Abstract
In vitro display technologies, best exemplified by phage and yeast display, were first described for the selection of antibodies some twenty years ago. Since that time a large number of antibodies, some with remarkable properties, have been selected and improved upon using these methods. The first antibodies derived using in vitro display methods are now in the clinic, with many more waiting in the wings. Here we discuss the scope of the technology, some of the powerful antibodies selected, and the future potential in a post-genomic world. Unique advantages offered by in vitro display technologies include the ability to carefully define selection conditions, allowing the derivation of antibodies recognizing predefined epitopes or conformations, the further improvement of selected antibodies, the potential for high throughput applications and the immediate availability of genes encoding the selected antibody. We anticipate that the high throughput potential of these technologies will soon lead to their use to select antibodies against all human proteins.
Introduction
For the past 35 years, hybridoma technology has enhanced our capacity for research and diagnostics by providing monoclonal antibody reagents allowing tracking, detection and quantitation of target molecules in cells and serum. Recently, a number of more advanced methods to harness the immune response have also been developed1,2,3 that significantly increase the number of antibody producing cells that can be screened. Alongside these “traditional” method of making monoclonal antibodies, a quiet revolution has been brewing in the generation of antibodies using in vitro display technologies, which offer a number of advantages, including a greater degree of control over the nature of the derived antibodies. The success of these technologies has relied upon many previous advances, including the conception and implementation of phage display4,5, the expression of antibody fragments in bacteria6 and PCR-mediated amplification of antibody genes and libraries7,8,9,10,11. The most popular technologies, antibody phage8,12,13 and yeast display14,15, which are complementary in their properties, can be used with naïve, immunized or synthetic repertoires.
As a direct consequence of genome sequencing, and the advent of high throughput biology, the demand for large numbers of renewable high quality affinity reagents, recognizing ever-greater numbers of proteins, for affinity reagent based proteomic scale experiments, is expected to increase dramatically. In vitro methods have the potential to deliver enormous improvements from parallelization, automation and miniaturization. In contrast, further advances in animal immunization technologies are expected to be slim. Furthermore, it is generally accepted that, irrespective of the source, there is an urgent need to improve antibody quality, as reflected by a raft of recent papers16,17,18,19,20,21,22 showing an alarmingly high proportion of commercial antibodies demonstrating poor specificity, or even failing to recognize their targets at all. Given that much of modern biological research relies on the fidelity of commercially supplied antibodies, there is an urgent need to resolve this problem. The high throughput potential of in vitro technologies make them ideal platforms for large scale projects to derive antibodies for all human proteins, which once completed are likely to have impacts perhaps as great as the completion of the human genome.
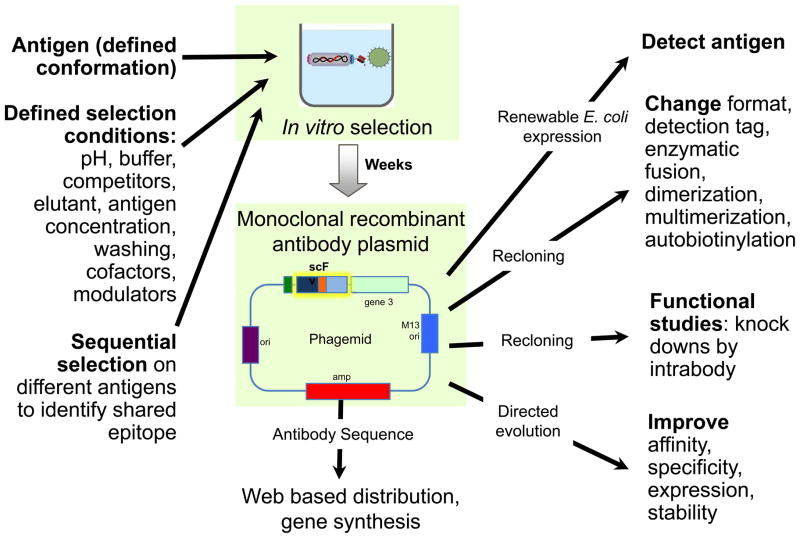
By carefully controlling selection and screening conditions, display technologies allow the generation of antibodies to defined antigen conformations or epitopes, for example, by the presentation of specific antigen conformations, or the inclusion of competitors to direct selection towards specific targets or epitopes (figure 1). Moreover, when variable regions from immunized sources are used with display technologies, specificities not detectable by traditional immunological techniques can often be selected23. During the process of in vitro antibody selection, the gene encoding the antibody is cloned at the same time as the antibody is selected, providing many advantages to the recombinant approach (Fig. 1). The availability of the antibody gene allows the creation of alternative constructs with added functionality by simple subcloning (see below). Libraries of mutagenized variants can be created and the same selection process repeated to yield variants that are improved, both in terms of specificity and affinity. The improvement of antibody affinity to picomolar levels24,25,26,27,28 has become relatively routine, with one study describing an antibody in the femtomolar range29. These affinities are far higher than those of antibodies obtained by immunization, which are limited to ~100 pM by the physiological mechanism of B cell activation30,31. In addition, antibody specificities can be broadened or narrowed by appropriate selection and screening.
Figure 1.
The additional capabilities of in vitro selection offer a new approach to antibody generation, allowing the selection of antibodies with particular properties by predefining panning conditions. Variations in salt and pH conditions, the conformational form of the target and the presence of closely related proteins help determine the biochemical properties, fine specificity, cross-reactivities and affinity of resulting binders. Further, the immediate availability of the antibody gene provides significant additional value. Complemented by a more rapid antibody generation cycle, this will broadly change the manner in which antibodies will be made and used for research in the near future.
As these in vitro methods are based on microbial systems, selection and screening are more amenable to automation than earlier hybridoma-based approaches. This provides the potential for high throughput binder generation32,33. In vitro methods also overcome immunological tolerance, allowing the selection of affinity reagents that recognize highly conserved targets such as ubiquitin34, histones35, hemoglobins36 and post-translational modifications37,38,39. While tricks can be used to overcome tolerance during immunization40,41, none are required to select antibodies against conserved proteins using in vitro display methods. Remarkably, the selection of hundreds of different antibodies from naïve human antibody repertoires against many different individual human targets has not been problematic32,42,43.
Most of the examples described below relate to antibody fragments. However, display technologies have allowed the development of alternative, non-antibody scaffolds and they too will provide affinity reagents with similar, or in some applications, superior properties to those described here. Selection platforms44,45,46 and different scaffold proteins47,48,49, including antibody fragments50, have been widely dealt with in previous reviews. Our goal here is not to reiterate the ways in which such libraries are made or used, but to illustrate how in vitro display methods have yielded antibodies with remarkable properties some of which have rarely, if ever, been obtained by immunization. This will be carried out by describing a number of different classes of unique and interesting antibodies, as well as outlining the enormous advantages provided by immediate access to cloned antibody genes.
Recognition of chemical modifications and small molecules
Monoclonal and polyclonal antibodies with specificities for small molecules have been obtained by traditional immunization51,52,53,54,55. However, the ability of display methods to tailor both affinity and specificity has led to significantly better antibodies than can be obtained by immunization (Table 1 and Fig. 2).
Table 1.
In vitro selected antibodies recognizing small molecules and modifications
| Targets | Notes | Refs |
|---|---|---|
| 6-monoacetylmorphine and morphine | Competition with morphine during panning to avoid crossreactivity | 56 |
| Fluorescein | Affinity matured to 48 fM by yeast display | 29 |
| testosterone, progesterone and 17β oestradiol | 57,58,59,60,61 | |
| Sulfotyrosine as a post-translational modification | Antibodies recognize all sulfo-tyrosinated proteins and peptides | 38,39 |
| Sulfur mustard–modified keratin | 62 | |
| Fluorogenic dyes | Antibody binding increases dye fluorescence up to 15,000 times by limiting conformational movement | 63 |
| Metallic gold | 64 |
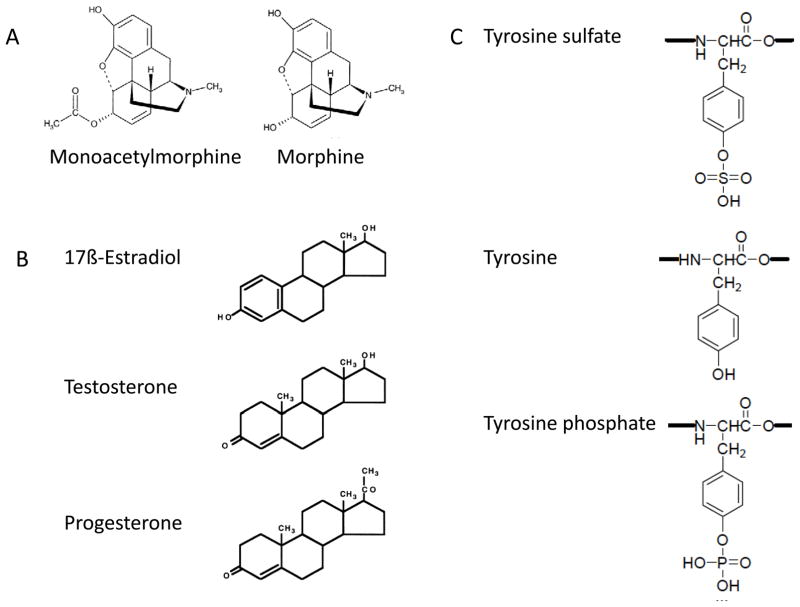
Figure 2.
In vitro selected antibodies can recognize minute differences in small molecules. A) Antibodies against 6-monoacetylmorphine, the major heroin metabolite, do not recognize the closely related morphine56. B) Many different antibodies have been selected and subsequently had both affinity and specificity matured to recognize each of the represented steroids (for references see Table 1). C) Antibodies against tyrosine sulfated modified proteins do not recognize proteins containing either tyrosine or tyrosine phosphate38,39.
Sulfotyrosine is a post-translational modification (PTM) predicted to occur in 30% of all secretory and membrane proteins65. Despite decades of immunization, it has proved impossible to generate antibodies recognizing this PTM by traditional means. This is probably due to the innate tolerance immune systems have for such ubiquitous protein modifications, as well as the presence of the recognized target in the secretory pathway, resulting in retention and an inability to secrete the antibody. Using phage display, two groups recently selected antibodies recognizing proteins containing sulfotyrosine (but not tyrosine phosphate), independently of protein context or sequence38,39. These antibodies recognized sulfotyrosinated proteins in western blotting, immunofluorescence, ELISA and immunoprecipitation, and recognition could be abolished by sulfatase treatment or preincubation with free tyrosine sulfate. This represents an enormous advance in the analysis of this modification, which has traditionally required thin-layer chromatography of radiolabeled protein hydrolysates66 or mass spectrometry (MS)67, with the presence of sulfate groups often inferred, rather than proven.
Specificity for protein sequences and conformations
Phage display allows the generation of antibodies against nearly any target, including toxins, pathogens, non-immunogenic, or highly conserved antigens. With respect to protein targets, antibodies have been selected with exquisite specificity, differentiating, for example, between chicken and quail lysozyme68 that differ by a single surface amino acid, and the SH2 domains of ABL1 and ABL269,70. Phage antibody libraries have been widely used to select antibodies against infectious agents. These include antibodies that discriminate between different strains of Hantavirus71, Dengue virus72, influenza73,74, Ebola75 and Venezuelan equine encephalitis virus76. Given that many of these viruses are classified serologically, the ability to select phage antibodies with similar specificities is not surprising, but unlike antibodies generated by immunization, these have the potential to be used therapeutically. Human antibodies, some of which are protective in animal models77,78,79, have also been selected against a number of bacterial biothreat targets, including Brucella melitensis80, Burkholderia mallei and Burkholderia pseudomallei81, anthrax toxins77,82,83 and spores84, and Botulinum toxin24,85. One library79 was generated from servicemen vaccinated against a plethora of different biothreat agents, reflecting the additional ability of display technologies to exploit antibodies generated during traditional immunization.
The in vitro nature of phage display technology has been exploited to target particular features of blood cells. In one study36, antibodies recognizing fetal hemoglobin but not adult hemoglobin, were selected by depleting high affinity cross-reactive antibodies followed by a selection against the fetal protein. Notably, the selected discriminatory antibody was of much lower affinity than cross-reactive antibodies, demonstrating the power of negative selections to favor clones with desirable binding specificities, even if their affinity is lower. Similar methods applied to cells have been used to select antibodies specifically recognizing fetal nucleated red blood cells95.
Protein allostery is a common means for the regulation of protein function, and many signaling proteins exist in alternative conformational states that mediate different cellular responses. Antibodies that recognize specific proteins conformers are powerful tools for probing the details of cell signaling. However, the generation of such antibodies by immunization is complicated by the difficulty of maintaining a particular protein conformation in an immunized animal. In contrast, in vitro selection technologies are ideally suited for these applications. Negative selections can be used to deplete non-specific binders, and affinity maturation strategies can be employed to fine-tune specificity. In one study, scFvs specific to the GTP-bound form of the small guanosine triphosphatase (GTPase) Rab6 were generated by performing selections against a GTP-locked mutant96. In another study97, small molecules were used to covalently lock caspase-1 in either the active or inactive form and the locked antigens were used to select Fabs that were highly selective for either the “on” or “off” form of the protease. The concept of using in vitro selections to generate conformation specific antibodies has also been combined with selections on whole cells in a powerful strategy that enables the probing the cell surfaces for conformational changes in response to various stimuli101.
Finally, phage display has enabled the generation of antibodies recognizing structured RNA molecules100, which are essentially non-immunogenic, and not as amenable to simple nucleotide probes. By ensuring a nuclease-free in vitro environment and selecting under conditions optimized for the structural stability of the RNA, high affinity Fabs were isolated against a structured domain from the Tetrahymena group I intron. These results hold great promise, as they establish general methods applicable to the generation of antibodies against other structured RNAs, and will be useful to decipher the biological roles of the vast numbers of noncoding RNAs found in metazoan transcriptomes.
Antibodies to cell surface receptors
Communication between cells is driven and controlled by interactions between cell surface receptors and the ligands they recognize. Antibodies can modify such interactions and many therapeutic antibodies exert their effect by interfering in communications at the cell surface using different mechanisms (Fig. 3). In vitro display technologies provide a powerful route to generating functional antibodies that interfere in normal or pathological extracellular signaling. Although it is usually difficult to select for function directly, display technologies have the ability to generate thousands of independent binders, each of which can then be screened for functional activity. For example, over 1200 different antibodies directed towards B Lymphocyte Stimulator (BlyS) were generated by phage display102. This large panel was subsequently screened in biochemical and cellular assays to identify antibodies that bound to BLyS, preventing its interaction with the receptor (Fig. 3a), and thereby blocking B cell activation. In some cases blocking antibodies with sub-nanomolar affinities were isolated directly from the naïve antibody-phage display library102. One of these antibodies, specific only for the secreted form of BlyS (BENLYSTA), was affinity matured103 and is close to approval for treatment of systemic lupus erythematosus. Similar results have been reported for the selection of phage antibodies against a panel of 28 different potentially therapeutic targets, with an average of 120 functionally active (i.e. antagonistic or agonistic) antibodies selected per target43.
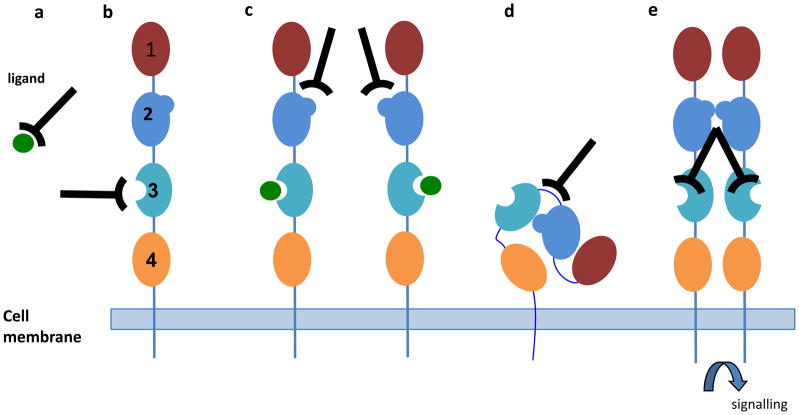
Figure 3.
Mechanisms for blocking or activating receptor signaling using antibodies. The EGF receptor is used to exemplify mechanisms by which antibodies can block signaling in different classes of receptor. The EGF receptor is a single trans-membrane domain with multiple extra-cellular domains (represented as different colored ovals) having different functional domains. In this example, binding of ligand (green circle) occurs at domain 3, receptor dimerization occurs through domain 2 and interactions between domains 2 and 4 stabilize the “closed” conformation of the receptor. Antibodies can block signaling by a. binding to the ligand and preventing interaction with receptor, b. binding the ligand binding site of the receptor and preventing interaction with ligand, c. preventing dimerization by binding the dimerization domain or sterically blocking the interaction d. stabilising the closed conformation of the receptor. e. Activation can occur by binding the ligand-binding site typically with bivalent antibodies.
An alternative strategy to block receptor signaling is to target the ligand binding sites on the receptors, thereby preventing the natural ligand from acting (Fig. 3b). This was used in a recent study, where antibodies were selected that prevented the interaction of insulin-like growth factor type 1 (IGF-1) with the IGF-1 receptor106. Several groups of receptor binders were generated that competed with ligand binding and blocked cell growth in vitro and in vivo. These antibodies were also found to reduce receptor expression by internalization and catabolism.
Studies on a panel of therapeutic antibodies targeting the EGF receptor (Erb-B1) have also shown competition with ligand binding. However, antibodies can also block receptor signaling by alternative mechanisms120. Erb-B1 has four extracellular domains, which adopt a mainly closed conformation in the absence of ligand, and a more extended conformation allowing dimerization, and subsequent phosphorylation of the intracellular domain, in the presence of ligand. Structural studies have shown that antibodies such as cetuximab stabilize the receptor in the closed conformation (Fig. 3d) while zalutumumab keeps intracellular domains apart preventing phosphorylation (Fig. 3c). Among the anti-Erb-B2 antibodies, Pertuzumab appears to work by preventing dimerization while trastuzumab (Herceptin) prevents receptor shedding and forms inactive tetramers120. While the original blocking antibodies in these examples were generated from mice, they demonstrate the therapeutic approaches that could benefit from human antibodies isolated directly from display technologies.
Antibodies that block Notch signaling reveal yet another mechanism of action. Following ligand binding, a conformational change occurs at the juxta-membrane negative regulatory region (NRR) exposing a protease cleavage site resulting in the release and translocation to the nucleus of the intracellular domain. In addition to generating antibodies that block the interaction with ligand, antibodies recognizing the NRR domains stabilized the “closed” confirmation of the Notch receptor (Fig. 3d) preventing the proteolytic cleavage and translocation of the intracellular domain107,108.
Dimeric antibodies targeting ligand-binding domains sometimes mimic the natural ligand, causing receptor activation rather than inhibition. This is the case for antibodies recognizing Met109, with monomeric antibodies being antagonistic. However, in the case of TRAIL receptor 1 (TRAIL-R1) and TRAIL receptor 2 (TRAIL-R2)119, an analysis of over 500 distinct selected antibodies, revealed some that were agonistic even as monomeric scFvs or Fabs. This is difficult to reconcile with the mode of action of TRAIL, which is a homotrimeric ligand that causes multimerization of the TRAIL receptor leading to apoptosis, particularly in tumor cells over-expressing the receptor.
Antibodies also have great potential in blocking protein interactions associated with viral entry into target cells, illustrated by antibodies selected from naïve antibody libraries against recombinant H5 hemagglutinin influenza ectodomain112,113. Structural analysis of one of the antibodies showed it bound to hemagglutinin at a highly conserved previously unrecognized pocket, found in many different influenza viruses. Binding prevents the structural reorganization required for membrane fusion, rendering the antibody neutralizing. Although antibodies have not been generated against this epitope by traditional immunization or infection, antibodies with similar VH gene usage and neutralizing activity have been selected from phage antibody libraries created from recently infected individuals23, showing that phage display can access the diversity of immune responses in ways not possible by traditional immunological means.
In vitro selection schemes have also been devised that allow the direct selection of antibodies mediating internalization117. This was carried out by incubating phage libraries with target cells and isolating those phage antibodies found within the cell after removing phage antibodies bound to the cell surface. The identification of the recognized antigen is usually carried out after selection. However, the use of mammalian cells transfected with the target of interest 118, or yeast displaying targets of interest on their surface114, provides a means of carrying this out on predetermined targets. This approach is particularly suitable for the selection of antibodies used for specific targeting of chemotherapeutics121,122.
In summary, antibodies and other binding molecules provide a means of modulating biological function by specifically interfering in protein interactions. In vitro display systems provide a means of presenting targets in appropriate conformations, including on cell surfaces, which facilitate rapid screening for potentially rare functional binders.
Improving antibody affinity and specificity
While initial leads can be used directly as affinity reagents, a major advantage of in vitro methods is that it is possible to further improve function by constructing secondary libraries that introduce additional mutations. The most prevalent application of secondary libraries is the improvement of affinity, and all three major display formats (phage, yeast and ribosome) have been applied to develop extremely tight affinities that exceed those possible with natural antibodies (Table 4). Both stepwise123 and computational124 methods have also been developed that are able to generate similarly high affinity antibodies, but they have not been as widely used as the in vitro display methods. There are many examples of in vitro affinity maturation, and here we highlight some key studies. In ribosome display, each selection cycles involves a PCR amplification step, which is ideal for introducing additional mutations by error prone PCR. This strategy has been used to simultaneously select and affinity mature anti-insulin antibodies with affinities in the sub-nanomolar range26. While yeast-displayed libraries are smaller than phage and ribosome libraries, they allow quantitative and exhaustive screening by fluorescence activated cell sorting (FACS). Coupled with sequential rounds of error-prone PCR, modest libraries of 105–107 unique clones were sufficient to affinity mature an anti-fluoroscein scFv into the low femtomolar range29.
Table 4.
Affinity and specificity maturation of antibodies by in vitro selection methods
| Target | Notes | Ref |
|---|---|---|
| Affinity maturation | ||
| HIV | CDRs targeted for mutation, 15pM affinity | 28 |
| c-erbB-2 | CDRs targeted for mutation, 13pM affinity | 27 |
| Insulin | Ribosome display, random errors, 82pM affinity | 26 |
| Fluorescein | Affinity matured to 48 fM by yeast display | 29 |
| Specificity modification, recognition specificities | ||
| CXCL10 & CXCL9 | Antibody selected against CXCL10 and evolved to also recognize CXCL9 | 125 |
| VEGF and Erb-B2 | Antigens are completely unrelated, and antibody binds with 3/0.2nM affinity to VEGF/Erb-B2 respectively | 126 |
| Botulinum toxin A, B, E and F | One antibody able to recognize all Botulinum types afflicting man was selected by yeast display. | 127 |
Specificity for a single antigen is generally the goal of antibody engineering. However, in certain applications, defined cross-reactivity is extremely useful. Species cross-reactivity allows better assessment of therapeutic efficacy and toxicity in animal models. Unfortunately, cross-reactive antibodies are often difficult to obtain by hybridoma methods, due to tolerance. In contrast, in vitro libraries are unaffected by immune tolerance and antibodies targeting conserved sites across species have proven to be the rule rather than the exception. In the case of highly conserved proteins, such as vascular endothelial growth factor (VEGF), human/mouse cross-reactive antibodies have been obtained directly from naïve libraries128,129. In the case of less conserved orthologs, such as BAFF/BLys receptor 3 (BR3), initial anti-human antibodies with weak cross-reactivity to the mouse protein have been obtained from naïve libraries and evolved to be highly cross-reactive130. Similar approaches have been used to generate antibodies recognizing two closely related chemokines (CXLC10 and CXCL9)125 thereby permitting neutralization of 2 human chemokines with a single antibody.
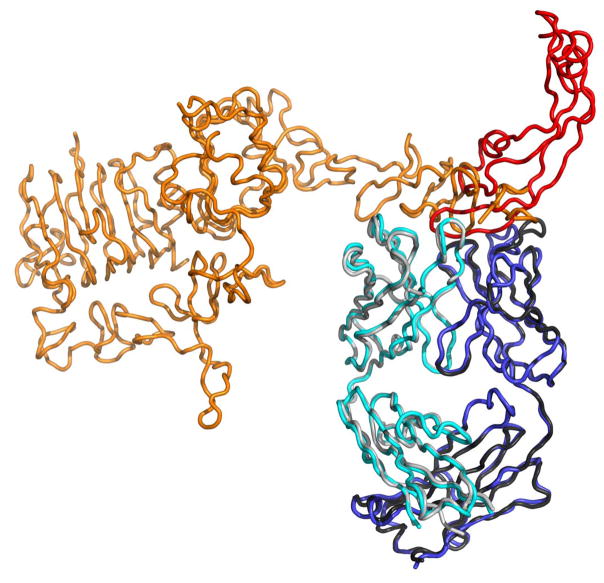
Most specificity engineering examples involve the improvement of pre-existing weak recognition, due to homology between the recognized targets. In perhaps the most extreme case of engineered cross-reactivity, Herceptin has been evolved to recognize a very different protein, VEGF, as well as its original target, Erb-B2126. After significant evolution the affinities for both targets were comparable to those of therapeutic antibodies (Kd = 3/0.2 nM for VEGF/Erb-B2). The antibody inhibited both VEGF and Erb-B2-mediated cell proliferation in vitro and tumor progression in mouse models. The structures of the bispecific Fab in complex with Erb-B2 or VEGF revealed a common paratope, with the Erb-B2 functional paratope located predominantly on VH, and that for VEGF on VL (Fig. 4). The ability to design antigen-binding sites with dual specificity against structurally unrelated antigens may be important in therapeutic strategies targeting two distinct signaling pathways with a single antibody.
Figure 4.
An engineered dual specificity synthetic Fab. The bH1 Fab binds to both Her2 (orange, PDB entry 3BDY) and VEGF (red, PDB entry 3BE1). The heavy and light chains of the Fab are colored cyan/grey or blue/black respectively, with the different colors derived from structures of bH1 binding to either Her2 or BEGF.
The ability to improve affinity and broaden specificity also has major implications for the development of antibodies against pathogens. For the effective inhibition of viral infection and bacterial toxins, antibodies would ideally recognize a variety of antigen subtypes with high affinity, to afford broad protection against pathogen variants. Furthermore, several studies have shown that multiple antibodies targeting distinct epitopes provide synergistic effects necessary for effective neutralization of pathogens131,132. In vitro antibody technologies provide an effective means for achieving these demanding criteria, as exemplified by a long-term study of neutralizing antibodies against the botulinum neurotoxin (BoNT). Phage antibody libraries from immunized mice and humans resulted in the isolation of three antibodies recognizing non-overlapping epitopes on BoNT133. The use of these three antibodies together as an oligoclonal” IgG provided strong synergy that dramatically increased toxin neutralization. A long series of affinity and specificity maturation cycles using yeast display, resulted in the final development of a remarkable antibody able to recognize Botulinum toxins A, B, E and F, all the serotypes afflicting man127.
Exploiting the recombinant nature of in vitro selected antibodies
All in vitro selection systems immediately provide the genes, and corresponding sequences, of antibodies selected against a particular target, providing ready access to additional antibody formats by simple sub-cloning. Functions adopted using this “gene-based” approach include dimerization134, multimerization135,136, and fusions to enzymes137, tags138 or fluorescent proteins139. Fusion to alkaline phosphatase is a particularly useful example of improved functionality. As this is a dimeric enzyme, fusing antibodies, either individually or as libraries, to alkaline phosphatase simultaneously provides dimerization and alkaline phosphatase activity, greatly facilitating screening32,137. Short peptides acting as in vivo biotinylation tags138, placed at the carboxy terminus of antibody fragments, allow stoichiometrically defined site specific antibody biotinylation, as well as a straightforward multimerization method140. Antibody fragments can also be transformed into full-length antibodies141, or scFv-Fc fusions, which are very similar in many aspects142. The use of engineered Fc regions can result in improved pharmacokinetics and effector functions (for reviews see 143,144), including bispecific IgG, in which engineering of two different Fc regions allow only heterologous pairing145,146.
Other approaches to generate bispecific antibodies build upon the observation that some scFv fragments form bivalent dimers (diabodies)147, trimers148,149 and even tetramers150 when the VH/VL linker is shortened. Various other bispecific antibody designs have also been created (see 151 for a review). Even more radically, completely novel biochemical entities have been added to antigen binding fragments. Fusions of scFv and Fab fragments to heterologous proteins, such as interleukins and cytokines152,153, apoptotic ligands, enzymes, toxins or RNases (see 154,155 for reviews) have allowed novel therapeutic paradigms. Many of the above candidate therapeutic antibody constructs arose from antibody genes initially isolated from mouse hybridomas, but this is expected to change as more human antibodies are made available from engineered repertoires.
Microinjected antibodies have been long used to knock out intracellular functions156. Antibody fragments can be expressed within target cells and targeted to various subcellular compartments141,157 by adding suitable signal sequences, allowing visualization or functional modification of proteins in different compartments. Removing the standard leader sequence results in cytoplasmic expression while the addition of a nuclear localization signal targets to the nucleus. The combination of a leader sequence and the endoplasmic reticulum (ER) retention sequence retains expressed antibodies in the ER and has been used to prevent the expression of membrane proteins by sequestration in the ER. These include human interleukin 2 receptor, the ErbB-2 receptor, s-amyloid precursor protein, vascular adhesion molecule 1 and many others158,159,160,161. The advantage of this strategy is that it requires antibodies that bind to any accessible epitope to provide the functional knockout, as opposed to the functional activity required of cytoplasmically expressed antibodies. Functional studies of membrane receptors or secreted proteins can thus be attempted by a single standardized subcloning step immediately after in vitro antibody selection, providing equivalence to RNAi knockdowns at the protein level.
While expression in the secretory pathway is straightforward, folding of antibody fragments in the cytoplasm is far more challenging, due to the absence of specific chaperones, and the reducing environment, which prevents disulfide bond formation162. Despite these problems, there are examples where cytoplasmic proteins have been targeted with intracellular scFvs96,163. The success of this approach has been improved by the creation of libraries of particularly stable scFvs164,165,166, preselecting antibodies for functional cytoplasmic expression167,168, or by using binder libraries based on molecular scaffolds that do not rely on disulphide bond formation, such as engineered ankyrin repeat proteins169,170. One major advantage of such protein based allosteric blockers is the ability to generate very specific binders, able to distinguish between closely related family members. While the need to genetically modify the target cell is a major disadvantage, this has been partly alleviated by fusion to internalizing sequences that allow antibodies to enter the cell from the outside171.
High throughput selection by in vitro display methods
The ease with which antibodies can be selected, screened and produced by in vitro display technologies, makes generation and screening of antibodies rapid and simple compared to hybridomas. Typically a panel of ELISA positive monoclonal antibody fragments can be generated within two weeks. Early experiments demonstrated the feasibility of semi-automated selection/screening of phage antibody libraries172,173,174 on small numbers of targets. More recently, selections on over 400 different antigens were successful with 54% of bacterial, and 88% of mammalian-produced antigens32 yielding antibodies, with the differences between the two protein classes probably due to the levels of correct folding. In a recent international comparative study antibodies were raised to 20 different human SH2 domains using hybridoma or phage display. Results from two of the participating phage display labs69,70, show that antibodies (some with sub-nanomolar affinities) were generated to all antigens, with 55% of positive antibodies specific for target SH2 domains when assessed against the entire SH2 panel. These antibodies were validated in a broad range of assays, including microarrays, immunoblots, immunofluorescence and immunoprecipitation.
The future vision of affinity reagents generated by display technologies
If antibodies selected by in vitro methods are so powerful, why are they not more widely perceived as valuable research reagents? Part of the answer lies in the difficult patent situation, which resulted in restriction of this technology to the high margin therapeutic markets for commercial use. It is perhaps significant in this regard that hybridoma technology was never patented, and achieved relatively wide acceptance within a short period. The situation for some of the core phage display patents is in the process of rapid change as most platform patents have either expired, or will do so over the next few years175, and it is possible that the technology will become more widely disseminated as a result.
Largely unnoticed by the research community, some commercial “monoclonal antibodies” are actually recombinant antibodies selected by phage display, reformatted to look like traditional murine antibodies by the fusion of Fc regions to human variable regions (e.g. the sulfotyrosine antibody described above38). ABDSerotec also sells a number of unmodified recombinant Fab fragments selected by phage display. It therefore seems that the most important impediments to widespread adoption are a lack of knowledge of the capabilities of this technology, coupled with limited expertise and library availability. Furthermore, the number of companies willing to carry out in vitro selection as a fee for service is vanishingly small compared to the 180 companies willing to generate antibodies by immunization16.
However, one commonly cited issue relates to the ability to express these antibodies. While the specificities obtained, and described herein, are remarkable, the expression and stability of antibody fragments varies enormously, from exceptionally stable scFv fragments used in clinical trials176 to other fragments that are poorly expressed. A typical selection almost always generates a number of different binders to any well-folded antigen. Among these there is usually at least one that is sufficiently stable and well produced for research use. Furthermore, it is expected that stability and expression levels will improve as libraries are based on more stable scaffolds177. The studies described above indicate that this goal can now be met in highly parallelized screening setups with low effort per antigen69,70, provided that libraries of sufficient diversity and optimized protocols are used. Furthermore, stability and expression screening can be easily included as part of the HT screening process. An additional issue with in vitro derived antibodies is that they are either not glycosylated if expressed in bacteria, or incorrectly glycosylated if expressed in standard yeast strains. If correct glycosylation is necessary, this can be overcome by expression in human cells or yeast modified to give human glycosylation patterns178.
Once an antibody is generated, it can be defined precisely by sequence and even “distributed” in this way. Gene synthesis is progressing at a remarkable pace, with the cost per base of synthesized genes falling dramatically. In fact, genes corresponding to the sequences of specific antibody fragments can now be synthesized for less than the cost of purchase of some antibodies from traditional vendors.
The present state of this field can be compared to the situation with sequencing technologies at the start of the human genome project. Just as enormous technical advances occurred in the human genome project once it was started and rigorous industrial processes were applied, so we anticipate dramatic improvement in all aspects of selection, screening, downstream use and distribution of in vitro derived affinity reagents once a proteomic scale project is initiated and financed.
In summary, in vitro display technologies permit the facile generation of antibodies by providing access to billions of potential binders in large “universal”, or immune, display libraries. The technologies facilitate production, screening and maturation of selected binders, allowing selection on target conformations and formats not possible by more traditional routes based on immunization. Furthermore, the easy availability of the gene sequence not only provides a definitive description of the product but also allows electronic sharing and recreation of the binding molecule through gene synthesis. Over the last 20 years display technologies have been applied successfully to the development of therapeutic antibody candidates. In the coming decade we expect to see increased realization of the benefits of this technology within the research and diagnostic markets as well.
Table 2.
In vitro selected antibodies recognizing protein sequences and conformations
| Target | Notes | Ref |
|---|---|---|
| Peptide MHC complexes (similar to T cell receptor recognition) | Similar antibodies obtained by immunization86,87 have lower affinities. | 88,89,90,91 |
| Fibronectin splice variants, EDA and EDB | Selection directed towards recognition of both human and mouse variants, allowing same antibody to be used in both models and clinical studies. | 92,93,94 |
| Fetal hemoglobin | 36 | |
| Fetal nucleated red cells | 95 | |
| GTP-bound Rab6 | Antibodies were used to track activated Rab6 in the cell as GFP fusions | 96 |
| Caspase 1 | Antibodies recognize either the “on” or “off” forms | 97 |
| Integral membrane proteins | CitS from Klebsiella pneumoniaei and KcsA from Streptomyces lividans. KcsA antibodies used as crystallization chaperones | 98,99 |
| RNA | Structured domain from Tetrahymena group I intron. Antibody used as crystallization chaperone | 100 |
| ABL1 versus ABL2 | Differ by only 11% | 69,70 |
| Chicken versus quail lysozyme | Differ by only four amino acids, of which only one surface exposed | 68 |
Table 3.
In vitro selected antibodies recognizing cell surface receptors
| Target | Notes/therapeutic indication | Ref |
|---|---|---|
| Blys | Systemic lupus erythematosis | 102 |
| Tumor necrosis factor a | Phage display was used to convert a murine mAb into a human antibody by guided selection. Rheumatoid arthritis, ankylosing spondylitis, chronic plaque psoriasis and Crohn’s disease, antibody developed by guided selection phage display | 104,105 |
| IGF-1 receptor | Blocking of ligand-binding site of receptor and receptor down-regulation by endocytosis. Potential application in cancer | 106 |
| Notch | Prevent proteolysis of juxtamembrane NRR domain | 107,108 |
| Met | Dimeric antibodies are agonistic, monomeric ones are antagonistic, and prospected for non-small cell lung cancer | 109 |
| MuSK | Agonsitic antibodies demonstrate that MuSK activation is capable of triggering a key event in neuromuscular junction formation | 110 |
| CD40 | Agonistic antibodies which activate normal human B suppress HIV-1 infection in vitro | 111 |
| Hemagglutinin | Antibodies recognize a previously unknown conserved conformational epitope. Isolated from both naïve and immunized libraries. | 23,112,113 |
| EphA2 and CD44 | Selected from phage antibody library on yeast displayed antigen, followed by selection for internalization on cells | 114 |
| CD166 | Internalizing antibodies selected directly for internalization on cancer cells (CD166 on prostate, ErbB2 and Transferrin receptor on breast, EGFR on A431). Antigen identified after selection. Potential utility for internalization of chemotherapeutics. | 115,116 |
| ErbB2 | 117 | |
| Transferrin receptor | 117 | |
| EGFR | 118 | |
| TRAIL-R | Over 500 different scFvs and Fabs isolated by phage display. | 119 |
Acknowledgments
ARMB is grateful to NIH (P50GM085273 & R01-HG004852-01A1), DOE (GTL) and DTRA for funding. SD gratefully acknowledges funding by the EU 7th framework programme (Projects: Affinomics and AffinityProteome). JMC is pleased to acknowledge funding by the Wellcome Trust.
References
- 1.Love JC, Ronan JL, Grotenbreg GM, van der Veen AG, Ploegh HL. A microengraving method for rapid selection of single cells producing antigen-specific antibodies. Nat Biotechnol. 2006 doi: 10.1038/nbt1210. [DOI] [PubMed] [Google Scholar]
- 2.Jin A, et al. A rapid and efficient single-cell manipulation method for screening antigen-specific antibody-secreting cells from human peripheral blood. Nat Med. 2009;15:1088–1092. doi: 10.1038/nm.1966. nm.1966 [pii] [DOI] [PubMed] [Google Scholar]
- 3.Reddy ST, et al. Monoclonal antibodies isolated without screening by analyzing the variable-gene repertoire of plasma cells. Nat Biotechnol. 2010;28:965–969. doi: 10.1038/nbt.1673. nbt.1673 [pii] [DOI] [PubMed] [Google Scholar]
- 4.Smith GP. Filamentous fusion phage: novel expression vectors that display cloned antigens on the virion surface. Science. 1985;228:1315–1317. doi: 10.1126/science.4001944. [DOI] [PubMed] [Google Scholar]
- 5.Scott JK, Smith GP. Searching for peptide ligands with an epitope library. Science. 1990;249:386–390. doi: 10.1126/science.1696028. [DOI] [PubMed] [Google Scholar]
- 6.Skerra A, Pluckthun A. Assembly of a functional immunoglobulin Fv fragment in Escherichia coli. Science. 1988;240:1038–1041. doi: 10.1126/science.3285470. [DOI] [PubMed] [Google Scholar]
- 7.Larrick JW, et al. Rapid cloning of rearranged immunoglobulin genes from human hybridoma cells using mixed primers and the polymerase chain reaction. Biochem Biophys Res Commun. 1989;160:1250–1256. doi: 10.1016/s0006-291x(89)80138-x. [DOI] [PubMed] [Google Scholar]
- 8.Marks JD, et al. By-passing immunization. Human antibodies from V-gene libraries displayed on phage. J Mol Biol. 1991;222:581–597. doi: 10.1016/0022-2836(91)90498-u. [DOI] [PubMed] [Google Scholar]
- 9.Orlandi R, Gussow DH, Jones PT, Winter G. Cloning immunoglobulin variable domains for expression by the polymerase chain reaction. Proc Natl Acad Sci U S A. 1989;86:3833–3837. doi: 10.1073/pnas.86.10.3833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huse WD, et al. Generation of a large combinatorial library of the immunoglobulin repertoire in phage lambda. Science. 1989;246:1275–1281. doi: 10.1126/science.2531466. [DOI] [PubMed] [Google Scholar]
- 11.Sastry L, et al. Cloning of the immunological repertoire in Escherichia coli for generation of monoclonal catalytic antibodies: construction of a heavy chain variable region-specific cDNA library. Proc Natl Acad Sci U S A. 1989;86:5728–5732. doi: 10.1073/pnas.86.15.5728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McCafferty J, Griffiths AD, Winter G, Chiswell DJ. Phage antibodies: filamentous phage displaying antibody variable domains. Nature. 1990;348:552–554. doi: 10.1038/348552a0. [DOI] [PubMed] [Google Scholar]
- 13.Breitling F, Dubel S, Seehaus T, Klewinghaus I, Little M. A surface expression vector for antibody screening. Gene. 1991;104:147–153. doi: 10.1016/0378-1119(91)90244-6. [DOI] [PubMed] [Google Scholar]
- 14.Boder ET, Wittrup KD. Yeast surface display for screening combinatorial polypeptide libraries. Nat Biotechnol. 1997;15:553–557. doi: 10.1038/nbt0697-553. [DOI] [PubMed] [Google Scholar]
- 15.Feldhaus MJ, et al. Flow-cytometric isolation of human antibodies from a nonimmune Saccharomyces cerevisiae surface display library. Nat Biotechnol. 2003 doi: 10.1038/nbt785. [DOI] [PubMed] [Google Scholar]
- 16.Bordeaux J, et al. Antibody validation. Biotechniques. 2010;48:197–209. doi: 10.2144/000113382. 000113382 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jositsch G, et al. Suitability of muscarinic acetylcholine receptor antibodies for immunohistochemistry evaluated on tissue sections of receptor gene-deficient mice. Naunyn Schmiedebergs Arch Pharmacol. 2009;379:389–395. doi: 10.1007/s00210-008-0365-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jensen BC, Swigart PM, Simpson PC. Ten commercial antibodies for alpha-1-adrenergic receptor subtypes are nonspecific. Naunyn Schmiedebergs Arch Pharmacol. 2009;379:409–412. doi: 10.1007/s00210-008-0368-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Spicer SS, Spivey MA, Ito M, Schulte BA. Some ascites monoclonal antibody preparations contain contaminants that bind to selected Golgi zones or mast cells. J Histochem Cytochem. 1994;42:213–221. doi: 10.1177/42.2.7507139. [DOI] [PubMed] [Google Scholar]
- 20.Pozner-Moulis S, Cregger M, Camp RL, Rimm DL. Antibody validation by quantitative analysis of protein expression using expression of Met in breast cancer as a model. Lab Invest. 2007;87:251–260. doi: 10.1038/labinvest.3700515. 3700515 [pii] [DOI] [PubMed] [Google Scholar]
- 21.Grimsey NL, et al. Specific detection of CB1 receptors; cannabinoid CB1 receptor antibodies are not all created equal! J Neurosci Methods. 2008;171:78–86. doi: 10.1016/j.jneumeth.2008.02.014. S0165–0270(08)00114–3 [pii] [DOI] [PubMed] [Google Scholar]
- 22.Saper CB. An open letter to our readers on the use of antibodies. J Comp Neurol. 2005;493:477–478. doi: 10.1002/cne.20839. [DOI] [PubMed] [Google Scholar]
- 23.Throsby M, et al. Heterosubtypic neutralizing monoclonal antibodies cross-protective against H5N1 and H1N1 recovered from human IgM+ memory B cells. PLoS ONE. 2008;3:e3942. doi: 10.1371/journal.pone.0003942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Razai A, et al. Molecular evolution of antibody affinity for sensitive detection of botulinum neurotoxin type A. J Mol Biol. 2005;351:158–169. doi: 10.1016/j.jmb.2005.06.003. [DOI] [PubMed] [Google Scholar]
- 25.Lee CV, et al. High-affinity human antibodies from phage-displayed synthetic Fab libraries with a single framework scaffold. J Mol Biol. 2004;340:1073–1093. doi: 10.1016/j.jmb.2004.05.051. [DOI] [PubMed] [Google Scholar]
- 26.Hanes J, Schaffitzel C, Knappik A, Pluckthun A. Picomolar affinity antibodies from a fully synthetic naïve library selected and evolved by ribosome display. Nat Biotechnol. 2000;18:1287–1292. doi: 10.1038/82407. [DOI] [PubMed] [Google Scholar]
- 27.Schier R, et al. Isolation of picomolar affinity anti-c-erbB-2 single-chain Fv by molecular evolution of the complementarity determining regions in the center of the antibody binding site. J Mol Biol. 1996;263:551–567. doi: 10.1006/jmbi.1996.0598. S0022–2836(96)90598–7 [pii] [DOI] [PubMed] [Google Scholar]
- 28.Yang WP, et al. CDR walking mutagenesis for the affinity maturation of a potent human anti-HIV-1 antibody into the picomolar range. J Mol Biol. 1995;254:392–403. doi: 10.1006/jmbi.1995.0626. S0022283685706262 [pii] [DOI] [PubMed] [Google Scholar]
- 29.Boder ET, Midelfort KS, Wittrup KD. Directed evolution of antibody fragments with monovalent femtomolar antigen-binding affinity. Proc Natl Acad Sci USA. 2000;97:10701–10705. doi: 10.1073/pnas.170297297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Foote J, Eisen HN. Breaking the affinity ceiling for antibodies and T cell receptors. Proc Natl Acad Sci U S A. 2000;97:10679–10681. doi: 10.1073/pnas.97.20.10679. 97/20/10679 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Batista FD, Neuberger MS. Affinity dependence of the B cell response to antigen: a threshold, a ceiling, and the importance of off-rate. Immunity. 1998;8:751–759. doi: 10.1016/s1074-7613(00)80580-4. S1074–7613(00)80580–4 [pii] [DOI] [PubMed] [Google Scholar]
- 32.Schofield DJ, et al. Application of phage display to high throughput antibody generation and characterization. Genome Biol. 2007;8:R254. doi: 10.1186/gb-2007-8-11-r254. gb-2007–8–11-r254 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dubel S, Stoevesandt O, Taussig MJ, Hust M. Generating recombinant antibodies to the complete human proteome. Trends Biotechnol. 2010;28:333–339. doi: 10.1016/j.tibtech.2010.05.001. S0167–7799(10)00082-X [pii] [DOI] [PubMed] [Google Scholar]
- 34.Koide A, Bailey CW, Huang X, Koide S. The fibronectin type III domain as a scaffold for novel binding proteins. J Mol Biol. 1998;284:1141–1151. doi: 10.1006/jmbi.1998.2238. [DOI] [PubMed] [Google Scholar]
- 35.Philibert P, et al. A focused antibody library for selecting scFvs expressed at high levels in the cytoplasm. BMC Biotechnol. 2007;7:81. doi: 10.1186/1472-6750-7-81. 1472–6750–7–81 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Parsons HL, et al. Directing phage selections towards specific epitopes. Protein Eng. 1996;9:1043–1049. doi: 10.1093/protein/9.11.1043. [DOI] [PubMed] [Google Scholar]
- 37.Lassen KS, Bradbury AR, Rehfeld JF, Heegaard NH. Microscale characterization of the binding specificity and affinity of a monoclonal antisulfotyrosyl IgG antibody. Electrophoresis. 2008;29:2557–2564. doi: 10.1002/elps.200700908. [DOI] [PubMed] [Google Scholar]
- 38.Kehoe JW, et al. Using Phage Display to Select Antibodies Recognizing Post-translational Modifications Independently of Sequence Context. Mol Cell Proteomics. 2006;5:2350–2363. doi: 10.1074/mcp.M600314-MCP200. [DOI] [PubMed] [Google Scholar]
- 39.Hoffhines AJ, Damoc E, Bridges KG, Leary JA, Moore KL. Detection and purification of tyrosine-sulfated proteins using a novel anti-sulfotyrosine monoclonal antibody. J Biol Chem. 2006;281:37877–37887. doi: 10.1074/jbc.M609398200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Grunewald J, et al. Mechanistic studies of the immunochemical termination of self-tolerance with unnatural amino acids. Proc Natl Acad Sci U S A. 2009;106:4337–4342. doi: 10.1073/pnas.0900507106. 0900507106 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dalum I, et al. Therapeutic antibodies elicited by immunization against TNF-alpha. Nat Biotechnol. 1999;17:666–669. doi: 10.1038/10878. [DOI] [PubMed] [Google Scholar]
- 42.Hust M, et al. A human scFv antibody generation pipeline for proteome research. J Biotechnol. 2010 doi: 10.1016/j.jbiotec.2010.09.945. S0168–1656(10)01879–1 [pii] [DOI] [PubMed] [Google Scholar]
- 43.Lloyd C, et al. Modelling the human immune response: performance of a 1011 human antibody repertoire against a broad panel of therapeutically relevant antigens. Protein Eng Des Sel. 2009;22:159–168. doi: 10.1093/protein/gzn058. gzn058 [pii] [DOI] [PubMed] [Google Scholar]
- 44.Paschke M. Phage display systems and their applications. Appl Microbiol Biotechnol. 2006;70:2–11. doi: 10.1007/s00253-005-0270-9. [DOI] [PubMed] [Google Scholar]
- 45.Zahnd C, Amstutz P, Pluckthun A. Ribosome display: selecting and evolving proteins in vitro that specifically bind to a target. Nat Methods. 2007;4:269–279. doi: 10.1038/nmeth1003. [DOI] [PubMed] [Google Scholar]
- 46.Chao G, et al. Isolating and engineering human antibodies using yeast surface display. Nat Protoc. 2006;1:755–768. doi: 10.1038/nprot.2006.94. [DOI] [PubMed] [Google Scholar]
- 47.Binz HK, Amstutz P, Pluckthun A. Engineering novel binding proteins from nonimmunoglobulin domains. Nat Biotechnol. 2005;23:1257–1268. doi: 10.1038/nbt1127. nbt1127 [pii] [DOI] [PubMed] [Google Scholar]
- 48.Binz HK, Pluckthun A. Engineered proteins as specific binding reagents. Curr Opin Biotechnol. 2005;16:459–469. doi: 10.1016/j.copbio.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 49.Skerra A. Alternative non-antibody scaffolds for molecular recognition. Curr Opin Biotechnol. 2007;18:295–304. doi: 10.1016/j.copbio.2007.04.010. S0958–1669(07)00080–8 [pii] [DOI] [PubMed] [Google Scholar]
- 50.Bradbury AR, Marks JD. Antibodies from phage antibody libraries. J Immunol Methods. 2004;290:29–49. doi: 10.1016/j.jim.2004.04.007. [DOI] [PubMed] [Google Scholar]
- 51.Wright K, Collins DC, Preedy JR. Comparative specificity of antisera raised against estrone, estradiol-17 and estriol using 6–0-carboxy-methyloxime bovine serum albumin derivatives. Steroids. 1973;21:755–769. doi: 10.1016/0039-128x(73)90140-2. [DOI] [PubMed] [Google Scholar]
- 52.Haning R, et al. The evolution of titer and specificity of aldosterone binding antibodies in hyperimmunized sheep. Steroids. 1972;20:73–88. doi: 10.1016/0039-128x(72)90119-5. [DOI] [PubMed] [Google Scholar]
- 53.Exley D, Johnson MW, Dean PD. Antisera highly specific for 17 -oestradiol. Steroids. 1971;18:605–620. doi: 10.1016/0039-128x(71)90073-0. [DOI] [PubMed] [Google Scholar]
- 54.Tateishi K, Hamaoka T, Takatsu K, Hayashi C. A novel immunization procedure for production of anti-testosterone and anti-5 alpha-dihydrotestosterone antisera of low cross-reactivity. J Steroid Biochem. 1980;13:951–959. doi: 10.1016/0022-4731(80)90170-3. [DOI] [PubMed] [Google Scholar]
- 55.Smith TW, Skubitz KM. Kinetics in interactions between antibodies and haptens. Biochemistry. 1975;14:1496–1502. doi: 10.1021/bi00678a023. [DOI] [PubMed] [Google Scholar]
- 56.Moghaddam A, et al. Identification of scFv antibody fragments that specifically recognise the heroin metabolite 6-monoacetylmorphine but not morphine. J Immunol Methods. 2003;280:139–155. doi: 10.1016/s0022-1759(03)00109-1. S0022175903001091 [pii] [DOI] [PubMed] [Google Scholar]
- 57.Dorsam H, et al. Antibodies to steroids from a small human naïve IgM library. FEBS Lett. 1997;414:7–13. doi: 10.1016/s0014-5793(97)00966-6. S0014–5793(97)00966–6 [pii] [DOI] [PubMed] [Google Scholar]
- 58.Pope A, et al. In vitro selection of a high affinity antibody to oestradiol using a phage display human antibody library. Immunotechnology. 1996;2:209–217. doi: 10.1016/s1380-2933(96)00051-6. S1380293396000516 [pii] [DOI] [PubMed] [Google Scholar]
- 59.Hemminki A, Niemi S, Hautoniemi L, Soderlund H, Takkinen K. Fine tuning of an anti-testosterone antibody binding site by stepwise optimisation of the CDRs. Immunotechnology. 1998;4:59–69. doi: 10.1016/s1380-2933(98)00002-5. S1380293398000025 [pii] [DOI] [PubMed] [Google Scholar]
- 60.Saviranta P, et al. Engineering the steroid-specificity of an anti-17beta-estradiol Fab by random mutagenesis and competitive phage panning. Protein Eng. 1998;11:143–152. doi: 10.1093/protein/11.2.143. [DOI] [PubMed] [Google Scholar]
- 61.Chames P, Coulon S, Baty D. Improving the affinity and the fine specificity of an anti-cortisol antibody by parsimonious mutagenesis and phage display. J Immunol. 1998;161:5421–5429. [PubMed] [Google Scholar]
- 62.Bikker FJ, Mars-Groenendijk RH, Noort D, Fidder A, van der Schans GP. Detection of sulfur mustard adducts in human callus by phage antibodies. Chem Biol Drug Des. 2007;69:314–320. doi: 10.1111/j.1747-0285.2007.00504.x. JPP504 [pii] [DOI] [PubMed] [Google Scholar]
- 63.Szent-Gyorgyi C, et al. Fluorogen-activating single-chain antibodies for imaging cell surface proteins. Nat Biotechnol. 2008;26:235–240. doi: 10.1038/nbt1368. nbt1368 [pii] [DOI] [PubMed] [Google Scholar]
- 64.Watanabe H, Nakanishi T, Umetsu M, Kumagai I. Human anti-gold antibodies: biofunctionalization of gold nanoparticles and surfaces with anti-gold antibodies. J Biol Chem. 2008;283:36031–36038. doi: 10.1074/jbc.M805547200. M805547200 [pii] [DOI] [PubMed] [Google Scholar]
- 65.Monigatti F, Gasteiger E, Bairoch A, Jung E. The Sulfinator: predicting tyrosine sulfation sites in protein sequences. Bioinformatics. 2002;18:769–770. doi: 10.1093/bioinformatics/18.5.769. [DOI] [PubMed] [Google Scholar]
- 66.Sako D, et al. A sulfated peptide segment at the amino terminus of PSGL-1 is critical for P-selectin binding. Cell. 1995;83:323–331. doi: 10.1016/0092-8674(95)90173-6. [DOI] [PubMed] [Google Scholar]
- 67.Rigby PW, Gething MJ, Hartley BS. Construction of intergeneric hybrids using bacteriophage P1CM: transfer of the Klebsiella aerogenes ribitol dehydrogenase gene to Escherichia coli. J Bacteriol. 1976;125:728–738. doi: 10.1128/jb.125.2.728-738.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ayriss J, Woods T, Bradbury A, Pavlik P. High-throughput screening of single-chain antibodies using multiplexed flow cytometry. J Proteome Res. 2007;6:1072–1082. doi: 10.1021/pr0604108. [DOI] [PubMed] [Google Scholar]
- 69.Pershad K, et al. Generating a panel of highly specific antibodies to 20 human SH2 domains by phage display. Protein Eng Des Sel. 2010;23:279–288. doi: 10.1093/protein/gzq003. gzq003 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mersmann M, et al. Towards proteome scale antibody selections using phage display. N Biotechnol. 2009 doi: 10.1016/j.nbt.2009.10.007. S1871–6784(09)01256–4 [pii] [DOI] [PubMed] [Google Scholar]
- 71.Velappan N, et al. Selection and characterization of scFv antibodies against the Sin Nombre hantavirus nucleocapsid protein. J Immunol Methods. 2007;321:60–69. doi: 10.1016/j.jim.2007.01.011. [DOI] [PubMed] [Google Scholar]
- 72.Cabezas S, et al. Phage-displayed antibody fragments recognizing dengue 3 and dengue 4 viruses as tools for viral serotyping in sera from infected individuals. Arch Virol. 2009;154:1035–1045. doi: 10.1007/s00705-009-0401-1. [DOI] [PubMed] [Google Scholar]
- 73.Lim AP, et al. Neutralizing human monoclonal antibody against H5N1 influenza HA selected from a Fab-phage display library. Virol J. 2008;5:130. doi: 10.1186/1743-422X-5-130. 1743–422X-5–130 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Okada J, et al. Monoclonal antibodies in man that neutralized H3N2 influenza viruses were classified into three groups with distinct strain specificity: 1968–1973, 1977–1993 and 1997–2003. Virology. 2010;397:322–330. doi: 10.1016/j.virol.2009.11.025. S0042–6822(09)00745–4 [pii] [DOI] [PubMed] [Google Scholar]
- 75.Meissner F, et al. Detection of antibodies against the four subtypes of ebola virus in sera from any species using a novel antibody-phage indicator assay. Virology. 2002;300:236–243. doi: 10.1006/viro.2002.1533. [DOI] [PubMed] [Google Scholar]
- 76.Kirsch MI, et al. Development of human antibody fragments using antibody phage display for the detection and diagnosis of Venezuelan equine encephalitis virus (VEEV) BMC Biotechnol. 2008;8:66. doi: 10.1186/1472-6750-8-66. 1472–6750–8–66 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Maynard JA, et al. Protection against anthrax toxin by recombinant antibody fragments correlates with antigen affinity. Nat Biotechnol. 2002;20:597–601. doi: 10.1038/nbt0602-597nbt0602-597[pii]. [DOI] [PubMed] [Google Scholar]
- 78.Mabry R, et al. Passive protection against anthrax by using a high-affinity antitoxin antibody fragment lacking an Fc region. Infect Immun. 2005;73:8362–8368. doi: 10.1128/IAI.73.12.8362-8368.2005. 73/12/8362 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wild MA, et al. Human antibodies from immunized donors are protective against anthrax toxin in vivo. Nat Biotechnol. 2003;21:1305–1306. doi: 10.1038/nbt891. [DOI] [PubMed] [Google Scholar]
- 80.Hayhurst A, et al. Isolation and expression of recombinant antibody fragments to the biological warfare pathogen Brucella melitensis. J Immunol Methods. 2003;276:185–196. doi: 10.1016/s0022-1759(03)00100-5. S0022175903001005 [pii] [DOI] [PubMed] [Google Scholar]
- 81.Zou N, Newsome T, Li B, Tsai S, Lo SC. Human single-chain Fv antibodies against Burkholderia mallei and Burkholderia pseudomallei. Exp Biol Med (Maywood) 2007;232:550–556. 232/4/550 [pii] [PubMed] [Google Scholar]
- 82.Steiniger SC, Altobell LJ, 3rd, Zhou B, Janda KD. Selection of human antibodies against cell surface-associated oligomeric anthrax protective antigen. Mol Immunol. 2007;44:2749–2755. doi: 10.1016/j.molimm.2006.11.011. [DOI] [PubMed] [Google Scholar]
- 83.Cirino NM, et al. Disruption of anthrax toxin binding with the use of human antibodies and competitive inhibitors. Infect Immun. 1999;67:2957–2963. doi: 10.1128/iai.67.6.2957-2963.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zhou B, Wirsching P, Janda KD. Human antibodies against spores of the genus Bacillus: a model study for detection of and protection against anthrax and the bioterrorist threat. Proc Natl Acad Sci U S A. 2002;99:5241–5246. doi: 10.1073/pnas.082121599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Garcia-Rodriguez C, et al. Molecular evolution of antibody cross-reactivity for two subtypes of type A botulinum neurotoxin. Nat Biotechnol. 2007;25:107–116. doi: 10.1038/nbt1269. [DOI] [PubMed] [Google Scholar]
- 86.Dadaglio G, Nelson CA, Deck MB, Petzold SJ, Unanue ER. Characterization and quantitation of peptide-MHC complexes produced from hen egg lysozyme using a monoclonal antibody. Immunity. 1997;6:727–738. doi: 10.1016/s1074-7613(00)80448-3. S1074–7613(00)80448–3 [pii] [DOI] [PubMed] [Google Scholar]
- 87.Krogsgaard M, et al. Visualization of myelin basic protein (MBP) T cell epitopes in multiple sclerosis lesions using a monoclonal antibody specific for the human histocompatibility leukocyte antigen (HLA)-DR2-MBP 85–99 complex. J Exp Med. 2000;191:1395–1412. doi: 10.1084/jem.191.8.1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Mutuberria R, et al. Isolation of human antibodies to tumor-associated endothelial cell markers by in vitro human endothelial cell selection with phage display libraries. J Immunol Methods. 2004;287:31–47. doi: 10.1016/j.jim.2004.0111. S0022175904000535 [pii] [DOI] [PubMed] [Google Scholar]
- 89.Cohen CJ, Denkberg G, Lev A, Epel M, Reiter Y. Recombinant antibodies with MHC-restricted, peptide-specific, T-cell receptor-like specificity: new tools to study antigen presentation and TCR-peptide-MHC interactions. J Mol Recognit. 2003;16:324–332. doi: 10.1002/jmr.640. [DOI] [PubMed] [Google Scholar]
- 90.Engberg J, Krogsgaard M, Fugger L. Recombinant antibodies with the antigen-specific, MHC restricted specificity of T cells: novel reagents for basic and clinical investigations and immunotherapy. Immunotechnology. 1999;4:273–278. doi: 10.1016/s1380-2933(98)00017-7. [DOI] [PubMed] [Google Scholar]
- 91.Stryhn A, et al. Shared fine specificity between T-cell receptors and an antibody recognizing a peptide/major histocompatibility class I complex. Proc Natl Acad Sci U S A. 1996;93:10338–10342. doi: 10.1073/pnas.93.19.10338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Villa A, et al. A high-affinity human monoclonal antibody specific to the alternatively spliced EDA domain of fibronectin efficiently targets tumor neo-vasculature in vivo. Int J Cancer. 2008;122:2405–2413. doi: 10.1002/ijc.23408. [DOI] [PubMed] [Google Scholar]
- 93.Pini A, et al. Design and use of a phage display library. Human antibodies with subnanomolar affinity against a marker of angiogenesis eluted from a two-dimensional gel. J Biol Chem. 1998;273:21769–21776. doi: 10.1074/jbc.273.34.21769. [DOI] [PubMed] [Google Scholar]
- 94.Schliemann C, Neri D. Antibody-based vascular tumor targeting. Recent Results Cancer Res. 2010;180:201–216. doi: 10.1007/978-3-540-78281-0_12. [DOI] [PubMed] [Google Scholar]
- 95.Huie MA, et al. Antibodies to human fetal erythroid cells from a nonimmune phage antibody library. Proc Natl Acad Sci USA. 2001;98:2682–2687. doi: 10.1073/pnas.051631798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Nizak C, et al. Recombinant antibodies to the small GTPase Rab6 as conformation sensors. Science. 2003;300:984–987. doi: 10.1126/science.1083911300/5621/984[pii]. [DOI] [PubMed] [Google Scholar]
- 97.Gao J, Sidhu SS, Wells JA. Two-state selection of conformation-specific antibodies. Proc Natl Acad Sci U S A. 2009;106:3071–3076. doi: 10.1073/pnas.0812952106. 0812952106 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Rothlisberger D, Pos KM, Pluckthun A. An antibody library for stabilizing and crystallizing membrane proteins - selecting binders to the citrate carrier CitS. FEBS Lett. 2004;564:340–348. doi: 10.1016/S0014-5793(04)00359-X. [DOI] [PubMed] [Google Scholar]
- 99.Uysal S, et al. Crystal structure of full-length KcsA in its closed conformation. Proc Natl Acad Sci U S A. 2009;106:6644–6649. doi: 10.1073/pnas.0810663106. 0810663106 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ye JD, et al. Synthetic antibodies for specific recognition and crystallization of structured RNA. Proc Natl Acad Sci U S A. 2008;105:82–87. doi: 10.1073/pnas.0709082105. 0709082105 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Eisenhardt SU, Schwarz M, Bassler N, Peter K. Subtractive single-chain antibody (scFv) phage-display: tailoring phage-display for high specificity against function-specific conformations of cell membrane molecules. Nat Protoc. 2007;2:3063–3073. doi: 10.1038/nprot.2007.455. nprot.2007.455 [pii] [DOI] [PubMed] [Google Scholar]
- 102.Edwards BM, et al. The remarkable flexibility of the human antibody repertoire; isolation of over one thousand different antibodies to a single protein, BLyS. J Mol Biol. 2003;334:103–118. doi: 10.1016/j.jmb.2003.09.054. [DOI] [PubMed] [Google Scholar]
- 103.Baker KP, et al. Generation and characterization of LymphoStat-B, a human monoclonal antibody that antagonizes the bioactivities of B lymphocyte stimulator. Arthritis Rheum. 2003;48:3253–3265. doi: 10.1002/art.11299. [DOI] [PubMed] [Google Scholar]
- 104.Osbourn J, Groves M, Vaughan T. From rodent reagents to human therapeutics using antibody guided selection. Methods. 2005;36:61–68. doi: 10.1016/j.ymeth.2005.01.006. [DOI] [PubMed] [Google Scholar]
- 105.Jespers LS, Roberts A, Mahler SM, Winter G, Hoogenboom HR. Guiding the selection of human antibodies from phage display repertoires to a single epitope of an antigen. Biotechnology (N Y) 1994;12:899–903. doi: 10.1038/nbt0994-899. [DOI] [PubMed] [Google Scholar]
- 106.Runnels HA, et al. Human monoclonal antibodies to the insulin-like growth factor 1 receptor inhibit receptor activation and tumor growth in preclinical studies. Adv Ther. 2010;27:458–475. doi: 10.1007/s12325-010-0026-5. [DOI] [PubMed] [Google Scholar]
- 107.Li K, et al. Modulation of Notch signaling by antibodies specific for the extracellular negative regulatory region of NOTCH3. J Biol Chem. 2008;283:8046–8054. doi: 10.1074/jbc.M800170200. M800170200 [pii] [DOI] [PubMed] [Google Scholar]
- 108.Wu Y, et al. Therapeutic antibody targeting of individual Notch receptors. Nature. 2010;464:1052–1057. doi: 10.1038/nature08878. nature08878 [pii] [DOI] [PubMed] [Google Scholar]
- 109.Martens T, et al. A novel one-armed anti-c-Met antibody inhibits glioblastoma growth in vivo. Clin Cancer Res. 2006;12:6144–6152. doi: 10.1158/1078-0432.CCR-05-1418. 12/20/6144 [pii] [DOI] [PubMed] [Google Scholar]
- 110.Xie MH, Yuan J, Adams C, Gurney A. Direct demonstration of MuSK involvement in acetylcholine receptor clustering through identification of agonist ScFv. Nat Biotechnol. 1997;15:768–771. doi: 10.1038/nbt0897-768. [DOI] [PubMed] [Google Scholar]
- 111.Ellmark P, Andersson H, Abayneh S, Fenyo EM, Borrebaeck CA. Identification of a strongly activating human anti-CD40 antibody that suppresses HIV type 1 infection. AIDS Res Hum Retroviruses. 2008;24:367–373. doi: 10.1089/aid.2007.0215. [DOI] [PubMed] [Google Scholar]
- 112.Sui J, et al. Structural and functional bases for broad-spectrum neutralization of avian and human influenza A viruses. Nat Struct Mol Biol. 2009;16:265–273. doi: 10.1038/nsmb.1566. nsmb.1566 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Sun L, et al. Generation, characterization and epitope mapping of two neutralizing and protective human recombinant antibodies against influenza A H5N1 viruses. PLoS ONE. 2009;4:e5476. doi: 10.1371/journal.pone.0005476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Zhou Y, Zou H, Zhang S, Marks JD. Internalizing cancer antibodies from phage libraries selected on tumor cells and yeast-displayed tumor antigens. J Mol Biol. 2010;404:88–99. doi: 10.1016/j.jmb.2010.09.006. S0022–2836(10)00965–4 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Roth A, et al. Anti-CD166 single chain antibody-mediated intracellular delivery of liposomal drugs to prostate cancer cells. Mol Cancer Ther. 2007;6:2737–2746. doi: 10.1158/1535-7163.MCT-07-0140. 6/10/2737 [pii] [DOI] [PubMed] [Google Scholar]
- 116.Liu B, et al. Recombinant full-length human IgG1s targeting hormone-refractory prostate cancer. J Mol Med. 2007;85:1113–1123. doi: 10.1007/s00109-007-0208-z. [DOI] [PubMed] [Google Scholar]
- 117.Poul MA, Becerril B, Nielsen UB, Morisson P, Marks JD. Selection of tumor-specific internalizing human antibodies from phage libraries. J Mol Biol. 2000;301:1149–1161. doi: 10.1006/jmbi.2000.4026. S0022–2836(00)94026–9 [pii] [DOI] [PubMed] [Google Scholar]
- 118.Heitner T, et al. Selection of cell binding and internalizing epidermal growth factor receptor antibodies from a phage display library. J Immunol Methods. 2001;248:17–30. doi: 10.1016/s0022-1759(00)00340-9. [DOI] [PubMed] [Google Scholar]
- 119.Dobson CL, et al. Human monomeric antibody fragments to TRAIL-R1 and TRAIL-R2 that display potent in vitro agonism. MAbs. 2009;1:552–562. doi: 10.4161/mabs.1.6.10057. 10057 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Peipp M, Dechant M, Valerius T. Effector mechanisms of therapeutic antibodies against ErbB receptors. Curr Opin Immunol. 2008;20:436–443. doi: 10.1016/j.coi.2008.05.012. S0952–7915(08)00089–7 [pii] [DOI] [PubMed] [Google Scholar]
- 121.Park JW, et al. Tumor targeting using anti-her2 immunoliposomes. J Control Release. 2001;74:95–113. doi: 10.1016/s0168-3659(01)00315-7. S0168365901003157 [pii] [DOI] [PubMed] [Google Scholar]
- 122.Nielsen UB, et al. Therapeutic efficacy of anti-ErbB2 immunoliposomes targeted by a phage antibody selected for cellular endocytosis. Biochim Biophys Acta. 2002;1591:109–118. doi: 10.1016/s0167-4889(02)00256-2. S0167488902002562 [pii] [DOI] [PubMed] [Google Scholar]
- 123.Wu H, et al. Stepwise in vitro affinity maturation of Vitaxin, an alphav beta3-specific humanized mAb. Proc Natl Acad Sci U S A. 1998;95:6037–6042. doi: 10.1073/pnas.95.11.6037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Lippow SM, Wittrup KD, Tidor B. Computational design of antibody-affinity improvement beyond in vivo maturation. Nat Biotechnol. 2007;25:1171–1176. doi: 10.1038/nbt1336. nbt1336 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Fagete S, et al. Specificity tuning of antibody fragments to neutralize two human chemokines with a single agent. MAbs. 2009;1:288–296. doi: 10.4161/mabs.1.3.8527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Bostrom J, et al. Variants of the antibody herceptin that interact with HER2 and VEGF at the antigen binding site. Science. 2009;323:1610–1614. doi: 10.1126/science.1165480. 323/5921/1610 [pii] [DOI] [PubMed] [Google Scholar]
- 127.Kalb SR, et al. Extraction of BoNT/A, /B, /E, and /F with a single, high affinity monoclonal antibody for detection of botulinum neurotoxin by Endopep-MS. PLoS ONE. 2010;5:e12237. doi: 10.1371/journal.pone.0012237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Fellouse FA, Wiesmann C, Sidhu SS. Synthetic antibodies from a four-amino-acid code: a dominant role for tyrosine in antigen recognition. Proc Natl Acad Sci U S A. 2004;101:12467–12472. doi: 10.1073/pnas.0401786101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Liang WC, et al. Cross-species vascular endothelial growth factor (VEGF)-blocking antibodies completely inhibit the growth of human tumor xenografts and measure the contribution of stromal VEGF. J Biol Chem. 2006;281:951–961. doi: 10.1074/jbc.M508199200. M508199200 [pii] [DOI] [PubMed] [Google Scholar]
- 130.Lee CV, et al. Synthetic anti-BR3 antibodies that mimic BAFF binding and target both human and murine B cells. Blood. 2006;108:3103–3111. doi: 10.1182/blood-2006-03-011031. blood-2006–03–011031 [pii] [DOI] [PubMed] [Google Scholar]
- 131.Volk WA, Bizzini B, Snyder RM, Bernhard E, Wagner RR. Neutralization of tetanus toxin by distinct monoclonal antibodies binding to multiple epitopes on the toxin molecule. Infect Immun. 1984;45:604–609. doi: 10.1128/iai.45.3.604-609.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Marks JD. Deciphering antibody properties that lead to potent botulinum neurotoxin neutralization. Mov Disord. 2004;19 (Suppl 8):S101–108. doi: 10.1002/mds.20023. [DOI] [PubMed] [Google Scholar]
- 133.Nowakowski A, et al. Potent neutralization of botulinum neurotoxin by recombinant oligoclonal antibody. Proc Natl Acad Sci U S A. 2002;99:11346–11350. doi: 10.1073/pnas.172229899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.de Kruif J, Logtenberg T. Leucine zipper dimerized bivalent and bispecific scFv antibodies from a semi-synthetic antibody phage display library. J Biol Chem. 1996;271:7630–7634. doi: 10.1074/jbc.271.13.7630. [DOI] [PubMed] [Google Scholar]
- 135.Hudson PJ, Kortt AA. High avidity scFv multimers; diabodies and triabodies. J Immunol Methods. 1999;231:177–189. doi: 10.1016/s0022-1759(99)00157-x. [DOI] [PubMed] [Google Scholar]
- 136.Dubel S, et al. Bifunctional and multimeric complexes of streptavidin fused to single chain antibodies (scFv) J Immunol Methods. 1995;178:201–209. doi: 10.1016/0022-1759(94)00257-w. 002217599400257W [pii] [DOI] [PubMed] [Google Scholar]
- 137.Griep RA, et al. pSKAP/S: An expression vector for the production of single-chain Fv alkaline phosphatase fusion proteins. Protein Expr Purif. 1999;16:63–69. doi: 10.1006/prep.1999.1041. [DOI] [PubMed] [Google Scholar]
- 138.Cloutier SM, et al. Streptabody, a high avidity molecule made by tetramerization of in vivo biotinylated, phage display-selected scFv fragments on streptavidin. Mol Immunol. 2000;37:1067–1077. doi: 10.1016/s0161-5890(01)00023-2. S0161589001000232 [pii] [DOI] [PubMed] [Google Scholar]
- 139.Casey JL, Coley AM, Tilley LM, Foley M. Green fluorescent antibodies: novel in vitro tools. Protein Eng. 2000;13:445–452. doi: 10.1093/protein/13.6.445. [DOI] [PubMed] [Google Scholar]
- 140.Thie H, Binius S, Schirrmann T, Hust M, Dubel S. Multimerization domains for antibody phage display and antibody production. N Biotechnol. 2009 doi: 10.1016/j.nbt.2009.07.005. S1871–6784(09)01102–9 [pii] [DOI] [PubMed] [Google Scholar]
- 141.Persic L, et al. An integrated vector system for the eukaryotic expression of antibodies or their fragments after selection from phage display libraries. Gene. 1997;187:9–18. doi: 10.1016/s0378-1119(96)00628-2. S0378–1119(96)00628–2 [pii] [DOI] [PubMed] [Google Scholar]
- 142.Hu S, et al. Minibody: A novel engineered anti-carcinoembryonic antigen antibody fragment (single-chain Fv-CH3) which exhibits rapid, high-level targeting of xenografts. Cancer Res. 1996;56:3055–3061. [PubMed] [Google Scholar]
- 143.Beck A, et al. Trends in glycosylation, glycoanalysis and glycoengineering of therapeutic antibodies and Fc-fusion proteins. Curr Pharm Biotechnol. 2008;9:482–501. doi: 10.2174/138920108786786411. [DOI] [PubMed] [Google Scholar]
- 144.Presta LG. Molecular engineering and design of therapeutic antibodies. Curr Opin Immunol. 2008;20:460–470. doi: 10.1016/j.coi.2008.06.012. S0952–7915(08)00117–9 [pii] [DOI] [PubMed] [Google Scholar]
- 145.Merchant AM, et al. An efficient route to human bispecific IgG. Nat Biotechnol. 1998;16:677–681. doi: 10.1038/nbt0798-677. [DOI] [PubMed] [Google Scholar]
- 146.Ridgway JB, Presta LG, Carter P. ‘Knobs-into-holes’ engineering of antibody CH3 domains for heavy chain heterodimerization. Protein Eng. 1996;9:617–621. doi: 10.1093/protein/9.7.617. [DOI] [PubMed] [Google Scholar]
- 147.Perisic O, Webb PA, Holliger P, Winter G, Williams RL. Crystal structure of a diabody, a bivalent antibody fragment. Structure. 1994;2:1217–1226. doi: 10.1016/s0969-2126(94)00123-5. [DOI] [PubMed] [Google Scholar]
- 148.Atwell JL, et al. scFv multimers of the anti-neuraminidase antibody NC10: length of the linker between VH and VL domains dictates precisely the transition between diabodies and triabodies. Protein Eng. 1999;12:597–604. doi: 10.1093/protein/12.7.597. [DOI] [PubMed] [Google Scholar]
- 149.Pei XY, Holliger P, Murzin AG, Williams RL. The 2.0-A resolution crystal structure of a trimeric antibody fragment with noncognate VH-VL domain pairs shows a rearrangement of VH CDR3. Proc Natl Acad Sci U S A. 1997;94:9637–9642. doi: 10.1073/pnas.94.18.9637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Le Gall F, Kipriyanov SM, Moldenhauer G, Little M. Di-, tri- and tetrameric single chain Fv antibody fragments against human CD19: effect of valency on cell binding. FEBS Lett. 1999;453:164–168. doi: 10.1016/s0014-5793(99)00713-9. S0014–5793(99)00713–9 [pii] [DOI] [PubMed] [Google Scholar]
- 151.Muller D, Kontermann RE. Bispecific antibodies for cancer immunotherapy: Current perspectives. BioDrugs. 2010;24:89–98. doi: 10.2165/11530960-000000000-00000. 2 [pii] [DOI] [PubMed] [Google Scholar]
- 152.Xiang J. Targeting cytokines to tumors to induce active antitumor immune responses by recombinant fusion proteins. Hum Antibodies. 1999;9:23–36. [PubMed] [Google Scholar]
- 153.Schliemann C, Neri D. Antibody-based targeting of the tumor vasculature. Biochim Biophys Acta. 2007;1776:175–192. doi: 10.1016/j.bbcan.2007.08.002. S0304–419X(07)00028–5 [pii] [DOI] [PubMed] [Google Scholar]
- 154.Deckert PM. Current constructs and targets in clinical development for antibody-based cancer therapy. Curr Drug Targets. 2009;10:158–175. doi: 10.2174/138945009787354502. [DOI] [PubMed] [Google Scholar]
- 155.Fuchs H, Bachran C. Targeted tumor therapies at a glance. Curr Drug Targets. 2009;10:89–93. doi: 10.2174/138945009787354557. [DOI] [PubMed] [Google Scholar]
- 156.Gawlitta W, Osborn M, Weber K. Coiling of intermediate filaments induced by microinjection of a vimentin-specific antibody does not interfere with locomotion and mitosis. Eur J Cell Biol. 1981;26:83–90. [PubMed] [Google Scholar]
- 157.Kontermann RE. Intrabodies as therapeutic agents. Methods. 2004;34:163–170. doi: 10.1016/j.ymeth.2004.04.002. S1046202304000702 [pii] [DOI] [PubMed] [Google Scholar]
- 158.Beerli RR, Wels W, Hynes NE. Intracellular expression of single chain antibodies reverts ErbB-2 transformation. J Biol Chem. 1994;269:23931–23936. [PubMed] [Google Scholar]
- 159.Richardson JH, Sodroski JG, Waldmann TA, Marasco WA. Phenotypic knockout of the high-affinity human interleukin 2 receptor by intracellular single-chain antibodies against the alpha subunit of the receptor. Proc Natl Acad Sci USA. 1995;92:3137–3141. doi: 10.1073/pnas.92.8.3137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Paganetti P, Calanca V, Galli C, Stefani M, Molinari M. beta-site specific intrabodies to decrease and prevent generation of Alzheimer’s Abeta peptide. J Cell Biol. 2005;168:863–868. doi: 10.1083/jcb.200410047. jcb.200410047 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Strebe N, et al. Functional knockdown of VCAM-1 at the posttranslational level with ER retained antibodies. J Immunol Methods. 2009;341:30–40. doi: 10.1016/j.jim.2008.10.012. S0022–1759(08)00323–2 [pii] [DOI] [PubMed] [Google Scholar]
- 162.Biocca S, Cattaneo A. Intracellular immunization: antibody targeting to subcellular compartments. Trends in Cell Biology. 1995;5:248–252. doi: 10.1016/s0962-8924(00)89019-4. [DOI] [PubMed] [Google Scholar]
- 163.Biocca S, Pierandrei-Amaldi P, Campioni N, Cattaneo A. Intracellular immunisation with cytosolic recombinant antibodies. BioTech. 1994;12:396–400. doi: 10.1038/nbt0494-396. [DOI] [PubMed] [Google Scholar]
- 164.Desiderio A, et al. A semi-synthetic repertoire of intrinsically stable antibody fragments derived from a single-framework scaffold. J Mol Biol. 2001;310:603–615. doi: 10.1006/jmbi.2001.4756. [DOI] [PubMed] [Google Scholar]
- 165.der Maur AA, et al. Direct in vivo screening of intrabody libraries constructed on a highly stable single-chain framework. J Biol Chem. 2002;277:45075–45085. doi: 10.1074/jbc.M205264200. [DOI] [PubMed] [Google Scholar]
- 166.Tanaka T, Chung GT, Forster A, Lobato MN, Rabbitts TH. De novo production of diverse intracellular antibody libraries. Nucleic Acids Res. 2003;31:e23. doi: 10.1093/nar/gng023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Auf der Maur A, Escher D, Barberis A. Antigen-independent selection of stable intracellular single-chain antibodies. FEBS Lett. 2001;508:407–412. doi: 10.1016/s0014-5793(01)03101-5. [DOI] [PubMed] [Google Scholar]
- 168.Visintin M, Tse E, Axelson H, Rabbitts TH, Cattaneo A. Selection of antibodies for intracellular function using a two-hybrid in vivo system. Proc Natl Acad Sci USA. 1999;96:11723–11728. doi: 10.1073/pnas.96.21.11723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Amstutz P, et al. Intracellular kinase inhibitors selected from combinatorial libraries of designed ankyrin repeat proteins. J Biol Chem. 2005;280:24715–24722. doi: 10.1074/jbc.M501746200. [DOI] [PubMed] [Google Scholar]
- 170.Kohl A, et al. Allosteric inhibition of aminoglycoside phosphotransferase by a designed ankyrin repeat protein. Structure (Camb) 2005;13:1131–1141. doi: 10.1016/j.str.2005.04.020. [DOI] [PubMed] [Google Scholar]
- 171.Rizk SS, et al. An engineered substance P variant for receptor-mediated delivery of synthetic antibodies into tumor cells. Proc Natl Acad Sci U S A. 2009;106:11011–11015. doi: 10.1073/pnas.0904907106. 0904907106 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Hallborn J, Carlsson R. Automated screening procedure for high-throughput generation of antibody fragments. Biotechniques. 2002;(Suppl):30–37. [PubMed] [Google Scholar]
- 173.Turunen L, Takkinen K, Soderlund H, Pulli T. Automated panning and screening procedure on microplates for antibody generation from phage display libraries. J Biomol Screen. 2009;14:282–293. doi: 10.1177/1087057108330113. 1087057108330113 [pii] [DOI] [PubMed] [Google Scholar]
- 174.Lou J, et al. Antibodies in haystacks: how selection strategy influences the outcome of selection from molecular diversity libraries. J Immunol Methods. 2001;253:233–242. doi: 10.1016/s0022-1759(01)00385-4. [DOI] [PubMed] [Google Scholar]
- 175.Storz U. In: Antibody Engineering. Kontermann R, Dubel S, editors. Springer-Verlag; 2010. pp. 517–581. [Google Scholar]
- 176.Kreitman RJ, et al. Phase I trial of recombinant immunotoxin anti-Tac(Fv)-PE38 (LMB-2) in patients with hematologic malignancies. J Clin Oncol. 2000;18:1622–1636. doi: 10.1200/JCO.2000.18.8.1622. [DOI] [PubMed] [Google Scholar]
- 177.Fellouse FA, et al. High-throughput generation of synthetic antibodies from highly functional minimalist phage-displayed libraries. J Mol Biol. 2007;373:924–940. doi: 10.1016/j.jmb.2007.08.005. S0022–2836(07)01062–5 [pii] [DOI] [PubMed] [Google Scholar]
- 178.Hamilton SR, Gerngross TU. Glycosylation engineering in yeast: the advent of fully humanized yeast. Curr Opin Biotechnol. 2007;18:387–392. doi: 10.1016/j.copbio.2007.09.001. S0958–1669(07)00113–9 [pii] [DOI] [PubMed] [Google Scholar]