Abstract
The key regulator of G2–M transition of the cell cycle is M-phase promoting factor (MPF), a complex composed of cdc2 and a B-type cyclin. Cyclin B1 nuclear localization involves phosphorylation within a region called the cytoplasmic retention signal, which also contains a nuclear export signal. The mechanism of MPF nuclear localization remains unclear since it contains no functional nuclear localization signal (NLS). We exploited the yeast two-hybrid screen to find protein(s) potentially mediating localization of cyclin B1 and identified a novel interaction between cyclin B1 and cyclin F. We found that cdc2, cyclin B1 and cyclin F form a complex that exhibits histone H1 kinase activity. Cyclin B1 and cyclin F also colocalize through immunofluorescence studies. Additionally, deletion analysis revealed that each putative NLS of cyclin F is functional. Taken together, the data suggest that the NLS regions of cyclin F regulate cyclin B1 localization to the nucleus. The interaction between cyclin B1 and cyclin F represents the first example of direct cyclin–cyclin binding, and elucidates a novel mechanism that regulates MPF localization and function.
Keywords: cyclin B1/cyclin F/cytoplasmic retention signal/M-phase promoting factor/nuclear localization signal
Introduction
The onset of mitosis is regulated by the activation of M–phase promoting factor (MPF) in both mitotic and meiotic cells (Draetta and Beach, 1988; Westendorf et al., 1989; Nurse, 1990; Hunt, 1991). MPF consists of two subunits, the serine/threonine kinase cdc2 and either cyclin B1 or cyclin B2 (Labbe et al., 1989; Draetta et al., 1989; Gautier et al., 1990). Cyclins were first identified as proteins whose levels oscillate during the embryonic cell cycle in invertebrate eggs (Evans et al., 1983). Cyclin B1 protein begins to accumulate during late S and G2 phases, peaks at late G2–M phase, begins to degrade at the start of metaphase, and is nearly completely degraded at the onset of anaphase (Clute and Pines, 1999). During interphase, cyclin B1 associates with its catalytic partner cdc2 through a conserved region of ∼150 amino acids known as the cyclin box (Minshull et al., 1989; Pines and Hunter, 1989; Westendorf et al., 1989; Nugent et al., 1991; Kobayashi et al., 1992). Most cyclins interact with a cyclin-dependent kinase (cdk) through the cyclin box, and to date, cyclins have not been found to interact directly with one another.
In late G2 phase, post-translational modification of an inactive cdc2–cyclin B1 complex occurs, involving both phosphorylation and dephosphorylation of tyrosine and threonine residues of cdc2 (Solomon et al., 1990; Krek and Nigg, 1991). cdc2 kinase activity is negatively regulated by phosphorylation at Thr14 by the Myt1 kinase and at Tyr15 by the Wee1/Mik1 or Myt1 protein kinases (Russell and Nurse, 1987; Gould and Nurse, 1989; Parker et al., 1992; Kornbluth et al., 1994; Mueller et al., 1995), and positively regulated by phosphorylation at Thr161 by cdc2-activating kinase (Gould et al., 1991; Solomon et al., 1992; Fesquet et al., 1993; Poon et al., 1993). Dephosphorylation of Thr14 and Tyr15 is controlled by the cdc25 phosphatase (Gautier et al., 1991; Millar et al., 1991; Honda et al., 1993), and in conjunction with Thr161 phosphorylation, is required for full MPF activity.
B-type cyclins also undergo phosphorylation in a variety of systems such as sea urchin eggs, yeast, starfish oocytes and human cells (Booher et al., 1989; Meijer et al., 1989; Pines and Hunter, 1989; Pondaven et al., 1990). Previous studies in our laboratory have shown that phosphorylation of cyclin B1 regulates its nuclear localization (Li et al., 1995, 1997). There are five sites of phosphorylation in Xenopus cyclin B1 that have been identified: Ser2, Ser94, Ser96, Ser101 and Ser113 (Izumi and Maller, 1991; Li et al., 1995). Four of these sites are located within the cytoplasmic retention signal (CRS) domain, a region consisting of residues 78–127 in Xenopus cyclin B1 that was initially believed to contain information necessary for retention of the protein in the cytoplasm (Pines and Hunter, 1994). Mutation of these Ser residues to Ala abrogates the ability of cyclin B1 to translocate to the nucleus and to mature oocytes. Incorporation of a nuclear localization signal (NLS) into the Ala mutant restores biological activity, suggesting that phosphorylation of cyclin B1 within the CRS is important for nuclear translocation (Li et al., 1997). In support of this hypothesis, mutation of these Ser residues to Glu results in localization of cyclin B1 to the nucleus and an acceleration of oocyte maturation (Li et al., 1997). Recently, several studies have shown that the CRS also contains a functional nuclear export signal (NES) that causes cyclin B1 to be exported from the nucleus to the cytoplasm during interphase by interaction with CRM1, and that this NES is inactivated by phosphorylation at one or more sites near the NES (Hagting et al., 1998; Toyoshima et al., 1998; Yang et al., 1998).
While it is clear that cyclin B1 is continuously exported from the nucleus during interphase, the mechanism of nuclear import of cyclin B1 is unclear and remains an intriguing issue. There are no consensus NLSs in either subunit of MPF, suggesting that an alternative mechanism of nuclear import may exist for MPF. Potentially, the CRS may itself be mediating nuclear localization, especially when in its phosphorylated state, which results in inactivation of the NES. However, previous studies in our laboratory have indicated that the phosphorylated CRS domain does not function as an NLS. Fusion of CRSWT, CRSAla or CRSGlu onto the N-terminus of pyruvate kinase, which is often used as a reporter for nuclear localization since its size excludes the possibility of passive diffusion, did not result in nuclear localization as discussed in Li et al. (1997). In addition, when the CRS of cyclin B1 was appended to cyclin A, the nuclear cyclin A protein was retained in the cytoplasm, although the phosphorylation state of the CRS was not determined (Pines and Hunter, 1994). It has also been suggested that a bipartite NLS may be formed when a cyclin binds to a cdk (Pines and Hunter, 1994). Another possibility may be that cyclins associate with transcription factors or other nuclear proteins as shown in the E2F interaction with cyclin A (Mudryj et al., 1991), or that cyclin B1 may enter the nucleus simply by association with cdc2, which has been speculated to contain a weak histidine-containing putative NLS (Boulikas, 1996). More recently, importin–β has been shown to interact directly with a truncated form of cyclin B1, but it was inconclusive how full-length cyclin B1 enters the nucleus (Moore et al., 1999). cdc2–cyclin B1 may also ‘piggyback’ into the nucleus by binding a third protein containing an NLS, as suggested by Maridor et al. (1993).
Using the CRS domain of Xenopus cyclin B1, we employed a yeast two-hybrid screen of a mouse embryonic library to identify proteins that may be involved in the nuclear localization of cyclin B1. Interestingly, we isolated mouse cyclin F. Cyclin F is the largest known protein of the cyclin family with a predicted mol. wt of 87 kDa, although it migrates as a 100–110 kDa protein (Bai et al., 1994). Examination of the amino acid sequence of cyclin F indicates the presence of a cyclin box, F–box, PEST sequences and two putative NLS regions. Cyclin F protein accumulates in interphase, reaches maximal levels at G2–M phase, and decreases at mitosis just prior to the destruction of cyclin B1 (Bai et al., 1994). Cyclins often complex to a cdk and act as the regulatory binding partner; however, neither cdc2 nor cdk2 has been found to interact with cyclin F (Bai et al., 1994), although cyclin F has been shown to interact with Skp1, a protein involved in proteolysis (Bai et al., 1996). The exact role of cyclin F in cell cycle regulation remains to be clearly defined. In this report, we describe the first known direct interaction between two cyclin proteins, show that the two putative NLS regions of cyclin F are functional, and propose a model by which cyclin F regulates the nuclear localization of cyclin B1.
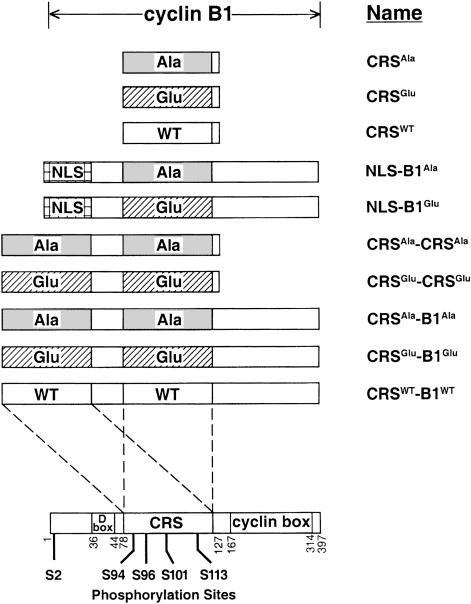
Results
Isolation of cyclin F from a two-hybrid screen
Previous work from our laboratory has shown that nuclear localization of cyclin B1 is regulated by phosphorylation in the CRS region (Li et al., 1997). Since cyclin B1 lacks a functional nuclear localization signal, another factor may be involved in the translocation of cyclin B1 to the nucleus. To investigate which protein(s) may be binding to the CRS region of cyclin B1 and affecting its localization, a yeast two-hybrid screen was employed. Full-length cyclin B1 could not be used as bait due to its toxicity in yeast. As previous experience suggested that a bait of at least 100 amino acids would be preferable for a successful two-hybrid screen (our unpublished data), another CRS region was appended to extend the length of each bait. With this size constraint in mind, we created two bait constructs of Xenopus cyclin B1: CRSAla-CRSAla, which is composed of two tandem CRS domains [amino acids (aa) 78–127] with each of the Ser residues mutated to Ala; and CRSGlu-CRSGlu, with the Ser residues mutated to Glu (Figure 1). The CRSAla-CRSAla construct prevents any phosphorylation in the CRS domain while the CRSGlu-CRSGlu construct mimics the phosphorylated state. The wild-type construct was not used in the screen as the phosphorylation state at the four phosphorylation sites within the CRS would be unpredictable. These two bait proteins were screened against a mouse embryonic cDNA library, resulting in 114 double transformants that grew on selective media after 3–4 days at 30°C (see Materials and methods). False positives were eliminated by screening against LexA-lamin, which was used to detect interactions specific to the bait (Vojtek and Hollenberg, 1995). After further characterization, 10 clones were identified and confirmed as valid interactions.
Fig. 1. Schematic representation of Xenopus cyclin B1-derived fusion proteins. The single and double CRS derivatives of cyclin B1 (aa 78–127) contained either all Ala or Glu mutations at residues 94, 96, 101 and 113. The NLS–B1 derivatives contained a nuclear localization signal appended to the N–terminus of cyclin B1. The CRS-B1 derivatives contained an additional CRS region appended to full-length cyclin B1.
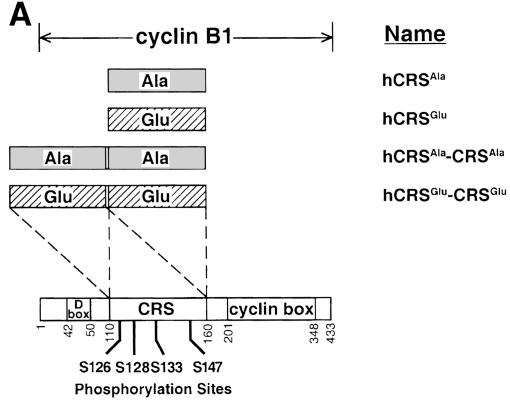
One of the clones identified was a 188 aa segment (aa 245–432) of mouse cyclin F, which interacted with both CRSAla-CRSAla and CRSGlu-CRSGlu baits. This segment encompasses the cyclin box and flanking regions of cyclin F (Figure 2B). To examine whether mouse cyclin F could interact with other species of cyclin B1, human CRS and CRS-CRS constructs were also made of each Ala and Glu mutant. Each of these hCRSAla, hCRSGlu, hCRSAla-CRSAla and hCRSGlu-CRSGlu derivatives (Figure 2A) interacted with the fragment of mouse cyclin F isolated from the two-hybrid screen (Table I). In addition, we obtained a full-length human cyclin F clone, which was tested for interaction with each of the Xenopus and human CRS and CRS-CRS mutants in the two-hybrid system; however, the protein proved to be toxic to the L40 yeast strain.
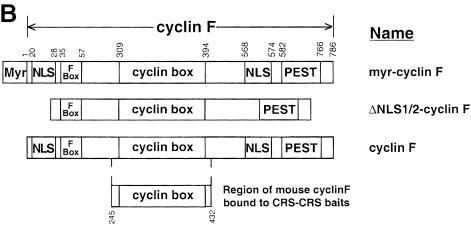
Fig. 2. Human cyclin B1 and cyclin F constructs. (A) The CRS region of human cyclin B1 was synthesized with the Ser residues at 126, 128, 133 and 147 all mutated to either Ala or Glu. (B) Myristylated human cyclin F (top), ΔNLS1/2-cyclin F (middle) and cyclin F (bottom) are shown. The 188 aa region of mouse cyclin F that interacted with the CRS of cyclin B1 corresponding to human cyclin F is indicated at the bottom.
Table I. Summary of two-hybrid interactions.
| LexA fusion | pVP16 | Mouse cyclin F (aa 245–432) |
|---|---|---|
| CRSAla | – | + |
| CRSGlu | – | + |
| CRSAla-CRSAla | – | + |
| CRSGlu-CRSGlu | – | + |
| hCRSAla | – | + |
| hCRSGlu | – | + |
| hCRSAla-CRSAla | – | + |
| hCRSGlu-CRSGlu | – | + |
| Lamin | nda | − |
and, not determined.
The potential for cyclin B1 and cyclin F interaction
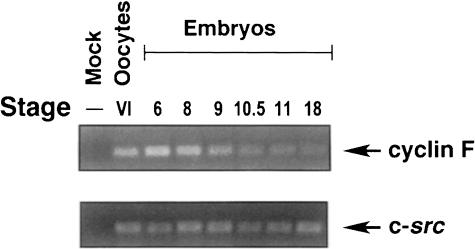
Both cyclin B1 and cyclin F proteins accumulate at G2 phase, thus providing an opportunity for interaction. Previously, it had been shown that cyclin F mRNA expression is present in various human tissues (Bai et al., 1994). We also wanted to determine whether cyclin F mRNA expression was present in Xenopus early embryonic development. Using quantitative RT–PCR, we examined cyclin F RNA expression in Stage VI oocytes, eggs and early embryos from different developmental stages after fertilization. As shown in Figure 3 (upper panel), the cyclin F RNA transcription levels remain constant to the end of blastulation (stage 9) and begin to decrease at the beginning of gastrulation (stages 10–11). c–src RNA expression was used as a positive control for RNA recovery by RT–PCR analysis (Figure 3, lower panel), since the level of c–src transcript remains constant throughout oogenesis and early embryogenesis (Rempel et al., 1995). These data indicate that cyclin F mRNA is present during meiotic maturation of oocytes and during early developmental stages and are consistent with the presence of cyclin F protein; however, in the absence of an antiserum against Xenopus cyclin F, we have not established whether this mRNA is translated.
Fig. 3. Expression of cyclin F RNA during Xenopus development. Total RNA isolated from stage VI oocytes or staged embryos was isolated and analyzed by RT–PCR. RNA (200 ng) was assayed for either the presence of cyclin F (upper panel) or, as an internal control, c–src RNA (lower panel).
Cyclin B1 and cyclin F interact in vitro and in vivo
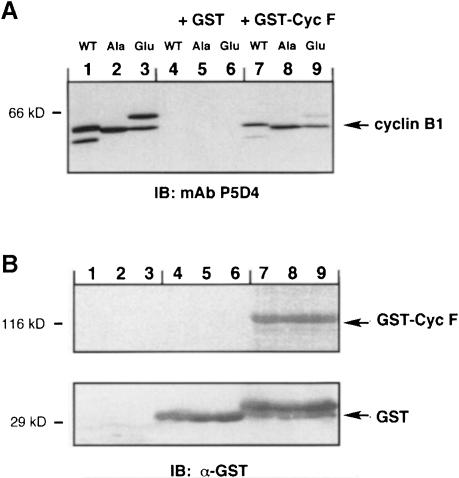
To test further for an interaction between cyclin B1 and cyclin F, a glutathione S-transferase (GST) fusion protein of human cyclin F was constructed and bacterially expressed. Cyclin B1 constructs, which contain a second CRS domain appended to the N-terminus of full-length Xenopus cyclin B1 to mimic the baits used in the two-hybrid screen (Figure 1), were in vitro translated. The Ser residues of each CRS domain were mutated to alanine (CRSAla-B1Ala), glutamic acid (CRSGlu-B1Glu) or unaltered (CRSWT-B1WT). Each Xenopus cyclin B1 construct was epitope-tagged to allow specific recognition by the monoclonal antibody (mAb) P5D4 (Kreis and Lodish, 1986). Incubation of GST–cyclin F with in vitro translated CRSAla-B1Ala, CRSGlu-B1Glu or CRSWT-B1WT showed that cyclin F bound to all three derivatives (Figure 4A, lanes 7, 8 and 9). As a negative control, each in vitro translated product was incubated with GST protein alone (Figure 4A, lanes 4, 5 and 6). Figure 4B confirms the presence of GST protein (lanes 4, 5 and 6) and GST–cyclin F (lanes 7, 8 and 9).
Fig. 4. In vitro binding of GST–cyclin F and Xenopus cyclin B1 fusion proteins. (A) Xenopus cyclin B1 derivatives were translated in reticulocyte lysates (lanes 1, 2 and 3) and then incubated with GST protein alone (lanes 4, 5 and 6) or with bacterially expressed GST–cyclin F (lanes 7, 8 and 9) bound to glutathione–agarose beads. The samples were analyzed by 7.5% SDS–PAGE and immunoblotted with mAb P5D4 directed against the epitope-tag VSV-G on each cyclin B1 derivative. (B) The membrane was re-probed with α–GST antibody, indicating the presence of GST protein (lanes 4, 5 and 6) and GST–cyclin F (lanes 7, 8 and 9). Molecular mass markers in kilodaltons are indicated.
The interaction between cyclin F and cyclin B1 was also tested in vivo by cotransfecting either CRSAla-B1Ala, CRSGlu-B1Glu or CRSWT-B1WT with cyclin F into COS–1 cells. Cell lysates were subjected to immunoprecipitation with cyclin F antibody, analyzed by SDS–PAGE, and immunoblotted with mAb P5D4 that recognized the epitope–tag of cyclin B1. All three cyclin B1 derivatives coimmunoprecipitated with cyclin F, confirming the interaction between cyclin B1 and cyclin F in vivo (data not shown). Similarly, immunoprecipitation with mAb P5D4 followed by immunoblotting with cyclin F antibody also demonstrated association between cyclin B1 and cyclin F (data not shown).
cdc2, cyclin B1 and cyclin F form a complex that exhibits histone H1 kinase activity
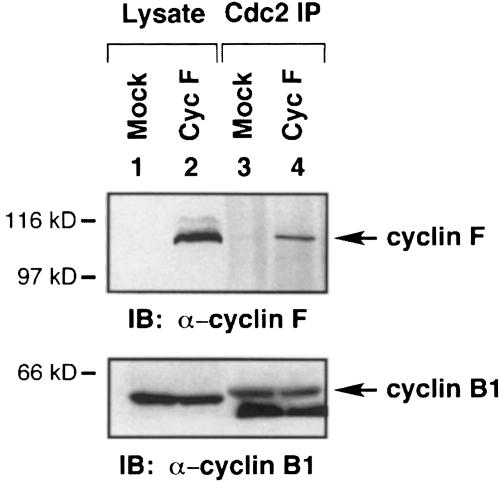
Since cyclin B1 is often complexed to cdc2, we were interested in determining whether cdc2 associated with the cyclin B1–cyclin F complex. Cyclin F was transfected into 293T cells, and lysates were then immunoprecipitated with α-cdc2 antibody. Immunoblot analysis demonstrated the presence of cyclin F in the α-cdc2 immunoprecipitate (Figure 5, upper panel, lane 4). Immunoblotting with cyclin B1 antibody indicated that cyclin B1 was present in each sample (Figure 5, lower panel). These data indicate that cdc2, cyclin B1 and overexpressed cyclin F are able to form a complex in vivo.
Fig. 5. cdc2, cyclin B1 and cyclin F form a complex. Cyclin F was transfected into 293T cells and then immunoprecipitated with α–cdc2 antibody. The samples were analyzed by 10% SDS–PAGE followed by immunoblotting with α–cyclin F antibody (upper panel) and α–cyclin B1 (lower panel). The lower band in the immunoprecipitated samples in lanes 3 and 4 of the lower panel represents the IgG band from the α–cdc2 antibody. Molecular mass markers are indicated.
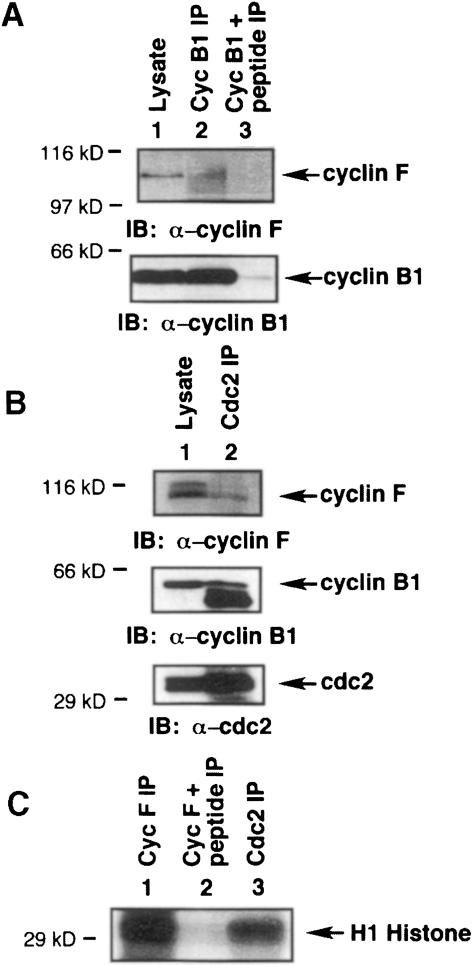
We then wished to determine whether we could detect an interaction between endogenous cyclin B1 and endogenous cyclin F. Using HeLa cells synchronized by double-thymidine block to G2–M phase, we immunoprecipitated with α–cyclin B1 antibody followed by immunoblotting with α–cyclin F antibody to show that cyclin F binds to cyclin B1 (Figure 6A, upper panel, lane 2). Immunoblotting with α–cyclin B1 confirmed the presence of cyclin B1 (Figure 6A, lower panel, lane 2). Addition of the blocking peptide to the α–cyclin B1 antiserum blocked the recovery of cyclin F in the cyclin B1 immunoprecipitate (Figure 6A, upper panel, lane 3). The lysate alone served as a positive control for the presence of cyclin B1 and cyclin F (Figure 6A, lane 1).
Fig. 6. Interaction of endogenous cyclin F with endogenous cyclin B1. (A) HeLa cells were synchronized by double-thymidine block to G2–M phase, and the lysate was immunoprecipitated with α–cyclin B1 antibody (lane 2) or α–cyclin B1 plus α–cyclin B1 blocking peptide (lane 3). Samples were analyzed by 10% SDS–PAGE followed by immunoblotting with α–cyclin F antibody (upper panel) and α–cyclin B1 antibody (lower panel). The lysate served as a positive control (lane 1). (B) Synchronized HeLa cells in G2–M were lysed and immunoprecipitated with α–cdc2 antibody (lane 2). Analysis by 10% SDS–PAGE followed by immunoblotting revealed the presence of cyclin F (upper panel), cyclin B1 (middle panel) and cdc2 (lower panel). The lysate served as a positive control (lane 1). The lower band in the cyclin B1 immunoblot of the α–cdc2 immunoprecipitate (lane 2) is due to the IgG band of the α–cdc2 antibody. (C) HeLa cells were lysed and immunoprecipitated with α–cyclin F (lane 1), α–cyclin F plus α–cyclin F blocking peptide (lane 2), or α–cdc2 antibody (lane 3). Samples were then assayed for histone H1 kinase activity. Molecular mass markers in all panels are indicated.
Next we investigated the interaction of endogenous cyclin F with endogenous MPF and whether this complex exhibited histone H1 kinase activity. Synchronized HeLa cells were harvested at G2–M phase and immunoprecipitated with α-cdc2 antibody. Figure 6B shows the presence of cyclin F (upper panel, lane 2), cyclin B1 (middle panel, lane 2) and cdc2 (lower panel, lane 2). The lysate alone was a positive control for the presence of all three proteins in Figure 6B, lane 1 of each panel. Thus, cdc2, cyclin B1 and cyclin F exist in a complex endogenously. To determine if cyclin F interacts with an active MPF complex, we performed a histone H1 kinase assay. HeLa cells were lysed, immunoprecipitated with α–cyclin F antibody, and then assayed for histone H1 kinase activity. As shown in Figure 6C, the α–cyclin F immunoprecipitate displayed kinase activity (lane 1), which was eliminated by inclusion of a blocking peptide to the α–cyclin F antibody (lane 2). The α-cdc2 immunoprecipitate served as a positive control (Figure 6C, lane 3). These results demonstrate that cyclin F can bind to the active MPF complex with retention of kinase activity, although we have not eliminated the possibility that the histone H1 kinase activity may reflect cyclin A–cdc2 complexes as well as cyclin B1–cdc2.
Cyclin B1 and cyclin F colocalize in immunofluorescence studies
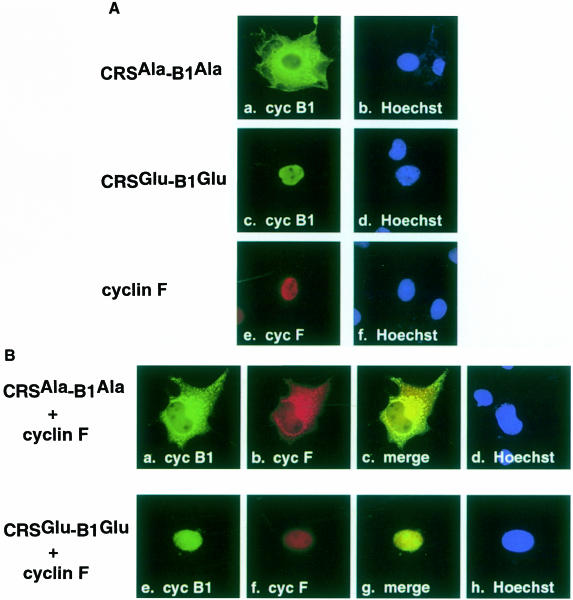
Cyclin F has been shown to localize predominantly to the nucleus in non-synchronized cells (Bai et al., 1994), whereas cyclin B1 has been shown to re-localize from the cytoplasm to the nucleus just prior to nuclear envelope breakdown (Pines and Hunter, 1991). We wished to examine whether overexpression of cyclin F would affect the localization of CRSAla-B1Ala or CRSGlu-B1Glu. In these experiments, COS–1 cells were transfected with cyclin F and/or cyclin B1 derivatives, and synchronized by double-thymidine block to enrich the G2 population of cells expressing cyclin F and cyclin B1. Indirect immunofluorescence experiments indicated that cyclin F is predomin- antly localized to the nucleus (Figure 7A, panel e). At the G2–M transition of the cell cycle, the CRSAla-B1Ala derivative is localized to the cytoplasm (Figure 7A, panel a) while the CRSGlu-B1Glu derivative is localized to the nucleus (panel c). When cotransfected, CRSAla-B1Ala (Figure 7B, panel a) and cyclin F (panel b) colocalize to the cytoplasm (panel c). In contrast, CRSGlu-B1Glu (Figure 7B, panel e) and cyclin F (panel f) mainly colocalize to the nucleus (panel g). These results demonstrate that cyclin F colocalizes with both derivatives of CRS-B1, correlating with the in vitro and in vivo binding data.
Fig. 7. (A) Individual localization of CRS-cyclin B1 and cyclin F in COS–1 cells synchronized in G2–M phase. Panels a and c are CRSAla-B1Ala and CRSGlu-B1Glu, respectively, fixed and stained with mAb P5D4 (green). Panel e shows cyclin F fixed and stained with α–cyclin F antibody (red). Hoechst dye 33342 reveals the nucleus in panels b, d and f (blue). (B) Colocalization of CRS-cyclin B1 and cyclin F. The CRSAla-B1Ala and cyclin F cotransfection (top row) shows that the cyclin B1 Ala mutant (green) and cyclin F (red) colocalize (yellow) in the cytoplasm. The CRSGlu-B1Glu and cyclin F cotransfection (bottom row) indicates that the cyclin B1 Glu mutant (green) and cyclin F (red) also colocalize (yellow), but in the nucleus.
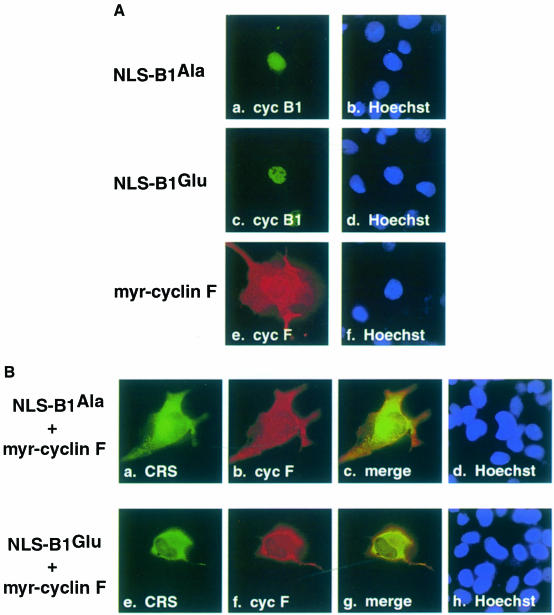
Myristylated cyclin F is able to recruit NLS–B1 to an abnormal subcellular compartment
The previous immunofluorescence results led us to question whether cyclin F was directing localization of cyclin B1 or vice versa. Thus, a myristylation signal was appended to cyclin F (myr-cyclin F; Figure 2B), in an attempt to target the normally nuclear protein to the plasma membrane. In addition, cyclin B1 Ala and Glu mutants with an NLS appended (NLS–B1Ala and NLS–B1Glu, shown in Figure 1) were used, ensuring that the cyclin B1 derivatives would be localized to the nucleus and thereby establishing a stringent criterion for the cyclin B1–cyclin F interaction. When transfected into COS–1 cells, we observed that myr-cyclin F was indeed localized to the plasma membrane and effectively excluded from the nucleus (Figure 8A, panel e). Both NLS–B1Ala and NLS–B1Glu localized to the nucleus in single transfections of COS–1 cells (Figure 8A, panels a and c). Cotransfection of NLS–B1Ala (Figure 8B, panel a) and myr-cyclin F (panel b) resulted in colocalization of the two proteins to the plasma membrane (panel c). Similarly, NLS–B1Glu (Figure 8B, panel e) and myr-cyclin F (panel f) colocalized to the plasma membrane (panel g). These results demonstrate that the interaction between cyclin B1 and cyclin F is strong, and that cyclin F can mislocalize a nuclear-targeted cyclin B1. The data also indicate that cyclin F is responsible for recruiting cyclin B1 and regulating its subcellular localization.
Fig. 8. (A) Individual localization of NLS–B1 and myr-cyclin F in COS–1 cells synchronized in G2–M phase. Panels a and c are NLS–B1 derivatives stained with mAb P5D4 (green). Panel e is myristylated cyclin F stained with α–cyclin F antibody (red). Hoechst dye shows the nucleus in panels b, d and f (blue). (B) Colocalization of NLS–B1 and myr-cyclin F. The NLS–B1Ala and myr-cyclin F cotransfection (top row) shows that the NLS–B1Ala mutant (green) and myr-cyclin F (red) colocalize (yellow). The NLS–B1Glu and myr-cyclin F cotransfection indicates that the NLS–B1Glu mutant (green) and myr-cyclin F (red) colocalize (yellow).
The two NLSs of cyclin F are functional
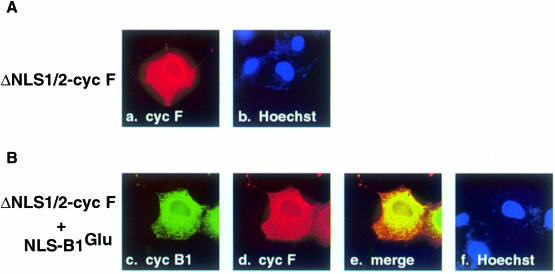
Cyclin F has two putative NLSs (Figure 2B). These NLS regions are rich in the basic residues Arg and Lys, and are likely to mediate localization of cyclin F to the nucleus. To determine whether one or both of these regions represented a functional NLS, we constructed mutants that lacked the first (ΔNLS1), second (ΔNLS2) or both (ΔNLS1/2) NLS region(s). Upon transfection of ΔNLS1-cyclin F into COS–1 cells, indirect immunofluorescence indicated that ∼50% more cells exhibited localization to the cytoplasm compared with wild-type in transfected cells (data not shown). The ΔNLS2-cyclin F mutant gave similar results (data not shown). The ΔNLS1/2-cyclin F mutant displayed localization to the cytoplasm in ∼90% of transfected cells. Figure 9A (panel a) shows a representative cell. These data provide evidence that each NLS of cyclin F is independent and functional, and that the two NLS regions appear to work in conjunction to localize properly the majority of cyclin F to the nucleus.
Fig. 9. ΔNLS1/2-cyclin F localizes to the cytoplasm and retains NLS–B1Glu in the cytoplasm. (A) ΔNLS1/2-cyclin F transfected cells were fixed and stained with an antibody directed against the epitope-tag of cyclin F (panel a, red). The nucleus is seen with Hoechst dye (panel b, blue) and indicates that ΔNLS1/2-cyclin F is completely excluded from the nucleus. (B) NLS–B1Glu (panel c, green) is cotransfected with ΔNLS1/2-cyclin F (panel d, red) and they colocalize (panel e, yellow).
ΔNLS1/2-cyclin F retains NLS–B1 in the cytoplasm
Since the ΔNLS1/2-cyclin F was mislocalized, we investigated whether it would behave as a dominant-negative protein, preventing localization of active cyclin B1 to the nucleus. ΔNLS1/2-cyclin F was cotransfected with the following cyclin B1 derivatives: CRSAla-B1Ala, NLS–B1Ala, CRSGlu-B1Glu or NLS–B1Glu. Transient transfections show that each cyclin B1 derivative predominantly colocalized with ΔNLS1/2-cyclin F to the cytoplasm (data not shown). Figure 9B shows the localization of NLS–B1Glu (panel c) and ΔNLS1/2-cyclin F (panel d). While NLS–B1Glu alone is ∼100% nuclear (Figure 8A, panel c), upon co-expression with ΔNLS1/2-cyclin F, both NLS–B1Glu and ΔNLS1/2-cyclin F exhibit cytoplasmic localization (Figure 9B, panel e). Thus, ΔNLS1/2-cyclin F recruits the nuclear NLS–B1Glu to the cytoplasm. Conversely, these results suggest that one role of wild-type cyclin F may be to localize cyclin B1 to the nucleus when the NES of cyclin B1 has been inactivated by phosphorylation.
Discussion
In this study, we found that a region containing the cyclin box of cyclin F interacts with both CRSAla-CRSAla and CRSGlu-CRSGlu mutants in a yeast two-hybrid screen. This interaction was confirmed in vitro and in vivo (data not shown) using full-length cyclin B1 constructs, and established the first known cyclin–cyclin association. The binding between cyclin B1 and cyclin F is strong, as shown by the ability of myr-cyclin F to retain nuclear-targeted cyclin B1 at the plasma membrane. We have also confirmed that cyclin B1 and cyclin F interact endogenously in coimmunoprecipitation experiments.
Both cyclin B1 and cyclin F accumulate at G2 phase (Bai et al., 1994), providing the opportunity for a potential interaction between the two cyclins. In addition, our immunofluorescence data consistently showed that ∼35% of cells expressing cyclin F protein exhibited perinuclear/cytoplasmic localization, and ∼65% of cells exhibited nuclear localization (our unpublished data), further providing the opportunity for cyclin F to bind to cyclin B1 in the cytoplasm. We have also shown that cyclin F mRNA is expressed in Xenopus oocytes and throughout early embryonic development, indicating that cyclin F is potentially present to interact with cyclin B1.
We have also found that cdc2 can be detected in the cyclin B1–cyclin F complex in vivo. Since it has been reported that cdc2 and cyclin F do not interact in vitro (Bai et al., 1994), it is likely that cyclin B1 is serving as the link between cyclin F and cdc2. The binding of cyclin F to cyclin B1 may allow cyclin F to interact indirectly with cdc2 and potentially help regulate its kinase activity in a surrogate manner. This is supported by the histone H1 kinase activity associated with the cyclin F immunoprecipitate, demonstrating that MPF maintains its kinase activity when complexed to cyclin F.
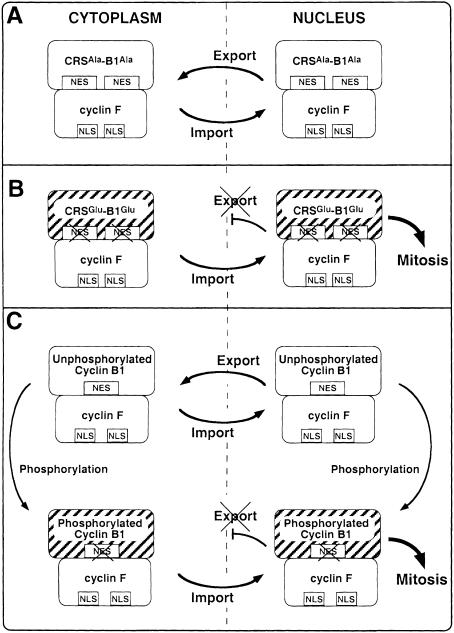
Immunofluorescence data show that cyclin B1 colocalizes with cyclin F, suggesting that cyclin F can bind indiscriminately to either CRSAla-B1Ala or CRSGlu-B1Glu, the unphosphorylated or phosphorylated forms, respectively, of cyclin B1. In Figure 10, a model is proposed to explain why both the Ala and Glu derivatives of cyclin B1 colocalize with cyclin F. Figure 10A shows that cyclin F transports CRSAla-B1Ala into the nucleus via the two NLS motifs of cyclin F, both of which have been shown in this study to be functional. Once in the nucleus, the functional NES of CRSAla-B1Ala shuttles the complex back to the cytoplasm. In these immunofluorescence studies, the CRSAla-B1Ala–cyclin F complex was not seen to colocalize to the nucleus at any time. A possible explanation is that the rate of nuclear export is faster than the rate of nuclear import. Thus, CRSAla-B1Ala and cyclin F are seen to colocalize to the cytoplasm.
Fig. 10. Model of cyclin B1–cyclin F colocalization. (A) CRSAla-B1Ala and cyclin F colocalize in the cytoplasm, undergo nuclear transport, and then re-localize to the cytoplasm due to the functional NES of CRSAla-B1Ala. (B) CRSGlu-B1Glu and cyclin F interact in the cytoplasm and then colocalize to the nucleus. The complex is then retained in the nucleus due to the inactive NES of CRSGlu-B1Glu and properly localized to trigger the onset of mitosis. (C) Upper panel: unphosphorylated cyclin B1 and cyclin F interact in the cytoplasm and enter the nucleus. Re-localization to the cytoplasm may then occur via the active NES of cyclin B1. Lower panel: phosphorylated cyclin B1 and cyclin F colocalize from the cytoplasm to the nucleus. MPF is active and ready to initiate mitosis. In each panel, cdc2 is complexed to cyclin B1, although it is not represented.
The model presented in Figure 10B explains the colocalization seen between CRSGlu-B1Glu and cyclin F. In this case, the CRSGlu-B1Glu–cyclin F complex traverses into the nucleus utilizing the two NLS regions of cyclin F. Recent studies have shown that phosphorylation of Ser residues within the CRS domain reduced the binding affinity for CRM1 and resulted in a decrease of nuclear export (Yang et al., 1998). Thus, the NES of CRSGlu-B1Glu is inactivated due to the mimicked phosphorylation by the Glu residues, and cyclin B1 is retained in the nucleus with cyclin F. The phosphorylated form of cyclin B1, which may also be complexed to cdc2, would then be properly localized in the nucleus to trigger the onset of mitosis by phosphorylating nuclear components necessary for mitosis. Therefore, the inactivation of the NES by phosphorylation of the Ser residues within cyclin B1 may be the trigger that initiates mitosis.
With regard to wild-type cyclin B1, the upper panel of Figure 10C depicts the localization of unphosphorylated cyclin B1 into the nucleus via the two NLS motifs of its binding partner cyclin F. In the nucleus, the functional NES of unphosphorylated cyclin B1 would result in export of the complex to the cytoplasm. With respect to the phosphorylated form of cyclin B1 complexed to cyclin F, we propose that the complex enters the nucleus via the NLS regions of cyclin F as shown in the lower panel of Figure 10C. Once in the nucleus, MPF is properly localized to initiate mitosis.
The ability of cyclin F to regulate the localization of cyclin B1 is clearly shown by the myr-cyclin F and NLS–B1 immunofluorescence studies. In addition, deletion of the two NLS regions of cyclin F resulted in its localization to the cytoplasm rather than to the nucleus. When this ΔNLS1/2-cyclin F was cotransfected with various cyclin B1 derivatives, the resulting complexes were colocalized to the cytoplasm. The ability of ΔNLS1/2cyclin F to retain the NLS–B1Glu derivative in the cytoplasm is especially notable, suggesting the possibility that cyclin F itself may contain an NES whose activity is manifested in the absence of its NLS activity. Alternatively, cyclin F may contain some type of CRS whose function becomes apparent upon inactivation of its NLS function. The presence of an NES in cyclin F may also be consistent with our data in the myr-cyclin F/NLS–B1 colocalization to the cytoplasm; the myristylation signal appended to cyclin F in conjunction with an NES overcomes the NLS appended to cyclin B1.
Our data support a model in which cyclin F mediates the nuclear localization of cyclin B1. Most nuclear import/export factors interact directly with their cargo, but importin–β uses the adaptor protein importin–α to interact with an NLS-containing protein (Gorlich et al., 1995a, b). Importin–α binds to importin–β to form a heterodimer, and during translocation through the nuclear pore complex (NPC), the heterodimer dissociates: importin–α and the NLS-containing protein enter and accumulate in the nucleoplasm as importin–β accumulates at the NPC (Gorlich et al., 1995b; Moroianu et al., 1995). It has recently been shown that importin–β can bind directly to a truncated form of Xenopus cyclin B1 (aa 121–397) and be imported into the nucleus without the presence of importin–α (Moore et al., 1999). However, this truncated cyclin B1 is lacking the CRS/NES region that has been shown to regulate cyclin B1 localization, and the import was not seen with full-length cyclin B1. Thus, the interaction found between cyclin F and the CRS region of cyclin B1 (aa 78–127) supports the possibility that importin–α binds to the NLS region(s) of cyclin F, which can then interact with importin–β to translocate cyclin B1 as well as cyclin F into the nucleus. Alternatively, the cyclin B1–cyclin F interaction could provide the link between both importin–β and importin–α. Whichever the case may be, the interaction of cyclin F would now provide a boost in cyclin B1 translocation to the nucleus since the truncated cyclin B1 protein exhibited slow nuclear import mediated by importin–β (Moore et al., 1999). Thus, at the G2–M transition of the cell cycle, the role of cyclin F may be to increase nuclear import of cyclin B1, possibly through an interaction with importin–α in addition to importin–β.
One of the few known roles of cyclin F is that it interacts with Skp1 through its F–box motif (Bai et al., 1996). Skp1 is involved in G1–S and G2–M transitions of the cell cycle, and is linked to ubiquitin-mediated proteolysis of many proteins such as Sic1p, Cln2p and Clb5p (Bai et al., 1996). Skp1 has been shown to act as a ubiquitin ligase when associated with Cdc53 and the F–box-containing protein Cdc4, forming a complex named SCFCdc4 (Skp1, Cdc53 and F–box protein Cdc4) (Skowyra et al., 1997). Skowyra et al. (1997) proposed a model in which an F–box protein specifically recruits phosphorylated substrates to the SCF complex and forms a ubiquitylation complex. Thus, cyclin F may be the F–box protein that completes the ubiquitin ligase complex, forming an SCFcyclinF and targeting phosphorylated cyclin B1 for ubiquitin-mediated degradation. However, this may not be the case as the timing of cyclin degradation suggests that cyclin F is degraded prior to cyclin B1 (Bai et al., 1994). Another role may be that cyclin F itself is destroyed by interaction with Skp1 and/or another F–box-containing protein that recognizes cyclin F. This implies that cyclin F degradation may be necessary for the degradation of cyclin B1. The destruction of cyclin B1 by the ubiquitylation pathway results in the inactivation of MPF, and the cell exits mitosis (Glotzer et al., 1991). Further experiments will be necessary to determine if cyclin F is indeed involved in the proteolysis of cyclin B1.
In conclusion, we have shown a novel interaction between two cyclins. The CRS region of cyclin B1 binds a region including the cyclin box of cyclin F and, through this interaction, cyclin F is able to colocalize with cyclin B1 and mediate nuclear localization of the phosphorylated form of cyclin B1. Since cyclin F has two putative NLSs and is present at approximately the same time as cyclin B1 during the cell cycle, the translocation of cyclin B1 into the nucleus is likely to be regulated by cyclin F. In this manner, one role of cyclin F in the cell cycle may be to regulate the nuclear translocation of cyclin B1 at the G2–M transition.
Materials and methods
Two-hybrid constructs
The two-hybrid plasmids pBTM116, pVP16 and pLexA-lamin (Vojtek et al., 1993) were generous gifts from S.Hollenberg and J.A.Cooper (Fred Hutchinson Cancer Research Center). Construction of CRSAla-CRSAla and CRSGlu-CRSGlu was achieved through digestion of the Bsu36I and BamHI sites from full-length cyclin B1 constructs made previously in our laboratory (Li et al., 1997). A linker oligonucleotide containing a stop codon was added after the last CRS domain of each construct. Each construct was then cut with EcoRI and BamHI and inserted into the pBTM116 vector, which was constructed by P.Bartel and S.Fields, at the corresponding sites. Human CRS regions were synthesized and inserted into the pBTM116 vector, the sequence was verified through dideoxy nucleotide sequencing. Human cyclin F (Bai et al., 1994), a generous gift from S.J.Elledge, was cloned into the pVP16 plasmid for two-hybrid analysis at the BamHI and NotI sites using an oligonucleotide linker.
Two-hybrid screen
The two-hybrid screen was performed according to previously published protocols (Vojtek et al., 1993; Vojtek and Hollenberg, 1995). Each LexA-CRSAla-CRSAla and LexA-CRSGlu-CRSGlu fusion construct was cotransformed with a 9.5 d.p.c. mouse embryo cDNA library fused to pVP16 into the L40 strain of Saccharomyces cerevisiae. The transformants were selected on –His plates for 3–4 days at 30°C with the addition of 5 mM 3–aminotriazole (3–AT) (Sigma) for the CRSAla-CRSAla transformants and 50 mM 3–AT for the CRSGlu-CRSGlu transformants. An interaction was scored positive for the ability of the yeast to grow beyond bait plus empty pVP16 vector and to overcome 3–AT inhibition. False positives were discriminated by testing cDNA-pVP16 against LexA-lamin. The remaining cDNA-pVP16 clones were then shuttled into Escherichia coli HB101 cells, rescued, and then sequenced as previously described (Vojtek and Hollenberg, 1995). The sequences were then entered into the NCBI BLAST program for identification.
RT–PCR assays
Total RNA from oocytes and staged embryos were prepared using TRIzol reagent as described by the manufacturer (Gibco-BRL, Life Technologies). RT–PCR was performed using 200 ng of total RNA, 50 pmol of sense primer (ATATGCACACGCTTTATTAGC) and 50 pmol of antisense primer (ATGGACGTGCTTGTCTGTGC) for the cyclin F gene to generate a 325 bp fragment, and 50 pmol of sense primer (TACATCACGTCTCGAACTC) and 50 pmol of antisense primer (ACAGCATATAACTGTACCAG) for the c–src gene to generate a 347 bp fragment.
In vitro binding assay
The cyclin F-pVP16 construct was cleaved by BamHI and NotI and inserted into pGEX–3X to generate GST–cyclin F. The GST–cyclin F fusion protein was produced by E.coli BL21 cells. The cells were induced with 1 mM isopropyl-β–d–thiogalactopyranoside (IPTG) for 3 h. The bacterial pellet was resuspended in 50 ml of lysis buffer [50 mM Tris pH 7.5, 150 mM NaCl, 10% glycerol, 1 mM dithiothreitol (DTT), 1 mM EDTA, 1 mM phenylmethylsulfonyl fluoride (PMSF), 10 μg/ml aprotinin, 10 μg/ml pepstatin and 10 μg/ml leupeptin] and sonicated five times for 15 s each. The pellet was resuspended in lysis buffer and 6 M urea, then slowly dialyzed. GST–cyclin F was then incubated with glutathione–agarose beads (Sigma) at 4°C and washed three times. Coupled in vitro transcription–translation (Promega) was performed according to the manufacturer's instructions for the full-length CRSAla-B1Ala, CRSGlu-B1Glu and CRSWT-B1WT derivatives. Each contained an epitope-tag derived from vesicular stomatitis virus glycoprotein (VSV–G) appended at the C–terminus, which was recognized by murine mAb P5D4 (Kreis and Lodish, 1986) as previously described (Li et al., 1997). The in vitro translated products were individually incubated with GST–cyclin F or GST proteins overnight at 4°C in binding buffer (20 mM Tris pH 7.5, 25 mM NaCl, 10% glycerol, 1 mM DTT, 1 mM EDTA, 1 mM PMSF, 10 μg/ml aprotinin, 10 μg/ml pepstatin and 10 μg/ml leupeptin). The reactions were then washed four times in binding buffer, boiled for 3 min in sample buffer, and analyzed on 7.5% SDS–PAGE followed by immunoblotting with mAb P5D4 (1:2000). Enhanced chemiluminescence (ECL) (Amersham) was then used to detect proteins. The membrane was then reprobed with GST antibody (Santa Cruz Biotechnology, Inc.) at 1:4000 and detected by ECL.
Coimmunoprecipitation
The cyclin F gene was cloned into pcDNA3 from the pAB23BXN vector. 293T cells were cultured in Dulbecco's modified Eagle's medium (DMEM), supplemented with 10% fetal bovine serum (FBS) and incubated at 37°C in 10% CO2. Sub-confluent cells were transfected with 20 μg of pcDNA3 or cyclin F-pcDNA3 by calcium phosphate precipitation (Chen and Okayama, 1987) in a 10 cm dish. Twenty-four hours after transfection, the cells were harvested and lysed in lysis buffer (20 mM Tris pH 8.0, 150 mM NaCl, 0.5% NP-40, 1 mM Na3VO4, 1 mM NaF, 1 mM benzamidine, 0.1 mM p–nitrophenylphosphate, 1 mM DTT, 1 mM PMSF, 10 μg/ml aprotinin, 10 μg/ml pepstatin and 10 μg/ml leupeptin). Lysates were pre-cleared with 40 μl of protein A–Sepharose beads and incubated with 2 μg of p34cdc2 antibody (Santa Cruz Biotechnology, Inc.) overnight at 4°C. Protein A–Sepharose beads were then added and the immunoprecipitated samples were washed four times, boiled for 3 min in sample buffer, and analyzed on 10% SDS–PAGE. The membrane was immunoblotted with α–cyclin F antibody (Santa Cruz Biotechnology, Inc.) at 1:8000 and reprobed with α–cyclin B1 antibody (Santa Cruz Biotechnology, Inc.) at 1:2000, followed by ECL (Amersham).
HeLa cells were cultured in 10% FBS in 5% CO2 to ∼60% confluency and then synchronized in G2–M phase by double-thymidine block as described elsewhere (Bai et al., 1994). Cells were harvested 9 h after release with 0.1% NP–40 lysis buffer (0.1% NP–40, 20 mM Tris–HCl pH 7.5, 100 mM NaCl, 1 mM DTT, 1 mM Na3VO4, 10 μg/ml aprotinin, 10 μg/ml leupeptin, 1 mM PMSF, 200 μM N–acetyl-leucinyl-leucinyl-norleucinal and 5 mM N–ethylmaleimide). Lysates were pre-cleared with protein A–Sepharose beads and then immunoprecipitated with 2 μg of α–cdc2, α–cyclin B1 or α–cyclin B1 antibody plus 10 μg of corresponding blocking peptide (Santa Cruz Biotechnology, Inc.). After washing four times, the samples were boiled for 3 min in sample buffer and analyzed by 10% SDS–PAGE. The gel was transferred to nitrocellulose membrane and then immunoblotted for cdc2, cyclin B1 or cyclin F, followed by ECL (Amersham).
Histone H1 kinase assay
HeLa cells cultured in 10% FBS were lysed in 0.1% NP–40 lysis buffer and then pre-cleared with protein A–Sepharose followed by immunoprecipitation with 2 μg of α–cdc2, α–cyclin F or α–cyclin F plus 10 μg of corresponding blocking peptide (Santa Cruz Biotechnology, Inc.). The immunoprecipitates were washed four times prior to the addition of 50 μl of kinase buffer (50 mM Tris–HCl pH 7.5, 10 mM MgCl2, 1 mM DTT, 20 mM EGTA, 50 mM ATP, 10 mCi of [γ–32P]ATP) and 74 μg/ml H1 histone (Boehringer Mannheim) for 10 min at 30°C. Sample buffer was added to stop the reaction and 50 μl of each sample were analyzed by 10% SDS–PAGE, followed by autoradiography.
Immunofluorescence microscopy
COS–1 cells were plated onto glass coverslips and cultured in DMEM supplemented with 10% FBS at 37°C in 5% CO2. Cells were transiently transfected by calcium phosphate precipitation with 10 μg of each DNA in a 60 mm dish. An epitope-tag consisting of 12 amino acids (EPDGAALEWHHL) was synthesized and appended to the C–terminus of cyclin F. The myristylated cyclin F construct was constructed by appending a 13 aa myristylation sequence from p60src (Cross et al., 1984; Pellman et al., 1985). The ΔNLS1/2–cyclin F construct was derived from the epitope-tagged cyclin F by deletion of residues 35–57 (ΔNLS1) and residues 568–574 (ΔNLS2). Each cyclin B1 derivative contained a VSV-G epitope-tag at the C–terminus. Sixteen hours after transfection, the cells were re-fed and then synchronized by double-thymidine block. Cells were treated with 2 mM thymidine for 15 h to arrest cells in S phase, rinsed three times, and released with fresh 10% FBS medium for 9 h. The cells were then retreated with 2 mM thymidine for 15 h. Five and a half hours after release >50% of the cells had a rounded mitotic phenotype. Cells were then fixed at this G2–M phase with 3% paraformaldehyde in phosphate-buffered saline (PBS) for 15 min and permeabilized with 0.1% Triton X–100, 0.2 M glycine and 2.5% FBS in PBS for 45 min. Primary antibody P5D4 (1:1000) was used to detect VSV–G-tagged cyclin B1 derivatives with FITC-conjugated goat anti-mouse secondary antibody (Boehringer Mannheim). To detect epitope-tagged cyclin F, affinity-purified rabbit antisera were made (Lenormand et al., 1999) and visualized with rhodamine-conjugated goat anti-rabbit secondary antibody (Boehringer Mannheim). Coverslips were mounted on slides with 100 mM Tris pH 8.0, 90% glycerol, 1 mg/ml phenylenediamine and 1 μg/ml Hoechst dye 33342 to detect DNA.
Acknowledgments
Acknowledgements
We thank S.Hollenberg and J.A.Cooper for the kind gifts of pBTM116, pVP16 and pLexA-lamin plasmids; P.Bartel and S.Fields for construction of the pBTM116 vector; V.Malhotra for providing mAb P5D4; and S.J.Elledge for kindly providing human cyclin F. We also thank K.C.Hart, R.W.Dellinger, A.N.Meyer, J.A.Tynan and other members of the D.J.D. laboratory for critical review of the manuscript. Support from NIH/NCI CA34456 is gratefully acknowledged. M.K. also gratefully acknowledges support from NIH/GM07313, and E.A.B. gratefully acknowledges support from NIH/NCI T32-CA09523.
References
- Bai C., Richman, R. and Elledge, S.J. (1994) Human cyclin F. EMBO J., 13, 6087–6098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai C., Sen, P., Hofmann, K., Ma, L., Goebl, M., Harper, J.W. and Elledge, S.J. (1996) SKP1 connects cell cycle regulators to the ubiquitin proteolysis machinery through a novel motif, the F–box. Cell, 86, 263–274. [DOI] [PubMed] [Google Scholar]
- Booher R.N., Alfa, C.E., Hyams, J.S. and Beach, D.H. (1989) The fission yeast cdc2/cdc13/suc1 protein kinase: regulation of catalytic activity and nuclear localization. Cell, 58, 485–497. [DOI] [PubMed] [Google Scholar]
- Boulikas T. (1996) Nuclear import of protein kinases and cyclins. J. Cell Biol., 60, 61–82. [DOI] [PubMed] [Google Scholar]
- Chen C. and Okayama, H. (1987) High-efficiency transformation of mammalian cells by plasmid DNA. Mol. Cell. Biol., 7, 2745–2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clute P. and Pines, J. (1999) Temporal and spatial control of cyclin B1 destruction in metaphase. Nature Cell Biol., 1, 82–87. [DOI] [PubMed] [Google Scholar]
- Cross F.R., Garber, E.A., Pellman, D. and Hanafusa, H. (1984) A short sequence in the p60src N terminus is required for p60src myristylation and membrane association and for cell transformation. Mol. Cell. Biol., 4, 1834–1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draetta G. and Beach, D. (1988) Activation of cdc2 protein kinase during mitosis in human cells: cell cycle-dependent phosphorylation and subunit rearrangement. Cell, 54, 17–26. [DOI] [PubMed] [Google Scholar]
- Draetta G., Luca, F., Westendorf, J., Bruzuela, L., Ruderman, J. and Beach, D. (1989) cdc2 protein kinase is complexed with both cyclin A and B: evidence for proteolytic inactivation of MPF. Cell, 56, 829–838. [DOI] [PubMed] [Google Scholar]
- Evans T., Rosenthal, E.T., Youngblom, J., Distel, D. and Hunt, T. (1983) Cyclin: a protein specified by maternal mRNA in sea urchin eggs that is destroyed at each cleavage division. Cell, 33, 389–396. [DOI] [PubMed] [Google Scholar]
- Fesquet D. et al. (1993)The MO15 gene encodes the catalytic subunit of a protein kinase that activates cdc2 and other cyclin-dependent kinases (CDKs) through phosphorylation of Thr 161 and its homologues. EMBO J., 12, 3111–3121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautier J., Minshull, J., Lohka, M., Glotzer, M., Hunt, T. and Maller, J.L. (1990) Cyclin is a component of maturation promoting factor from Xenopus.Cell, 60, 487–494. [DOI] [PubMed] [Google Scholar]
- Gautier J., Solomon, M.J., Booher, R.N., Bazan, J.F. and Kirschner, M.W. (1991) cdc25 is a specific tyrosine phosphatase that directly activates p34cdc2.Cell, 67, 197–211. [DOI] [PubMed] [Google Scholar]
- Glotzer M., Murray, A.W. and Kirschner, M.W. (1991) Cyclin is degraded by the ubiquitin pathway. Nature, 349, 132–138. [DOI] [PubMed] [Google Scholar]
- Gorlich D., Kostka, S., Kraft, R., Dingwall, C., Laskey, R.A., Hartmann, E. and Prehn, S. (1995a) Two different subunits of importin cooperate to recognize nuclear localization signals and bind them to the nuclear envelope. Curr. Biol., 5, 383–392. [DOI] [PubMed] [Google Scholar]
- Gorlich D., Vogel, F., Mills, A.D., Hartmann, E. and Laskey, R.A. (1995b) Distinct functions for the two importin subunits in nuclear protein import. Nature, 377, 246–248. [DOI] [PubMed] [Google Scholar]
- Gould K.L. and Nurse, P. (1989) Tyrosine phosphorylation of the fission yeast cdc2+ protein kinase regulates entry into mitosis. Nature, 342, 39–45. [DOI] [PubMed] [Google Scholar]
- Gould K.L., Moreno, S., Owen, D.J., Sazer, S. and Nurse, P. (1991) Phosphorylation at Thr161 is required for Schizosaccharomyces pombe p34cdc2 function. EMBO J., 10, 3297–3309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagting A., Karlsson, C., Clute, P., Jackman, M. and Pines, J. (1998) MPF localization is controlled by nuclear export. EMBO J., 17, 4127–4138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda R., Ohba, Y., Nagata, A., Okayama, H. and Yasuda, H. (1993) Dephosphorylation of human p34cdc2 kinase on both Thr-14 and Tyr–15 by human cdc25B phosphatase. FEBS Lett., 318, 331–334. [DOI] [PubMed] [Google Scholar]
- Hunt T. (1991) Cyclins and their partners: from a simple idea to complex reality. Semin. Cell Biol., 2, 213–222. [PubMed] [Google Scholar]
- Izumi T. and Maller, J.L. (1991) Phosphorylation of Xenopus cyclins B1 and B2 is not required for cell cycle transitions. Mol. Cell. Biol., 11, 3860–3867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi H., Stewart, E., Poon, R., Adamczewski, J.P., Gannon, J. and Hunt, T. (1992) Identification of the domains in cyclin A required for binding to and activation of, p34cdc2 and p32cdk2 protein kinase subunits. Mol. Biol. Cell, 3, 1279–1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornbluth S., Sebastian, B., Hunter, T. and Newport, J. (1994) Membrane localization of the kinase which phosphorylates p34cdc2 on threonine 14. Mol. Biol. Cell, 5, 273–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreis T.E. and Lodish, H.F. (1986) Oligomerization is essential for transport of vesicular stomatitis viral glycoprotein to the cell surface. Cell, 46, 929–937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krek W. and Nigg, E.A. (1991) Differential phosphorylation of vertebrate p34cdc2 kinase at the G1/S and G2/M transitions of the cell cyclin: identification of major phosphorylation sites. EMBO J., 10, 305–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labbe J.-C., Capony, J.-P., Caput, D., Cavadore, J.-C., Derancourt, J., Kaghdad, M., Lelias, J.-M., Picard, A. and Doree, M. (1989) MPF from starfish oocytes at first meiotic metaphase is a heterodimer containing one molecule of cdc2 and one molecule of cyclin B. EMBO J., 8, 3053–3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenormand J.-L., Dellinger, R.W., Knudsen, K., Subramani, S. and Donoghue, D.J. (1999) Speedy: a novel cell cycle regulator of the G2/M transition. EMBO J., 18, 1869–1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Meyer, A.N. and Donoghue, D.J. (1995) Requirement for phosphorylation of cyclin B1 for Xenopus oocyte maturation. Mol. Biol. Cell, 6, 1111–1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Meyer, A.N. and Donoghue, D.J. (1997) Nuclear localization of cyclin B1 mediates its biological activity and is regulated by phosphorylation. Proc. Natl Acad. Sci. USA, 94, 502–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maridor G., Gallant, P., Golsteyn, R. and Nigg, E.A. (1993) Nuclear localization of vertebrate cyclin A correlates with its ability to form complexes with cdk catalytic subunits. J. Cell Sci., 106, 535–544. [DOI] [PubMed] [Google Scholar]
- Meijer L., Arion, D., Golsteyn, R., Pines, J., Brizuela, L., Hunt, T. and Beach, D. (1989) Cyclin is a component of the sea urchin egg M–phase specific histone H1. EMBO J., 8, 2275–2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar J.B., McGowan, C.H., Lenaers, G., Jones, R. and Russell, P. (1991) p80cdc25 mitotic inducer is the tyrosine phosphatase that activated p34cdc2 kinase in fission yeast. EMBO J., 10, 4301–4309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minshull J., Pines,J., Golsteyn,R., Standart,N., Mackie,S., Colman,A., Blow,J., Ruderman,J.V., Wu,M. and Hunt,T. (1989) The role of cyclin synthesis, modification and destruction in the control of cell division. J. Cell Sci., Suppl., 12, 77–97. [DOI] [PubMed] [Google Scholar]
- Moore J.D., Yang, J., Truant, R. and Kornbluth, S. (1999) Nuclear import of cdk/cyclin complexes: identification of distinct mechanisms for import of cdk2/cyclin E and cdc2/cyclin B1. J. Cell Biol., 144, 213–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moroianu J., Hijikata, M., Blobel, G. and Radu, A. (1995) Mammalian karyopherin α1β and α2β heterodimers: α1 or α2 subunit binds nuclear localization signal and β subunit interacts with peptide repeat-containing nucleoporins. Proc. Natl Acad. Sci. USA, 92, 6532–6536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mudryj M., Devoto, S.H., Hiebert, S.W., Hunter, T., Pines, J. and Nevins, J.R. (1991) Cell cycle regulation of the E2F transcription factor involves an interaction with cyclin A. Cell, 65, 1243–1253. [DOI] [PubMed] [Google Scholar]
- Mueller P.R., Coleman, T.R., Kumagai, A. and Dunphy, W.G. (1995) Myt1: a membrane-associated inhibitory kinase that phosphorylates cdc2 on both threonine-14 and tyrosine-15. Science, 270, 86–90. [DOI] [PubMed] [Google Scholar]
- Nugent J.H., Alfa, C.E., Young, T. and Hyams, J.S. (1991) Conserved structural motifs in cyclins identified by sequence analysis. J. Cell Sci., 99, 669–674. [DOI] [PubMed] [Google Scholar]
- Nurse P. (1990) Universal control mechanism regulating onset of M–phase. Nature, 344, 503–508. [DOI] [PubMed] [Google Scholar]
- Parker L.L., Atherton-Fessler, S. and Piwnica-Worms, H. (1992) p107wee1 is a dual-specificity kinase that phosphorylates p34cdc2 on tyrosine 15. Proc. Natl Acad. Sci. USA, 89, 2917–2921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellman D., Gerber, E.A., Cross, F.R. and Hanafusa, H. (1985) An N–terminal peptide from p60src can direct myristylation and plasma membrane localization when fused to heterologous proteins. Nature, 314, 374–377. [DOI] [PubMed] [Google Scholar]
- Pines J. and Hunter, T. (1989) Isolation of a human cyclin cDNA: evidence for cyclin mRNA and protein regulation in the cell cycle and for interaction with p34cdc2.Cell, 58, 833–846. [DOI] [PubMed] [Google Scholar]
- Pines J. and Hunter, T. (1991) Human cyclins A and B1 are differentially located in the cell and undergo cell cycle-dependent nuclear transport. J. Cell Biol., 115, 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pines J. and Hunter, T. (1994) The differential localization of human cyclins A and B is due to a cytoplasmic retention signal in cyclin B. EMBO J., 13, 3772–3781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pondaven P., Meijer, L. and Beach, D. (1990) Activation of M-phase-specific histone H1 kinase by modification of the phosphorylation of its p34cdc2 and cyclin components. Genes Dev., 4, 9–17. [DOI] [PubMed] [Google Scholar]
- Poon R.Y., Yamashita, K., Adamczewski, J.P., Hunt, T. and Shuttleworth, J. (1993) The cdc2-related protein p40MO15 is the catalytic subunit of a protein kinase that can activate p32cdk2 and p34cdc2. EMBO J., 12, 3123–3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rempel R.E., Sleight, S.B. and Maller, J.L. (1995) Maternal Xenopus Cdk2–cyclin E complexes function during meiotic and early embryonic cell cycles that lack a G1 phase. J. Biol. Chem., 270, 6843–6855. [DOI] [PubMed] [Google Scholar]
- Russell P. and Nurse, P. (1987) Negative regulation of mitosis by wee1+, a gene encoding a protein kinase homolog. Cell, 49, 559–567. [DOI] [PubMed] [Google Scholar]
- Skowyra D., Craig, K.L., Tyers, M., Elledge, S.J. and Harper, J.W. (1997) F–box proteins are receptors that recruit phosphorylated substrates to the SCF ubiquitin-ligase complex. Cell, 91, 209–219. [DOI] [PubMed] [Google Scholar]
- Solomon M.J., Glotzer, M., Lee, T.H., Philippe, M. and Kirschner, M.W. (1990) Cyclin activation of p34cdc2.Cell, 63, 1013–1024. [DOI] [PubMed] [Google Scholar]
- Solomon M.J., Lee, T. and Kirschner, M.W. (1992) Role of phosphorylation in p34cdc2 activation: identification of an activating kinase. Mol. Biol. Cell, 3, 13–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyoshima F., Moriguchi, F., Wada, A., Fukuda, M. and Nishida, E. (1998) Nuclear export of cyclin B1 and its possible role in the DNA damage-induced G2 checkpoint. EMBO J., 17, 2728–2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vojtek A.B. and Hollenberg, S.M. (1995) Ras–Raf interaction: two-hybrid analysis. Methods Enzymol., 255, 331–342. [DOI] [PubMed] [Google Scholar]
- Vojtek A.B., Hollenberg, S.M. and Cooper, J.A. (1993) Mammalian Ras interacts directly with the serine/threonine kinase Raf. Cell, 74, 205–214. [DOI] [PubMed] [Google Scholar]
- Westendorf J.M., Swenson, K.I. and Ruderman, J.V. (1989) The role of cyclin B in meiosis I. J. Cell Biol., 108, 1431–1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Bardes, E.S.G., Moore, J.D., Brennan, J., Powers, M.A. and Kornbluth, S. (1998) Control of cyclin B1 localization through regulated binding of the nuclear export factor CRM1. Genes Dev., 12, 2131–2143. [DOI] [PMC free article] [PubMed] [Google Scholar]