Abstract
Cdc45p assembles at replication origins before initia tion and is required for origin firing in Saccharomyces cerevisiae. A heat-inducible cdc45 degron mutant was constructed that promotes rapid degradation of Cdc45p at the restrictive temperature. Consistent with a role in initiation, loss of Cdc45p in G1 prevents all detectable DNA replication without preventing subsequent entry into mitosis. Loss of Cdc45p activity during S-phase blocks S-phase completion but not activation of replication checkpoints. Using density substitution, we show that after allowing replication fork establishment, Cdc45p inactivation prevents the subsequent progression of individual replication forks. This provides the first direct functional evidence that Cdc45p plays an essential role during elongation. Thus, like the large T antigen in SV40 replication, Cdc45p plays a central role in both initiation and elongation phases of chromosomal DNA replication.
Keywords: cell cycle/DNA polymerase/DNA replication/pre-replicative complex/S-phase
Introduction
Considerable progress has been made in understanding the earliest events in the replication of budding yeast chromosomes. These include origin recognition by the origin recognition complex (ORC) and pre-replicative complex (pre-RC) assembly by Cdc6p and the Mcm2–7 proteins (Bell and Stillman, 1992; Diffley and Cocker, 1992; Diffley et al., 1994; Cocker et al., 1996; Aparicio et al., 1997; Donovan et al., 1997; Liang and Stillman, 1997; Tanaka et al., 1997). Several factors have been instrumental in facilitating the analysis of replication origin function. The development and application of sensitive assays such as genomic footprinting (Diffley and Cocker, 1992; Diffley et al., 1994; Santocanale and Diffley, 1996) and chromatin immunoprecipitation (CHIP) (Aparicio et al., 1997; Tanaka et al., 1997) has allowed examination of protein complexes at individual origins. Determination of the order in which pre-RC components assemble has been made possible primarily because the very rapid degradation of Cdc6p (Piatti et al., 1995; Drury et al., 1997) has provided the means to efficiently and rapidly eliminate this central pre-RC component from cells. Finally, using two-dimensional gel electrophoresis to examine replication intermediates, it has been possible to demonstrate directly that mutants in pre-RC components are defective in establishing bidirectional replication forks at individual origins (Yan et al., 1993; Fox et al., 1995; Liang et al., 1995; Zou et al., 1997).
It should also be possible to examine individual replication forks during elongation. Indeed, CHIP has been used to demonstrate that Cdc45p and Mcm proteins redistribute from origins to distal sequences after origin firing, consistent with the possibility that they travel with or near replication forks (Aparicio et al., 1997). However, functional assays that test whether particular proteins are required for ongoing DNA synthesis at individual replication forks are lacking. At present, elongation is generally studied in vivo by examining global DNA synthesis defects in conditional mutants, either by monitoring S-phase progression using flow cytometry or incorporation of radiolabelled precursors into DNA, or by examining the size distribution of total nascent DNA. Because these approaches are not designed to examine the progression of individual replication forks, they cannot unambiguously determine which stage of replication is defective in the mutant. For example, because eukaryotic chromosomes are replicated from multiple origins that fire throughout S-phase (Ferguson et al., 1991; Fangman and Brewer, 1992; Brewer et al., 1993), a defect in S-phase progression might be caused by a defect in elongation but might also be caused by defects in origin firing. Similarly, the accumulation of small nascent DNA might be due to defects in processing Okazaki fragments throughout elongation, defects in establishing the elongation machinery at replication origins or even degradation of nascent DNA.
SV40 has provided a very important model for eukaryotic DNA replication and the cell-free SV40 replication system has allowed the identification and characterization of many replication proteins (Challberg and Kelly, 1989; Weinberg et al., 1990; Waga and Stillman, 1994). However, because the viral-encoded large T antigen has essential roles both in initiation as an origin recognition factor and during elongation as the replicative helicase (Fanning and Knippers, 1992), many cellular factors involved in the initiation and elongation phases of chromosomal DNA replication have yet to be fully characterized.
Based on examination of mutant phenotypes, a number of yeast proteins have been shown to have essential roles in initiation; however, any role for these proteins in elongation has been more difficult to demonstrate. For example, Cdc45p associates with origins during G1- or early S-phase (Aparicio et al., 1997, 1999; Zou and Stillman, 1998) and two-dimensional gel electrophoresis has shown that the cold-sensitive cdc45-1 mutant is defective in initiating DNA replication (Zou et al., 1997). Thus, it seems clear that Cdc45p has some essential role during initiation. There is circumstantial evidence suggesting that Cdc45p may also have a role during elongation. After initiation, CHIP analysis has shown that Cdc45p can be cross-linked near replication forks during S-phase (Aparicio et al., 1997). Recently, it has also been shown that a topoisomerase 1 (top1), cdc45 double mutant transiently accumulates small amounts of low molecular weight DNA intermediates at the restrictive temperature (Reid et al., 1999). However, even this double mutant shows little or no defect in DNA replication; significant S-phase delay and accumulation of small DNA molecules requires the presence of a third mutation in the DPB11 gene (Reid et al., 1999). Arguing against a role in elongation, reciprocal shift experiments with the cold-sensitive cdc45-1 mutant indicated that Cdc45p plays no role in replication after hydroxyurea (HU) arrest, indicating that it might not be required after initiation (Owens et al., 1997).
In this paper we describe a strategy for studying elongation using a new class of thermolabile mutants and classical density-transfer technology. We have used this to demonstrate that in addition to its role in initiation, Cdc45p has an essential role throughout S-phase in the elongation phase of DNA replication.
Results
Analysis of a thermolabile ‘degron’ mutant shows that Cdc45p is required for an early event in DNA replication
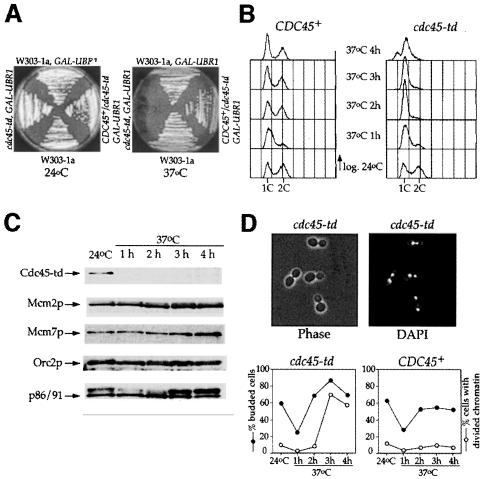
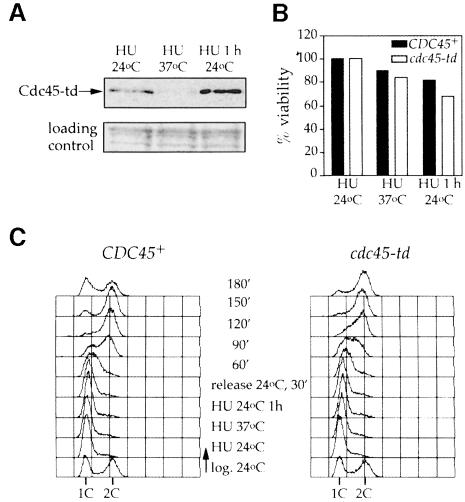
Temperature-sensitive mutants can be extremely useful in determining the function of proteins in vivo; however, there can also be significant problems in interpreting results of experiments with such mutants. As they have usually been selected simply for viability at one temperature but not another, they can be significantly defective in function at permissive temperatures and not completely defective at restrictive temperatures. In addition, they often exhibit reduced levels of not only the mutant protein, but also other wild-type proteins that interact with the mutant protein even at permissive temperatures. For example, levels of Mcm5p are reduced in mcm3, mcm4 and mcm7 mutants at permissive temperature (S.Donovan and J.F.X.Diffley, unpublished observations). To circumvent these problems in our examination of Cdc45p, we have constructed a heat-inducible degron mutant, cdc45-td, by fusing a ‘degron cassette’ to the N-terminus of Cdc45p (Dohmen et al., 1994). We have used a modification of this method in which UBR1, the E3 ubiquitin ligase required for N-end rule degradation, is overexpressed from the GAL1,10 promoter, which increases the efficiency of degradation at 37°C (Labib et al., 2000; see Materials and methods). The cdc45-td mutant grows well at 24°C with no apparent defects in growth rate but arrests in the first cell cycle after shift to 37°C in the presence of high levels of UBR1 (Figure 1A and B).
Fig. 1. Characterization of the cdc45-td mutant. (A) cdc45-td mutant cells grow normally at 24°C but show a temperature-sensitive phenotype at 37°C. (B) Logarithmic phase cultures of cdc45-td mutant and parental wild-type cells growing at 24°C in YP plus raffinose were shifted to 37°C in YP medium containing galactose. Samples were collected at the time points indicated and analysed by flow cytometry to follow cell cycle progression. (C) Immunoblot analysis of Cdc45-td and other proteins during the course of the experiment. (D) Cells from the experiment shown in (B) were fixed, stained with DAPI and analysed by fluorescence microscopy. Budding index and the proportion of cells with divided chromatin are indicated.
The cdc45-td mutant arrests cell growth because Cdc45p is rapidly degraded at 37°C. Figure 1C shows that Cdc45-td is completely degraded within the first hour when the culture is shifted to 37°C. In other experiments we have found that Cdc45-td is completely degraded within 20 min when UBR1 is overexpressed prior to temperature shift (see supplementary material, available in The EMBO Journal Online). Several factors, including Mcm proteins (Hopwood and Dalton, 1996; Dalton and Hopwood, 1997; Zou et al., 1997; Zou and Stillman, 1998; Kukimoto et al., 1999), ORC (Zou et al., 1997) and DNA polymerase α (Mimura and Takisawa, 1998; Kukimoto et al., 1999), have been shown to interact with Cdc45p in vivo. At least some of these proteins including ORC and Mcm2p are not present in large excess in cells (Rowley et al., 1995; Lei et al., 1996). Figure 1C shows that although Cdc45-td is rapidly degraded after the temperature shift, the levels of Mcm2p, Mcm7p, Orc2p and the B subunit of DNA polymerase α-primase (p86/91) are not affected even after 4 h at 37°C. This result is not surprising since proteins such as cyclins, which are rapidly degraded by the proteosome after polyubiquitylation, do not target tightly bound proteins like cyclin-dependent kinases (Cdks) for degradation. We have constructed degron mutants in several multi-subunit protein complexes including the MCM complex, ORC and DNA polymerase α-primase. In each case we have found that only the degron-fused subunit is degraded; other tightly associated subunits remain stable after temperature shift (Labib et al., 2000; E.A.Noton, C.Desdouets and J.F.X.Diffley, unpublished data). Thus, specific degradation appears to be a general feature of degron mutants. In addition, the degron mutant is recessive (Figure 1A; see supplementary material), which might not be predicted if it acted by targeting another protein for degradation. Finally, the degron was designed so that recognition by UBR1 should occur at both permissive and restrictive temperatures (Dohmen et al., 1994). Polyubiquitylation, however, should only occur at the restrictive temperature when the degron unfolds. The fact that the cdc45-td mutant is temperature sensitive argues that it is Cdc45-td itself that is acting as the acceptor for polyubiquitylation. Taken together, these results strongly argue that the cdc45-td mutant blocks entry into S-phase by specifically eliminating the Cdc45 protein from cells.
To further characterize this mutant, we studied the progression of cdc45-td and isogenic wild-type cells through the cell cycle, analysing their DNA content by flow cytometry after shifting logarithmic cultures to the restrictive temperature. Figure 1B shows that in contrast to wild-type cells, cdc45-td mutant cells accumulate with 1C DNA content after shifting to the non-permissive temperature, indicating that Cdc45p is required for entry into S-phase. This is in agreement with work published previously (Hennessy et al., 1991; Hopwood and Dalton, 1996; Hardy, 1997; Owens et al., 1997; Zou et al., 1997), and is consistent with the idea that Cdc45p plays an essential role in the initiation of DNA replication.
In addition to their failure to enter S-phase, Figure 1B shows that cdc45-td mutant cells undergo cell division at late time points and cells with less than 1C DNA content begin to accumulate. This phenotype is strikingly similar to that of cells lacking Cdc6p (Piatti et al., 1995) and indicates that cdc45-td mutant cells undergo mitosis in the absence of DNA replication. A time course shows that nuclear division without DNA replication occurs in most cdc45-td cells 2–3 h after shift to 37°C (Figure 1D). Therefore, loss of Cdc45p from cells prevents S-phase entry but does not prevent the subsequent entry into mitosis.
Cdc45p is required for entry into and progression through S-phase
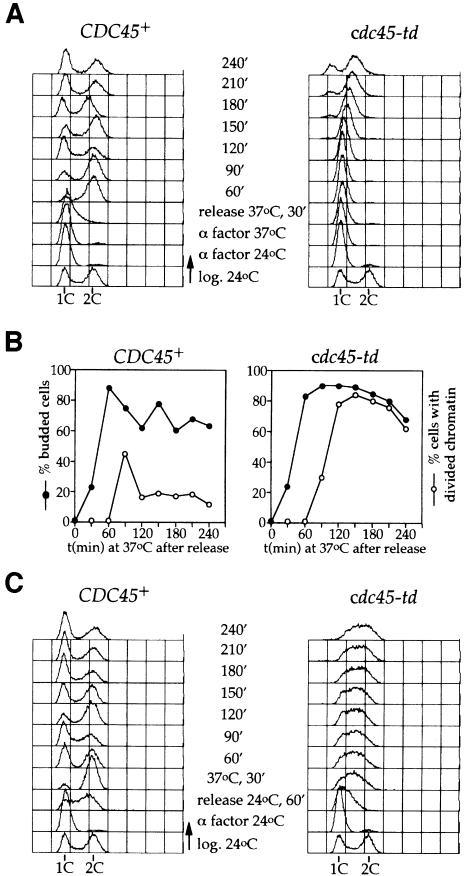
We next examined the requirement of Cdc45p for the entry into S-phase in synchronous cultures. Cells were first arrested in G1 with α factor and then released from the α factor arrest after shifting to the restrictive temperature. Figure 2A shows that, after release from the arrest, wild-type cells enter S-phase at ∼30 min and finish S-phase by 60 min. In contrast, cdc45-td mutant cells undergo little or no apparent increase in DNA content after release from the block with α factor at 37°C, indicating that Cdc45p is required to enter S-phase, consistent with the results of Owens et al. (1997). In agreement with the data obtained with asynchronous cultures (Figure 1C), cdc45-td mutant cells divide without replicating their DNA and accumulate at late time points with less than 1C DNA content at 37°C (Figure 2A). Figure 2B shows that cells with divided nuclei first appear in the wild-type strain (CDC45+) ∼90 min after release from α factor. Although the cdc45-td mutant cells do not progress through S-phase, the right panel in Figure 2B shows that they undergo nuclear division 90 min after release from the α factor block, at the same time as the wild-type cells.
Fig. 2. Cdc45p is required for progression through S-phase. (A) Cycling cultures of cdc45-td and parental wild-type strains were arrested in G1 with α factor at 24°C in YP plus raffinose. They were shifted to 37°C in YP plus galactose and held in α factor for 45 min. Cells were then released from the block at 37°C and samples were collected at the time points indicated and analysed by flow cytometry. (B) Budding index and percentage of cells with divided chromatin corresponding to the experiment shown in (A). (C) Logarithmic cultures of cdc45-td mutant and parental wild-type cells were arrested in G1 with α factor at 24°C in YP plus raffinose. They were released from the arrest at 24°C in YP plus galactose, and after 60 min the cultures were shifted to 37°C. Samples were taken for flow cytometry analysis.
We were interested in determining whether Cdc45p is only required for entry into S-phase or whether it is also required for S-phase progression after the G1–S transition. For this, we synchronized cells in G1 with α factor at the permissive temperature (24°C). We then released these cells from the arrest at 24°C. After 60 min both wild-type and mutant cells had entered S-phase. At this point, cultures were shifted to 37°C to degrade Cdc45-td. FACS analysis in Figure 2C shows that under these conditions, wild-type cells rapidly finish S-phase and enter a new cell cycle, soon losing synchrony. However, cdc45-td mutant cells do not increase in number (data not shown) and, although they have clearly entered S-phase, they do not complete S-phase even 4 h after the shift to 37°C (Figure 2C). In this experiment, where Cdc45-td is degraded during S-phase, there is no chromatin division even at late time points and cells remain as large, budded cells with a single intact nucleus (data not shown), indicating that the replication checkpoint is activated and remains active when Cdc45-td is degraded. This is in contrast to Figure 2A, where Cdc45p was degraded prior to entry into S-phase. Taken together, these experiments show that Cdc45p is required not only for the entry into S-phase, but also for progression through S-phase. The experiment in Figure 2C argues that degradation of Cdc45p after cells enter S-phase is sufficient to activate the S-phase checkpoint.
Cdc45p is required to complete DNA replication after replication fork progression is arrested with HU
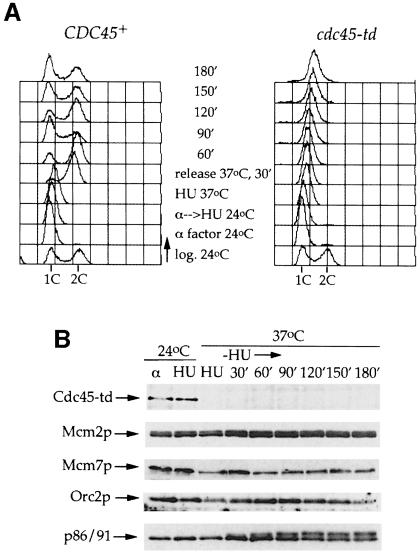
Figure 2C shows that Cdc45p is required for progression through S-phase. However, this experiment does not distinguish an essential role in origin firing during S-phase from an essential role after initiation, in the elongation phase of DNA replication. To begin to investigate the possibility that Cdc45p is also involved in elongation, we asked whether Cdc45p is required to complete DNA replication after S-phase arrest with HU. Such ‘reciprocal shift’ analysis has proved powerful in establishing the order of gene function during the yeast cell cycle (Hartwell, 1976). HU inhibits ribonucleotide reductase causing an arrest in early S-phase. When cells synchronized in G1 with α factor are released into HU, replication initiates from early firing origins and replication forks stall (Bousset and Diffley, 1998; Santocanale and Diffley, 1998). The firing of late origins is prevented under these conditions by the Mec1/Rad53 checkpoint pathway (Santocanale and Diffley, 1998); however, new origin firing is not required to complete S-phase after release from HU (Bousset and Diffley, 1998). In Figure 3A, cultures of wild-type and cdc45-td mutant cells were first synchronized in G1 with α factor at 24°C, and released into HU for 90 min at 24°C. Under these conditions, early firing origins ARS305 and ARS306 have initiated efficiently (data not shown; Figure 6). Cultures were then shifted to 37°C and released from HU at 37°C. Under these conditions, wild-type cells finished S-phase rapidly and entered the next cell cycle after release from HU. However, cdc45-td cells did not complete S-phase, even 3 h after release from HU at 37°C. In fact, this experiment shows that the cdc45-td mutant synthesizes very little DNA after release from HU at the restrictive temperature. If instead of shifting to 37°C, cultures are maintained at 24°C throughout the course of the experiment, both wild-type and cdc45-td mutant cells complete S-phase and divide with similar kinetics (data not shown). Figure 3B shows that Cdc45-td is rapidly degraded after shift to 37°C in HU and is absent for the remainder of the experiment at 37°C while levels of other associated proteins (ORC, DNA polymerase α-primase and Mcm proteins) are not affected (Figure 3B). The cdc45-td cells remain as large, budded cells with single undivided nuclei in this experiment (data not shown), indicating that as in Figure 2C, the DNA replication checkpoint pathway remains active. Since new origin firing is not required to complete S-phase after release from HU (Bousset and Diffley, 1998), these results strongly suggest a role for Cdc45p in the elongation of DNA replication.
Fig. 3. Cdc45p is necessary to complete S-phase after an HU block. (A) Logarithmic phase cultures of cdc45-td and parental wild-type cells growing in YP plus raffinose were synchronized in G1 with α factor at 24°C and then released for 90 min into fresh medium at 24°C containing 0.2 M HU. The cells arrested with HU were shifted to 37°C in the presence of galactose and held in HU for 45 min to degrade Cdc45-td. They were then released from the arrest at 37°C. Samples were taken at the time points indicated and analysed by flow cytometry. (B) Immunoblot analysis of Cdc45-td and other proteins during the course of the experiment.
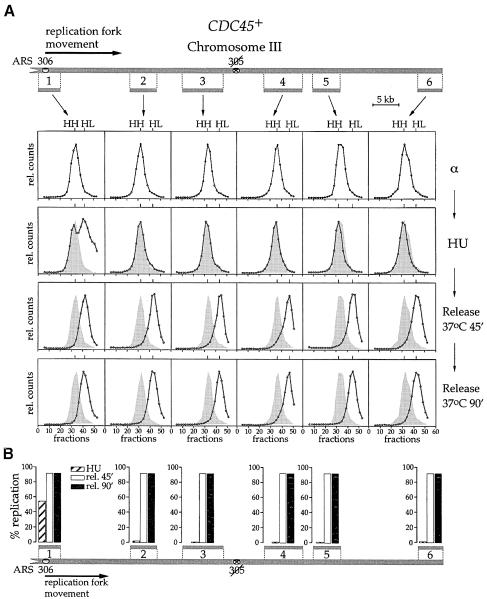
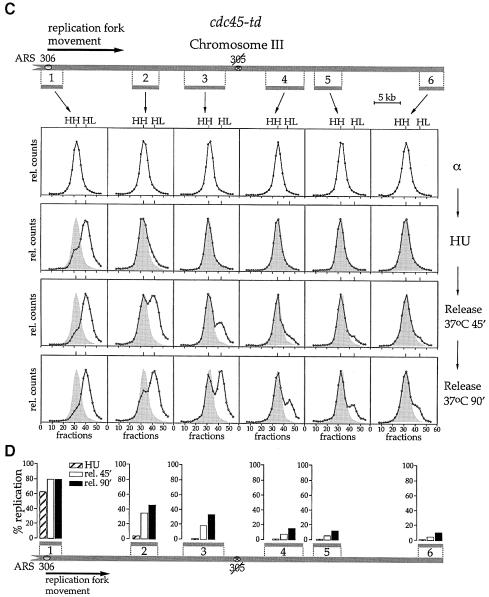
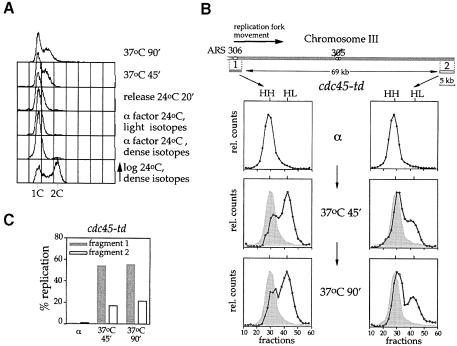
Fig. 6. Cdc45p is required for elongation after the stalling of replication forks. See text for experimental details. Data are shown for both CDC45+ (A) and cdc45-td strains (C). Progression of individual replication forks from ARS306 to the end of the replicon was analysed by dense-isotope transfer (see Materials and methods) using specific probes recognizing the ClaI fragments 1–6. The relative amounts of radioactivity in the hybridized DNA are plotted against the fraction number. The position of unreplicated heavy–heavy (HH) and fully replicated heavy–light (HL) peaks is indicated. At later time points the position of the initial HH peak is shown for comparison (hatched shadow). The lower panels (B and D) show a quantitation of the data.
The block to S-phase progression is reversible
Figure 3 shows that, when Cdc45-td is degraded in HU-arrested cells, these cells cannot finish S-phase when released from the HU arrest at the non-permissive temperature. We were interested in determining whether resynthesis of Cdc45p prior to release from HU could restore the ability of these cells to complete S-phase. Figure 4A shows that after degradation of Cdc45-td at 37°C, a period of 1 h at 24°C is sufficient to restore starting levels of Cdc45-td. Moreover, Figure 4B shows that cdc45-td mutant cells do not lose viability when plated at 24°C after shifting to 37°C. Finally, Figure 4C shows that if cells are released from the HU arrest after allowing resynthesis of Cdc45-td, they finish S-phase with kinetics only slightly slower than wild-type cells. Together, these results show that the block to S-phase completion after Cdc45-td degradation is reversible. This implies that the newly re-synthesized Cdc45p can enter the nucleus and execute its essential function in DNA replication even during S-phase.
Fig. 4. The cdc45-td arrest is reversible. (A) Cdc45-td is degraded when cells arrested with HU at 24°C in raffinose are shifted to 37°C in the presence of galactose for 1 h. After returning to 24°C in YPD plus HU, Cdc45p is remade. As a loading control, the membrane was stained with Ponceau S and scanned prior to incubation with antibody. The region of the membrane around the molecular weight of Cdc45-td is shown. (B) Viability of CDC45+ and cdc45-td strains during the course of the experiment. (C) Cells were subsequently released from HU arrest at 24°C, and DNA content was determined by flow cytometry at all stages of the experiment.
Re-establishment of individual replication forks after HU arrest requires Cdc45p
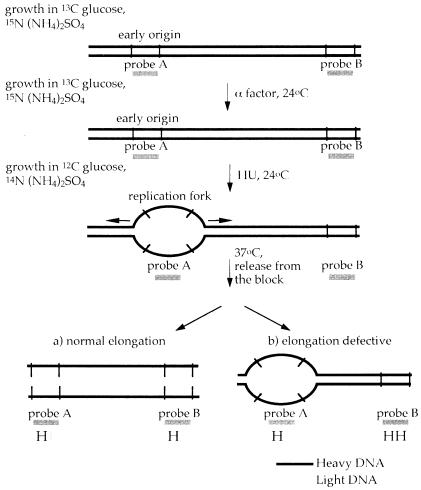
To establish firmly a post-initiation role for Cdc45p after release from HU, it is necessary to follow the progression of individual replication forks. We chose to examine two well characterized replicons. The first is on the left arm of chromosome III (Newlon et al., 1993). Normally, there are two active origins, ARS305 and ARS306, in this region. To increase the size of this replicon, we have deleted ARS305. Therefore, ARS306 replicates the left arm of chromosome III in these cells. The second replicon is on chromosome VI, where a fork from ARS607 replicates the remainder of the right arm of this chromosome in most cell cycles (Friedman et al., 1997; Yamashita et al., 1997). Thus, both regions replicated from single forks are ∼70 kbp. This is large enough to allow initiation to occur and proceed for a measurable distance prior to inactivation of Cdc45-td. Although there are other ARSs on both these chromosome arms, they are either inactive [ARS301–304 (Dubey et al., 1991; Vujcic et al., 1999); ARS608 (Friedman et al., 1997; Yamashita et al., 1997)] or weakly active [ARS609 (Friedman et al., 1997; Yamashita et al., 1997)] in their normal chromosomal location. We developed a strategy to determine whether forks from ARS306 and ARS607 could complete normal elongation after release from HU. This approach utilizes density transfer as shown schematically in Figure 5. In these experiments, cultures of either wild-type or cdc45-td mutant cells are grown in the presence of ‘heavy’ isotopes (13C and 15N) for a number of generations to ensure that both strands of the parental DNA are fully substituted. Cells are arrested in G1 with α factor, transferred to medium containing light isotopes in the presence of α factor and released into HU. Cultures are then shifted to 37°C in HU and released from the HU block at 37°C. DNA is isolated, digested with restriction enzymes and fractionated on CsCl gradients. The position of specific DNA sequences in the gradients is then determined by DNA slot blot hybridization. Using this technique, replication is seen as transfer of specific restriction fragments from the heavy–heavy (HH) peak, in which both DNA strands are substituted with dense isotopes, to the heavy–light (HL) peak, in which only the parental strand contains the dense isotopes. Replication of different sequences on each chromosome can then be examined by using specific probes. Figure 5 shows that if elongation is normal after release from HU at the restrictive temperature, then not only will the origin-containing fragment become HL, but also an origin distal fragment in the replicon will shift to the HL position. Alternatively, if elongation is defective, the origin-distal fragment will remain HH after release from HU.
Fig. 5. An assay to examine replication fork progression. DNA substituted with dense isotopes is shown as thick black lines while DNA made in the presence of light isotopes is shown as thinner grey lines. Vertical lines represent restriction sites and the positions of probes for DNA blot hybridization are shown. See text for details.
In Figure 6 we used probes to examine six different ClaI restriction fragments along the left arm of chromosome III. When cells were arrested in G1 with α factor, all six restriction fragments along chromosome III, in both the wild-type and mutant strains, were found entirely in the HH peak (Figure 6A and C). After release from the arrest in α factor into HU in the presence of light isotopes, most of the ARS306 restriction fragment (fragment 1, Figure 6) shifted to the HL peak in both wild-type and mutant cells. This is consistent with the fact that early firing origins like ARS306 fire efficiently in the HU arrest. There was a fraction that did not appear to shift to the HL position. This could be due to incomplete origin firing; however, based on the facts that pre-RCs are quantitatively lost from early origins under these conditions and there are many replication intermediates in HU much smaller than the full-length restriction fragment (Santocanale and Diffley, 1998), we think that this is likely to be due to the stalling of forks close to the origin. The regions corresponding to the other five probes of the ARS306 replicon (fragments 2–6, Figure 6) remained completely in the HH peak in HU, consistent with the fact that forks stall in the vicinity of the origin in the presence of HU.
After release from HU at 37°C, ARS306 was shifted almost entirely to the HL peak by the first time point (45 min) in both wild-type and cdc45-td mutant cells. In wild-type cells, all of the chromosome III restriction fragments tested had completely shifted to the HL peak by 45 min and remained in the HL peak at 90 min, consistent with the flow cytometry experiment in Figure 3A showing that the entire genome replicates in ∼30 min after release from HU at 37°C. In the cdc45-td mutant, restriction fragment 2 is partially shifted to the HL peak at 45 min after release from HU. A smaller fraction of restriction fragment 3 is also shifted to the HL peak. By 90 min after release at 37°C, slightly more of fragments 2 and 3 are shifted to the HL peak, while fragments 4–6 remain almost entirely in the HH peak. Therefore, in the cdc45-td mutant, replication forks resume a small amount of DNA synthesis after release from HU but the vast majority stop soon thereafter. In contrast to the wild-type cells, replication forks do not reach the end of the replicon in this mutant. These results are quantified at the bottom of each panel (Figure 6).
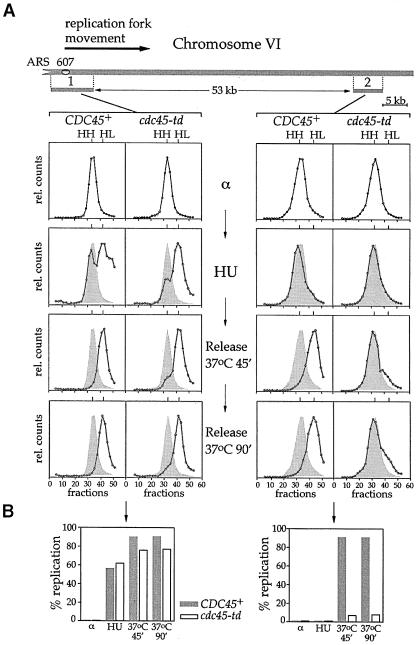
We also examined the progression of a replication fork on chromosome VI, from the ARS607 origin to the end of the right arm of the chromosome. In this case, we examined two sequences of this replicon: one corresponding to the origin itself and the other corresponding to a sequence near the end of the chromosome. The results with chromosome VI (Figure 7) are in agreement with those obtained for chromosome III. All the DNA is in the HH peak in cells arrested in α factor, and after release into HU, the DNA shifts to the HL peak in the region of the ARS607 origin (fragment 1) in both the wild type and the cdc45-td mutant, consistent with the fact that early origins fire in HU. After release at the non-permissive temperature, the restriction fragment near the end of chromosome VI (fragment 2) is transferred to the HL peak in wild-type cells, indicating that it has been fully replicated, while it remains almost entirely in the HH peak in cdc45-td cells. Therefore, Cdc45p is necessary for replication fork progression after initiation from two different origins (ARS306 and ARS607), indicating strongly that Cdc45p plays a general role in replication fork progression.
Fig. 7. (A) Progression of individual replication forks on chromosome VI from ARS607 was analysed by density substitution experiments, as described in Figure 6. Percentages of replication of fragments corresponding to the origin (1) and to the end of the replicon (2) are indicated. (B) Quantitation of the same data.
Cdc45p is required for normal replication fork progression
The experiments shown above demonstrate for the first time that Cdc45p has an essential post-initiation role in DNA replication in promoting replication fork progression after HU arrest. They do not distinguish between an essential role in ongoing DNA synthesis and an essential role in replication fork recovery after stalling. We have used density transfer again to address this issue. First, we fully substituted the DNA with dense isotopes in the cdc45-td mutant. Cells were then arrested in α factor at 24°C and transferred to medium containing light isotopes plus α factor, as shown in Figures 6 and 7. Cells were subsequently released into light medium without α factor at 24°C for 20 min to allow a significant fraction to enter S-phase. At this point, the culture was transferred to 37°C, still in light medium, to promote inactivation of the Cdc45-td protein. For this experiment to work, Cdc45p must be inactivated after ARS306 has fired but before the replication forks from ARS306 reach the end of the replicon. This is technically difficult because origin firing after release from α factor is not synchronous, particularly in minimal medium, and because inactivation of the thermolabile Cdc45 protein is not instantaneous after temperature shift. The deletion of ARS305 increases the size of the terminal replicon on chromosome III, increasing the likelihood of inactivating Cdc45p before forks reach the end of the chromosome. Figure 8A shows that shifting to 37°C 20 min after release from α factor prevents completion of S-phase. A population of cells (∼50%) remain with a 1C DNA content under these conditions. These are either cells that do not release from the α factor arrest in minimal medium or that released but did not initiate replication as in Figure 2A. The second population of cells (∼50%) arrest with a DNA content between 1C and 2C. These cells have therefore initiated DNA replication from at least some origins but were unable to complete S-phase. Wild-type cells rapidly complete S-phase when shifted to 37°C during S-phase (Figure 2C). This is consistent with the idea that Cdc45p is essential for S-phase progression as well as S-phase entry as in Figure 2C. When the replication of individual sequences was examined as described previously for Figures 6 and 7, we found that ∼50% of the ARS306-containing ClaI fragment was replicated 45 min after shift to 37°C. This proportion did not change by 90 min, consistent with the FACS analysis showing that 50% of the cells did not enter S-phase. Approximately 17% of the ClaI fragment at the end of the ARS306 replicon was replicated by the first time point after temperature shift. This presumably represents forks that have reached the end of the chromosome before Cdc45p was inactivated. Most importantly, this proportion did not increase significantly even after an additional 45 min of incubation (21% at 90 min). Therefore, the inactivation of Cdc45p prevents active replication forks from ARS306 from reaching the end of the replicon. From these experiments we conclude that Cdc45p is essential for normal replication fork progression.
Fig. 8. Cdc45p is required for elongation during an unperturbed S-phase. See text for details. DNA content was determined by flow cytometry at the times indicated (A), and the replication of particular regions of chromosome III was determined by dense-isotope transfer, as described above (B). Percentages of replication of the different regions during the experiment are indicated (C).
Discussion
In this paper we have described a systematic approach to study the genetic requirements for the elongation phase of DNA replication. Using this approach we have demonstrated that Cdc45p, a factor previously shown only to be required for the initiation of DNA replication, plays an essential role during the elongation phase of DNA replication. There are two key facets to this approach. First, we used a modified version of the heat-inducible degron method to deplete Cdc45p from cells (Dohmen et al., 1994; Labib et al., 2000). The degron is useful because it acts in a defined way to eliminate the protein from cells rather than inactivate it in some undefined way as is the case for most conventional conditional mutants. We showed that this degradation is specific; Cdc45p was degraded after shifting to the restrictive temperature while other associated proteins including ORC, DNA polymerase α-primase and the Mcm2–7 complex were unaffected by Cdc45p depletion. The second facet of this approach involved the use of density substitution to examine the progression of individual replication forks after Cdc45p inactivation. We showed that when Cdc45p was eliminated from cells after initiation was allowed to occur, DNA synthesis from individual replication forks was prevented. Cdc45p inactivation blocked replication fork progression either after HU arrest or during the normal progression of forks. Previous experiments showed that the cold-sensitive cdc45-1 mutant could complete S-phase at the restrictive temperature; however, they were significantly slower than wild-type cells (Owens et al., 1997). At the time this was attributed to defects in initiation; however, based on our results, we suggest that it might also have been due to defects in elongation. We anticipate that the approach described in this paper will be useful to examine the roles of many other proteins during elongation.
Less surprisingly, the cdc45-td mutant also shows a defect in entry into S-phase. This is consistent with previous experiments in both budding yeast and Xenopus showing that Cdc45p is essential for initiating DNA replication (Hennessy et al., 1991; Hopwood and Dalton, 1996; Hardy, 1997; Owens et al., 1997; Zou et al., 1997; Mimura and Takisawa, 1998). This block to S-phase entry at the restrictive temperature does not prevent subsequent entry into mitosis. In fact, nuclear division occurs in cdc45-td mutant cells that have not replicated their DNA at approximately the same time as it does in wild-type cells that have replicated their DNA (Figure 2B). The inability to restrain mitosis in the absence of DNA replication is unlikely to be due to defects in the checkpoint machinery since cdc45-td mutants efficiently activate S-phase checkpoints if Cdc45p is depleted during, instead of prior to, S-phase entry (Figures 2C and 3A). Instead, our results, together with the results of others, suggest that any tight conditional mutant that is defective in initiating DNA replication cannot produce the signal, probably the replication fork, which is required to activate the S-phase checkpoint (Kelly et al., 1993; Li and Deshaies, 1993; Piatti et al., 1995; Toyn et al., 1995; Tavormina et al., 1997) and, in the absence of S-phase checkpoint activation, cells cannot subsequently prevent entry into mitosis.
The biochemical function of Cdc45p during initiation and elongation is presently unclear. Its primary amino acid sequence has not, thus far, been helpful; it does not contain any characterized protein motifs and the only proteins with significant similarity to budding yeast Cdc45p in database searches are Cdc45p homologues in other organisms. However, its association with other replication proteins suggests a possible role. Cdc45p has previously been shown to have genetic interactions with the Mcm proteins (Hennessy et al., 1991; Hopwood and Dalton, 1996; Dalton and Hopwood, 1997; Zou et al., 1997; Zou and Stillman, 1998; Kukimoto et al., 1999) and a subcomplex of Mcm proteins has been shown to possess intrinsic DNA helicase activity (Ishimi, 1997; You et al., 1999). Cdc45p also shows genetic interactions with Dpb11 (Kamimura et al., 1998), a protein previously shown to interact with DNA polymerase ε (Araki et al., 1995). In addition, Cdc45p is required to load DNA polymerase α-primase onto chromatin in budding yeast (Aparicio et al., 1999) and Xenopus egg extracts (Mimura and Takisawa, 1998), and in Xenopus, physical interactions between Cdc45p and DNA polymerase α-primase have been detected (Mimura and Takisawa, 1998). We note that our density-substitution experiments show that origin-distal restriction fragments remain almost completely unreplicated after Cdc45p inactivation. This indicates that both the leading and lagging strand DNA synthesis machinery is inactivated after Cdc45p inactivation. Taken with the genetic interactions, this might suggest that Cdc45p plays an essential role in co-ordinating the DNA helicase with leading and lagging strand DNA polymerase activity during elongation. While this is a useful hypothesis, it is likely that elucidation of the precise function of Cdc45p in elongation awaits the dissection of soluble cell-free replication systems.
Resynthesis of Cdc45p after depletion in HU-arrested cells allows the resumption of S-phase and restores cell viability. At present we do not know whether stalled replication forks resume DNA synthesis after Cdc45p resynthesis or if the resumption of S-phase requires the activation of late-firing replication origins. Since Cdc45p depletion during S-phase leads to activation of S-phase checkpoints (Figure 2; data not shown), which have been shown to prevent activation of late firing origins, we think it is unlikely that newly synthesized Cdc45p promotes late origin firing without the resumption of DNA synthesis from the stalled replication forks. Cdks actively prevent assembly of new pre-RCs in HU-arrested cells (Noton and Diffley, 2000). Taken together, these experiments demonstrate that newly synthesized Cdc45p can execute an essential DNA replication function after Cdk activation, under conditions where new pre-RCs cannot form. Thus, although Cdc45p may or may not associate with origins during G1-phase prior to Cdk activation (Aparicio et al., 1997, 1999; Zou and Stillman, 1998), it behaves differently from other pre-RC components like Cdc6p and Mcm2–7, which are unable to execute their essential function if synthesized in the presence of Cdk activity (Dahmann et al., 1995; Piatti et al., 1996; Tanaka et al., 1997; Labib et al., 1999, 2000).
Proteins such as SV40 large T antigen and the Escherichia coli dnaB protein first associate with their respective replication origins prior to initiation and subsequently play essential roles in both the initiation and elongation phases of DNA replication. Our results show that in eukaryotic cells there is at least one protein, Cdc45p, which plays a similar dual role in chromosome replication.
Materials and methods
Strains and media
The strains used in this study are all based on W303-1a (MATa ade2-1 ura3-1 his3-11,15 trp1-1 leu2-3,112 can1-100). The cdc45-td mutant was constructed by a modification of the heat-inducible degron method (Dohmen et al., 1994; Labib et al., 2000). In this approach, we used GAL-controlled expression of the UBR1 gene to enhance inactivation of the temperature-sensitive degron fusion. The resulting strain was YJT18 (cdc45-td ubr1Δ::GAL-UBR1::HIS3, W303-1a background). YKL83 (ubr1Δ::GAL-UBR1::HIS3, W303-1a background) was used as a control.
For the density-transfer experiments, the strains were YJT27 (cdc45-td ΔARS305::kanMX, ade2-1::ADE2, W303-1a background) and YJT28 (ΔARS305::kanMX, ade2-1::ADE2, W303-1a background). To delete the ARS305 origin, a fragment from the plasmid pUG6, including the kanMX cassette, was amplified by PCR using the oligonucleotides 5′-TGAGCTATTCGCGACGCCCGACGCCGTAATAACTACTTTCG ACAGCAGCTGAAGCTTCGTACGC and 5′-TGAAAGTGTTTGATT GTATATCGATATATCAGAAACATACATATGCATAGGCCACTA GTGGATCTG. This fragment was transformed into the CDC45+ and cdc45-td strains, and transformants were selected as described previously (Güldener et al., 1996).
Strains were grown in the rich medium YP (1% yeast extract, Difco, plus 2% bacto peptone, Difco) unless otherwise indicated. This medium was supplemented with glucose, raffinose or galactose to a final concentration of 2%.
Cell cycle synchronization, FACS analysis
Cell cycle blocks with α factor and HU were as described previously (Diffley et al., 1994; Donovan et al., 1997). Samples for flow-cytometric analysis were collected and processed as described previously (Labib et al., 1999). They were analysed using a Becton Dickinson FACScan.
Immunoblotting
Protein extracts were prepared as described previously (Foiani et al., 1994). Immunoblot analysis of Cdc45-td was performed using monoclonal antibody 12CA5 (antiCdc45-HA was used at 3.2 µg/ml); p86/91 was detected using monoclonal antibody 6D2 (dilution 1:10 000 of ascites fluid) (Foiani et al., 1994); Mcm2p and Mcm7p were detected using polyclonal antibodies (Santa Cruz Biotechnology SC-6680 and SC6688) and JAB12 polyclonal antibody (1:500) was used to detect Orc2p (1:500) (Donovan et al., 1997). Horseradish peroxidase (HRP)-coupled anti-mouse antibodies were used with the monoclonal antibodies 12CA5 and SD2; HRP-coupled anti-goat was used with the anti-Mcm2 and -7 antibodies and HRP-coupled protein A was used with JAB12. Immunoreactive bands were visualized with enhanced chemiluminescence (ECL; Amersham) according to the manufacturer’s instructions.
Dense-isotope substitution experiments
For density-transfer assays, the procedure was basically as described previously (McCarroll and Fangman, 1988). YJT27 and YJT28 cells were grown at 24°C for eight generations in heavy minimal medium containing 0.1% 13C glucose and 0.01% 15N (NH4)2SO4 as carbon and nitrogen source, respectively. They were then arrested in G1 with α factor and transferred afterwards to light minimal medium [2% 12C glucose and 0.01% 14N (NH4)2SO4] in the presence of α factor. Cells were then washed and released into minimal medium containing 0.2 M HU. After most cells had budded, cultures were incubated at 37°C for 45 min in minimal medium containing 0.2 M HU. Cells were then washed and released from the block at 37°C. Samples were taken for analysis after the indicated time points. The DNA was digested with ClaI and separated on CsCl gradients (McCarroll and Fangman, 1988). The gradients were fractionated and every second fraction of a refractive index between 1.407 and 1.403 was blotted and hybridized with specific probes (see below) labelled with [α-32P]dCTP. A Molecular Dynamics PhosphorImager was used for quantification. Extent of replication was calculated from the equation: % replication = 100 [0.5 HL/HH + 0.5 HL]. Dense isotopes were purchased from CK Gas Products Ltd, UK. The specific probes recognizing the ClaI fragments from chromosome III indicated in Figure 6 were the following: probe 1, nucleotides 73001–73958; probe 2, nucleotides 55449–56401; probe 3, nucleotides 45020–46048; probe 4, nucleotides 27947–28804; probe 5, nucleotides 21295–22320; probe 6, nucleotides 2062–3100. In Figure 7, the probes recognizing the ClaI fragments from chromosome VI were probe 1, nucleotides 198945–199832 and probe 2, nucleotides 260048–261088. The numbers of the probes correspond to the numbers of the DNA fragments in figures.
Supplementary material
Supplementary material to this paper is available in The EMBO Journal Online.
Acknowledgments
Acknowledgements
We thank Kristine Bousset for advice and suggestions and the other members of our laboratory for helpful discussions. We thank Lee Johnston and Alexander Varshavsky for plasmids. This work was supported by the Imperial Cancer Research Fund. J.A.T. is a recipient of a postdoctoral fellowship from the Human Frontier Science Program.
References
- Aparicio O.M., Weinstein,D.M. and Bell,S.P. (1997) Components and dynamics of DNA replication complexes in S.cerevisiae: redistribution of MCM complexes and Cdc45p during S phase. Cell, 91, 59–69. [DOI] [PubMed] [Google Scholar]
- Aparicio O.M., Stout,A.M. and Bell,S.P. (1999) Differential assembly of Cdc45p and DNA polymerases at early and late origins of DNA replication. Proc. Natl Acad. Sci. USA, 96, 9130–9135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araki H., Leem,S.H., Phongdara,A. and Sugino,A. (1995) Dpb11, which interacts with DNA polymerase II (ε) in Saccharomyces cerevisiae, has a dual role in S-phase progression and at a cell-cycle checkpoint. Proc. Natl Acad. Sci. USA, 92, 11791–11795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell S.P. and Stillman,B. (1992) ATP-dependent recognition of eukaryotic origins of DNA replication by a multiprotein complex. Nature, 357, 128–134. [DOI] [PubMed] [Google Scholar]
- Bousset K. and Diffley,J.F.X. (1998) The Cdc7 protein kinase is required for origin firing during S phase. Genes Dev., 12, 480–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewer B.J., Diller,J.D., Friedman,K.L., Kolor,K.M., Raghuraman,M.K. and Fangman,W.L. (1993) The topography of chromosome replication in yeast. Cold Spring Harb. Symp. Quant. Biol., 58, 425–434. [DOI] [PubMed] [Google Scholar]
- Challberg M.D. and Kelly,T.J. (1989) Animal virus DNA replication. Annu. Rev. Biochem., 58, 671–717. [DOI] [PubMed] [Google Scholar]
- Cocker J.H., Piatti,S., Santocanale,C., Nasmyth,K. and Diffley,J.F.X. (1996) An essential role for the Cdc6 protein in forming the pre-replicative complexes of budding yeast. Nature, 379, 180–182. [DOI] [PubMed] [Google Scholar]
- Dahmann C., Diffley,J.F.X. and Nasmyth,K.A. (1995) S-phase-promoting cyclin-dependent kinases prevent re-replication by inhibiting the transition of origins to a pre-replicative state. Curr. Biol., 5, 1257–1269. [DOI] [PubMed] [Google Scholar]
- Dalton S. and Hopwood,B. (1997) Characterization of Cdc47p-minichromosome maintenance complexes in Saccharomyces cerevisiae: identification of Cdc45p as a subunit. Mol. Cell. Biol., 17, 5867–5875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diffley J.F.X. and Cocker,J.H. (1992) Protein–DNA interactions at a yeast replication origin. Nature, 357, 169–172. [DOI] [PubMed] [Google Scholar]
- Diffley J.F.X., Cocker,J.H., Dowell,S.J. and Rowley,A. (1994) Two steps in the assembly of complexes at yeast replication origins in vivo.Cell, 78, 303–316. [DOI] [PubMed] [Google Scholar]
- Dohmen R.J., Wu,P. and Varshavsky,A. (1994) Heat-inducible degron: a method for constructing temperature-sensitive mutants. Science, 263, 1273–1276. [DOI] [PubMed] [Google Scholar]
- Donovan S., Harwood,J., Drury,L.S. and Diffley,J.F.X. (1997) Cdc6-dependent loading of Mcm proteins onto pre-replicative chromatin in budding yeast. Proc. Natl Acad. Sci. USA, 94, 5611–5616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drury L.S., Perkins,G. and Diffley,J.F.X. (1997) The Cdc4/34/53 pathway targets Cdc6p for proteolysis in budding yeast. EMBO J., 16, 5966–5976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubey D.D., Davis,L.R., Greenfeder,S.A., Ong,L.Y., Zhu,J.G., Broach,J.R., Newlon,C.S. and Huberman,J.A. (1991) Evidence suggesting that the ARS elements associated with silencers of the yeast mating-type locus HML do not function as chromosomal DNA replication origins. Mol. Cell. Biol., 11, 5346–5355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fangman W.L. and Brewer,B.J. (1992) A question of time: replication origins of eukaryotic chromosomes. Cell, 71, 363–366. [DOI] [PubMed] [Google Scholar]
- Fanning E. and Knippers,R. (1992) Structure and function of simian virus 40 large tumor antigen. Annu. Rev. Biochem., 61, 55–85. [DOI] [PubMed] [Google Scholar]
- Ferguson B.M., Brewer,B.J. and Fangman,W.L. (1991) Temporal control of DNA replication in yeast. Cold Spring Harb. Symp. Quant. Biol., 56, 293–302. [DOI] [PubMed] [Google Scholar]
- Foiani M., Marini,F., Gamba,D., Lucchini,G. and Plevani,P. (1994) The B subunit of the DNA polymerase α-primase complex in Saccharomyces cerevisiae executes an essential function at the initial stage of DNA replication. Mol. Cell. Biol., 14, 923–933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox C.A., Loo,S., Dillin,A. and Rine,J. (1995) The origin recognition complex has essential functions in transcriptional silencing and chromosomal replication. Genes Dev., 9, 911–924. [DOI] [PubMed] [Google Scholar]
- Friedman K.L., Brewer,B.J. and Fangman,W.L. (1997) Replication profile of Saccharomyces cerevisiae chromosome VI. Genes Cells, 2, 667–678. [DOI] [PubMed] [Google Scholar]
- Güldener U., Heck,S., Fiedler,T., Beinhauer,J. and Hegemann,J.H. (1996) A new efficient gene disruption cassette for repeated use in budding yeast. Nucleic Acids Res., 24, 2519–2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy C.F. (1997) Identification of Cdc45p, an essential factor required for DNA replication. Gene, 187, 239–246. [DOI] [PubMed] [Google Scholar]
- Hartwell L.H. (1976) Sequential function of gene products relative to DNA synthesis in the yeast cell cycle. J. Mol. Biol., 104, 803–817. [DOI] [PubMed] [Google Scholar]
- Hennessy K.M., Lee,A., Chen,E. and Botstein,D. (1991) A group of interacting yeast DNA replication genes. Genes Dev., 5, 958–969. [DOI] [PubMed] [Google Scholar]
- Hopwood B. and Dalton,S. (1996) Cdc45p assembles into a complex with Cdc46p/Mcm5p, is required for minichromosome maintenance and is essential for chromosomal DNA replication. Proc. Natl Acad. Sci. USA, 93, 12309–12314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishimi Y. (1997) A DNA helicase activity is associated with an MCM4, -6 and -7 protein complex. J. Biol. Chem., 272, 24508–24513. [DOI] [PubMed] [Google Scholar]
- Kamimura Y., Masumoto,H., Sugino,A. and Araki,H. (1998) Sld2, which interacts with Dpb11 in Saccharomyces cerevisiae, is required for chromosomal DNA replication. Mol. Cell. Biol., 18, 6102–6109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly T.J., Nurse,P. and Forsburg,S.L. (1993) Coupling DNA replication to the cell cycle. Cold Spring Harb. Symp. Quant. Biol., 58, 637–644. [DOI] [PubMed] [Google Scholar]
- Kukimoto I., Igaki,H. and Kanda,T. (1999) Human CDC45 protein binds to minichromosome maintenance 7 protein and the p70 subunit of DNA polymerase α. Eur. J. Biochem., 265, 936–943. [DOI] [PubMed] [Google Scholar]
- Labib K., Diffley,J.F.X. and Kearsey,S.E. (1999) G1-phase and B-type cyclins exclude the DNA-replication factor Mcm4 from the nucleus. Nature Cell Biol., 1, 415–422. [DOI] [PubMed] [Google Scholar]
- Labib K., Tercero,J.A. and Diffley,J.F.X. (2000) DNA replication fork progression requires uninterrupted MCM2–7 function. Science, in press. [DOI] [PubMed] [Google Scholar]
- Lei M., Kawasaki,Y. and Tye,B.K. (1996) Physical interactions among MCM proteins and effects of MCM dosage on DNA-replication in Saccharomyces cerevisiae.Mol. Cell. Biol., 16, 5081–5090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J.J. and Deshaies,R.J. (1993) Exercising self-restraint: discouraging illicit acts of S and M in eukaryotes. Cell, 74, 223–226. [DOI] [PubMed] [Google Scholar]
- Liang C. and Stillman,B. (1997) Persistent initiation of DNA replication and chromatin-bound MCM proteins during the cell cycle in cdc6 mutants. Genes Dev., 11, 3375–3386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang C., Weinreich,M. and Stillman,B. (1995) ORC and Cdc6p interact and determine the frequency of initiation of DNA replication in the genome. Cell, 81, 667–676. [DOI] [PubMed] [Google Scholar]
- McCarroll R.M. and Fangman,W.L. (1988) Time of replication of yeast centromeres and telomeres. Cell, 54, 505–513. [DOI] [PubMed] [Google Scholar]
- Mimura S. and Takisawa,H. (1998) Xenopus Cdc45-dependent loading of DNA polymerase α onto chromatin under the control of S-phase cdk. EMBO J., 17, 5699–5707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newlon C.S., Collins,I., Dershowitz,A., Deshpande,A.M., Greenfeder, S.A., Ong,L.Y. and Theis,J.F. (1993) Analysis of replication origin function on chromosome III of Saccharomyces cerevisiae.Cold Spring Harb. Symp. Quant. Biol., 58, 415–423. [DOI] [PubMed] [Google Scholar]
- Noton E.A. and Diffley,J.F.X. (2000) CDK inactivation is the only essential function of the APC/C and the mitotic exit network proteins for origin resetting during mitosis. Mol. Cell, 5, 85–95. [DOI] [PubMed] [Google Scholar]
- Owens J.C., Detweiler,C.S. and Li,J.J. (1997) CDC45 is required in conjunction with CDC7/DBF4 to trigger the initiation of DNA replication. Proc. Natl Acad. Sci. USA, 94, 12521–12526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piatti S., Lengauer,C. and Nasmyth,K. (1995) Cdc6 is an unstable protein whose de novo synthesis in G1 is important for the onset of S phase and for preventing a ‘reductional’ anaphase in the budding yeast Saccharomyces cerevisiae.EMBO J., 14, 3788–3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piatti S., Bohm,T., Cocker,J.H., Diffley,J.F.X. and Nasmyth,K. (1996) Activation of S-phase promoting CDKs in late G1 defines a ‘point of no return’ after which Cdc6 synthesis cannot promote DNA replication in yeast. Genes Dev., 10, 1516–1531. [DOI] [PubMed] [Google Scholar]
- Reid R.J., Fiorani,P., Sugawara,M. and Bjornsti,M.A. (1999) CDC45 and DPB11 are required for processive DNA replication and resistance to DNA topoisomerase I-mediated DNA damage. Proc. Natl Acad. Sci. USA, 96, 11440–11445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowley A., Cocker,J.H., Harwood,J. and Diffley,J.F.X. (1995) Initiation complex assembly at budding yeast replication origins begins with the recognition of a bipartite sequence by limiting amounts of the initiator, ORC. EMBO J., 14, 2631–2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santocanale C. and Diffley,J.F.X. (1996) ORC- and Cdc6-dependent complexes at active and inactive chromosomal replication origins in Saccharomyces cerevisiae.EMBO J., 15, 6671–6679. [PMC free article] [PubMed] [Google Scholar]
- Santocanale C. and Diffley,J.F.X. (1998) A Mec1- and Rad53-dependent checkpoint controls late-firing origins of DNA replication. Nature, 395, 615–618. [DOI] [PubMed] [Google Scholar]
- Tanaka T., Knapp,D. and Nasmyth,K. (1997) Loading of an Mcm protein onto DNA-replication origins is regulated by Cdc6p and CDKs. Cell, 90, 649–660. [DOI] [PubMed] [Google Scholar]
- Tavormina P.A., Wang,Y. and Burke,D.J. (1997) Differential requirements for DNA replication in the activation of mitotic checkpoints in Saccharomyces cerevisiae.Mol. Cell. Biol., 17, 3315–3322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyn J.H., Johnson,A.L. and Johnston,L.H. (1995) Segregation of unreplicated chromosomes in Saccharomyces cerevisiae reveals a novel G1/M-phase checkpoint. Mol. Cell. Biol., 15, 5312–5321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vujcic M., Miller,C.A. and Kowalski,D. (1999) Activation of silent replication origins at autonomously replicating sequence elements near the HML locus in budding yeast. Mol. Cell. Biol., 19, 6098–6109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waga S. and Stillman,B. (1994) Anatomy of a DNA replication fork revealed by reconstitution of SV40 DNA replication in vitro.Nature, 369, 207–212. [DOI] [PubMed] [Google Scholar]
- Weinberg D.H., Collins,K.L., Simancek,P., Russo,A., Wold,M.S., Virshup,D.M. and Kelly,T.J. (1990) Reconstitution of simian virus 40 DNA replication with purified proteins. Proc. Natl Acad. Sci. USA, 87, 8692–8696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita M., Hori,Y., Shinomiya,T., Obuse,C., Tsurimoto,T., Yoshikawa,H. and Shirahige,K. (1997) The efficiency and timing of initiation of replication of multiple replicons of Saccharomyces cerevisiae chromosome VI. Genes Cells, 2, 655–665. [DOI] [PubMed] [Google Scholar]
- Yan H., Merchant,A.M. and Tye,B.-K. (1993) Cell cycle-regulated nuclear localisation of MCM2 and MCM3, which are required for the initiation of DNA synthesis at chromosomal replication origins in yeast. Genes Dev., 7, 2149–2160. [DOI] [PubMed] [Google Scholar]
- You Z., Komamura,Y. and Ishimi,Y. (1999) Biochemical analysis of the intrinsic Mcm4–Mcm6–Mcm7 DNA helicase activity. Mol. Cell. Biol., 19, 8003–8015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou L. and Stillman,B. (1998) Formation of a preinitiation complex by S-phase cyclin CDK-dependent loading of Cdc45p onto chromatin. Science, 280, 593–596. [DOI] [PubMed] [Google Scholar]
- Zou L., Mitchell,J. and Stillman,B. (1997) CDC45, a novel yeast gene that functions with the origin recognition complex and Mcm proteins in initiation of DNA replication. Mol. Cell. Biol., 17, 553–563. [DOI] [PMC free article] [PubMed] [Google Scholar]