K.F. wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Dr Chyung-Ru Wang, Department of Micro-biology and Immunology, Northwestern University, 320 E Superior St, Searle 3-401, Chicago, IL 60611; e-mail: chyung-ru-wang@northwestern.edu.
References
- 1.Bendelac A, Savage PB, Teyton L. The biology of NKT cells. Annu Rev Immunol. 2007;25:297–336. doi: 10.1146/annurev.immunol.25.022106.141711. [DOI] [PubMed] [Google Scholar]
- 2.Godfrey DI, MacDonald HR, Kronenberg M, Smyth MJ, Van Kaer L. NKT cells: what's in a name? Nat Rev Immunol. 2004;4(3):231–237. doi: 10.1038/nri1309. [DOI] [PubMed] [Google Scholar]
- 3.Matsuda JL, Naidenko OV, Gapin L, et al. Tracking the response of natural killer T cells to a glycolipid antigen using CD1d tetramers. J Exp Med. 2000;192(5):741–754. doi: 10.1084/jem.192.5.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Behar SM, Cardell S. Diverse CD1d-restricted T cells: diverse phenotypes, and diverse functions. Semin Immunol. 2000;12(6):551–560. doi: 10.1006/smim.2000.0273. [DOI] [PubMed] [Google Scholar]
- 5.Gapin L, Matsuda JL, Surh CD, Kronenberg M. NKT cells derive from double-positive thymocytes that are positively selected by CD1d. Nat Immunol. 2001;2(10):971–978. doi: 10.1038/ni710. [DOI] [PubMed] [Google Scholar]
- 6.Bendelac A. Positive selection of mouse NK1+ T cells by CD1-expressing cortical thymocytes. J Exp Med. 1995;182(6):2091–2096. doi: 10.1084/jem.182.6.2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coles MC, Raulet DH. NK1.1+ T cells in the liver arise in the thymus and are selected by interactions with class I molecules on CD4+CD8+ cells. J Immunol. 2000;164(5):2412–2418. doi: 10.4049/jimmunol.164.5.2412. [DOI] [PubMed] [Google Scholar]
- 8.Stanic AK, Bezbradica JS, Park JJ, et al. NF-kappa B controls cell fate specification, survival, and molecular differentiation of immunoregulatory natural T lymphocytes. J Immunol. 2004;172(4):2265–2273. doi: 10.4049/jimmunol.172.4.2265. [DOI] [PubMed] [Google Scholar]
- 9.Nichols KE, Hom J, Gong SY, et al. Regulation of NKT cell development by SAP, the protein defective in XLP. Nat Med. 2005;11(3):340–345. doi: 10.1038/nm1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gadue P, Morton N, Stein PL. The Src family tyrosine kinase Fyn regulates natural killer T cell development. J Exp Med. 1999;190(8):1189–1196. doi: 10.1084/jem.190.8.1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eberl G, Lowin-Kropf B, MacDonald HR. Cutting edge: NKT cell development is selectively impaired in Fyn-deficient mice. J Immunol. 1999;163(8):4091–4094. [PubMed] [Google Scholar]
- 12.Chung B, Aoukaty A, Dutz J, Terhorst C, Tan R. Signaling lymphocytic activation molecule-associated protein controls NKT cell functions. J Immunol. 2005;174(6):3153–3157. doi: 10.4049/jimmunol.174.6.3153. [DOI] [PubMed] [Google Scholar]
- 13.Benlagha K, Kyin T, Beavis A, Teyton L, Bendelac A. A thymic precursor to the NK T cell lineage. Science. 2002;296(5567):553–555. doi: 10.1126/science.1069017. [DOI] [PubMed] [Google Scholar]
- 14.Pellicci DG, Hammond KJ, Uldrich AP, Baxter AG, Smyth MJ, Godfrey DI. A natural killer T (NKT) cell developmental pathway iInvolving a thymus-dependent NK1.1(-)CD4(+) CD1d-dependent precursor stage. J Exp Med. 2002;195(7):835–844. doi: 10.1084/jem.20011544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Engel I, Hammond K, Sullivan BA, et al. Co-receptor choice by V alpha14i NKT cells is driven by Th-POK expression rather than avoidance of CD8-mediated negative selection. J Exp Med. 2010;207(5):1015–1029. doi: 10.1084/jem.20090557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang L, Carr T, Xiong Y, et al. The sequential activity of Gata3 and Thpok is required for the differentiation of CD1d-restricted CD4+ NKT cells. Eur J Immunol. 2010;40(9):2385–2390. doi: 10.1002/eji.201040534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Godfrey DI, Stankovic S, Baxter AG. Raising the NKT cell family. Nat Immunol. 2010;11(3):197–206. doi: 10.1038/ni.1841. [DOI] [PubMed] [Google Scholar]
- 18.Savage AK, Constantinides MG, Han J, et al. The transcription factor PLZF directs the effector program of the NKT cell lineage. Immunity. 2008;29(3):391–403. doi: 10.1016/j.immuni.2008.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kovalovsky D, Uche OU, Eladad S, et al. The BTB-zinc finger transcriptional regulator PLZF controls the development of invariant natural killer T cell effector functions. Nat Immunol. 2008;9(9):1055–1064. doi: 10.1038/ni.1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mycko MP, Ferrero I, Wilson A, et al. Selective requirement for c-Myc at an early stage of V(alpha)14i NKT cell development. J Immunol. 2009;182(8):4641–4648. doi: 10.4049/jimmunol.0803394. [DOI] [PubMed] [Google Scholar]
- 21.Dose M, Sleckman BP, Han J, Bredemeyer AL, Bendelac A, Gounari F. Intrathymic proliferation wave essential for Valpha14+ natural killer T cell development depends on c-Myc. Proc Natl Acad Sci U S A. 2009;106(21):8641–8646. doi: 10.1073/pnas.0812255106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bezbradica JS, Hill T, Stanic AK, Van Kaer L, Joyce S. Commitment toward the natural T (iNKT) cell lineage occurs at the CD4+8+ stage of thymic ontogeny. Proc Natl Acad Sci U S A. 2005;102(14):5114–5119. doi: 10.1073/pnas.0408449102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Egawa T, Eberl G, Taniuchi I, et al. Genetic evidence supporting selection of the Valpha14i NKT cell lineage from double-positive thymocyte precursors. Immunity. 2005;22(6):705–716. doi: 10.1016/j.immuni.2005.03.011. [DOI] [PubMed] [Google Scholar]
- 24.Hu T, Simmons A, Yuan J, Bender TP, Alberola-Ila J. The transcription factor c-Myb primes CD4+CD8+ immature thymocytes for selection into the iNKT lineage. Nat Immunol. 2010;11(5):435–441. doi: 10.1038/ni.1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Matsuda JL, Gapin L, Sidobre S, et al. Homeostasis of V alpha 14i NKT cells. Nat Immunol. 2002;3(10):966–974. doi: 10.1038/ni837. [DOI] [PubMed] [Google Scholar]
- 26.Townsend MJ, Weinmann AS, Matsuda JL, et al. T-bet regulates the terminal maturation and homeostasis of NK and Valpha14i NKT cells. Immunity. 2004;20(4):477–494. doi: 10.1016/s1074-7613(04)00076-7. [DOI] [PubMed] [Google Scholar]
- 27.Yokoyama WM. Betting on NKT and NK cells. Immunity. 2004;20(4):363–365. doi: 10.1016/s1074-7613(04)00085-8. [DOI] [PubMed] [Google Scholar]
- 28.Gallant S, Gilkeson G. ETS transcription factors and regulation of immunity. Arch Immunol Ther Exp (Warsz) 2006;54(3):149–163. doi: 10.1007/s00005-006-0017-z. [DOI] [PubMed] [Google Scholar]
- 29.Hromas R, Orazi A, Neiman RS, et al. Hematopoietic lineage- and stage-restricted expression of the ETS oncogene family member PU.1. Blood. 1993;82(10):2998–3004. [PubMed] [Google Scholar]
- 30.Oikawa T, Yamada T. Molecular biology of the Ets family of transcription factors. Gene. 2003;303:11–34. doi: 10.1016/s0378-1119(02)01156-3. [DOI] [PubMed] [Google Scholar]
- 31.Barton K, Muthusamy N, Fischer C, et al. The Ets-1 transcription factor is required for the development of natural killer cells in mice. Immunity. 1998;9(4):555–563. doi: 10.1016/s1074-7613(00)80638-x. [DOI] [PubMed] [Google Scholar]
- 32.Lacorazza HD, Miyazaki Y, Di Cristofano A, et al. The ETS protein MEF plays a critical role in perforin gene expression and the development of natural killer and NK-T cells. Immunity. 2002;17(4):437–449. doi: 10.1016/s1074-7613(02)00422-3. [DOI] [PubMed] [Google Scholar]
- 33.Walunas TL, Wang B, Wang CR, Leiden JM. Cutting edge: the Ets1 transcription factor is required for the development of NK T cells in mice. J Immunol. 2000;164(6):2857–2860. doi: 10.4049/jimmunol.164.6.2857. [DOI] [PubMed] [Google Scholar]
- 34.Bassuk AG, Barton KP, Anandappa RT, Lu MM, Leiden JM. Expression pattern of the Ets-related transcription factor Elf-1. Mol Med. 1998;4(6):392–401. [PMC free article] [PubMed] [Google Scholar]
- 35.Huang X, Brown C, Ni W, Maynard E, Rigby AC, Oettgen P. Critical role for the Ets transcription factor ELF-1 in the development of tumor angiogenesis. Blood. 2006;107(8):3153–3160. doi: 10.1182/blood-2005-08-3206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sarafova S, Siu G. A potential role for Elf-1 in CD4 promoter function. J Biol Chem. 1999;274(23):16126–16134. doi: 10.1074/jbc.274.23.16126. [DOI] [PubMed] [Google Scholar]
- 37.Rellahan BL, Jensen JP, Howcroft TK, Singer DS, Bonvini E, Weissman AM. Elf-1 regulates basal expression from the T cell antigen receptor zeta-chain gene promoter. J Immunol. 1998;160(6):2794–2801. [PubMed] [Google Scholar]
- 38.Serdobova I, Pla M, Reichenbach P, et al. Elf-1 contributes to the function of the complex interleukin (IL)-2-responsive enhancer in the mouse IL-2 receptor alpha gene. J Exp Med. 1997;185(7):1211–1221. doi: 10.1084/jem.185.7.1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Finco TS, Justice-Healy GE, Patel SJ, Hamilton VE. Regulation of the human LAT gene by the Elf-1 transcription factor. BMC Mol Biol. 2006;7(1):4. doi: 10.1186/1471-2199-7-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Garrett-Sinha LA, Dahl R, Rao S, Barton KP, Simon MC. PU. 1 exhibits partial functional redundancy with Spi-B, but not with Ets-1 or Elf-1. Blood. 2001;97(9):2908–2912. doi: 10.1182/blood.v97.9.2908. [DOI] [PubMed] [Google Scholar]
- 41.Mandal M, Chen XR, Alegre ML, et al. Tissue distribution, regulation and intracellular localization of murine CD1 molecules. Mol Immunol. 1998;35(9):525–536. doi: 10.1016/s0161-5890(98)00055-8. [DOI] [PubMed] [Google Scholar]
- 42.Goossens PL, Jouin H, Marchal G, Milon G. Isolation and flow cytometric analysis of the free lymphomyeloid cells present in murine liver. J Immunol Methods. 1990;132(1):137–144. doi: 10.1016/0022-1759(90)90407-m. [DOI] [PubMed] [Google Scholar]
- 43.Zimmer MI, Colmone A, Felio K, Xu H, Ma A, Wang CR. A cell-type specific CD1d expression program modulates invariant NKT cell development and function. J Immunol. 2006;176(3):1421–1430. doi: 10.4049/jimmunol.176.3.1421. [DOI] [PubMed] [Google Scholar]
- 44.Geng Y, Laslo P, Barton K, Wang CR. Transcriptional regulation of CD1D1 by Ets family transcription factors. J Immunol. 2005;175(2):1022–1029. doi: 10.4049/jimmunol.175.2.1022. [DOI] [PubMed] [Google Scholar]
- 45.Benlagha K, Wei DG, Veiga J, Teyton L, Bendelac A. Characterization of the early stages of thymic NKT cell development. J Exp Med. 2005;202(4):485–492. doi: 10.1084/jem.20050456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Anderson MK, Hernandez-Hoyos G, Diamond RA, Rothenberg EV. Precise developmental regulation of Ets family transcription factors during specification and commitment to the T cell lineage. Development. 1999;126(14):3131–3148. doi: 10.1242/dev.126.14.3131. [DOI] [PubMed] [Google Scholar]
- 47.Randi AM, Sperone A, Dryden NH, Birdsey GM. Regulation of angiogenesis by ETS transcription factors. Biochem Soc Trans. 2009;37(Pt 6):1248–1253. doi: 10.1042/BST0371248. [DOI] [PubMed] [Google Scholar]
- 48.Oettgen P. The role of ets factors in tumor angiogenesis. J Oncol. 2010;2010:767384. doi: 10.1155/2010/767384. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.