Abstract
The genome sequence of the enteric bacterial pathogen Campylobacter jejuni NCTC 11168 (11168-GS) was published in 2000, providing a valuable resource for the identification of C. jejuni-specific colonization and virulence factors. Surprisingly, the 11168-GS clone was subsequently found to colonize 1-day-old chicks following oral challenge very poorly compared to other strains. In contrast, we have found that the original clinical isolate from which 11168-GS was derived, 11168-O, is an excellent colonizer of chicks. Other marked phenotypic differences were also identified: 11168-O invaded and translocated through tissue culture cells far more efficiently and rapidly than 11168-GS, was significantly more motile, and displayed a different morphology. Serotyping, multiple high-resolution molecular genotyping procedures, and subtractive hybridization did not yield observable genetic differences between the variants, suggesting that they are clonal. However, microarray transcriptional profiling of these strains under microaerobic and severely oxygen-limited conditions revealed dramatic expression differences for several gene families. Many of the differences were in respiration and metabolism genes and operons, suggesting that adaptation to different oxygen tensions may influence colonization potential. This correlates biologically with our observation that anaerobically priming 11168-GS or aerobically passaging 11168-O caused an increase or decrease, respectively, in colonization compared to the parent strain. Expression differences were also observed for several flagellar genes and other less well-characterized genes that may participate in motility. Targeted sequencing of the sigma factors revealed specific DNA differences undetected by the other genomic methods. These observations highlight the capacity of C. jejuni to adapt to multiple environmental niches, the likelihood that this adaptation involves genetic evolution, and provides the first whole-genome molecular exploration of the effect of laboratory culture and storage on colonization and virulence properties of this pathogen.
Campylobacter jejuni is the most common cause of human acute bacterial enteritis in industrialized countries like the United States and United Kingdom (4, 10, 19). It is estimated that nearly 1 in 100 individuals in both the United States and the United Kingdom develop campylobacter-related illness each year (79), with acute symptoms ranging from watery to bloody diarrhea, fever, nausea, and vomiting. Although the vast majority of cases are self-limiting, severe consequences of this infection can arise, including bacteremia, arthropathies, and polyneuropathies like Guillain-Barré Syndrome (51, 65, 74).
Human campylobacteriosis is considered primarily a food-borne disease. C. jejuni is a common gut commensal in most food-producing animals and birds, and fecal contamination of meat during processing is a well-recognized route of transmission to humans. Epidemiological studies indicate that a major source of human infection is the consumption and handling of meat from broiler chickens (19). C. jejuni colonizes the cecum and colon of such birds readily and in extremely high numbers (54). Thus, strategies for the control and prevention of poultry colonization at the farm level are considered an important approach to the reduction or elimination of campylobacteriosis in humans.
The mechanisms by which C. jejuni colonizes the intestinal tract are poorly understood. Models of colonization in orally challenged chickens have enabled some colonization factors to be identified by using defined mutants; these factors include flagellin (78), superoxide dismutase (60), and GalE (20). However, such studies are highly dependent on the presumed properties of genes and thus do not generally lead to the identification of C. jejuni-specific colonization factors. Recently, with the advent of genome sequencing and the development of strategies, such as signature-tagged mutagenesis, for the mass screening of mutants, the identification of bacterial factors important for virulence has become more rapid and independent of known gene functions (9, 48, 62).
In 2000, the genome sequence of C. jejuni NCTC 11168 was completed (58). This appeared to provide a unique database for postgenomic investigations, including the potential for identification of C. jejuni-specific colonization factors. However, initial studies demonstrated that the genome-sequenced variant of NCTC 11168, unlike other C. jejuni strains, was deficient in its ability to colonize 1-day-old chicks (1) or SCID mice (28) following oral challenge. Previous reports have indicated that the virulence properties of some bacteria, including campylobacters, may be reduced by laboratory subculture over time (7, 17, 40, 52, 72). However, none of these studies identified discernible molecular changes or differences that might lend insight into possible mechanisms underlying this phenomenon.
C. jejuni NCTC 11168 was originally isolated from the feces of a diarrheic patient in 1977 by Martin Skirrow (http://cphl.phls.org.uk/divisions/cdmssd/nctc/qcrc.htm). Fortuitously, this original clinical isolate (strain number 5636/77), archived by Skirrow in 1977 and frozen ever since, was donated to the campylobacter strain collection of the Veterinary Laboratories Agency (VLA) in 1999. In stark contrast to the genome-sequenced variant, we have found that the original isolate is an excellent colonizer of chickens. To identify other potential phenotype differences and to explore possible mechanisms underlying the colonization disparity, we undertook a detailed phenotypic, genotypic, and transcriptional comparison of the genome-sequenced and original variants of C. jejuni NCTC 11168 (henceforth designated 11168-GS and 11168-O, respectively). Our findings include a likely role for bacterial metabolism and adaptation to different oxygen tensions in colonization potential. This study also highlights the remarkable capacity of C. jejuni to evolve rapidly as a result of storage, culture, and passage conditions. Finally, our work demonstrates the potential of microarray-based transcriptional profiling in screening for differences in bacterial variants that have adapted to new environments, probably via small but accumulative vertical evolutionary events, without having incurred more readily identifiable genomic changes such as gene loss, gene gain, or horizontal gene transfer.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
C. jejuni strain 11168-O (the original clinical isolate) was isolated from human diarrheic feces in 1977 by M. Skirrow and designated strain 5636/77 at that time. This strain was stored frozen, without known subculture, at −70°C to −80°C first at Worcester Royal Infirmary, then at VLA Luddington, and finally at VLA Weybridge. This strain was accessed into the National Collection of Type Cultures (NCTC) by Skirrow in 1977, where it was subcultured to provide a stock which was then freeze-dried. The strain was also probably distributed to a number of other international laboratories at that time. The genome-sequenced C. jejuni strain NCTC 11168 (11168-GS) was kindly provided by Brendan Wren. This clone was used for the Sanger Centre C. jejuni sequencing project and was previously obtained from the NCTC. The subculture history of this variant is unknown.
To avoid damage due to laboratory passage (particularly for 11168-O), C. jejuni strains were subcultured on plates no more than twice prior to initiating any experiment. The C. jejuni strains were either cultured on 10% (vol/vol) sheep blood agar plates with actidione (250 μg/ml) and Skirrow's supplement (10 μg of vancomycin/ml, 2.5 IU of polymyxin B/ml, and 5 μg of trimethoprim/ml) (BASA) at 42°C in a microaerobic environment (7.5% [vol/vol] CO2, 7.5% [vol/vol] O2, and 85% [vol/vol] N2) for 24 to 48 h or cultured on Mueller Hinton (MH) agar and in MH broth with 10 μg of vancomycin/ml and 5 μg of trimethoprim/ml at 37°C with an Oxoid CampyGen pack to generate microaerobic conditions (6% O2 and 12% CO2). To generate severely O2-limited conditions, bacteria were cultured with Oxoid AnaeroGen packs. According to the manufacturer's specifications, these packs result in an O2 environment of <1% within 30 min. Escherichia coli strain TOPO 10F′ (Invitrogen) was grown on Luria-Bertani agar or in Luria-Bertani broth at 37°C under atmospheric conditions. For one experiment, 11168-O was passaged aerobically on BASA plates. The plates were not dried prior to inoculation and were placed in an unsealed gas jar with a moist tissue at the bottom and incubated at 37°C for 24 to 48 h. This process was repeated 13 times. As a control, 11168-O was also passaged 13 times microaerobically, again on undried BASA plates at 37°C for 24 to 48 h. A CampyPak (BBL, Becton Dickinson) was used in a sealed gas jar to generate the microaerobic environment.
Chicken colonization model.
A 1-day-old chick model of colonization (78) was used to determine colonization potential. Briefly, eggs from specific-pathogen-free chickens (Lohmann's) were hatched in isolators. Groups of chicks (n = 10) were maintained in separate isolators with unlimited food and water. At 1 day old, chicks were each dosed orally by gavage with approximately 102 to 109 CFU of C. jejuni in 0.1 ml of phosphate-buffered saline (PBS). Bacteria were grown overnight on blood agar plates under microaerobic conditions at 42°C. Chicks were killed 5 days after dosing, and bacterial colonization levels were determined by plating out dilutions of cecal contents. The detection limit for colonization was 102 CFU/g of cecal content. Further colonization studies were carried out in which birds were dosed with (i) 11168-GS grown as above and then placed in anaerobic conditions at 37°C for 24 h by using an anaerobic cabinet and (ii) 11168-O that had been subcultured 13 times under aerobic conditions at 37°C.
In vitro phenotypic studies.
The morphologies of the two strains were compared by transmission electron microscopy (TEM). Briefly, bacteria were grown under microaerobic conditions, suspended in water, allowed to sediment onto Formvar-carbon-coated grids, negatively stained with 2% (wt/vol) phosphotungstic acid, and examined with a Phillips CM 10 TEM. Motility was measured by swarming on motility agar plates. Each strain was grown as described above, and the concentration was adjusted to approximately 108 CFU/ml in PBS. One microliter of this suspension was then stabbed into a 0.4% MH agar plate. Following microaerobic growth at 42°C for 24 to 48 h, the diameter of each halo of growth was measured.
Bacterial invasion into INT407 cells was studied by a gentamicin protection assay (6, 16, 42, 76) with confluent INT407 monolayers seeded 24 h prior to infection. The starting bacterial inoculum for each strain was ∼4 × 108 to 6 × 108 CFU. Invasion was assayed in minimum essential medium (MEM) without serum, with triplicate wells seeded for each strain for each time point. Bacteria were allowed to invade for 30 min or 2 h, then the cells were washed five times with prewarmed MEM prior to the addition of 250 μg of gentamicin/ml in MEM. After 2 h, cells were washed five times again with MEM and lysed with water and a 25-gauge syringe. For experiments in which the bacteria were first centrifuged onto the INT407 cells prior to initiating invasion, centrifugation was carried out at 450 × g for 15 min at room temperature. Translocation assays were carried out by using 18-day differentiated Caco-2 cells, grown in transwells (Corning Costar Corp.) with a pore size of 3 μm, as previously described (6, 42). Separate transwells were used for each time point, and the assay was performed in triplicate for each strain at each time point. The starting bacterial inoculum for each strain was ∼2.5 × 108 CFU. Translocation was performed in MEM plus 10% fetal bovine serum and allowed to proceed for 2 or 6 h. The entire basolateral fraction was collected, and bacteria were enumerated by serial dilution plate counts. Prior to initiating each assay, bacteria were inoculated in biphasic MH flasks (5 ml of broth over 10 ml of agar) at an optical density at 600 nm (OD600) of 0.002 and grown at 37°C microaerobically for ∼16 h to an OD600 of ∼0.3 before starting the experiment.
Electron microscopy of Caco-2 sections.
Polarized Caco-2 cells infected with C. jejuni were washed gently three times with PBS and fixed at 4°C for 90 min with 2% glutaraldehyde in 0.1 M sodium phosphate buffer, pH 7.4. Samples were postfixed in 1% OsO4 (Polysciences) for 90 min at 4°C and stained with 0.25% uranyl acetate (Polysciences) at 4°C for 1 h. After dehydration with a series of ethanol and water solutions, samples were embedded in firm Spurr's resin (Polysciences). Samples were then sectioned, stained with uranyl acetate and lead citrate, and examined on a Philips 201c electron microscope.
Molecular genotyping.
Amplified fragment length polymorphism (AFLP) was performed according to the method of Duim et al. (14). The final products were separated on a 7.3% denaturing acrylamide sequencing gel with an ABI 377 automated DNA sequencer. Pulsed-field gel electrophoresis (PFGE) was performed by adjusting a bacterial suspension to an OD500 of 0.5 in PBS prior to preparation of agar plugs. Cells were lysed by two consecutive 24-h incubations in lysis buffer and proteinase K at 55°C prior to digestion of genomic DNA with SmaI for 48 h. The DNA fragments were separated by using a Chef-DR 111 system (Bio-Rad) with a ramped pulse of 10 to 35 s for 21 h at 200 V and 14°C. Fla typing was performed as described previously (53).
Microarray-based genotyping (genomotyping) studies were performed by using a C. jejuni DNA microarray consisting of amplicons representing the largest nonoverlapping, nonhomologous regions of 99% of the open reading frames (ORFs) in the 11168-GS genome (44). Each array contains two spots per gene. Four hybridizations were performed, each of which used 11168-GS as a reference. Two hybridizations used 11168-GS as the test DNA (as a control for labeling and dye variability); two hybridizations used 11168-O as the test DNA. Hybridizations were performed essentially as described previously (44). Briefly, 500 ng of genomic DNA was incubated with amino-allyl dUTP (Sigma) and Klenow fragment (exo-) (NEB) at 37°C for 2 h to generate labeled probes. The probes were coupled to monofunctional Cy dyes (Amersham), with Cy3 for the reference dye and Cy5 for the test dye. Arrays were hybridized at 55°C for 16 h and scanned with an Axon 4000A scanner and GenePix software. Data were analyzed by using GACK (39) and Cluster (15).
Gene expression analyses.
After a single passage on MH plates, bacteria were inoculated into MH broth at an OD600 of 0.002 and grown microaerobically with the Oxoid CampyGen system at 37°C for ∼14 h with shaking at 200 rpm to mid-log phase (OD600 of ∼0.3). Bacteria were diluted back to an OD600 of 0.1, split into microaerobic or severely O2-limited cultures (with the CampyGen or AnaeroGen Oxoid system, respectively), and shaken at 200 rpm at 37°C in an airtight 2.5-liter BBL Campyjar. Starting cultures were 25 ml in a 150-ml flask. At various time points, 3-ml samples were harvested from the same flask to determine the OD600 and number of CFU per milliliter and to prepare RNA, after which the flask was returned to the jar with a fresh pack for further incubation. Flasks were exposed to atmospheric conditions for <30 s at each time point. For RNA preparation, samples were harvested into a 1/10 volume of 5% phenol-95% ethanol, spun, and frozen at −80°C. The samples were resuspended in 0.4 mg of lysozyme/ml, incubated at room temperature for 5 min, and lysed in a 20× volume of Trizol reagent (Invitrogen). Following chloroform extraction, the samples were combined with an equal volume of 70% ethanol, loaded onto an RNeasy Mini column (Qiagen), and processed according to manufacturer's procedures (including on-column DNase treatment). RNA was confirmed as DNA-free by reverse transcription (RT)-PCR. For hybridizations, RT was performed with 0.6 pmol of C. jejuni gene-specific primer mix (Sigma-Genosys) and 1.5 μg of C. jejuni RNA per hybridization. cDNA labeling and array hybridization were performed essentially as described above for genomic DNA labeling and in reference 44, except that the Klenow labeling and probe-generating reactions were performed for 16 h at 37°C prior to dye coupling. For the RNA-based hybridizations, reference cDNA was generated by performing RT reactions on a pool of RNA comprised of equimolar aliquots of RNA from each sample in the experiment. Probes generated from this reference cDNA were labeled with Cy3 and hybridized against probes generated from individual samples labeled with Cy5. Data were entered into the Stanford Microarray Database, and data were retrieved only for those spots exhibiting a regression correlation of >0.6 and which contained >75% good data. To identify spots with the most pronounced differences between the variants, we employed SAM (significance analysis of microarrays), a statistical program which uses repeated permutations of the data to determine whether the expression of any gene (or spot) is significantly related to the response (71). Spots were identified on the basis of an overall (i.e., over the four time points) >1.5× signal intensity difference and a low false-discovery rate. Because expression levels change over time, for a given spot, the signal intensity difference is likely to be much greater than 1.5× between the variants at certain time points and slightly less than 1.5× different at others (see Fig. 5C and D and 6C and D for examples). The spots identified were confirmed as significant by using ORF order and Cluster analyses to investigate operon organization and coexpression with known genes of likely similar function. The original data for the spots identified were mean centered and hierarchically clustered with Cluster and visualized by Treeview (15). Quantitative real-time RT-PCR (QRT-PCR) was performed with a Bio-Rad iCycler, primers designed to produce 70-bp amplimers, and recombinant Tth enzyme, buffer, and SYBR Green (Perkin-Elmer/Molecular Probes). Standard curves were generated for each primer pair in every reaction mixture. Reaction conditions were as follows: (i) RT, 50°C for 5 min and 60°C for 30 min; (ii) PCR, 50 cycles of 94°C for 10 s, 50°C for 30 s, and 60°C for 30 s; (iii) melt curve, 100 to 60°C in 1°C increments every 30 s.
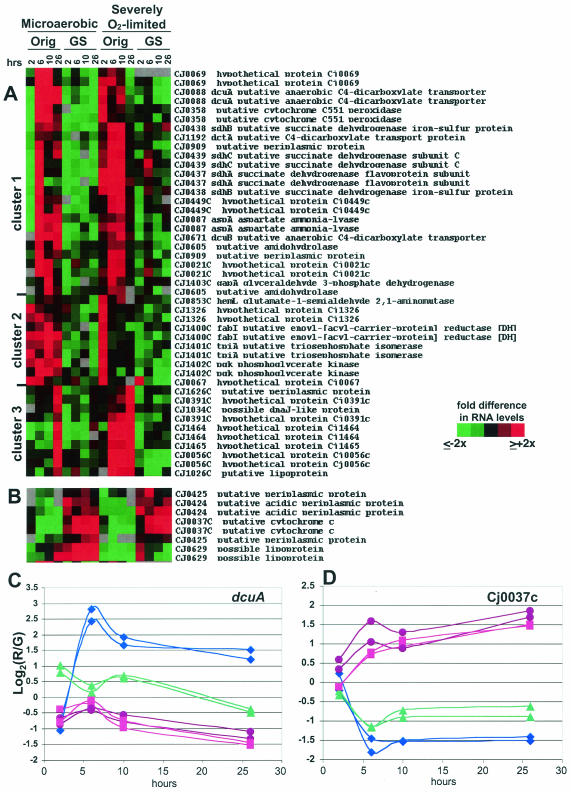
FIG. 5.
11168-O and 11168-GS cultured as described in the legend to Fig. 4 were harvested for RNA and gene expression analyses. SAM analysis was used to identify genes with statistically significant differences under both conditions. Panel A shows genes expressed more highly in 11168-O than in 11168-GS. Panel B shows genes expressed more highly in 11168-GS than in 11168-O. For both panels A and B, red indicates expression higher than the mean for each gene and green indicates lower expression than the mean (see key on figure). Panels C and D show numerical data, in log2 format, for both spots representing dcuA (C) and Cj0037c (D). For both panels C and D, the y axis is the log2 of the red/green (R/G) ratio relative to the mean for that gene; the actual difference (n-fold) from the mean for any particular data point is 2A, where A is the number on the y axis (i.e., if the y axis value is 1.5, the actual R/G level, or difference [n-fold] from the mean, is 21.5, or ∼3.7). Also for panels C and D, purple circles are microaerobic 11168-GS; blue diamonds are microaerobic 11168-O; pink squares are severely O2-limited 11168-GS; and green triangles are severely O2-limited 11168-O.
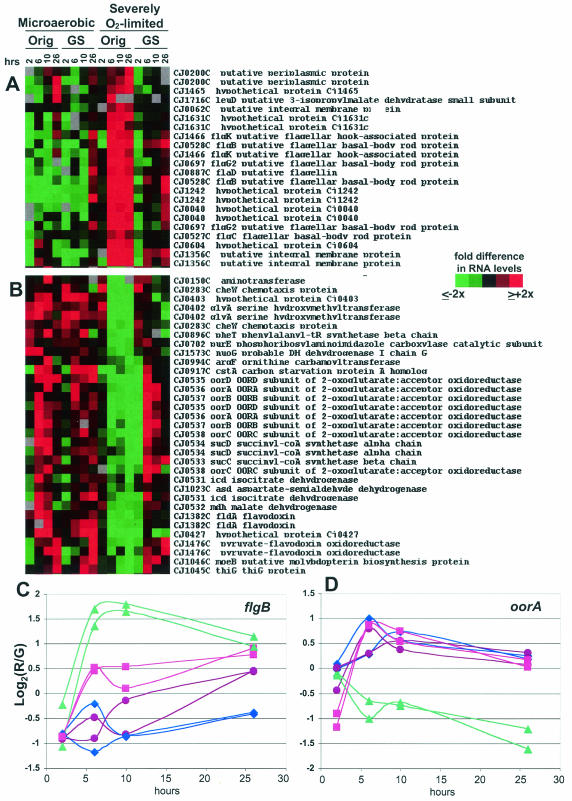
FIG. 6.
11168-O and 11168-GS cultured as described in the legend to Fig. 4 were harvested for RNA and gene expression analyses. SAM analysis was used to identify genes with statistically significant differences between the variants only under anaerobic conditions. Panel A shows genes expressed more highly in 11168-O (anaerobic) than in 11168-GS (anaerobic). Panel B shows genes expressed more highly in 11168-GS (anaerobic) than in 11168-O (anaerobic). For both panels A and B, red indicates expression higher than the mean for each gene and green indicates lower expression than the mean (see key on figure). Panels C and D show numerical data, in log2 format, for both spots representing flgB (C) and oorA (D). For both panels C and D, the y axis is the log2 of the red/green (R/G) ratio relative to the mean for that gene; the actual difference (n-fold) from the mean for any particular data point is 2A, where A is the number on the y axis (i.e., if the y axis value is 1.5, the actual R/G level, or difference [n-fold] from the mean, is 21.5, or ∼3.7). Also for panels C and D, purple circles are microaerobic 11168-GS; blue diamonds are microaerobic 11168-O; pink squares are severely O2-limited 11168-GS; and green triangles are severely O2-limited 11168-O.
DNA sequencing.
The three sigma factor genes of C. jejuni (rpoD, rpoN, and fliA) were amplified from 11168-O with primers derived from the NCTC 11168 genome sequence (58). The amplicons were cloned into the pCR-2.1TOPO vector (Invitrogen) and sequenced with M13 forward and reverse primers. Two separate clones for each sigma factor were sequenced at least three times in both directions. Sequencing was carried out with BigDye Terminator mix (Applied Biosystems), and the products were separated with an ABI 3700 DNA sequencer (Applied Biosystems). Contigs were assembled by using the SeqMan program (Lasergene; DNAstar).
Subtractive hybridization.
The method used for subtractive hybridization was similar to that described previously (1), except that both primary and secondary hybridizations were carried out at 58°C. Subtracted products were cloned into pCR-2.1TOPO, and a selection, based on the dot blot results, were sequenced with M13 forward and reverse primers.
RESULTS
Significant differences in chick colonization of the 11168 variants.
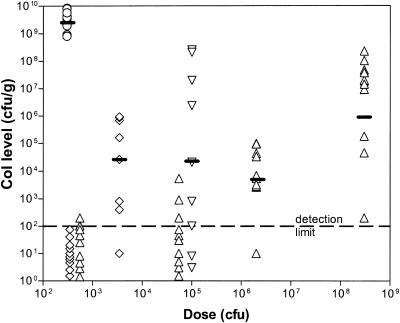
To investigate the colonization potential of the variants, 1-day-old chicks were dosed with approximately 102 to 109CFU of C. jejuni and colonization levels were determined 5 days later (Fig. 1). The resultant data clearly show that 11168-O colonizes much more efficiently than 11168-GS. For instance, a relatively low dose of 3 × 102 CFU of 11168-O maximally colonized all birds (108 to 1010 CFU/g of cecal contents), with a geometric mean level of 3 × 109 CFU/g. In contrast, a similar dose of 11168-GS (5 × 102 CFU) only detectably colonized 3 of 10 birds, all at a very low level of 102 CFU/g, and even a very high dose of 3 × 108 CFU only gave a geometric mean colonization level of 106 CFU/g. 11168-O that had been passaged aerobically 13 times was significantly attenuated for colonization, although it still colonized slightly better than 11168-GS. Specifically, a dose of 4 × 102 CFU of the aerobically passaged 11168-O did not colonize to a detectable level, and a dose of 3.5 × 103 CFU gave a mean colonization level of 3 × 104 CFU/g. Conversely, exposing 11168-GS to anaerobic conditions for 24 h prior to inoculation increased its colonization potential, although not to the level of 11168-O grown microaerobically. A dose of 105 CFU of the anaerobically primed 11168-GS colonized 8 of 10 birds to a geometric mean level of 2.5 × 105 CFU/g, whereas doses of 5.5 × 104 and 2 × 106 CFU of 11168-GS grown microaerobically colonized 7 of 10 and 9 of 10 birds to mean levels of 7 × 100 and 5 × 103 CFU/g, respectively.
FIG. 1.
Differences in chick colonization of C. jejuni strain 11168 variants. One-day-old chicks were inoculated with various doses of bacteria (x axis). Numbers of CFU per gram of cecal content were calculated for each chick by serial dilution and plotted on the y axis. Chicks were dosed with C. jejuni as follows: open circles are 11168-O, microaerobically grown; upright open triangles are 11168-GS, microaerobically grown; open diamonds are 11168-O, aerobically passaged 13 times; inverted open triangles are 11168-GS, anaerobically primed for 24 h. The dashes indicate the geometric means for each strain at the given dose.
11168-GS and 11168-O exhibit dramatic differences in morphology, motility, invasion, and translocation.
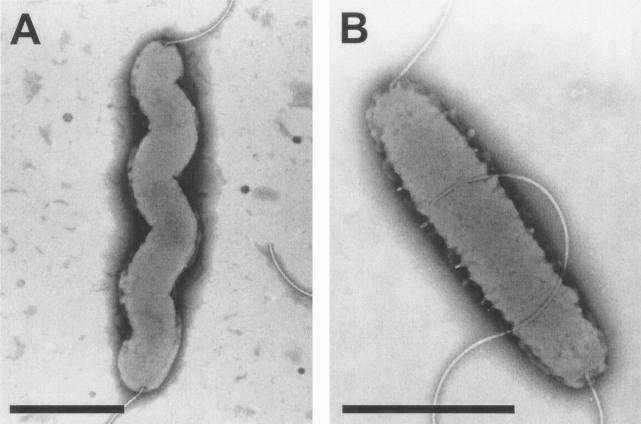
To determine whether other colonization- and virulence-associated attributes of C. jejuni might also be altered in 11168-GS compared to 11168-O, we next performed a detailed phenotypic characterization of the two strains. The distinct morphologies of the variants are shown in Fig. 2. 11168-O has the classic spiral shape generally expected of C. jejuni, whereas 11168-GS has lost this shape and appears as a straight rod. Both variants have polar flagella. However, 11168-GS was significantly impaired for motility both in soft agar and in liquid broth culture as observed by light microscopy, whereas 11168-O was highly motile in soft agar and exhibited extremely rapid darting motility from a liquid broth culture (Table 1). Two virulence-associated phenotypes were also assayed: (i) invasion into INT407 cells and (ii) translocation through differentiated Caco-2 cells. 11168-O invaded INT407 cells far more rapidly and at significantly higher numbers than 11168-GS (P < 0.05). After just 30 min, a 280-fold difference in invasion was observed, and after 2 h of invasion, a 3-fold difference was still apparent (Table 1). Invasion assays in which the bacteria were first centrifuged onto the INT407 monolayers also resulted in 11168-O invading at a significantly higher frequency than 11168-GS (P < 0.05) (data not shown). Similarly, 11168-O rapidly translocated through differentiated Caco-2 monolayers while 11168-GS was essentially unable to translocate (Table 1). Interestingly, thin sectioning of the infected Caco-2 monolayers followed by electron microscopy indicated that 11168-O bacterial cells were readily observed both inside and between the Caco-2 cells (Fig. 3). In these sections, which were taken 2 h after inoculation (insufficient time for >1 round of intracellular replication), multiple bacteria were often found within a single cell, suggesting that several invasion events may have occurred at once and/or at one location. Furthermore, bacteria were often observed at likely intracellular sites very near the intercellular spaces. Very few 11168-GS bacteria were observed between the Caco-2 cells (data not shown); the number of internalized bacteria observed by electron microscopy is shown in Table 1.
FIG. 2.
Morphology of the 11168 variants, as demonstrated by negative staining. (A) 11168-O; (B) 11168-GS. Bars, ∼1 μm.
TABLE 1.
Invasion, translocation, and motility phenotypes of the 11168 variants
| Phenotype | Time | Result for strain:
|
Fold difference | |
|---|---|---|---|---|
| 11168-O | 11168-GS | |||
| Invasion in INT407 cellsa | 30 min | 2,800 (793)f | 10 (0) | 280 |
| 2 h | 14,000 (5,291) | 4,533 (642) | 3 | |
| Translocation in Caco-2 cellsb | 2 h | 33,000 (19,798) | 25 (7) | 1,320 |
| 6 h | 68,500 (4,949) | 0 (0) | ≥68,500 | |
| No. of bacteria/cell in Caco-2 cellsc | 2 h | 1.48 | 0.36 | 4.1 |
| 6 h | 1.04 | 0.31 | 3.6 | |
| Motility | ||||
| Swarm agard | 24 h | 21.0 mm (2.7) | 0 (0) | NAg |
| Liquid culturee | 14 h | +++ | − | NA |
Total number of bacteria internalized (similar inocula).
Total number of bacteria harvested from the basolateral side of transwell (similar inocula).
Number of bacteria/cell observed inside or between cells, enumerated by random observation of 26 to 50 cells in 2 TEM fields.
Halo size on swarm agar plates.
Early-log-phase cultures assayed by light microscopy. +++, very rapid, darting motility; −, apparent Brownian motion only.
Numbers in parenthesis indicate standard deviations.
NA, not applicable.
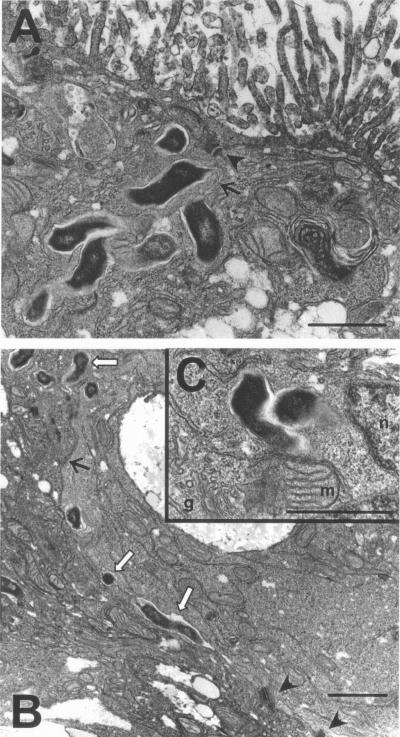
FIG. 3.
TEMs of 11168-O invading and translocating through polarized differentiated Caco-2 cells. After 2 h of infection, cells were fixed, thin sectioned, and prepared for electron microscopy. Arrowheads point out desmosomes connecting the cells; line arrows show the line of the intercellular space. Bacteria are the dark, electron-dense structures. In panel B, the hollow arrows point to bacteria that are in the intercellular space. Other bacteria in panels A and B could either be between the cells or internalized near the intercellular space. Panel C shows bacteria clearly inside cells near several organelles (g, Golgi; m, mitochondria; n, nucleus). Bars, ∼10 μm.
11168-GS and 11168-O appear identical by current molecular genotyping techniques.
We next undertook extensive serotyping and molecular genotyping analyses of the 11168 variants, with the dual goals of (i) exploring the genetic relationship between these strains and (ii) identifying possible genetic bases underlying the dramatic phenotype differences observed. Results of these studies are shown in Table 2. Both variants were found to be biotype 1, Penner serotype 2, Fla-type 1.1, and sequence type 43 (ST43) by multilocus sequence typing. Neither strain contained a plasmid. Two well-established molecular genotyping techniques, AFLP and PFGE, also yielded identical profiles for the two variants. Genomotyping studies with an ORF-specific C. jejuni DNA microarray followed by GACK analysis of the resultant data (39) also did not identify any genes that were significantly different between the two strains. Furthermore, cluster analyses comparing the microarray profiles of 11168-GS and 11168-O to those of a panel of other isolates (44) resulted in the 11168 variants definitively clustering together and away from the other isolates. Finally, subtractive hybridization, which was recently used to identify genes present in another highly colonizing C. jejuni strain compared to 11168-GS (1), did not reveal any differences between the 11168 variants. Together, these studies indicate that 11168-O and 11168-GS appear clonal by all typing methods employed and that 11168-GS is not likely to have acquired or lost large pieces of DNA or been subject to horizontal gene transfer during passage before, through, or after accession into the NCTC.
TABLE 2.
11168-O and 11168-GS appear identical by multiple serotyping and molecular genotyping methods
| Method | Result for:
|
||
|---|---|---|---|
| 11168-O | 11168-GS | ||
| Biotype | 1 | 1 | |
| Penner serotype | 2 | 2 | |
| Fla type | 1.1 | 1.1 | |
| MLSTa | ST43 | ST43 | |
| Plasmid analysis | No plasmids | No plasmids | |
| AFLP | Identical profile
|
||
| PFGE | Identical profile
|
||
| Subtractive hybridization | No differences
|
||
| Microarray genomotyping | |||
| 11168-O vs 11168-GSb | No variant genes
|
||
| 11168-O and 11168-GS vs clinical isolatesc | 11168 strains cocluster away from other isolates | ||
Transcriptional profiling of 11168-O versus 11168-GS reveals multiple differences in respiration and metabolism and flagellar and motility gene expression.
Because we were unable to detect genetic differences by the typing techniques above, we next used the C. jejuni DNA microarray to explore gene expression patterns in the 11168 variants. Initial microarray experiments tracking expression profiles of the variants over several growth stages indicated that a surprising number of genes and operons exhibited significant expression differences between the variants, with a large proportion of those genes being involved in aerobic and anaerobic respiration and metabolism (data not shown). C. jejuni is considered an obligate microaerophile; however, it must be able to survive within a variety of environmental niches, including the microaerobic and often severely oxygen (O2)-limited environment of the avian and mammalian gut. Thus, we expanded the initial experiment to investigate expression differences between the two variants over a time course of exposure to both microaerobic and severely O2-limited culture conditions. To cover growth stages ranging from early log to stationary, samples were collected 2, 6, 10, and 26 h after dilution back to an OD600 of 0.1. Microaerobic and severely O2-limited conditions were established by using the Oxoid CampyGen and AnaeroGen systems, respectively (see Materials and Methods).
Growth data and growth stage and gene expression controls.
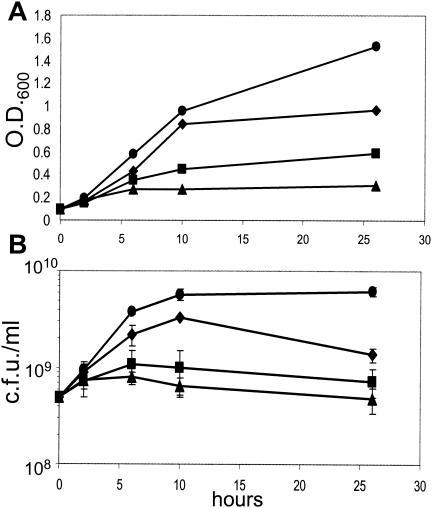
OD600 and CFU/ml data indicated that during microaerobic culture, 11168-GS grew faster, reached a higher number of CFU per milliliter at stationary phase, and maintained a high level of viability for a longer period of time than 11168-O (Fig. 4). Under severely O2-limited conditions, neither variant grew well, as expected given previous reports on the inability of C. jejuni to grow during anoxia (64). Nevertheless, both variants retained a significant proportion of viable cells at the 26-h time point under both conditions.
FIG. 4.
Growth data for 11168-O and 11168-GS cultured for 26 h microaerobically or under severe O2 limitation. At the 0-h time point, overnight mid-log cultures were diluted back to an OD600 of 0.1 and samples were taken for CFU analysis. The OD600 (A) and number of CFU per milliliter (B) were determined at 2, 6, 10, and 26 h after back dilution. Circles are microaerobic 11168-GS; diamonds are microaerobic 11168-O; squares are severely O2-limited 11168-GS; and triangles are severely O2-limited 11168-O.
Because the experiment was performed as a time course, known growth stage-dependent genes could be monitored to determine the relative likelihood that expression changes in interesting genes might be due to growth stage rather than valid differences between the variants. Separate pairwise whole-genome comparisons of the variants under each condition revealed that despite the growth differences noted above, the two variants exhibited remarkably similar growth stage transitions (data not shown). For instance, in both variants, expression of ribosomal and DNA synthesis genes peaked at 2 h and declined steadily thereafter; conversely, groEL, groES, and several heat shock genes were expressed at low levels during exponential growth but increased dramatically at the 26-h time point. Genes identified as growth stage dependent in other experiments (E. C. Gaynor and S. Falkow, unpublished data) were also similar; for instance, expression of the surface antigens peb1 and pebC peaked at the 6- and 10-h time points in both variants under both conditions. Thus, the differences in gene expression between the variants were considered to be valid and not due to growth stage discrepancies. We also performed QRT-PCR on several of the genes highlighted in Fig. 5 and 6 (i.e., dcuA, Cj1464, and Cj0037c), using RNA from both strains under each condition tested. In every case, both microarray analyses and QRT-PCR yielded nearly identical strain-to-strain and condition-to-condition differences in gene expression levels (data not shown). It is also of note that many of the genes identified as exhibiting differential expression between the variants fall into discrete coregulated operons. Together, these observations provide a high degree of confidence in the validity of the microarray expression trends described below.
Gene expression data.
Using the time points as technical replicates, we first employed SAM (see Materials and Methods) to identify genes that differed significantly between the variants regardless of culture condition (Fig. 5). Whole-genome analyses indicated that some differences observed only under severe O2 limitation were not identified by this method; thus, we also used SAM to identify genes with significant differences specifically under these conditions (Fig. 6). In both Fig. 5 and 6, panel A shows genes with higher expression in 11168-O compared to 11168-GS, with the log2 of the red/green ratios shown numerically for one example in panel C. Panel B shows genes with lower expression in 11168-O than in 11168-GS, with the log2 of the red/green ratios shown numerically for one example in panel D. Where possible, both array spots for a given gene are shown (i.e., when both spots passed the filtering and SAM criteria).
(i) Respiration and metabolism clusters. (a) TCA cycle, C4-dicarboxylate, and electron transport chain genes.
The tricarboxylic acid (TCA) cycle plays a dual role in cell metabolism: it generates biosynthetic starting compounds that are used in amino acid metabolism and it provides a metabolic energy source by creating intermediates that feed into the electron transport chain. In facultative anaerobes like E. coli, the TCA cycle is primarily oxidative during aerobic growth while during anaerobiosis it branches into oxidative (biosynthetic) and reductive (energy-generating) arms. Half of the TCA cycle is comprised of C4-dicarboxylic intermediates (e.g., fumarate, succinate, malate, and aspartate [during anaerobiosis]); the other half is comprised of classic biosynthetic starting compounds (e.g., α-ketoglutarate, succinyl-coenzyme A [succinyl-CoA], and oxaloacetate). The genome sequence of C. jejuni suggests that it contains a complete TCA cycle and a complex, branched electron transport chain that is likely to allow both aerobic and anaerobic respiration with a variety of electron acceptors (38, 58, 66). Apart from the genome information and a few studies on enzyme activity (64, 67), little is known about the TCA cycle in C. jejuni.
Several genes likely to be involved in uptake or metabolism of C4-dicarboxylate TCA cycle intermediates were expressed at significantly higher levels in 11168-O than in 11168-GS under both culture conditions (Fig. 5A, cluster 1). One of these genes, dcuA, is also shown in Fig. 5C, where a >9-fold difference in signal intensity is observed at the 6-h time point for 11168-O (microaerobic) compared to 11168-GS (either condition). dcuA and dcuB have been characterized in other bacteria as mediating anaerobic succinate-fumarate antiport and succinate uptake for use as a carbon source, whereas dctA catalyzes aerobic uptake of succinate (34). aspA, in a two-gene operon with dcuA, encodes an aspartate ammonia-lyase that catalyzes the conversion of aspartate to fumarate, which can feed into the anaerobic reductive branch of the TCA cycle. Up-regulation of several of these genes in 11168-O, although faster under severely O2-limited conditions, was somewhat higher during microaerobic growth. This cluster also contains an operon (sdhABC) annotated as encoding succinate dehydrogenase, an enzyme that has been well-characterized in E. coli and several other bacteria as a key component of the aerobic oxidative TCA cycle that converts succinate to fumarate and also connects the TCA cycle with the aerobic respiratory chain (8, 64). In those bacteria, succinate dehydrogenase is repressed during anaerobiosis so that the reverse reaction, reduction of fumarate to succinate by fumarate reductase, can prevail. In our experiments, expression of the frdCAB operon likely to encode fumarate reductase was modestly higher in 11168-O than in 11168-GS but only at the 6- and 10-h time points under microaerobic conditions (data not shown). Other interesting genes in cluster 1 of Fig. 5A include Cj0358, which has been suggested to be an oxygen-independent terminal oxidase in the C. jejuni electron transport chain (73), and Cj0069 and Cj0021c, which are annotated as hypothetical proteins but contain conserved domains potentially involved in glutathione synthesis and coenzyme modification and conversion of tyrosine to fumarate and acetoacetate, respectively.
In contrast, several other TCA cycle and putative electron transport chain genes exhibited significantly lower expression levels in 11168-O than in 11168-GS but only under severely O2-limited conditions (Fig. 6B). Eight of these genes are contained in a single operon (Cj0531 to Cj0538). The first four of these genes encode homologs of enzymes that participate in the more biosynthetic intermediate-generating half of the TCA cycle: the conversion of isocitrate to α-ketoglutarate (icd), the interconversion of succinate and succinyl-CoA (sucCD), and the interconversion of malate and oxaloacetate (mdh). Several other genes in the Fig. 6B cluster are likely to be involved in general biosynthetic processes, e.g., glyA, pheT, purE, moeB, and thiG. This cluster also contains genes likely to encode two oxygen-labile oxidoreductases that have been characterized in Helicobacter pylori as mediating electron transport to NADP (29, 30) (see also Fig. 6D). OOR, a 2-oxoglutarate-acceptor oxidorecuctase, interconverts 2-oxoglutarate and succinyl-CoA using a ferredoxin; POR, a pyruvate-flavodoxin oxidoreductase, acts together with the fldA flavodoxin (Fig. 6B) to convert pyruvate to acetyl-CoA.
(b) Gluconeogenesis genes.
C. jejuni cannot use glucose as a primary carbon source; however, genome sequence and experimental data indicate that C. jejuni possesses an Embden-Meyerhof (glycolysis and gluconeogenesis) pathway that is likely to be largely gluconeogenic (49, 58, 73). C. jejuni requires gluconeogenesis to generate glucose-derived polysaccharides and can also incorporate glycolysis end products like pyruvate into the TCA cycle via anaplerotic enzymes like pyruvate carboxylase (49, 73). A four-gene operon (Cj1403c to Cj1400c) likely to encode three key bidirectional enzymes of the glycolysis-gluconeogenesis pathway (gapA, pgk, and tpiA) and one fatty acid synthesis gene (fabI) were expressed at significantly higher levels in 11168-O than in 11168-GS (Fig. 5A). Most of these genes fall into cluster 2 of Fig. 5A and were expressed even more highly under microaerobic conditions. The expression of other genes in this pathway (e.g., pgm, eno) (73) was similar in both variants (data not shown).
(ii) Flagellar export, motility, and other gene clusters.
A number of likely flagellar export apparatus genes were expressed significantly more highly in 11168-O under severely O2-limited conditions than in any of the other samples (Fig. 6A). This cluster contains several putative hook and basal body structures and one putative flagellin. flgB, for example, exhibits an ∼7-fold-higher signal intensity in 11168-O at the 6-h time point under severely O2-limited conditions than in either strain cultured microaerobically (Fig. 6C). Other flagellar genes and the major flagellins flaA and flaB were expressed at similar levels in the two variants (data not shown). Figure 6A also contains one spot representing Cj1465; the three other spots representing Cj1464 and Cj1465 are found in Fig. 5A, cluster 3. As with the flagellar genes in Fig. 6A, these genes were expressed much more highly in 11168-O during severe O2 limitation and fall into a likely operon (Cj1462 to Cj1466) with flgK (Cj1466), flgI (Cj1462, a putative flagellar P-ring protein), and Cj1463. Although flgI and Cj1463 did not pass the SAM cutoff employed, both spots for each gene exhibited expression profiles similar to those of the other genes in the operon (data not shown). Cj1463, Cj1464, and Cj1465 have no homology to any other proteins and no conserved domains. Several other genes also found in cluster 3 of Fig. 5A also have no known homologies or conserved domains (e.g., Cj0056c and Cj0931c) but were strikingly coexpressed with Cj1464 and Cj1465. Other interesting genes with higher expression in 11168-O under severely O2-limited conditions (Fig. 6A) include Cj0062c, which was recently shown to play a role in motility (27), Cj1631c, which contains an EF hand Ca2+ binding domain, and Cj0604, a close homolog of the recently identified ppk2 gene from Pseudomonas aeruginosa (31, 80), a polyphosphate kinase that preferentially synthesizes GTP from polyphosphate.
Finally, a number of genes were expressed at significantly higher levels in 11168-GS than in 11168-O under both culture conditions (Fig. 5B and D). Apart from the putative cytochrome C (Cj0037c), which exhibits a >10-fold-higher signal intensity in 11168-GS (microaerobic) than 11168-O (microaerobic) at the 6- and 26-h time points (Fig. 5D), none of these genes have homology to any other translated ORFs in the National Center for Biotechnology Information microbial genome database.
DNA sequencing of the three major sigma factors reveals amino acid differences between the two variants.
Although many of the genes exhibiting significant expression differences are functionally related, they are physically scattered throughout the 11168 genome, suggesting that the two variants may possess differences in global regulatory molecules. To investigate this, and to explore the relative frequency with which single-nucleotide polymorphisms (SNPs) may occur between the otherwise clonal variants, we sequenced the three C. jejuni sigma factors from 11168-O and compared them with the known 11168-GS sequences. We found that each sigma factor contains SNPs resulting in at least one amino acid change in 11168-GS relative to 11168-O (Table 3). There are two amino acid changes in the RpoN sequence. The serine at amino acid 84 in 11168-O is replaced by a glycine in 11168-GS, and the lysine at amino acid 411 in 11168-O is replaced by an asparagine in 11168-GS. RpoD and FliA each have a single amino acid change. In RpoD, the alanine at amino acid 40 in 11168-O is replaced by a threonine in 11168-GS, and in FliA, the threonine at amino acid 91 in 11168-O is replaced by a methionine in 11168-GS.
TABLE 3.
Sigma factor differences in the 11168 variants
| Sigma factor | Position (aa)a | Amino acid for:
|
|
|---|---|---|---|
| 11168-O | 11168-GS | ||
| RpoN | 84 | Serine | Glycine |
| 411 | Lysine | Asparagine | |
| RpoD | 40 | Alanine | Threonine |
| FliA | 91 | Threonine | Methionine |
aa, amino acid.
DISCUSSION
The culture and storage conditions for any organism studied in a laboratory clearly have the potential to affect its normal biological processes. For organisms such as bacterial pathogens, where gaining an increased understanding of the pathogen's virulence is often the research focus, this is a particularly acute issue. Investigations of clinical versus sequenced and/or extensively passaged laboratory strains of several well-studied bacterial pathogens such as E. coli O157:H7 versus K-12, Mycobacterium tuberculosis CDC1551 versus H37Rv, and Mycobacterium bovis BCG vaccine strains have identified large genetic polymorphisms that are readily discernible by many genotyping techniques (3, 12, 22, 25, 45). In these instances, gene acquisition, gene loss, and/or horizontal gene transfer likely account for many of the phenotype differences, although subsequent whole-genome sequence analyses of several of these strains also identified SNPs throughout the genome (18, 59). Genetic diversity among strains, isolates, and serotypes of C. jejuni can also be detected by molecular genotyping methods (13, 41, 43, 44), as can the presence of plasmids (2) and instances of horizontal recombination (5). Other studies specifically investigating the effects of laboratory passage or culture condition on virulence phenotypes have been undertaken for several pathogenic bacteria; examples include H. pylori (40), Burkholderia pseudomallei (72), and Actinobacillus actinomycetemcomitans (17). C. jejuni has also been shown to be susceptible to genetic and phenotypic changes as a consequence of environmental conditions (7, 26, 52, 77). Although a few of these studies identified large-scale genetic differences between laboratory-passaged, clinical, and environmental isolates, most were unable to identify changes in specific genes and/or biological processes that might account for the gain or loss of virulence-associated phenotypes, particularly when those phenotypes arose as a result of vertical evolution rather than gene loss, gene gain, or horizontal transfer.
In this work, we report that the genome-sequenced variant of C. jejuni NCTC 11168 (11168-GS) and the original clinical isolate from which it was derived (11168-O) exhibit dramatic differences in numerous virulence-associated phenotypes, including colonization, invasion, translocation, and motility. Despite these differences, these clonally derived strains appear indistinguishable by multiple high-resolution molecular genotyping techniques, although targeted sequencing detected SNPs in each sigma factor. Unlike previous studies on C. jejuni attenuation or gain of virulence associated phenotypes due to culture condition, the availability of the precise DNA sequence of 11168-GS, together with technical advances such as microarray technology, has allowed us to take a whole-genome approach that includes transcriptional profiling as a platform for exploring the molecular and biological bases of the phenotypic alterations. The identification of several unexpected differences in gene expression has provided a heretofore unavailable window of insight into what might have changed in these variants and perhaps also how and why these changes occurred.
In nature, C. jejuni encounters a range of environments. Only in the intestinal tracts of temperate animal species does this organism naturally grow. However, long-term survival can occur in such diverse environments as fecal matter, water, and retail meats. These environments encompass a range of nutrient availability and oxygen tension, from largely aerobic to microaerobic to largely anaerobic. C. jejuni possesses genes and pathways that can allow survival and often growth under many different oxygen tensions (58, 64, 66). Efficient regulation of these genes and pathways is thus vital to the success of this organism in traversing these different niches. As 11168-O became 11168-GS, it was frozen, passaged in the laboratory, freeze-dried, and then passaged in the laboratory again. Our genotyping data strongly suggest that the original and genome-sequenced variants are clonal and that 11168-GS evolved vertically from 11168-O. Thus, it is likely that selective pressure to survive and thrive under laboratory growth and culture conditions (possibly including exposure to a largely aerobic environment) resulted in small genetic changes that allowed more robust in vitro growth (Fig. 4) while concomitantly compromising the strain's ability to readapt readily to the environment of the intestinal tract (Fig. 1).
Indeed, our gene expression and animal data suggest that the ability of C. jejuni to shift its metabolism and respiration strategies to accommodate the lower-oxygen microaerobic and largely anaerobic environments encountered in the gut is likely to be important for in vivo survival and colonization. Figure 6 clearly illustrates that many genes exhibit differential expression in 11168-O but not 11168-GS as a result of severe O2 limitation, suggesting that 11168-O may be better able to sense and respond to such an environment than 11168-GS. Many of the affected genes are involved in respiration and metabolism. For instance, the expression of several TCA cycle enzymes likely to be involved in generating biosynthetic starting compounds (e.g., icd, mdh, and sucCD) was lower in 11168-O cultured under severe O2 limitation than in any of the other samples, as was expression of several putative biosynthesis genes (Fig. 6B). In contrast, a number of genes likely to be involved in transport and metabolism of C4-dicarboxylates were expressed much more highly in 11168-O than 11168-GS under both conditions tested (Fig. 5A); many of these genes have also been characterized as participating in anaerobic metabolism (e.g., dcuA, dcuB, aspA, and Cj0358). It is thus possible that, upon sensing oxygen depletion, 11168-O (but not 11168-GS) efficiently shifts the TCA cycle towards transport, production, and utilization of C4-dicarboxylate intermediates at the expense of other TCA intermediates and biosynthetic processes. This may have important implications for energy generation during intestinal colonization. The putative OOR and POR complexes and the POR-associated flavodoxin also exhibited a relative down-regulation in 11168-O cultured under severe O2 limitation (Fig. 6B and D). Both enzymes are oxygen labile in other bacteria; if transcription of their genes is closely linked to protein levels in the cell, 11168-O may down-regulate these genes by sensing and responding either to their presence or to the O2 environment. Finally, several genes likely to operate in both directions of the gluconeogenesis-glycolysis pathway were preferentially up-regulated in 11168-O under microaerobic conditions (Fig. 5A). Glycolysis-gluconeogenesis and TCA cycle enzymes have been shown to participate in colonization and virulence in several other bacteria (47, 68, 69); as such, exploration of these pathways in C. jejuni should yield new insight into colonization requirements for this pathogen.
The chick colonization data presented in Fig. 1 also support our hypothesis that adaptation to different O2-containing environments is a crucial and heretofore unappreciated colonization determinant for C. jejuni. Previous work indicated that aerobically adapted C. jejuni can colonize mice; however, in that study, not every animal was colonized and colonization levels were not presented (35). In our experiments, repeated aerobic passage of 11168-O significantly attenuated its ability to colonize chicks, whereas anaerobically priming 11168-GS for 24 h prior to infection significantly increased its colonization potential (Fig. 1). The fact that neither strain could be induced to mimic the other completely in colonization potential indicates that the changes incurred at the genetic level as 11168-O became 11168-GS are more significant than can be rescued solely by preadapting each strain to a different oxygen tension. Indeed, targeted sequencing demonstrated that the 11168-GS variant has undergone genetic changes in all three sigma factors, possibly as a result of selective pressure (Table 3). The functional consequences of these changes remain to be established. However, these investigations will need to be closely monitored, since as the evidence in this study demonstrates, genetic manipulations to test the outcome of single amino acid changes in a highly plastic organism like C. jejuni may well invoke additional undocumented genetic alterations. Furthermore, given the abundance of differences between these variants and the relative frequency with which SNPs have already been identified, it is possible that other regulatory molecules such as two-component systems might also be altered, that specific promoter regions might have undergone change, and/or that a combination of SNPs are responsible for the transcriptional and phenotype variations presented. Significant further work will be required to dissect this issue.
As noted in Results, expression patterns for the sdh and frd operons are somewhat paradoxical given the abundance of other data suggesting that 11168-GS is better adapted for higher-O2 laboratory culture and 11168-O is better adapted for the very low-O2 environment of the intestinal tract. In E. coli, succinate dehydrogenase is only present in aerobically growing cells; during anaerobiosis, the sdh genes are strongly repressed by ArcA (32, 57) while fumarate reductase expression is enhanced by the fumarate nitrate reductase regulator FNR (63). In C. jejuni, literally nothing is known about the regulation and function of the sdh and frd genes. Several putative arcA-like two-component regulators are present in the C. jejuni genome, but a functional homolog has not yet been identified. One FNR-CRP (catabolite repressing protein) homolog is also present in the C. jejuni genome (Cj0466); however, its deletion did not impair in vitro survival under either microaerobic or severely O2-limited conditions (J. K. MacKichan and S. Falkow, unpublished data). Furthermore, although C. jejuni possesses fumarate reductase activity (67), and fumarate reductase is essential for H. pylori colonization of the mouse stomach (21), a C. jejuni frdA mutant was not defective for colonizing mice (J. K. MacKichan, S. Falkow, C. Chang, and J. Miller, unpublished data). It is also possible that the gene expression differences observed are due to the specific culture conditions employed in our experiments. Unraveling the regulation and function of the C. jejuni sdh and frd operons will require significant biochemical and transcriptional analyses and should, in the future, lend considerable insight into the roles of these very similar enzyme complexes in C. jejuni biology and colonization potential.
Another striking phenotype change in 11168-GS compared to 11168-O is the near-complete loss of motility. This is likely to contribute to the colonization and translocation differences observed as well as to the marked difference in invasion speed and efficiency, although 11168-O still invaded more efficiently than 11168-GS when the bacteria were centrifuged onto the monolayer (Table 1) (G. Manning and D. G. Newell, unpublished data). Phase variation in flagellar and other genes has previously been shown to affect C. jejuni motility and shape (11, 37, 46, 55, 56). However, all genes thus far identified as phase variable affect flagellar biosynthesis; as both 1168 variants have full-length polar flagella (Fig. 2) and similar flaA and flaB expression profiles, it seems unlikely that phase variation is responsible for the motility differences. Both the FliA and RpoN sigma factors have been implicated as important regulators of C. jejuni motility (23, 24, 27, 55, 75), and each contains 1 to 2 amino acid differences in the variants. Complete regulons for these sigma factors have not been identified for C. jejuni; however, several flagellar export genes shown to be part of the H. pylori RpoN regulon (36) were identified in our experiments as exhibiting significantly higher expression in 11168-O cultured under severely O2-limited conditions than in any other samples (Fig. 6A). As noted in Results, this cluster also contains several uncharacterized genes that, by virtue of their coexpression with known motility-related genes, might lend insight into their function. For instance, Cj1631c contains a likely calcium-binding domain, and Cj0604 is a likely polyphosphate kinase. Both calcium and inorganic polyphosphate are known to participate in motility in other bacteria (50, 61, 70), but their roles in C. jejuni have not yet been established. It will also be interesting to explore the function of genes with no database homologies, such as Cj1464 and Cj1465, and genes that are strikingly coexpressed with them, such as Cj0056c, in the context of their coexpression with known motility genes.
Is there a connection between the motility and morphology phenotypes and respiration and metabolism gene expression differences between these variants? Mechanical energy for flagellar rotation is provided by a proton motive force across the membrane that can be generated by the electron transport chain. Many genes exhibiting expression differences between the variants in our study are involved in metabolism and respiration; at least one, Cj0358, has been directly implicated in energy generation via the electron transport chain (64). Furthermore, in 11168-O, up-regulation of flagellar export genes occurred specifically under severely O2-limited conditions, suggesting that a low-oxygen environment may trigger these expression changes. It was recently found that the aerobically passaged 11168-O strain is, like 11168-GS, both markedly attenuated for motility and appears as a straight rod by electron microscopy (Manning and Newell, unpublished). Preliminary data also suggest that severe O2 limitation enhances 11168-O motility and can induce a small fraction of 11168-GS within a population to become motile (Gaynor and Falkow, unpublished). These observations highlight the remarkable vertical plasticity of C. jejuni and are consistent with the existence of intermediate NCTC 11168 variants such as the motile strain used by Jagannathan et al. for motility studies (33). We are currently pursuing the intriguing possibility of a genetic link between adaptation to different oxygen tensions, morphological change, and motility in C. jejuni and are exploring these phenomena in a controlled stepwise evolutionary manner by assaying phenotype and transcriptional profiles as adaptation to a higher O2 tension occurs. Studies such as these may also lend insight into whether genes exhibiting significantly higher expression in 11168-GS, such as a putative cytochrome c (Cj0037c) and coexpressed periplasmic and membrane-bound genes, might functionally interact to enhance the growth capacity of C. jejuni in laboratory and/or higher-O2 conditions.
In summary, C. jejuni is a remarkably malleable zoonotic bacterium that, despite its stringent in vitro growth conditions, is able to survive a diverse range of ecological niches. As it adapts to new environments, it is also likely to undergo some degree of genetic evolution, resulting in changes in gene expression and phenotype to accommodate its surroundings. Evidence of its large-scale genetic plasticity has recently been provided by several whole-genome DNA comparisons (13, 44), which demonstrated that different C. jejuni isolates exhibit marked variation in genes modifying the capsule, lipooligosaccharide, and flagella. One interpretation of those studies is that a possible defense mechanism for C. jejuni may be a constant remodeling of surface structures, via genome evolution, selective pressure, and/or natural selection, to survive a variety of natural host environments and bottlenecks. Here, we have used whole-genome approaches, only recently made accessible by virtue of the 11168 genome sequence and advances such as microarray technology, to gain insight into how and why two supposedly isogenic C. jejuni strains, one of which descended directly from the other via clonal propagation, might exhibit such different colonization and other virulence-related phenotypes. Unlike the studies mentioned above, these strains do not contain large variant regions of the genome or genes present in one strain but not the other, nor are the differences observed in surface structure genes. Rather, these strains contain very subtle genetic changes, detectable only by direct sequencing, but which clearly engender marked differences in transcription (primarily affecting respiration, metabolism, motility, and unknown genes) and phenotype. Because the 11168 variants are likely clonal, one important outcome of this study is that the NCTC 11168 genome sequence can be considered an accurate scaffold for exploring colonization and other virulence-associated phenotypes, especially if used in conjunction with the 11168-O variant. Furthermore, the use of microarray technology to explore not only genomic but also gene expression differences between isolates (or indeed clones) should be considered a powerful new tool for identifying possible mechanisms underlying phenotypic variation or change. Finally, this work underscores the need to sequence multiple isolates of the same pathogenic bacterial species and to appreciate that even inadvertent in vitro manipulations have the potential for altering the genome and biology of pathogens studied in the laboratory.
Acknowledgments
We thank Nafisa Ghori (Stanford) and Mary Bagnall and the members of TMB4 (VLA) for expert electron microscopy, Anne Ridley (VLA) for subtyping analyses, and Inna Bilis and Amita Kawale (Stanford) for technical assistance. We also thank Brendan Wren and Martin Skirrow for strains; Elizabeth Joyce, Denise Monack, Corrie Detweiler, and Michael Grigg for critical input on the manuscript; Elizabeth Joyce, David Schneider, and members of the Schneider lab for invaluable assistance with QRT-PCR procedures; and members of the Falkow and Newell laboratories for helpful discussions.
E.C.G. is funded by the Life Sciences Research Foundation and the Burroughs Wellcome Fund, S.F. is funded by grants from the NIH, and G.M., S.C., and D.G.N. are funded by the Department of Environmental Food and Rural Affairs (DEFRA), United Kingdom.
REFERENCES
- 1.Ahmed, I. H., G. Manning, T. M. Wassenaar, S. Cawthraw, and D. G. Newell. 2002. Identification of genetic differences between two Campylobacter jejuni strains with different colonization potentials. Microbiology 148:1203-1212. [DOI] [PubMed] [Google Scholar]
- 2.Bacon, D. J., R. A. Alm, D. H. Burr, L. Hu, D. J. Kopecko, C. P. Ewing, T. J. Trust, and P. Guerry. 2000. Involvement of a plasmid in virulence of Campylobacter jejuni 81-176. Infect. Immun. 68:4384-4390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Behr, M. A., M. A. Wilson, W. P. Gill, H. Salamon, G. K. Schoolnik, S. Rane, and P. M. Small. 1999. Comparative genomics of BCG vaccines by whole-genome DNA microarray. Science 284:1520-1523. [DOI] [PubMed] [Google Scholar]
- 4.Blaser, M. J., J. G. Wells, R. A. Feldman, R. A. Pollard, and J. R. Allen. 1983. Campylobacter enteritis in the United States. A multicenter study. Ann. Intern. Med. 98:360-365. [DOI] [PubMed] [Google Scholar]
- 5.Boer, P., J. A. Wagenaar, R. P. Achterberg, J. P. Putten, L. M. Schouls, and B. Duim. 2002. Generation of Campylobacter jejuni genetic diversity in vivo. Mol. Microbiol. 44:351-359. [DOI] [PubMed] [Google Scholar]
- 6.Bras, A. M., and J. M. Ketley. 1999. Transcellular translocation of Campylobacter jejuni across human polarised epithelial monolayers. FEMS Microbiol. Lett. 179:209-215. [DOI] [PubMed] [Google Scholar]
- 7.Cawthraw, S. A., T. M. Wassenaar, R. Ayling, and D. G. Newell. 1996. Increased colonization potential of Campylobacter jejuni strain 81116 after passage through chickens and its implication on the rate of transmission within flocks. Epidemiol. Infect. 117:213-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cecchini, G., I. Schroder, R. P. Gunsalus, and E. Maklashina. 2002. Succinate dehydrogenase and fumarate reductase from Escherichia coli. Biochim. Biophys. Acta 1553:140-157. [DOI] [PubMed] [Google Scholar]
- 9.Chiang, S. L., J. J. Mekalanos, and D. W. Holden. 1999. In vivo genetic analysis of bacterial virulence. Annu. Rev. Microbiol. 53:129-154. [DOI] [PubMed] [Google Scholar]
- 10.CPHLS. 2000. Common gastrointestinal infections, England and Wales. Commun. Dis. Rep. Rev. 10:9-12. [Google Scholar]
- 11.Diker, K. S., G. Hascelik, and M. Akan. 1992. Reversible expression of flagella in Campylobacter spp. FEMS Microbiol. Lett. 78:261-264. [DOI] [PubMed] [Google Scholar]
- 12.Dobrindt, U., F. Agerer, K. Michaelis, A. Janka, C. Buchrieser, M. Samuelson, C. Svanborg, G. Gottschalk, H. Karch, and J. Hacker. 2003. Analysis of genome plasticity in pathogenic and commensal Escherichia coli isolates by use of DNA arrays. J. Bacteriol. 185:1831-1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dorrell, N., J. A. Mangan, K. G. Laing, J. Hinds, D. Linton, H. Al-Ghusein, B. G. Barrell, J. Parkhill, N. G. Stoker, A. V. Karlyshev, P. D. Butcher, and B. W. Wren. 2001. Whole genome comparison of Campylobacter jejuni human isolates using a low-cost microarray reveals extensive genetic diversity. Genome Res. 11:1706-1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Duim, B., T. M. Wassenaar, A. Rigter, and J. Wagenaar. 1999. High-resolution genotyping of Campylobacter strains isolated from poultry and humans with amplified fragment length polymorphism fingerprinting. Appl. Environ. Microbiol. 65:2369-2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eisen, M. B., P. T. Spellman, P. O. Brown, and D. Botstein. 1998. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. USA 95:14863-14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Elsinghorst, E. A. 1994. Measurement of invasion by gentamicin resistance. Methods Enzymol. 236:405-420. [DOI] [PubMed] [Google Scholar]
- 17.Fine, D. H., D. Furgang, H. C. Schreiner, P. Goncharoff, J. Charlesworth, G. Ghazwan, P. Fitzgerald-Bocarsly, and D. H. Figurski. 1999. Phenotypic variation in Actinobacillus actinomycetemcomitans during laboratory growth: implications for virulence. Microbiology 145(Pt 6):1335-1347. [DOI] [PubMed] [Google Scholar]
- 18.Fleischmann, R. D., D. Alland, J. A. Eisen, L. Carpenter, O. White, J. Peterson, R. DeBoy, R. Dodson, M. Gwinn, D. Haft, E. Hickey, J. F. Kolonay, W. C. Nelson, L. A. Umayam, M. Ermolaeva, S. L. Salzberg, A. Delcher, T. Utterback, J. Weidman, H. Khouri, J. Gill, A. Mikula, W. Bishai, W. R. Jacobs, Jr., J. C. Venter, and C. M. Fraser. 2002. Whole-genome comparison of Mycobacterium tuberculosis clinical and laboratory strains. J. Bacteriol. 184:5479-5490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Friedman, C. R., J. Neimann, H. C. Wegener, and R. Tauxe. 2001. Epidemiology of Campylobacter jejuni infection in the United States and other industrialized nations, p. 121-138. In I. Nachamkin and M. J. Blaser (ed.), Campylobacter. ASM Press, Washington, D.C.
- 20.Fry, B. N., S. Feng, Y. Y. Chen, D. G. Newell, P. J. Coloe, and V. Korolik. 2000. The galE gene of Campylobacter jejuni is involved in lipopolysaccharide synthesis and virulence. Infect. Immun. 68:2594-2601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ge, Z., Y. Feng, C. A. Dangler, S. Xu, N. S. Taylor, and J. G. Fox. 2000. Fumarate reductase is essential for Helicobacter pylori colonization of the mouse stomach. Microb. Pathog. 29:279-287. [DOI] [PubMed] [Google Scholar]
- 22.Gordon, S. V., R. Brosch, A. Billault, T. Garnier, K. Eiglmeier, and S. T. Cole. 1999. Identification of variable regions in the genomes of tubercle bacilli using bacterial artificial chromosome arrays. Mol. Microbiol. 32:643-655. [DOI] [PubMed] [Google Scholar]
- 23.Guerry, P., R. A. Alm, M. E. Power, S. M. Logan, and T. J. Trust. 1991. Role of two flagellin genes in Campylobacter motility. J. Bacteriol. 173:4757-4764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guerry, P., S. M. Logan, S. Thornton, and T. J. Trust. 1990. Genomic organization and expression of Campylobacter flagellin genes. J. Bacteriol. 172:1853-1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hahm, B. K., Y. Maldonado, E. Schreiber, A. K. Bhunia, and C. H. Nakatsu. 2003. Subtyping of foodborne and environmental isolates of Escherichia coli by multiplex-PCR, rep-PCR, PFGE, ribotyping and AFLP. J. Microbiol. Methods 53:387-399. [DOI] [PubMed] [Google Scholar]
- 26.Hanninen, M. L., M. Hakkinen, and H. Rautelin. 1999. Stability of related human and chicken Campylobacter jejuni genotypes after passage through chick intestine studied by pulsed-field gel electrophoresis. Appl. Environ. Microbiol. 65:2272-2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hendrixson, D. R., B. J. Akerley, and V. J. DiRita. 2001. Transposon mutagenesis of Campylobacter jejuni identifies a bipartite energy taxis system required for motility. Mol. Microbiol. 40:214-224. [DOI] [PubMed] [Google Scholar]
- 28.Hodgson, A. E., B. W. McBride, M. J. Hudson, G. Hall, and S. A. Leach. 1998. Experimental campylobacter infection and diarrhoea in immunodeficient mice. J. Med. Microbiol. 47:799-809. [DOI] [PubMed] [Google Scholar]
- 29.Hughes, N. J., P. A. Chalk, C. L. Clayton, and D. J. Kelly. 1995. Identification of carboxylation enzymes and characterization of a novel four-subunit pyruvate:flavodoxin oxidoreductase from Helicobacter pylori. J. Bacteriol. 177:3953-3959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hughes, N. J., C. L. Clayton, P. A. Chalk, and D. J. Kelly. 1998. Helicobacter pylori porCDAB and oorDABC genes encode distinct pyruvate:flavodoxin and 2-oxoglutarate:acceptor oxidoreductases which mediate electron transport to NADP. J. Bacteriol. 180:1119-1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ishige, K., H. Zhang, and A. Kornberg. 2002. Polyphosphate kinase (PPK2), a potent, polyphosphate-driven generator of GTP. Proc. Natl. Acad. Sci. USA 99:16684-16688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Iuchi, S., and E. C. Lin. 1988. arcA (dye), a global regulatory gene in Escherichia coli mediating repression of enzymes in aerobic pathways. Proc. Natl. Acad. Sci. USA 85:1888-1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jagannathan, A., C. Constantinidou, and C. W. Penn. 2001. Roles of rpoN, fliA, and flgR in expression of flagella in Campylobacter jejuni. J. Bacteriol. 183:2937-2942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Janausch, I. G., E. Zientz, Q. H. Tran, A. Kroger, and G. Unden. 2002. C4-dicarboxylate carriers and sensors in bacteria. Biochim. Biophys. Acta 1553:39-56. [DOI] [PubMed] [Google Scholar]
- 35.Jones, D. M., E. M. Sutcliffe, R. Rios, A. J. Fox, and A. Curry. 1993. Campylobacter jejuni adapts to aerobic metabolism in the environment. J. Med. Microbiol. 38:145-150. [DOI] [PubMed] [Google Scholar]
- 36.Josenhans, C., E. Niehus, S. Amersbach, A. Horster, C. Betz, B. Drescher, K. T. Hughes, and S. Suerbaum. 2002. Functional characterization of the antagonistic flagellar late regulators FliA and FlgM of Helicobacter pylori and their effects on the H. pylori transcriptome. Mol. Microbiol. 43:307-322. [DOI] [PubMed] [Google Scholar]
- 37.Karlyshev, A. V., D. Linton, N. A. Gregson, and B. W. Wren. 2002. A novel paralogous gene family involved in phase-variable flagella-mediated motility in Campylobacter jejuni. Microbiology 148:473-480. [DOI] [PubMed] [Google Scholar]
- 38.Kelly, D. J. 2001. The physiology and metabolism of Campylobacter jejuni and Helicobacter pylori. Symp. Ser. Soc. Appl. Microbiol. 30:16S-24S. [DOI] [PubMed]
- 39.Kim, C. C., E. A. Joyce, K. Chan, and S. Falkow. 2002. Improved analytical methods for microarray-based genome-composition analysis. Genome Biol. 3:1-17. [DOI] [PMC free article] [PubMed]
- 40.Kim, S. S., H. S. Lee, Y. S. Cho, Y. S. Lee, C. S. Bhang, H. S. Chae, S. W. Han, I. S. Chung, and D. H. Park. 2002. The effect of the repeated subcultures of Helicobacter pylori on adhesion, motility, cytotoxicity, and gastric inflammation. J. Korean Med. Sci. 17:302-306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kokotovic, B., and S. L. On. 1999. High-resolution genomic fingerprinting of Campylobacter jejuni and Campylobacter coli by analysis of amplified fragment length polymorphisms. FEMS Microbiol. Lett. 173:77-84. [DOI] [PubMed] [Google Scholar]
- 42.Konkel, M. E., D. J. Mead, S. F. Hayes, and W. Cieplak, Jr. 1992. Translocation of Campylobacter jejuni across human polarized epithelial cell monolayer cultures. J. Infect. Dis. 166:308-315. [DOI] [PubMed] [Google Scholar]
- 43.Lam, K. M., R. Yamamoto, and A. J. DaMassa. 1995. DNA diversity among isolates of Campylobacter jejuni detected by PCR-based RAPD fingerprinting. Vet. Microbiol. 45:269-274. [DOI] [PubMed] [Google Scholar]
- 44.Leonard, E. E., II, T. Takata, M. J. Blaser, S. Falkow, L. S. Tompkins, and E. C. Gaynor. 2003. Use of an open-reading frame-specific Campylobacter jejuni DNA microarray as a new genotyping tool for studying epidemiologically related isolates. J. Infect. Dis. 187:691-694. [DOI] [PubMed] [Google Scholar]
- 45.Mahairas, G. G., P. J. Sabo, M. J. Hickey, D. C. Singh, and C. K. Stover. 1996. Molecular analysis of genetic differences between Mycobacterium bovis BCG and virulent M. bovis. J. Bacteriol. 178:1274-1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Matz, C., A. H. van Vliet, J. M. Ketley, and C. W. Penn. 2002. Mutational and transcriptional analysis of the Campylobacter jejuni flagellar biosynthesis gene flhB. Microbiology 148:1679-1685. [DOI] [PubMed] [Google Scholar]
- 47.McKinney, J. D., K. Honer zu Bentrup, E. J. Munoz-Elias, A. Miczak, B. Chen, W. T. Chan, D. Swenson, J. C. Sacchettini, W. R. Jacobs, Jr., and D. G. Russell. 2000. Persistence of Mycobacterium tuberculosis in macrophages and mice requires the glyoxylate shunt enzyme isocitrate lyase. Nature 406:735-738. [DOI] [PubMed] [Google Scholar]
- 48.Mecsas, J. 2002. Use of signature-tagged mutagenesis in pathogenesis studies. Curr. Opin. Microbiol. 5:33-37. [DOI] [PubMed] [Google Scholar]
- 49.Mendz, G. L., G. E. Ball, and D. J. Meek. 1997. Pyruvate metabolism in Campylobacter spp. Biochim. Biophys. Acta 1334:291-302. [DOI] [PubMed] [Google Scholar]
- 50.Michiels, J., C. Xi, J. Verhaert, and J. Vanderleyden. 2002. The functions of Ca(2+) in bacteria: a role for EF-hand proteins? Trends Microbiol. 10:87-93. [DOI] [PubMed] [Google Scholar]
- 51.Nachamkin, I. 2002. Chronic effects of Campylobacter infection. Microbes Infect. 4:399-403. [DOI] [PubMed] [Google Scholar]
- 52.Newell, D. G., H. McBride, F. Saunders, Y. Dehele, and A. D. Pearson. 1985. The virulence of clinical and environmental isolates of Campylobacter jejuni. J. Hyg. (London) 94:45-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Newell, D. G., J. E. Shreeve, M. Toszeghy, G. Domingue, S. Bull, T. Humphrey, and G. Mead. 2001. Changes in the carriage of Campylobacter strains by poultry carcasses during processing in abattoirs. Appl. Environ. Microbiol. 67:2636-2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Newell, D. G., and J. A. Wagenaar. 2000. Poultry infections and their control at the farm level, p. 497-509. In I. Nachamkin and M. J. Blaser (ed.), Campylobacter. American Society for Microbiology, Washington, D.C.
- 55.Nuijten, P. J., F. J. van Asten, W. Gaastra, and B. A. van der Zeijst. 1990. Structural and functional analysis of two Campylobacter jejuni flagellin genes. J. Biol. Chem. 265:17798-17804. [PubMed] [Google Scholar]
- 56.Park, S. F., D. Purdy, and S. Leach. 2000. Localized reversible frameshift mutation in the flhA gene confers phase variability to flagellin gene expression in Campylobacter coli. J. Bacteriol. 182:207-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Park, S. J., C. P. Tseng, and R. P. Gunsalus. 1995. Regulation of succinate dehydrogenase (sdhCDAB) operon expression in Escherichia coli in response to carbon supply and anaerobiosis: role of ArcA and Fnr. Mol. Microbiol. 15:473-482. [DOI] [PubMed] [Google Scholar]
- 58.Parkhill, J., B. W. Wren, K. Mungall, J. M. Ketley, C. Churcher, D. Basham, T. Chillingworth, R. M. Davies, T. Feltwell, S. Holroyd, K. Jagels, A. V. Karlyshev, S. Moule, M. J. Pallen, C. W. Penn, M. A. Quail, M. A. Rajandream, K. M. Rutherford, A. H. van Vliet, S. Whitehead, and B. G. Barrell. 2000. The genome sequence of the food-borne pathogen Campylobacter jejuni reveals hypervariable sequences. Nature 403:665-668. [DOI] [PubMed] [Google Scholar]
- 59.Perna, N. T., G. Plunkett III, V. Burland, B. Mau, J. D. Glasner, D. J. Rose, G. F. Mayhew, P. S. Evans, J. Gregor, H. A. Kirkpatrick, G. Posfai, J. Hackett, S. Klink, A. Boutin, Y. Shao, L. Miller, E. J. Grotbeck, N. W. Davis, A. Lim, E. T. Dimalanta, K. D. Potamousis, J. Apodaca, T. S. Anantharaman, J. Lin, G. Yen, D. C. Schwartz, R. A. Welch, and F. R. Blattner. 2001. Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature 409:529-533. [DOI] [PubMed] [Google Scholar]
- 60.Purdy, D., S. Cawthraw, J. H. Dickinson, D. G. Newell, and S. F. Park. 1999. Generation of a superoxide dismutase (SOD)-deficient mutant of Campylobacter coli: evidence for the significance of SOD in Campylobacter survival and colonization. Appl. Environ. Microbiol. 65:2540-2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rashid, M. H., N. N. Rao, and A. Kornberg. 2000. Inorganic polyphosphate is required for motility of bacterial pathogens. J. Bacteriol. 182:225-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sassetti, C. M., D. H. Boyd, and E. J. Rubin. 2001. Comprehensive identification of conditionally essential genes in mycobacteria. Proc. Natl. Acad. Sci. USA 98:12712-12717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sawers, G. 1999. The aerobic/anaerobic interface. Curr. Opin. Microbiol. 2:181-187. [DOI] [PubMed] [Google Scholar]
- 64.Sellars, M. J., S. J. Hall, and D. J. Kelly. 2002. Growth of Campylobacter jejuni supported by respiration of fumarate, nitrate, nitrite, trimethylamine-N-oxide, or dimethyl sulfoxide requires oxygen. J. Bacteriol. 184:4187-4196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Skirrow, M., and M. J. Blaser. 2001. Clinical aspects of Campylobacter infection, p. 69-88. In I. Nachamkin and M. J. Blaser (ed.), Campylobacter. ASM Press, Washington, D.C.
- 66.Smith, M. A., M. Finel, V. Korolik, and G. L. Mendz. 2000. Characteristics of the aerobic respiratory chains of the microaerophiles Campylobacter jejuni and Helicobacter pylori. Arch. Microbiol. 174:1-10. [DOI] [PubMed] [Google Scholar]
- 67.Smith, M. A., G. L. Mendz, M. A. Jorgensen, and S. L. Hazell. 1999. Fumarate metabolism and the microaerophily of Campylobacter species. Int. J. Biochem. Cell Biol. 31:961-975. [DOI] [PubMed] [Google Scholar]
- 68.Somerville, G. A., M. S. Chaussee, C. I. Morgan, J. R. Fitzgerald, D. W. Dorward, L. J. Reitzer, and J. M. Musser. 2002. Staphylococcus aureus aconitase inactivation unexpectedly inhibits post-exponential-phase growth and enhances stationary-phase survival. Infect. Immun. 70:6373-6382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Thongboonkerd, V., J. Luengpailin, J. Cao, W. M. Pierce, J. Cai, J. B. Klein, and R. J. Doyle. 2002. Fluoride exposure attenuates expression of Streptococcus pyogenes virulence factors. J. Biol. Chem. 277:16599-16605. [DOI] [PubMed] [Google Scholar]
- 70.Tisa, L. S., and J. Adler. 1992. Calcium ions are involved in Escherichia coli chemotaxis. Proc. Natl. Acad. Sci. USA 89:11804-11808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tusher, V. G., R. Tibshirani, and G. Chu. 2001. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. USA 98:5116-5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ulett, G. C., B. J. Currie, T. W. Clair, M. Mayo, N. Ketheesan, J. Labrooy, D. Gal, R. Norton, C. A. Smith, J. Barnes, J. Warner, and R. G. Hirst. 2001. Burkholderia pseudomallei virulence: definition, stability and association with clonality. Microbes Infect. 3:621-631. [DOI] [PubMed] [Google Scholar]
- 73.Velayudhan, J., and D. J. Kelly. 2002. Analysis of gluconeogenic and anaplerotic enzymes in Campylobacter jejuni: an essential role for phosphoenolpyruvate carboxykinase. Microbiology 148:685-694. [DOI] [PubMed] [Google Scholar]
- 74.Wassenaar, T. M., and M. J. Blaser. 1999. Pathophysiology of Campylobacter jejuni infections of humans. Microbes Infect. 1:1023-1033. [DOI] [PubMed] [Google Scholar]
- 75.Wassenaar, T. M., N. M. Bleumink-Pluym, D. G. Newell, P. J. Nuijten, and B. A. van der Zeijst. 1994. Differential flagellin expression in a flaA flaB+ mutant of Campylobacter jejuni. Infect. Immun. 62:3901-3906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wassenaar, T. M., N. M. Bleumink-Pluym, and B. A. van der Zeijst. 1991. Inactivation of Campylobacter jejuni flagellin genes by homologous recombination demonstrates that flaA but not flaB is required for invasion. EMBO J. 10:2055-2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wassenaar, T. M., B. Geilhausen, and D. G. Newell. 1998. Evidence of genomic instability in Campylobacter jejuni isolated from poultry. Appl. Environ. Microbiol. 64:1816-1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wassenaar, T. M., B. A. van der Zeijst, R. Ayling, and D. G. Newell. 1993. Colonization of chicks by motility mutants of Campylobacter jejuni demonstrates the importance of flagellin A expression. J. Gen. Microbiol. 139(Pt 6):1171-1175. [DOI] [PubMed] [Google Scholar]
- 79.World Health Organization. 2000. Annual report. World Health Organization, Copenhagen, Denmark.
- 80.Zhang, H., K. Ishige, and A. Kornberg. 2002. A polyphosphate kinase (PPK2) widely conserved in bacteria. Proc. Natl. Acad. Sci. USA 99:16678-16683. [DOI] [PMC free article] [PubMed] [Google Scholar]