Abstract
Binding of the 50S ribosomal subunit to the 30S initiation complex and the subsequent transition from the initiation to the elongation phase up to the synthesis of the first peptide bond represent crucial steps in the translation pathway. The reactions that characterize these transitions were analyzed by quench-flow and fluorescence stopped-flow kinetic techniques. IF2-dependent GTP hydrolysis was fast (30/s) followed by slow Pi release from the complex (1.5/s). The latter step was rate limiting for subsequent A-site binding of EF-Tu⋅GTP⋅Phe-tRNAPhe ternary complex. Most of the elemental rate constants of A-site binding were similar to those measured on poly(U), with the notable exception of the formation of the first peptide bond which occurred at a rate of 0.2/s. Omission of GTP or its replacement with GDP had no effect, indicating that neither the adjustment of fMet-tRNAfMet in the P site nor the release of IF2 from the ribosome required GTP hydrolysis.
Keywords: A-site binding/fast kinetics/IF2-dependent GTPase/phosphate release/translation initiation
Introduction
Initiation of protein synthesis in bacteria involves several steps that ultimately bring together the 70S ribosome, the initiator fMet-tRNAfMet and the translation initiation region of the mRNA to form a functional complex, ready to enter the elongation phase of translation. During the transition from the 30S to the 70S initiation complex and the subsequent passage from the initiation to the elongation phase of translation, initiation factors IF1 and IF3 are ejected from the ribosome, IF2 hydrolyzes GTP and dissociates from the ribosome, the acceptor end of initiator fMet-tRNAfMet becomes substrate for the ribosomal peptidyltransferase and the first dipeptide is formed (for reviews, see Hershey, 1987; Gualerzi and Pon, 1990; Gualerzi et al., 2000).
Among the three factors (IF1, IF2 and IF3) that kinetically govern initiation, IF2 is the one that assists the initiation process throughout most of the steps. IF2 binds to the 30S subunit and promotes first the codon-independent and then the initiation codon-dependent binding of fMet-tRNAfMet (Wintermeyer and Gualerzi, 1983; Canonaco et al., 1986; Gualerzi and Wintermeyer, 1986; La Teana et al., 1996); subsequently, IF2 stimulates the association of 30S and 50S subunits (Grunberg-Manago et al., 1975) and finally allows the adjustment of fMet-tRNAfMet in the bona fide P-site where it can form the first peptide bond with the aminoacyl-tRNA (aa-tRNA) bound to the second codon triplet and brought to the A-site by EF-Tu⋅GTP (La Teana et al., 1996).
As in the case of the translation factors EF-Tu, EF-G and RF3, IF2 is a GTPase that is activated by the interaction with the 50S ribosomal subunit. All translation factors appear to bind to overlapping sites on the ribosome. This overlap is the probable basis for the mutual incompatibility between the binding of EF-Tu and EF-G and for a similar incompatibility believed to exist between IF2 and the two elongation factors (Parmeggiani and Sander, 1981; Luchin et al., 1999, and references cited therein).
Stimulation by IF2 of fMet-tRNAfMet binding to the ribosomes is accomplished by an increase in the rate of codon–anticodon interaction and by a decrease in the dissociation rate of the ribosome–tRNA complex. Since the affinity of IF2 for aa-tRNAs bearing a blocked α-amino group is at least two orders of magnitude greater than that displayed for non-modified aa-tRNAs, IF2 strongly favors, both in vitro and in vivo, binding of fMet-tRNAfMet to the ribosome. In contrast to EF-Tu, whose interaction with aa-tRNAs depends on the presence of GTP, the interaction of IF2 with fMet-tRNAfMet is not affected by nucleotides (Petersen et al., 1979); only the affinity of IF2 for the 30S subunit is somewhat influenced by the nature of the guanosine nucleotide bound to the factor (Pon et al., 1985). Nonetheless, since the cellular concentration of GTP is at least one order of magnitude higher than the Kd of the IF2⋅GTP complex, IF2 is likely to perform all of its functions on the 30S subunit in the GTP-bound state. The GTP molecule is hydrolyzed only during formation of the 70S initiation complex. Although the release of IF2 from the 70S initiation complex (Hershey, 1987; Luchin et al., 1999) and/or the adjustment of the initiator tRNA in the P-site (La Teana et al., 1996, and references therein) are thought to require (or, at least, to be accelerated by) GTP hydrolysis, the role of the IF2-dependent GTPase remained elusive.
Here, the kinetics of IF2-dependent GTP hydrolysis and of the steps that mark the transition from the initiation to the elongation phase of translation were studied by quench-flow and fluorescence stopped-flow techniques. Using ribosomes programmed with a model mRNA (022mRNA; La Teana et al., 1993) with AUG and UUU as first and second codon, respectively, we investigated the kinetics of the following steps: (i) IF2-dependent GTPase; (ii) Pi release; (iii) binding of ternary complex EF-Tu⋅GTP⋅Phe-tRNAPhe to 70S initiation complexes; and (iv) the formation of the first dipeptide, fMetPhe. The last two steps are used as indicator reactions of the transition of the 70S initiation complex to its elongation-competent state. A kinetic mechanism for the transition from the initiation to the elongation phase of protein synthesis is derived from the data.
Results
Rapid kinetics of IF2-dependent GTP hydrolysis
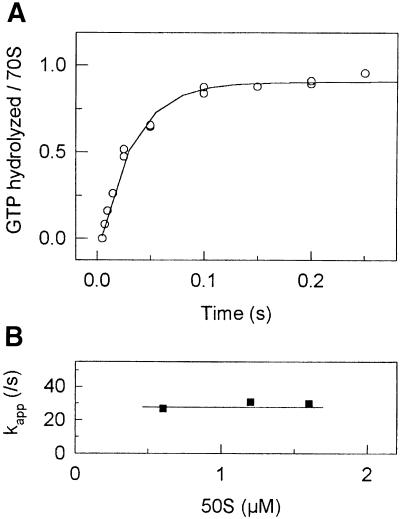
GTP hydrolysis during 70S initiation complex formation was studied by quench-flow measurements. Two solutions, one containing 30S subunits, 022mRNA, IF1, IF2, IF3, f[3H]Met-tRNAfMet and [γ-32P]GTP, and another containing 50S subunits, were mixed rapidly and the time course of GTP hydrolysis was measured. Rapid hydrolysis of GTP was observed with an apparent rate constant of kapp = 30 ± 5/s, reflecting single-round GTP hydrolysis by IF2 after joining of 50S to the 30S initiation complex (Figure 1A). Subsequent slow multiple turnover GTP hydrolysis was observed due to the uncoupled GTP hydrolysis by IF2 on the ribosomes; the rate constant of the multiple turnover reaction was 0.15 ± 0.02/s (not shown).
Fig. 1. Single round of IF2-dependent GTP hydrolysis. (A) Time course of GTP hydrolysis in the presence of 0.6 µM 50S. The data were fitted by a single exponential function with kapp = 30 ± 5/s. (B) Dependence of kapp upon 50S subunit concentration.
GTP hydrolysis by IF2 requires binding of 50S to the 30S initiation complex; if the latter reaction limited the rate of GTP hydrolysis, there should be a delay in the time course of GTP cleavage. Figure 1A shows that the reaction progressed in a clearly exponential fashion, and that there is no significant delay phase. Furthermore, the rate of GTP hydrolysis by IF2 did not change upon addition of increasing concentrations of 50S subunits (Figure 1B). Thus, joining of 50S subunits to the 30S initiation complex occurred much more rapidly than the subsequent hydrolysis of GTP.
Pi release from IF2
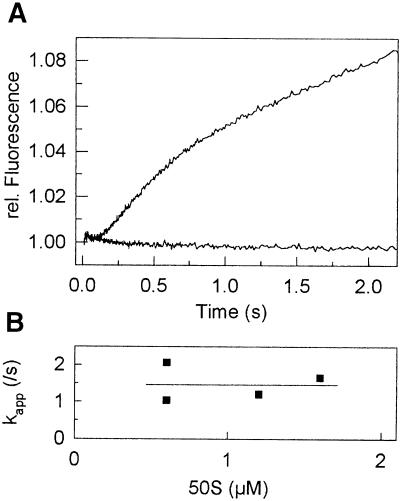
Pi release was studied according to the method of Brune et al. (1994), using a fluorescent derivative of phosphate-binding protein (PBP) from Escherichia coli, PBP-MDCC. Binding of Pi to MDCC-labeled PBP is rapid (kon = 108/M/s) and tight (Kd = 0.1 µM), and the formation of the complex strongly increases the fluorescence of MDCC (Brune et al., 1994). After a short lag phase (∼200 ms), which included the time required for GTP hydrolysis and possibly other steps, a rapid increase in fluorescence was observed, reflecting Pi dissociation after the first round of GTP hydrolysis, followed by a further slower increase due to multiple rounds of GTP hydrolysis (Figure 2A). In the first round, Pi was released at a rate of 1.5 ± 0.5/s, independently of the concentration of 50S subunits (Figure 2B), suggesting that this value represented the rate constant of Pi release (Scheme 1). The rate of the turnover reaction (0.15 ± 0.05/s) was the same as measured in the GTPase reaction (not shown). The molecular basis for the 200 ms delay is not known, because only ∼30 ms are required to complete the GTPase reaction (1/kGTPase = 1/30 s).
Fig. 2. Pi release following GTP hydrolysis in IF2. (A) Time course of Pi release in the presence of 0.6 µM 50S subunits. The data were fitted by a function comprising the sum of two exponential terms [delay and kapp(fast)] and a linear term [kapp(slow)]. The parameters of the fit: kapp(fast) = 1.5 ± 0.5/s, kapp(slow) = 0.15 ± 0.10/s. Lower curve, control. (B) Dependence of kapp(fast) on 50S subunit concentration.
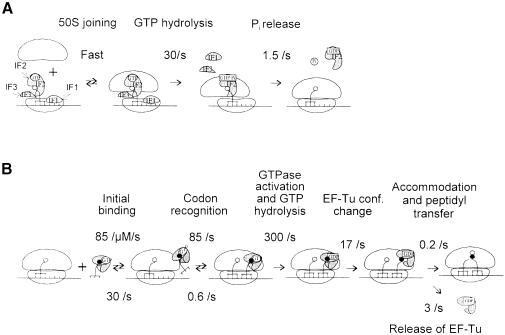
Scheme 1. Transition from initiation to elongation. (A) Kinetic mechanism of transition from the 30S to 70S initiation complex. (B) EF-Tu-dependent binding of aa-tRNA to the ribosomal A-site.
Binding of the ternary complex EF-Tu⋅GTP⋅Phe-tRNAPhe to the 70S initiation complex
If IF2-dependent GTP hydrolysis is required for the release of IF2 from the ribosome upon formation of the 70S initiation complex, it is possible that the presence of IF2 on the ribosome, or its slow release, may interfere with the interaction of ternary complex with the A-site. On the other hand, a failure of IF2 to place the initiator fMet-tRNAfMet correctly into the P-site in the absence of GTP should result in a reduced efficiency and/or a slower formation of dipeptide (i.e. fMetPhe in the case of 022mRNA). Thus, the ability of the 70S initiation complex to bind EF-Tu⋅GTP⋅Phe-tRNAPhe was used to study the completion of initiation and the transition to the elongation state of the ribosome. The 30S initiation complex was formed in the presence of GTP or GDP, or in the absence of nucleotide. The ternary complex EF-Tu⋅GTP⋅Phe-tRNAPhe was purified from unbound GTP by gel filtration (Rodnina et al., 1994a). The extent of ternary complex binding was measured by nitrocellulose filtration, and the kinetics by stopped-flow, monitoring the fluorescence change of proflavin attached to the D loop of tRNAPhe (Rodnina et al., 1994a).
The multistep process of aa-tRNA binding to the A-site of the ribosome is depicted in Scheme 1. In the first step, a ternary complex of aa-tRNA with elongation factor Tu (EF-Tu) and GTP forms a labile complex with the ribosome (initial binding, rate constants of the forward and backward reactions are defined as k1 and k–1, respectively) (Rodnina et al., 1996). Subsequent codon recognition (k2 and k–2) triggers GTP hydrolysis (k3) (Rodnina et al., 1995) which results in a large-scale conformational change of EF-Tu to the GDP-bound form (k4) (Dell et al., 1990; Abel et al., 1996; Polekhina et al., 1996). As a consequence, aa-tRNA is released from EF-Tu and enters the A-site on the 50S ribosomal subunit (accommodation) to take part in peptide bond formation (k5), whereas EF-Tu⋅GDP dissociates from the ribosome (k6) (Rodnina et al., 1995).
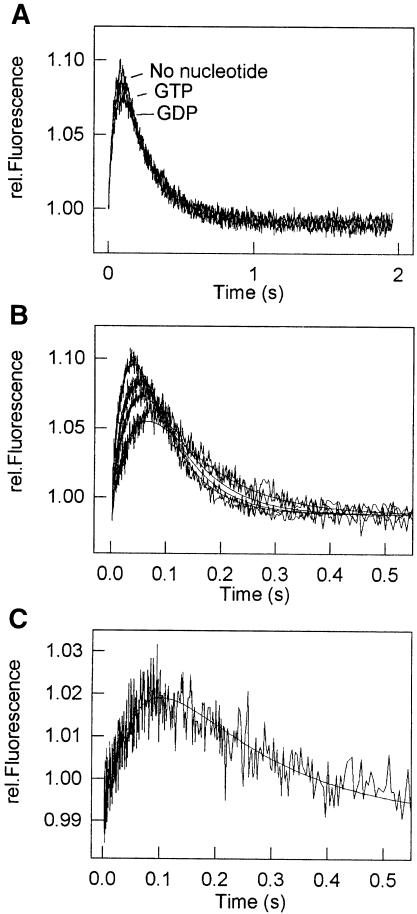
In the stopped-flow experiment, the 30S initiation complex was pre-mixed with 50S subunits in the pre-mixing chamber of the stopped-flow apparatus to form the 70S initiation complex which was then mixed rapidly with ternary complex in the second mixing chamber, and the proflavin fluorescence was monitored. As shown in Figure 3A, the signal changed in a biphasic way: a rapid increase in the fluorescence was followed by a slower decrease. Qualitatively, the same result had been observed previously with poly(U)-programmed ribosomes (Rodnina et al., 1994a); therefore, the assignment of the respective fluorescence changes to particular steps was taken from the poly(U) system (Pape et al., 1998, and references cited therein). The increase in proflavin fluorescence reflects the steps of initial codon-independent binding of the ternary complex to the ribosome as well as the codon recognition (Scheme 1), as established in experiments with non-programmed ribosomes (Rodnina et al., 1996) and with poly(U)-programmed ribosomes but in the presence of a non-hydrolyzable GTP analog in the ternary complex (Rodnina et al., 1994a). Further steps of EF-Tu activation for the GTPase reaction and GTP hydrolysis do not change the fluorescence of proflavin. The fluorescence decrease reflects the coupled conformational change of EF-Tu and aa-tRNA during the transition of the factor from the GTP to the GDP form and aa-tRNA release from EF-Tu. In the presence of poly(U) and AcPhe-tRNAPhe in the P-site, this step is partly masked by the increase of fluorescence of the A-site-bound tRNA due to its accommodation and peptide bond formation (Rodnina et al., 1994a; Pape et al., 1998); in the mRNA system, the latter two reactions do not change proflavin fluorescence (Figure 3A). Quantitative differences between the mRNA and poly(U) systems in both apparent rate constants and amplitudes are attributed to the different buffer conditions (Pape et al., 1998), and to the presence of a different tRNA in the P-site (Rodnina et al., 1994a).
Fig. 3. Binding of ternary complex to the 70S initiation complex. (A) Time course of EF-Tu⋅GTP⋅[14C]Phe-tRNAPhe binding in the presence of GTP or GDP, or in the absence of nucleotides. According to the nitrocellulose filtration, binding of [14C]Phe-tRNAPhe to the ribosome was 98, 95 and 87% of the added complex in the presence of GTP and GDP, or in the absence of nucleotide, respectively. (B) Time courses of ternary complex binding to 0.3 (lowest curve), 0.5, 0.7 and 1.0 µM (highest curve) 70S initiation complexes monitored by fluorescence changes of Prf16/17 in tRNA. (C) Time course of ternary complex binding to 0.7 µM 70S initiation complexes monitored by fluorescence changes of mant-GTP. Experiments were also performed at 0.3, 0.5 and 1.0 µM ribosomes (not shown).
To study the effect of the nucleotide in IF2 on the transition from the initiation to the elongation step, experiments were performed in the presence of GTP or GDP, or without nucleotide during initiation. The presence or the nature of the nucleotide did not significantly influence the extent of f[3H]Met-tRNAfMet binding to the 30S subunits (Pon et al., 1985; La Teana et al., 1996; and data not shown). The time course of ternary complex binding to the ribosomes monitored by proflavin fluorescence was not affected by the presence or absence of nucleotide during formation of the initiation complex (Figure 3A). Also, the efficiency of ternary complex binding measured by nitrocellulose filtration did not depend on the presence of GTP (not shown). Thus, the 70S initiation complexes formed with or without GTP did not differ with respect to the binding of the ternary complex.
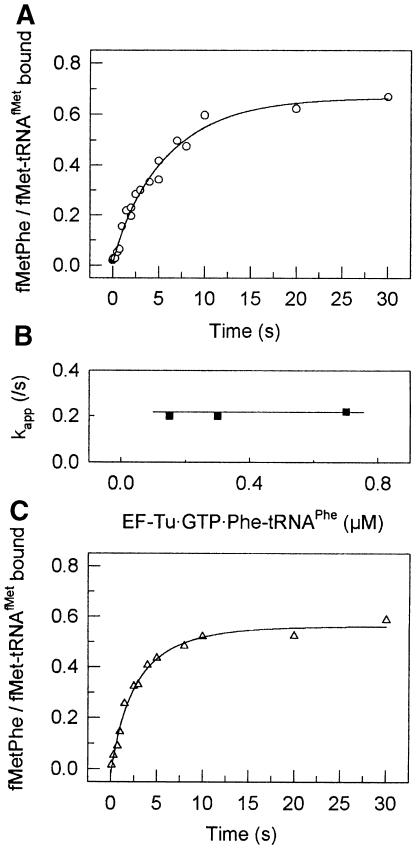
To determine the rate constants of the A-site binding to the first elongation codon, experiments were performed as described before (Pape et al., 1998). In brief, the time course of A-site binding was measured at different concentrations of 70S initiation complex. Fluorescence changes of proflavin in position 16/17 of the tRNA were used to monitor the conformational changes during initial binding of the EF-Tu ternary complex, codon recognition and release of aa-tRNA from EF-Tu followed by accommodation in the A-site (Figure 3B). Fluorescence changes of the GTP derivative, mant-GTP, were used to monitor the GTPase activation of EF-Tu and the dissociation of EF-Tu⋅mant-GDP from the ribosome (Figure 3C). The rate constants of initial binding of the ternary complex to the ribosome prior to codon recognition were determined separately from the ribosome titration experiments analogous to those shown in Figure 3B using non-programmed ribosomes (not shown). The value for the rate constant of dipeptide formation was taken from Figure 5A (see below). Elemental rate constants of A-site binding were determined by numerical integration using all available data (see Materials and methods). The results of the calculations are shown in Table I. The values of the elemental rate constants of A-site binding on 022mRNA were similar to those found with poly(U)-programmed ribosomes with two exceptions. For the rate constants k1, k–1, k2, k3 and k6, the differences from the previously reported values (Pape et al., 1998) were well within the range expected for the present ionic conditions (7 mM Mg2+). Two rate constants, k4 and, especially, k5, were lower than in the poly(U) system.
Fig. 5. Kinetics of fMetPhe formation. (A) Time course of dipeptide formation in the presence of GTP in the 30S initiation complex. The value of the apparent rate constant was determined by numerical integration (Materials and methods). (B) Dependence of kapp upon 50S subunit concentration. (C) Time course of dipeptide formation in the presence of GDP in the 30S initiation complex.
Table I. Elemental rate constants of EF-Tu-dependent aa-tRNA binding to the A site of 70S initiation complexes.
| Step | Rate constant (/s) | |
|---|---|---|
| Initial binding | k1 | 85 ± 15a |
| k–1 | 30 ± 10b | |
| Codon recognition | k2 | 85 ± 10 |
| k–2 | 0.6c | |
| GTPase activation and GTPhydrolysisd | k3 | 300 ± 100 |
| GTP–GDP conformation changeof EF-Tu | k4 | 17 ± 5 |
| Aa-tRNA accommodation andpeptide bond formationd | k5 | 0.2 ± 0.1 |
| Dissociation of EF-Tu | k6 | 3 ± 1 |
a/µM/s.
bDetermined independently.
cCalculated by extrapolation of previously reported data (Pape et al., 1998).
dGrouped for the analysis, because the former reaction is rate limiting and the latter follows instantaneously.
Transition from initiation to elongation
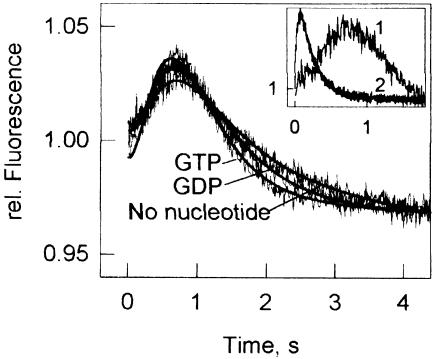
To study the transition from the 30S initiation complex to the 70S initiation complex competent in ternary complex binding, stopped-flow experiments were performed using a different mode of pre-mixing than that of Figure 3A. The 30S initiation complexes were mixed rapidly with 50S subunits and ternary complex without prior incubation, and the proflavin fluorescence was monitored. In this case, the time course of ternary complex binding showed a clear delay phase (Figure 4). The duration of the delay was not affected by the nucleotide present during the 30S initiation step.
Fig. 4. Transition from initiation to elongation monitored by ternary complex binding. The 30S initiation complexes were prepared in the presence of GTP or GDP, or in the absence of nucleotide. Smooth lines show time curves simulated according to Scheme 1 using the following elemental rate constants: kGTP = 30/s, kPi = 1.5/s, k1 = 85/s, k–1 = 25/s, k2 = 85/s, k3 = 290/s, k4= 17/s, k5 = 0.2/s (c.f. Figures 1 and 2 and Table I). The data obtained in the presence of GDP or in the absence of the nucleotide were fitted by numerical integration using a similar model, except that the first two steps (kGTP and kPi) were replaced by a single step; the value of the respective rate constant, ktransition, was determined to be 1.0 ± 0.5/s. The inset illustrates the delay phase of the ternary complex binding (curve 1) compared with the rapid interaction with the pre-formed 70S initiation complex (curve 2, from Figure 3A); both with GTP.
The data were evaluated quantitatively by numerical integration (Materials and methods). For the fitting, the model presented below (Scheme 1A) was used. In the first step, the 30S initiation complex binds the 50S subunit. As shown above, this step was fast compared with the subsequent reactions and was therefore not considered in the kinetic model (Materials and methods). The next step of GTP hydrolysis by IF2 proceeded with a rate constant of 30/s (cf. Figure 1) followed by Pi release, 1.5/s. When GDP or no nucleotide was present during the formation of the 30S initiation complex, these two steps were omitted from the calculations. Instead, to account for the delay in the ternary complex binding, a step was introduced which may include the dissociation of the initiation factors, final positioning of fMet-tRNAfMet in the P-site or some other as yet unidentified reactions; kinetically, all these presumed processes were grouped into a single step limiting the rate of ternary complex binding. After the completion of the transition from the initiation to the elongation, the binding of the ternary complex took place (Scheme 1B). To describe the A-site binding reactions, the values for the rate constants for the present ionic conditions were used (Table I).
The results of fitting are shown as smooth lines in Figure 4. Scheme 1 fully accounts for the observed kinetic behavior in the presence of GTP, Pi release from IF2 being the rate-limiting step for the subsequent steps of A-site binding. The fitting yields a unique solution for the rate constant of the rate-limiting step completing the initiation in the presence of GDP or in the absence of the nucleotide. At ∼1 ± 0.5/s, this value is very similar in the presence of GTP or GDP, or in the absence of the nucleotide.
Dipeptide formation
To test whether GTP hydrolysis is required for proper positioning of fMet-tRNAfMet in the P-site, we analyzed the formation of the dipeptide fMetPhe after rapid mixing of 30S initiation complex and ternary complex with 50S subunits. The time course of dipeptide formation measured by quench-flow is shown in Figure 5. The rate of dipeptide formation was similar in the presence of GTP and GDP, 0.20 ± 0.04/s (Figure 5A) and 0.18 ± 0.03/s (Figure 5C), respectively, and did not increase further upon increasing the concentration of ternary complex (Figure 5B). The rate of fMetPhe formation was 35-fold lower than that of AcPhePhe formation under similar ionic conditions (Pape et al., 1998), but still compatible with the 0.3/s rate of initiation measured in vivo (Kennell and Riezman, 1977).
Dipeptide formation started after a lag period of ∼750 ms, the time required for the transition from initiation to elongation and for the binding of the ternary complex. Replacement of GTP by GDP during the 30S initiation complex formation did not affect the duration of this lag significantly, indicating that GTP hydrolysis by IF2 did not accelerate the steps that led to production of the transpeptidation-competent complex. These results, together with the finding that there is no influence of the presence of GTP on the rate of dipeptide formation obtained with a standard experimental approach by La Teana et al. (1996), contradict the premise that GTP hydrolysis by IF2 is required for the adjustment of the fMet-tRNA in the P-site and for the formation of the first peptide bond (for references, see La Teana et al., 1996).
Discussion
Kinetic mechanism of transition from initiation to elongation
The multistep pathway of 30S initiation complex formation has been studied extensively (Gualerzi and Pon, 1990; Gualerzi et al., 2000). On the other hand, comparatively little is known concerning the later steps of initiation that characterize the transitions from the 30S to the 70S initiation complex and from the initiation to the elongation phase of protein synthesis. These transitions are of crucial importance insofar as they entail essential events such as the ejection of the initiation factors IF1 and IF3, the triggering of the GTPase activity of IF2, the adjustment of the fMet-tRNAfMet in the P-site, the release of IF2 from the ribosome and the formation of the first peptide bond (initiation dipeptide) between fMet-tRNAfMet and the aa-tRNA brought to the A-site by EF-Tu⋅GTP in response to the second codon of the mRNA.
The present results provide the first description of these events based on a rapid kinetic analysis of individual steps, which are the IF2-dependent hydrolysis of GTP, the release of Pi from the complex, the entry of the EF-Tu⋅GTP⋅aa-tRNA complex into the A-site of the 70S initiation complex and the formation of the first peptide bond (Scheme 1). Our results show that GTP is hydrolyzed rapidly by IF2 on the ribosome, triggered by the addition of 50S subunits to the 30S initiation complex containing IF2. The reaction has a rate constant of ∼30/s and the time course of the GTP hydrolysis appears to follow a single exponential, with no lag in the onset of the reaction observed in the beginning. This means that after the mixing with the 30S initiation complex, the 50S subunit associates very rapidly to form a 70S complex and this is followed by fast GTP hydrolysis by IF2. The kinetics of Pi release from the complex, which occurs following an ∼200 ms delay phase, are slower (1.5/s) than GTP hydrolysis, and GTP hydrolysis accounts for only ∼30 ms (1/30/s) of the 200 ms lag phase of Pi release, indicating that IF2 stays in the GDP⋅Pi form for the rest of the delay time. Most probably, during this time, the factor remains bound to the ribosome at a site overlapping the binding site for EF-Tu⋅GTP⋅aa-tRNA, as during the same period of time the binding of the ternary complex is inhibited.
The binding of the EF-Tu⋅GTP⋅aa-tRNA complex is also delayed in the absence of nucleotide, or in the presence of GDP in initiation, as the rate constant of the rate-limiting step that determines the onset of the A-site binding in this situation, ∼1/s, is similar to that observed in the presence of GTP, 1.5/s. This indicates that there is a conformational rearrangement that takes place after joining of the 50S subunit with the 30S initiation complex and that determines the rate of transition to the state which is competent for binding of the ternary complex. IF2 is expelled from the 70S ribosome only after this transition has taken place, regardless of whether it has GDP⋅Pi, GDP or no nucleotide bound. The same transition seems to limit the rate of Pi release from IF2.
The elemental rate constants of EF-Tu⋅GTP⋅Phe-tRNAPhe binding to the 70S initiation complex up to the GTP hydrolysis step were found to be essentially the same as estimated for the poly(U) system at 7 mM Mg2+ in the absence of initiation factors (Pape et al., 1998). From the Mg2+ dependence of the rate constants measured by stopped-flow and quench-flow techniques at 5 and 10 mM Mg2+, the following values are estimated for 7 mM Mg2+: k1 = 85 /µM/s, k–1 = 30/s, k2 = 90/s and k3 = 250/s; these values are almost identical to the values measured in the present study (Table I). Rate constants of steps following GTP hydrolysis are different. The present value of k4 (17/s), the rate constant of the conformational change of EF-Tu, is three times lower than the (Mg2+-independent) value measured for poly(U)-programmed ribosomes (60/s; Pape et al., 1998). While this difference may reflect another step where the ribosome has to adapt to elongation, it has to be interpreted with caution, because similar differences in k4 were observed previously, for instance when different tRNAs were present in the P-site (Rodnina et al., 1994a, 1995), or when paromomycin was bound to the decoding center (Pape et al., 2000). Thus, the transition of EF-Tu from the GTP to the GDP form seems to be influenced by the ribosome, indicating that the conformational change of EF-Tu is coupled to a rearrangement of the ribosome that may be affected in various ways.
A large difference is observed between the present value of k5 (0.2/s), the rate constant of fMetPhe formation, and that of AcPhePhe formation in the poly(U) system (7/s; Pape et al., 1998). As the rate of peptide bond formation is determined by the rate of aa-tRNA accommodation in the A-site (Pape et al., 1998), the different values of k5 probably mean that the accommodation of Phe-tRNAPhe is slower in the initiation complex with fMet-tRNAfMet in the P-site compared with the ribosome complex programmed with poly(U) and containing AcPhe-tRNAPhe in the P-site. As there is evidence suggesting that the accommodation step is coupled to a structural transition of 16S rRNA (Pape et al., 2000), the present results indicate that the same, or a related, transition of 16S rRNA may be required for the ribosome to perform the transition from initiation to elongation.
Does IF2 function as a switch operated by GTP hydrolysis?
IF2 is a large multidomain GTPase that belongs to the same subfamily of G proteins as EF-Tu, EF-G and RF3 (Bourne et al., 1991). Generally, G proteins are active, i.e. they interact with their respective effectors, in the GTP-bound form and are inactivated by GTP hydrolysis whereby they are switched into the inactive GDP-bound form. Whereas many GTPases follow this switch mechanism, recent evidence suggests that the functional mechanism of at least one translation factor, EF-G, is different. EF-G promotes the translocation of tRNAs on the ribosome, and kinetic measurements show that translocation is accelerated by GTP hydrolysis and that GTP hydrolysis precedes rather than follows translocation, in contrast to what is expected for a switch mechanism (Rodnina et al., 1997).
The present data show that IF2 also does not follow a switch mechanism. First, the formation of the 30S initiation complex is independent of the nucleotide in IF2 (La Teana et al., 1996), indicating that there is little affinity difference between the GTP- and GDP-bound forms of the protein in regulating the binding to its target (Pon et al., 1985). Secondly, GTP is hydrolyzed immediately after joining the 50S subunit, which is expected to provide the element(s) required for the GTPase activation in translation factors. Finally, the transition from initiation to elongation is not affected by the nucleotide in IF2. Hence, none of the functional steps of initiation are regulated by GTP binding or hydrolysis.
GTP hydrolysis in G proteins leads to conformation changes of the G domain, notably in the switch 1 and 2 regions that are important for the interactions with effectors of the GTPases (Wittinghofer, 1998). Structural rearrangements that are induced by the cleavage of the bond between β and γ phosphate are likely to occur stepwise, via an intermediate that retains both GDP and Pi. Until now, the kinetics of Pi release have been measured for a number of GTPases and ATPases, including Ras (Nixon et al., 1995), transducin (Ting and Ho, 1991), myosin (Brune et al., 1994; Lionne et al., 1995; White et al., 1997; Suzuki et al., 1998), kinesin (Gilbert et al., 1995; Moyer et al., 1998), dynamin (Vernon et al., 1999), tubulin (Vandecandelaere et al., 1999) and EF-G (Rodnina et al., 1999). Whereas Pi is released instantaneously after GTP hydrolysis by Ras, it has been shown for transducin, myosin, kinesin and dynamin that Pi release is slow compared with the hydrolysis of ATP, and the ADP⋅Pi form of the latter proteins is crucial for their function. Interestingly, a significant delay between GTP hydrolysis and release of Pi was also observed in EF-G (Rodnina et al., 1999; A.Savelsbergh, M.V.Rodnina and W.Wintermeyer, unpublished), supporting the notion that the functional cycle of the factor differs from that of conventional switch GTPases (Rodnina et al., 1997). With a long delay between GTP hydrolysis and Pi release, IF2 seems to be yet another G protein that does not follow the switch mechanism.
Is there a function for GTP hydrolysis in IF2?
Two main functions have been attributed to the GTPase activity of IF2: the adjustment of the initiator tRNA in the ribosomal P-site and the release of IF2 from the ribosome. Previous work, carried out primarily with Bacillus stearothermophilus components, has shown that the presence of GTP is not mandatory to place fMet-tRNAfMet into the puromycin-reactive site unless the initiation complex is formed under suboptimal conditions (e.g. when there is no proper codon for the initiator tRNA) (La Teana et al., 1996). Since these studies were carried out using standard techniques which allow the investigation in a time range which is at the borderline of the rates (≈3.2 s/initiation event) found in vivo by Kennell and Riezman (1977), the conclusion that IF2-dependent GTP hydrolysis is not required for the adjustment of the initiator tRNA in the P-site could be limited to those physiological situations characterized by similar initiation rates.
The second role attributed to IF2-dependent GTP hydrolysis is that of allowing the dissociation of IF2 from the ribosome at the end of the initiation phase. This view is supported by early data as well as by a recent study in which the dominant lethal effect of IF2 mutations impairing the GTPase activity was attributed to the failure of this factor to dissociate from the ribosomes (Luchin et al., 1999), thereby inhibiting the binding of other translation factors (EF-G, EF-Tu and possibly RF3) or their function, as all translational GTPases may require the same ribosomal contacts, possibly with protein L7/L12 (Savelsbergh et al., 2000), to elicit their GTPase activity. Indeed, early experiments have also shown that IF2 may prevent the interaction of EF-Tu with the ribosome in the absence of GTP hydrolysis (Lockwood et al., 1974; Beaudry et al., 1979). Recently, extending the ‘molecular mimicry’ hypothesis, it has been suggested that in addition to IF1 (Moazed et al., 1995), IF2 also (in a binary complex with IF1) interacts with the A-site, mimicking the structure of EF-G which in turn mimicks that of the EF-Tu⋅GTP⋅aa-tRNA complex (Brock et al., 1998). Thus, it is thought that IF2 and EF-Tu interfere in binding, so that failure of IF2 to be released from the ribosome should prevent the binding of the EF-Tu ternary complex to the A-site (Luchin et al., 1999).
Although we cannot exclude that under our in vitro assay conditions a certain step of initiation may have become so unphysiologically slow and rate-limiting as to render the effect of GTP hydrolysis on the physiological rate-limiting step no longer detectable, our data indicate that: (i) the subunits associate properly into the 70S initiation complex without GTP; (ii) the positioning of the P-site-bound fMet-tRNAfMet is correct with respect to the peptidyltransferase center in the absence of GTP in the initiation complex; and (iii) the relative positioning of the tRNAs in the P- and the A-sites is the same in the presence and absence of GTP. Furthermore, our data indicate that IF2-dependent GTPase activity is not implicated in the ejection of IF2 from the complete 70S initiation complex.
These conclusions are in obvious conflict with current hypotheses concerning the role of the IF2-dependent GTPase in translation initiation. A possible reason for this discrepancy is that many of the conclusions reported in the earlier literature were based on results obtained with non-hydrolyzable GTP analogs, such as GDPCP and GDPNP, used to assess the possible role of GTP hydrolysis in the processes under study. It has become clear, however, that these analogs impose different structural constraints on IF2 which lead to the loss of functions not necessarily related to the lack of GTP hydrolysis (La Teana et al., 1996). Concerning the conflict of our data with the more recent results of Luchin et al. (1999), we would like to stress that, although we have obtained similar dominant lethal mutants of IF2 from B.stearothermophilus, we were unable to confirm that mutants lacking the GTPase activity remain preferentially associated with the ribosomes. Instead, we show that there is no relationship between the dominant lethal phenotype and the residual GTPase activity of mutant IF2 (Gualerzi et al., 2000; J.Tomšic, A.Smorlesi, R.Spurio, C.L.Pon and C.O.Gualerzi, in preparation).
Materials and methods
Buffers and reagents
Buffer A: 25 mM Tris–HCl pH 7.5, 70 mM NH4Cl, 30 mM KCl and 7 mM MgCl2. Buffer B: 20 mM Tris–HCl pH 7.5, 0.1 mM EDTA, 5 mM 2-mercaptoethanol, 10% glycerol, 0.1 mM benzamide and 0.1 mM phenylmethylsulfonyl fluoride (PMSF). GTP, GDP, GDPNP, dithiothreitol (DTT), pyruvate kinase and phosphoenolpyruvate were from Roche Diagnostics. The purity of GTP was analyzed by HPLC and was found to be >95%. Chemicals were of analytical grade and purchased from Merck; radioactive compounds were from ICN or Amersham. All kinetic experiments were performed at 20°C.
Ribosomes, factors, tRNAs and mRNA
Escherichia coli MRE600 ribosomal subunits, tRNAPhe and tRNAfMet were kindly provided by Y.P.Semenkov and S.V.Kirillov (Gatchina, Russia). Initiation factors were prepared essentially as described (Gualerzi et al., 1991), except that for IF2 the purification was modified in that IF2-containing fractions from phosphocellulose chromatography were purified further by FPLC on MonoS using a linear gradient from 50 to 400 mM NH4Cl in buffer B. IF2-containing fractions were identified by SDS–PAGE, pooled and dialyzed against buffer B containing 200 mM NH4Cl. The protein concentration was determined both colorimetrically and from its molar extinction coefficient at 280 nm (Hershey et al., 1977).
EF-Tu, fluorescent tRNAPhe (Prf16/17) and purified ternary complex, EF-Tu⋅GTP⋅[14C]Phe-tRNAPhe, were prepared as described (Rodnina et al., 1994a). f[3H]Met-tRNAfMet was prepared as in Rodnina et al. (1994b).
The 022mRNA (120 nucleotides) (La Teana et al., 1993) contained an appropriately spaced four nucleotide Shine–Dalgarno sequence followed by an AUG initiation codon and UUU as a first elongation codon. The rest of the translation initiation region of this mRNA also resembled the consensus sequence of natural mRNAs (La Teana et al., 1993).
GTPase activity
The 30S initiation complexes were prepared in buffer A using 0.6 µM 30S subunits, 0.9 µM of each of the initiation factors and of f[3H]Met-tRNAfMet, 1.8 µM 022mRNA and 72 µM [γ-32P]GTP (∼1000 d.p.m./pmol). After incubation at 37°C for 15 min, the complexes were stored on ice. Quench-flow experiments were performed using a KinTek RQF-3 apparatus. The 30S initiation complexes (final concentration 0.3 µM) were mixed rapidly with 50S subunits (26 µl each) at varying concentrations, as indicated (Figure 1). The reactions were quenched by 1 M HClO4 and 3 mM KH2PO4, and [32P]phosphate was determined by molybdate extraction as described (Rodnina et al., 1999). Time courses were evaluated by fitting a single exponential function. Time constants were reproducible within 20%.
Pi release
Pi release was monitored by the fluorescence change of phosphate-binding protein labeled with MDCC (Brune et al., 1994) in an SX-18MV stopped-flow spectrometer (Applied Photophysics). The fluorescence of MDCC was excited at 425 nm and monitored after passing a KV450 filter (Schott). The 30S initiation complexes were prepared as described above, except that non-radioactive GTP was used. The concentrations after mixing of 60 µl each of 30S initiation complexes and 50S subunits were the same as in the GTPase experiment (see above). The rate constant of Pi binding to MDCC-PBP, k1 = 96/µM/s, was determined in separate experiments. The concentration of MDCC-PBP (2.5 µM) was chosen such that the rate of Pi binding to MDCC-PBP (k1′ = k1⋅[MDCC-PBP] = 240/s) was much faster than the rate of GTP hydrolysis in IF2 (30/s) and thus the uptake of Pi liberated from the factor by MDCC-PBP was practically instantaneous. To minimize phosphate contamination, all solutions and the stopped-flow apparatus were pre-incubated with 600 µM 7-methylguanosine and 0.3 U/ml purine nucleoside phosphorylase (Brune et al., 1994). Rate constants were determined by fitting a function that included the delay before the onset of Pi release, an exponential term for Pi release and a slope for the linear turnover Pi release. The calculations were performed using the TableCurve software (Jandel Scientific).
A-site binding
Fluorescence stopped-flow measurements were performed on an SFM-3 apparatus (Biologic) or on an SX-18MV spectrometer (Applied Photophysics) and the data evaluated as described previously (Rodnina et al., 1994a; Pape et al., 1998). The fluorescence of proflavin was excited at 436 nm and measured after passing a KV500 filter (Schott). The fluorescence of mant-GTP was excited at 360 nm and measured after passing a KV408 filter (Schott).
The experiments of Figure 3A were performed by rapidly mixing equal volumes (60 µl each) of the purified ternary complex containing fluorescent tRNA and the 70S initiation complex. Final concentrations of ternary complex, 50S subunits and 30S initiation complex were 0.1, 0.3 and 0.39 µM, respectively. From experiment to experiment, the reproducibility of the rate constants given was about ±10%, that of the amplitudes about ±15%; within one experiment, the reproducibility from shot to shot was ∼5% for both parameters.
To determine independently the rate constants of initial binding of the ternary complex to the ribosome, time courses of EF-Tu⋅GTP⋅Phe-tRNAPhe(Prf16/17) (0.1 µM) binding were measured in the presence of 0.3–1.0 µM non-programmed ribosomes, and the rate constants, k1 and k–1, were calculated from the concentration dependence of the kapp of the reaction. The labile binding of the ternary complex to the ribosomes in the absence of mRNA or in the presence of a non-matching codon in the A-site reflects the initial codon-independent step of A-site binding that precedes codon recognition (Rodnina et al., 1996). The rate constants of the reaction, k1 and k–1, were shown to be independent of the mRNA (Pape et al., 1998). The calculations were performed by numerical intergration as described (Pape et al., 1998, 2000), and gave values of k1 = 85 ± 15/s and k–1 = 30 ± 10/s.
The experiments of Figure 3B and C were carried out with 0.1 µM ternary complex and 0.3–1.0 µM 70S initiation complex. Rate constants were calculated from the combined sets of time courses for proflavin and mant-GTP fluorescence changes at four different ribosome concentrations. The data were fitted by numerical integration to a kinetic model depicted in Scheme 1 using Scientist for Windows (MicroMath Scientific Software) as described (Pape et al., 1998).
For the calculations, independently determined values of k1 and k–1 were used (see above). The dissociation rate constant of the codon recognition complex, k–2, is known to be small compared with the rate constant of the forward reactions (k2 and k3) (Eccleston et al., 1985; Pape et al., 1998). For the present purpose, the value of k–2 was estimated as 0.6/s by extrapolation of the previously obtained data (Pape et al., 1998) to the present ionic conditions. The rate of dipeptide formation, k5, was determined separately from the data of Figure 6 (see below). The values of k1, k–1, k–2 and k5 were fixed during the calculations of the remaining constants. The fitting yielded a unique solution for the rate constants k2, k3, k4 and k6. For values that were measured directly, the given standard deviation (Table I) was calculated from the results of several experiments. For values calculated by global fitting, the standard deviation for a given parameter was determined for the case when all other parameters, except the fixed ones, are allowed to change, i.e. if a given parameter was set to a value that is outside the range of the standard deviation, there was no fit satisfying all data sets.
The experiments of Figure 4 were performed by mixing 30S initiation complexes, 50S subunits and the ternary complex containing fluorescent labeled tRNA. Final concentrations of ternary complex, 50S subunits and the 30S initiation complex were 0.17, 0.6 and 0.3 µM, respectively. Time courses were fitted by numerical integration using Scientist for Windows. A model was used where Equation 2 was combined with the A-site binding model (scheme 1):
which denotes GTP hydrolysis in IF2 upon joining of 50S subunits to the 30S initiation complex (kGTP), followed by Pi release from IF2 after GTP hydrolysis (kPi) resulting in an elongation-competent state of the 70S initiation complex (B, see model 1).
To account for the delay in A-site binding in the experiments with GDP or in the absence of nucleotide in the initiation complex, the upper equation in model (2) was simplified to the following expression:
where ktransition includes all the steps that take place after the joining of the 50S subunit and lead to the formation of the 70S initiation complex competent in A-site binding.
Dipeptide formation
The 30S initiation complexes were prepared as described above. Quench-flow experiments were performed in a KinTech RQF-3 apparatus. One sample syringe was loaded with pre-formed 30S initiation complex (0.3 µM) and EF-Tu⋅GTP⋅[14C]Phe-tRNAPhe (0.6 µM) in the presence of either GTP or GDP (2 mM), and the other sample syringe was loaded with 50S subunits. Reactions were performed at 20°C and quenched with 0.2 M KOH. The quenched reaction mixtures were incubated for 15 min at 37°C, neutralized with acetic acid, centrifuged for 5 min at 12 000 r.p.m., and peptides were analyzed by HPLC. Since time courses of dipeptide formation were sigmoidal in the beginning, the data were analyzed by numerical integration using Scientist for Windows.
Acknowledgments
Acknowledgements
We thank Martin Webb for the E.coli strain overproducing mutant PBP, and Yuri Semenkov and Stanislav Kirillov for ribosomal subunits and tRNAs. C.O.G. was the recipient of an Alexander von Humboldt Research Prize. The work was supported by grants from the Deutsche Forschungsgemeinschaft, the EC Biotechnology Program, the Alfried Krupp von Bohlen und Halbach-Stiftung and the Fonds der Chemischen Industrie to W.W. and M.V.R., and from the Italian CNR (Progetto Strategico ST74), MURST-CNR 95/95 and EC Biotechnology Program to C.O.G.
References
- Abel K., Yoder,M.D., Hilgenfeld,R. and Jurnak,F. (1996) An α to β conformational switch in EF-Tu. Structure, 4, 1153–1159. [DOI] [PubMed] [Google Scholar]
- Beaudry P., Sander,G., Grunberg-Manago,M. and Douzou,P. (1979) Cation-induced regulatory mechanism of GTPase activity dependent on polypeptide initiation factor 2. Biochemistry, 18, 202–207. [DOI] [PubMed] [Google Scholar]
- Bourne H.R., Sanders,D.A. and McCormick,F. (1991) The GTPase superfamily: conserved structure and molecular mechanism. Nature, 349, 117–127. [DOI] [PubMed] [Google Scholar]
- Brock S., Szkaradkiewicz,K. and Sprinzl,M. (1998) Initiation factors of protein biosynthesis in bacteria and their structural relationship to elongation and termination factors. Mol. Microbiol., 29, 409–417. [DOI] [PubMed] [Google Scholar]
- Brune M., Hunter,J.L., Corrie,J.E. and Webb,M.R. (1994) Direct, real-time measurement of rapid inorganic phosphate release using a novel fluorescent probe and its application to actomyosin subfragment 1 ATPase. Biochemistry, 33, 8262–8271. [DOI] [PubMed] [Google Scholar]
- Canonaco M.A., Calogero,R.A. and Gualerzi,C.O. (1986) Mechanism of translation initiation in prokaryotes. Evidence for a direct effect of IF2 on the activity of the 30S ribosomal subunit. FEBS Lett., 207, 198–204. [DOI] [PubMed] [Google Scholar]
- Dell V.A., Miller,D.L. and Johnson,A.E. (1990) Effects of nucleotide- and aurodox-induced changes in elongation factor Tu conformation upon its interactions with aminoacyl transfer RNA. A fluorescence study. Biochemistry, 29, 1757–1763. [DOI] [PubMed] [Google Scholar]
- Eccleston J.F., Dix,D.B. and Thompson,R.C. (1985) The rate of cleavage of GTP on the binding of Phe-tRNA⋅elongation factor Tu⋅GTP to poly(U)-programmed ribosomes of Escherichia coli.J. Biol. Chem., 260, 16237–16241. [PubMed] [Google Scholar]
- Gilbert S.P., Webb,M.R., Brune,M. and Johnson,K.A. (1995) Pathway of processive ATP hydrolysis by kinesin. Nature, 373, 671–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunberg-Manago M., Dessen,P., Pantaloni,D., Godefroy-Colburn,T., Wolfe,A.D. and Dondon,J. (1975) Light-scattering studies showing the effect of initiation factors on the reversible dissociation of Escherichia coli ribosomes. J. Mol. Biol., 94, 461–478. [DOI] [PubMed] [Google Scholar]
- Gualerzi C.O. and Pon,C.L. (1990) Initiation of mRNA translation in prokaryotes. Biochemistry, 29, 5881–5889. [DOI] [PubMed] [Google Scholar]
- Gualerzi C.O. and Wintermeyer,W. (1986) Prokaryotic initiation factor 2 acts at the level of the 30S ribosomal subunit. A fluorescence stopped-flow study. FEBS Lett., 202, 1–6. [Google Scholar]
- Gualerzi C.O., Severini,M., Spurio,R., La Teana,A. and Pon,C.L. (1991) Molecular dissection of translation initiation factor IF2. Evidence for two structural and functional domains. J. Biol. Chem., 266, 16356–16362. [PubMed] [Google Scholar]
- Gualerzi C.O., Brandi,L., Caserta,E., La Teana,A., Spurio,R., Tomsic,J. and Pon,C.L. (2000) Translation initiation in bacteria. In Garrett,R.A., Douthwaite,S.R., Liljas,A., Matheson,A.T., Moore,P.B. and Noller,H.F. (eds), The Ribosome:Structure, Function, Antibiotics and Cellular Interactions. ASM Press, Washington, DC, pp. 477–494. [Google Scholar]
- Hershey J.W.B. (1987) Protein synthesis. In Neidhardt,F.C., Ingraham,J.L., Brooks Low,K., Magasanik,B., Schaechter,M. and Umbarger,H.E. (eds), Escherischia coli and Salmonella typhimurium. Cellular and Molecular Biology. American Society for Microbiology, Washington, DC, pp. 613–647. [Google Scholar]
- Hershey J.W., Yanov,J., Johnston,K. and Fakunding,J.L. (1977) Purification and characterization of protein synthesis initiation factors IF1, IF2 and IF3 from Escherichia coli.Arch. Biochem. Biophys., 182, 626–638. [DOI] [PubMed] [Google Scholar]
- Kennell D. and Riezman,H. (1977) Transcription and translation initiation frequencies of the Escherichia coli lac operon. J. Mol. Biol., 114, 1–21. [DOI] [PubMed] [Google Scholar]
- La Teana A., Pon,C.L. and Gualerzi,C.O. (1993) Translation of mRNAs with degenerate initiation triplet AUU displays high initiation factor 2 dependence and is subject to initiation factor 3 repression. Proc. Natl Acad. Sci. USA, 90, 4161–4165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Teana A., Pon,C.L. and Gualerzi,C.O. (1996) Late events in translation initiation. Adjustment of fMet-tRNA in the ribosomal P-site. J. Mol. Biol., 256, 667–675. [DOI] [PubMed] [Google Scholar]
- Lionne C., Brune,M., Webb,M.R., Travers,F. and Barman,T. (1995) Time resolved measurements show that phosphate release is the rate limiting step on myofibrillar ATPases. FEBS Lett., 364, 59–62. [DOI] [PubMed] [Google Scholar]
- Lockwood A.H., Maitra,U., Brot,N. and Weissbach,H. (1974) The role of ribosomal proteins L7 and L12 in polypeptide chain initiation in Escherichia coli.J. Biol. Chem., 249, 1213–1218. [PubMed] [Google Scholar]
- Luchin S., Putzer,H., Hershey,J.W., Cenatiempo,Y., Grunberg-Manago,M. and Laalami,S. (1999) In vitro study of two dominant inhibitory GTPase mutants of Escherichia coli translation initiation factor IF2. Direct evidence that GTP hydrolysis is necessary for factor recycling. J. Biol. Chem., 274, 6074–6079. [DOI] [PubMed] [Google Scholar]
- Moazed D., Samaha,R.R., Gualerzi,C. and Noller,H.F. (1995) Specific protection of 16S rRNA by translational initiation factors. J. Mol. Biol., 248, 207–210. [DOI] [PubMed] [Google Scholar]
- Moyer M.L., Gilbert,S.P. and Johnson,K.A. (1998) Pathway of ATP hydrolysis by monomeric and dimeric kinesin. Biochemistry, 37, 800–813. [DOI] [PubMed] [Google Scholar]
- Nixon A.E., Brune,M., Lowe,P.N. and Webb,M.R. (1995) Kinetics of inorganic phosphate release during the interaction of p21ras with the GTPase-activating proteins, p120-GAP and neurofibromin. Biochemistry, 34, 15592–8. [DOI] [PubMed] [Google Scholar]
- Pape T., Wintermeyer,W. and Rodnina,M.V. (1998) Complete kinetic mechanism of elongation factor Tu-dependent binding of aminoacyl-tRNA to the A site of the E.coli ribosome. EMBO J., 17, 7490–7497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pape T., Wintermeyer,W. and Rodnina,M.V. (2000) Conformational switch in the decoding region of 16S rRNA during aminoacyl-tRNA selection on the ribosome. Nature Struct. Biol., 7, 104–107. [DOI] [PubMed] [Google Scholar]
- Parmeggiani A. and Sander,G. (1981) Properties and regulation of the GTPase activities of elongation factors Tu and G and of initiation factor 2. Mol. Cell. Biochem., 35, 129–158. [DOI] [PubMed] [Google Scholar]
- Petersen H.U., Roll,T., Grunberg-Manago,M. and Clark,B.F. (1979) Specific interaction of initiation factor IF2 of E.coli with formylmethionyl-tRNAfMet. Biochem. Biophys. Res. Commun., 91, 1068–1074. [DOI] [PubMed] [Google Scholar]
- Polekhina G., Thirup,S., Kjeldgaard,M., Nissen,P., Lippmann,C. and Nyborg,J. (1996) Helix unwinding in the effector region of elongation factor EF-Tu-GDP. Structure, 4, 1141–1151. [DOI] [PubMed] [Google Scholar]
- Pon C.L., Paci,M., Pawlik,R.T. and Gualerzi,C.O. (1985) Structure–function relationship in Escherichia coli initiation factors. Biochemical and biophysical characterization of the interaction between IF-2 and guanosine nucleotides. J. Biol. Chem., 260, 8918–8924. [PubMed] [Google Scholar]
- Rodnina M.V., Fricke,R. and Wintermeyer,W. (1994a) Transient conformational states of aminoacyl-tRNA during ribosome binding catalyzed by elongation factor Tu. Biochemistry, 33, 12267–12275. [DOI] [PubMed] [Google Scholar]
- Rodnina M.V., Semenkov,Y.P. and Wintermeyer,W. (1994b) Purification of fMet-tRNA (fMet) by fast protein liquid chromatography. Anal. Biochem., 219, 380–381. [DOI] [PubMed] [Google Scholar]
- Rodnina M.V., Fricke,R., Kuhn,L. and Wintermeyer,W. (1995) Codon-dependent conformational change of elongation factor Tu preceding GTP hydrolysis on the ribosome. EMBO J., 14, 2613–2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodnina M.V., Pape,T., Fricke,R., Kuhn,L. and Wintermeyer,W. (1996) Initial binding of the elongation factor Tu⋅GTP⋅aminoacyl-tRNA complex preceding codon recognition on the ribosome. J. Biol. Chem., 271, 646–652. [DOI] [PubMed] [Google Scholar]
- Rodnina M.V., Savelsbergh,A., Katunin,V.I. and Wintermeyer,W. (1997) Hydrolysis of GTP by elongation factor G drives tRNA movement on the ribosome. Nature, 385, 37–41. [DOI] [PubMed] [Google Scholar]
- Rodnina M.V., Savelsbergh,A., Matassova,N.B., Katunin,V.I., Semenkov,Y.P. and Wintermeyer,W. (1999) Thiostrepton inhibits the turnover but not the GTPase of elongation factor G on the ribosome. Proc. Natl Acad. Sci. USA, 96, 9586–9590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savelsbergh A., Mohr,D., Wilden,B., Wintermeyer,W. and Rodnina,M.V. (2000) Stimulation of the GTPase activity of translation elongation factor G by ribosomal protein L7/12. J. Biol. Chem., 275, 890–894. [DOI] [PubMed] [Google Scholar]
- Suzuki Y., Yasunaga,T., Ohkura,R., Wakabayashi,T. and Sutoh,K. (1998) Swing of the lever arm of a myosin motor at the isomerization and phosphate-release steps. Nature, 396, 380–383. [DOI] [PubMed] [Google Scholar]
- Ting T.D. and Ho,Y.-K. (1991) Molecular mechanism of GTP hydrolysis by bovine transducin: pre-steady-state kinetic analysis. Biochemistry, 30, 8996–9007. [DOI] [PubMed] [Google Scholar]
- Vandecandelaere A., Brune,M., Webb,M.R., Martin,S.R. and Bayley,P.M. (1999) Phosphate release during microtubule assembly: what stabilizes growing microtubules? Biochemistry, 38, 8179–8188. [DOI] [PubMed] [Google Scholar]
- Vernon G.G., Brune,M., Webb,M.R. and Woolley,D.M. (1999) Real-time visual detection of Pi released by flagellar dynein ATPase. Pflugers Arch., 437, 771–775. [DOI] [PubMed] [Google Scholar]
- White H.D., Belknap,B. and Webb,M.R. (1997) Kinetics of nucleoside triphosphate cleavage and phosphate release steps by associated rabbit skeletal actomyosin, measured using a novel fluorescent probe for phosphate. Biochemistry, 36, 11828–11836. [DOI] [PubMed] [Google Scholar]
- Wintermeyer W. and Gualerzi,C.O. (1983) Effect of E.coli initiation factors on the kinetics of NAc-Phe-tRNA binding to 30 ribosomal subunits. A fluorescence stopped flow study. Biochemistry, 22, 690–694. [DOI] [PubMed] [Google Scholar]
- Wittinghofer F. (1998) Ras signalling—caught in the act of the switch-on. Nature, 394, 317. [DOI] [PubMed] [Google Scholar]