Abstract
Background
We have previously demonstrated that alcohol exposure at early neurulation induces growth retardation, neural tube abnormalities, and alteration of DNA methylation. To explore the global gene expression changes which may underline these developmental defects, microarray analyses were performed in a whole embryo mouse culture model that allows control over alcohol and embryonic variables.
Result
Alcohol caused teratogenesis in brain, heart, forelimb, and optic vesicle; a subset of the embryos also showed cranial neural tube defects. In microarray analysis (accession number GSM9545), adopting hypothesis-driven Gene Set Enrichment Analysis (GSEA) informatics and intersection analysis of two independent experiments, we found that there was a collective reduction in expression of neural specification genes (neurogenin, Sox5, Bhlhe22), neural growth factor genes [Igf1, Efemp1, Klf10 (Tieg), and Edil3], and alteration of genes involved in cell growth, apoptosis, histone variants, eye and heart development. There was also a reduction of retinol binding protein 1 (Rbp1), and de novo expression of aldehyde dehydrogenase 1B1 (Aldh1B1). Remarkably, four key hematopoiesis genes (glycophorin A, adducin 2, beta-2 microglobulin, and ceruloplasmin) were absent after alcohol treatment, and histone variant genes were reduced. The down-regulation of the neurospecification and the neurotrophic genes were further confirmed by quantitative RT-PCR. Furthermore, the gene expression profile demonstrated distinct subgroups which corresponded with two distinct alcohol-related neural tube phenotypes: an open (ALC-NTO) and a closed neural tube (ALC-NTC). Further, the epidermal growth factor signaling pathway and histone variants were specifically altered in ALC-NTO, and a greater number of neurotrophic/growth factor genes were down-regulated in the ALC-NTO than in the ALC-NTC embryos.
Conclusion
This study revealed a set of genes vulnerable to alcohol exposure and genes that were associated with neural tube defects during early neurulation.
Background
Children born to women who drink heavily during pregnancy are at risk for various developmental disorders, collectively called Fetal Alcohol Spectrum Disorder (FASD). Fetal Alcohol Syndrome (FAS) is a severe form of FASD in which the affected child is diagnosed with growth retardation, abnormal central nervous system development (typically including microencephaly), and a characteristic pattern of abnormal facial features [1-4]; organ dysmorphology, particularly of the eye and heart, may be evident in FAS cases as well [5,6]. Disruption of complex molecular cascades that regulate embryonic morphogenesis likely are responsible for the teratogenic effects of alcohol. Potential mechanisms include metabolic stress, reduced signaling by transcription factors, retinoic acid or growth factors, disrupted cell-cell interactions, impaired cell proliferation, and apoptosis [7-16]. Several of these mechanisms may have direct roles in causing the cell death and growth retardation in multiple systems, including brain and head (for review see [17]).
Expression of a number of genes during development was reported to be affected by alcohol in different experimental paradigms, including homeobox genes such as Msx2 [18] and sonic hedgehog [19,20], neurotrophic molecules (e.g. ADNP gene [21]), fetal liver kinase 1 (Flk1) [22]), retinol-related genes (e.g. Crabp1 and Fabp4; [20]), nucleotide excision repair gene, (Ercc6l) [23], stress-related genes (e.g. heat shock protein 47 [24]), and differentiation and apoptosis genes such as Timp4, Bmp15, Rnf25, Akt1, Tulp4, Dexras1 [25]. These altered genes suggest potential mechanisms for the abnormal development in FASD. However, the wide-ranging developmental abnormalities in FASD are likely a consequence of the interaction of multiple genes. Examination of global gene expression provides a holistic view of genes that potentially interact and collaboratively contribute to the abnormal development. Alcohol exposure induced changes in a group of cellular adhesion genes (e.g. L1cam and integrin) in neuroblastoma cells [26]. A brief ethanol exposure (3 h) at gestation day 8 (E8) in mouse embryos altered expression of genes of metabolic, cell programming and cytoskeletal signaling pathways [27]. An earlier alcohol exposure at E6-E8 also altered a set of genes related to PLUNC, neurofilament, and pale ear [28].
In animal models of prenatal alcohol exposure, sources of variability include the pattern, concentration, amount, and developmental stage of alcohol exposure, maternal stress, embryonic growth and maturation of embryos between litters and even within a given litter and within inbred strains of mice [29]. Control of all these variables in rapidly developing embryos is virtually unattainable in vivo. To limit these variables, a whole embryonic culture [30,31] was adopted, including stage alignment based on somite number, in which the pattern, amount and concentration of alcohol and embryonic staging were controlled. Inbred C57BL/6 mice, with known susceptibility to ethanol teratogenesis [32,33], were used for this study.
Differences in the dose and timing of alcohol exposure are known contributors to variation in the phenotypic spectrum in FASD. Understanding the pattern of gene alterations that co-vary with different outcomes produced by different alcohol doses or developmental timing of exposure would provide valuable insights into mechanisms underlying this phenotypic variability. As development is highly dynamic throughout gestation, we asked how alcohol exposure might affect genome-wide gene expression at the critical stage of neurulation (E8-10), when the nervous system (and other major organs) are actively forming in mouse. We have shown that at this key stage, neural tube formation was highly sensitive to the alcohol insult [29]. DNA methylation was altered, with the degree of change commensurate with severity of neural tube defect [34]. In the current study, in an initial experiment, cluster analysis indicated distinct differences in gene expression not only between control- and alcohol-treated embryos, but also between two phenotypic subsets of alcohol-treated embryos discernable at the end of alcohol treatment, one group which had a closed neural tube (ALC-NTC) and the other group with an open neural tube (ALC-NTO). A second study with a larger set of arrays was then performed in which alcohol-treated embryos of both neural tube phenotypes were specifically compared. We report here the correlation of alcohol-induced embryonic growth retardation and neural tube abnormalities with changes in expression in networks of genes known to regulate embryonic growth, organ development, and neural specification processes.
Results
Embryonic Growth Retardation/Abnormalities
As was seen in our previous report [29], the size and somite number varied (from 1-6) among embryos within a litter at the time of harvesting from the mother. We selected embryos of similar developmental stages (3-5 somites) and randomly assigned them to the two treatment groups (alcohol or control). The alcohol concentration profile of the culture media over the 46 hours was similar to that in our previous report [29]. The concentration of ethanol in the medium was ~88 mM at the start of each day (when first added to the media) and declined to ~44 mM by the end of each day. Among all cultured embryos, more than 95% maintained active heartbeats and blood circulation over this time, and only those were used for analysis. Development of the heart, caudal neural tube, brain vesicles, optic system, and limb buds in the embryos were significantly compromised in the alcohol treated group (Table 1). Brain vesicle development was retarded and the brain vesicles were smaller in size in the alcohol group. The significant effects in multiple organs and regions and in total scores (Table 1) demonstrated that alcohol treatment resulted in retardation of the overall growth and interfered with development of several specific structures, including brain, heart, and limb development, in this embryonic culture model.
Table 1.
Embryonic dysmorphology after alcohol exposure, scored according to Maele-Fabry et al,1992.
| Region | Control | Alcohol | ALC-NTC | ALC-NTO |
|---|---|---|---|---|
| Allantois | 3 ± 0 | 2.80 ± 0.08 | 2.86 ± 0.10 | 2.70 ± 0.14 |
| Branchial bars | 2.77 ± 0.09 | 2.15 ± 0.21 | 2.17 ± 0.26 | 2.11 ± 0.39 |
| Brain: Forebrain | 4.76 ± 0.10 | 3.81 ± 0.27* | 4.57 ± 0.14 | 2.29 ± 0.29** ^^ |
| Brain: Midbrain | 4.52 ± 0.11 | 3.71 ± 0.27 | 4.50 ± 0.14 | 2.14 ± 0.14** ^^ |
| Brain: Hindbrain | 4.71 ± 0.10 | 3.86 ± 0.24* | 4.50 ± 0.14 | 2.57 ± 0.30** ^^ |
| Caudal Neural Tube | 4.76 ± 0.12 | 4.11 ± 0.19* | 4.09 ± 0.26* | 4.14 ± 0.26* |
| Flexion | 4.80 ± 0.09 | 4.33 ± 0.19 | 4.59 ± 0.19 | 3.81 ± 0.36* |
| Heart | 4.80 ± 0.10 | 4.10 ± 0.16** | 4.15 ± 0.19* | 4.00 ± 0.31* |
| Limb: Forelimb | 2.01 ± 0.06 | 1.51 ± 0.13** | 1.48 ± 0.18* | 1.57 ± 0.20 |
| Limb: Hindlimb | 0.53 ± 0.10 | 0.20 ± 0.08* | 0.21 ± 0.09 | 0.19 ± 0.14 |
| Mandibular process | 2.08 ± 0.11 | 1.99 ± 0.09 | 2.12 ± 0.08 | 1.71 ± 0.18 |
| Maxillary process | 2.41 ± 0.14 | 2.06 ± 0.16 | 2.21 ± 0.18 | 1.76 ± 0.30 |
| Olfactory system | 0.47 ± 0.08 | 0.26 ± 0.08 | 0.29 ± 0.11 | 0.20 ± 0.13 |
| Optic system | 3.59 ± 0.14 | 2.87 ± 0.14** | 3.02 ± 0.17* | 2.57 ± 0.20** |
| Otic system | 3.95 ± 0.12 | 3.68 ± 0.11 | 3.88 ± 0.10 | 3.29 ± 0.18* ^ |
| Somites | 4.81 ± 0.09 | 4.38 ± 0.16 | 4.50 ± 0.17 | 4.14 ± 0.34 |
| Total score | 53.97 ± 0.66 | 45.83 ± 1.54** | 49.14 ± 1.54** | 39.23 ± 1.72** ^^ |
* P-Value <0.05, ** <0.01; compared to control
^ P-Value <0.05, ^^ <0.01; compared to NTC
Control n = 21; Alcohol (all alcohol-treated, n = 21; ALC-NTC, n = 14; ALC-NTO, n = 7)
The overall growth retardation was accompanied by varying degrees of abnormality in organ system development (Figure 1). These abnormalities included an increased size of the heart and ventricular chambers, reduced size of lung buds, flattened forebrain, small/slanted eyes, abnormal tail morphology, abnormal limb web, and unfinished turning of neural axis. A reduced blood/vascular system was also evident by less vascularization in yolk sac (Table 1), and lower red coloration apparent in many blood vessels of yolk sacs and embryos in the alcohol-treated than the control embryos (Figure 2).
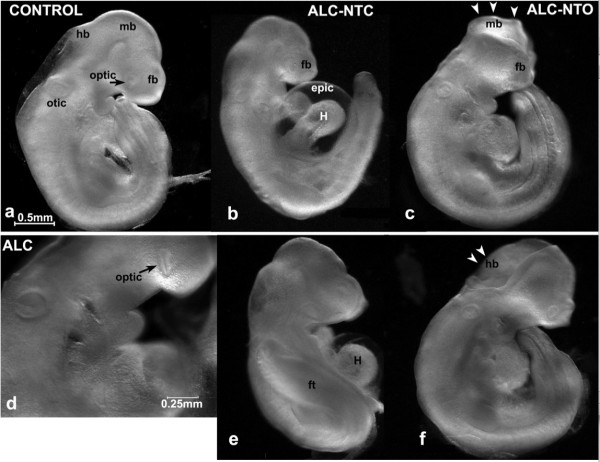
Figure 1.
Alcohol causes dysmorphology of growing embryos. Control embryos (a), Alcohol-treated (b-f). There are many dysmorphologies including microencephaly of forebrain (b, c, f), failure of closure of midbrain (mb; c) or hindbrain (hb; f), dysmorphic optical vesicle (optic; d), flex tail (ft; e) in caudal neural tube, delay formation of heart (H) chamber (b) and occasional detachment of epicardium (epic; b and e), neural tube opening at midbrain (mb; c, arrowheads) and hindbrain (hb; f, arrowheads) in the alcohol group. Majority of the brain vesicles in alcohol-treated group were closed (ALC-NTC; b, e). Approximately 30% of the embryos were found with a neural tube opening (ALC-NTO), usually in the head fold. Scale bars: a, b, c, e, f = 0.05mm; d = 0.25 mm.
Figure 2.
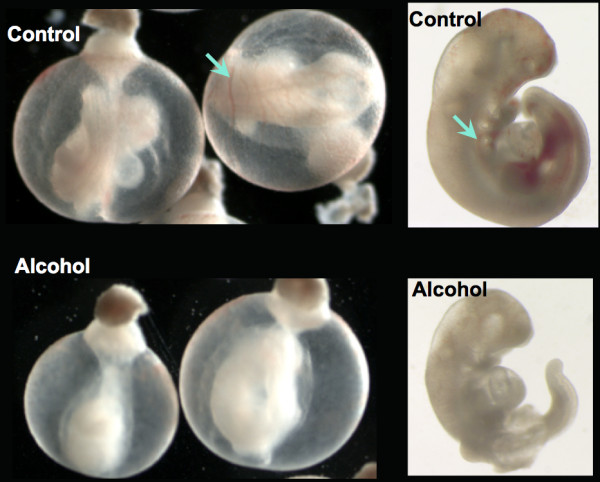
The red blood vessels were less distinguishable in the yolk sac (arrow, left) and embryo (arrow, right) in the alcohol-treated group as compared with those of the Control. All embryos examined for red blood vessels had active heart beat at the termination of experiment.
Among 127 samples of alcohol-treated embryos, 34 (27%) had various degrees of incomplete neural tube closing (Figure 1); this compares to 3 (2%) out of the 139 controls. These openings in the neural tube mostly occurred in the head fold, although delayed or incomplete neural tube closure in midbrain and hindbrain was also seen. The abnormalities and developmental delays are clearly more severe in ALC-NTO than in ALC-NTC subgroups, particularly in development of the neural axis including hindbrain, midbrain, forebrain, otic vesicle.
Differences in Gene Expression
At the end of the culture period, the total RNA extracted from alcohol-treated embryos was approximately half that of controls: controls = 2.8 ± 0.5 (n = 13), ALC-NTC = 1.6 ± 0.5 (n = 13, P < 0.05 compared with control), ALC-NTO = 1.2 ± 0.5 (n = 8, P < 0.05 compared with control). In Experiment 1, 14,243 out of 22,690 probe sets (62.7%) were present in at least half of the samples in either control or alcohol treated groups. Hierarchical clustering by arrays (Figure 3. Exp 1) clearly separated the samples into three groups, control, ALC/NTC, and ALC/NTO, rather than just two (ALC vs. control). In Experiment 2, 26,674 out of 45101 probe sets (59.1%) were present in at least half of the samples in either control or alcohol treated group. Again, the hierarchical cluster analysis (Figure 3. Exp 2) separated the samples into the same three groups, control, ALC/NTC, and ALC/NTO.
Figure 3.
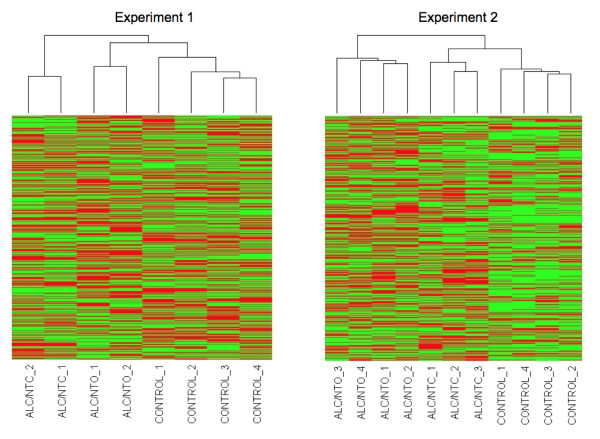
Hierarchical clustering by arrays in Experiment 1 and Experiment 2.
In Experiment 1, 850 probe sets (6% of the probe sets that were present) were differentially expressed in alcohol-treated embryos as a group (p ≤ 0.05). In Experiment 2, which had more power due to the larger number of arrays and also examined twice as many probe sets, 2519 probe sets (9.4% of the probe sets that were present) were differentially expressed in alcohol-treated embryos considered as a group (p ≤ 0.05). These relaxed stringencies were employed to reduce false negatives when comparing genes across the two experiments. The probe sets on the Mouse Genome 430A GeneChip were a subset of those on the Mouse Genome 430 2.0 GeneChip. Comparing this common subset across the two experiments, 87 probe sets were significant in both experiments and consistent in direction; because there are 13810 genes present in both experiments, the null expectation is that only 17 genes would be expected to be in common with the same direction of change. 49 probe sets were lower in alcohol-treated embryos and 38 were higher (Table 2). Among these were genes for alcohol metabolism, epigenetics (histone and histone variants), hematopoiesis, neurotrophic factors, retinol metabolism, cell cycle, cell adhesion, homeobox genes, and oncogenes.
Table 2.
Genes with changed expression†
| Gene symbol | UniGene | Source.id | Exp. 1 Fold Change | Exp. 2 Fold Change | Category/Function | Description |
|---|---|---|---|---|---|---|
| AI415282 | Mm.254704 | 1415793_at | -1.1 | -1.6 | expressed sequence AI415282 | |
| Atp6ap2 | Mm.25148 | 1439456_x_at | -1.2 | -1.2 | Energy | ATPase, H+ transporting, lysosomal protein 2 |
| BC008163 | Mm.11473 | 1425328_at | -1.2 | -1.2 | CDNA sequence BC008163 | |
| Cask | Mm.253779 | 1427692_a_at | -1.5 | -1.1 | MAGUK family | calcium/calmodulin-dependent serine protein kinase |
| Clk1 | Mm.1761 | 1426124_a_at | -1.6 | -1.3 | Cell cycle | CDC-like kinase 1 |
| Clk4 | Mm.239354 | 1427663_a_at | -1.7 | -1.3 | Cell cycle | CDC like kinase 4 |
| Cri1 | Mm.44244 | 1448406_at | -1.2 | -1.2 | DNA transcription/differentiation | CREBBP/EP300 inhibitory protein 1 |
| Cyr61 | Mm.1231 | 1416039_x_at | -1.4 | -1.6 | extracellular matrix | Cysteine rich protein 61 |
| Dach2 | Mm.79760 | 1449823_at | -1.3 | -1.3 | Myogenin | Dachshund 2 (Drosophila) |
| Ebf1 | Mm.255321 | 1416302_at | -1.5 | -1.6 | hematopoiesis | early B-cell factor 1 |
| Ebf2 | Mm.319947 | 1449101_at | -1.2 | -1.4 | hematopoiesis | early B-cell factor 2 |
| Ebf3 | Mm.30282 | 1428349_s_at | -1.5 | -1.3 | hematopoiesis | early B-cell factor 3 |
| Edil3 | Mm.41716 | 1433474_at | -1.5 | -1.4 | Homeobox | EGF-like repeats and discoidin I-like domains 3 |
| Efemp1 | Mm.44176 | 1427183_at | -1.4 | -1.9 | Neurotrophin | EGF-containing fibulin-like extracellular matrix protein 1 |
| Foxd1 | Mm.347441 | 1418876_at | -1.4 | -1.2 | Homeobox | forkhead box D1 |
| Gypc | Mm.292145 | 1423878_at | -1.2 | -1.4 | hematopoiesis | glycophorin C |
| Hist1h3a | Mm.221301 | 1422948_s_at | -1.6 | -1.3 | Epigenetic | histone 1, H3a |
| Hist1h4i | Mm.14775 | 1424854_at | -1.7 | -1.5 | Epigenetic | Histone 1, H4i |
| Hist3h2a | Mm.212549 | 1435866_s_at | -2.1 | -1.7 | Epigenetic | histone 3, H2a |
| Igf1 | Mm.268521 | 1419519_at | -1.4 | -1.3 | Neurotrophin | insulin-like growth factor 1 |
| Lgals1 | Mm.43831 | 1419573_a_at | -1.4 | -1.7 | Angiogenesis/neural development | Lectin, galactose binding, soluble 1 |
| Mageh1 | Mm.6890 | 1422498_at | -1.3 | -1.3 | Oncogene | Melanoma antigen, family H, 1 |
| Myct1 | Mm.33762 | 1452072_at | -1.3 | -1.3 | Oncogene | myc target 1 |
| Napb | Mm.274308 | 1423172_at | -1.6 | -1.4 | Synapsis | N-ethylmaleimide sensitive fusion protein beta |
| Ndrg1 | Mm.30837 | 1423413_at | -1.9 | -1.8 | Cell cycle | N-myc downstream regulated gene 1 |
| Peli1 | Mm.28957 | 1417371_at | -1.2 | -1.1 | Kinase | Pellino 1 |
| Pim1 | Mm.328931 | 1435872_at | -1.3 | -1.3 | hematopoiesis | proviral integration site 1 |
| Ppox | Mm.300006 | 1416618_at | -1.3 | -1.3 | hematopoiesis | protoporphyrinogen oxidase |
| Ppp1r14a | Mm.2343 | 1418086_at | -1.1 | -1.3 | signal transduction | Protein phosphatase 1, regulatory subunit 14A |
| Ptx3 | Mm.276776 | 1418666_at | -1.5 | -1.7 | plasma proteins | pentaxin related gene |
| Rab11a | Mm.1387 | 1449256_a_at | -1.1 | -1.2 | Oncogene | RAB11a, member RAS oncogene family |
| Rbp1 | Mm.302504 | 1448754_at | -1.2 | -1.2 | Retinol metabolism | retinol binding protein 1, cellular |
| Rpl13a | Mm.180458 | 1433928_a_at | -1.1 | -1.1 | Synthesis | ribosomal protein L13a |
| Rpl17 | Mm.276337 | 1453752_at | -1.3 | -1.2 | Synthesis | ribosomal protein L17 |
| Skil | Mm.15406 | 1422054_a_at | -1.6 | -1.4 | Oncogene | SKI-like |
| Sncg | Mm.282800 | 1417788_at | -4.4 | -1.5 | Oncogene | synuclein, gamma |
| Stmn2 | Mm.29580 | 1423281_at | -1.7 | -1.8 | Neural specification | Stathmin-like 2 |
| Stmn3 | Mm.2319 | 1460181_at | -1.7 | -1.7 | Neural specification | Stathmin-like 3 |
| Syap1 | Mm.44207 | 1416472_at | -1.2 | -1.1 | Synapsis | Synapse associated protein 1 |
| Timp3 | Mm.4871 | 1419089_at | -1.3 | -1.3 | Tissue inhibitor of metalloproteinase 3 | |
| Ube2b | Mm.280233 | 1423107_at | -1.1 | -1.1 | Epigenetic | ubiquitin-conjugating enzyme E2B, RAD6 homology |
| Vcam1 | Mm.76649 | 1448162_at | -1.3 | -1.3 | Cell adhesion | Vascular cell adhesion molecule 1 |
| 1110008H02Rik | Mm.28311 | 1436506_a_at | -1.3 | -1.3 | Energy | RIKEN cDNA 1110008H02 gene |
| 2010011I20Rik | Mm.30013 | 1424695_at | -1.2 | -1.6 | RIKEN cDNA 2010011I20 gene | |
| 2310034L04Rik | Mm.41891 | 1426416_a_at | -1.2 | -1.2 | RIKEN cDNA 2310034L04 gene | |
| 5033414D02Rik | Mm.275511 | 1460361_at | -1.2 | -1.3 | RIKEN cDNA 5033414D02 gene | |
| 5230400G24Rik | Mm.139176 | 1451572_a_at | -1.3 | -1.1 | RIKEN cDNA 5230400G24 gene | |
| 5730420B22Rik | Mm.28129 | 1427050_at | -1.4 | -1.4 | RIKEN cDNA 5730420B22 gene | |
| A630082K20Rik | Mm.293175 | 1427359_at | -1.7 | -1.3 | RIKEN cDNA A630082K20 gene | |
| Acsl6 | Mm.267478 | 1451257_at | 1.2 | 1.4 | Lipid metabolism | acyl-CoA synthetase long-chain family member 6 |
| Atp1a1 | Mm.280103 | 1451071_a_at | 1.2 | 1.2 | Energy | ATPase, Na+/K+ transporting, alpha 1 polypeptide |
| AW547365 | Mm.270088 | 1433645_at | 1.2 | 1.2 | Membrane Transport | expressed sequence AW547365 |
| C78212 | Mm.27090 | 1435369_at | 1.2 | 1.3 | Expressed sequence C78212 | |
| Cad | Mm.305535 | 1452830_s_at | 1.2 | 1.3 | Amino acid metabolism | carbamoyl-phosphate synthetase 2, aspartate transcarbamylase, and dihydroorotase |
| Cdv3 | Mm.261025 | 1415704_a_at | 1.2 | 1.2 | Lipid metabolism | carnitine deficiency-associated gene expressed in ventricle 3 |
| Clstn1 | Mm.38993 | 1421861_at | 1.2 | 1.2 | Cell adhesion | Calsyntenin 1 |
| Cpd | Mm.276736 | 1434547_at | 1.2 | 1.2 | Protease activity | carboxypeptidase D |
| E130306I01Rik | Mm.277582 | 1424419_at | 1.1 | 1.3 | RIKEN cDNA E130306I01 gene | |
| Emb | Mm.274926 | 1415856_at | 1.3 | 1.2 | Cell adhesion | embigin |
| Exosc2 | Mm.150972 | 1426630_at | 1.1 | 1.2 | RNA degradation | exosome component 2 |
| Hmga2 | Mm.157190 | 1450780_s_at | 1.2 | 1.1 | high mobility group AT-hook 2 | |
| Hsd11b2 | Mm.5079 | 1416761_at | 1.4 | 1.4 | Steroid Metabolism | Hydroxysteroid 11-beta dehydrogenase 2 |
| Ide | Mm.28366 | 1423120_at | 1.2 | 1.2 | Protease activity | Insulin degrading enzyme |
| Ifrg15 | Mm.253335 | 1418116_at | 1.1 | 1.1 | interferon alpha responsive gene | |
| Ipo11 | Mm.132208 | 1428096_at | 1.2 | 1.2 | Nuclear Protein Transport | importin 11 |
| Itga6 | Mm.225096 | 1422445_at | 1.1 | 1.2 | Cell adhesion | integrin alpha 6 |
| Klf16 | Mm.41513 | 1416350_at | 1.3 | 1.3 | Alcohol metabolism | Kruppel-like factor 16 |
| Ndufs1 | Mm.290791 | 1425143_a_at | 1.1 | 1.1 | Energy | NADH dehydrogenase (ubiquinone) Fe-S protein 1 |
| Phf13 | Mm.25582 | 1455175_at | 1.2 | 1.1 | Alcohol metabolism | PHD finger protein 13 |
| Podxl | Mm.89918 | 1448688_at | 1.3 | 1.3 | hematopoiesis, kinase | Podocalyxin-like |
| Psmd3 | Mm.12194 | 1448479_at | 1.1 | 1.2 | Proteasome | Proteasome (prosome, macropain) 26 S subunit, non-ATPase, 3 |
| Ptcd1 | Mm.332840 | 1454970_at | 1.4 | 1.3 | pentatricopeptide repeat domain 1 | |
| Rhou | Mm.168257 | 1449027_at | 1.2 | 1.2 | Signal transduction | ras homolog gene family, member U |
| Rpo1-4 | Mm.135581 | 1417775_at | 1.2 | 1.1 | Synthesis | RNA polymerase 1-4 |
| Saa2 | Mm.200941 | 1419075_s_at | 1.7 | 1.7 | Lipid metabolism | serum amyloid A 2 |
| Slc27a4 | Mm.330113 | 1424441_at | 1.2 | 1.3 | Lipid metabolism | solute carrier family 27 (fatty acid transporter), member 4 |
| Trp53bp1 | Mm.215389 | 1433659_at | 1.1 | 1.2 | Cell cycle | transformation related protein 53 binding protein 1 |
| Ttr | Mm.2108 | 1454608_x_at | 2 | 1.6 | Retinol | Transthyretin |
| Ube2j1 | Mm.259095 | 1417723_at | 1.2 | 1.3 | Epigenetic | ubiquitin-conjugating enzyme E2, J1 |
| 0610007A15Rik | Mm.28122 | 1452132_at | 1.8 | 1.5 | RIKEN cDNA 0610007A15 gene | |
| 1110060D06Rik | Mm.319964 | 1430291_at | 1.3 | 1.3 | Adult male corpora quadrigemina cDNA, RIKEN full-length enriched library, clone:B230210C03 product:u | |
| 1300001I01Rik | Mm.214574 | 1428106_at | 1.2 | 1.2 | RIKEN cDNA 1300001I01 gene | |
| 1700017B05Rik | Mm.22712 | 1429758_at | 1.3 | 1.3 | RIKEN cDNA 1700017B05 gene | |
| 1700054N08Rik | Mm.157746 | 1451483_s_at | 1.4 | 1.2 | RIKEN cDNA 1700054N08 gene | |
| 4632417K18Rik | Mm.1643 | 1422628_at | 1.2 | 1.1 | RIKEN cDNA 4632417K18 gene | |
| 4930485D02Rik | Mm.293449 | 1424810_at | 1.3 | 1.3 | RIKEN cDNA 4930485D02 gene | |
| 5930416I19Rik | Mm.143908 | 1452313_at | 1.2 | 1.1 | RIKEN cDNA 5930416I19 gene |
†P < 0.05
Furthermore, in Experiment 2 (which had more power to detect differences), a number of genes in addition to the above list were present in the controls but were absent in the alcohol treated samples (Table 3). Notably, glycophorin A (Gypa) and beta-2 microglobulin (B2m) genes were absent in ALC-NTO, and ceruloplasmin (Cp), adducin 2 (Add2), B2 m, and ceruloplasmin (Cp) genes were absent in ALC-NTC. All of these are critical in hematopoiesis and/or red blood cell function [35-39]. In contrast, the aldehyde dehydrogenase 1 family, B1 (Aldh1b1), which catalyzes oxidation of retinaldehyde, was present only in the alcohol-treated embryos with open neural tubes (ALC-NTO) (Table 3 last row). No gene was found to be absent in Control but present in ALC-NTC. Another retinol regulating gene, cellular retinol binding protein 1 (Crbp1), was reduced by alcohol exposure (Table 2).
Table 3.
Genes in Experiment 2 that are turned on or off by alcohol treatment.
| Gene Symbol | Genbank | p-value | Change in Alcohol-treated | Description |
|---|---|---|---|---|
| Alc-NTC | ||||
| Add2* | NM_013458 | 0.0196 | Off | Adducin 2 (beta) |
| B2m* | NM_009735 | 0.0446 | Off | Beta-2 microglobulin |
| Cfi | NM_007686 | 0.0494 | Off | Complement component factor i |
| Cp* | NM_007752 | 0.0225 | Off | Ceruloplasmin |
| Fbxo2 | NM_176848 | 0.0290 | Off | F-box only protein 2 |
| Gch1 | NM_008102 | 0.0017 | Off | GTP cyclohydrolase 1 |
| Gfi1b | NM_008114 | 0.0007 | Off | Growth factor independent 1B |
| Nppb | NM_008726 | 0.0103 | Off | Natriuretic peptide precursor type B |
| Pitpnm1 | NM_008851 | 0.0272 | Off | Phosphatidylinositol membrane-associated 1 |
| Ppgb | NM_008906 | 0.0044 | Off | Protective protein for beta-Galactosidase |
| Tacr2 | NM_009314 | 0.0108 | Off | Tachykinin receptor 2 |
| ALC-NTO | ||||
| Acbd5 | NM_028793 | 0.0292 | Off | Acyl-Coenzyme A binding domain containing 5 |
| B2m* | NM_009735 | 0.0446 | Off | Beta-2 microglobulin |
| Fbxo2 | NM_176848 | 0.0290 | Off | F-box only protein 2 |
| Frmd3 | NM_172869 | 0.0004 | Off | FERM domain containing 3 |
| Gypa* | NM_010369 | 0.00001 | Off | Glycophorin A |
| Mlr1 | BB298201 | 0.0295 | Off | Mblk1-related protein-1 |
| Ogn | NM_008760 | 0.0363 | Off | Osteoglycin |
| Pdcd4 | BG230003 | 0.0468 | Off | Programmed cell death 4 |
| Sqstm1 | NM_011018 | 0.0357 | Off | Sequestosome 1 |
| Aldh1b1 | NM_028270 | 0.0048 | On | Aldehyde dehydrogenase 1 family, member B1 |
Off = present in ≥ 75% of arrays in control and in no arrays of alcohol-treated samples.
On = not present in any control, present in ≥ 75% of arrays from alcohol-treated.
Only named genes are shown. * = hematopoiesis gene.
P-value: as compared with Control.
Gene Set Enrichment Analysis (GSEA) Analyses
Four GSEA analyses were conducted within each experiment: control versus all alcohol-treated (ALC), control versus ALC-NTC, control versus ALC-NTO, and ALC-NTC versus ALC-NTO. As 415 GO gene sets and 191 stem cell related gene set were pre-selected, there were totally 4 × (415+191) = 2424 GSEA tests. We found 15 gene sets that were significant at 5% and shared the same enrichment direction in both experiments. By chance, one would expect only 2424 × (0.05 × 0.05 × 0.5) = 3; therefore, the FDR is 3/15 = 20%. The significant gene sets common to the two experiments are outlined below.
a. Early Developmental Biology Gene Sets
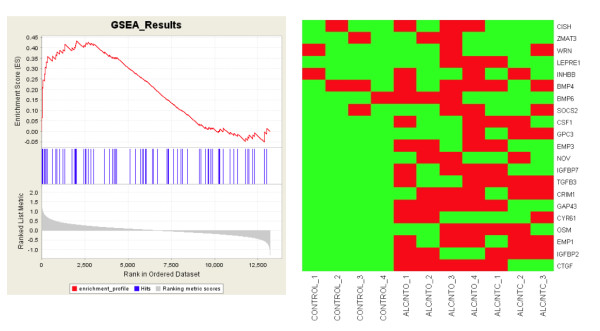
GSEA analysis using the GO biological function categories selected as being related to development (Additional files 1 and 2.) identified 20 enriched sets in Experiment 2. Of these 20 sets, 9 were also identified by Experiment 1 (Table 4). Included in these shared gene sets are multiple GO categories related to growth, eye and heart development, and epigenetics. When comparing the control embryos to all alcohol treated embryos, there were 7 GO categories that were enriched in the control groups (i.e., down-regulated in the alcohol-treated groups): five growth-related GO sets, one epigenetics (histone and chromatin regulator) GO set, and one angiogenesis GO set (Table 4). No gene set was enriched in the alcohol-treated group. An example of gene enrichment analysis is shown in Figure 4 for GO:0040007, Growth. This gene set contained 75 genes. The GSEA p-values for this enrichment score were 0.010 in Experiment 1 and 0.005 in Experiment 2.
Table 4.
GSEA for Early Developmental Biology GO sets.
| Comparison | Keyword | Gene Set | Gene Set Description | Size | p-value Exp. 2 |
Significant Genes | p-value Exp. 1 |
|---|---|---|---|---|---|---|---|
| Control vs ALC-NTO/ALC-NTC (see legends) |
Growth, Growth Regulation* |
GO:0016049 GO:0040007 |
^Cell growth ^Growth |
47 75 |
0.002 0.005 |
(Ctgf, Igfbp2, Emp1, Osm, Cyr61, Gap43, Crim1, Tgfb3, Igfbp7, Nov, Emp3), Gpc3, Csf1, Socs2, Bmp6, Bmp4, Inhbb, Lepre1, Wrn, Wig1, Cish |
0.010 0.010 |
|
GO:0001558 GO:0040008 |
Regulation of cell growth Regulation of growth |
39 56 |
0.002 0.016 |
(Ctgf, Igfbp2, Osm, Cyr61, Gap43, Crim1, Igfbp7, Nov), Gpc3, Csf1, Socs2, | 0.006 0.017 |
||
| GO:0005520 | Insulin-like growth factor binding | 14 | 0.000 | Ctgf, Igfbp2, Cyr61, Crim1, Igfbp7, Nov | 0.012 | ||
| Heart* | GO:0001525 | Angiogenesis | 53 | 0.022 | Ctgf, Anxa2, Cyr61, Thbs1, Vegfa, Tie1, Elk3, Flt1, Crhr2, Vegfc, Kdr, Bmp4, Adra2b, Tnfrsf12a | 0.022 | |
| Eye# | GO:0001654 | Eye Development | 26 | 0.040 | Mab21l1, Neurod1, Neurod4, Ntrk2, Fkbp8, Bmpr1b, Crb1, Stat3, Tspan5, Pax6, Bmp4, Map3k1 | 0.004 | |
| Epigenetic factor~,^ | GO:0006334 | Nucleosome modeling | 30 | 0.021 | Hist3h2b, a; Hist1h3f; Hist1h1c; Hist1h2b, c; Hist1h3a; H1f0; Smarca2; Nap1l3 | 0.033 | |
| ALC-NTO vs ALC-NTC | Growth, Growth retardation | GO:0007173 | Epidermal growth factor receptor (EGFR) signaling pathway | 5 | 0.019 |
Pde6g, Egfr, Hbegf (Enriched in ALC-NTC) |
0.023 |
^= Gene set reduced in ALC-NTO as compared with Control.
*= Gene sets reduced in ALC (all alcohol group) and ALC-NTC as compared with Control.
~= Gene set reduced in ALC as compared with Control
#= Gene set reduced in ALC-NTC as compared with Control.
( ) = genes in Cell growth related GO set, which is included in Growth GO set; or genes in Regulation of Cell growth related GO set, which is included in Regulation of Growth GO set.
Figure 4.
Illustration of Gene Set Enrichment Analysis (GSEA) informatics with neurotrophic factor related gene set. (Left panel) Profile of the running enrichment score (ES) and positions of a prominent neurotrophic factor related gene set, GO:0040007: Growth, on the rank ordered list GSEA output for the comparison ALC vs. CONTROL. This test is a one-way test, i.e. whether gene expression is higher in control than in ALC. The x-axis lists all the genes ranked based on their associations with phenotype, i.e. the comparison ALC vs. CONTROL. The blue vertical bars indicate candidate genes in the target gene set. The ES profile records the cumulative score of the gene ranks from the target gene set. If a majority of gene ranks from the candidate gene set are high (i.e. toward the start of ranking) compared to the rest of genes, the cumulative ranking score (profile) will have a high peak, suggesting a significant enrichment of this gene set. The statistical significances (p-value) were calculated based on the height of this peak through a permutation test (p-value = 0.010 in Experiment 1 and 0.005 in Experiment 2). (Right panel) The significant genes (enriched in control) are determined by the position of the peak of the profile. There are 21 candidate genes up to this peak position which are claimed as significant. They are plotted in the Heatmap (green means high expression level, and red means low expression level) in Experiment 2.
The growth-related genes represented the largest group of affected genes. There were 5 GO sets of growth-associated genes (Table 4). Many of these genes, identified by GSEA in both experiments, were also identified in Experiment 2 at the single gene level; e.g. the Growth gene set (GO:0040007): Ctgf, Igfbp2, Emp1, Osm, Cyr61, Gap43, Crim1, Tgfb3, Nov, Socs2, and Wrn were significantly reduced in Experiment 2, and Igfbp7, Emp3, Bmp4, Bmp6, Inhbb, Wig1, and Cish were reduced but did not reach the criteria for significance. The additional growth genes in Epidermal growth factor receptor (EGFR) signaling pathway GO group appear to be reduced to a greater extent in ALC-NTO than in ALC-NTC (Table 4).
b. Stem Cell Related Gene Sets
Three gene sets were enriched in the control embryos compared to the combined alcohol-treated embryos (i.e., down-regulated in the alcohol-treated group): TGF-Beta activin-responsive genes (important for maintenance of pluripotency and embryonic stem cells [40]), extracellular matrix (ECM) molecules, and ECM protease inhibitors (Table 5). Three gene sets were down-regulated in the ALC-NTC subgroup (i.e., enriched in the control group): other related growth factors (Neural specification), other regulators of cell differentiation, and ECM protease inhibitors (Table 5). Two gene sets were down-regulated in the ALC-NTO group (enriched in the control group): other related growth factor and other ECM molecules (Table 5). There were no significant gene sets in comparisons between ALC-NTC and ALC-NTO. No gene set was enriched in any alcohol-treated group.
Table 5.
GSEA for Stem Cell Related Gene Sets
| Comparison | Gene Set | Size | p-value Exp. 2 |
Significant Genes | p-value Exp. 1 |
|---|---|---|---|---|---|
| Control vs ALC | Other ECM Molecules~^ | 9 | 0.002 | Ctgf, Thbs2, Tgfbi, Ecm1 | 0.016 |
| ECM Protease Inhibitors~,# | 7 | 0.004 | Thbs1, Timp3 | 0.006 | |
| TGF-β Activin-responsiv~ e | 16 | 0.010 | Junb, Fos, Tgfbi, Pdgfb, Tgfb1i1, Igf1 | 0.014 | |
| Other Regulators of Cell Differentiation (Neural Specification) # | 17 | 0.003 | Elavl3, Neurod1, Neurod4, Nhlh1, Neurog1, Nhlh2, Neurog3, Spock2, Neurog2 | 0.004 | |
|
Other Related growth Factor^ Other Related growth Factor# |
7 | 0.005 0.040 |
Ctgf, Igf1 Ctgf Hgf Igf1 |
0.015 0.008 |
~= Gene set is reduced in ALC (all alcohol group) as compared with Control.
^= Gene set is reduced in ALC-NTO as compared with Control.
#= Gene set is reduced in ALC-NTC as compared with Control.
ECM: extracellular matrix
Validation by Quantitative RT-PCR
Quantitative RT-PCR (qRT-PCR) was used to verify some of the genes that were significantly affected by alcohol, including a sample of genes from the functional gene sets for neural specification and trophic factors identified in GSEA (Tables 6 and 7). These studies used independent embryos subjected to identical ethanol exposure. The qRT-PCR verified that all 11 down-regulated neural specification genes (Table 6) and neurotrophic/growth factor genes (Table 7) tested differed in the same direction. One gene (Mylc2) that did not differ in the microarray experiments was also tested and the lack of difference was confirmed.
Table 6.
RT-PCR confirmation of differences in gene expression: Neural specification genes from Experiment 1.
| Microarray | qRT-PCR | |||||
|---|---|---|---|---|---|---|
| Gene | GenBank | Fold Change* | p-value* | Fold Change | p-value | Description |
| Bhlhb5 | NM_021560 | -1.6 | 0.0021 | -2.3 | 0 | basic helix-loop-helix domain, class B5 |
| Ngn2 | BC055743 | -2 | 0.0015 | -2 | 0 | neurogenin 2 |
| Ngn1 | NM_010896 | -1.3 | 0.047 | -1.7 | 0.021 | neurogenin 1 |
| Sox5 | AI528773 | -1.4 | 0.015 | -1.7 | 0.005 | SRY-box containing gene 5 |
| Mylc2 | NM_023402 | -1 | 0.8 | -1 | 0.19 | myosin light chain |
*Control versus alcohol-treated
Table 7.
RT-PCR confirmation of differences in gene expression: Growth/neurotrophic factor genes from Experiment 2.
| ALC-NTC | ALC-NTO | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Microarray | qRT-PCR | Microarray | qRT-PCR | ||||||
| Gene symbol | GenBank | Fold Change* | p-value* | Fold Change* | p-value* | Fold | p-value* | Fold Change* | p-value* |
| Ctgf | NM_010217 | -1.7 | 0.01 | -1.2 | 0.300 | -2.2 | 0.004 | -2.0 | 0.048 |
| Edil3 | NM146015 | -1.6 | 0.00 | -1.4 | 0.019 | -1.9 | 0.025 | Not tested | |
| Efemp1 | NM_146015 | -1.5 | 0.01 | -2.3 | 0.002 | -2.3 | 0.001 | -1.8 | 0.010 |
| Igf1 | NM_184052 | -1.4 | 0.02 | -2.6 | 0.001 | -1.3 | 0.060 | Not tested | |
| Igfbp2 | NM_008342 | -2.0 | 0.02 | -1.6 | 0.084 | -1.5 | 0.035 | -2.0 | 0.045 |
| Ntf3 | NM_008742 | -1.2 | 0.30 | -1.7 | 0.060 | -1.8 | 0.030 | -1.9 | 0.038 |
| Tieg1 | NM_013692 | -1.2 | 0.36 | -1.3 | 0.015 | -1.5 | 0.010 | -1.2 | 0.053 |
*Control versus alcohol-treated
Fold change: positive = ratio of alcohol-treated to control, negative = ratio of control to alcohol-treated. p-value: t-test of control vs. alcohol treated
Discussion
1. Developmental Deficits and Correlation with Gene Expression Profiles
The abnormal embryonic development resulting from the alcohol treatment at this specific stage of development (Figure 1; Table 1) was consistent with our previous report [29] and those of others [41,42]. Two different facets of abnormal development could be identified: growth delay and frank teratogenesis. Delays in growth were also evident by the significant reductions in the total RNA per embryo and in the delayed morphological staging (Table 1). The affected structures were derived from each of the three germ layers, i.e., neural tube and brain vesicles (ectoderm), somites and cardiovascular system (mesoderm/endoderm), and involved a wide range of tissues and organs (e.g., heart, head, limbs). Alterations in all of these have been observed in FAS cases. The teratogenic consequences were evident as dysmorphology of various organs (central nervous system, eye, and heart) that involved pathogenic effects beyond just the observed delay of the normal course of development. Examples include enlarged heart primordium and abnormally enlarged ventricular chambers, detached pericardial sac, small forebrain, flat telencephalic vesicle, failure in neural tube closure, and small and irregularly shaped eyes.
Neural tube defect
We observed in Experiment 1 that gene expression profiles from alcohol treatment of embryos in this controlled culture system yielded two distinguishable patterns; comparison to the morphological data revealed that these were correlated with two different phenotypes: open (ALC-NTO) and closed neural tubes (ALC-NTC). The phenotypes and correlated gene expression differences were reproduced in Experiment 2. The embryos with open neural tubes (ALC-NTO) had more severe delays in brain and otic development than those with closed neural tubes (ALC-NTC) (Table 1). These different phenotypes are consistent with our previous in vivo observation in a liquid diet model of prenatal alcohol exposure in C57BL/6 mice, which resulted in partial penetration of incomplete neural tube closure (as late as embryonic day 15) and a cascade of deficits in midline structural development [43]. Finding this difference in development in experimentally controlled culture conditions indicates either a stochastic event or that an extremely sensitive gene-environment interaction is involved, e.g. different outcomes based on small differences in developmental stage at the time of exposure or small differences in tissue concentrations of alcohol across embryos. We have recently found greater DNA hypermethylation in ALC-NTO than in ALC-NTC embryos, particularly in genes on chromosomes 7, 10, and X. Remarkably, there was a >10 fold increase in the number of hypermethlyated genes on chromosomes 10 and X in ALC-NTO than ALC-NTC [34].
Both the ALC-NTC and the ALC-NTO embryos demonstrated lower expression of genes in sets related to cell growth, growth factors, heart (angiogenesis), and eye (in NTC vs. Control) (Table 4; Table 7). The ALC-NTC and ALC-NTO embryos also differed in other sets of functionally related genes. The histone gene set was selectively reduced in ALC-NTO compared to controls. The epidermal growth factor signaling pathway genes were lower in ALC-NTO than ALC-NTC (Table 4). At the single gene analysis level, Experiment 2 showed a greater number of neurotrophic/growth factor genes were down-regulated in ALC-NTO than in ALC-NTC groups, particularly in the TGFβ, NTF3, S100, and EGF families. These differences in gene expression between the ALC-NTO and ALC-NTC embryos appear to be correlated with the more severe teratogenic trajectory of the ALC-NTO group, but causal relationships have yet to be established.
The neural tube abnormality may either be a delay in neural tube closure or a neural tube defect. In either case, a delay in closing of the neural tube is associated with deficits in midline brain development due to disruption of the timing of critical events of early brain development. At more mature stages, such midline deficits include craniofacial abnormalities, corpus callosum, olfactory bulb, cerebellum, and raphe neuron formation [43-50].
2. Patterns of Gene Expression
A. Temporal patterns
Green and colleagues [27] reported that a 3 to 4 h binge-like alcohol exposure, with blood alcohol concentration 300 to 400 mg/dL at E8, produced a major abnormality in craniofacial and eye development in C57BL/6 mice at E15 or E17 (effects in the C57BL/6J substrain were greater than in the C57BL/6N substrain). Alterations of gene expression were reported to occur within hours of alcohol exposure at E8; these genes included metabolic and cellular gene, down-regulated ribosome and proteasome pathways; upregulated glycolysis and pentose phosphate, tight junction, and Wnt signaling pathways, as well as other cellular profile genes. In another study, a comparable high dose of alcohol exposure at an earlier stage, E6-E8, produced growth retardation, abnormal tail torsion, open neural tube, reduction of somite number, and other malformations [28]. The altered gene expression at E10 included cytoskeletal (Neurofilament), signal transduction (Zinc finger protein, MAP kinase related, Transcription factor Nf2l2), and metabolic genes (lactate dehydrogenase, Aldolase 1). In the current study, a similar dose of alcohol exposure at the stage of neurulation (E8-10) produced a major neural and cardiovascular retardation and other organ system abnormalities. The trends of gene expression are consistent with the observed developmental delay and growth retardation in FASD. Among the genes with reduced expression in the alcohol-treated embryos were those involved in growth retardation, neural development, heart and hematopoiesis, and epigenetics. Among the identified functionally related gene sets, the most notable effect was the down regulation of growth-related genes, which represented the largest group of affected genes (Table 4). These genes provide plausible candidates for mechanistic links to the observed embryonic growth retardation.
B. Neural specification genes
Expression of neural specification genes (Table 5 and 7) and neurotrophic/growth factor genes (Table 4 and 7) was also reduced by the ethanol exposure. These participate in neuronal specification, neural stem cell differentiation, and neural fate determination [51-55]. Suppression of these genes predicts a downstream reduction in the early formation of neural cells. Null neurog 1 (Ngn1) or neurog 2 (Ngn2) leads to sensory abnormality [56,57]). These differential expression of neuronal specification/patterning genes together with neurotrophic genes supports the dysmorphism and developmental delay of neural tube and fore-to mid-brain formation. The Igf1 and EGF genes were also identified by a microarray study with 3 h alcohol treatment [27] indicating they are altered early after ethanol exposure. The down-regulation of these neural specification and neural trophic/growth factor genes may play a major role in the neurodevelopmental deficit observed in the current study and featured in FASD.
C. Genes related to other organ defects
Although heterogeneity of tissue arising from use of whole embryos might have masked some changes in specific tissues, two functional gene sets, optic vesicle and the heart (Table 4), were identified and specifically linked to our observed developmental delay and abnormalities. Also, the collective down-regulation of key hematopoiesis genes that were either absent (Table 3) or reduced (Table 2) is consistent with the reduced blood circulation observed in the embryos.
D. Histone variants
Many histone genes related to epigenetic regulation of transcription were affected by ethanol (Table 4). The reduction of many histone variants would alter chromatin organization, affecting transcription at a global level [58,59]; this may be an important effect of the alcohol that leads to the reduction of total RNA and induced growth retardation. Modification of epigenetic processes is a potential mechanism by which alcohol may alter gene expression during development, and may be an important candidate mechanism for the pathophysiology of fetal alcohol syndrome.
E. Alcohol delayed or induced gene expression
Other genes that were present in the control group but absent in the alcohol-treated group (Table 3) likely reflect a delay in onset or a strong inhibition of normal expression at this stage of development. Among them, four hematopoiesis genes [glycophorin A (Gypa), adducin 2 (Add2), beta-2 microglobulin (B2m), and ceruloplasmin (Cp)] associated with blood cell formation were absent in the alcohol-treated groups; these genes are key components in the pathway of white and red blood cell formation [36,38,60-62]. The absence of these genes is in agreement with the low circulating blood cells seen in alcohol treated embryos (Figure 2). The expression of aldehyde dehydrogenase 1B1 (Aldh1b1) was induced in both of our experiments by alcohol treatment during this period of early neurulation (Table 2 last row). Because Aldh1b1 encodes an efficient enzyme for breakdown of acetaldehyde formed during metabolism of ethanol, this up-regulation is likely a detoxification response to the high level of ethanol in the environment. However, the metabolism of other substrates of this enzyme (e.g., retinoic acid, corticosteroids, biogenic amines, neurotransmitters, and lipids) that are required for normal development may be adversely affected by this increase in Aldh1b1 expression [63,64].
Conclusion
In summary, alcohol exposure during the period of early neurulation at ~E8-E10, is predominantly inhibitory to gene expression, particularly the neural developmental genes. We found major reductions in gene sets involved in neurospecification, neural growth factors, cell growth and hematopoiesis. These effects on gene expression parallel the growth delay and developmental abnormalities including brain, neural tube, eye, heart, blood cells, and embryonic vascularization which are major targets in FASD. Our study, in conjunction with others that use different developmental periods of alcohol exposure, provides an important portfolio of alcohol-induced changes in gene expression associated with altered development. Together, these gene profiles should contribute to the generation of testable new hypotheses concerning the mechanistic path from gene expression changes to embryonic structural deficits, and for causal mechanisms of alcohol-induced teratogenesis (e.g., brain growth retardation, neural tube midline deficit, craniofacial dysmorphology) in fetal alcohol spectrum disorder. Two such hypotheses emerge from the current study. The first is that alcohol causes a delay in development of the nervous system by inhibiting specific sets of genes involved in neural development (Ngn, Nhlh, Sox, Igf, Ntf, and Egf). The second is that neural tube defects are mediated by the inhibition of genes in the epidermal growth factor signaling pathway and genes encoding histone variants.
Methods
Embryonic Culture
All experimental procedures were approved by the Institutional Animal Care and Use Committee of the Indiana University School of Medicine (Indianapolis, IN) and are in accordance with the guidelines of the Institutional Animal Care and Use Committee of the National Institute on Drug Abuse, National Institutes of Health, and the Guide for the Care and Use of Laboratory Animals [65]. Two-month-old C57BL/6 mice (~20 g) were purchased from Harlan, Inc. (Indianapolis, IN). Upon arrival, breeder mice were individually housed and acclimated for at least one week before mating began. The mice were maintained on a reverse 12 h light-dark cycle (lights on: 19:00 - 07:00) and provided with laboratory chow and water ad libitum. Two females were placed with one male for two hours between 08:00 and 10:00. When a vaginal plug was detected after the mating period, it was designated as embryonic day 0 (E0). On E8.25 at 15:00, dams were sacrificed using CO2 gas. The embryos were treated at this stage, which is the beginning of neurulation. The window of 46 hrs treatment covered the stages of the formation of the major organs, neural specification and patterning. These stages are known to be vulnerable to alcohol [66].
The technique for whole embryo culture was based on the methods described by New [31]. The gravid uterus was removed and placed in sterile PBS (0.1 M phosphate buffer containing saline) at 37°C. The embryo in the visceral yolk sac along with a small piece of the ectoplacental cone (hereafter called embryo, unless otherwise stated) was carefully removed from the deciduas tissues and the Reichert's membrane in PBS containing 4% fetal bovine serum (Sigma, St Louise. MO). After removal, three embryos bearing 3-5 somites (E8.25) were incubated in a culture bottle in 20 mL of medium which consisted of 70% immediately centrifuged heat-inactivated rat serum (Harlan Sprague-Dawley, Inc, Indianapolis, IN) and 30% phosphate buffered saline (137 mM NaCl, 2.7 mM KCl, 0.5 mM MgCl2, 8 mM Na2HPO4, 1.47 mM KH2PO4, 0.9 mM CaCl2, 5.6 mM glucose, 0.33 mM sodium pyruvate, pH7.4) supplemented with 20 units/ml penicillin and 20 units/ml streptomycin (Sigma, St. Louis, MO), and gassed with 5% O2, 5% CO2, and 90% N2 in a rotating culture system (B.T.C. Precision Incubator Unit, B.T.C. engineering, Cambridge, England, 36 rpm) for 2 h. After 2 h, treatment was initiated by transferring embryos into the same medium with or without 88 mM ethanol in isotonic buffer. The bottles were gassed for an additional 20 h with 5% O2, 5% CO2, and 90% N2, and then between 22 h and 46 h with 20% O2, 5% CO2, and 75% N2. The culture medium in alcohol and control cultures was replaced with fresh medium (with or without ethanol, respectively) 22 h after the start of the treatment. In this culture system, it was previously determined that the media alcohol concentration declined from 88 mM to 44 mM over the course of the experiment. Alcohol concentrations in this range (44-88 mM) have been commonly used in whole embryo cultures to generate FAS-related structural malformations [41,42,67] in multiple strains of mice [29], and are comparable to blood alcohol concentrations produced by in vivo doses of acute ethanol injections that produce teratogenic effects in mice during this embryonic period [68]. This level, though high, is within the range attained by human alcoholics [69,70].
All cultures were terminated 46 hrs from the beginning of treatment. The concentration of ethanol in the medium was assayed at three time points on each day (0 [initial], 12, and 22 hours on the first day; at 0 [after media change], 12, and 24 hours on the second day) in a separate group of embryos not used for the analyses, to avoid the potential confounding effects of drawing samples from the cultures. Media samples from alcohol- or vehicle-treated cultures were assayed in duplicate for alcohol concentrations using an Analox alcohol analyzer (Analox Instruments USA, Lunenburg, MA).
At the end of culture, viability was confirmed by observing the blood circulation of the yolk sac and the beating heart. Cultured embryos were quickly immersed in 0.7 ml TRIzol (Invitrogen, Carlsbad, CA) and homogenized for extracting total RNA for the RT-PCR and microarray processes (see microarray section, below), or fixed in 4% paraformaldehyde in PBS for the evaluation of the developmental status.
Whole embryos were used because the dysmorphology is observed throughout tissue derived from the three germ layers and in various developing organs (e.g., head fold, caudal neural tube, heart, lung bud, somites, and limbs). Also, dissection of the millimeter size embryos would unavoidably introduce another source of variability: whole embryos yield sufficient total RNA for single embryo analysis, whereas dissected tissues yield too little RNA and would require pooling or amplification for microarray analysis. Although this limits the resolution of genes contributing to different topographic changes, we thought that obtaining a complete gene expression profile in parallel with this widespread alcohol-induced teratogenesis in the embryo would be informative.
Embryonic dysmorphology
The analysis of embryo dysmorphology was performed as described by van Maele-Fabry et al. [71] and in our previous report [29]. The morphological features of the developing embryo, including the allantois, flexion, heart, caudal neural tube, hind-brain, midbrain, forebrain, otic system, optic system, branchial bars, maxillary process, mandibular process, forelimb, hindlimb, and somites, were examined and scored for any malformations using the ordinal scales of our previous report [29]. Scores for each of the above features were typically not normally distributed, so they were analyzed statistically by the non-parametric Mann-Whitney U test. The number of somites was normally distributed, so those data were analyzed by Student's t-test, using StatView software (SAS Institute, Inc. Cary, NC).
Gene expression analyses
Two microarray experiments were performed. In Experiment 1, total RNA was isolated from individual whole embryos (4 vehicle control, 4 alcohol treated). Each embryo was immediately immersed in 700 ml TRIzol (Invitrogen) and homogenized using a Polytron. Extraction followed the TRIzol protocol. Ethanol precipitated RNA was resuspended in DEPC water. RNA was cleaned up using RNeasy mini-kit (Qiagen, Valencia, CA) The quality of RNA was assessed by the Agilent Bioanalyzer (Agilent Technologies, Waldbronn, Germany)and by spectrophotometry from 220 nm to 350 nm; concentration was determined from A260. Typical total RNA yields were 5-10 μg/embryo. Microarray analysis was performed at the Center for Medical Genomics at the Indiana University School of Medicine. Labeling and hybridization to Affymetrix Mouse Genome 430A GeneChips® (Affymetrix, Santa Clara, CA) were carried out following the manufacturer's suggested procedure. Fragmented biotinylated RNA from each embryo was separately hybridized to its own GeneChip for 17 hours at 42°C. The microarray analysis revealed striking differences among the 4 alcohol treated samples, which segregated as two separate pairs rather than one set of four; subsequently, it was noted that one pair of embryos had an open neural tube (ALC-NTO) and the other pair had the neural tube closed (ALC-NTC). All 4 control embryos had closed neural tubes.
Experiment 2 was designed to follow-up these initial results and provide an independent test of the gene expression correlations with the two neural tube phenotypes. Total RNA was isolated from individual embryos (4 vehicle control, 7 alcohol treated: 4 ALC-NTO, 3 ALC-NTC). RNA extraction and microarray analysis was as described above, except that Affymetrix Mouse Genome 430 2.0 GeneChips® (Affymetrix, Santa Clara, CA) were used.
The Mouse Genome 430A chip contains over 22,600 probe sets representing transcripts and variants from over 14,000 well-characterized mouse genes. The newer Mouse Genome 430 2.0 Array contains all of the probe sets present on the earlier 430A chip plus additional probe sets for a total of approximately 45,000 probe sets that analyze the expression of over 39,000 transcripts and variants from over 34,000 well characterized mouse genes. The differences in feature size and probe set content make direct comparisons inappropriate, due to scanning and scaling issues, but because the probe sets on the 430A are present on the 430 2.0 array, those can be compared at the level of gene lists.
The data from independent arrays (each with RNA from a single embryo) for each of the treatments were extracted using the Affymetrix Microarray Suite 5.0 (MAS5) algorithm. Data for both experiments have been deposited in GEO/NCBI and have been assigned series accession number GSE9545 and sample numbers GSM241642 through GSM241660.
To minimize false positive results, only genes detected ("present" by the MAS5 algorithm) on at least half of all individual arrays in at least one experimental condition were retained for further analysis. This avoids data that primarily represent "noise" [72,73].
To detect differentially expressed genes, control samples were compared to ALC-NTC samples, or ALC-NTO samples, or their combination, using a Welch's t-test on the log-transformed signals. To see genes that were similarly affected in both experiments, we intersected the gene lists. To avoid missing genes that met a stringent significance threshold in one experiment but were just beyond that threshold in the second, we chose p ≤ 0.05 as the threshold for each experiment. Given that the two experiments were independent, the probability that a gene overlaps by chance and differs in the same up/down direction in both experiments is (0.05)*(0.05)/2 = 0.00125. False discovery rate (FDR) was calculated based on the number of genes expected to be significant and in the same direction in both experiments under the null hypothesis/the number of such genes actually found.
Hierarchical clustering with average linkage function was used to construct a dendrogram based upon all genes that were present on at least half of the arrays in an experimental group.
Gene Set Enrichment Analysis (GSEA) [74,75] was carried out to identify groups of related genes that were differentially expressed. GSEA analyses were conducted for 4 different comparisons: control vs. ALC, control vs. ALC/NTC, control vs. ALC/NTO, and ALC/NTC vs. ALC/NTO. The top ranked genes in a significant gene set, in the region up to the maximum score, were considered significant. To reduce multiple testing issues, the GSEA in this study was conducted using two gene set databases designed to test the hypotheses that groups of genes related to Early Development or Stem Cells were differentially affected by alcohol.
(a) Early Developmental Biology Gene Sets (Additional file 1): 415 GO categories that were defined by 29 key words were selected (identified gene sets, Additional file 2).
(b) Stem Cell Related Gene Sets: 191 GO categories related to stem cells, neurogenesis, osteogenesis, extracellular matrix, developmental signal transduction pathway, cell cycle, growth factor, TGFβ/BMP signaling, Wnt signaling, and notch signaling were developed by Superarray Bioscience http://www.superarray.com. The gene set information is listed in Additional file 3 (shown with consent of Superarray Bioscience, Frederick, MD).
Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
A number of differentially expressed genes detected in Experiment 1 were selected for qRT-PCR validation based on their biological significance. To test selected genes from the neural specification gene group, the total RNA of each embryo was isolated using the RNeasy mini kit (Qiagen, Valencia, CA) as described above. Vector NTI Advance 9.0 software (Invitrogen, Frederick, MD) was used to design the primers for qRT-PCR (Table 8); if possible, at least one primer in each pair spanned an exon-intron boundary. The number of embryos used in the control group varied from 7 to 9 for different genes, and the number used in the alcohol treated group varied from 9 to 11. The cDNA templates were generated from 50 ng total RNA (TaqMan Reverse Transcription Reagents, Applied Biosystems, Foster City, CA) from each individual embryo, and added to PCR reactions that contained 0.1 μM of forward and reverse primers and SYBR Green PCR Master Mix (Applied Biosystems). Triplicate qRT-PCR were performed for each sample in at least 3 experiments (n = 9). The cycle threshold (Ct) for each cDNA template was determined on the ABI Prism 7700 Sequence Detection System. The Ct refers to the cycle number at which the fluorescence of the amplified product reached an arbitrary threshold that was within the exponential phase of amplification. To correct for sample-to-sample variation, Gapdh served as an internal reference. Relative values of expression of neural specific genes were determined for each sample using the ΔΔCt method [76], and these values were normalized to the Ct values of Gapdh. The average Gapdh Ct values for alcohol treatment and control were the same in each tested sample, making it an appropriate control gene to normalize the expression of the candidate genes of interest.
Table 8.
Primers for qRT-PCR
| Primer | Sequence | RefSeq ID |
|---|---|---|
| Bhlhb5-f | CCTATTCAACAGCGTCTCGTCC | NM_021560 |
| Bhlhb5-r | GCTTCTCACTTTCCTCTAGCTTTGG | |
| Ctgf-f | AGATTGGAGTGTGCACTGCCAAAG | NM_010217 |
| Ctgf-r | TCCAGGCAAGTGCATTGGTATTTG | |
| Dll 1-f | ATAGCGACTGAGGTGTAAGATGGAAGC | NM_007865 |
| Dll 1-r | CTTCGCCTGAACCTGGTTCTCAG | |
| Efemp1-f | TCTACCTACGACAAACAAGCCCTGTG | NM_146015 |
| Efemp1-r | AGAGCTTGTGCGGAAGGTTCCTATAC | |
| Gapdh-f | TCCTGGTATGACAATGAATACGGC | NM_008084 |
| Gapdh-r | TCTTGCTCAGTGTCCTTGCTGG | |
| Igf1-f | ACTGACATGCCCAAGACTCAGAAGTC | NM_184052 |
| Igf1-r | TGCCTCCGTTACCTCCTCCTGTTC | |
| Igfbp2-f | CACAGCAGGTTGCAGACAGTGATG | NM_008342 |
| Igfbp2-r | CAGCTCCTTCATGCCTGACTTGAG | |
| Ntf3-f | TGGTTACTTCTGCCACGATCTTACAGG | NM_008742 |
| Ntf3-r | CTCCTTTGATCCATGCTGTTGCC | |
| Mylc2a-f | GGAAGAGTTCAAGCAGCTTCTC | NM_023402 |
| Mylc2a-r | ACTTGTAGTCAATGTTGCCGGC | |
| Neurog1-f | TCCCTCGGCTTCAGAAGACTTCAC | NM_010896 |
| Neurog1-r | AGGCCTAGTGGTATGGGATGAAACAG | |
| Neurog2-f | GCGTAGGATGTTCGTCAAATCTG | BC055743 |
| Neurog2-r | TCCGCGCTGGAGGACATC | |
| Sox5 f | AATATGAGTGGAGATTCTGACGGAAGC | AI528773 |
| Sox5 | GGCATTCATTGGACGCTTTATGTG | |
| Tieg1-f | CAGTCCCAGCATTTTGTTTAACGC | NM_013692 |
| Tieg1-r | GCAGCATCGGAGAAAGATTTGAAG | |
| Edil3-f | GCTCTCAGGCTGTTCAGAACCTTTG | AF031524 |
| Edil3-r | GGCTTTCCTTGGTTCCCAAGTAAAC |
Primers are named according to the gene with-f for forward primer and -r for reverse primer.
After Experiment 2, we decided to test the three groups (control, ALC/NTO, ALC/NTC) as pools, and chose growth/neurotrophic genes. A separate experiment was carried out with embryonic treatments identical to those used in Experiment 1. Whole embryos were homogenized in TRIzol (Invitrogen) using a Mini-Bead-Beater-8 (Bipspec products, INC, Bartlesville, OK), and total RNA isolation was as described above. Two different pools were created for each condition: Control1 (n = 12), ALC/NTC1 (n = 16), ALC/NTO1 (n = 5), Control2 (n = 5), ALC/NTC2 (n = 9), ALC/NTO2 (n = 6). The relative quantification of expression of each RNA pool was performed using the ABI Prism 7700 Sequence Detection System and calculated using the standard curve method (Applied Biosystems, User Bulletin #2; http:////www.appliedbiosystems.com). In each experiment, a relative expression level was determined for the two pools from each group in triplicate; 3-4 repeat experiments were performed, resulting in 18-24 values from each group. The treatment groups were compared with one way ANOVA followed by Student's t test.
Authors' contributions
FCZ involved in the overall design and coordination of the study. CG involved in the design of the experiment. QZ and LL carried out the statistical analysis, and YL the informatics. TL performed RTPCR analysis. HJE and JNM were involved in experimental design and carried out the microarray study. All authors participated in the writing of the manuscript. All authors read and approved the manuscript.
Supplementary Material
Keyword and GO categories. List of keywords used and the GO categories identified by these key words for Early Development.
GO gene sets selected by keywords. List of gene sets used in GSEA analyses, derived from GO categories selected by key words for Early Development. Title of each set followed by Entrez Gene Ids of the genes in the set.
GO categories related to Stem Cells. Gene sets used in GSEA analysis based on GO categories related to Stem Cells. Title of each set followed by Entrez Gene Ids of the genes in the set.
Contributor Information
Feng C Zhou, Email: imce100@iupui.edu.
Qianqian Zhao, Email: qizhao@iupui.edu.
Yunlong Liu, Email: yunliu@iupui.edu.
Charles R Goodlett, Email: goodlett@iupui.edu.
Tiebing Liang, Email: tliang@iupui.edu.
Jeanette N McClintick, Email: jnmcclin@iupui.edu.
Howard J Edenberg, Email: edenberg@iupui.edu.
Lang Li, Email: lali@iupui.edu.
Acknowledgements
This study was supported by NIAAA P50 AA07611 to the Alcohol Research Center at Indiana University School of Medicine (Center PI, David W. Crabb; FAS project, Feng C. Zhou), and in part by AA016698 to FCZ. Microarray studies were carried out in the Center for Medical Genomics at Indiana University School of Medicine, which is partially supported by the Indiana Genomic Initiative at Indiana University (INGEN®); INGEN® is supported in part by the Lilly Endowment, Inc. We thank Ms. Li-jun Ni for her technical assistance in embryonic culture, and Ronald Jerome and Chunxiao Zhu for technical assistance in preparation of microarray.
References
- Jones KL, Smith DW. Recognition of the fetal alcohol syndrome in early infancy. Lancet. 1973;2(7836):999–1001. doi: 10.1016/S0140-6736(73)91092-1. [DOI] [PubMed] [Google Scholar]
- Sampson PD, Streissguth AP, Bookstein FL, Little RE, Clarren SK, Dehaene P, Hanson JW, Graham JM Jr. Incidence of fetal alcohol syndrome and prevalence of alcohol-related neurodevelopmental disorder. Teratology. 1997;56(5):317–326. doi: 10.1002/(SICI)1096-9926(199711)56:5<317::AID-TERA5>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Clarren SK, Alvord EC jr, Sumi SM, Streissguth AP, Smith DW. Brain malformations related to prenatal exposure to ethanol. J Pediatr. 1978;92(1):64–67. doi: 10.1016/S0022-3476(78)80072-9. [DOI] [PubMed] [Google Scholar]
- Kalter H. Teratology in the 20th century: environmental causes of congenital malformations in humans and how they were established. Neurotoxicol Teratol. 2003;25(2):131–282. doi: 10.1016/S0892-0362(03)00010-2. [DOI] [PubMed] [Google Scholar]
- Stromland K, Hellstrom A. Fetal alcohol syndrome--an ophthalmological and socioeducational prospective study. Pediatrics. 1996;97(6 Pt 1):845–850. [PubMed] [Google Scholar]
- Burd L, Deal E, Rios R, Adickes E, Wynne J, Klug MG. Congenital heart defects and fetal alcohol spectrum disorders. Congenit Heart Dis. 2007;2(4):250–255. doi: 10.1111/j.1747-0803.2007.00105.x. [DOI] [PubMed] [Google Scholar]
- Climent E, Pascual M, Renau-Piqueras J, Guerri C. Ethanol exposure enhances cell death in the developing cerebral cortex: role of brain-derived neurotrophic factor and its signaling pathways. J Neurosci Res. 2002;68(2):213–225. doi: 10.1002/jnr.10208. [DOI] [PubMed] [Google Scholar]
- Miller MW, Kuhn PE. Cell cycle kinetics in fetal rat cerebral cortex: effects of prenatal treatment with ethanol assessed by a cumulative labeling technique with flow cytometry. Alcohol Clin Exp Res. 1995;19(1):233–237. doi: 10.1111/j.1530-0277.1995.tb01497.x. [DOI] [PubMed] [Google Scholar]
- Ikonomidou C, Bittigau P, Ishimaru MJ, Wozniak DF, Koch C, Genz K, Price MT, Stefovska V, Horster F, Tenkova T. et al. Ethanol-induced apoptotic neurodegeneration and fetal alcohol syndrome. Science. 2000;287(5455):1056–1060. doi: 10.1126/science.287.5455.1056. [DOI] [PubMed] [Google Scholar]
- Light KE, Belcher SM, Pierce DR. Time course and manner of Purkinje neuron death following a single ethanol exposure on postnatal day 4 in the developing rat. Neuroscience. 2002;114(2):327–337. doi: 10.1016/S0306-4522(02)00344-5. [DOI] [PubMed] [Google Scholar]
- Holownia A, Ledig M, Menez JF. Ethanol-induced cell death in cultured rat astroglia. Neurotoxicol Teratol. 1997;19(2):141–146. doi: 10.1016/S0892-0362(96)00226-7. [DOI] [PubMed] [Google Scholar]
- Kotch LE, Sulik KK. Patterns of ethanol-induced cell death in the developing nervous system of mice; neural fold states through the time of anterior neural tube closure. Int J Dev Neurosci. 1992;10(4):273–279. doi: 10.1016/0736-5748(92)90016-S. [DOI] [PubMed] [Google Scholar]
- Ewald SJ, Shao H. Ethanol increases apoptotic cell death of thymocytes in vitro. Alcohol Clin Exp Res. 1993;17(2):359–365. doi: 10.1111/j.1530-0277.1993.tb00776.x. [DOI] [PubMed] [Google Scholar]
- Sulik KK. Genesis of alcohol-induced craniofacial dysmorphism. Exp Biol Med (Maywood) 2005;230(6):366–375. doi: 10.1177/15353702-0323006-04. [DOI] [PubMed] [Google Scholar]
- Kilburn BA, Chiang PJ, Wang J, Flentke GR, Smith SM, Armant DR. Rapid induction of apoptosis in gastrulating mouse embryos by ethanol and its prevention by HB-EGF. Alcohol Clin Exp Res. 2006;30(1):127–134. doi: 10.1111/j.1530-0277.2006.00008.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SM. Alcohol-induced cell death in the embryo. Alcohol Health Res World. 1997;21(4):287–297. [PMC free article] [PubMed] [Google Scholar]
- Goodlett CR, Horn KH, Zhou FC. Alcohol teratogenesis: mechanisms of damage and strategies for intervention. Exp Biol Med (Maywood) 2005;230(6):394–406. doi: 10.1177/15353702-0323006-07. [DOI] [PubMed] [Google Scholar]
- Rifas L, Towler DA, Avioli LV. Gestational exposure to ethanol suppresses msx2 expression in developing mouse embryos. Proc Natl Acad Sci USA. 1997;94(14):7549–7554. doi: 10.1073/pnas.94.14.7549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahlgren SC, Thakur V, Bronner-Fraser M. Sonic hedgehog rescues cranial neural crest from cell death induced by ethanol exposure. Proc Natl Acad Sci USA. 2002;99(16):10476–10481. doi: 10.1073/pnas.162356199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada Y, Nagase T, Nagase M, Koshima I. Gene expression changes of sonic hedgehog signaling cascade in a mouse embryonic model of fetal alcohol syndrome. The Journal of craniofacial surgery. 2005;16(6):1055–1061. doi: 10.1097/01.scs.0000183470.31202.c9. discussion 1062-1053. [DOI] [PubMed] [Google Scholar]
- Poggi SH, Goodwin K, Hill JM, Brenneman DE, Tendi E, Schinelli S, Abebe D, Spong CY. The role of activity-dependent neuroprotective protein in a mouse model of fetal alcohol syndrome. Am J Obstet Gynecol. 2003;189(3):790–793. doi: 10.1067/S0002-9378(03)00834-2. [DOI] [PubMed] [Google Scholar]
- Xu Y, Xiao R, Li Y. Effect of ethanol on the development of visceral yolk sac. Hum Reprod. 2005;20(9):2509–2516. doi: 10.1093/humrep/dei075. [DOI] [PubMed] [Google Scholar]
- Xu Y, Chen X, Li Y. Ercc6l, a gene of SNF2 family, may play a role in the teratogenic action of alcohol. Toxicol Lett. 2005;157(3):233–239. doi: 10.1016/j.toxlet.2005.02.013. [DOI] [PubMed] [Google Scholar]
- Lee IJ, Soh Y, Song BJ. Molecular characterization of fetal alcohol syndrome using mRNA differential display. Biochem Biophys Res Commun. 1997;240(2):309–313. doi: 10.1006/bbrc.1997.7655. [DOI] [PubMed] [Google Scholar]
- Hard ML, Abdolell M, Robinson BH, Koren G. Gene-expression analysis after alcohol exposure in the developing mouse. J Lab Clin Med. 2005;145(1):47–54. doi: 10.1016/j.lab.2004.11.011. [DOI] [PubMed] [Google Scholar]
- Miller MW, Mooney SM, Middleton FA. Transforming growth factor beta1 and ethanol affect transcription and translation of genes and proteins for cell adhesion molecules in B104 neuroblastoma cells. J Neurochem. 2006;97(4):1182–1190. doi: 10.1111/j.1471-4159.2006.03858.x. [DOI] [PubMed] [Google Scholar]
- Green ML, Singh AV, Zhang Y, Nemeth KA, Sulik KK, Knudsen TB. Reprogramming of genetic networks during initiation of the Fetal Alcohol Syndrome. Dev Dyn. 2007;236(2):613–631. doi: 10.1002/dvdy.21048. [DOI] [PubMed] [Google Scholar]
- Da Lee R, Rhee GS, An SM, Kim SS, Kwack SJ, Seok JH, Chae SY, Park CH, Yoon HJ, Cho DH. et al. Differential gene profiles in developing embryo and fetus after in utero exposure to ethanol. J Toxicol Environ Health A. 2004;67(23-24):2073–2084. doi: 10.1080/15287390490515001. [DOI] [PubMed] [Google Scholar]
- Ogawa T, Kuwagata M, Ruiz J, Zhou FC. Differential teratogenic effect of alcohol on embryonic development between C57BL/6 and DBA/2 mice: a new view. Alcohol Clin Exp Res. 2005;29(5):855–863. doi: 10.1097/01.ALC.0000163495.71181.10. [DOI] [PubMed] [Google Scholar]
- Cockroft D. In: Postimplantaion Mammalian Embryos: A Practice Approach. AC, DC, editor. New Yolk: Oxford University Press; 1990. Dissection and culture of postimplantation embryos; pp. 15–40. [Google Scholar]
- New DA. Whole-embryo culture and the study of mammalian embryos during organogenesis. Biol Rev Camb Philos Soc. 1978;53(1):81–122. doi: 10.1111/j.1469-185X.1978.tb00993.x. [DOI] [PubMed] [Google Scholar]
- Boehm SL, Lundahl KR, Caldwell J, Gilliam DM. Ethanol teratogenesis in the C57BL/6J, DBA/2J, and A/J inbred mouse strains. Alcohol. 1997;14(4):389–395. doi: 10.1016/S0741-8329(97)87950-5. [DOI] [PubMed] [Google Scholar]
- Gilliam DM, Irtenkauf KT. Maternal genetic effects on ethanol teratogenesis and dominance of relative embryonic resistance to malformations. Alcohol Clin Exp Res. 1990;14(4):539–545. doi: 10.1111/j.1530-0277.1990.tb01196.x. [DOI] [PubMed] [Google Scholar]
- Liu Y, Balaraman Y, Wang G, Nephew KP, Zhou FC. Alcohol exposure alters DNA methylation profiles in mouse embryos at early neurulation. Epigenetics. 2009;4(7) doi: 10.4161/epi.4.7.9925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vickers MA, Hoy T, Lake H, Kyoizumi S, Boyse J, Hewitt M. Estimation of mutation rate at human glycophorin A locus in hematopoietic stem cell progenitors. Environmental and molecular mutagenesis. 2002;39(4):333–341. doi: 10.1002/em.10076. [DOI] [PubMed] [Google Scholar]
- Yenerel MN, Sundell IB, Weese J, Bulger M, Gilligan DM. Expression of adducin genes during erythropoiesis: a novel erythroid promoter for ADD2. Experimental hematology. 2005;33(7):758–766. doi: 10.1016/j.exphem.2005.03.015. [DOI] [PubMed] [Google Scholar]
- Ortega F, Gonzalez M, Moro MJ, Gascon A, Duarte I, Martin M, Hernandez J, Jimenez-Galindo R, Portero JA, Sanz M. et al. [Prognostic effect of beta 2-microglobulin in multiple myeloma] Medicina clinica. 1992;99(17):645–648. [PubMed] [Google Scholar]
- Mzhel'skaya TI. Biological functions of ceruloplasmin and their deficiency caused by mutation in genes regulating copper and iron metabolism. Bulletin of experimental biology and medicine. 2000;130(8):719–727. doi: 10.1007/BF02766076. [DOI] [PubMed] [Google Scholar]
- Barnes G, Frieden E. Ceruloplasmin receptors of erythrocytes. Biochem Biophys Res Commun. 1984;125(1):157–162. doi: 10.1016/S0006-291X(84)80348-4. [DOI] [PubMed] [Google Scholar]
- Jiang J, Ng HH. TGFbeta and SMADs talk to NANOG in human embryonic stem cells. Cell Stem Cell. 2008;3(2):127–128. doi: 10.1016/j.stem.2008.07.011. [DOI] [PubMed] [Google Scholar]
- Wilkemeyer MF, Chen SY, Menkari CE, Sulik KK, Charness ME. Ethanol antagonist peptides: structural specificity without stereospecificity. J Pharmacol Exp Ther. 2004;309(3):1183–1189. doi: 10.1124/jpet.103.063818. [DOI] [PubMed] [Google Scholar]
- Chen SY, Charness ME, Wilkemeyer MF, Sulik KK. Peptide-mediated protection from ethanol-induced neural tube defects. Dev Neurosci. 2005;27(1):13–19. doi: 10.1159/000084528. [DOI] [PubMed] [Google Scholar]
- Zhou FC, Sari Y, Powrozek T, Goodlett CR, Li T-K. Moderate alcohol exposure compromises neural tube midline development in prenatal brain. Developmental Brain Research. 2003;144:43–55. doi: 10.1016/S0165-3806(03)00158-5. [DOI] [PubMed] [Google Scholar]
- Bookstein FL, Sampson PD, Connor PD, Streissguth AP. Midline corpus callosum is a neuroanatomical focus of fetal alcohol damage. Anat Rec. 2002;269(3):162–174. doi: 10.1002/ar.10110. [DOI] [PubMed] [Google Scholar]
- Sowell ER, Mattson SN, Thompson PM, Jernigan TL, Riley EP, Toga AW. Mapping callosal morphology and cognitive correlates: Effects of heavy prenatal alcohol exposure. Neurology. 2001;57(2):235–244. doi: 10.1212/wnl.57.2.235. [DOI] [PubMed] [Google Scholar]
- Eriksen JL, Gillespie RA, Druse MJ. Effects of in utero ethanol exposure and maternal treatment with a 5-HT(1A) agonist on S100B-containing glial cells. Brain Res Dev Brain Res. 2000;121(2):133–143. doi: 10.1016/S0165-3806(00)00029-8. [DOI] [PubMed] [Google Scholar]
- Coulter CL, Leech RW, Schaefer GB, Scheithauer BW, Brumback RA. Midline cerebral dysgenesis, dysfunction of the hypothalamic-pituitary axis, and fetal alcohol effects. Arch Neurol. 1993;50(7):771–775. doi: 10.1001/archneur.1993.00540070083022. [DOI] [PubMed] [Google Scholar]
- Sulik KK, Johnston MC, Daft PA, Russell WE, Dehart DB. Fetal alcohol syndrome and DiGeorge anomaly: critical ethanol exposure periods for craniofacial malformations as illustrated in an animal model. Am J Med Genet Suppl. 1986;2:97–112. doi: 10.1002/ajmg.1320250614. [DOI] [PubMed] [Google Scholar]
- Johnson VP, Swayze VW, Sato Y, Andreasen NC. Fetal alcohol syndrome: craniofacial and central nervous system manifestations. Am J Med Genet. 1996;61(4):329–339. doi: 10.1002/(SICI)1096-8628(19960202)61:4<329::AID-AJMG6>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- Zhou FC, Sari Y, Powrozek T, Goodlett CR, Li TK. Moderate alcohol exposure compromises neural tube midline development in prenatal brain. Brain Res Dev Brain Res. 2003;144(1):43–55. doi: 10.1016/S0165-3806(03)00158-5. [DOI] [PubMed] [Google Scholar]
- Kele J, Simplicio N, Ferri AL, Mira H, Guillemot F, Arenas E, Ang SL. Neurogenin 2 is required for the development of ventral midbrain dopaminergic neurons. Development. 2006;133(3):495–505. doi: 10.1242/dev.02223. [DOI] [PubMed] [Google Scholar]
- Lee J, Wu Y, Qi Y, Xue H, Liu Y, Scheel D, German M, Qiu M, Guillemot F, Rao M. et al. Neurogenin3 participates in gliogenesis in the developing vertebrate spinal cord. Dev Biol. 2003;253(1):84–98. doi: 10.1006/dbio.2002.0868. [DOI] [PubMed] [Google Scholar]
- Korzh V, Sleptsova I, Liao J, He J, Gong Z. Expression of zebrafish bHLH genes ngn1 and nrd defines distinct stages of neural differentiation. Dev Dyn. 1998;213(1):92–104. doi: 10.1002/(SICI)1097-0177(199809)213:1<92::AID-AJA9>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- Kageyama R, Ohtsuka T, Hatakeyama J, Ohsawa R. Roles of bHLH genes in neural stem cell differentiation. Experimental cell research. 2005;306(2):343–348. doi: 10.1016/j.yexcr.2005.03.015. [DOI] [PubMed] [Google Scholar]
- Lee JE. NeuroD and neurogenesis. Dev Neurosci. 1997;19(1):27–32. doi: 10.1159/000111182. [DOI] [PubMed] [Google Scholar]
- Fode C, Gradwohl G, Morin X, Dierich A, LeMeur M, Goridis C, Guillemot F. The bHLH protein NEUROGENIN 2 is a determination factor for epibranchial placode-derived sensory neurons. Neuron. 1998;20(3):483–494. doi: 10.1016/S0896-6273(00)80989-7. [DOI] [PubMed] [Google Scholar]
- Ma Q, Anderson DJ, Fritzsch B. Neurogenin 1 null mutant ears develop fewer, morphologically normal hair cells in smaller sensory epithelia devoid of innervation. J Assoc Res Otolaryngol. 2000;1(2):129–143. doi: 10.1007/s101620010017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403(6765):41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- Berger SL. Histone modifications in transcriptional regulation. Curr Opin Genet Dev. 2002;12(2):142–148. doi: 10.1016/S0959-437X(02)00279-4. [DOI] [PubMed] [Google Scholar]
- Nehls V, Drenckhahn D, Joshi R, Bennett V. Adducin in erythrocyte precursor cells of rats and humans: expression and compartmentalization. Blood. 1991;78(7):1692–1696. [PubMed] [Google Scholar]
- Bernier GM. beta 2-Microglobulin: structure, function and significance. Vox sanguinis. 1980;38(6):323–327. doi: 10.1111/j.1423-0410.1980.tb04500.x. [DOI] [PubMed] [Google Scholar]
- Chang YZ, Qian ZM, Wang K, Zhu L, Yang XD, Du JR, Jiang L, Ho KP, Wang Q, Ke Y. Effects of development and iron status on ceruloplasmin expression in rat brain. J Cell Physiol. 2005;204(2):623–31. doi: 10.1002/jcp.20321. [DOI] [PubMed] [Google Scholar]
- Duester G. Genetic dissection of retinoid dehydrogenases. Chemico-biological interactions. 2001;130-132(1-3):469–480. doi: 10.1016/S0009-2797(00)00292-1. [DOI] [PubMed] [Google Scholar]
- Alnouti Y, Klaassen CD. Tissue distribution, ontogeny, and regulation of aldehyde dehydrogenase (aldh) enzymes mRNA by prototypical microsomal enzyme inducers in mice. Toxicol Sci. 2008;101(1):51–64. doi: 10.1093/toxsci/kfm280. [DOI] [PubMed] [Google Scholar]
- National-Academy-of-Sciences. Guide for the care and use of laboratory animals. 7. Washington. D.C.: National Academy Press; 2010. [Google Scholar]
- Dunty WC jr, Chen SY, Zucker RM, Dehart DB, Sulik KK. Selective vulnerability of embryonic cell populations to ethanol-induced apoptosis: implications for alcohol-related birth defects and neurodevelopmental disorder. Alcohol Clin Exp Res. 2001;25(10):1523–1535. doi: 10.1111/j.1530-0277.2001.tb02156.x. [DOI] [PubMed] [Google Scholar]
- Wilkemeyer M, Chen SY, Menkari CE, Brenneman DE, Sulik KK, Charness ME. Differential effects of ethanol antagonism an neuroprotection in napvsipq prevention of ethanol-induced developmental toxicity. PNAS. 2003. in press . [DOI] [PMC free article] [PubMed]
- Webster W, Walsh D, Lipson A, McEwen S. Teratogenesis after acute alcohol exposure in inbred and outbred mice. Neurobehav Toxicol. 1980;2:227–243. [Google Scholar]
- Hoffman F. In: A Handbook of Drug and Alcohol Abuse. Hoffman F, Hoffman A, editor. New York: Oxford University Press; 1975. Generalized depressants of the central nervous system; pp. 95–128. [Google Scholar]
- Lindblad B, Olsson R. Unusually high levels of blood alcohol? Jama. 1976;236(14):1600–1602. doi: 10.1001/jama.236.14.1600. [DOI] [PubMed] [Google Scholar]
- van Maele-Fabry G, Delhaise F, Picard JJ. Evolution of the developmental scores of sixteen morphological features in mouse embryos displaying 0 to 30 somites. The International journal of developmental biology. 1992;36(1):161–167. [PubMed] [Google Scholar]
- McClintick JN, Jerome RE, Nicholson CR, Crabb DW, Edenberg HJ. Reproducibility of oligonucleotide arrays using small samples. BMC Genomics. 2003;4(1):4. doi: 10.1186/1471-2164-4-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClintick JN, Edenberg HJ. Effects of filtering by Present call on analysis of microarray experiments. BMC Bioinformatics. 2006;7:49. doi: 10.1186/1471-2105-7-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag S, Lehar J, Puigserver P, Carlsson E, Ridderstrale M, Laurila E. et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34(3):267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Keyword and GO categories. List of keywords used and the GO categories identified by these key words for Early Development.
GO gene sets selected by keywords. List of gene sets used in GSEA analyses, derived from GO categories selected by key words for Early Development. Title of each set followed by Entrez Gene Ids of the genes in the set.
GO categories related to Stem Cells. Gene sets used in GSEA analysis based on GO categories related to Stem Cells. Title of each set followed by Entrez Gene Ids of the genes in the set.