Abstract
2-C-methyl-d-erythritol 4-phosphate is the first committed intermediate in the biosynthesis of the isoprenoid precursors isopentenyl diphosphate and dimethylallyl diphosphate. Supplementation of the growth medium with 2-C-methyl-d-erythritol has been shown to complement disruptions in the Escherichia coli gene for 1-deoxy-d-xylulose 5-phosphate synthase, the enzyme that synthesizes the immediate precursor of 2-C-methyl-d-erythritol 4-phosphate. In order to be utilized in isoprenoid biosynthesis, 2-C-methyl-d-erythritol must be phosphorylated. We describe the construction of Salmonella enterica serovar Typhimurium strain RMC26, in which the essential gene encoding 1-deoxy-d-xylulose 5-phosphate synthase has been disrupted by insertion of a synthetic mevalonate operon consisting of the yeast ERG8, ERG12, and ERG19 genes, responsible for converting mevalonate to isopentenyl diphosphate under the control of an arabinose-inducible promoter. Random mutagenesis of RMC26 produced defects in the sorbitol phosphotransferase system that prevented the transport of 2-C-methyl-d-erythritol into the cell. RMC26 and mutant strains of RMC26 unable to grow on 2-C-methyl-d-erythritol were incubated in buffer containing mevalonate and deuterium-labeled 2-C-methyl-d-erythritol. Ubiquinone-8 was isolated from these cells and analyzed for deuterium content. Efficient incorporation of deuterium was observed for RMC26. However, there was no evidence of deuterium incorporation into the isoprenoid side chain of ubiquinone Q8 in the RMC26 mutants.
With more than 33,000 different compounds known to date, isoprenoids are among the most diverse groups of compounds found in nature (9). Isoprenoid molecules are important for a wide variety of cellular functions, including electron transport (quinones), stabilization of membranes (sterols), cell wall biosynthesis (dolichols), signaling (prenylated proteins and hormones), photosynthesis (chlorophylls), photoprotection (carotenoids), and protein synthesis (modified tRNAs). Despite a high degree of structural diversity, all isoprenoids are derived from two simple five-carbon molecules, isopentenyl diphosphate (IPP) and dimethylallyl diphosphate (DMAPP). Although all higher isoprenoid molecules are derived from IPP and DMAPP, these fundamental building blocks are produced by two different biosynthetic pathways.
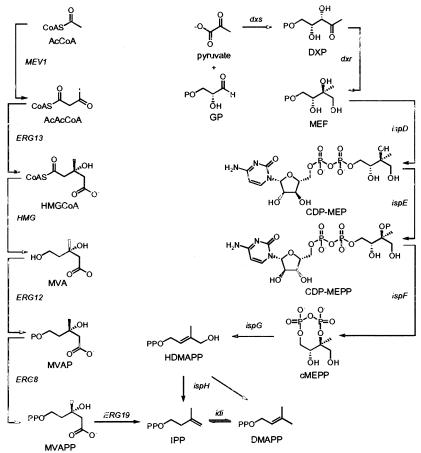
In Archaea, Eukarya, and some gram-positive bacteria, IPP and DMAPP are synthesized from acetyl coenzyme A by the mevalonate (MVA) pathway (Fig. 1). The first two steps of the MVA pathway condense three molecules of acetyl coenzyme A to form 3-hydroxy-3-methylglutaryl-coenzyme A (HMG-CoA) (24, 33). HMG-CoA is then reduced to MVA, which is converted to MVA diphosphate in two steps (5, 8). IPP obtained by an ATP-dependent decarboxylation of MVA diphosphate is then isomerized to DMAPP (1, 15). The 2-C-methyl-d-erythritol 4-phosphate (MEP) pathway is orthogonal to the MVA pathway (Fig. 1) and is found in most bacteria and plant chloroplasts. The first step in the MEP pathway is the condensation of d-glyceraldehyde 3-phosphate and pyruvate to give 1-deoxy-d-xylulose 5-phosphate (DXP) (30). MEP synthase then catalyzes an NADPH-dependent rearrangement and reduction of DXP to give MEP (27). DXP is also a precursor for biosynthesis of vitamins B1 and B6. Thus, MEP is the first committed precursor for biosynthesis of IPP and DMAPP. MEP is converted to 2-C-methyl-d-erythritol 2,4-cyclodiphosphate (cMEPP) by the consecutive actions of 4-diphosphocytidyl-2-C-methyl-d-erythritol synthase, 4-diphosphocytidyl-2-C-methyl-d-erythritol 2-phosphate synthase, and cMEPP synthase (22, 28, 29, 31, 39). A reductive ring-opening of cMEPP produces 1-hydroxy-2-methyl-2-buten-4-yl diphosphate (4, 11, 21). In the final step, the protein encoded by ispH (formerly lytB) converts 1-hydroxy-2-methyl-2-buten-4-yl diphosphate to a mixture of IPP and DMAPP (2, 3, 13, 32, 38). The segregation of the MVA and MEP pathways makes the latter an attractive target for development of antibiotics and herbicides.
FIG. 1.
MVA (left) and MEP (right) pathways.
Disruptions of the genes encoding each of the proteins in the MEP pathway are lethal (3, 4, 10, 11, 13, 20, 32). Disruptions in dxs can be complemented by supplementation of the growth medium with deoxyxylulose (DX) or 2-C-methyl-d-erythritol (ME), while disruptions in dxr can be complemented with ME (6, 16, 19, 37, 40, 43). In order to be utilized in the MEP pathway, exogenous DX and ME must be transported into the cell and phosphorylated. DX is converted to DXP by d-xylulokinase, the enzyme that normally phosphorylates d-xylulose (46), although at this point the transport mechanism for uptake of DX is not known. Likewise, the mechanism for uptake of ME had not been established.
Work from our laboratory (20) and by Boronat and colleagues (10) demonstrated the ability of a plasmid-encoded MVA operon to restore cell viability to strains of Escherichia coli that had been disrupted in dxs. The bacterium was blocked in de novo biosynthesis of IPP and DMAPP by the MEP pathway but was able to synthesize both molecules from MVA. To identify genes involved in the MEP pathway, the chromosomal copy of dxs in Salmonella enterica serovar Typhimurium was disrupted with our synthetic MVA operon (20). For the chromosomal disruption, the araBAD promoter from pBAD was inserted upstream of the MVA pathway genes with a kanamycin resistance (Kanr) cassette appended downstream to allow easy detection of insertion mutants. To facilitate a correct insertion, the MVA operon was flanked by regions of homology to dxs. Boronat and colleagues (10) have described a related construct in which the synthetic MVA operon was inserted into the tryptophan biosynthetic operon in E. coli and chromosomal dxs was disrupted with a chloramphenicol acetyltransferase (CAT) cassette. We now report work that implicates the sorbitol phosphotransferase system (PTS) in the transport and phosphorylation of ME (for a review of PTS, see reference 45).
MATERIALS AND METHODS
Materials.
Restriction endonucleases, Klenow fragment, and T7 DNA polymerase were purchased from New England Biolabs. Ampicillin, kanamycin (KAN), and chloramphenicol (CAM) were purchased from Sigma. Shrimp alkaline phosphatase was purchased from U.S. Biochemical Corp. Ampligase was purchased from Epicentre Technologies Corporation. KlenTaq-LA DNA polymerase mix was purchased from Clontech. Taq/TaqStart DNA polymerase, E. coli DH5α competent cells, and agarose were purchased from GIBCO. Oligonucleotides used in this study are listed in Table 1 and were custom synthesized by the Protein/DNA Core Facility of the Utah Regional Cancer Center.
TABLE 1.
Oligonucleotide sequences used in this study
| Oligonucleotide | Sequencea |
|---|---|
| crcDXSARA | 5′ CAATGAATCACGCAGGCGATCTCGCCGCAGCCGAACGACC 3′ |
| crcKANDXS | 5′ TGCAGGTCGAGGGGGGGCCTGAGGGGCGAAAGTCGTAAA 3′ |
| sISPA1 | 5′ CGTTAGACTTGGGCGTTGAG 3′ |
| asARAC1 | 5′ CTTTTGAGCACCACCCGGAT 3′ |
| asYAJO1 | 5′ AGTTAATGCCGCCCTCAAGG 3′ |
| sKAN1 | 5′ GGCAGAGCATTACGCTGACT 3′ |
| SygbPFRT2 | 5′ GGGCAAACTCATCGATAAACCATCCAGGGATTTAACATGGCAGCCACTTTATTGGACGTTTGCGCCGTGGTGCCGGCGAAGCCACTGGAGCACCTCAA 3′ |
| ASygbPFRT2 | 5′ CCAAACGCGTGTACGTCAAAACCGTGTCCAATTCGCATTATGCTTTCTCCTGGTGGATGGTTCGGGTCAGATAAACGGGGAGAGCCTGAGCAAA 3′ |
| sychBcamPCR | 5′ ATGATGACCCATTGGCCTTCTCCGGCAAAATTAAATCTGTTTTTATATATCACCGGACGAAGCCACTGGAGCACCTCAA 3′ |
| asychBcamPCR | 5′ TTAGAGTAACTCTCGATGCAATGGGGAGAGGTTGACACCCTTCGCCACAAAAGCATTGAGCGGGGAGAGCCTGAGCAAA 3′ |
| sychBCamseq2 | 5′ GGTGGCTATGACAGCAAAA 3′ |
| asychBCamseq2 | 5′ GGATCCTTACTCGTCTTCATTCAC 3′ |
| sygbBcamPCR | 5′ ATGCGAATTGGACACGGTTTTGACGTACACGCGTTTGGCGGAGAAGGGCCGATTATTATTAAGCCACTGGAGCACCTCAA 3′ |
| asygbBcamPCR | 5′ TCATTTCGCCGCCTTCATCAGCAACGCGACCGCTTCGCAGGCAATGCCTTCTCCGCGTCCCGGGGAGAGCCTGAGCAAA 3′ |
| sygbBCamseq2 | 5′ TAAATCTTCAACCACCACAAAATCTTCC 3′ |
| asygbBCamseq2 | 5′ GGATCCAATTTTATCTGACCCGAA 3′ |
| sgcpEcam2 | 5′ TTATTTTTCAACCTGCAGCACGTCAATCCGACGCGCTTCAAGCCACTGGAGCACCTCAA 3′ |
| asgcpEcam2 | 5′ ATGCATAACCAGGCTCCGATTCAACGTAGAAAATCGACACGGGGAGAGCCTGAGCAAA 3′ |
| sgcpEseq | 5′ CCAGGCAGATAATCGTTCATG 3′ |
| sgcpEseq | 5′ GTATCAAGGAAAACCTGTCG 3′ |
| smud33 | 5′ ACCTTTGGTCACCAACGCTTTTCC 3′ |
| smud54 | 5′ CCGAATAATCCAATGTCC 3′ |
| smud59 | 5′ CAGATCCCGAATAATCCAATGTCCT 3′ |
| smud86 | 5′ GCAAGCCCCACCAAATCTAATCCCA 3′ |
| sgutEFRT2 | 5′ ACTTGAGCAGAAAGTTCATCTGACAGGAGCGGTATCATGACACGGGTTCGCATTGAGAAAGGCGCCGGCGGCTGGGGCAAGCCACTGGAGCACCTCAA 3′ |
| asgutEFRT2 | 5′ ATGGTGGTTTGATAAATCACGCTCATGTCTGAACCTCTTATTGATAGATAAAGCCGGAAACAAACCAGGCGATAAGTACGGGGAGAGCCTGAGCAAA 3′ |
| SgutEseq | 5′ CATATGTCACCGATCTCACCACCGCT 3′ |
| ASgutEseq | 5′ GGATCCGCTCATGTCTGAACCTCTTA 3′ |
| Ssr1E | 5′ CCATGGCACGGGTTCGCATTGAGAAAGGC 3′ |
| STYasgutEPCR | 5′ CGGATTCTTAATGATGATGATGATGATGTTGATAGATAAAGCCGGAAA CAAACC 3′ |
| SsrlACam | 5′ AGAAAACTAAAAACCCTACAGGAGAGAACAATGATCGAAAAAGCCACTGGAGCACCTCAA 3′ |
| ASsrlACam | 5′ CAATGCGAACCCGTGTCATGATACCGCTCCTGTCAGATGACGGGGAGAGCCTGAGCAAA 3′ |
| SsrlBCam | 5′ GGCTTTATCTATCAATAAGAGGTTCAGACATGAGCGTGATAAGCCACTGGAGCACCTCAA 3′ |
| ASsrlBCam | 5′ CAACGGCAACTTGATTCATGTTTTTTCTCCTTAAGCGACACGGGGAGAGCCTGAGCAAA 3′ |
| SsrlAX | 5′ TTGCGATCACAATAACACT 3′ |
| ASsrlAX | 5′ TGTTAGTTGCGCCAGTTT 3′ |
| SsrlBX | 5′ CAGATGCTGATTACGTTTCG 3′ |
| ASsrlBX | 5′ GATTAAAGCGGGAAATCTGC 3′ |
Italics indicate homology to CAT.
Growth media and supplements.
Luria-Bertani (LB) was used with or without supplementation for all growth conditions (41). KAN was used at a concentration of 40 μg/ml, CAM was used at a concentration of 20 μg/ml, and ampicillin was used at a concentration of 100 μg/ml. ME and [1,1-2H2]2-C-methyl-d-erythritol ([1,1-2H2]ME) were synthesized as described by Duvold et al. (16) and used at a concentration of 50 μg/ml. MVA was prepared by hydrolysis of 1 volume of 1 M MVA lactone with 1.02 volumes of 1 M KOH followed by incubation of the mixture at 37°C for 30 min. DX was prepared as previously described (7) and used at a concentration of 2 mM. l-arabinose (Ara) was used at a concentration of 0.02%.
General methods.
Minipreparations of plasmid DNA for restriction analysis were made using QIAprep Spin Miniprep kits (Qiagen). Large-scale plasmid preparations were made using Qiagen Plasmid Midi Prep kits. PCR products were purified by QIAquick PCR purification kits (Qiagen) or on agarose gels (GIBCO) with either soaking of the band overnight in TE or purification using QIAquick gel extraction kits (Qiagen). Genomic DNA was isolated using Easy DNA kits (Invitrogen). Restriction digests, ligations, transformations, and electroporations were performed as described by Sambrook et al. (41). PCRs were carried out using a Perkin-Elmer GeneAmp PCR system 2400 DNA thermal cycler and KlenTaq LA polymerase mix (Clontech), unless otherwise noted. Chain reaction cloning (CRC) was performed as described by Pachuk et al. (34). E. coli DH5α (GIBCO) was used as a host for plasmid constructs. S. enterica serovar Typhimurium strain TR6579 was used to shuttle plasmids constructed in E. coli to S. enterica serovar Typhimurium. Transductions were mediated by the high-frequency P22 mutant HT105/1 int-201 as previously described (42). Phage P22 lysates were prepared as previously described (14). Random mutagenesis was performed as described by Hughes and Roth (23) using TE3461 (17) for Mud-Cam insertions by selecting on LB-CAM-MVA-Ara or LB-CAM. λ-Red-mediated recombination was performed as described by Price-Carter et al. (36). DNA was sequenced at the Health Sciences Center Sequencing Facility, Eccles Institute of Human Genetics, University of Utah. Sequence searches were performed at the National Center for Biotechnology Information and the WU GSC BLAST server, Genome Sequencing Center, Washington University School of Medicine.
Plasmid and strain construction.
Plasmids and bacterial strains used in this study are listed in Table 2. All parental S. enterica serovar Typhimurium strains and phages used were gifts of J. Roth (University of California—Davis).
TABLE 2.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Description | Reference or source |
|---|---|---|
| E. coli | ||
| DH5α | Host for cloning vectors | GIBCO |
| S. typhimurium | ||
| TR6579 | Host for cloning vectors | J. R. Roth |
| TT22236 | LT2/pTP223 | J. R. Roth |
| RMC25 | LT2 dxs::MVA operon | This work |
| RMC26 | LT2 dxs::MVA operon | This work |
| RMC28 | RMC26/pTP223 | This work |
| CR4 | RMC26 ispD::Cam | This work |
| TE3461 | Mud-cam source | J. R. Roth |
| CR5 | RMC26 srlE::Cam | This work |
| CR24 | LT2 srlE::Cam | This work |
| CR29 | CR5 pBAD/srlE | This work |
| RMC78 | RMC26 srlA::Cam | This work |
| RMC79 | RMC26 srlB::Cam | This work |
| Plasmids | ||
| pBAD/gIII | Source of araC and PBAD | Invitrogen |
| pGEM-T Easy | TA cloning vector; Ampr | Promega |
| pGEM-T | TA cloning vector; Ampr | Promega |
| pTP223 | λ-Red genes | J. R. Roth |
| pFCO1 | ERG genes inserted in pBluescript SK+ | 20 |
| pUC4K | Contains Kanr cassette | Pharmacia |
| pFCO1/kan | pFCO1 with Kanr cassettes from pUC4K | This work |
| pCAT53-3 | MVA operon | This work |
| pRMC13 | Serovar Typhimurium dxs in pGEM-T | This work |
| pRMC14 | Plasmid-encoded dxs::MVA operon | This work |
| pBAD/myc-his A | Arabinose-induced expression vector; Ampr | Invitrogen |
| pBAD/srlE | srlE cloned into pBAD/myc-his A | This work |
Construction of pCAT53-3.
The SalI fragment from pUC4K containing the gene conferring Kanr was ligated into the XhoI site of pFCO1 (20) to form pFCO1/kan. Transformants bearing the ligated products were selected on LB-KAN. The PBAD promoter and araC gene were excised from pBAD/gIII (Invitrogen) by digestion with NsiI, treatment with Klenow to blunt end, and digestion with SacI, followed by ligation of the resulting fragment into pFCO1/kan, which had also been digested with SapI, treated with Klenow, and digested with SacI to form pCAT53-3.
Construction of pRMC14.
S. enterica serovar Typhimurium dxs was PCR amplified from genomic template, ligated directly into pGEM-T (Promega), digested with Bsu36I and EcoRV and subsequently treated with Klenow to give a 4-kb fragment. The 7-kb Klenow-treated BsrBI/PspOMI fragment from pCAT53-3 was ligated into the 4-kb fragment from pRMC13 to create pRMC14 using CRC. The bridge oligonucleotides crcDXSARA and crcKANDXS were used with thermal cycler settings of denaturation at 94°C for 30 s, followed by an annealing cycle at 52°C for 30 s and ligation at 66°C for 1 min for 30 cycles.
Construction of RMC26.
An ∼8-kb BsrBI/SpeI fragment from pRMC14 was electroporated into TT22236 containing the λ-Red plasmid pTP223 (35). Linear recombinants were selected by plating to LB-KAN-ME. The correct insertion was initially verified phenotypically by growth on LB-KAN-ME and LB-KAN-MVA-Ara and by lack of growth on LB-KAN. Subsequent verification was accomplished by PCR using primers nested in ispA (sISPA1) and araC (asARAC1) for one end of the insertion and primers nested in yajO (asYAJO1) and the Kan cassette (sKAN1) for the other end of the insert followed by sequencing of the PCR products. The genes ispA and yajO flank dxs. The strain was designated RMC25. The insertion was backcrossed into LT2 to result in RMC26, with the disruption designated dxs::MVAoperon.
Random mutagenesis of RMC26.
Mutagenesis experiments were conducted using TE3461 as a donor strain in a standard P22 transduction with RMC26 as the recipient, resulting in random Mud-Cam insertions (17, 23). Mutants were selected by plating transduction crosses directly to LB-CAM-MVA-Ara. After incubation for 2 days at 37°C, the plates were replica printed to LB-CAM-ME and LB-CAM-MVA-Ara. Colonies exhibiting growth on MVA-Ara but not on ME were then streaked to green-CAM-MVA-Ara plates. Nonlysogenic colonies (white colonies on green plates) were cross streaked against P22 (H5) to ensure phage sensitivity. When phage-sensitive mutants were verified, single colonies were streaked to LB-CAM-MVA-Ara plates. When the cells grew, the plates were replica plated as described previously. Cells exhibiting the proper phenotype were used to inoculate liquid cultures, and the insertion point of the Cam cassette was determined by single-primer (semirandom) PCR of the genomic DNA. Semirandom PCR (23) was performed using Taq/TaqStart and the primers smud33, smud54, smud59, and smud86. Thermal cycler settings were as follows: an initial denaturation at 94°C for 30 s, followed by 20 cycles of high-stringency PCR in which the samples were heated to 94°C (with no hold time) and cooled to 53°C (no hold time), followed by a 1-min elongation step at 72°C. Thirty cycles of low-stringency PCR ensued, where the samples were heated to 94°C (no hold time) and cooled to 40°C (no hold time), followed by a 1-min extension at 72°C. Thirty additional cycles of high-stringency PCR followed as described above.
Insertions identified by semirandom PCR were verified using standard two-primer PCR. In the mutants, all MEP pathway genes were probed for insertions using PCR.
Construction of directed knockouts of srlE, srlA, srlB, ispD, ispE, ispF, and ispG in RMC26.
Disruptions of srlE, srlA, srlB, and MEP pathway genes from ispD through ispG were constructed by insertion of a CAT cassette using λ-Red-mediated linear recombination as described for disrupting dxs with the MVAoperon. In all cases, the insertion of the CAT cassette also resulted in a partial gene deletion. All primers were designed to PCR amplify the CAT cassette of pACYC184 while incorporating approximately 40 bp of homology to the target at either end of the amplificate. The amplificate was then gel purified and used to directly transform the recipient cells to CAM resistance. The srlE and MEP pathway disruptions were constructed in RMC28 using the primers sgutEFRT2 and asgutEFRT2, SygbPFRT2 and ASygbPFRT2, sygbBcamPCR and asygbBcamPCR, sychBcamPCR and asychBcamPCR, and sgcpEcam2 and asgcpEcam2 to yield CR3 (srlE::Cam), CR2 (ispD::Cam), CT10 (ispE::Cam), CT11 (ispF::Cam), and CR20 (ispG::Cam), respectively. When a correct insertion was verified by phenotype (growth on MVA-Ara, no growth on ME), PCR analysis, and sequencing, most disruptions were backcrossed into RMC26. The primers SgutEseq and ASgutEseq (srlE::Cam), sygbPseq and asygbPseq (ispD::Cam), sychBCamseq2 and asychBCamseq2 (ispE::Cam), sygbBCamseq2 and asygbBCamseq2 (ispF::Cam), and sgcpEseq and asgcpEseq (ispG::Cam) with homology to regions flanking the disruptions were used for sequencing. The CAT insertions in srlA and srlB were initially constructed in TT22236. After verification of the correct recombination event by sequencing, the srlA and srlB insertions were crossed into RMC26 by standard P22 transduction, resulting in strains RMC78 and RMC79, respectively. The primers used to construct the srlA insertion were SsrlACam and ASsrlBCam. Primers SsrlBCam and ASsrlBCam were used for the srlB insertion. The srlA insertion was verified using primers SsrlAX and ASsrlAX. The analogous primers SsrlBX and ASsrlBX were used to sequence the srlB insertion. Strains RMC26, CR5, RMC78, and RMC79 were plated to LB-KAN-MVA-Ara and LB-KAN-ME to verify phenotype.
Partial purification of ubiquinone Q8.
Ubiquinone-8 (Q8) was purified as described by Charon et al. (12) with minor modifications. Mass spectrometry was performed using positive ion atmospheric pressure chemical ionization mass spectrometry (APCI) and positive ion liquid secondary ion mass spectrometry (LSIMS). APCI was performed by direct injection of the sample dissolved in 1:4:1:1 chloroform-methanol-n-butanol-water. A mobile phase of 1:1 water-acetonitrile was used at a flow rate of 1 ml/min. LSIMS was performed using m-nitrobenzyl alcohol as the matrix and argon.
RESULTS
Disruption of chromosomal dxs in S. enterica serovar Typhimurium.
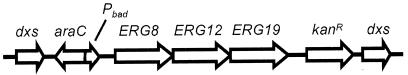
A dxs mutant of S. enterica serovar Typhimurium was constructed by disruption of the gene with a synthetic operon containing the three genes from Saccharomyces cerevisiae responsible for converting MVA to IPP (20) under the control of the PBAD promoter (10) (Fig. 2). pCAT53-3 was constructed from plasmid pFCO1 by adding a Kanr cassette to the 3′ end of yeast ERG8 (phosphomevalonate kinase), ERG12 (MVA kinase), and ERG19 (MVA diphosphate decarboxylase) and adding the araC gene and PBAD promoter to the 5′ end. The modified operon (MVAoperon) was excised from pCAT53-3 and used to disrupt plasmid-encoded dxs using CRC to form pRMC14. The traditional blunt-end ligation of the MVA operon into dxs was attempted but was not successful. The ligation most likely failed due to the size of the MVAoperon (∼7 kb). A stable integration into the S. enterica serovar Typhimurium chromosome was generated by λ-Red-mediated homologous recombination (23). Selection was performed using KAN resistance as well as ME auxotrophy. Homologous recombination was facilitated using pTP223, which carries the phage λ genes encoding a 5′-3′ exonuclease (exo) to facilitate interstrand invasion of DNA (bet) and a host cell RecBCD-inactivating protein (gam). This construct synthesizes IPP from MVA, when supplemented with Ara, or from ME.
FIG. 2.
Diagram of the MVA operon inserted into dxs. araC and PBAD are upstream of the yeast ERG genes, and the Kanr cassette is located downstream.
Random mutagenesis of RMC26.
Mutagenesis was performed by phage-mediated random “hopping” of a Mud-Cam element, which contains a CAT cassette, into the RMC26 genome. Mutants that exhibited the desired phenotype of growth on MVA and no growth on ME were isolated, and the disrupted gene was identified by semirandom PCR using primers nested in the CAT cassette. A total of nine mutants exhibiting this phenotype were isolated, out of approximately 200,000 screened.
Disruption of srlE, srlA, srlB, and MEP pathway genes (ispD and ispG) in RMC26 and LT2.
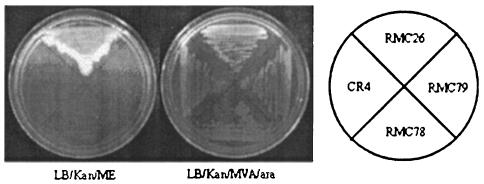
BLAST searches on the sequences obtained from the semirandom PCR products of the nine mutants returned four sequences from srlE. The gene encoding the SrlE protein in RMC28 was disrupted by homologous recombination and subsequently transduced into RMC26 to give CR5. In addition to exhibiting the phenotype of growth on MVA-Ara and no growth on ME, CR5 was viable when supplemented with DX. Restoration of growth by DX supplementation indicated that the endogenous MEP pathway remained intact and suggested that the mechanism for DX utilization is different from that for ME. The srlE::CAT disruption was also successfully crossed into wild-type LT2. The resulting strain, CR24, was viable on LB-CAM, indicating that srlE is not an essential gene. The srlA and srlB genes were also disrupted in RMC26, and the resulting strains, RMC78 and RMC79, exhibited the phenotype of growth on MVA-Ara and no growth on ME. Figure 3 displays the growth patterns of RMC26, CR5, RMC78, and RMC79 when they were plated to LB-KAN-MVA-Ara and LB-KAN-ME. Each of the strains was viable when the medium was supplemented with MVA-Ara, but only RMC26 was viable with ME supplementation, indicating that SrlA, SrlB, and SrlE are all involved in ME utilization. All of the directed knockouts in RMC26 of MEP pathway genes resulted in the desired phenotype of growth on MVA-Ara and no growth on ME (unpublished results).
FIG. 3.
Genetic evidence that SrlE, SrlA, and SrlB are involved in the utilization of exogenous ME. Growth patterns of RMC26, CR5, RMC78, and RMC79 on LB-KAN-ME (left) and LB-KAN-MVA-Ara (right) are shown.
Q8 isolation and analysis.
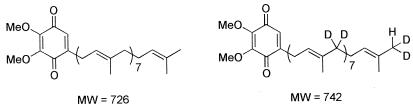
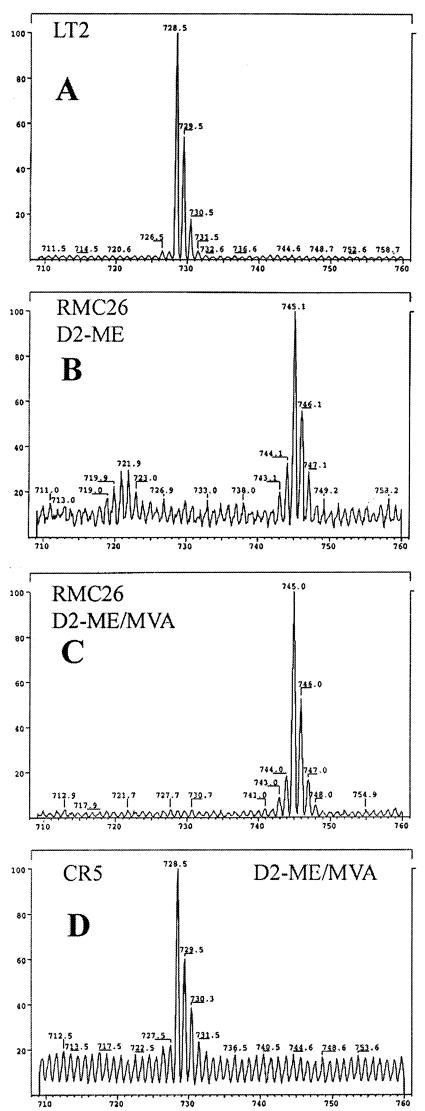
Feeding experiments with [1,1-2H2]ME established that SrlE is required for uptake and utilization of ME by the cell. Q8 was partially purified from the following cultures: LT2 grown in LB, RMC26 grown in LB-KAN-[1,1-2H2]ME, RMC26 grown in LB-KAN-[1,1-2H2]ME-MVA-Ara, and CR5 grown in LB-CAM-[1,1-2H2]ME-MVA-Ara. LT2 grown in LB was used as a control for Q8 isolation. Q8 derived from unlabeled isoprene units has a molecular weight (MW) of 726, whereas Q8 derived from [1,1-2H2]ME has a MW of 742 (Fig. 4). Q8 was isolated from LT2 and analyzed by positive-ion APCI and positive-ion LSIMS. In APCI, a peak at m/z 727 was observed corresponding to M + H+ for Q8 (MW = 726). The LSIMS spectrum had a peak at m/z 729, reflecting reduction of the quinone to the corresponding quinol during mass spectral analysis, as was previously reported (12).
FIG. 4.
Q8 derived from MVA (left) and from [1,1-2H2]ME (right). MW.
Purified samples of Q8 from the following incubations were analyzed by LSIMS to demonstrate that ME utilization is compromised when srlE is disabled: (i) LT2 was incubated in LB; (ii) RMC26 was incubated with [1,1-2H2]ME; (iii) RMC26 was incubated with [1,1-2H2]ME and MVA-Ara; and (iv) CR5 was incubated with [1,1-2H2]ME and MVA-Ara. The LSIMS spectra are shown in Fig. 5. APCI spectra for Q8 isolated from incubation ii had a peak at m/z 743, and the LSIMS peak was at m/z 745, indicating complete incorporation of deuterium into the isoprenoid side chain of Q8. Incubation iii also gave an APCI peak at 743 and an LSIMS peak at 745, indicating that deuterium-labeled IPP was incorporated exclusively into the Q8 side chain. Q8 isolated from incubation iv gave an APCI peak at 727 and an LSIMS peak at 729, demonstrating no deuterium incorporation into Q8 when srlE is disrupted.
FIG. 5.
LSIMS spectra of Q8 isolated from feeding experiments: (A) wild-type LT2 without supplementation; (B) RMC26 supplemented with [1,1-2H2]ME; (C) RMC26 supplemented with [1,1-2H2]ME-MVA-Ara; (D) CR5 supplemented with [1,1-2H2]ME-MVA-Ara.
Characterization of the remaining insertion mutants.
Four of the isolated insertion mutations in RMC26 mapped to srlE. The remaining five have been resistant to semirandom PCR and subsequent sequencing. None of the remaining five mutants have insertions in the known genes of the MEP pathway as probed by two-primer PCR. Initial attempts to complement these mutations with an S. enterica serovar Typhimurium genomic library have been unsuccessful.
DISCUSSION
Random mutagenesis of RMC26 generated four mutants with the desired phenotype that had disruptions in srlE, which encodes a protein in the sorbitol PTS. PCR using genomic DNA from the mutants as a template confirmed an insertion in the srlE region. Insertion of a CAT cassette into srlE produced a MVA-Ara-dependent phenotype in RMC26, confirming its involvement in ME utilization. During PTS-mediated phosphorylation, a phosphate group from PEP is first transferred to the N1 position of a histidine residue of EI and subsequently transferred to the N1 position of a histidine in HPr, with neither of these steps being sugar specific. The phosphate is then transferred from HPr to a histidine N3 of EIIA in the sugar-specific transport complex, followed by transfer to a cysteine residue of EIIB and finally to the sugar molecule (45). Because of the coordinated mechanism of the EIIA, EIIB, and EIIC subunits in a PTS system, srlA and srlB were also implicated along with srlE in ME utilization. Insertions of CAT cassettes into srlA and srlB gave a MVA-Ara-dependent phenotype similar to that seen for srlE, confirming the role of all three genes in ME utilization.
Incorporation of ME and DX into isoprenoids was reported before many of the steps in the MEP pathway had been discovered (6, 19, 37, 40, 43). It is now clear that both compounds must be phosphorylated before they are utilized as substrates in the MEP pathway. The sorbitol PTS provides a mechanism for import and phosphorylation of ME. In vitro experiments have demonstrated that d-xylulokinase (encoded by xylB), the enzyme responsible for phosphorylating d-xylulose, also phosphorylates DX (46), although the transport mechanism for uptake by cells has not been reported. The viability of CR5 when DX is present in the growth medium indicates that the mechanism for utilization of DX in the MEP pathway is different from that for ME. One could presumably identify the gene (or genes) responsible for transporting exogenous DX by performing a similar set of experiments described in this work except using DX instead of ME in the screening process.
Involvement of the srlE gene product in the utilization of exogenously supplied ME for isoprenoid biosynthesis was verified biochemically by analysis of Q8. S. enterica serovar Typhimurium strain RMC26 readily incorporated the deuterium label from ME into the isoprenoid side chain of Q8. Interestingly, RMC26, which synthesizes isoprenoids from either ME or MVA, had a substantial preference for utilization of ME when incubated with a mixture of MVA-Ara and deuterium-labeled ME. In contrast, incubation of CR5, where SrlE in the sorbitol PTS system is disabled, with the same mixture of substrates produced Q8 with no deuterium in the side chain. Thus, a functional copy of the SrlE protein is required for ME utilization.
The sorbitol PTS complex also appears to be responsible for phosphorylation of ME. In particular, the SrlE protein is the EIIB subunit of the sorbitol PTS and is analogous to protein that transfers phosphate to the sugar in the glucose PTS (18). Although d-sorbitol and ME are polyhydroxyl compounds, the absolute stereochemistries of their chiral centers do not precisely map onto one another, and it is not clear why the operon responsible for the utilization of sorbitol is also responsible for the transport and phosphorylation of ME.
In some cases overproduction of enzymes in the MEP pathway stimulates the synthesis of isoprenoids (25, 26). For example, higher levels of lycopene are obtained when isopentenyl diphosphate isomerase is overexpressed (25). The level of lycopene can also be increased when DXP synthase and MEP synthase are overproduced (44). It might be possible to further stimulate the synthesis of lycopene from ME by overproduction of the SrlA, SrlE, and SrlB proteins along with DXP synthase and MEP synthase.
It is notable that no insertions were seen in known biosynthetic genes in the MEP pathway even though directed knockouts of the genes in RMC26 gave strains with the same phenotype as CR5 (unpublished results). We were able to specifically disrupt each of the MEP pathway genes beyond MEP synthase in RMC26, with each exhibiting the phenotype of growth on MVA-Ara and no growth on ME. This would imply that we should be able to identify mutations that render each of the genes inactive, yet all of the isolated mutants appear to be involved in ME uptake and phosphorylation. In addition to the four srlE mutants, the five unknown mutants do not have an absolute requirement for the MVAoperon and readily lose it through recombination when crossed with a tightly linked marker, such as thiI::Tn10d-Tet. If the random insertions were in MEP pathway genes, the MVA operon would be essential to provide cellular isoprenoids, as demonstrated by the inability of CR4 to lose the MVA operon in the same cross. If it is assumed that there are 1,000 genes encoded by the S. enterica serovar Typhimurium chromosome, with five being potential MEP pathway targets in our screen (ispD through ispH) and five belonging to the proposed ME transport system (srlA, srlB, srlE, and the genes encoding HPr and EI), an estimated 100 mutants exhibiting the phenotype of growth on MVA-Ara and no growth on ME would be expected for completely random insertions. Our finding of only nine mutants out of 200,000 is low. We have no evidence to explain this discrepancy.
Acknowledgments
We thank John Roth for helpful discussions, Lisa Eubanks for providing DXP synthase, and David Fox for MEP synthase.
R.M.C. was an NIH trainee supported by grant GM 08537. This project was supported by NIH grant GM 25521.
REFERENCES
- 1.Agranoff, B. W., H. Eggerer, U. Henning, and F. Lynen. 1959. Isopentenyl pyrophosphate isomerase. J. Am. Chem. Soc. 81:1254-1255. [PubMed] [Google Scholar]
- 2.Altincicek, B., E. C. Duin, A. Reichenberg, R. Hedderich, A. K. Kollas, M. Hintz, S. Wagner, J. Wiesner, E. Beck, and H. Jomaa. 2002. LytB protein catalyzes the terminal step of the 2-C-methyl-D-erythritol-4-phosphate pathway of isoprenoid biosynthesis. FEBS Lett. 532:437-440. [DOI] [PubMed] [Google Scholar]
- 3.Altincicek, B., A. Kollas, M. Eberl, J. Wiesner, S. Sanderbrand, M. Hintz, E. Beck, and H. Jomaa. 2001. LytB, a novel gene of the 2-C-methyl-D-erythritol 4-phosphate pathway of isoprenoid biosynthesis in Escherichia coli. FEBS Lett. 499:37-40. [DOI] [PubMed] [Google Scholar]
- 4.Altincicek, B., A. K. Kollas, S. Sanderbrand, J. Wiesner, M. Hintz, E. Beck, and H. Jomaa. 2001. GcpE is involved in the 2-C-methyl-D-erythritol 4-phosphate pathway of isoprenoid biosynthesis in Escherichia coli. J. Bacteriol. 183:2411-2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Amdur, B. H., H. Rilling, and K. Bloch. 1957. The enzymatic conversion of mevalonic acid to squalene. J. Am. Chem. Soc. 79:2646-2647. [Google Scholar]
- 6.Arigoni, D., S. Sagner, C. Latzel, W. Eisenreich, A. Bacher, and M. H. Zenk. 1997. Terpenoid biosynthesis from 1-deoxy-D-xylulose in higher plants by intramolecular skeletal rearrangement. Proc. Natl. Acad. Sci. USA 94:10600-10605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blagg, B. S., and C. D. Poulter. 1999. Synthesis of 1-deoxy-D-xylulose and 1-Deoxy-D-xylulose-5-phosphate. J. Org. Chem. 64:1508-1511. [DOI] [PubMed] [Google Scholar]
- 8.Bloch, K., S. Chaykin, A. H. Phillips, and A. de Waard. 1959. Mevalonic acid pyrophosphate and isopentenyl pyrophosphate. J. Biol. Chem. 234:2595-2604. [PubMed] [Google Scholar]
- 9.Bochar, D. A., J. A. Freisen, C. V. Stauffacher, and V. W. Rodwell. 1999. Biosynthesis of mevalonic acid from acetyl-CoA, p. 15-44. In D. Cane (ed.), Comprehensive natural products chemistry, vol. 2. Pergamon Press, Oxford, United Kingdom.
- 10.Campos, N., M. Rodriguez-Concepcion, S. Sauret-Gueto, F. Gallego, L. M. Lois, and A. Boronat. 2001. Escherichia coli engineered to synthesize isopentenyl diphosphate and dimethylallyl diphosphate from mevalonate: a novel system for the genetic analysis of the 2-C-methyl-D-erythritol 4-phosphate pathway for isoprenoid biosynthesis. Biochem. J. 353:59-67. [PMC free article] [PubMed] [Google Scholar]
- 11.Campos, N., M. Rodriguez-Concepcion, M. Seemann, M. Rohmer, and A. Boronat. 2001. Identification of gcpE as a novel gene of the 2-C-methyl-D-erythritol 4-phosphate pathway for isoprenoid biosynthesis in Escherichia coli. FEBS Lett. 488:170-173. [DOI] [PubMed] [Google Scholar]
- 12.Charon, L., J. F. Hoeffler, C. Pale-Grosdemange, L. M. Lois, N. Campos, A. Boronat, and M. Rohmer. 2000. Deuterium-labelled isotopomers of 2-C-methyl-D-erythritol as tools for the elucidation of the 2-C-methyl-D-erythritol 4-phosphate pathway for isoprenoid biosynthesis. Biochem J. 346:737-742. [PMC free article] [PubMed] [Google Scholar]
- 13.Cunningham, F. X., Jr., T. P. Lafond, and E. Gantt. 2000. Evidence of a role for LytB in the nonmevalonate pathway of isoprenoid biosynthesis. J. Bacteriol. 182:5841-5848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Davis, R. W., D. Bolstein, and J. R. Roth. 1980. Advanced bacterial genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, New York.
- 15.Dhe-Paganon, S., J. Magrath, and R. H. Abeles. 1994. Mechanism of mevalonate pyrophosphate decarboxylase: evidence for a carbocationic transition state. Biochemistry 33:13355-13362. [DOI] [PubMed] [Google Scholar]
- 16.Duvold, T., P. Cali, J.-M. Bravo, C. Pale-Grosdemange, and M. Rohmer. 1997. Incorporation of 2-C-methyl-D-erythritol, a putative isoprenoid intermediate in the mevalonate-independent pathway, into ubiquinone and menaquinone of Escherichia coli. Tetrahedron Lett. 38:4769-4772. [Google Scholar]
- 17.Elliot, T. 1993. Transport of 5-aminolevulinic acid by the dipeptide permease in Salmonella typhimurium. J. Bacteriol. 175:325-331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Erni, B., H. Trachsel, P. W. Postma, and J. P. Rosenbusch. 1982. Bacterial phosphotransferase system. Solubilization and purification of the glucose-specific enzyme II from membranes of Salmonella typhimurium. J. Biol. Chem. 257:13726-13730. [PubMed] [Google Scholar]
- 19.Giner, J. L., B. Jaun, and D. Arigoni. 1998. Biosynthesis of isoprenoids in Escherichia coli: the fate of the 3-H and 4-H atoms of 1-deoxy-D-xylulose. J. Chem. Soc. Chem. Commun. 17:1857-1858. [Google Scholar]
- 20.Hahn, F. M., L. M. Eubanks, C. A. Testa, B. S. Blagg, J. A. Baker, and C. D. Poulter. 2001. 1-Deoxy-D-xylulose 5-phosphate synthase, the gene product of open reading frame (ORF) 2816 and ORF 2895 in Rhodobacter capsulatus. J. Bacteriol. 183:1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hecht, S., W. Eisenreich, P. Adam, S. Amslinger, K. Kis, A. Bacher, D. Arigoni, and F. Rohdich. 2001. Studies on the nonmevalonate pathway to terpenes: the role of the GcpE (IspG) protein. Proc. Natl. Acad. Sci. USA 98:14837-14842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Herz, S., J. Wungsintaweekul, C. A. Schuhr, S. Hecht, H. Lüttgen, S. Sagner, M. Fellermeier, W. Eisenreich, M. H. Zenk, A. Bacher, and F. Rohdich. 2000. Biosynthesis of terpenoids: YgbB protein converts 4-diphosphocytidyl-2-C-methyl-D-erythritol 2-phosphate to 2-C-methyl-D-erythritol 2,4-cyclodiphosphate. Proc. Natl. Acad. Sci. USA 97:2486-2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hughes, K. T., and J. R. Roth. 1988. Transitory cis complementation: a method for providing transposition functions to defective transposons. Genetics 119:9-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jordan, P. M., and P. N. Gibbs. 1983. Mechanism of action of beta-oxoacyl-CoA thiolase from rat liver cytosol. Direct evidence for the order of addition of the two acetyl-CoA molecules during the formation of acetoacetyl-CoA. Biochem. J. 213:153-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kajiwara, S., P. D. Fraser, K. Kondo, and N. Misawa. 1997. Expression of an exogenous isopentenyl diphosphate isomerase gene enhances isoprenoid biosynthesis in Escherichia coli. Biochem. J. 324:421-426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim, S.-W., and J. D. Keasling. 2001. Metabolic engineering of the nonmevalonate isopentenyl diphosphate synthesis pathway in Escherichia coli enhances lycopene production. Biotechnol. Bioeng. 72:408-415. [DOI] [PubMed] [Google Scholar]
- 27.Koppisch, A. T., D. T. Fox, B. S. Blagg, and C. D. Poulter. 2002. E. coli MEP synthase: steady-state kinetic analysis and substrate binding. Biochemistry 41:236-243. [DOI] [PubMed] [Google Scholar]
- 28.Kuzuyama, T., M. Takagi, Kaneda, K., T. Dairi, and H. Seto. 2000. Formation of 4-(cytidine 5′-diphospho)-2-C-methyl-D-erythritol from 2-C-methyl-D-erythritol 4-phosphate by 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase. Tetrahedron Lett. 41:703-706. [Google Scholar]
- 29.Kuzuyama, T., M. Takagi, K. Kaneda, H. Watanabe, T. Dairi, and H. Seto. 2000. Studies on the nonmevalonate pathway: conversion of 4-(cytidine 5′-diphospho)-2-C-methyl-D-erythritol to its 2-phospho derivative by 4-(cytidine 5′-diphospho)-2-C-methyl-D-erythritol kinase. Tetrahedron Lett. 41:2925-2928. [Google Scholar]
- 30.Lange, B. M., M. R. Wildung, D. McCaskill, and R. Croteau. 1998. A family of transketolases that directs isoprenoid biosynthesis via a mevalonate-independent pathway. Proc. Natl. Acad. Sci. USA 95:2100-2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lüttgen, H., F. Rohdich, S. Herz, J. Wungsintaweekul, S. Hecht, C. A. Schuhr, M. Fellermeier, S. Sagner, M. H. Zenk, A. Bacher, and W. Eisenreich. 2000. Biosynthesis of terpenoids: YchB protein of Escherichia coli phosphorylates the 2-hydroxy group of 4-diphosphocytidyl-2-C-methyl-D-erythritol. Proc. Natl. Acad. Sci. USA 97:1062-1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McAteer, S., A. Coulson, N. McLennan, and M. Masters. 2001. The lytB gene of Escherichia coli is essential and specifies a product needed for isoprenoid biosynthesis. J. Bacteriol. 183:7403-7407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miziorko, H. M., and M. D. Lane. 1977. 3-Hydroxy-3-methylgutaryl-CoA synthase. Participation of acetyl-S-enzyme and enzyme-S-hydroxymethylgutaryl-SCoA intermediates in the reaction. J. Biol. Chem. 252:1414-1420. [PubMed] [Google Scholar]
- 34.Pachuk, C. J., M. Samuel, J. A. Zurawski, L. Snyder, P. Phillips, and C. Satishchandran. 2000. Chain reaction cloning: a one-step method for directional ligation of multiple DNA fragments. Gene 243:19-25. [DOI] [PubMed] [Google Scholar]
- 35.Poteete, A. R., and A. C. Fenton. 2000. Genetic requirements of phage lambda red-mediated gene replacement in Escherichia coli K-12. J. Bacteriol. 182:2336-2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Price-Carter, M., J. Tingey, T. A. Bobik, and J. R. Roth. 2001. The alternative electron acceptor tetrathionate supports B12-dependent anaerobic growth of Salmonella enterica serovar Typhimurium on ethanolamine or 1,2-propanediol. J. Bacteriol. 183:2463-2475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Putra, S. R., L. M. Lois, N. Campos, A. Boronat, and M. Rohmer. 1998. Incorporation of [2,3-13C2]- and [2, 4-13C2]-D-1-deoxyxylulose into ubiquinone of Escherichia coli via the mevalonate-independent pathway for isoprenoid biosynthesis. Tetrahedron Lett. 39:23-26. [Google Scholar]
- 38.Rohdich, F., S. Hecht, K. Gartner, P. Adam, C. Krieger, S. Amslinger, D. Arigoni, A. Bacher, and W. Eisenreich. 2002. Studies on the nonmevalonate terpene biosynthetic pathway: metabolic role of IspH (LytB) protein. Proc. Natl. Acad. Sci. USA 99:1158-1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rohdich, F., J. Wungsintaweekul, M. Fellermeier, S. Sagner, S. Herz, K. Kis, W. Eisenreich, A. Bacher, and M. H. Zenk. 1999. Cytidine 5′-triphosphate-dependent biosynthesis of isoprenoids: YgbP protein of Escherichia coli catalyzes the formation of 4-diphosphocytidyl-2-C-methylerythritol. Proc. Natl. Acad. Sci. USA 96:11758-11763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sagner, S., C. Latzel, W. Eisenreich, A. Bacher, and M. H. Zenk. 1998. Differential incorporation of 1-deoxy-D-xylulose into monoterpenes and carotenoids in higher plants. J. Chem. Soc. Chem. Commun. 2:221-222. [Google Scholar]
- 41.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 42.Schmieger, H. 1972. Phage P22 mutants with increased or decreased transduction abilities. Mol. Gen. Genet. 119:75-88. [DOI] [PubMed] [Google Scholar]
- 43.Schwender, J., J. Zeidler, R. Groner, C. Muller, M. Focke, S. Braun, F. W. Lichtenthaler, and H. K. Lichtenthaler. 1997. Incorporation of 1-deoxy-D-xylulose into isoprene and phytol by higher plants and algae. FEBS Lett. 414:129-134. [DOI] [PubMed] [Google Scholar]
- 44.Seon-Won, K., and J. D. Keasling. 2001. Metabolic engineering of the nonmevalonate isopentenyl diphosphate pathway in Escherichia coli enhances lycopene production. Biotechnol. Bioeng. 72:408-415. [DOI] [PubMed]
- 45.Tchieu, J. H., V. Norris, J. S. Edwards, and M. H. Saier, Jr. 2001. The complete phosphotranferase system in Escherichia coli. J. Mol. Microbiol. Biotechnol. 3:329-346. [PubMed] [Google Scholar]
- 46.Wungsintaweekul, J., S. Herz, S. Hecht, W. Eisenreich, R. Feicht, F. Rohdich, A. Bacher, and M. H. Zenk. 2001. Phosphorylation of 1-deoxy-D-xylulose by D-xylulokinase of Escherichia coli. Eur. J. Biochem. 268:310-316. [DOI] [PubMed] [Google Scholar]