The PNC is a nuclear body that forms in malignant cells. We characterize a newly identified complex in the PNC; determine the dynamics of PNC proteins; and describe the recruitment, localization, and sedimentation properties of PNC components.
Abstract
The perinucleolar compartment (PNC) forms in cancer cells and is highly enriched with a subset of polymerase III RNAs and RNA-binding proteins. Here we report that PNC components mitochondrial RNA–processing (MRP) RNA, pyrimidine tract–binding protein (PTB), and CUG-binding protein (CUGBP) interact in vivo, as demonstrated by coimmunoprecipitation and RNA pull-down experiments. Glycerol gradient analyses show that this complex is large and sediments at a different fraction from known MRP RNA–containing complexes, the MRP ribonucleoprotein ribozyme and human telomerase reverse transcriptase. Tethering PNC components to a LacO locus recruits other PNC components, further confirming the in vivo interactions. These interactions are present both in PNC-containing and -lacking cells. High-resolution localization analyses demonstrate that MRP RNA, CUGBP, and PTB colocalize at the PNC as a reticulated network, intertwining with newly synthesized RNA. Furthermore, green fluorescent protein (GFP)–PTB and GFP-CUGBP show a slower rate of fluorescence recovery after photobleaching at the PNC than in the nucleoplasm, illustrating the different molecular interaction of the complexes associated with the PNC. These findings support a working model in which the MRP RNA–protein complex becomes nucleated at the PNC in cancer cells and may play a role in gene expression regulation at the DNA locus that associates with the PNC.
INTRODUCTION
The perinucleolar compartment (PNC) is a unique and dynamic nuclear body associated with the nucleolus. It is an irregularly shaped structure that ranges from 0.25 to 4 μm in size (Huang et al., 1997; Pollock and Huang, 2009). Electron microscopic (EM) analyses demonstrate that PNCs are structurally distinct from the nucleolus and composed of multiple electron-dense strands between 80 and 180 nm in diameter (Huang et al., 1998). The structure directly associates with DNA, where it nucleates on an unidentified locus (Norton et al., 2009). The PNC is of particular interest because of its association with cancer. PNCs form in malignant cells during transformation and correlate with metastasis, and a high PNC prevalence indicates worse disease prognosis (Huang et al., 1997; Kamath et al., 2005; Norton et al., 2008). The role of the PNC in malignancy and metastasis, however, is not yet known.
The PNC is associated with an unidentified DNA locus (Norton et al., 2009) and is enriched with several small, noncoding RNAs including mitochondrial RNA–processing (MRP) RNA, RNase P RNA, Alu RNA, signal recognition particle RNA, and human Y (hY) RNAs (Matera et al., 1995; Lee et al., 1996; Wang et al., 2003). However, not all RNA polymerase (Pol) II transcripts are detected in the PNC. In situ hybridization experiments did not show other RNA Pol III transcripts, including tRNA, U6, or 5S rRNA, in the PNC (Matera et al., 1995; Pollock and Huang, 2009). The PNC is also enriched with RNA-binding proteins, including pyrimidine tract–binding protein (PTB), CUG-binding protein (CUGBP), Raver1, Raver2, nucleolin, and KH-type splicing regulatory protein (Ghetti et al., 1992; Timchenko et al., 1996; Huang et al., 1998; Huttelmaier et al., 2001; Hall et al., 2004; Kopp and Huang, 2005). While the proteins that enrich to the PNC are known to interact primarily with RNA Pol II transcripts, no Pol II RNAs have been identified at the PNC (Hall et al., 2004; Kopp and Huang, 2005), and RNA Pol II inhibition does not disrupt PNC structure (Huang et al., 1998). Why these proteins and RNAs enrich at the PNC and what their role in malignancy may be remain unknown.
In this study we begin to explore the function of the PNC by analyzing the molecular interactions among MRP RNA, PTB, and CUGBP associated with the PNC. We observe that MRP RNA reciprocally coprecipitates CUGBP and PTB proteins, and PNC components are able to recruit each other in vivo. These interactions are unique from known MRP RNA–containing complexes judged from localization, coprecipitation, and glycerol gradient analyses. They colocalize in subdomains of the PNC as examined by high-resolution light microscopy. Furthermore, fluorescence recovery after photobleaching (FRAP) analyses indicate that PNC enriching proteins have different dynamics at the PNC as compared with those distributed in the nucleoplasm and presumably involved in pre-mRNA metabolism. These results indicate that MRP RNA forms a previously uncharacterized complex that nucleates at the PNC in PNC-containing cells. The newly characterized ribonucleoproteins (RNPs) may play a role in gene expression regulation in malignant cells.
RESULTS
Colocalization of CUGBP, PTB, and MRP RNA at the PNC with Optical Microscope eXperimental (OMX) microscopy
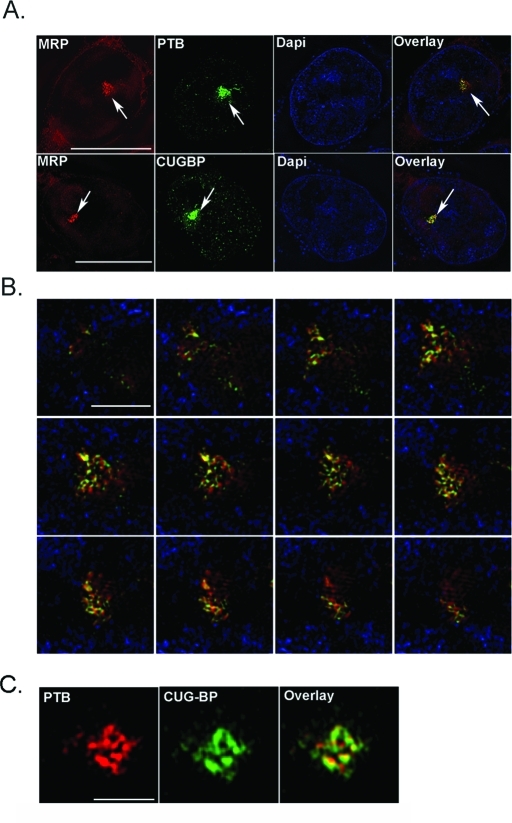
Previous localization studies have detected a set of proteins and Pol III RNAs enriched at the PNC (Ghetti et al., 1992; Matera et al., 1995; Lee et al., 1996; Timchenko et al., 1996; Huang et al., 1998; Wang et al., 2003; Hall et al., 2004). The distribution of these proteins and RNAs within the PNC at a higher resolution, however, has not been previously studied. Here we examined the intra-PNC localization of three PNC enriching components—MRP RNA, CUGBP, and PTB—using the DeltaVision OMX (Applied Precision, Issaquah, WA) system, a super-resolution fluorescence microscopy based on the structure illumination technique (Davis, 2009). The OMX system has a resolution of 100 nm at X and Y. MRP RNA and CUGBP or PTB were simultaneously detected within subregions of the PNC through immunolabeling of the proteins and in situ hybridization to MRP RNA in HeLa cells. The detection signals were examined through z-sections of PNCs in 250-nm intervals using the OMX system. The results show that PTB and MRP RNA and, similarly, CUGBP and MRP RNA are largely colocalized at the PNC in a reticulated meshwork (Figure 1, A and B). Using coimmunolabeled cells, we also observed that PTB colocalizes mostly with CUGBP at the PNC (Figure 1C). These findings are consistent with our earlier EM observation that the PNC is composed of a densely stained filamentous structure forming a reticulated meshwork (Huang et al., 1997). The close proximity of these components also suggests that they may be interacting with each other at the PNC.
FIGURE 1:
PNC components colocalize to the PNC at high resolution. (A) Super-resolution OMX microscopy shows fluorescence localization of MRP RNA with CUGBP and PTB proteins in HeLa cells. Scale bar = 10 μm. Arrows mark PNCs. (B) Z-section of the PNC of a single HeLa cell shows colocalization of MRP (in red) and PTB (in green), with 4′,6-diamino-2-phenylindole in blue. Scale bar = 1 μm. (C) PTB (red) colocalization with CUGBP (green) is also shown in high resolution at the PNC. Scale bar = 1 μm.
MRP RNA is in a complex with CUGBP and PTB in vivo
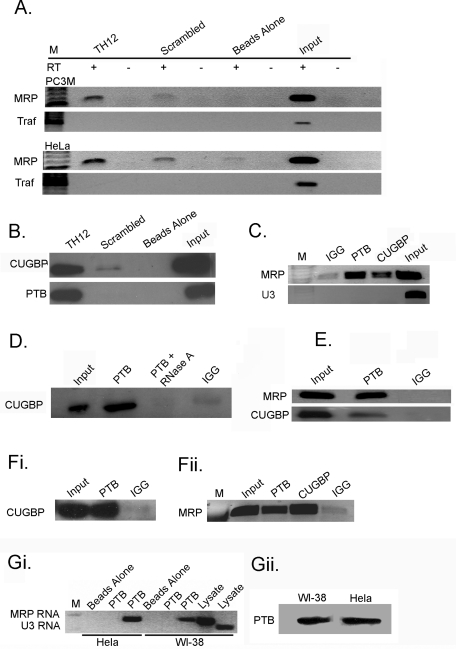
To determine whether PNC-associated MRP RNA, CUGBP, or PTB are in a previously uncharacterized complex, we performed RNA pull-down and reciprocal immunoprecipitation (IP) experiments. For RNA pull-downs, a 2′-O-methylated biotinylated oligo, TH12 (Matera et al., 1995), was used to pull down MRP RNA and interacting proteins. If CUGBP or PTB proteins interact with MRP RNA, we would expect that they would be pulled down along with the RNA. The TH12 oligo was shown to in situ hybridize specifically to MRP RNA (Matera et al., 1995). To test whether TH12 specifically pulls down MRP RNA, the oligo was linked to magnetic beads and incubated with HeLa or PC3M nuclear lysate, and the precipitates were examined for MRP RNA using reverse transcriptase (RT) PCR (Figure 2A). A scrambled oligo with the same CG content as TH12 and beads alone were utilized as negative controls. The TH12 oligo robustly pulled down MRP RNA, compared with scrambled oligo and beads-alone controls. As an additional specificity evaluation, we tested for a pre-mRNA with the closest sequence similarity to the oligo (87.5% identity), tumor necrosis factor receptor–associated factor (TRAF) 4 RNA, in the precipitates through RT-PCR, and the results are negative (Figure 2A). These observations demonstrate that the TH12 oligo specifically pulls down MRP RNA.
FIGURE 2:
MRP RNA coprecipitates with CUGBP and PTB. (A) RNA pull-down assays were completed in PC3M and HeLa cells using a TH12 oligo specific for MRP RNA and scrambled TH12 oligo and beads-alone controls, and RT-PCR was utilized to amplify MRP and TRAF RNAs. (B) Anti-CUGBP and anti-PTB antibodies were used to detect proteins from RNA pull-down assays in HeLa cells. (C) HeLa cell lysates were subjected to IP using anti-PTB, anti-CUGBP, and anti-IgG antibodies and analyzed by RT-PCR for MRP and U3 RNA, (D) or Western blot with an anti-CUGBP antibody, (E) and under cross-linking conditions. (Fi) CUGBP and (Fii) MRP RNA are precipitated in α-amanitin–treated HeLa cells. (Gi) HeLa and WI-38 cells were immunoprecipitated with PTB or beads alone and analyzed by RT-PCR. In lanes 2 and 5 PTB is precipitated and U3 RNA is amplified, and in lanes 3 and 6 MRP RNA is amplified. (Gii) PTB protein was precipitated from HeLa and WI-38 cells and blotted with anti-PTB.
To determine whether MRP RNA is interacting with PNC proteins, the protein pull-downs from the TH12 oligo were blotted against specific anti-PTB or anti-CUGBP antibodies. The results show that both CUGBP and PTB proteins are pulled down with MRP RNA, while neither protein is specifically pulled down with the scrambled oligo or beads-alone control (Figure 2B). These results suggest that MRP RNA interacts with both PTB and CUGBP proteins in vivo. To confirm these results, reciprocal precipitations were performed. Specific antibodies against PTB and CUGBP were used to precipitate their respective proteins, and MRP RNA was detected using RT-PCR from the precipitates. Both PTB and CUGBP antibodies robustly coprecipitate MRP RNA, while immunoglobulin G (IgG) control does not (Figure 2C). To further confirm the specificity of the coimmunoprecipitation, an abundant and small noncoding nucleolar RNA, U3 RNA, was also examined in the precipitates, and the result shows that the U3 RNA was not coprecipitated with either of the proteins. These results demonstrate that MRP RNA interacts with PTB and CUGBP specifically in vivo.
To determine whether PTB and CUGBP are in the same complex, PTB, CUGBP, and IgG antibodies were used for IPs. Samples were subsequently analyzed via Western blot with a CUGBP-specific antibody. Our results show that CUGBP is coprecipitated with PTB (Figure 2D), suggesting that these proteins interact with each other in addition to MRP RNA. When the nuclear lysate is treated with RNase A for 30 min before IP, PTB no longer precipitates CUGBP, which demonstrates the RNA-dependent nature of this interaction (Figure 2D). To determine the specificity of the interactions, we performed the precipitation experiments in cross-linked samples, and similar coprecipitations were observed (Figure 2E). To further evaluate whether the PTB–CUGBP interactions are mediated by pre-mRNA or by MRP RNA, because both proteins are primarily known for their roles in pre-mRNA processing, we treated cells with α-amanitin, which specifically blocks RNA Pol II transfection. Coimmunoprecipitations were performed in cells treated with α-amanitin (10 μg/ml for 16 h), in which most of the RNA Pol II is degraded (Aeby et al., 2010). The results show that the coprecipitation appears to be enhanced (Figure 2F, i and ii), suggesting the interactions are primarily mediated by RNAs other than pre-mRNA. This finding is consistent with our previous report that the colabeling of PTB and CUGBP in the PNC enlarges during RNA Pol II transcription inhibition (Huang et al., 1998).
To study whether the observed interactions were specific to PNC-containing cells, we examined the PTB–MRP interaction in WI-38 cells, which contain little to no PNCs. The results show that PTB also specifically precipitates MRP RNA in WI-38 cells (Figure 2Gi). This observation suggests that the interaction observed between MRP RNA and PTB is not specific to PNC-containing cells and may represent a ubiquitous function that is altered in malignancy. Interestingly, the amount of MRP RNA precipitated by PTB in HeLa cells appears to be greater than in WI-38 cells, even though the amount of protein precipitated in each cell line is similar (Figure 2Gii). This suggests that while the interaction between PTB and MRP RNA is not specific to PNC-containing cells, it could be more abundant in HeLa cells.
MRP RNA forms a PNC-associated complex independent of previously identified interactions
MRP RNA has been previously shown to be the RNA component of two RNPs. A ribozyme, the MRP RNP, contains 10 proteins in addition to MRP RNA and has been previously detected in mitochondria and nucleoli, with the majority being localized in the nucleolus (Reimer et al., 1988; Li et al., 1994). MRP RNA forms a second complex with the human telomerase reverse transcriptase (hTERT) protein, which can be found in the cytoplasm, nucleus, nucleolus, and telomere (Yan et al., 2004). If the PNC-associated MRP RNA is in one of these complexes, we would expect a coenrichment of these proteins in the PNC with MRP RNA. To evaluate this possibility, we compared the cellular localization of components of the known RNP complexes with the PNC.
To determine whether PNC-associated MRP RNA is in the MRP RNP complex, HeLa cells were transfected with a green fluorescent protein (GFP)–tagged protein subunit of the MRP RNP complex, Rpp20. The localization of GFP-Rpp20 was compared with MRP RNA localization, as detected by in situ hybridization with an MRP RNA oligo. Microscopy analysis of these cells shows that Rpp20 is not enriched at the PNC like MRP RNA (Figure 3A), and similar results are observed with two additional MRP RNP subunits (unpublished data). These findings suggest that MRP RNA is enriched at the PNC independent of MRP RNP protein subunits, and therefore PNC-associated MRP RNA is unlikely to be in the form of MRP RNP particles. To determine whether MRP RNA is in a complex with hTERT in the PNC, PTB and hTERT were immunolabeled simultaneously in HeLa cells. Colocalization analyses show that hTERT is not correspondingly enriched at the PNC (Figure 3A). These results suggest that MRP RNA is enriched at the PNC neither as part of the MRP RNP or in complex with hTERT. These findings are consistent with previous observations that hY RNAs enriched at the PNC are not in the form of their functional Ro RNPs (Matera et al., 1995).
FIGURE 3:
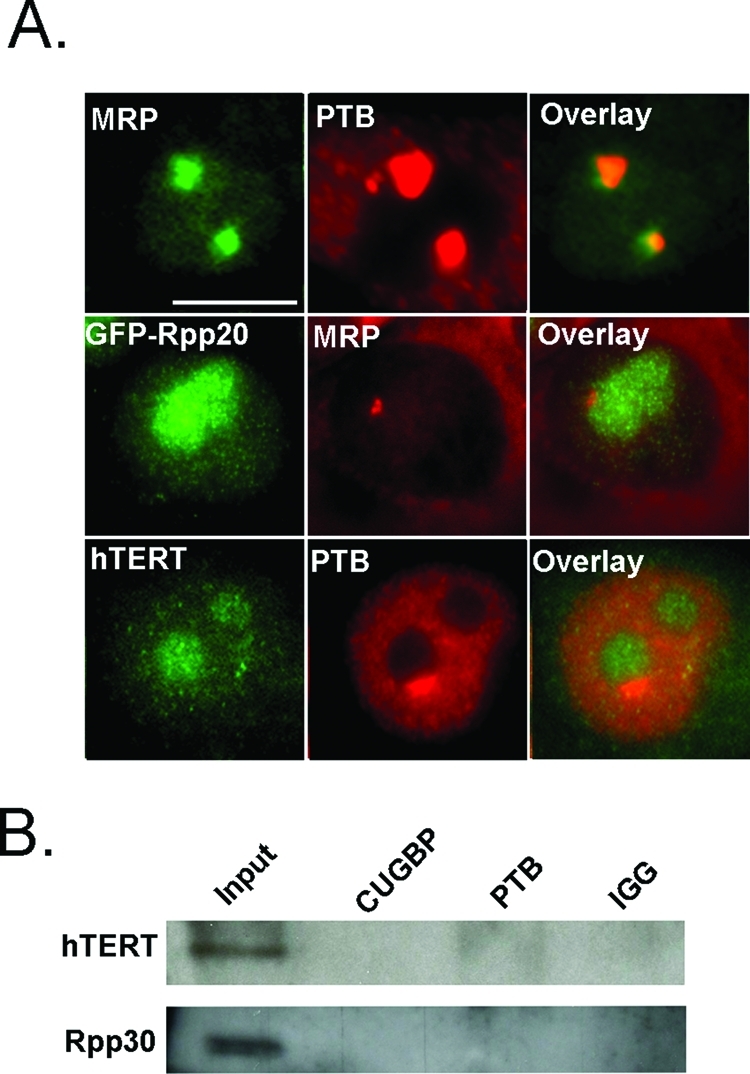
MRP RNA forms a PNC-associated complex independent of the MRP RNP and hTERT complexes. (A, top) Fluorescence microscopy was used to examine normal subcellular localization of MRP (green) and PTB (red), (A, middle) GFP-Rpp20 (green) and MRP (red), (A, bottom) and hTERT (green) and PTB (red) in HeLa cells. (B) HeLa cells are precipitated with anti-CUGBP and anti-PTB antibodies and blotted with anti-hTERT and anti-Rpp30 antibodies. Scale bar = 10 μm.
Furthermore, to biochemically analyze that the interaction between MRP RNA and CUGBP or PTB proteins is independent of previously characterized MRP RNA complexes, PTB and CUGBP immunoprecipitates were blotted with hTERT and Rpp30 antibodies. The results show that while Rpp30 and hTERT are detected in the input, neither the Rpp30 nor the hTERT protein is robustly precipitated by PTB or CUGBP antibodies (Figure 3B), confirming that the interactions between MRP RNA and PTB and CUGBP proteins are distinct from the previously described MRP RNA complexes.
Sedimentation properties of MRP RNA, CUGBP, and PTB
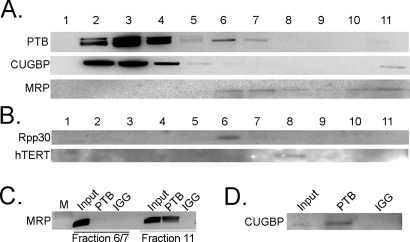
To further characterize the properties of the newly identified MRP–CUGBP–PTB complex, we analyzed their sedimentation properties using glycerol gradient. If these three components coprecipitate to form a complex, we would expect them to be detected in overlapping gradient fractions. Cell lysates were fractionated in a 5–45% step glycerol gradient. MRP RNA is detected in fractions 6, 7, and 11 and to some extent in 8, 9, and 10, while CUGBP and PTB are detected in fractions 2, 3, 4, 5, 6, 7, and 11 (Figure 4A). To determine what MRP RNA–containing fractions are likely to be involved in PTB and CUGBP interactions versus previously described MRP RNP and hTERT complexes, gradient fractions were blotted with hTERT and Rpp30 antibodies. hTERT is observed only in fraction 8, where MRP RNA is also detected, but not with PTB or CUGBP (Figure 4B). Rpp30 is detected in fractions 6 and 7, but not fraction 11. This finding indicates that fractions 6 and 7 contain MRP RNA, which is consistent with the size of the MRP RNP ribozyme (Figure 4B).
FIGURE 4:
The MRP RNA–CUGBP–PTB complex is detected in fraction 11. (A) PTB and CUGBP proteins are detected in fractions of a 5–45% glycerol gradient with anti-PTB and anti-CUGBP antibodies, and MRP RNA is detected with Northern blot. (B) Fractions are blotted with anti-Rpp30 and anti-hTERT antibodies. (C) PTB is precipitated from fractions 6/7 and 11, and MRP is detected with RT-PCR. (D) PTB is precipitated from fraction 11, and anti-CUGBP is used to blot. Ten percent of material used in IP was loaded as input.
The lack of MRP RNA in fractions 2, 3, and 4 suggests that it is unlikely that PTB or CUGBP forms complexes with the RNA in those fractions. Coimmunoprecipitation of these fractions between PTB and CUGBP failed to show interactions between the two (unpublished data), indicating the MRP RNA–PTB or –CUGBP complexes are not in these fractions. To determine whether the interactions of the MRP RNA–CUGBP–PTB complex are specific to fractions 6, 7, or 11, PTB was immunoprecipitated from pooled fractions 6 and 7 and fraction 11, and the precipitates were examined for MRP RNA with RT-PCR. The results show that while MRP RNA is present in both groups, PTB precipitates MRP RNA only in fraction 11 (Figure 4C). Additionally, PTB is able to precipitate CUGBP in fraction 11 (Figure 4D). This suggests that the newly characterized complex is specific to fraction 11, which also suggests that this complex may be of large size.
PNC components recruit each other
To further determine whether these interactions take place in vivo at the PNC, we utilized a protein tethering system recently used to study the assembly of the Cajal nuclear body (Kaiser et al., 2008). In this system, GFP-tagged PNC proteins of interest are fused with the Escherichia coli lac repressor (LacI) and transfected into HeLa cells whose genome is inserted with 256 Lac operator (LacO) repeats at chromosome 7 (Belmont et al., 1998; Kaiser et al., 2008). GFP-tagged LacI-PNC proteins were tethered onto the LacO locus, and the localization of other endogenous PNC components was evaluated. If PNC components are interacting, we expect that a protein immobilized at the LacO site should be able to recruit additional PNC components.
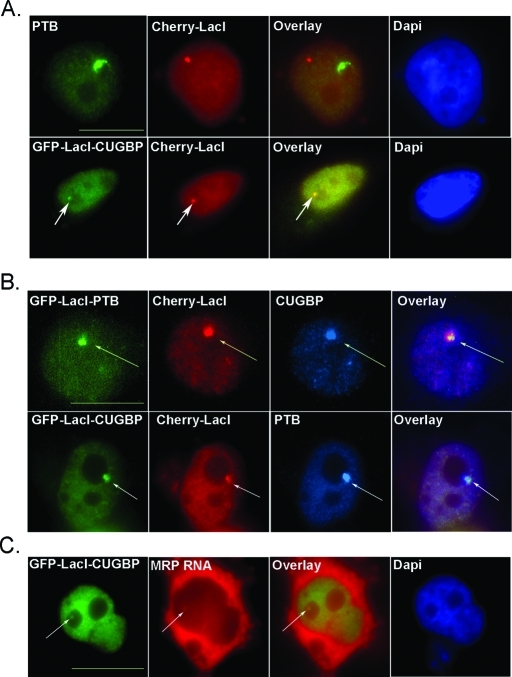
To show that the tethering system was functional, HeLa cells were transfected with either GFP-LacI-CUGBP and Cherry-LacI or Cherry-LacI alone. Cherry-LacI was used to visualize the LacO site and determine whether recruitment was occurring at this site. We found that transfected GFP-LacI-CUGBP successfully colocalized with Cherry-LacI at the LacO site, and additionally Cherry-LacI, when transfected alone, did not colocalize with endogenous PTB at the PNC (Figure 5A). To determine whether CUGBP and PTB are able to recruit each other, HeLa cells were cotransfected with Cherry-LacI and GFP-LacI-PTB or GFP-LacI-CUGBP and immunostained for endogenous CUGBP or PTB, respectively. Immobilization of CUGBP was able to efficiently recruit PTB and form a de novo PNC-like structure (Figure 5B). In 40% (n = 100) of cells transfected with GFP-LacI-CUGBP, colocalization of endogenous PTB was detected. Similarly, transfected GFP-LacI-PTB is able to recruit CUGBP, detected by immunostaining in 35% (n = 100) of transfected cells (Figure 5B). Additionally, to determine whether an immobilized PNC protein can recruit PNC-associating RNAs, we transfected cells with GFP-LacI-CUGBP, chosen for its greater transfection efficiency, and visualized MRP RNA with in situ hybridization. Our results show that immobilized CUGBP is able to recruit MRP RNA in 25% of cells (Figure 5C). These results support our hypothesis that PNC components are interacting with each other in vivo. Additionally, the recruitment efficiencies of PTB, CUGBP, and MRP RNA are similar to what has been observed in some components of the Cajal body (Kaiser et al., 2008), suggesting that like the Cajal body, the PNC components are able to self-organize, and the moderate efficiency of this organization may reflect the multifunctional nature of the components.
FIGURE 5:
Immobilized PNC proteins are able to recruit additional PNC components. Immunofluorescence microscopy of HeLa cells expressing GFP-LacI protein and/or Cherry-LacI is represented. (A, top) HeLa cells are transfected with Cherry-LacI (red) and endogenous PTB is stained (green). (A, bottom, marked with arrows) Colocalization of transfected GFP-LacI-CUGBP (green) and Cherry-LacI (red) is visualized. (B) HeLa cells are cotransfected with Cherry-LacI (red) and either GFP-LacI-PTB or GFP-LacI-CUGBP (green) and immunostained for endogenous CUGBP or PTB (blue), respectively. (C) HeLa cells were transfected with GFP-LacI-CUGCP (green) and endogenous MRP RNA was in situ labeled (red) to visualize colocalization. Scale bar = 10 μm. Arrows point to colocalization of transfected proteins with endogenous PNC components.
CUGBP and PTB behave differently at the PNC compared with nucleoplasm
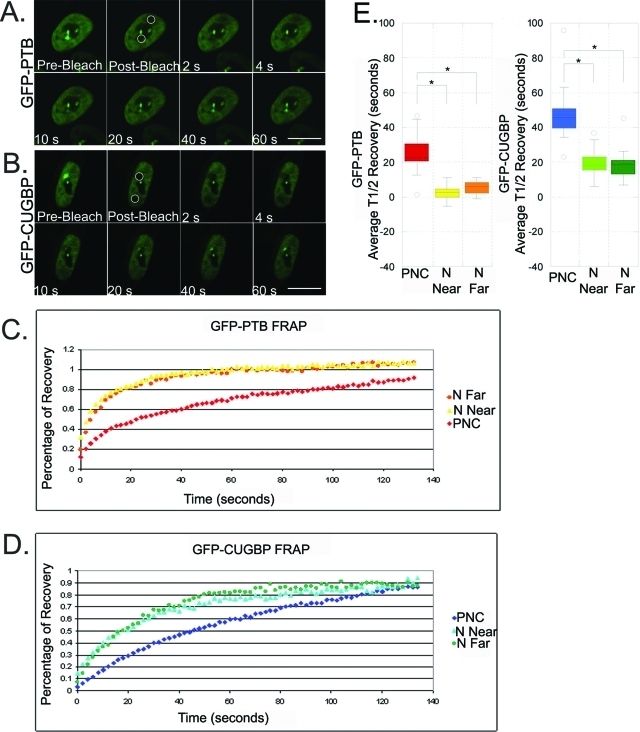
To begin analyzing the in vivo characteristic of this complex, we examined and compared the dynamics of these proteins in the PNC versus in the nucleoplasm using FRAP because the association with the PNC represents those in the unknown complex and the nucleoplasmic pool represents predominantly those involved in pre-mRNA processing. To carry out our FRAP analysis, HeLa cells containing PNCs were transiently transfected with GFP-CUGBP; GFP-PTB; or an empty vector, GFP-C1. After 24 h, two regions of interest (ROI) within the same cells were bleached and the recovery of fluorescence was recorded. One ROI covered the entire PNC including surrounding nucleoplasm, and another ROI of equal size contained nucleoplasm further away from the PNC. The two photobleached areas were further divided into three equally sized regions for quantification, which included the PNC, the nucleoplasm surrounding the PNC (N Near), and the nucleoplasm further away from the PNC (N Far).
The results of FRAP experiments for both GFP-PTB and GFP-CUGBP at the PNC and the nucleoplasm are shown in Figure 6. GFP-C1 recovery was immediate as a result of diffusion (unpublished data). In contrast, the fluorescence recovery of GFP-PTB in the PNC was significantly slower than recovery in either area of the nucleoplasm (Figure 6, A, C, and E). Additionally, the half-times of recovery for GFP-PTB in both regions of nucleoplasm were statistically indistinguishable from one another. The fluorescence of recovery for GFP-CUGBP was also much slower in the PNC (Figure 6, B, D, and E). As with GFP-PTB, we could not differentiate between the half-times of recovery for GFP-CUGBP in either part of the nucleoplasm, but both were significantly faster than in the PNC. These results demonstrate that both GFP-PTB and GFP-CUGBP exchange with different dynamics in the PNC versus in the nucleoplasm, further supporting the idea that the two proteins engage in different molecular complexes when associated with the PNC opposed to those distributed in the nucleoplasm. The compartmentalization of a subset of PTB and CUGBP to the PNC may represent a concentration of a specific function at this site. Together with the finding that PNC-associated RNAs are not associated with their characterized functional complexes at the PNC, these observations further support the presence of uncharacterized RNA–protein interactions in the PNC. The slower recovery times of PTB and CUGBP at the PNC indicate higher affinity interactions in association with the PNC as compared with the predominantly Pol II RNA–protein interactions in the nucleoplasm.
FIGURE 6:
The dynamics of GPF-CUGBP and GFP-PTB differ between the PNC and nucleoplasm. (A) Select images are shown from a representative FRAP time series acquisition of transiently transfected GFP-PTB and (B) GFP-CUGBP in HeLa cells (n = 18). Circles in the prebleached image demarcate the two bleached regions subject to quantification in each cell. Scale bar = 10 μm. (C) FRAP studies on cells that were transfected with GFP-PTB or (D) GFP-CUGBP were averaged, and percentage recovery was plotted over time. (E) Box-and-whisker plots depict the distribution of the half-recovery times for GFP-PTB and GFP-CUGBP FRAP studies. At the PNC, GFP-PTB (t1/2 = 24.9) and GFP-CUGBP (t1/2 = 48.3) recovered more slowly than nucleoplasmic GFP-PTB (N Near t1/2 = 2.6, and N Far t1/2 = 5.3) and GFP-CUGBP (N Near t1/2 = 20.0, and N Far t1/2 = 19.0). Three ROI were quantified for each study: the PNC, nucleoplasm near the PNC (N Near), and nucleoplasm further from the PNC (N Far). Each study was corrected for photofading and normalized to the prebleached image before averaging. *p < 0.001; Student’s t test.
The PNC-associated complex intertwines with newly synthesized RNA
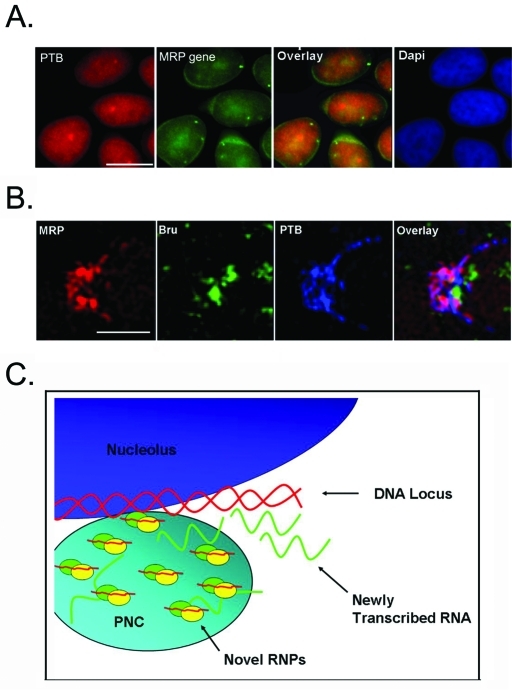
Previous studies have shown that PNCs rapidly incorporate newly synthesized RNA when cells are pulse labeled with bromouridine (BrU), and PNCs disassemble within 3 h of RNA Pol III transcription inhibition but not after RNA Pol II transcription inhibition (Huang et al., 1998; Wang et al., 2003). These findings suggest that PNCs are associated with newly synthesized Pol III RNA. However, in situ hybridization to the MRP RNA gene did not show a spatial association of the gene to the PNC (Figure 7A). It is therefore not clear whether the enrichment of MRP RNA represents newly synthesized RNA rapidly recruited to the PNC. To evaluate this possibility, HeLa cells were triple labeled with BrU after 5 min labeling, MRP RNA, and PTB. The results show that while PTB and MRP RNA are mostly colocalized in the PNC, the majority of the PNC-associated newly synthesized RNA, labeled with anti-BrU antibody, does not colocalize with, but rather intertwines around, the tight PTB-MRP labeling (Figure 7B). A prolonged BrU labeling for 15 min did not show differences in the relationship between BrU and PTB-MRP labeling (unpublished data). These results indicate that a separate population of newly synthesized RNA might associate with the PNC, but likely not as part of the MRP RNA, PTB, and CUGBP complex.
FIGURE 7:
A working model of the PNC. (A) Fluorescence microscopy was used to show localization of the PNC, labeled with PTB (red) and the MRP DNA locus (green). Scale bar = 10 μm. (B) High-resolution microscopy was used to visualize the PNC of cells triple labeled with MRP (red), BrU (green), and PTB (blue). Scale bar = 1 μm. (C) We propose that the novel RNPs associated with the PNC may be involved in processing the newly synthesized RNA at a discreet DNA locus/loci.
DISCUSSION
Although the PNC was discovered nearly two decades ago, its structure and function are not yet fully understood. PNC formation is associated with the malignant phenotype (Huang et al., 1997; Norton et al., 2008), and the PNC is enriched with a subset of Pol III, noncoding RNAs, and RNA-binding proteins that have been implicated mainly in pre-mRNA processing (reviewed in Pollock and Huang, 2009). However, the molecular relationships between these components and how they relate to PNC function have not been revealed. In this report, we show that MRP RNA, CUGBP, and PTB proteins localize to specific regions of the PNC. MRP RNA forms a previously unknown complex with CUGBP and PTB at the PNC. This newly identified RNP may play an important role in PNC function.
This is the first description that MRP RNA forms a complex with PTB and CUGBP. These interactions were tested both in vitro and in vivo through multiple approaches. Localization studies show that neither hTERT nor the protein subunits of the MRP RNP are coenriched in the PNC with MRP RNA, and coprecipitation studies show neither CUGBP nor PTB precipitates hTERT or MRP RNP protein subunits. Furthermore, glycerol gradient analyses show a separation of hTERT or MRP RNP proteins from PTB or CUGBP in MRP RNA–containing fractions, demonstrating that MRP RNA–PTB or –CUGBP interactions are distinct from previously characterized MRP RNA–containing complexes. However, it is unclear whether MRP RNA binds PTB or CUGBP directly. PTB preferentially binds pyrimidine-rich sequences (Auweter et al., 2007), and sequence analysis of MRP RNA identified a pyrimidine-rich region (CTCTGTTCCTCTCCTTTCCGCCT) in the P3 loop of the RNA. However, many attempts at in vitro gel shift assays have failed to detect a direct interaction (unpublished data). It is not clear whether the failure is due to the lack of direct interactions between MRP RNA and PTB or to the lack of the proper in vivo modifications needed for interactions in the in vitro conditions. As the gradient analyses indicate that the MRP RNA–PTB–CUGBP complex is a large complex with a high sediment index, the interactions among these factors can be more complexed. Future studies will analyze the composition of the complex to understand its functional role in cells.
While PTB and CUGBP have not previously been shown to interact with each other, they have been functionally linked in the alternative splicing of α-actinin RNA (Gromak et al., 2003). It was reported that in the α-actinin gene, smooth muscle exon splicing was associated with the displacement of PTB by binding of CUGBP at adjacent sites. This antagonistic binding was used as a form of alternative splicing regulation (Gromak et al., 2003). Here we detect coprecipitation between PTB and CUGBP, PTB and MRP RNA, as well as CUGBP and MRP RNA. However, it is not clear whether all three are in the same complex all of the time. There are at least two possibilities. CUGBP and PTB could be competitively binding MRP RNA as a means of regulating its function, similar to their behavior in alternative splicing. On the other hand, MRP RNA, CUGBP, and PTB could be forming a complex that acts to regulate other RNA or performs unknown functions. Evidence supporting the later includes the coprecipitation of PTB with CUGBP, the colocalization of CUGBP and PTB with MRP RNA at a high resolution, and cosedimentation at the same fraction in glycerol gradients. However, the RNase sensitivity of CUGBP and PTB interactions suggests either that these proteins bind on distant regions of RNA where they do not have direct protein–protein interactions or that they antagonistically bind MRP RNA. Future analysis of the binding sites and competitive binding assays between MRP RNA, PTB, and CUGBP should help clarify these questions.
Several lines of evidence demonstrate that the newly described interaction between PTB or CUGBP and MRP RNA is associated with the PNC. The FRAP studies show that the RNA-binding proteins in the PNC exchange in a different dynamic as compared with their counterparts in the nucleoplasm, demonstrating different molecular interactions for both PTB and CUGBP in the PNC versus in the nucleoplasm. Second, these components colocalize with each other in the PNC as detected at 100-nm resolution through the use of OMX microscopy. Third, our earlier finding that reduction of PTB proteins in HeLa cells significantly reduces the size and number of PNCs as measured by the localization of MRP RNA and CUGBP further supports the association of these complexes with the PNC (Wang et al., 2003). Finally, tethering of PNC components can recruit each other in vivo at the perinucleolar region further demonstrate they are par to the PNC. However, it does not preclude the existence of these complexes outside the PNC. In fact, the MRP RNA and PTB interaction can also be detected in WI-38 cells, which have little to no PNCs. Therefore these novel interactions may represent a constitutive cellular function that undergoes alteration to nucleate at the PNC in cancer cells. The identification of the complex in the future will help us analyze its function and its significance.
Although MRP RNA is associated with the PNC in complex with PTB and CUGBP, two observations suggest that it does not represent the newly synthesized RNA from RNA Pol III at the PNC. First, the localization of the gene that codes for MRP RNA is not physically close to the PNC. Second, simultaneous detection of BrU-labeled newly synthesized RNA and MRP RNA shows that MRP RNA does not colocalize with BrU labeling in the PNC as examined using the superresolution light microscopy (OMX). These findings make it less likely that the RNP containing MRP RNA, PTB, and CUGBP represents a posttranscriptional processing of a newly synthesized MRP RNA. This complex may instead play a yet to be identified role in regulating specific gene expression. A previous study from our lab shows that the PNC associates with an unknown, active gene locus and that disruption of DNA base pairing or RNA Pol III transcription inhibition blocks the nucleation of PTB, CUGBP, and MRP RNA upon this locus (Wang et al., 2003; Norton et al., 2009). The highly reticulated fibrous distribution of these components resembles chromatin fibrils, supporting the association of this complex with chromatin or RNA that is newly synthesized from the locus.
On the basis of previous studies and our observations reported here, we propose a working model regarding the novel RNA–protein complexes and their functional relationship to the PNC. In this model (Figure 7C), the novel RNPs nucleate upon DNA locus or loci, forming the PNC. These RNPs may interact directly with DNA or with newly synthesized RNA at the PNC and play a role in the regulation of gene expression at the PNC-associated DNA locus or loci. These functions may take place in PNC-lacking cells but most likely undergo significant increases in PNC-containing cells. We are currently working to identify the DNA locus that the PNC associates with and therein to determine the identity of the newly transcribed RNAs. Studies are also underway to identify the complete novel RNP complex so that we will be able to functionally analyze its role at the PNC and ultimately the function of PNCs in cancer cells.
MATERIALS AND METHODS
Cell culture and transfection
HeLa and HeLa-LacO (Kaiser et al., 2008) cells were cultured in DMEM (Invitrogen), supplemented with 10% fetal bovine serum (FBS) (Gemini Science, Tucson, AZ) and 5% penicillin/streptomycin (Invitrogen, Carlsbad, CA). PC3M cells were cultured in RPMI 1640 media (Invitrogen), supplemented with 10% FBS and 5% penicillin/streptomycin. For transfection, HeLa-LacO cells were seeded onto glass coverslips in 35-mm Petri dishes and grown for 24 h at 37ºC. Plasmids were transiently transfected into cells with Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions or via electroporation using standard procedures (Wang et al., 2003) and incubated for 18–24 h before experimentation.
Plasmids
pEGFP-C1-LacI-NLS plasmids were kindly provided by Miroslav Dundr of Rosalind Franklin University of Medicine and Science (Chicago, IL) (Kaiser et al., 2008). To make LacI fusion proteins, PTB was PCR amplified and subcloned into the plasmid as a Kpn1-BamH1 fragment. CUGBP was PCR amplified and subcloned in as an EcoR1-BamH1 fragment. The plasmid GFP-CUGBP was a generous gift from Marion Hamshere of the University of Nottingham (Nottingham, UK) (Fardaei et al., 2001). The GFP-PTB plasmid was created and characterized in our lab (Huang et al., 1997).
Pull-down and IP
To prepare nuclear extract, cells were washed with 1× phosphate-buffered saline (PBS), resuspended in hypotonic buffer (10 mM Tris-HCl, 3 mM MgCl2, 5 mM NaCl), and disrupted by Dounce homogenization with 0.5% Triton. Nuclei were separated through centrifugation (600 × g for 10 min) and suspended in IPP 150 buffer (0.05% NP40, 10 mM Tris-HCl, 150 mM NaCl, 1× protease inhibitor).
For pull-downs, 10 μl TH12 and control oligos (100 uM) (Keck Foundation, Yale University, New Haven, CT) were incubated with 50 μl streptavidin Dynabeads (Invitrogen) overnight in 500 μl IPP 150 buffer at 4°C. Beads with linked oligos were then incubated with prepared nuclear extract for 2 h and subsequently washed with IPP 150 buffer. Pulled-down proteins and RNAs were analyzed with Western blot and RT-PCR, respectively.
For IP, 10 μl antibody was incubated with 50 μl protein A Dynabeads (Invitrogen) overnight at 4°C in IPP 150 buffer. Then 500 μl nuclear buffer was incubated with the prepared beads for 1 h at 4°C. Samples were washed with IPP 150 buffer and analyzed with RT-PCR and Western blot. Antibodies used for IP and Western blot include anti-Rpp30 (Jiang et al., 2001), anti–PTB SH54 (Huang et al., 1997), hnRNP I (Santa Cruz Biotechnology, Santa Cruz, CA), anti–CUGBP 3B1 (Santa Cruz), and anti-hTERT (Rockland Immunochemicals, Gilbertsville, PA).
In situ hybridization and immunolabeling
Immunolabeling and in situ hybridization to RNA experiments were carried out as described in Wang et al. (2003), with the following modifications. In situ hybridization was performed with the biotinylated TH12 MRP RNA–specific oligo 5′-GUAA CUAG AGGG AGCU BBB-3′. Cells were fixed with 4% formaldehyde in PBS for 12 min and washed three times with PBS. Coverslips were equilibrated with wash solution (4× SSC and 0.1% Tween-20 in PBS) for 10 min, and hybridization was completed as previously described. In situ hybridization to DNA is performed according to the protocol described in Live Cell Imaging: A Laboratory Manual (Goldman and Spector, 2005). A Bac-containing MRP RNA gene was used to prepare the hybridization probes through nick translation. Cells were immunolabeled with SH54 to mark the PNC before the DNA in situ hybridization.
RT-PCR
Total RNA was isolated using TRIzol reagent (Invitrogen) according to the manufacturer’s instructions. cDNA was synthesized with Superscript Reverse Transcriptase III (Invitrogen), and DNA was PCR amplified with gene-specific primers.
Western blot
Proteins were prepared with Laemmli buffer, boiled for 10 min, and run on a 12% SDS–PAGE gel. Proteins were transferred to a polyvinylidene difluoride membrane (Bio-Rad, Hercules, CA) and blocked in 5% milk buffer for 30 min. Primary antibody incubations were overnight at 4ºC, followed by 1-h room temperature incubations with TrueBlot horseradish peroxidase–linked anti–mouse secondary antibody (eBiosciences, San Diego, CA). Blots were developed using SuperSignal West Pico Chemiluminescent Substrate (Thermo Scientific, Waltham, MA).
FRAP
The FRAP experiments were conducted using a Zeiss LSM 510 confocal microscope equipped with a Hamamatsu CCD camera. A 63× oil lens with a numerical aperture (NA) of 1.4 was used for image acquisition. Fluorescence intensities were measured in three ROI—the PNC, the nucleoplasm near the PNC (N Near), and the nucleoplasm far from the PNC (N Far)—in each frame. The average intensities of the areas of interest in images including before, immediately after, and the series of time points after bleaching were measured under the same conditions for each data set. The relative fluorescence intensity (RFI) in the FRAP analysis was calculated as (ROIB/ROIBEN) / (ROIPB/ROIPEN). ROIB is the average intensity of the bleached area at various time points after bleaching. ROIBEN is the average intensity of the entire nucleus at the corresponding time points. ROIPB is the average intensity of the bleached area before bleaching. ROIPEN is the average intensity of the entire nucleus before bleaching. When ROIB/ROIBEN = ROIPB/ROIPEN, namely, when the RFI is 1, fluorescence recovery of the bleached zone reaches 100%. The ROIB of the images acquired immediately after bleaching equals 0. Using the equation, we have taken into consideration the overall fluorescence change during subsequent image acquisitions.
BrU incorporation
HeLa cells were grown to confluence on glass coverslips, and BrU incorporation was performed as described previously (Wang et al., 2008). MRP RNA in situ hybridization and PTB immunolabeling were prepared as described above.
Microscopy analysis
To obtain fluorescent images, glass coverslips (prepared as described above) were analyzed with a Nikon Eclipse E800 microscope with a 60× objective lens equipped with a SenSys cooled CCD camera (Photometrics, Tucson, AZ). Images were captured using MetaMorph image acquisition software (Universal Imaging, Sunnyvale, CA). For DeltaVision OMX microscopic analyses, HeLa cells were plated to 80% confluence on a coverslip, and RNA and proteins were visualized by in situ hybridization and immunolabeling as described above. Images of cells were obtained with the OMX structured illumination microscope at Applied Precision or at Andrew Belmont’s laboratory. The standard configuration (100×, 1.4 NA, objective lens, 2× EM-CCD cameras, 405- and 488-nm lasers 100×) was used in the image acquisition and the z-sections were acquired in 250-nm intervals.
Glycerol gradient sedimentation
The nuclear lysate of HeLa cells was prepared as previously described and layered on a 5–45% glycerol gradient containing IPP 150 buffer. The gradient was run for 18 h at 38,000 rpm at 4°C in a Beckman SW41 rotor and fractioned into 11 samples. Glycerol gradient IPs were performed under the same conditions as previously described IPs, with fractioned samples used as lysate.
Acknowledgments
We would to thank the generous colleagues who provided us with reagents, Mirek Dundr and Sidney Altman. We will also like to thank Andrew Belmont and Andy (Wei Cheng) Wu, of Applied Precision, for allowing us to use the OMX system for data acquisition. Thanks are also directed to Xavier Darzacq, who hosted S. H. for the work in sedimentation property characterization. This work is supported by a National Cancer Institute Training Program in Oncogenesis and Developmental Biology grant (T32 CA-080621) to C. P., a National Institutes of Health grant (R01 GM-078555) to S. H., École Normale Supérieure, a grant from Fondation Pierre Gilles de Gennes, and a Partners University Fund program grant to O. B. and S. H.
Abbreviations used:
- BrU
bromouridine
- CUGBP
CUG-binding protein
- EM
electron microscopy
- FBS
fetal bovine serum
- FRAP
fluorescence recovery after photobleaching
- GFP
green fluorescent protein
- hTERT
human telomerase reverse transcriptase
- IgG
immunoglobulin G
- IP
immunoprecipitation
- MRP
mitochondrial RNA processing
- NA
numerical aperture
- OMX
Optical Microscope, eXperimental
- PBS
phosphate-buffered saline
- PNC
perinucleolar compartment
- Pol
polymerase
- PTB
pyrimidine tract–binding protein
- RFI
relative fluorescence intensity
- RNP
ribonucleoprotein
- ROI
regions of interest
- TRAF
tumor necrosis factor receptor–associated factor.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E10-09-0768) on January 13, 2011.
REFERENCES
- Aeby E, Ullu E, Yepiskoposyan H, Schimanski B, Roditi I, Muhlemann O, Schneider A. tRNASec is transcribed by RNA polymerase II in Trypanosoma brucei but not in humans. Nucleic Acids Res. 2010;38:5833–5843. doi: 10.1093/nar/gkq345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auweter SD, Oberstrass FC, Allain FH. Solving the structure of PTB in complex with pyrimidine tracts: an NMR study of protein-RNA complexes of weak affinities. J Mol Biol. 2007;367:174–186. doi: 10.1016/j.jmb.2006.12.053. [DOI] [PubMed] [Google Scholar]
- Belmont AS, Li G, Sudlow G, Robinett C. Visualization of large-scale chromatin structure and dynamics using the lac operator/lac repressor reporter system. Methods Cell Biol. 1998;58:203–222. doi: 10.1016/s0091-679x(08)61957-3. [DOI] [PubMed] [Google Scholar]
- Davis I. The ‘super-resolution’ revolution. Biochem Soc Trans. 2009;37:1042–1044. doi: 10.1042/BST0371042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fardaei M, Larkin K, Brook JD, Hamshere MG. In vivo co-localisation of MBNL protein with DMPK expanded-repeat transcripts. Nucleic Acids Res. 2001;29:2766–2771. doi: 10.1093/nar/29.13.2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghetti A, Pinol-Roma S, Michael WM, Morandi C, Dreyfuss G. hnRNP I, the polypyrimidine tract-binding protein: distinct nuclear localization and association with hnRNAs. Nucleic Acids Res. 1992;20:3671–3678. doi: 10.1093/nar/20.14.3671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman R, Spector D, editors. Live Cell Imaging: A Laboratory Manual. New York: Cold Spring Harbor Laboratory Press; 2005. [Google Scholar]
- Gromak N, Matlin AJ, Cooper TA, Smith CW. Antagonistic regulation of α-actinin alternative splicing by CELF proteins and polypyrimidine tract binding protein. RNA. 2003;9:443–456. doi: 10.1261/rna.2191903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall MP, Huang S, Black DL. Differentiation-induced colocalization of the KH-type splicing regulatory protein with polypyrimidine tract binding protein and the c-src pre-mRNA. Mol Biol Cell. 2004;15:774–786. doi: 10.1091/mbc.E03-09-0692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S, Deerinck TJ, Ellisman MH, Spector DL. The dynamic organization of the perinucleolar compartment in the cell nucleus. J Cell Biol. 1997;137:965–974. doi: 10.1083/jcb.137.5.965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S, Deerinck TJ, Ellisman MH, Spector DL. The perinucleolar compartment and transcription. J Cell Biol. 1998;143:35–47. doi: 10.1083/jcb.143.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huttelmaier S, Illenberger S, Grosheva I, Rudiger M, Singer RH, Jockusch BM. Raver1, a dual compartment protein, is a ligand for PTB/hnRNPI and microfilament attachment proteins. J Cell Biol. 2001;155:775–786. doi: 10.1083/jcb.200105044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang T, Guerrier-Takada C, Altman S. Protein-RNA interactions in the subunits of human nuclear RNase P. RNA. 2001;7:937–941. doi: 10.1017/s1355838201010299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser TE, Intine RV, Dundr M. De novo formation of a subnuclear body. Science. 2008;322:1713–1717. doi: 10.1126/science.1165216. [DOI] [PubMed] [Google Scholar]
- Kamath RV, et al. Perinucleolar compartment prevalence has an independent prognostic value for breast cancer. Cancer Res. 2005;65:246–253. [PubMed] [Google Scholar]
- Kopp K, Huang S. Perinucleolar compartment and transformation. J Cell Biochem. 2005;95:217–225. doi: 10.1002/jcb.20403. [DOI] [PubMed] [Google Scholar]
- Lee B, Matera AG, Ward DC, Craft J. Association of RNase mitochondrial RNA processing enzyme with ribonuclease P in higher ordered structures in the nucleolus: a possible coordinate role in ribosome biogenesis. Proc Natl Acad Sci USA. 1996;93:11471–11476. doi: 10.1073/pnas.93.21.11471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li K, Smagula CS, Parsons WJ, Richardson JA, Gonzalez M, Hagler HK, Williams RS. Subcellular partitioning of MRP RNA assessed by ultrastructural and biochemical analysis. J Cell Biol. 1994;124:871–882. doi: 10.1083/jcb.124.6.871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matera AG, Frey MR, Margelot K, Wolin SL. A perinucleolar compartment contains several RNA polymerase III transcripts as well as the polypyrimidine tract-binding protein, hnRNP I. J Cell Biol. 1995;129:1181–1193. doi: 10.1083/jcb.129.5.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norton JT, Pollock CB, Wang C, Schink JC, Kim JJ, Huang S. Perinucleolar compartment prevalence is a phenotypic pancancer marker of malignancy. Cancer. 2008;113:861–869. doi: 10.1002/cncr.23632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norton JT, Wang C, Gjidoda A, Henry RW, Huang S. The perinucleolar compartment is directly associated with DNA. J Biol Chem. 2009;284:4090–4101. doi: 10.1074/jbc.M807255200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollock C, Huang S. The perinucleolar compartment. J Cell Biochem. 2009;107:189–193. doi: 10.1002/jcb.22107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reimer G, Raska I, Scheer U, Tan EM. Immunolocalization of 7-2-ribonucleoprotein in the granular component of the nucleolus. Exp Cell Res. 1988;176:117–128. doi: 10.1016/0014-4827(88)90126-7. [DOI] [PubMed] [Google Scholar]
- Timchenko LT, Miller JW, Timchenko NA, DeVore DR, Datar KV, Lin L, Roberts R, Caskey CT, Swanson MS. Identification of a (CUG)n triplet repeat RNA-binding protein and its expression in myotonic dystrophy. Nucleic Acids Res. 1996;24:4407–4414. doi: 10.1093/nar/24.22.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Norton JT, Ghosh S, Kim J, Fushimi K, Wu JY, Stack MS, Huang S. Polypyrimidine tract-binding protein (PTB) differentially affects malignancy in a cell line-dependent manner. J Biol Chem. 2008;283:20277–20287. doi: 10.1074/jbc.M803682200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Politz JC, Pederson T, Huang S. RNA polymerase III transcripts and the PTB protein are essential for the integrity of the perinucleolar compartment. Mol Biol Cell. 2003;14:2425–2435. doi: 10.1091/mbc.E02-12-0818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan P, Benhattar J, Seelentag W, Stehle JC, Bosman FT. Immunohistochemical localization of hTERT protein in human tissues. Histochem Cell Biol. 2004;121:391–397. doi: 10.1007/s00418-004-0645-5. [DOI] [PubMed] [Google Scholar]