Abstract
Electrostatic interactions play an important role in both packaging of DNA inside bacteriophages and its release into bacterial cells. While at physiological conditions DNA strands repel each other, the presence of polyvalent cations such as spermine and spermidine in solutions leads to the formation of DNA condensates. In this study, we discuss packaging of DNA into bacteriophages P4 and Lambda under repulsive and attractive conditions using a coarse-grained model of DNA and capsids. Packaging under repulsive conditions leads to the appearance of the coaxial spooling conformations; DNA occupies all available space inside the capsid. Under the attractive potential both packed systems reveal toroidal conformations, leaving the central part of the capsids empty. We also present a detailed thermodynamic analysis of packaging and show that the forces required to pack the genomes in the presence of polyamines are significantly lower than those observed under repulsive conditions. The analysis reveals that in both the repulsive and attractive regimes the entropic penalty of DNA confinement has a significant non-negligible contribution into the total energy of packaging. Additionally we report the results of simulations of DNA condensation inside partially packed Lambda. We found that at low densities DNA behaves as free unconfined polymer and condenses into the toroidal structures; at higher densities rearrangement of the genome into toroids becomes hindered, and condensation results in the formation of non-equilibrium structures. In all cases packaging in a specific conformation occurs as a result of interplay between bending stresses experienced by the confined polymer and interactions between the strands.
Keywords: bacteriophage, packaging, molecular dynamics, confinement, DNA condensation
INTRODUCTION
The phenomenon of DNA compaction is highly important for all living organisms (van Holde, 1989). It is a vital step in the lifecycle of bacteriophages, in which genome packaging is not a spontaneous process but is aided by an ATP- driven motor (Smith et al., 2001). The molecular motors responsible for DNA translocation are located in a unique vertex of capsids. These motors are capable of developing forces as high as 100 pN (Rickgauer et al., 2008), which can be measured experimentally by optical tweezers applied to a single bacteriophage system. The compacted genome is stored under pressure of several tens of atmospheres (Grayson et al., 2006) and organized into well ordered structures (Jiang et al., 2006; Lander et al., 2006), the specific conformation of which depends on the size and the shape of the capsids (Ali et al., 2006; Comolli et al., 2008; Earnshaw and Harrison, 1977; Earnshaw et al., 1978; Petrov et al., 2007a). The high pressure inside tightly packed bacteriophages is the result of complex interactions between DNA strands that includes not only electrostatic and van der Waals interactions but also the hydration energies associated with the displacement of water molecules from the polyelectrolyte’s surface (Podgornik et al., 1989; Rau et al., 1984). The latter component dominates at separation distances of 5–15 Å as revealed by osmotic pressure experiments measured for ordered DNA arrays (Parsegian et al., 1995; Rau and Parsegian, 1992). These studies also showed that mono- and divalent cations have only secondary effects on the magnitude of the osmotic pressure (Parsegian et al., 2000). A substantial contribution to the pressure of DNA packed inside bacteriophages also arises from the entropic penalty of DNA confinement into a small closed space (Locker et al., 2007; Petrov and Harvey, 2007).
In the presence of multivalent cations such as Co(NH3)63+, spermine, or spermidine, DNA spontaneously collapses and forms precipitates (Bloomfield, 1991; Bloomfield, 1997). Most frequently, DNA condensates form regular toroidal structures (Hud and Vilfan, 2005), although the formation of rod-like shape conformations has also been reported (Sarkar et al., 2009).
Polycations had been originally thought to play an active role in packing DNA into bacteriophages in vivo and in vitro. However, it was later demonstrated that the polycations are not necessarily required for this purpose (Hafner et al., 1979; Hamada et al., 1986) and packaging is primarily assisted by an ATP-driven motor (Smith et al., 2001). Thus, polycations are not essential for the packing mechanism, and their presence in vivo at physiological concentrations would only have a peripheral effect on the thermodynamics of packaging (Black, 1989). The formation of toroidal condensates in the presence of polycations has been observed in vitro inside partially packed bacteriophages Lambda (Evilevitch, 2006). Recently, a detailed study of DNA condensation in vitro inside bacteriophage T5 revealed that toroidal condensates are twisted and DNA curvature results in variations in the DNA helical pitch (Leforestier and Livolant, 2009).
Bacteriophage Lambda has been a model system for studying viral packaging process at the molecular level (Evilevitch, 2006; Evilevitch et al., 2003; Evilevitch et al., 2005; Fuller et al., 2007b). The wild type genome contains a 48.5 kilobase-pair (kbp) dsDNA genome, which is stored inside an icosahedral T=7 capsid. Packaging of DNA causes a significant capsid expansion during maturation, and the volume of the protein shell nearly doubles. The structure of the mature capsid has been recently obtained at subnanometer resolution using modern cryo-EM techniques (Lander et al., 2008). The mature capsid gains additional stability due to the presence of gpD proteins bound to its surface. It has also been shown that the motor of bacteriophage Lambda is capable of developing the forces above 50 pN; however, a force of ~25 pN was found to be sufficient to overcome the internal pressure of 30 atm and package the entire wild-type Lambda genome (Fuller et al., 2007b). Unlike Lambda, bacteriophage P4 is a parasite phage, which only exists in concurrence with phage P2 (Dokland et al., 1992). The 11.6 kbp genome of bacteriophage P4 does not contain genes that code for structural proteins; it uses the proteins generated by P2 to build its own T=4 capsid. When packaging is completed, the genome of P4 cyclizes due to the presence of cohesive ends forming a knotted structure (Arsuaga et al., 2002b; Arsuaga et al., 2005; Micheletti et al., 2008).
In recent years, DNA packaging in bacteriophages (Petrov and Harvey, 2008) as well as more general problem of a polymer confinement in a sphere (Morrison and Thirumalai, 2009) have been extensively studied by theoretical and computational approaches. The first group of studies uses a continuum representation of DNA and the apparatus of statistical mechanics. These methods have been able to reflect the basic physics of DNA confinement and to estimate thermodynamic properties such as forces and free energy (Grayson et al., 2006; Kindt et al., 2001; Purohit et al., 2003; Purohit et al., 2005; Tzlil et al., 2003). The continuum approach has also been successful in explaining the role of the osmotic pressure during genome ejection (Evilevitch et al., 2008). The main disadvantage of the continuum approach is the lack of predicting power regarding the conformation of the packed genome. The second group of studies consists of the simulations of packaging performed within the framework of coarse-grain representations of DNA and capsids by means of various computational techniques, which include Molecular Dynamics (LaMarque et al., 2004; Locker et al., 2007; Petrov and Harvey, 2007), Stochastic Rotational Dynamics (Ali et al., 2004; Ali et al., 2006), and Langevin Dynamics (Forrey and Muthukumar, 2006a). These methods have been able to reveal information regarding forces, thermodynamics and kinetics of packaging, as well as to demonstrate the dependence of DNA conformation on capsid shape and size (Ali et al., 2006; Petrov et al., 2007a). For the detailed progress achieved by computational techniques see surveys (Harvey et al., 2009; Marenduzzo, 2008; Petrov and Harvey, 2008) and references therein. Computer simulations have also been used to model DNA condensation. Models that account for the presence of condensing agents in both explicit (Stevens, 2001) and implicit (Sarkar et al., 2009) ways have successfully predicted the formation of toroidal and rod-like shape condensates.
In this study we extend our Molecular Dynamics (MD) studies to characterize packaging of DNA genomes inside bacteriophages P4 and Lambda at different ionic conditions. We perform the packaging simulations in two distinct regimes: the repulsive (in sodium/magnesium buffers) and attractive (done upon addition of condensing agents). These two regimes can be viewed as two limiting cases for in vivo systems. We provide detailed thermodynamic analyses of the packaging in both regimes, and estimate individual components of free energy. Further, we compare energetics of packaging in different regimes as well as discuss the role of the cations and their effect on genome conformation. Additionally, we simulate condensation of partially packed DNA at different volume fractions and discuss the phase transition and the morphology of the observed condensates.
METHODS
Models for P4 and Lambda
The 11.6 kbp genome of P4 (Dokland et al., 1992) and 37.800 kbp genome of a Lambda mutant (78% of 48.5 kbp wild-type Lambda genome) (Lander et al., 2008) were represented using a coarse-grained “beads on a string” model (Locker and Harvey, 2006), in which each bead represents six base pairs and is connected to adjacent beads via a set of harmonic bond- and angular constraints. The P4 capsid was modeled as a regular non-expandable icosahedron with a side of 450 Å in diameter (Dokland et al., 1992), whose faces were filled out with a number of soft spheres with a radius of 8 Å. A few spheres were removed from a single (unique) vertex of the icosahedron to mimic the packaging portal and enable DNA packaging. In contrast with ε15 (Jiang et al., 2006), the packaging portal of P4 is very small, so no explicit portal model was used for P4. The capsid of bacteriophage Lambda also has icosahedral symmetry (Dokland and Murialdo, 1993). Since both longitudinal and equatorial cross-sections of the Lambda capsid closely resemble regular circles (Lander et al., 2008), the capsid was modeled as a sphere of radius R. To define the capsid’s boundary, a dummy atom was placed at the origin and all DNA atoms were subjected to a semiharmonic restraint (i.e. if a DNA pseudoatom exceeds the capsid boundary R by a distance x, a repulsive force directed to the origin and proportional to x pulls it back). Bacteriophage Lambda undergoes significant expansion upon DNA injection. To account for this, the radius of the capsid was linearly expanded from 210 Å to 290 Å between 20 and 40% of total Lambda genome packed (Dokland and Murialdo, 1993; Fuller et al., 2007b).
Force-field
The DNA elastic parameters were chosen to match the known elastic moduli for stretching and bending; the former is computed from the average and variance in rise per basepair of free dsDNA reported in the nucleic acid database, and the latter is derived from the experimentally observed persistence length of DNA (500 Å). Specifically, the numerical values of constants are kb=3.5 kcal/(mol Å2), b0=19.9 Å, kθ=22.4 kcal/(mol rad2), θ0=π rad, kDNA-DNA= 11.0 kcal/(mol Å2), d0,DNA-DNA=25.0 Å, kDNA-Capsid=8.8 kcal/(mol Å2), d0,DNA-Capsid=20.5 Å, kDNA-Sphere=8.8 kcal/(mol Å2). A cutoff distance of 50 Å was used for calculating the volume exclusion terms.
DNA-DNA interactions, which are highly important during the packaging process, will be considered in two distinct regimes in the current study. The first regime is repulsive and it corresponds to the case when only mono- and divalent ions are present in the solvent. This case has been widely considered in our previous studies (Petrov et al., 2007a). For the repulsive regime the functional form of DNA-DNA interactions was empirically derived from the experimental data of Rau and Parsegian and modeled as a function of distance, r, by a modified Debye-Hückel function (Petrov and Harvey, 2007):
| (1) |
where Lb = 7.135 Å is the Bjerrum length and 0.59 is the conversion factor to kcal/mol. Two sets of parameters were used to model the packaging in two repulsive buffers. The first set (the effective charge, qeff= −12.6 e per pseudoatom, the effective screening constant, κeff = 0.31 Å−1, and DNA radius, a = 12.5 Å) corresponds to the buffer containing 10 mM MgCl2, 100 mM NaCl and 10 mM TrisCl and the second set (qeff = −6.63 e, κeff = 0.21 Å−1, a = 10. Å) fits the experimental data obtained for the buffer containing 10 mM MgCl2, and 10 mM TrisCl.
The interaction between DNA strands in the attractive regime, i.e. in the presence of polycations, which induce DNA condensation, is described by the following empirical relationship:
| (2) |
with A1 =11 cal/(mol*bp); A2=12 cal/(mol*bp); b1=30.5 Å; b2=37.5 Å c1= 2.6 Å; c2=2.2 Å. The parameters have been derived to match the data for the attractive interactions in the presence of cobalt haxamine occurring in the range ~25–34 Å with a minimum of ~120 cal/(mol*bp) at ~27.7 Å as estimated in the theoretical study by Tzlil et al. (Tzlil et al., 2003) and the repulsive interactions in the range 35–50 Å as experimentally observed by the osmotic pressure data obtained in the presence of polycations by Rau and Parsegian (Rau and Parsegian, 1992). We point out that later experimental data (Raspaud et al., 2005) suggest that DNA condensed in the presence of spermine result in slightly larger interstrand distances (28.5–29.0 Å) and the values of the attractive energy per nucleotide are somewhat lower (~40–50 cal/(mol*bp)). This experimental work also shows a significant dependence of the attractive potential on the concentration of sodium ions, which often coexist in viral systems. Thus, our attractive parameters do not correspond to a specific buffer composition but are instead aimed at describing the most characteristic features of the attractive regime at the phenomenological level.
Molecular Dynamics protocol
All MD trajectories for packaging DNA inside bacteriophages were generated using the freely available MD package YUP (Tan et al., 2006) designed for molecular modeling of coarse-grained systems. For each MD trajectory initial velocities of the first ten DNA beads originally placed inside the capsid were randomly drawn from the Boltzmann distribution. Because DNA inside each bacteriophage is packed at a unique conformation (and kinetically trapped there at high packing densities), independent MD trajectories were generated for each packaging mode for a given system (40 for P4 and 32 for Lambda) to collect statistically representative data for the thermodynamic functions. All packaging simulations were performed with a time step of 1ps in the repulsive regime and 0.5 ps in the attractive regime. Packaging was performed at 300K by coupling the systems to a Berendsen thermostat (Berendsen et al., 1984). DNA was driven into the capsid using harmonic restrains between the first five incoming DNA pseudoatoms and five fixed pseudoatoms lying on the axis of DNA outside the capsid (“studs”). These restraints have a force constant of 0.01 pN/Å. Each step in the packaging was achieved by changing the restraints to move the DNA pseudoatoms forward 10 Å and followed by extensive equilibration, thus ratcheting the DNA into the capsid. The length of packing trajectories was 40 µs and 250 µs for P4 and Lambda, respectively. This is several orders of magnitude faster than the packaging time in vivo or in vitro but sufficient for equilibration at every point along the trajectories, since the model is coarse-grained and does not contain explicit solvent. For packaging in both the repulsive and attractive regimes the reference state was assumed to be the same (random-coil DNA outside the capsid). Thus, in our simulations attraction between DNA strands was only allowed inside the capsids. To determine the packaging forces, conformations obtained at intervals of ten percent along the packing trajectories were taken as starting points for a series of new MD runs. Each of these was equilibrated for an additional 20 ns at 300K before the force measurement runs. The packed DNA is under pressure and tries to leave the capsid. This causes the springs connecting the DNA tail beads and the stud atoms to be stretched from their equilibrium positions. These displacements generate a force acting on the DNA tail atoms that is equal to and opposite the force developed by the motor, which pushes DNA out of the capsid. To collect statistically uncorrelated data, the restraining force was measured at 500 ps intervals along new MD trajectories and a total of 1000 independent data points were collected. All reported thermodynamic properties were averaged over independent MD trajectories (40 for P4 and 32 for Lambda).
DNA condensation studies
Intermediate DNA conformations at 10, 30 and 78% of the total length of the Lambda genome packed in the repulsive regime have also been used to study DNA condensation in a confined space. These conformations were taken as starting points for new MD runs, in which DNA-DNA interactions were switched to the attractive regime. No additional packaging was done during these simulations. To relax the DNA conformation in the new force field, and to avoid the numerical instability of the MD algorithm, the new simulations were started at 10 K, and the systems were gradually preheated to 300 K during the first 1000 steps. The intermediate data points were then collected every 2000 steps along free MD trajectories.
Electron density maps
Density maps were reconstructed by averaging 32 individual trajectories generated for the Lambda mutant and 40 for the bacteriophage P4 according to previously described procedures (Petrov et al., 2007b). The positions of the pseudoatoms were extracted from the trajectory files and were used to generate cylindrical segments with a length of 20 Å and a radius of 10 Å, which connect two adjacent pseudoatoms. These cylinders were uniformly filled with 2000 points (“atoms”). The sets of such atoms were converted to corresponding values of single particle density maps with a voxel size of 3 Å using Spider (Frank et al., 1996). The individual densities were superimposed, and the average density map was visualized using Chimera (Pettersen et al., 2004).
RESULTS AND DISCUSSION
The experimental studies of packaging are normally performed in the repulsive regime, i.e., in the presence of buffers containing sodium and magnesium ions (Fuller et al., 2007a; Rickgauer et al., 2008; Smith et al., 2001). Additionally, the effect of the condensing agents on the conformation of DNA in partially packed phages has been studied (Evilevitch, 2006; Leforestier and Livolant, 2009). We performed simulations of genome packaging inside bacteriophages P4 and Lambda in both repulsive and attractive regimes. These allow us to extract the information about the structure and thermodynamics under two different conditions, and to draw fundamental conclusions regarding the behavior of DNA in confined spaces. By switching the potential for the partially packed systems from the repulsive to the attractive, we simulated the process of DNA condensation in confined spaces. Thus, this manuscript contains three main parts. In the first part we describe the structural aspects of packaging simulations of bacteriophages P4 and the 38.7 kbp Lambda mutant (further referred to as bacteriophage Lambda) in the repulsive and the attractive regimes. In the second part we provide a thermodynamic analysis of the packaging process. Finally, in the last part we discuss simulations of DNA condensation in the partially packed bacteriophage Lambda.
DNA conformations inside bacteriophages
Packaging in the repulsive regime
Bacteriophage P4 contains a relatively small genome (only 11.6 kbp) and it was used as a model system to test the packaging parameters in our simulations. We have previously shown (Arsuaga et al., 2002a; Rollins et al., 2008) that in the repulsive regime DNA packs itself in the coaxial spool motif (i.e. the inner and outer layers form DNA spools, whose axes are not parallel but are instead rotated relative to each other by some angle). This motif was confirmed in the current study, in which we performed 40 independent trajectory runs. A representative conformation of P4 is shown in Figure 1a and Figure 1S (Supporting Material). This conformation also reveals a characteristic feature of any packaging simulation performed under repulsive conditions: DNA occupies the entire volume of the capsid and arranges itself to maximize the distance between the strands. The behavior of DNA along the packaging trajectory undergoes the same standard set of stages that we have previously reported (Petrov et al., 2007a). At the beginning of packaging (0%–25%, for P4) DNA density is low and DNA explores most of the space inside the capsid. This can be seen in movies of the MD trajectories, given at http://rumour.biology.gatech.edu/Publications/showcase.html. At the intermediate densities (25–70%) one can observe the appearance of the global pattern (coaxial spooling) and the motion of DNA strands becomes somewhat restricted; finally, at high densities (>70%) newly packaged DNA reptates along previously packed DNA layers, while the global conformation remains practically unchanged, simply growing more compact.
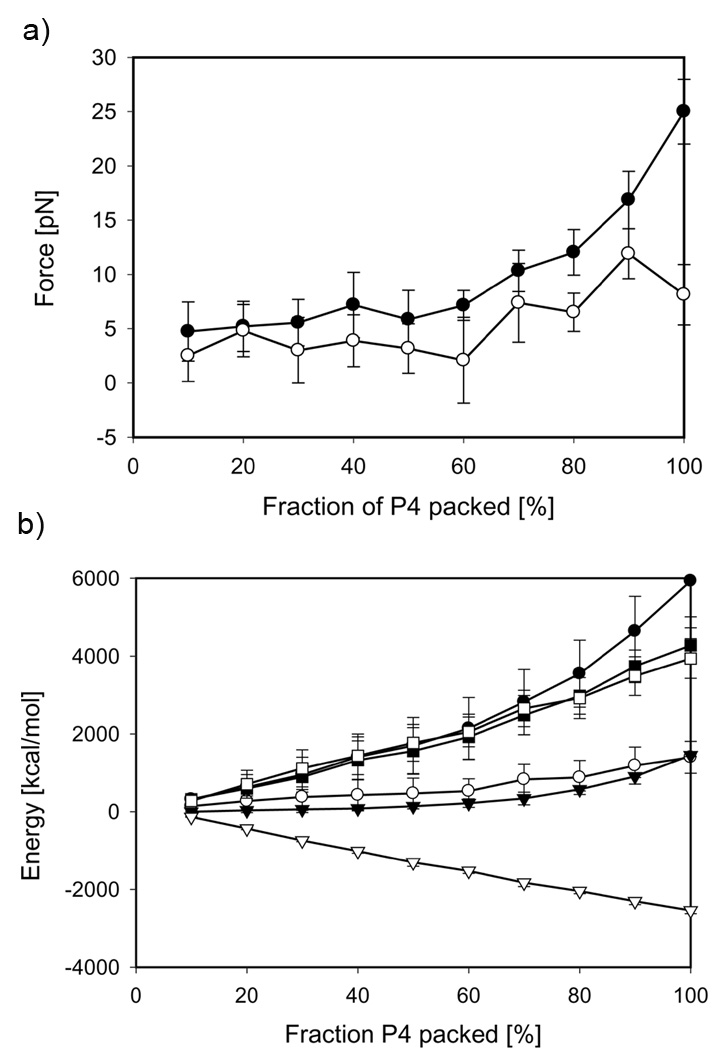
Figure 1.
DNA conformations inside bacteriophage P4 at 100% of genome packed in a) repulsive regime, b) attractive regime and inside bacteriophage Lambda at 78% of wild-type genome length packed in c) repulsive regime, d) attractive regime Side and top views are shown.
Packaging of DNA inside bacteriophage Lambda (Figure 1c, and Figure 2S, Supporting Material) occurs in a similar fashion, except that fraction of DNA at which the ordering and the appearance of the global conformation occurs is smaller (~15%–20%; here and after the percent fractions of genome Lambda relative to the wild-type 48.5 kbp genome are reported). This happens because the Lambda capsid expands upon packaging and the volume of the capsid nearly doubles (Dokland and Murialdo, 1993). In our protocol we used a linear expansion of the capsid’s radius from 210 Å to 290 Å in the range of 20%–40% of Lambda genome packed (Fuller et al., 2007b). Thus, before the capsid’s expansion DNA reaches critical density and the global conformation is formed. During the next stage of packaging (20–40%) the capsid expands and DNA density slightly decreases. This, however, does not affect the global conformation, which remains preserved. We point out that during this stage new portions of DNA wrap around the previously packaged DNA (the “inside-out” model). This does not agree with the proposed inverse-spool model (Purohit et al., 2003; Purohit et al., 2005), which assumes that the initial portion of DNA (head) arranges itself in the outer layers in a highly ordered fashion, whereas the final portion of the genome is predominantly organized in the inner layers (the “outside-in” model). Since we have previously observed “inside-out” packaging in the simulations of bacteriophage ε15 (Petrov et al., 2007b), which does not expand upon packaging, we can probably rule out the hypothesis that the formation of the observed “inside-out” conformation in the repulsive regime is affected by capsid expansion. This is also supported by an observation of the late stages of packaging: after the capsid’s expansion is completed at 40%, DNA continues wrapping around the outermost layer of the capsid. Additionally, we note that Lambda capsid in this series of the simulations was modeled as a “teflon” sphere, thus the interactions between DNA and the capsid were neglected. We believe that the presence of weak repulsive interactions between the capsid and DNA would favor the “inside-out” model, whereas weak attractive interactions would result in the “outside-in” inverse-spool model. Similar to P4 and the other systems we have simulated, the internal motion of DNA diminishes with increasing packing density. During this last stage we also observed that the coaxial spooling motif becomes distorted. In a few analyzed trajectories (Figure 3S, Supporting Material), the distortion was so pronounced that the global DNA conformation changed to a folded toroid (Hud, 1995; Petrov and Harvey, 2007). This is in agreement with the recent morphological analysis of the individual packaging conformations of T5 mutant (Leforestier and Livolant, 2010), in which locally ordered hexagonal structures organized into domains coexist with local defects, due to DNA bending or supertwisting. Additionally, this study provided experimental evidence for multiple DNA conformations inside the capsids (Leforestier and Livolant, 2010). Our results indicate that there is no substantial difference in the internal energy between coaxially spooled and folded-toroidal conformations of fully packed Lambda; this also supports the idea of multiple conformations inside large bacteriophages.
Packaging in the attractive regime
It has been previously shown both experimentally and theoretically that in the presence of multivalent cations free DNA condenses into toroidal or rod-like structures. The diameter of such toroidal condensates was found to vary between 1000 Å and 2000 Å (Hud and Vilfan, 2005), which is larger than the dimensions of bacteriophages P4 and Lambda. It has also been shown that simulations of DNA packaging into a capsid whose linear dimensions are nearly two times smaller than the DNA persistence length, under attractive conditions, lead to the formation of the folded toroid conformations (Forrey and Muthukumar, 2006b).
The analysis of the attractive packaging trajectories for P4 shows that at the very beginning of the packaging, DNA acts much as it does in the repulsive regime; it bends and explores all space inside the capsid, pushing against its walls. This stage is very short, however, and after as little as three turns of DNA get packed, it collapses on itself and forms a stable toroidal condensate. After nucleation has occurred, new portions of ejected DNA collapse on the growing toroid almost immediately (Figure 1b, and Figure 1S, Supporting Material). Along the course of packaging the genome arranges itself into several nearly ideal hexagonally packed coaxial layers. The DNA density of completely packed bacteriophage P4 is relatively low (0.32) and the toroidal condensate remains hollow until almost the entire genome is packed. Only the very last portion of the genome occupies a fraction of the inner part of the toroid in a disordered fashion. By looking at the final conformations one can clearly see that a substantial amount of space inside the capsid remains unoccupied. This is a fundamental difference from the repulsive regime, in which the genome distributes itself throughout the entire volume of the capsid.
Figure 1b also reveals that all toroidal condensates inside P4 contain a single nearly ideal hexagonal lattice region (~300°), separated by a crossover region (~60°). Crossover regions are necessary components of toroidal condensates (Hud and Vilfan, 2005). They appear because when a single DNA forms multiple loops organized in a number of layers, the layers must be connected. Well defined hexagonal domains and crossover regions have been observed experimentally for free and confined toroidal condensates (Hud and Vilfan, 2005; Leforestier and Livolant, 2009). These studies suggest the existence of multiple crossover regions. Possible reasons for not having multiple crossover domains in our simulations are a) a much smaller toroidal circumference in P4 compared to the experimental systems and b) the use of the pair-wise additive force field, which tends to favor hexagonal packaging. (It can be shown that the addition of a point defect in the hexagonal lattice treated in the framework of the pair-wise additive potential favors the preserving of the hexagonal symmetry and not the formation of the pentagonal arrangement).
Packaging of DNA into Lambda under attractive conditions is quite similar to that of P4 (Figure 1d and Figure 2S, Supporting Material). After the nucleation stage, the toroidal structure grows by forming multiple loops organized into several layers. Between 20% and 40% of the genome packed, when capsid expansion occurs, the incoming DNA collapses onto the outer surface of the growing toroid. This pattern also continues until the expansion is completed. The final conformation (containing 78% of the Lambda genome) is a toroidal condensate with an empty core, which contains a single hexagonal domain and a single crossover region. Thus, both repulsive and attractive regimes reveal that Lambda is packed in the “inside-out” fashion. This mechanism is not well pronounced in P4, because of much smaller genome.
Density maps
The current resolution of the experimental cryo-EM methods is not sufficient to provide the detailed information about conformation of DNA inside bacteriophages. From the individual images, one can see the global packaging pattern (Comolli et al., 2008; Jiang et al., 2006), not the distinct positions of the individual strands. The reconstructed EM-densities lack information about single DNA conformations (Petrov and Harvey, 2007). They are obtained by superposing a large number of single individual images, and represent an average, most probable location of the genome inside the capsid.
Thus, following the experimentalists’ footsteps (Jiang et al., 2006; Lander et al., 2006), we averaged the individual conformations and reconstructed the electron density maps. These maps generated for bacteriophages P4 and Lambda are shown in Figures 2a and 2b, respectively. Both maps reveal a similar pattern. The density in the outer regions is ordered and forms distinct layers, while the density in the inner regions is nearly uniformly distributed inside the capsid. Such a pattern, which has previously been reported for other phages (Jiang et al., 2006; Lander et al., 2006), appears because of the following reasons. In the repulsive mode DNA is stored under high pressure, and outer layers, by pushing against the capsid walls are always positioned as close as possible to the protein shell. This leads to the formation of a well defined outer layer of DNA followed by a region where, on average, very little DNA can be found because the next layer must be separated from the first by a layer of water molecules. Each consecutive layer of the observed density is less pronounced due to the intrinsic disorder of DNA stored inside under large bending stresses, resulting in a nearly uniform distribution of density in the inner regions.
Figure 2.
Cross-sectional views of the reconstructed density maps for DNA packaged inside bacteriophages a) P4 and b) Lambda.
Despite the similarity, the density maps depicted in Figure 2 are quantitatively different. The density obtained for bacteriophage Lambda reveals high organization and contains at least 5 distinct radial shells. The distribution of DNA inside bacteriophage P4 is more uniform. Its density exhibits only 2–3 distinct layers. We believe that this difference is because of two factors. First, the volume fraction of DNA inside fully packed P4 (0.32) is less than that of the Lambda mutant (0.4); therefore, the former system is intrinsically more highly disordered. Second, the diameter of bacteriophage P4 is smaller than that of Lambda, so DNA in the outer layers of P4 experiences higher bending stresses compared to Lambda, and it has smaller tendency to get aligned in a regular “parallel” fashion.
Thermodynamics of DNA packaging
The advantages of the computational methodology allowed us to not only reveal information about experimentally measurable forces developed by ATP-driven motors and to estimate the energetic cost of packaging but also to evaluate the individual components of the energy terms (Petrov and Harvey, 2007). The packing energies are calculated as the difference between the energies in the packaged state and those in the unconfined state. We note that in both the attractive and the repulsive regimes the reference state is free uncondensed DNA.
Thermodynamics of P4 packaging
The forces required to hold DNA inside the capsid have been evaluated at every 10% along the packaging course from independent equilibrium trajectories (Figure 3a). As expected, the forces for the repulsive mode are higher than those in the attractive regime. At the final stages, the forces reach the values of 25 and 8 pN, respectively. Surprisingly, however, the forces obtained for the attractive regime are positive in the entire range of the genome fraction packed, which indicates that even in the presence of condensing agents inside the capsid an external energy generated by a motor is required to bring DNA into a confined state. This penalty is associated with a significant entropic cost of DNA confinement into the capsid.
Figure 3.
Thermodynamic analysis of P4 genome packaging a) Packaging forces and b) free energy components: the Helmholtz free energy, ΔA (circles), the internal energy, ΔU (triangles) and entropy, −TΔS (squares). Filled and open symbols represent repulsive and attractive regimes, respectively. Data points correspond to the mean values of respective thermodynamic properties extracted from forty independent MD trajectories, and the error bars represent standard deviations.
Since the volume of P4 capsid does not change over the course of the simulation, the integral of the forces computed at the equilibrium conditions over the entire genome length yields the Helmholtz free energy, ΔA (Figure 3b). The energetic penalty of packing the full P4 genome is 5800 kcal/mol and 1400 kcal/mol for the repulsive and attractive conditions, respectively.
The internal energy of the P4 systems, ΔU, which consists of the bending and angular elastic terms, DNA-DNA interactions and the DNA-capsid term, is known directly from the output of the MD simulations (Figure 4S, Supporting Material). The internal energy of P4 systems packed in the repulsive regime grows monotonically to 1450 kcal/mol, whereas ΔU of packing in the attractive regime steadily decreases to −2540 kcal/mol (Figure 3b). Decomposition of the internal energy into its individual components revealed that the interaction between DNA strands is the major component of the internal energy: 1130 kcal/mol and −2410 kcal/mol for the repulsive and attractive modes, respectively.
The difference between the Helmholtz free energy, ΔA, and the internal free energy, ΔU, represents the entropic term, −TΔS, associated with DNA confinement into the capsid from a free state. The entropic penalty of packaging the P4 genome in the absence and in the presence of the condensing agents is 4300 kcal/mol and 3950 kcal/mol, respectively (Figure 3b). The significance of this result is twofold. First, our analysis shows that for P4 the entopic penalty associated with DNA confinement is the largest component of the Helmholtz free energy. This is contrary to our previous findings for the φ29 and ε15 systems (Petrov and Harvey, 2007; Petrov et al., 2007b), for which the enthalpic term contributed 60–65% of the total free energy in the repulsive mode. This difference appears because a) P4 is smaller than φ29 and ε15; and b) the DNA volume fraction of fully packed P4 (0.31) is significantly smaller than that of φ29 (0.46) and ε15 (0.52). Thus, the DNA-DNA repulsion for P4 is much weaker than that for φ29 and ε15. Second, the entropic penalties independently obtained for two P4 systems confined to the same volume from identical free states by using two different force fields (repulsive and attractive) are very similar. This strongly supports our earlier observation that the entropic penalty associated with DNA confinement upon its packaging into bacteriophages is substantial (Locker et al., 2007; Petrov and Harvey, 2007). We also point out that the negative change in the entropy of confinement (positive −TΔS) corresponds to ordering of DNA molecule. This is contrary to the entropy of the whole system, which increases due to disordering of the water molecules (Jeembaeva et al., 2010).
Thermodynamics of Lambda packaging
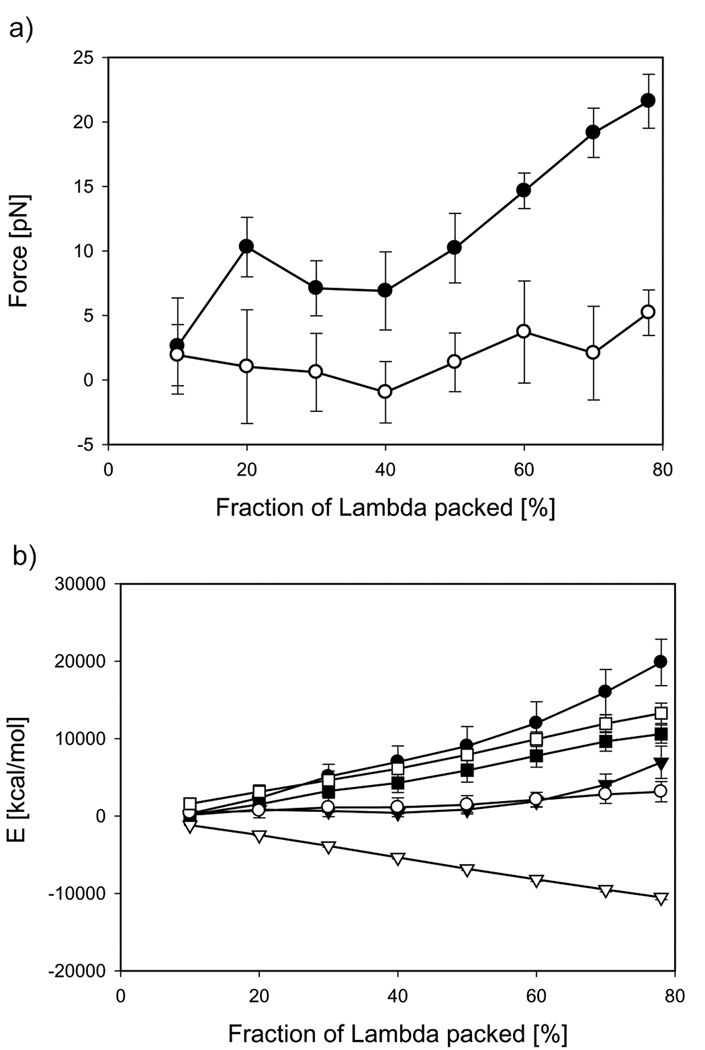
A substantial difference between the packaging protocol used for bacteriophage P4 and that for bacteriophage Lambda is the presence of the controlled capsid expansion between 20% and 40% along the packaging trajectory for the latter system. Thus, the packaging process is associated not only with the enthalpic and entropic penalties but also with the expansion work done by DNA. The range of capsid expansion is distinctly noticeable on the force curve measured in the repulsive mode (Figure 4a): the force rises during the first 20% of packaging, reaching 11 pN, and then steeply decreases due to a substantial volume increase. Further packaging results in a monotonic increase in the force up to 22 pN. A similar dependence of the force on the amount of the genome packed has also been observed experimentally by Fuller et al. (Fuller et al., 2007b). Their packaging experiments performed in 10 mM MgCl2 +20 mM Tris-HCl buffer revealed a peak around 30% of the genome packed, which reflected the capsid expansion. The experimental force reached a value of ~13 pN at a packing density of 78%, which corresponds to the amount of DNA packaged into the Lambda mutant. We note that our calculations done for Lambda in the repulsive regime were parameterized against osmotic pressure data obtained in similar conditions (10 mM MgCl2 +10 mM Tris-HCl). The capsid expansion has a less pronounced effect on the force in the attractive regime, with only a slight decrease in force over the expansion range (Figure 4a). During the rest of the packaging the force reaches a final value of 5 pN. Again, the positive value of the force indicates that work needs to be done by the motor to translocate DNA into the capsid. This is contrary to the expectations that the attraction between DNA strands would drive the packaging.
Figure 4.
Thermodynamic analysis of Lambda genome packaging a) Packaging forces and b) free energy components: the Gibbs free energy, ΔG (circles), the internal energy, ΔU (triangles) and entropy, −TΔS (squares). Filled and open symbols represent repulsive and attractive regimes, respectively. Data points correspond to the mean values of respective thermodynamic properties extracted from thirty two independent MD trajectories, and the error bars represent standard deviations.
To estimate the expansion work done by DNA, the pressure inside the capsids was also calculated from the data generated by the independent MD trajectories used for the force analysis. The pressure exerted on the Lambda capsid walls by confined DNA reached the values of 23.2 atm and 1.3 atm for the repulsive and attractive modes, respectively. Pressure data at the intermediate densities are summarized in Table 1. This table also contains information about the cumulative expansion work pΔV; as expected this work is significantly larger for the repulsive mode (2400 kcal/mol) than for the attractive one (370 kcal/mol).
Table 1.
Pressure, volume and the expansion work done by DNA upon packaging into bacteriophage Lambda in the repulsive and attractive regimes
| % | R [Å] | V [Å3]*10−7 | P [atm] | pΔV [kcal/mol] |
P [atm] | pΔV [kcal/mol] |
|---|---|---|---|---|---|---|
| Repulsive mode | Attractive mode | |||||
| 10 | 210 | 3.87 | 0.71 | 0 | 0.15 | 0 |
| 20 | 210 | 3.87 | 3.23 | 0 | 0.97 | 0 |
| 30 | 250 | 6.54 | 3.01 | 1216 | 0.16 | 352.2 |
| 40 | 290 | 10.2 | 1.40 | 2400 | 0.02 | 370.2 |
| 50 | 290 | 10.2 | 2.90 | 2400 | 0.11 | 370.2 |
| 60 | 290 | 10.2 | 6.21 | 2400 | 0.38 | 370.2 |
| 70 | 290 | 10.2 | 12.5 | 2400 | 0.79 | 370.2 |
| 78 | 290 | 10.2 | 23.5 | 2400 | 1.34 | 370.2 |
Due to the capsid expansion, the integration of the packing force over the DNA contour length equal to the Gibbs free energy. The rest of the thermodynamic analysis for Lambda is similar to that for P4 and is shown in Figure 4b. For the repulsive and attractive modes, the maximum values of the total free energies are 19800 kcal/mol and 3200 kcal/mol and those for the internal energies are 6900 kcal/mol and −10500 kcal/mol, respectively. Thus, the respective entropic penalties of confinement for bacteriophage Lambda obtained by subtracting the enthalpic and expansion work contributions from the Gibbs free energy are 10500 kcal/mol and 13300 kcal/mol. These data again demonstrate that a) the entropic penalties estimated from two sets of independent simulations substantial, and their magnitudes are comparable; and b) the Gibbs free energies are positive in both regimes, and, therefore, in both regimes work needs to be done by a motor to pack DNA inside the bacteriophage. The individual components of the internal energy are shown in Figure 5S, Supporting Material.
DNA condensation inside bacteriophage Lambda
Recently, studies on DNA condensation have been performed not only in the bulk solution but also in the confined spaces inside bacteriophages. It has been shown that partially packed genomes inside Lambda and T5 rearrange into distinct toroidal structures upon addition of condensing agents (Evilevitch, 2006; Leforestier and Livolant, 2009). The study by Leforestier et al. revealed quantitative information about the organization of toroids and the defects that appeared upon condensation. Inspired by these experiments, in this section we report the results of simulations of DNA condensation inside the partially packed Lambda capsid, and address the question how confinement affects DNA condensation.
We extracted DNA conformations generated inside the Lambda capsid in the repulsive regime from the packing trajectories at 10%, 30% and 78% genome packed, which correspond to DNA volume fractions of 0.13, 0.24 and 0.39, respectively, and used them as starting structures for condensation simulations. The DNA-DNA potential was switched to the attractive regime, and the MD simulations were initiated.
Analysis of the MD trajectories (http://rumour.biology.gatech.edu/Publications/showcase.html) shows that in all three cases DNA collapses on itself during the first 50 ns of the MD runs. After this initial stage (Figure 5a), the system containing 10% of the DNA genome (DNA volume fraction 0.13) undergoes a significant structural rearrangement and compaction, and turns into a regular toroidal conformation (Figure 5b). The condensed toroid has a single well-defined hexagonal region adjacent to a crossover region, as did the structure generated by sequential packaging the first ten percent of the Lambda genome in the attractive regime (discussed above).
Figure 5.
DNA conformations inside bacteriophage Lambda at 10%, 30% and 78% of wild-type genome length a) initial structures prepacked in the repulsive regime, b) final structures obtained by condensation in the attractive regime. Side and top views are shown.
Condensation of the system containing 30% of the Lambda genome undergoes the transition in a similar way. The initial conformation relaxes into a toroidal structure; nevertheless, a few strands form a loop below the toroidal plane, which stays there until the end of the MD trajectory (Figure 5b). It is possible that secondary relaxation processes occur on time scales much longer than those used in our simulations, in which case we are observing the appearance of non-equilibrium kinetic conformational traps, which arise as a result of fast DNA condensation.
The formation of non-equilibrium structures is more pronounced during condensation of 78% of the genome inside Lambda phage (Figure 5b). The genome packed under the repulsive regime in a distorted spooled conformation shrinks upon changing the DNA-DNA interaction potential, but it remains topologically trapped in the same state and cannot rearrange itself into a toroidal structure. Thus, the high DNA density favors the formation of the condensed non-equilibrium domains. In the absence of confinement DNA would condense into toroidal structures with a diameter of greater than 1000 Å (Hud and Vilfan, 2005). The DNA bending stresses inside the Lambda capsid, whose diameter (580 Å) are comparable to the DNA persistence length, is another factor that hinders relaxation of the genome into a regular toroidal conformation. The experimental studies performed by Leforestier et al. (Leforestier and Livolant, 2009) on bacteriophage T5 use DNA chains up to 55 kbp in length, which corresponds to the volume fraction up to 0.18. Additionally, the diameter of T5 is compared with the smaller toridal condensates formed in the unconstrained bulk conditions (1000 Å). At these conditions the formation of regular toroidal structures is favored.
It has been recently demonstrated by small angle X-ray scattering experiments that upon addition of condensing agents to partially packed Lambda with the genome fraction greater than 85%, the DNA-DNA separation decreases from ~30 Å to ~27.5 Å (Qui et al., 2009). This fact led the authors of the study to conclude that DNA at high densities inside Lambda condenses into conformations other than toroidal spools. As shown in Table 2, the distance between DNA strands increases with the amount of DNA packed from 26.5 Å to 27.3 Å. This suggests that this parameter can be used as a measure of non-ideality in DNA condensates. Thus, our simulations done at high packing densities agree with the experimental data and suggest that the confinement affects the conformation of the condensed DNA, resulting in the formation of non-equilibrium condensates at high densities.
Table 2.
Average separation distance between DNA strands in the repulsive and attractive regimes at different packing densities.
| Lambda genome, [%] |
Separation distance,[Å] | |
|---|---|---|
| Repulsive | Attractive | |
| 10% | 33.1 | 26.2 |
| 30% | 32.8 | 26.4 |
| 78% | 30.8 | 27.2 |
CONCLUSIONS
Our study reveals fundamental differences in conformations of DNA packed into bacteriophages P4 and Lambda in the repulsive and attractive modes. In the repulsive regime the genomes form concentrically spooled structures and occupy all available space inside the capsids. Packaging in the attractive mode leads to formation of regular toroidal condensates. The compaction in a specific conformation occurs as a result of interplay between bending stresses experienced by the confined polymer and interactions between the strands. Packaging in both regimes is not spontaneous and requires assistance by an external motor. Detailed thermodynamic analysis of packaging shows that confinement is accompanied by large entopic penalties, and these are weakly dependent on the packaging mode. Further, at the low packing densities the confined genomes condense into toroidal conformations upon changing the DNA-DNA interaction potential from the repulsive to the attractive regime. Rearrangement of the genome into toroids becomes hindered at higher DNA densities; condensation of the tightly packed genome leads to the formation of non-equilibrium kinetically trapped condensates. These results are supported by the experimental observations.
Supplementary Material
ACKNOWLEDGMENTS
We would like to thank Donald C. Rau, Dr. Nicholas V. Hud, Dr. Alex Evilevitch, Dr., Dr. Robert K.-Z. Tan, Dr. Xiangyun Qui, Dr. Rebecca C. Locker, Dr. Gabriel C. Lander for helpful discussions. We also acknowledge Mr. Alexandre Lomsadze for technical assistance with calculations. The research is supported by National Institutes of Health Grant R01 GM70875.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supplementary data: Supplementary data is available online at
REFERENCES
- Ali I, Marenduzzo D, Yeomans JM. Dynamics of polymer packaging. Journal of Chemical Physics. 2004;121:8635–8641. doi: 10.1063/1.1798052. [DOI] [PubMed] [Google Scholar]
- Ali I, Marenduzzo D, Yeomans JM. Polymer packaging and ejection in viral capsids: Shape matters. Physical Review Letters. 2006;96 doi: 10.1103/PhysRevLett.96.208102. [DOI] [PubMed] [Google Scholar]
- Arsuaga J, Tan RKZ, Vazquez M, Sumners DW, Harvey SC. Investigation of viral DNA packaging using molecular mechanics models. Biophysical Chemistry. 2002a;101:475–484. doi: 10.1016/s0301-4622(02)00197-7. [DOI] [PubMed] [Google Scholar]
- Arsuaga J, Vazquez M, Trigueros S, Sumners D, Roca J. Knotting probability of DNA molecules confined in restricted volumes: DNA knotting in phage capsids. Proceedings of the National Academy of Sciences of the United States of America. 2002b;99:5373–5377. doi: 10.1073/pnas.032095099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arsuaga J, Vazquez M, McGuirk P, Trigueros S, Sumners DW, Roca J. DNA knots reveal a chiral organization of DNA in phage capsids. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:9165–9169. doi: 10.1073/pnas.0409323102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berendsen HJC, Postma JPM, Vangunsteren WF, Dinola A, Haak JR. Molecular-Dynamics With Coupling To An External Bath. Journal Of Chemical Physics. 1984;81:3684–3690. [Google Scholar]
- Black LW. DNA packaging in dsDNA bacteriophages. Annual Review of Microbiology. 1989;43:267–292. doi: 10.1146/annurev.mi.43.100189.001411. [DOI] [PubMed] [Google Scholar]
- Bloomfield VA. Condensation of DNA by multivalent cations: Considerations on mechanism. Biopolymers. 1991;31:1471–1481. doi: 10.1002/bip.360311305. [DOI] [PubMed] [Google Scholar]
- Bloomfield VA. DNA condensation by multivalent cations. Biopolymers. 1997;44:269–282. doi: 10.1002/(SICI)1097-0282(1997)44:3<269::AID-BIP6>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- Comolli LR, Spakowitz AJ, Siegerist CE, Jardine PJ, Grimes S, Anderson DL, Bustamante C, Downing KH. Three-dimensional architecture of the bacteriophage phi 29 packaged genome and elucidation of its packaging process. Virology. 2008;371:267–277. doi: 10.1016/j.virol.2007.07.035. [DOI] [PubMed] [Google Scholar]
- Dokland T, Murialdo H. Structural transitions during maturation of bacteriophage-lambda capsids. Journal of Molecular Biology. 1993;233:682–694. doi: 10.1006/jmbi.1993.1545. [DOI] [PubMed] [Google Scholar]
- Dokland T, Lindqvist BH, Fuller SD. Image reconstruction from cryoelectron micrographs reveals the morphopoietic mechanism in the P2–P4 bacteriophage system. Embo Journal. 1992;11:839–846. doi: 10.1002/j.1460-2075.1992.tb05121.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earnshaw WC, Harrison SC. DNA arrangement in isometric phage heads. Nature. 1977;268:598–602. doi: 10.1038/268598a0. [DOI] [PubMed] [Google Scholar]
- Earnshaw WC, King J, Harrison SC, Eiserling FA. Strucural organization of DNA packaged within heads of T4 wild-type, isometric and giant bacteriophages. Cell. 1978;14:559–568. doi: 10.1016/0092-8674(78)90242-8. [DOI] [PubMed] [Google Scholar]
- Evilevitch A. Effects of condensing agent and nuclease on the extent of ejection from phage lambda. Journal of Physical Chemistry B. 2006;110:22261–22265. doi: 10.1021/jp060573j. [DOI] [PubMed] [Google Scholar]
- Evilevitch A, Lavelle L, Knobler CM, Raspaud E, Gelbart WM. Osmotic pressure inhibition of DNA ejection from phage. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:9292–9295. doi: 10.1073/pnas.1233721100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evilevitch A, Gober JW, Phillips M, Knobler CM, Gelbart WM. Measurements of DNA lengths remaining in a viral capsid after osmotically suppressed partial ejection. Biophysical Journal. 2005;88:751–756. doi: 10.1529/biophysj.104.045088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evilevitch A, Fang LT, Yoffe AM, Castelnovo M, Rau DC, Parsegian VA, Gelbart WM, Knobler CM. Effects of salt concentrations and bending energy on the extent of ejection of phage genomes. Biophysical Journal. 2008;94:1110–1120. doi: 10.1529/biophysj.107.115345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forrey C, Muthukumar M. Langevin dynamics simulations of genome packing in bacteriophage. Biophys J. 2006a;91:25–41. doi: 10.1529/biophysj.105.073429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forrey C, Muthukumar M. Langevin dynamics simulations of genome packing in bacteriophage. Biophysical Journal. 2006b;91:25–41. doi: 10.1529/biophysj.105.073429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank J, Radermacher M, Penczek P, Zhu J, Li YH, Ladjadj M, Leith A. SPIDER and WEB: Processing and visualization of images in 3D electron microscopy and related fields. Journal Of Structural Biology. 1996;116:190–199. doi: 10.1006/jsbi.1996.0030. [DOI] [PubMed] [Google Scholar]
- Fuller DN, Rickgauer JP, Jardine PJ, Grimes S, Anderson DL, Smith DE. Ionic effects on viral DNA packaging and portal motor function in bacteriophage phi 29. Proceedings of the National Academy of Sciences of the United States of America. 2007a;104:11245–11250. doi: 10.1073/pnas.0701323104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller DN, Raymer DM, Rickgauer JP, Robertson RM, Catalano CE, Anderson DL, Grimes S, Smith DE. Measurements of single DNA molecule packaging dynamics in bacteriophage lambda reveal high forces, high motor processivity, and capsid transformations. Journal of Molecular Biology. 2007b;373:1113–1122. doi: 10.1016/j.jmb.2007.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grayson P, Evilevitch A, Inamdar MM, Purohit PK, Gelbart WM, Knobler CM, Phillips R. The effect of genome length on ejection forces in bacteriophage lambda. Virology. 2006;348:430–436. doi: 10.1016/j.virol.2006.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafner EW, Tabor CW, Tabor H. Mutants of Escherichia coli that do not contain 1,4-diaminobutane (putrescine) or spermidine. Journal of Biological Chemistry. 1979;254:2419–2426. [PubMed] [Google Scholar]
- Hamada K, Fujisawa H, Minagawa T. A defined in vitro system for packaging of bacteriophage T3 DNA. Virology. 1986;151:119–123. doi: 10.1016/0042-6822(86)90109-1. [DOI] [PubMed] [Google Scholar]
- Harvey SC, Petrov AS, Devkota B, Boz MB. Viral assembly: a molecular modeling perspective. Physical Chemistry Chemical Physics. 2009;11:10553–10564. doi: 10.1039/b912884k. [DOI] [PubMed] [Google Scholar]
- Hud NV. Double stranded DNA organization in bacteriophage heads: an alternative toroid-based model. Biophysical Journal. 1995;69:1355–1362. doi: 10.1016/S0006-3495(95)80002-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hud NV, Vilfan ID. Toroidal DNA condensates: Unraveling the fine structure and the role of nucleation in determining size. Annual Review of Biophysics and Biomolecular Structure. 2005;34:295–318. doi: 10.1146/annurev.biophys.34.040204.144500. [DOI] [PubMed] [Google Scholar]
- Jeembaeva M, Jonsson B, Castelnovo M, Evilevitch A. DNA Heats Up: Energetics of Genome Ejection from Phage Revealed by Isothermal Titration Calorimetry. Journal of Molecular Biology. 2010;395:1079–1087. doi: 10.1016/j.jmb.2009.11.069. [DOI] [PubMed] [Google Scholar]
- Jiang W, Chang J, Jakana J, Weigele P, King J, Chiu W. Structure of epsilon15 bacteriophage reveals genome organization and DNA packaging/injection apparatus. Nature. 2006;439:612–616. doi: 10.1038/nature04487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kindt J, Tzlil S, Ben-Shaul A, Gelbart WM. DNA packaging and ejection forces in bacteriophage. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:13671–13674. doi: 10.1073/pnas.241486298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaMarque JC, Le TVL, Harvey SC. Packaging double-helical DNA into viral capsids. Biopolymers. 2004;73:348–355. doi: 10.1002/bip.10529. [DOI] [PubMed] [Google Scholar]
- Lander GC, Evilevitch A, Jeembaeva M, Potter CS, Carragher B, Johnson JE. Bacteriophage lambda stabilization by auxiliary protein gpD: Timing, location, and mechanism of attachment determined by cryo-EM. Structure. 2008;16:1399–1406. doi: 10.1016/j.str.2008.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander GC, Tang L, Casjens SR, Gilcrease EB, Prevelige P, Poliakov A, Potter CS, Carragher B, Johnson JE. The structure of an infectious P22 virion shows the signal for heedful DNA packaging. Science. 2006;312:1791–1795. doi: 10.1126/science.1127981. [DOI] [PubMed] [Google Scholar]
- Leforestier A, Livolant F. Structure of toroidal DNA collapsed inside the phage capsid. Proceedings of the National Academy of Sciences of the United States of America. 2009;106 doi: 10.1073/pnas.0901240106. 9157-9152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leforestier A, Livolant F. The Bacteriophage Genome Undergoes a Succession of Intracapsid Phase Transitions upon DNA Ejection. Journal of Molecular Biology. 2010;396:384–395. doi: 10.1016/j.jmb.2009.11.047. [DOI] [PubMed] [Google Scholar]
- Locker CR, Harvey SC. A model for viral geome packing. Multiscale Modeling and Simulation. 2006;5:1264–1279. [Google Scholar]
- Locker CR, Fuller SD, Harvey SC. DNA organization and thermodynamics during viral packing. Biophysical Journal. 2007;93:2861–2869. doi: 10.1529/biophysj.106.094771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marenduzzo D. Computer simulations of DNA packing inside bacteriophages: Elasticity, electrostatics and entropy. Computational and Mathematical Methods in Medicine. 2008;9:317–325. [Google Scholar]
- Micheletti C, Marenduzzo D, Orlandini E, Sumners DW. Simulations of knotting in confined circular DNA. Biophysical Journal. 2008;95:3591–3599. doi: 10.1529/biophysj.108.137653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison G, Thirumalai D. Semiflexible chains in confined spaces. Physical Review E. 2009;79 doi: 10.1103/PhysRevE.79.011924. [DOI] [PubMed] [Google Scholar]
- Parsegian VA, Rand RP, Rau DC. Macromolecules and water: Probing with osmotic stress. Energetics Of Biological Macromolecules. 1995:43–94. doi: 10.1016/0076-6879(95)59039-0. [DOI] [PubMed] [Google Scholar]
- Parsegian VA, Rand RP, Rau DC. Osmotic stress, crowding, preferential hydration, and binding: A comparison of perspectives. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:3987–3992. doi: 10.1073/pnas.97.8.3987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrov AS, Harvey SC. Structural and thermodynamic principles of viral packaging. Structure. 2007;15:21–27. doi: 10.1016/j.str.2006.11.013. [DOI] [PubMed] [Google Scholar]
- Petrov AS, Harvey SC. Packaging double-helical DNA into viral capsids: Structures, forces, and energetics. Biophysical Journal. 2008;95:497–502. doi: 10.1529/biophysj.108.131797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrov AS, Boz MB, Harvey SC. The conformation of double-stranded DNA inside bacteriophages depends on capsid size and shape. Journal of Structural Biology. 2007a;160:241–248. doi: 10.1016/j.jsb.2007.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrov AS, Lim-Hing K, Harvey SC. Packaging of DNA by Bacteriophage Epsilon15: Structure, Forces and Thermodynamics. Structure. 2007b;15:807–812. doi: 10.1016/j.str.2007.05.005. [DOI] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera: A visualization system for exploratory research and analysis. Journal Of Computational Chemistry. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Podgornik R, Rau DC, Parsegian VA. The action of interhelical forces on the organization of DNA double helices: fluctuation-enhanced decay of electrostatic double-layer andd hydration forces. Macromolecules. 1989;22:1780–1786. [Google Scholar]
- Purohit PK, Kondev J, Phillips R. Force steps during viral DNA packaging? Journal of the Mechanics and Physics of Solids. 2003;51:2239–2257. [Google Scholar]
- Purohit PK, Inamdar MM, Grayson PD, Squires TM, Kondev J, Phillips R. Forces during bacteriophage DNA packaging and ejection. Biophysical Journal. 2005;88:851–866. doi: 10.1529/biophysj.104.047134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qui X, Rau DC, Parsegian VA, Fang LT, Knobler CM, Gelbart WM. Charting the structure and energetrics of packaged DNA in bacteriophages lambda. Biophysical Journal. 2009;Vol. 96:422a. Biophysical Society Meeting Abstracts. [Google Scholar]
- Raspaud E, Durand D, Livolant F. Interhelical spacing in liquid crystalline spermine and spermidine-DNA precipitates. Biophysical Journal. 2005;88:392–403. doi: 10.1529/biophysj.104.040113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rau DC, Parsegian VA. Direct measurment of the intermolecular forces between counterion-condensed DNA double helices: Evidence for long-range attractive hydration forces. Biophysical Journal. 1992;61:246–259. doi: 10.1016/S0006-3495(92)81831-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rau DC, Lee B, Parsegian VA. Measurment of the repulsive force between poly-electrolyte molecules in ionic solution: hydration forces between parallel DNA double helicies. Proceedings of the National Academy of Sciences of the United States of America-Biological Sciences. 1984;81:2621–2625. doi: 10.1073/pnas.81.9.2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rickgauer JP, Fuller DN, Grimes S, Jardine PJ, Anderson DL, Smith DE. Portal motor velocity and internal force resisting viral DNA packaging in bacteriophage phi 29. Biophysical Journal. 2008;94:159–167. doi: 10.1529/biophysj.107.104612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rollins GC, Petrov AS, Harvey SC. The role of DNA twist in the packaging of viral genomes. Biophysical Journal. 2008;94:L38–L40. doi: 10.1529/biophysj.107.126698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar T, Petrov AS, Vitko JR, Santai CT, Harvey SC, Mukerji I, Hud NV. Integration Host Factor (IHF) Dictates the Structure of Polyamine-DNA Condensates: Implications for the Role of IHF in the Compaction of Bacterial Chromatin. Biochemistry. 2009;48:667–675. doi: 10.1021/bi8019965. [DOI] [PubMed] [Google Scholar]
- Smith DE, Tans SJ, Smith SB, Grimes S, Anderson DL, Bustamante C. The bacteriophage phi 29 portal motor can package DNA against a large internal force. Nature. 2001;413:748–752. doi: 10.1038/35099581. [DOI] [PubMed] [Google Scholar]
- Stevens MJ. Simple simulations of DNA condensation. Biophysical Journal. 2001;80:130–139. doi: 10.1016/S0006-3495(01)76000-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan RKZ, Petrov AS, Harvey SC. YUP: A molecular simulation program for coarse-grained and multiscaled models. Journal of Chemical Theory and Computation. 2006;2:529–540. doi: 10.1021/ct050323r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzlil S, Kindt JT, Gelbart WM, Ben-Shaul A. Forces and pressures in DNA packaging and release from viral capsids. Biophysical Journal. 2003;84:1616–1627. doi: 10.1016/S0006-3495(03)74971-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Holde KE. Chromatin. New York: Springer-Verlag; 1989. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.