Abstract
Centromere-binding protein B (CENP-B) is a widely conserved DNA binding factor associated with heterochromatin and centromeric satellite repeats1. In fission yeast, CENP-B homologs have been shown to silence Long Terminal Repeat (LTR) retrotransposons by recruiting histone deacetylases2. However, CENP-B factors also have unexplained roles in DNA replication3, 4. Here, we show that a molecular function of CENP-B is to promote replication fork progression through the LTR. Mutants have increased genomic instability caused by replication fork blockage that depends on the DNA binding factor Switch Activating Protein 1 (Sap1), which is directly recruited by the LTR. The loss of Sap1-dependent barrier activity allows the unhindered progression of the replication fork, but results in rearrangements deleterious to the retrotransposon. We conclude that retrotransposons influence replication polarity through recruitment of Sap1 and transposition near replication fork blocks, while CENP-B counteracts this activity and promotes fork stability. Our results may account for the role of LTR in fragile sites, and for the association of CENP-B with pericentromeric heterochromatin and tandem satellite repeats.
In fission yeast, CENP-B proteins are encoded by three homologs, autonomously replicating sequence binding protein 1 (abp1), cenp-B homolog 1 (cbh1) and cbh2, and were previously characterized as DNA binding factors at origins of replication and centromeric repeats, respectively3, 5. abp1 mutants grow slowly, while double mutants with cbh1 or cbh2 have severely stunted growth, abnormal mitosis and morphological defects, and triple deletion mutants are inviable6, 7. As a result, double Δabp1Δcbh1 mutants form microcolonies on solid media (Fig. 1a and Supplementary Table 1) and exhibit high levels of cell death (Supplementary Fig. 1). We observed the spontaneous appearance of faster growing cells in a culture of Δabp1Δcbh1 that grew at rates similar to the Δabp1 single mutant, lacked morphological defects (Fig. 1a and Table 1) and showed lower levels of cell death (Supplementary Fig. 1). Genetic analysis revealed the presence of a single essential locus that also suppressed the lethality of the triple mutantΔabp1Δcbh1Δcbh2 (not shown). We performed whole genome resequencing in the mutant strain8 and isolated a missense mutation in the coding sequence of the DNA binding factor Sap1 (sap1E101D, henceforth called sap1-c; Supplementary Fig. 2) that cosegregated with suppression of slow growth in Δabp1Δcbh1 and resulted in lethality in a WT background. Sap1 is a protein with essential roles in chromosome stability9. Sap1 has been implicated in a programmed replication fork block in the rDNA monomer that ensures directional replication to prevent mitotic recombination between rDNA repeats10–12.
Figure 1.
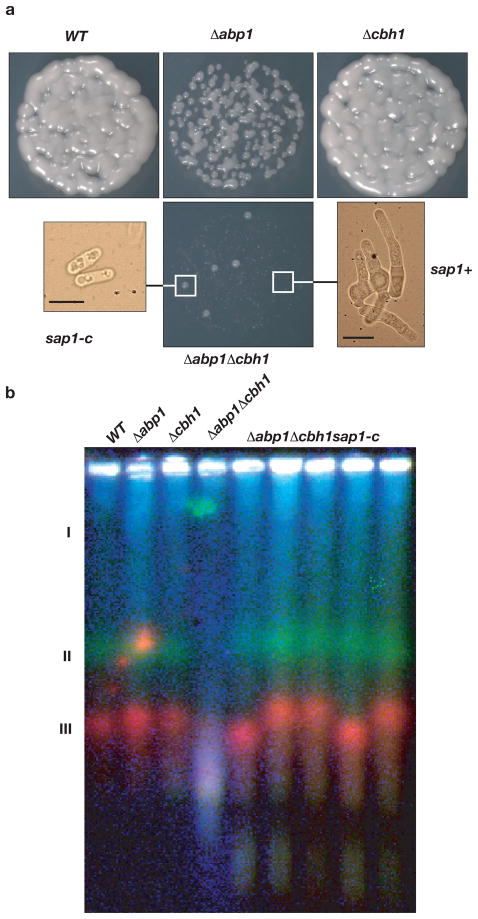
DNA damage in CENP-B mutants is suppressed by sap1 mutation. a, Images of 103 plated cells of WT, Δabp1 and Δabp1Δcbh1 with Δabp1Δcbh1sap1-c colonies. Microscopy image inserts: Images showing branched phenotype in Δabp1Δcbh1 background (right) and Δabp1Δcbh1sap1-c mutant (left). Scale bar is 10 μm. b, Pulsed Field Gel blot analysis of wild type (WT), CENP-B mutants (Δabp1, Δcbh1, Δabp1Δcbh1) and 5 CENP-B/sap1-c mutant isolates (Δabp1Δcbh1sap1-c). The position of the three chromosomes is indicated on the right. The image is a false-colored composite of hybridizations for all three chromosomes.
To test the effects of CENP-B and sap1-c mutations on genome integrity, we examined chromosomes by Pulsed Field Gel Electrophoresis (PFGE). While singleΔabp1 and Δcbh1 mutants had wild-type chromosome lengths, the double Δabp1Δcbh1 mutant had a smear of DNA fragments indicating double strand breaks (DSB) in all three chromosomes (Fig. 1b). Treatment of the Δabp1Δcbh1 sample plugs with the restriction enzyme NotI allowed migration of the chromosomes into the gel, and detection of telomeric and centromeric sequences (Supplementary Fig. 3), suggesting the presence of scattered unresolved replication or recombination intermediates that interfere with the migration of full length chromosomes, but not with NotI digested DNA, into the pulsed field gel. This indicates that Abp1 and Cbh1 have roles in the maintenance of genome integrity.
Surprisingly, the sap1-c mutation restored genome integrity to all chromosomes with Chromosome 3 exhibiting size variability in several isolates of Δabp1Δcbh1sap1-c mutant (Fig. 1b). In fission yeast, Chromosome 3 harbors the ribosomal DNA (rDNA) repeats. Temperature sensitive (ts) alleles of sap1 exhibit changes in the size of Chromosome 3 attributable to loss of fork barrier activity and an increase in mitotic recombination at rDNA12, and the changes in chromosome 3 size in the sap1-c mutants are associated with altered rDNA copy number (Supplementary Fig. 4). The ts alleles sap1-1 and sap1–4812 suppressed slow growth in the Δabp1Δcbh1 double mutant, mimicking sap1-c (Supplementary Fig. 5). Consistent with a reduction in fork barrier activity, a probe containing a canonical Sap1 binding sequence had reduced electrophoretic mobility shift (EMSA) in crude extracts from sap1-c mutants (Supplementary Fig. 6). We conclude that the suppression of the Δcbh1Δabp1 phenotype is not specific to the sap1-c mutation but a result of defective function of Sap1, and therefore that the loss of genome integrity in Δabp1Δcbh1 mutants is a consequence of Sap1 activity.
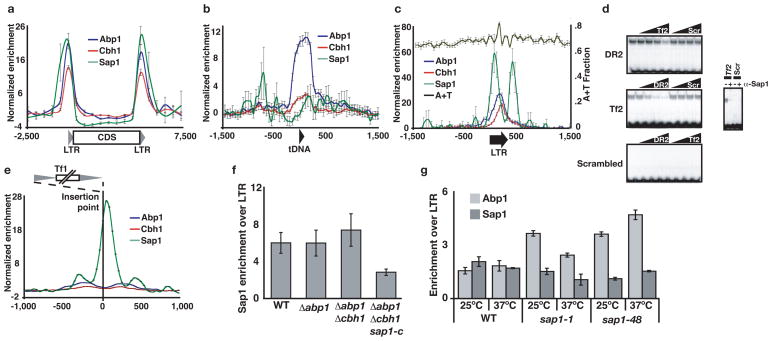
Blocked replication forks are potential sources of genome instability because they can lead to collapse of the replisome and DSB formation13. The fact that Sap1 activity leads to DNA damage in the absence of Abp1/Cbh1 suggests that the function of CENP-B is to manage Sap1-arrested replication forks. In the absence of Sap1 loss of replication fork blockage would render Abp1/Cbh1 activity unnecessary and lead to increased genome stability in Δabp1Δcbh1 mutants. This model predicts that CENP-B and Sap1 would colocalize to the regions where they acted on the replication fork, and that these regions would engage in homologous recombination (HR) and degrade to DSB in the absence of CENP-B. To test this hypothesis, we performed Chromatin Immunoprecipitation of Sap1, Abp1 and Cbh1 followed by High Throughput sequencing (ChIP-seq). Abp1 has previously been shown to localize and recruit Cbh1 to the LTR of Tf1 and Tf2 retrotransposons, where they play a role in their transcriptional silencing2. We demonstrated a strong colocalization of Sap1 with Abp1 and Cbh1 at these LTR as well as at solo LTR scattered throughout the genome (Fig. 2a, c and Supplementary Fig. 7a, b) and at the mating type locus (Supplementary Fig. 8), where Sap1 and Abp1 have been described to regulate mating type switching14, 15. Both Sap1 and Abp1/Cbh1 also localized to genomic regions independently of each other, suggesting that they do not form a stable complex or mediate their mutual recruitment. In particular, Abp1 exhibited binding to tRNA genes (Fig. 2b and Supplementary Fig.7b), known to be potent replication pause sites13, 16. Abp1 and Cbh1 co-localize to a highly A/T rich region located in positions 100–150 of the LTR (Fig. 2c and Supplementary Fig. 7a, b). The localization of Sap1 within the LTR was concentrated in the first 50bp of sequence (Fig. 2c), coinciding with a predicted Sap1 binding site17 (Supplementary Fig. 7a, c). We tested this sequence by EMSA and detected specific binding in WT extracts (Fig. 2d) as well as decreased binding and altered mobility in extracts from Δabp1Δcbh1sap1-c mutants (Supplementary Fig. 7d). Interestingly, solo LTR and full length Tf2 insertions were associated with a prominent peak of Sap1 binding located outside the 3′ end of the transposon sequence (Fig. 2c). These observations indicate that Sap1 binding precedes and possibly guides Tf element integration. To test this prediction, we plotted the average enrichment of Sap1, Abp1 and Cbh1 around more than 70,000 de novo Tf1 integration sites recently reported18 and observed a dramatic association of these integration sites with a peak of Sap1 binding immediately downstream of the insertion site (Fig. 2e and Supplementary Fig. 8), with no appreciable CENP-B enrichment. These results strongly suggest that Sap1 binding sequences determine the targeting and orientation of Tf retroelement transposition.
Figure 2.
Sap1 and CENP-B colocalize at the LTR of retrotransposons in vivo. Average genome-wide enrichment by ChIP-seq of Sap1, Abp1 and Cbh1 on a, all Tf2 elements, b, euchromatic tRNA and c, solo LTR. Error bars represent Standard Error. d, Left panel: Competition EMSA. Right panel: Inactivation by incubation with anti-Sap1 serum9. e, Average Sap1, Abp1 and Cbh1 enrichment around Tf1 de novo insertion points18. f, ChIP of Sap1 with LTR of Tf2 in CENP-B and sap1-c mutants and g, of Abp1 with LTR of Tf2 in and sap1 ts mutants. Error bars represent standard deviation for triplicates.
To evaluate the mutual influence of Sap1 and Abp1/Cbh1 on LTR binding we performed ChIP analysis of Sap1 in Δabp1Δcbh1 mutants and of Abp1 in ts sap1 mutants that affect DNA binding activity12. Sap1 binding to the LTR was unaffected inΔabp1 andΔcbh1 mutants, and was slightly increased in Δabp1Δcbh1 double mutants (Fig. 2f), but consistently reduced (2 fold) in Δabp1Δcbh1sap1-c. Conversely, Abp1 binding to the LTR was increased between 2 and 3 times at the permissive temperature in sap1-1 and sap1–48 mutants (Fig. 2g). These results indicate that Sap1 and Abp1/Cbh1 bind to the LTR independently of each other and mutually counteract their recruitment, and that the sap1-c mutation impairs its binding to the LTR in vivo as well as in vitro (Fig. S4).
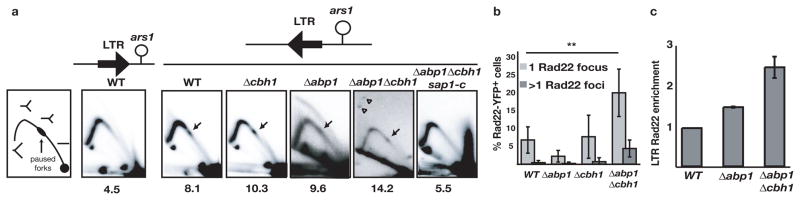
A failure of replication fork stability at LTR, which are distributed throughout the genome, would explain the widespread DNA damage in Δabp1Δ cbh1 mutants. We assessed the behavior of the replication fork as it traversed the LTR using 2D agarose gel electrophoresis. Sap1 dependent programmed fork blocks are directional and only hinder fork progression in one orientation10, 11. We cloned a full length LTR and its first 50 base pairs (containing the Sap1 binding site) in a plasmid in both orientations with respect to the replication origin ars1. 2D gel electrophoresis in a WT strain transformed with this episomal system showed a modest accumulation of fork signal at the location of the cloned LTR (Fig. 3a) but only when the Sap1 binding site was proximal to the origin, and not in the opposite orientation (Supplementary Fig. 9). The Sap1 binding site was sufficient for this blocking activity, with the same orientation requirement (Supplementary Fig. 9). We next assayed the LTR for pausing activity in Δabp1, Δcbh1 and sap1-c mutants (Fig. 3a). Strikingly, the paused fork signal was consistently enhanced and always at the same location in Δabp1 and Δcbh1 mutants, while theΔabp1Δcbh1 double mutant exhibited additional signals outside the replication arc suggestive of recombination intermediates19. The fork blocking activity of the LTR disappeared in Δabp1Δcbh1sap1-c mutants. Unresolved fork blocks can collapse and undergo HR for fork recovery. We confirmed the presence of HR in the Δabp1Δcbh1 double mutants by measuring the increase in the formation of Rad22 (homologous to Rad52 in S cerevisiae) foci in a Rad22-YFP strain20 (Fig. 3b), and we observed that Δabp1Δcbh1 double mutant cells accumulated the HR protein Rad22 at the LTR (Fig. 3c). Consistently, the recombination factor Rhp51 (Rad51 homolog) was essential for viability of Δabp1Δcbh1 double mutants (Supplementary Fig. 10), indicating that HR is necessary for recovery from fork stalling at LTR. These results indicate that Abp1/Cbh1 counteract Sap1 barrier activity and stabilize the replication fork at LTR. This results in loss of genome integrity and HR at the LTR in Δabp1Δcbh1 mutants.
Figure 3.
CENP-B promotes replication fork progression through the Sap1 dependent barrier present at the LTR and prevents HR. a, 2D gel electrophoresis of a plasmid fragment containing the Tf2 LTR oriented towards (left) and away (right) from the ars1 origin. Arrows indicate paused replication intermediates and open arrows recombination intermediates. The percentage of signal over the LTR is indicated below each panel. b, Quantification of Rad22 -GFP foci (N >400 nuclei for all mutants). Error bars depict standard errors. c, Rad22 -YFP ChIP with LTR in WT, Δabp1 and Δabp1Δcbh1 mutants. Error bars represent standard deviation for triplicates.
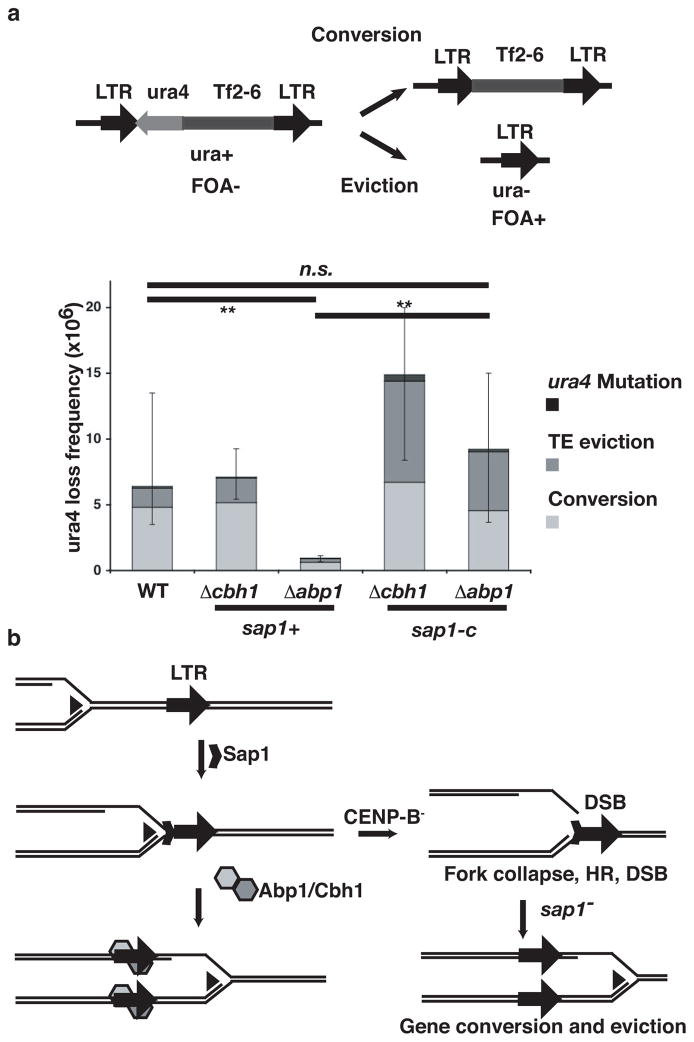
The Sap1 binding sequence is conserved in Tf1 and Tf2 retrotransposon LTR (Supplementary Fig. 6c) suggesting that it plays a role in the retrotransposon life cycle. We assayed the effect of sap1 and abp/cbh1 on Tf2 stability by measuring the frequencies of loss of a ura4 reporter transgene inserted in the Tf2-6 transposon21. Mutation of abp1 resulted in a dramatic decrease of Tf2 ectopic recombination, which returned to normal levels when sap1 was also mutated (Fig. 4). In the presence of sap1+ there is a preference for gene conversion, which normally constitutes the majority of ectopic recombination events22, but in Δabp1sap1-c and Δcbh1sap1-c mutants the proportion of eviction and conversion events is similar (Fig. 4). Therefore, we propose that the LTR recruits Sap1 in order to control the direction of transposon replication and increase transposon persistence in the genome, perhaps by coordinating lagging strand synthesis, which prevents single strand annealing from complementary direct repeats (Supplementary Fig. 11a,b). CENP-B counteracts this activity, possibly by promoting replication fork progression through the Sap1 dependent barrier. Thus, CENP-B and Sap1 promote genome and transposon integrity, respectively, in a “tug-of-war” between transposon and host. Abp1 stimulates fork progression by recruiting the fork restart protein MCM104 which has primase activity, and the histone deacetylase Mst1, which has roles in replication fork stability, also interacts directly with Cbh123 (Supplementary Fig. 11a). In S cerevisiae the histone deacetylase (HDAC) Sir2 silences and inhibits recombination in repetitive DNA24. CENP-B factors recruit the HDACs Clr3 and Clr6, which carry out LTR silencing2. The result of these functions would be to preserve genome integrity at LTRs by preventing DNA damage and recombination. This novel role of CENP-B may not be limited to LTR and tDNA, as mutation of the replication fork blocking factor reb1, which is specific to rDNA repeats, also suppresses the slow growth of the abp1 mutant25. Similarly, our ChIP-seq data indicates that Sap1 may also be implicated in the functionality of the replication terminator RTS1 (Supplementary Fig 8) in collaboration with Rtf1. In this manner, the function and regulation of the Sap1 bound regions is determined by the binding in their vicinity of different factors affecting replication fork progression.
Figure 4.
CENP-B and Sap1 have opposite effects on Tf2 stability. a, ectopic recombination fluctuation assay. Two potential mechanisms of ura4 loss from the marked Tf2-6::ura4 are indicated, gene conversion and eviction by LTR recombination. Columns represent total median ura4 loss frequency in WT, Δabp1, Δcbh1, Δabp1sap1-c and Δcbh1sap1-c mutants, error bars represent 95% confidence intervals. Colors indicate distribution of mode of ectopic recombination events in the ura4- colonies obtained from WT (n=93), Δabp1 (n=88), Δcbh1 (n=94), Δabp1sap1-c (n=91), and Δcbh1sap1-c (n=89) mutants. b, Model for the interactions between Abp1, Cbh1, Sap1 and the replication fork at the LTR.
Because of their repetitive nature, transposons have a close relationship with replication and recombination. For example, the IS608 transposon of E coli is targeted to the lagging strand and always replicated in the same direction26. This might prevent recombination between tandemly arranged copies. We have shown that retrotransposons influence DNA replication via recruitment of directional fork blocking factor Sap1 and that activity of CENP-B is required for replication fork management Additionally, retrotransposition is targeted to the genomic localization of Sap1. These mechanisms influence the replicative dynamics of the host genome. The genomes of eukaryotes show widespread colonization by retrotransposons, and pericentromeric satellite repeats are often of transposon origin27. When such sequences are arranged as tandem repeats, control of replication direction by CENP-B would prevent chromosome breaks and preserve genome integrity. This mechanism accounts for the role of other regulators of fork progression in inter-LTR recombination28, 29. In contrast, when flanked by LTR in opposite orientations, fragile sites fail to replicate and result in chromosome breaks13, 30.
Methods summary
ChIP was performed using tagged TAP-Abp1 and TAP-Cbh1 strains with an Anti-Calmodulin Binding Protein antibody (Millipore) and a polyclonal serum against the native Sap1 protein9. High throughput sequencing was performed on the illumina G2 genome analyzer, and analyzed for polymorphism detection or statistical analysis of enrichment. 2D gel electrophoresis was performed as described11, see supplementary information for construction of the episomal system. EMSA was performed as described previously17.
Full methods and any associated references are available in the Supplementary Information.
Supplementary Material
Acknowledgments
We thank the Martienssen lab, Victoria Aranda, Eva Mejia-Ramirez and Francisco Antequera for technical advice and discussions, and Robin Allshire, Eishi Noguchi, Matthew O’Connell, Peter Espenshade and the National BioResource Project (Taro Nakamura, Japan) for strains. This work was supported by NIH grant RO1GM076396 to R.A.M., by Cancer Research UK grant C9546/A6517 to J.B., by l’Agence Nationale de la Recherche grant ANR-06-BLAN-0271 to B.A., by a C.J. Martin Fellowship to D.V.I and by a Postdoctoral Fellowship from the Spanish Ministry of Education to M.Z. The authors declare no competing interests.
Footnotes
Author Contributions M.Z., B.A. and R.A.M. designed the experiments presented and wrote the paper. M.Z. performed and analyzed the experiments. M.W.V. provided bioinformatic analysis. D.G. and D.V.I. provided strains. S.W. and J.B. performed additional experiments.
Author Information The sequences from the ChIP-seq experiments are available at the Sequence Read Archive (www.ncbi.nln.nih.gov/sra) with accession number SRA024710.2. Reprints and permissions information is available at www.nature.com/reprints. The authors declare no competing financial interests. Correspondence and requests for materials should be addressed to R.A.M. (martiens@cshl.edu).
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
References
- 1.Okada T, et al. CENP-B controls centromere formation depending on the chromatin context. Cell. 2007;131:1287–300. doi: 10.1016/j.cell.2007.10.045. [DOI] [PubMed] [Google Scholar]
- 2.Cam HP, Noma K, Ebina H, Levin HL, Grewal SI. Host genome surveillance for retrotransposons by transposon-derived proteins. Nature. 2008;451:431–6. doi: 10.1038/nature06499. [DOI] [PubMed] [Google Scholar]
- 3.Murakami Y, Huberman JA, Hurwitz J. Identification, purification, and molecular cloning of autonomously replicating sequence-binding protein 1 from fission yeast Schizosaccharomyces pombe. Proc Natl Acad Sci U S A. 1996;93:502–7. doi: 10.1073/pnas.93.1.502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Locovei AM, Spiga MG, Tanaka K, Murakami Y, D’Urso G. The CENP-B homolog, Abp1, interacts with the initiation protein Cdc23 (MCM10) and is required for efficient DNA replication in fission yeast. Cell Div. 2006;1:27. doi: 10.1186/1747-1028-1-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee JK, Huberman JA, Hurwitz J. Purification and characterization of a CENP-B homologue protein that binds to the centromeric K-type repeat DNA of Schizosaccharomyces pombe. Proc Natl Acad Sci U S A. 1997;94:8427–32. doi: 10.1073/pnas.94.16.8427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baum M, Clarke L. Fission yeast homologs of human CENP-B have redundant functions affecting cell growth and chromosome segregation. Mol Cell Biol. 2000;20:2852–64. doi: 10.1128/mcb.20.8.2852-2864.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Irelan JT, Gutkin GI, Clarke L. Functional redundancies, distinct localizations and interactions among three fission yeast homologs of centromere protein-B. Genetics. 2001;157:1191–203. doi: 10.1093/genetics/157.3.1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Irvine DV, et al. Mapping epigenetic mutations in fission yeast using whole-genome next-generation sequencing. Genome Res. 2009;19:1077–83. doi: 10.1101/gr.089318.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Lahondes R, Ribes V, Arcangioli B. Fission yeast Sap1 protein is essential for chromosome stability. Eukaryot Cell. 2003;2:910–21. doi: 10.1128/EC.2.5.910-921.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mejia-Ramirez E, Sanchez-Gorostiaga A, Krimer DB, Schvartzman JB, Hernandez P. The mating type switch-activating protein Sap1 Is required for replication fork arrest at the rRNA genes of fission yeast. Mol Cell Biol. 2005;25:8755–61. doi: 10.1128/MCB.25.19.8755-8761.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Krings G, Bastia D. Sap1p binds to Ter1 at the ribosomal DNA of Schizosaccharomyces pombe and causes polar replication fork arrest. J Biol Chem. 2005;280:39135–42. doi: 10.1074/jbc.M508996200. [DOI] [PubMed] [Google Scholar]
- 12.Noguchi C, Noguchi E. Sap1 promotes the association of the replication fork protection complex with chromatin and is involved in the replication checkpoint in Schizosaccharomyces pombe. Genetics. 2007;175:553–66. doi: 10.1534/genetics.106.065334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Szilard RK, et al. Systematic identification of fragile sites via genome-wide location analysis of gamma-H2AX. Nat Struct Mol Biol. 2010;17:299–305. doi: 10.1038/nsmb.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aguilar-Arnal L, Marsellach FX, Azorin F. The fission yeast homologue of CENP-B, Abp1, regulates directionality of mating-type switching. Embo J. 2008;27:1029–38. doi: 10.1038/emboj.2008.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Arcangioli B, Klar AJ. A novel switch-activating site (SAS1) and its cognate binding factor (SAP1) required for efficient mat1 switching in Schizosaccharomyces pombe. Embo J. 1991;10:3025–32. doi: 10.1002/j.1460-2075.1991.tb07853.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Deshpande AM, Newlon CS. DNA replication fork pause sites dependent on transcription. Science. 1996;272:1030–3. doi: 10.1126/science.272.5264.1030. [DOI] [PubMed] [Google Scholar]
- 17.Ghazvini M, Ribes V, Arcangioli B. The essential DNA-binding protein sap1 of Schizosaccharomyces pombe contains two independent oligomerization interfaces that dictate the relative orientation of the DNA-binding domain. Mol Cell Biol. 1995;15:4939–46. doi: 10.1128/mcb.15.9.4939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guo Y, Levin HL. High-throughput sequencing of retrotransposon integration provides a saturated profile of target activity in Schizosaccharomyces pombe. Genome Res. 2010;20:239–48. doi: 10.1101/gr.099648.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Segurado M, Gomez M, Antequera F. Increased recombination intermediates and homologous integration hot spots at DNA replication origins. Mol Cell. 2002;10:907–16. doi: 10.1016/s1097-2765(02)00684-6. [DOI] [PubMed] [Google Scholar]
- 20.Meister P, et al. Nuclear factories for signalling and repairing DNA double strand breaks in living fission yeast. Nucleic Acids Res. 2003;31:5064–73. doi: 10.1093/nar/gkg719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sehgal A, Lee CY, Espenshade PJ. SREBP controls oxygen-dependent mobilization of retrotransposons in fission yeast. PLoS Genet. 2007;3:e131. doi: 10.1371/journal.pgen.0030131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kupiec M, Petes TD. Allelic and ectopic recombination between Ty elements in yeast. Genetics. 1988;119:549–59. doi: 10.1093/genetics/119.3.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gomez EB, Nugent RL, Laria S, Forsburg SL. Schizosaccharomyces pombe histone acetyltransferase Mst1 (KAT5) is an essential protein required for damage response and chromosome segregation. Genetics. 2008;179:757–71. doi: 10.1534/genetics.107.085779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang J, DM Association of the RENT complex with nontranscribed and coding regions of rDNA and a regional requirement for the replication fork block protein Fob1 in rDNA silencing. Genes Dev. 2003;17:2162–76. doi: 10.1101/gad.1108403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Roguev A, et al. Conservation and rewiring of functional modules revealed by an epistasis map in fission yeast. Science. 2008;322:405–10. doi: 10.1126/science.1162609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ton-Hoang B, et al. Single-Stranded DNA Transposition Is Coupled to Host Replication. Cell. 2010;142:398–408. doi: 10.1016/j.cell.2010.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wong LH, Choo KH. Evolutionary dynamics of transposable elements at the centromere. Trends Genet. 2004;20:611–6. doi: 10.1016/j.tig.2004.09.011. [DOI] [PubMed] [Google Scholar]
- 28.Putnam CD, Hayes TK, Kolodner RD. Specific pathways prevent duplication-mediated genome rearrangements. Nature. 2009;460:984–9. doi: 10.1038/nature08217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wallis JW, Chrebet G, Brodsky G, Rolfe M, Rothstein R. A hyper-recombination mutation in S. cerevisiae identifies a novel eukaryotic topoisomerase. Cell. 1989;58:409–19. doi: 10.1016/0092-8674(89)90855-6. [DOI] [PubMed] [Google Scholar]
- 30.Lemoine FJ, Degtyareva NP, Lobachev K, Petes TD. Chromosomal translocations in yeast induced by low levels of DNA polymerase a model for chromosome fragile sites. Cell. 2005;120:587–98. doi: 10.1016/j.cell.2004.12.039. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.