Nuclear targeting of intact proteasome particles was tested in the Xenopus egg extract system. Both the 26S proteasome holoenzyme and the 20S core particle were targeted to the nuclear envelope but could not enter the nucleus. A novel proteolytically active 20S+ particle was actively imported into the nucleoplasm in a Ran-independent fashion.
Abstract
The 26S proteasome is a conserved 2.5 MDa protein degradation machine that localizes to different cellular compartments, including the nucleus. Little is known about the specific targeting mechanisms of proteasomes in eukaryotic cells. We used a cell-free nuclear reconstitution system to test for nuclear targeting and import of distinct proteasome species. Three types of stable, proteolytically active proteasomes particles were purified from Xenopus egg cytosol. Two of these, the 26S holoenzyme and the 20S core particle, were targeted to the nuclear periphery but did not reach the nucleoplasm. This targeting depends on the presence of mature nuclear pore complexes (NPCs) in the nuclear envelope. A third, novel form, designated here as 20S+, was actively imported through NPCs. The 20S+ proteasome particle resembles recently described structural intermediates from other systems. Nuclear import of this particle requires functional NPCs, but it is not directly regulated by the Ran GTPase cycle. The mere presence of the associated “+” factors is sufficient to reconstitute nuclear targeting and confer onto isolated 20S core particles the ability to be imported. Stable 20S+ particles found in unfertilized eggs may provide a means for quick mobilization of existing proteasome particles into newly formed nuclear compartments during early development.
INTRODUCTION
The distinctive feature of all eukaryotic cells is compartmentalization, which is achieved by a set of membrane boundaries and their associated translocation machineries. The nuclear border consists of the two concentric membranes of the nuclear envelope, which converge at the sites of nuclear pore complexes (NPCs; Hetzer and Wente, 2009). NPCs are huge proteinaceous assemblies that form gated channels for selective bidirectional traffic between the cytoplasm and the nucleus (Weis, 2003; Fahrenkrog et al., 2004). Most nuclear transport pathways are mediated by specific shuttling receptors and are regulated by the small GTPase Ran (Gorlich et al., 1996). Importin β, the prototypic nuclear import receptor, ferries proteins into the nucleus through the use of adaptor molecules, such as importin α, or by direct binding of some cargoes. Distinct transport pathways are defined by combinations of specific shuttling receptors and their adaptors (Fried and Kutay, 2003; Fahrenkrog et al., 2004; Harel and Forbes, 2004; Pemberton and Paschal, 2005).
The 26S proteasome is a highly conserved 2.5 MDa supramolecular assembly that is responsible for the degradation of most cytosolic, nuclear, and endoplasmic reticulum-associated proteins (ER) (Glickman and Ciechanover, 2002; Finley, 2009). The proteasome holoenzyme is a molecular machine composed of a catalytic 20S core particle (20S CP) connected to one or two ATP-dependent regulatory 19S complexes (19S RP) (Pickart and Cohen, 2004). Polyubiquitinated proteins are targeted to the 19S RP, which is responsible for their ATP-dependent unfolding and threading through a narrow channel into the internal cavity of the 20S CP (Glickman and Ciechanover, 2002; Pickart and Cohen, 2004). The 20S CP is composed of four stacked heptameric rings, each made up of seven related but nonidentical α or β subunits. The β subunits contain three types of functional peptidase sites that carry out the degradation of substrates inside the internal cavity of the core particle (Glickman, 2000; Glickman and Ciechanover, 2002; Finley, 2009). Although in most studies the 26S proteasome holoenzyme seems to be the active and primary form, it may exist in a dynamic equilibrium with its subcomplexes (20S CP and 19S RP) in different cell types or in response to changes in growth conditions (Godon et al., 1998; Bajorek et al., 2003; Liu et al., 2006; Tonoki et al., 2009; Tsvetkov et al., 2009).
Proteasomes have been localized to many different compartments within eukaryotic cells, including the cytoplasm, specific intranuclear locations (the nucleolus, promyelocytic leukemia [PML]-nuclear bodies, and chromatin), the nuclear envelope, the outer side of the endoplasmic reticulum, and cytoskeletal elements (Olink-Coux et al., 1994; Palmer et al., 1996; Arcangeletti et al., 1997; Reits et al., 1997; Wilkinson et al., 1998; Wojcik, 1999; Mattsson et al., 2001; Gordon, 2002; Wojcik and DeMartino, 2003; Girao et al., 2005; Adori et al., 2006; von Mikecz et al., 2008). Little is known about the dynamic distribution and regulated activity of proteasomes in the context of these different locations. Equally unknown is the identity of most of the cellular factors responsible for targeting and/or anchoring proteasomes to specific locations. Nuclear import of newly synthesized proteasome subunits has been investigated in some detail in yeast. These studies suggest that yeast 20S CP components are imported into the nucleus as inactive precursor complexes by the importin α/β pathway, possibly aided by additional factors such as the maturation factor Ump1 (Lehmann et al., 2002). Additional evidence suggests that components of the 19S regulatory complex are imported separately and independently from the 20S CP (Wendler et al., 2004; Isono et al., 2007). Thus in yeast the 26S proteasome may be assembled inside the nucleus from different, independently imported modules. The difficulty in assessing the dynamic distribution of proteasomes in living cells stems from the high abundance of these particles and their affinity to many different cellular substructures. Because proteasomes are essential, long-lived complexes, de novo synthesis of proteasomal subunits occurs in addition to stable pools of 20S CP and 26S particles and adds to this complexity. Although most studies have focused on de novo synthesized subunits, work in digitonin-permeabilized mammalian cells has suggested that intact proteasome particles may be imported into nuclei under some conditions (Wang et al., 1997; Mayr et al., 1999). At a width of 15–20 nm, import of intact 20S or even 26S proteasome complexes is potentially possible, given that the gated channel of the NPC has been shown to expand and to accommodate cargoes with a diameter of up to ∼39 nm (Pante and Kann, 2002).
In many arrested cells, such as oocytes and unfertilized eggs of various animal species, proteasomes exhibit distinct cytoplasmic staining patterns but are excluded from meiotic spindles and chromatin. Dramatic changes are observed after fertilization and in the early cleavage divisions, with rapid accumulation of proteasomes inside newly formed nuclei (Ryabova et al., 1994; reviewed by Wojcik and DeMartino, 2003). To address the dynamic distribution of intact proteasomes across the nucleocytoplasmic border, we chose to use a cell-free system derived from Xenopus egg extracts. These extracts have been widely used in experimental cell biology because of their ability to maintain specific cell-cycle states and to recapitulate in vitro many of the events in early embryonic development (Forbes et al., 1983; Lohka and Masui, 1983; Murray and Kirschner, 1989; Finlay and Forbes, 1990; Hirano and Mitchison, 1991; Murray et al., 1996; Harel et al., 2003b). Specifically, we used interphase extracts to reconstitute nuclei with functional NPCs. We then followed the targeting and import of distinct proteasome species into the newly formed nuclear compartments.
RESULTS
Proteasomes associate with functional nuclei reconstituted in vitro from Xenopus egg extract
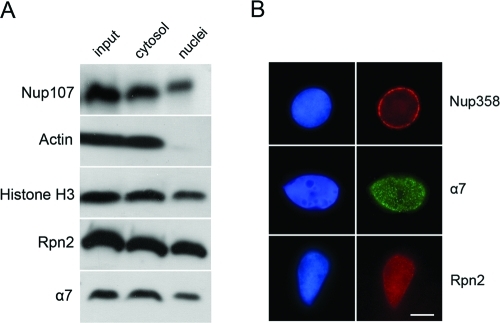
Amphibian eggs contain a large reservoir of nuclear components, which are stored for use during the rapid cell divisions that follow fertilization. This forms the basis for the use of cell-free systems to study nuclear assembly and dynamics (Forbes et al., 1983; Lohka and Masui, 1983; Murray and Kirschner, 1989). We used Xenopus egg extracts to assemble functional nuclei around templates of demembranated sperm chromatin. A typical assembly mixture containing cytosol, membrane vesicles, and chromatin templates leads to robust assembly of nuclei within 1 h of incubation. Nuclear envelopes replete with functional NPCs are formed around decondensed chromatin, and endogenous nuclear proteins from the extract are gradually imported into the reconstituted nuclei. Following assembly, we monitored the redistribution of proteasome subunits between the nuclear and cytoplasmic compartments. Nuclei were separated from cytosol by centrifugation through a sucrose cushion and the resulting fractions were immunoblotted with various antibodies. Actin served as a cytosolic marker and was completely excluded from nuclei, while significant amounts of both histone H3 and the nucleoporin Nup107 were detected in the nuclear fraction. Similarly, α7 (a 20S CP subunit) and Rpn2 (a component of the 19S RP) were detected in both the cytosolic and nuclear fractions (Figure 1A). Next, nuclear targeting of proteasomes was visualized by immunofluorescence staining (Figure 1B). Successful reconstitution of nuclei was confirmed with an antibody directed against Nup358, the major component of the cytoplasmic filaments of the NPC. In parallel, immunostaining with the proteasome markers α7 and Rpn2 indicated that a portion of these endogenous components in Xenopus egg extract associates with the reconstituted nuclei. The staining pattern suggests that these 20S CP and 19S RP components are not merely associated with nuclear envelopes but also reach the nuclear interior.
FIGURE 1:
Nuclear targeting of proteasomes. (A) In vitro reconstituted nuclei were separated from the surrounding cytosol by centrifugation through a sucrose cushion, as described in Materials and Methods. Total reaction, cytosolic, and nuclear fractions were immunoblotted with specific markers. Note that actin is exclusively cytosolic, while a fraction of the histone and nucleoporin markers are recruited to nuclei within the time of incubation. The α7 and Rpn2 proteasome subunits are also significantly recruited to the nuclear fraction. (B) Reconstituted nuclei were fixed and processed for indirect immunofluorescence. DNA was stained with Hoechst 33258 (left column). Anti-Nup358 exhibits the typical punctuate rim staining of NPC components. Both α7 and Rpn2 antibodies exhibit punctuate staining throughout the nucleus. Scale bar, 10 μm.
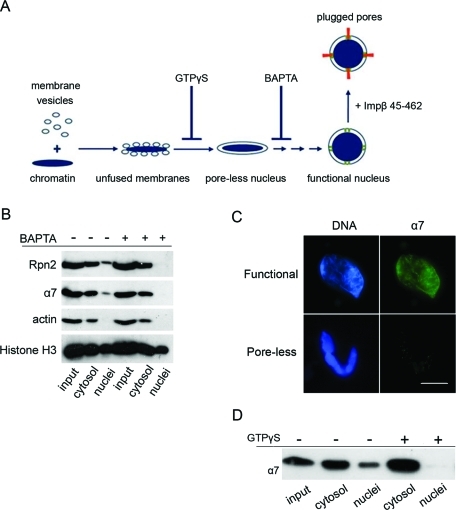
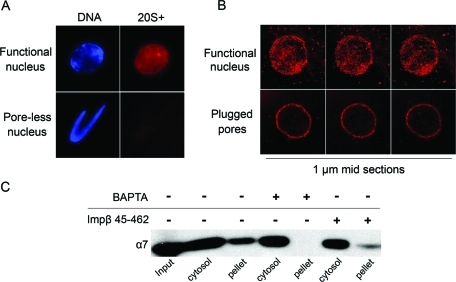
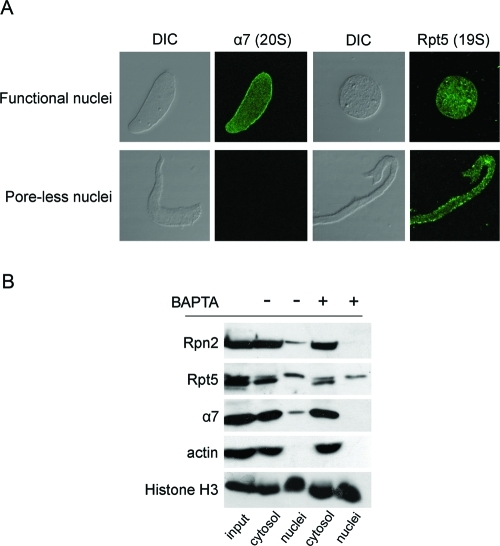
The observed nuclear localization of proteasomes could be the result of an interaction with chromatin prior to nuclear assembly or of specific targeting to nuclei after they are fully formed. To differentiate between these possibilities, we used known inhibitors that interfere with the early stages of nuclear assembly (Macaulay and Forbes, 1996; Harel and Forbes, 2004). As shown schematically in Figure 2A, nuclear assembly is a sequential process that can be blocked at distinct steps by the nonhydrolyzable GTP analog, GTPγS, and the calcium chelator BAPTA, which results in aborted assembly intermediates lacking mature NPCs and the majority of nuclear pore subunits (Harel et al., 2003a). The proteasomal α7 and Rpn2 subunits did not associate with BAPTA-inhibited intermediates, which contain fully sealed nuclear membranes with no NPCs (Figure 2B). This result was corroborated by immunofluorescence staining with anti-α7 (Figure 2C). An earlier stage in assembly is represented by GTPγS-inhibited intermediates, which contain unfused membrane vesicles docked onto chromatin. Again the α7 proteasomal marker was absent from these isolated intermediates (Figure 2D). Taken together, our results suggest that endogenous proteasomes are partially localized to newly formed nuclei. This nuclear targeting requires mature nuclear envelopes containing NPCs.
FIGURE 2:
Proteasome targeting requires mature nuclear envelopes and NPCs. (A) Schematic representation of major steps and known chemical inhibitors of the nuclear assembly pathway (reviewed by Harel and Forbes, 2004). The addition of GTPγS at t = 0 of nuclear reconstitution results in unfused membrane vesicles docked on the surface of chromatin. BAPTA inhibits a subsequent step, resulting in aborted assembly intermediates with sealed double nuclear membranes, but no NPCs (poreless nuclei). If nuclear assembly is allowed to proceed normally and recombinant Impβ 45–462 is later added, mature NPCs are formed, but nuclear transport is blocked (plugged pores). (B) Separation of nuclear and cytosolic fractions was carried out as in Figure 1A and compared between normal nuclei (left three columns) and BAPTA-inhibited intermediates (right three columns). Proteasome markers were missing in BAPTA-inhibited nuclear intermediates. (C) Normal (functional) nuclei and BAPTA-inhibited intermediates were immunostained with anti-α7 as in Figure 1B. Scale bar, 10 μm. (D) Separated nuclear and cytosolic fractions were compared between normal assembly and GTPγS-inhibited reactions. The α7 proteasomal marker is absent from the aborted assembly intermediates.
Biochemical purification of functional proteasome species from Xenopus egg extract
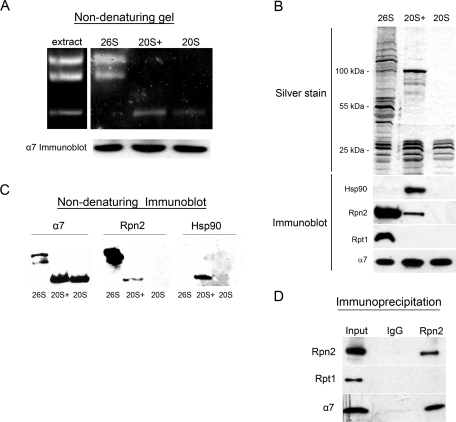
Having found that endogenous proteasome subunits redistribute to newly formed nuclei (Figures 1 and 2), we asked whether they enter as mature, functional proteasome complexes. Proteasomes in dividing cells are usually found as a mixture of 20S CPs and 26S holoenzymes. These two molecular species are easily resolved by nondenaturing PAGE (“native gels”) (Rechsteiner et al., 1993; Bajorek et al., 2003). Indeed, we found two proteasome species in Xenopus interphase egg extract: the slower migrating 26S holoenzyme doublet, and a faster migrating, proteolytically active form that appeared to correspond to the 20S CP (Figure 3A, extract). These functional proteasomes serve as the source for purifying proteasome complexes from egg extract (see Materials and Methods, Supplemental Figure S1, and Peters et al., 1991). Isolated 26S proteasome particles retained their proteolytic activity (Figure 3A) and contained all the known bona fide proteasome subunits, as determined by mass spectrometry analysis (Supplemental Table S1). However, these isolated 26S particles did not accumulate inside in vitro reconstituted nuclei (unpublished data; see Figure 4). This initial observation raised the possibility that fully assembled 26S proteasome holoenzymes cannot be imported through NPCs. We next attempted to purify the 20S CP using a series of biochemical purification steps based on established protocols from budding yeast (Supplemental Figure S1 and Materials and Methods). This process resulted in the isolation of the faster migrating form, as determined by native gel analysis (Figure 3A, right). However, when analyzed by denaturing gel electrophoresis, the 20S CP sample contained several additional protein bands with apparent molecular masses above the 20S α and β subunits (Figure 3B, silver staining, middle column). Mass spectrometry analysis identified the 19S subunits Rpn1 and Rpn2, as well as Hsp90 and importin β, among these components (Supplemental Table S2). More stringent conditions, involving salt treatment and hydroxyapetite chromatography, stripped off these proteins and yielded typical “naked” 20S core particles (Supplemental Figure S1 and Figure 3, A and B, 20S lanes).
FIGURE 3:
Biochemical purification of three distinct species of proteasome particles. (A) Migration of endogenous proteasomes (extract) and purified particles (designated 26S, 20S+, and 20S) in native gels. Active particles were visualized by the Suc-LLVY-AMC peptide, which becomes fluorescent upon proteolytic cleavage in the internal cavity of the proteasome core particle. The slower migrating top two bands correspond to single- and double-capped 26S forms known from yeast extracts and other systems. The 20S+ particle shows higher peptidase activity than an equivalent sample of the 20S CP. (B) Silver staining (top) and immunoblot analysis (bottom) of samples of the three purified particle preparations, following denaturing SDS–PAGE. The blot was probed with antibodies directed against α7 (a 20S CP subunit), Rpn2 and Rpt1 (19S subunits), and Hsp90, which is only detected in the 20S+ particle. (C) Identical samples of the three purified particle preparations (26S, 20S+, and 20S) were loaded for the in-gel peptidase activity assay shown in (A) and in three native gels used for immunoblotting. Equal loading based on anti-α7 reactivity in a denaturing immunoblot is shown in the bottom strip of (A). The native blots were probed against α7, Rpn2, and Hsp90, all of which were detected in the single band of the 20S+ particle. (D) Immunoprecipitation out of Xenopus egg cytosol was performed with affinity-purified antibodies generated against the central PC-repeat domain of xRpn2, or control rabbit IgG. Rpn2 and α7 were coimmunoprecipitated, but Rpt1 was missing, indicating the presence of an endogenous particle in the extract containing the 20S CP together with an exposed Rpn2 subunit.
FIGURE 4:
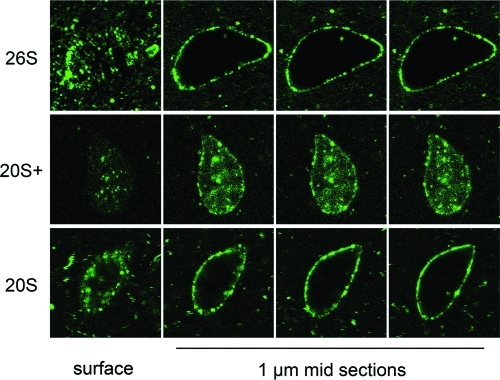
Nuclear targeting and import of fluorescently labeled proteasomes. Purified 26S, 20S+, and 20S particles were fluorescently labeled and added into nuclear reconstitution reactions after the assembly of functional nuclei had been completed. Samples were fixed and analyzed by confocal microscopy. For each of the three particles a representative nucleus is shown, starting with a surface (top) view section on the left, and followed by three consecutive 1-μm-thick confocal sections through the middle of the nucleus. The labeled 26S and 20S particles accumulate at the nuclear envelope, while the 20S+ particle reaches the nucleoplasm.
These results suggest that core particles that are not incorporated into 26S holoenzymes may associate with other factors to yield a stable complex. Specifically, in Xenopus egg extract we report the presence of a 20S-associated molecular species, which we refer to here as 20S+. This particle is not easily discerned from the 20S CP by native gel separation (Figure 3A). Immunoblotting purified proteasome samples identified both Rpn2 and Hsp90 in the 20S+ sample, comigrating as a ∼100 kDa band (Figure 3B). By contrast, only Rpn2 was detected in 26S complexes. Other 19S regulatory complex subunits were missing from the purified 20S+ particle, as demonstrated for the Rpt1 ATPase that was detected only in the 26S holoenzyme (Figure 3B).
To confirm that the components in the 20S+ preparation were present as a stable complex, we performed nondenaturing immunoblot analysis. The three purified proteasome preparations (26S, 20S+, and 20S) were resolved by nondenaturing (native) gels. Loading was normalized based on a denaturing (SDS–PAGE) immunoblot with anti-α7, a subunit that is present in all three particles (Figure 3A, α7 immunoblot). We note that the in-gel activity of the 20S+ form appears to be stronger than that of comparable 20S CPs. All bands showing peptidase activity in the native gel were identified by the same anti-α7 antibody in the nondenaturing immunoblot (Figure 3C, left). Anti-Rpn2 reacted with both 26S bands and 20S+, indicating that these complexes contain Rpn2 as a stably comigrating component. No Rpn2 was identified in the 20S band (Figure 3C, middle). On the basis of the signal intensities of anti-Rpn2 relative to anti-α7 in each species, it may be concluded that Rpn2 is found at lower levels in 20S+ (this is also seen for Rpn2 in the denaturing immunoblot in Figure 3B). Anti-Hsp90 reacted only with 20S+ (Figure 3C, right) and is missing from 26S and 20S. Most importantly, the alignment of the three nondenaturing immunoblots shows that α7, Rpn2, and Hsp90 are all present in the same native complex, which corresponds to the single proteolytically active band of 20S+. In addition, coimmunoprecipitation performed with anti-Rpn2 in unfractionated egg cytosol demonstrated that Rpn2 and the 20S CP subunit α7 are found in a common complex, which lacks Rpt1 (Figure 3D). Together, our results suggest that Rpn2 and Rpn1 (Supplemental Table S2) may associate with the 20S CP independently of other 19S RP subunits and that they can form a stable particle form that also contains Hsp90 (see Discussion).
Thus it is possible to biochemically obtain three distinct types of proteasome complexes from egg extract, although it is difficult to distinguish between them solely by in-gel peptidase activity in native gel electrophoresis.
26S and 20S particles are targeted to the nuclear periphery while 20S+ is imported through nuclear pores
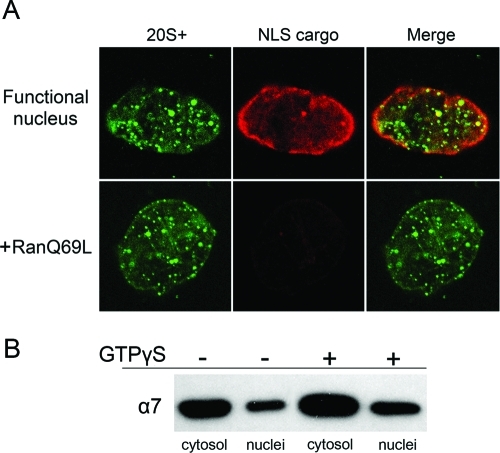
With three proteasome species in hand, we could label the purified particles and introduce them into reconstitution reactions to test their nuclear targeting capacity. Each of the purified particles (26S, 20S+, and 20S) was fluorescently labeled and added into in vitro reconstitution reactions after nuclei had fully formed. A large number of nuclei were examined, in multiple reactions, by epifluorescence and confocal microscopy. Each of the three proteasome species displayed a consistent targeting behavior, demonstrated by a series of optical sections shown for representative nuclei in each category in Figure 4. Both the 26S holoenzyme and the 20S CP strongly accumulated at the nuclear periphery, giving a punctuate nuclear rim but no intranuclear staining. By contrast, the 20S+ particle accumulated inside the nucleus, implying that it was imported through NPCs (Figure 4, middle row).
Labeled 20S+ particles were not targeted to BAPTA-inhibited (poreless) nuclear intermediates (Figure 5A). This result confirms our conclusion from Figure 2 that targeting depends on the presence of NPCs rather than being a general feature of nuclear membranes. Similarly, labeled 26S and 20S particles did not stain BAPTA-inhibited nuclear intermediates (Supplemental Figure S2). These results indicate that all three proteasome species can accumulate at the nuclear envelope if nuclear pores are present, and a unique feature of 20S+ enables it to be imported and reach the nucleoplasm.
FIGURE 5:
Nuclear targeting of the 20S+ particle requires functional NPCs. (A) Labeled 20S+ particles were added into normal reconstitution reactions (functional nuclei), or reactions in which NPC assembly was inhibited by BAPTA (poreless nuclei). Samples were fixed and analyzed by epifluorescence microscopy; 20S+ staining was not observed in BAPTA-inhibited intermediates. (B) Normal reconstituted reactions were split in two and 8 μM Impβ 45–462 was added to plug the channel of preassembled NPCs in one sample. Labeled 20S+ particles were then added to both samples. Consecutive confocal sections through the middle of representative nuclei are shown. The 20S+ accumulated inside normal nuclei, but only stained the nuclear envelope when pores were plugged. (C) Nuclear and cytosolic fractions were separated and immunoblotted with the α7 proteasomal marker, as in Figure 2. Normal nuclei were compared with BAPTA-inhibited nuclei [corresponding to poreless intermediates in (A)], and Impβ 45–462 was added after assembly [plugged pores in (B)]. The α7 subunit is not detected in the nuclear fraction of BAPTA-inhibited intermediates and a smaller portion of α7 cofractionates with plugged-pore nuclei as compared with normal nuclei.
Next we assembled normal nuclei with functional NPCs and subsequently added the recombinant Impβ 45–462 protein. This dominant negative form of importin β is known to irreversibly bind phenylalanine–glycine (FG) repeat nucleoporins in preexisting NPCs and block the central pore channel, thus preventing nuclear transport (Figure 2A; Kutay et al., 1997; Jaggi et al., 2003). When labeled 20S+ particles were introduced to nuclei with plugged pores, we observed strong accumulation at nuclear rims but no intranuclear staining (Figure 5B). This finding suggests that 20S+ particles were no longer able to translocate through the pore channel but could still bind to the cytoplasmic side of the NPC. To complement the experiments performed with fluorescently labeled proteasome particles, we went back to the separation of nuclei from cytosol (Figures 1A and 2B). Normal nuclei, BAPTA-inhibited intermediates, and nuclei with plugged pores were probed for the endogenous α7 subunit, which is present in all three types of proteasome particles. The results confirm that proteasomes do not cofractionate with nuclei when NPC assembly is inhibited by BAPTA and that a smaller portion of endogenous proteasomes cofractionates with nuclei after plugging the NPCs with Impβ 45–462 (Figure 5C).
20S+ is imported in a Ran-independent manner
The purified 20S+ particle was the only one out of the three proteasome species that was imported through NPCs. This suggests that a unique feature of 20S+ (e.g., the presence of associated copurifying proteins) confers this ability. Shuttling transport receptors of the importin β superfamily all share an N-terminal Ran-binding domain. For import receptors, the binding of RanGTP induces cargo release within the target compartment, the nucleus (Strom and Weis, 2001; Fried and Kutay, 2003). Mutant forms of the Ran GTPase, which are locked in the GTP-bound form, disrupt nuclear import pathways by binding the receptors and dissociating import complexes in the cytoplasm. To ask whether the import of the 20S+ particle is mediated by import receptors in one of the canonical pathways, we used the RanQ69L mutant, preloaded with GTP (Klebe et al., 1995). Nuclei were reconstituted in vitro and import of labeled 20S+ particles was followed simultaneously with a classical nuclear localization signal (NLS) substrate, which is imported by the importin α/β pathway. Control nuclei exhibited simultaneous accumulation of the NLS substrate (red) and 20S+ particles (green) in the nucleoplasm (Figure 6A, top). By contrast, nuclei that were preincubated with RanQ69L-GTP showed a complete block in classical NLS import but accumulated 20S+ particles to normal levels (Figure 6A, bottom). The same issue was also addressed by the nuclear–cytosolic fractionation approach and the addition of GTPγS at a late stage of reconstitution. In this case, unlike the addition at t = 0 (Figure 2, A and D), nuclear membrane fusion and NPC assembly are completed, but subsequent GTPase functions including the Ran cycle are blocked. Immunoblot analysis of the separated fractions did not show a reduction in the nuclear fraction of the α7 proteasomal marker following GTPγS addition (Figure 6B). Thus nuclear import of the 20S+ particle does not appear to be regulated by the Ran GTPase cycle.
FIGURE 6:
20S+ import is not blocked by RanGTP. (A) Reconstituted nuclei were preincubated with buffer (control) or 10 μM RanQ69L-GTP for 15 min, before the addition of fluorescently labeled 20S+ particles and TRITC-NLS-BSA import substrate. Both 20S+ and the NLS cargo accumulated in control nuclei (top row; confocal midsection). The import of the NLS cargo was blocked in the presence of RanQ69L-GTP, while 20S+ particles still accumulated inside the nucleus. (B) Nuclear and cytosolic fractions were separated and immunoblotted with anti-α7 as in Figure 5C, comparing normal nuclei with the addition of 2 mM GTPγS after 1 h of assembly. Both reactions were fractionated after 20 min of incubation. The inhibition of GTPase functions after nuclear assembly did not affect the nuclear/cytoplasmic ratio of the α7 proteasomal marker.
Reconstitution of an import-compatible proteasome particle
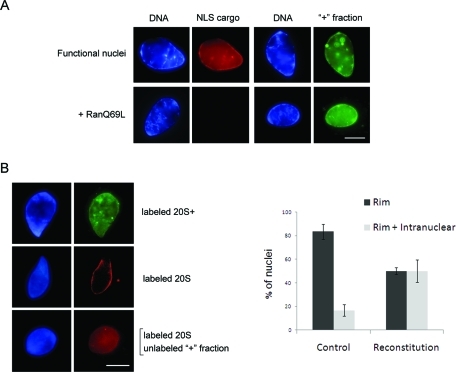
We found one species of proteasome particle, containing the 20S CP and additional factors (“20S+”), that was readily imported into reconstituted nuclei. To functionally define the factors required for nuclear targeting, we collected proteins dissociated from 20S+ core particles and further purified them through an additional size exclusion step by gel filtration chromatography (see Materials and Methods). This isolated “+” fraction still contained multiple components including Rpn1, Rpn2, Hsp90, and importin β, but no 20S CP subunits (Supplemental Table S3 and Supplemental Figure S1C). We first tested whether this isolated + fraction is capable of nuclear import on its own. Fluorescently labeled + fraction components strongly accumulated inside reconstituted nuclei and did so in a RanGTP-independent manner (Figure 7A). This behavior mirrored the targeting behavior of the 20S+ particle (Figures 4 and 6).
FIGURE 7:
Reconstitution of an import-compatible form of the proteasome. (A) The purified + fraction was fluorescently labeled and tested in an import assay with reconstituted nuclei, as in Figure 6. The labeled + fraction accumulated inside nuclei (right). The addition of RanQ69L-GTP blocked the import of TRITC-NLS-BSA cargo, but did not affect the nuclear accumulation of the + fraction. (B) Preincubation of labeled proteasome particles with cytosol prior to their addition into nuclear reconstitution reactions did not affect their targeting properties. The 20S+ particles were efficiently imported (top row), while 20S particles remained perinuclear (middle row) in the majority of nuclei. By contrast, when labeled 20S particles were preincubated with the unlabeled + fraction, about half of the nuclei exhibited substantial intranuclear accumulation together with nuclear rim staining (bottom row). The histogram on the right compiles the results of three separate experiments, in which individual nuclei were scored as showing either nuclear rim or rim + intranuclear staining. Labeled 20S particles were preincubated with cytosol (control) or with cytosol supplemented with the concentrated + fraction (reconstitution). Error bars represent the SE. Scale bars, 10 μm.
Next we asked whether the isolated + fraction could interact with the 20S CP and reconstitute an import-compatible particle. To this end, we incubated the unlabeled, concentrated + fraction with labeled 20S particles prior to their addition into the import assay. In the absence of the additional + components, 20S particles did not reach the nucleoplasm in the majority of nuclei tested, reiterating our earlier observations. By contrast, when preincubated with the concentrated + fraction, a dramatic shift in proteasome distribution was observed. About half of the nuclei showed clear intranuclear staining of labeled proteasome particles (Figure 7B). This finding suggests that components of the unlabeled + fraction were able to interact with the 20S particles and drive their import into the nucleoplasm.
Different 19S subunits show distinct nuclear targeting properties
The Rpn1 and Rpn2 subunits can be targeted to the nucleoplasm through mature NPCs, as part of the 20S+ particle. Other 19S components are missing from this particle, which raises the question whether they might be separately targeted to the nuclear compartment. To begin to address this question, we used a monoclonal antibody directed against the Rpt5 ATPase and checked the localization of the endogenous protein in reconstituted nuclei by immunofluorescence. Anti-Rpt5 produced a strong intranuclear staining pattern in normal nuclei, which was similar to the result obtained with the anti-α7 20S CP subunit (Figure 8A; see also Figure 1B for the endogenous α7 and Rpn2). Surprisingly, anti-Rpt5 also strongly stained the poreless assembly intermediates formed in the presence of BAPTA (Figure 8A). This was in sharp contrast to the lack of staining of poreless nuclei by anti-α7 (Figures 8A and 2C) or by labeled 20S+ particles (Figure 5A). This contrast suggests that at least the Rpt5 ATPase subunit of the 19S regulatory particle is able to reach the nuclear interior by a separate targeting mechanism from that of 20S+. Confirming this observation, immunoblots showed that α7 and Rpn2 were found only in normal nuclei, whereas Rpt5 was detected in the nuclear fraction of both normally assembled and poreless nuclei (Figure 8B).
FIGURE 8:
Different 19S subunits show distinct nuclear targeting properties. (A) Normal (functional) nuclei and BAPTA-inhibited (poreless) nuclei were immunostained with anti-α7 and anti-Rpt5 and analyzed by confocal microscopy. Differential interference contrast (DIC) images and confocal midsections are shown for all nuclei. The 20S CP subunit α7 was missing from poreless nuclei, but anti-Rpt5 strongly stained these nuclear assembly intermediates. (B) Nuclear and cytosolic fractions were separated and compared between normal and BAPTA-inhibited nuclei, as in Figure 2B. Immunoblotting of the separate fractions confirmed that Rpt5 was present in the nuclear fraction in both normal and poreless nuclei, while α7 and Rpn2 only associated with normal nuclei.
Taken together, our results suggest that, of the 19S RP subunits, at least Rpn1 and Rpn2 can enter the nucleus as part of the 20S+ particle, which is imported through active, unplugged nuclear pores. Other 19S subunits may be separately targeted, as exemplified for the Rpt5 ATPase, which is found in newly formed nuclear compartments even in the absence of mature pores. We propose that the import of the 20S+ particle may form the basis for the assembly of the full proteasome holoenzyme inside the nucleus.
DISCUSSION
In this study, we use the Xenopus nuclear reconstitution system to follow the dynamic distribution of proteasomes across the nucleocytoplasmic border. We demonstrate the existence of stable proteasome particles in egg cytosol and their redistribution following nuclear assembly. We purify three distinct populations of proteolytically active proteasomes from Xenopus eggs and provide evidence that intact particles of one specific type can be imported through NPCs into the nucleoplasm.
It is not surprising to find active proteasome particles in extracts made from unfertilized eggs, since proteasome activity is essential for cell-cycle transitions in early development, long before the activation of the zygotic transcription program (Newport and Kirschner, 1982; Murray et al., 1996). Three distinct populations of proteasomes were obtained from egg cytosol using biochemical purification methods. Two of these, the 26S holoenzyme and the 20S CP, correspond to typical proteasome conformations identified in many other systems (Peters et al., 1993, 1994; Liu et al., 2006). A third form, designated here as 20S+, contains the core 20S particle and at least two 19S base subunits. This 20S+ species lacks the majority of the 19S RP subunits but contains additional copurifying proteins, such as Hsp90 and importin β. Similar, though nonidentical, “20S+”-like species have been purified directly from extracts obtained from mammalian cells (Andersen et al., 2009; Hendil et al., 2009) or from yeast (Rosenzweig et al., 2008; Zmijewski et al., 2009). One possible interpretation of these data is that the isolated 20S+ particle is an incomplete purification of the 20S CP. This finding would indicate that Rpn1 and Rpn2 interact with the 20S CP more strongly than other 19S subunits, and it supports recently published data describing putative assembly intermediates of the 26S holoenzyme in mammalian cells that contain Rpn1 and Rpn2 yet lack most other 19S RP subunits (Andersen et al., 2009; Hendil et al., 2009). Detailed analysis of similar structural complexes in budding yeast indicated that Rpn2 and Rpn1 interface directly with the alpha ring surface of the 20S CP, forming a stable unit (Rosenzweig et al., 2008). Thus an alternative interpretation is that the 20S+ particle represents a stable proteasome species with its own composition and unique properties. It is important to note that the 20S+ particle appears to be naturally present in egg cytosol, based on our immunoprecipitation results (Figure 3D). This particle, containing an exposed Rpn2 subunit, can be isolated from cytosol by anti-Rpn2 antibodies. Presumably Rpn2 is masked by other, more peripheral subunits in the full 26S holoenzyme.
Why would such 20S+ particles be present in unfertilized eggs? One possibility is that they serve as a reservoir for rapid entry into newly formed nuclei after fertilization. Indeed when we fluorescently labeled purified 20S+ particles and introduced them into reconstitution reactions, they strongly accumulated inside newly formed nuclei (Figure 4). By contrast, labeled 26S and 20S particles accumulated at the nuclear periphery but did not reach the nucleoplasm. This observation suggests that 20S+ may serve as a basis for forming a nuclear pool of 26S holoenzyme by reassociating inside the nucleus with other 19S subunits. In this scenario, the remainder of the 19S RP subunits would be independently imported for assembly onto the 20S+ template. We found evidence that at least one 19S subunit, the Rpt5 ATPase, is targeted to the nuclear compartment before the formation of mature NPCs (Figure 8) and, therefore, by a separate mechanism from 20S+ import.
Preexisting 26S components coming together within the nucleus may represent a distinct pathway from the assembly of new proteasomes from de novo synthesized subunits. The assembly of the base subcomplex of the 19S RP from newly synthesized subunits involves four chaperones, each of which interacts with one or two of the six Rpt ATPases, leading to assembled 26S by more than one pathway. According to one model, pairs of ATPases, each aided by their dedicated chaperones, dock onto the 20S surface accompanied by Rpn2 and Rpn1 recruitment to form the base subcomplex (Funakoshi et al., 2009; Kaneko et al., 2009; Murata et al., 2009). A somewhat distinct model described how the 20S itself may serve as a template for base assembly. Aided by chaperones, Rpn2 and three ATPases attach to the 20S to form a base precursor, which later recruits Rpn1 and the three remaining Rpts (Kusmierczyk and Hochstrasser, 2008; Park et al., 2009; Roelofs et al., 2009). A separate observation found a stable association of the 20S CP with Rpn2 that also included other base and lid components yet lacked the ATPases (Hendil et al., 2009). Further work in the Xenopus extract system and nuclear targeting data on additional 19S subunits is needed before a detailed assembly model can be proposed and compared with the de novo synthesis scenario.
In vivo, nuclear assembly of the proteasome holoenzyme from its components may be required for the resumption of the cell cycle at the earliest stages of embryonic development. Intriguingly, high cellular mobility and rapid nuclear import of proteasomes was recently reported in fission yeast (Cabrera et al., 2010). Since this was observed in live photobleaching experiments, it presumably reflects the mobilization of preexisting proteasome particles. Thus nuclear import of intact proteasome particles may also occur alongside the import of de novo synthesized subunits in diverse eukaryotic systems.
As mentioned earlier, labeled 26S and 20S CPs accumulate at the nuclear periphery and exhibit punctate rim staining (Figure 4). Because these labeled particles do not stain poreless BAPTA-inhibited intermediates, their targeting to normal nuclei suggests docking onto mature NPCs. Interestingly, plugging NPCs with Impβ 45–462 also causes labeled 20S+ particles to produce a punctuate nuclear rim staining. This finding implies that the import of the 20S+ particle may be a two-step process consisting of docking on the cytoplasmic face of the NPC, followed by translocation into the nucleoplasm. Taken together, our results suggest that all three species of proteasome particles can dock onto NPCs, perhaps through a common constituent of the 20S CP. Further work is needed to identify the potential docking sites for proteasomes, with the best candidates being non-FG domains of cytoplasmic-facing nucleoporins.
What mediates the passage of the large 20S+ particle through the NPC and into the nucleoplasm? The usual suspects would be import receptors of the importin β superfamily, which deliver their cargoes into the nucleus and release them upon binding nuclear RanGTP. Ran imposes directionality on the transport process by binding the receptor molecules and causing irreversible cargo release on the nuclear side of the NPC (Stewart, 2007). Indeed importin β was identified in multiple preparations of the purified 20S+ particle and the isolated + fraction (Supplemental Tables S2 and S3). However, our use of the dominant negative RanQ69L mutant demonstrates that 20S+ import is not directly regulated by the Ran GTPase cycle. This does not completely rule out the involvement of importin β in 20S+ import, although it probably precludes a simple mechanism based on the recognition of classical nuclear localization signals through importin α. If 20S+ import was mediated by this classical pathway, the RanQ69L mutant would be expected to release importins from the particle in the cytoplasm and block import. Importin β, or other known import receptors, may still be needed for translocation through the NPC, whereas a different means is used to trigger cargo release. Large cargoes have been suggested to rely on the recruitment of multiple transport receptor molecules in gaining access through the permeability barrier of the NPC (Ribbeck and Gorlich, 2002). Premature release of some receptor molecules from a large particle could result in pore clogging and may explain the need for a Ran-independent release mechanism. A similar mechanism was previously proposed for Ran-independent import of the ∼1 MDa U5 snRNP by the snurportin1–importin β heterodimer, based on work in digitonin-permeabilized cells (Huber et al., 2002). Subsequent work has suggested a different interpretation, involving a reduced energy requirement but still depending on Ran (Mitrousis et al., 2008). Overall, the examination of large versus small cargoes in the canonical transport pathways has pointed to cargo size and shape, the number of receptors bound to a specific cargo, and the affinities of the receptor–cargo complex for different nucleoporins as the main factors affecting the translocation through the NPC (Lyman et al., 2002; Ribbeck and Gorlich, 2002). It is unlikely that the large 20S+ particle breaks the general rules of passage through the selective molecular sieve of the NPC. It is possible, however, that this proteasome species forms atypical interactions with transport receptors, such as RanGTP-insensitive interactions, or that it interacts directly with nucleoporins.
The nuclear effector of the Wnt signaling cascade, β-catenin, has been proposed to shuttle in and out of the nucleus via direct interactions of its arm repeats with the NPC (Fagotto et al., 1998; Yokoya et al., 1999; Henderson and Fagotto, 2002). Nuclear export of β-catenin was later shown to be enhanced by interactions with RanGTP and RanBP3 (Hendriksen et al., 2005). Both Rpn1 and Rpn2 contain extensive PC repeat domains that are evolutionarily related to the arm/HEAT repeats of importin β-like receptors and other proteins (Kajava, 2002). Moreover, yeast Rpn1 and Rpn2 were recently shown to form curved α-helical solenoid structures that resemble the overall structure of importin β (Effantin et al., 2009). It is tempting to speculate that the PC repeat domains of these proteins mediate direct interactions with nuclear transport receptors or with the NPC itself.
Hsp90, which is consistently detected in preparations of the 20S+ particle and in the isolated + fraction (Supplemental Tables S2 and S3), is another potential interaction partner that is likely involved in stabilizing or modulating the proteasome for nuclear import. Hsp90 is an abundant component of the 20S+ particle, but it is not detected in the 26S proteasome or in the “naked” 20S CP (Figure 3, B and C). One hypothesis might be that Hsp90 blocks binding surfaces on the 20S CP to halt the normally rapid process of holoenzyme assembly and to allow subsequent passage through the NPC.
A clear disadvantage of the in vitro nuclear reconstitution system, as well as digitonin-permeabilized cells, is that additional factors may be recruited to the isolated proteasome particles after they are introduced into the system. Specific import factors could be recruited to the 20S+ particle at the nuclear envelope or at the NPC itself. Nevertheless, our results demonstrate that an import-compatible form of the proteasome can be reconstituted with the aid of the isolated + fraction. Thus some components of this functionally defined set of proteins are sufficient to confer nuclear import capability on preexisting 20S core particles. An important future challenge is to define specific roles for each of the interaction partners within the 20S+ sphere of interactions.
Nuclear import of intact proteasome particles was first suggested to occur by work in digitonin-permeabilized mammalian cells (Wang et al., 1997; Mayr et al., 1999). Proteasomes considered to be 20S particles were purified by a standard protocol from human erythrocytes but were not further characterized in these studies. It is interesting to consider the possibility that these particles were in fact more similar to the 20S+ species described in our study. The import of proteasomes from the archaeon Thermoplasma acidophilum, observed by Wang et al. (1997), remains unexplained but may reflect interactions with import factors derived from the permeabilized mammalian cells. Interestingly, when authentic 20S core particles were purified from budding yeast and tested in digitonin-permeabilized mammalian cells, they were not targeted to the nucleus (Lehmann et al., 2002).
In summary, we have demonstrated differential nuclear targeting and import of three distinct types of proteasome particles purified from the cytosol of unfertilized eggs. Import of the novel 20S+ particle hints at a mechanism allowing for the quick mobilization of existing proteasome particles into newly formed nuclear compartments. This may be typical of early developmental stages, with a large pool of existing particles and rapid formation of new nuclei, but could also work alongside de novo synthesis of proteasomal subunits in other cellular scenarios.
MATERIALS AND METHODS
Antibodies
Commercially obtained antibodies included monoclonal anti-α7 (PW8110; Biomol, Plymouth Meeting, PA), monoclonal anti-Rpn2 (PW9270, Biomol), monoclonal anti-Rpt1 (PW8825, Biomol), monoclonal anti-Rpt5 (PW8770, Biomol), rabbit anti-Hsp90 (Ab13495; Abcam, Cambridge, UK), rabbit anti–histone H3 (9715; Cell Signaling, Beverly, MA) and goat anti-actin (#sc1615; Santa Cruz Biotechnology, Santa Cruz, CA). Rabbit immunoglobulin (IgG; Calbiochem, La Jolla, CA) was used as a control in immunoprecipitation experiments. Affinity purified anti-xNup107 was a gift from Ulrike Kutay (ETH Zurich, Switzerland) and was previously described (Rotem et al., 2009). Rabbit polyclonal antibodies against Xenopus laevis Rpn2 and Nup358 were generated for this study and affinity purified as described (Rotem et al., 2009).
Recombinant protein expression and purification
The coding sequence for xRpn2 aa 312–486 (LOC 734707) was inserted into pET28A and expressed as a soluble hexahistidine-T7 tagged protein in the Escherichia coli strain BL21 (DE3) Rosetta. The purified protein was used to immunize two rabbits, and antibodies were affinity purified as described (Rotem et al., 2009). Similarly, the coding sequence for xNup358 aa: SFQPVPSPSKSPTKLNHSRVSVGTDEESDVTQEEERDGQYFEPVV (from a partial clone, GenBank accession no. CA983354; see also Walther et al., 2002) was inserted into pET28A and used to produce and purify polyclonal antibodies. The expression of Impβ 45–462 has been described (Harel et al., 2003a). Histidine-tagged proteins were purified on Ni-NTA resin (Qiagen, Chatsworth, CA) according to standard procedures. Expression, purification, and loading of RanQ69L with GTP were performed as by Rotem et al. (2009).
Egg extracts and nuclear reconstitution
Preparation of demembranated sperm chromatin, Xenopus egg extracts, and fractionation into cytosolic and membrane fractions were performed as previously described (Harel et al., 2003b; Rotem et al., 2009). For biochemical purification of cytosolic components, the crude soluble fraction of egg extract was diluted in two volumes of buffer A (25 mM Tris, pH 7.4, 10 mM MgCl2, 10% glycerol, 1 mM dithiothreitol) and centrifuged for 1 h at 270,000g to remove membranes and large aggregates. Nuclei were reconstituted by mixing Xenopus egg membrane vesicles and cytosolic fractions at a 1:20 ratio, with an ATP-regeneration system and sperm chromatin, as described (Harel et al., 2003b). The nuclear assembly inhibitors, 2 mM GTPγS (Roche Applied Science, Mannheim, Germany) and 5 mM BAPTA (Calbiochem), were added to this initial reconstitution mixture and incubated for 60 min at room temperature. When inhibition or analysis was required after the assembly of functional NPCs, nuclear morphology was monitored by staining with Hoechst 33258 (Sigma-Aldrich, St. Louis, MO) after 50–60 min of assembly. Samples were also removed for import analysis with tetramethyl rhodamine isothiocyanate (TRITC)-labeled NLS-bovine serum albumin (BSA) substrate and for staining with mAb414 (Covance, Princeton, NJ) (Harel et al., 2003b). Once nuclear and NPC assembly had been confirmed, the reconstitution reactions were diluted by the addition of two volumes of 1× Egg lysis buffer + sucrose (ELBS; 10 mM HEPES, pH 7.6, 250 mM sucrose, 50 mM KCl, 2.5 mM MgCl2) and import substrates or labeled proteasome particles were added for an additional incubation of 30 min. To inhibit nuclear import or plug existing NPCs, 10 μM RanQ69L-GTP or 8 μM Impβ 45–462 was added at the time of dilution (60 min) and incubated with the reconstituted nuclei for 15 min before the addition of import substrate or labeled proteasome particles. For the reconstitution of an import-compatible form of the proteasome, labeled particles were preincubated with cytosol, or with cytosol supplemented with the concentrated + fraction for 20 min in the dark at room temperature, prior to their addition to assembled nuclei. The overall dilution factor of labeled proteasomes in relation to the nuclei was kept constant in these experiments and in the normal import assays. To quantify the reconstitution effect, 20–40 nuclei in each category in three separate experiments were randomly chosen and individually scored as having either an exclusive rim staining pattern or nuclear rim + intranuclear accumulation. For the separation of nuclear and cytosolic fractions, 50 μl reactions were overlaid on 300 μl cushions of 0.5 M sucrose in ELBS and centrifuged in a horizontal rotor at 11,000g, 4°C, for 2 min. Complete recovery of nuclei and nuclear intermediates was verified by Hoechst 33258 staining and visual inspection. Normalized samples of the total reaction, cytosolic, and nuclear fractions were immunoblotted with specific markers.
Purification of proteasome particles
Amphibian oocytes and eggs are abundant in proteasomes (Peters et al., 1993; Peters et al., 1994; King et al., 1995). Purification protocols for Xenopus proteasome particles were developed on the basis of yeast protocols (Glickman et al., 1998; Glickman and Coux, 2001) and peptidase activity was monitored with the fluorigenic peptide N-succinyl-Leu-Leu-Val-Tyr-7-amino-4-methylcoumarin (Suc-LLVY-AMC; Biotest, Dreieich, Germany) throughout the procedure. For the purification of 26S holoenzyme, the clarified crude soluble fraction of egg extract was filtered through a 0.45 μm filter (Millipore, Billerica, MA) and loaded on a 28 ml DEAE Affi-Gel Blue column (Bio-Rad, Richmond, CA), washed extensively in buffer A and eluted with 150 mM NaCl in buffer A. ATP (0.5 mM; Sigma-Aldrich), was added to the starting material and all subsequent buffers. The resulting material was diluted in buffer A, loaded on a 16 ml Q-Sepharose FF anion-exchange column (GE Healthcare, Waukesha, WI), and resolved on a 300 ml linear gradient of 0–1 M NaCl in buffer A. Peak activity fractions were pooled, concentrated, and loaded on a Superose-6 gel filtration column (GE Healthcare) and 0.5 ml fractions were collected and concentrated in Amicon Ultra-4 microconcentrators (10000 MWCO, Millipore). For the purification of 20S or 20S+ particles, ATP was omitted from the buffers, and peak activity fractions eluted from the Superose-6 column were pooled and exchanged into phosphate buffer. KCl was added to a final concentration of 0.5 M and incubated with the sample for 1 h at 30°C (20S+ purification) or 1 M KCl for 2 h (20S purification). The samples were then loaded on a 1 ml ceramic hydroxyapatite (CHT) type I column (BioRad), which was subsequently eluted with a 20 ml linear gradient of 0–0.4 M potassium phosphate, pH 7.6. Active fractions of 0.5 ml were collected and dialyzed against 1× phosphate-buffered saline (PBS), 5% glycerol. Purified proteasome particle preparations were either used immediately or quick-frozen in liquid nitrogen and stored at −80° C for future use.
For the purification of + fraction components, the major silver-staining band of ∼100 kDa eluting off the CHT hydroxyapatite column after 1 M KCl treatment of active particle fractions, was followed. Fractions that contained this band but did not show peptidase activity or the presence of α7 by immunoblotting were pooled, concentrated, and loaded on an additional Superose-6 column. Three peak fractions from the gel filtration column were concentrated, exchanged into 1× PBS, and analyzed by mass spectrometry and functional assays.
Native and denaturing PAGE, immunoblotting, and immunoprecipitation
SDS–PAGE and immunoblotting were performed using polyvinylidene fluoride (PVDF; Millipore) and standard techniques. Silver staining of 10% SDS–PAGE gels was performed with Silver Stain Plus (Bio-Rad). Native 4% PAGE was performed as previously described (Glickman et al., 1998) and gels were incubated for 15 min at 30°C with 0.1 mM Suc-LLVY-AMC to visualize proteasome bands upon exposure to UV light (380 nm). An amount of 10 μl of unfractionated egg cytosol was loaded for the extract sample and samples of the three purified particle preparations were normalized according to the staining intensity of 20S CP bands on denaturing gels and of a denaturing immunoblot with anti-α7. Immunoprecipitation out of Xenopus egg cytosol was performed essentially as described by Shah et al. (1998), including coupling of the antibodies to protein A-Sepharose (GE Healthcare) by dimethylpimelimidate (Sigma-Aldrich). Affinity-purified antibodies or microgram-equivalent amounts of control rabbit IgG were used and all dilutions and washes were in 1× PBS. Immunoprecipitated proteins were eluted from the beads by the addition of 100 mM glycine, pH 2.5, and analyzed by immunoblotting.
Fluorescent labeling of purified proteasome particles
Purified proteasome particles were concentrated to ∼1 mg/ml and labeled on exposed amine groups with Oregon Green 488 succinimidyl ester (Molecular Probes, Eugene, OR), or TRITC (Sigma-Aldrich), for 1 h at 4°C. Unreacted dye was quenched with 0.1 M ethanolamine, pH 8.0, for 2 h and removed by extensive dialysis. The conjugated dye was distributed over multiple subunits in each particle type, as determined by SDS–PAGE, and the labeled species remained stable as high-molecular-weight particles and retained their peptidase activity.
Microscopy
Indirect immunofluorescence staining was performed as previously described (Harel et al., 2003b; Rotem et al., 2009) with Alexa Fluor 488-donkey anti–mouse IgG and Alexa Fluor 568-goat anti–rabbit IgG (Invitrogen) as secondary antibodies. For the direct visualization of TRITC-NLS-BSA and labeled proteasome particles, nuclei were fixed in 3% paraformaldehyde and stained with Hoechst 33258. Images were acquired on an Olympus BX61TRF epifluorescence microscope equipped with a DP70 digital camera, or a Zeiss LSM 510 META confocal microscope. Figures were prepared using LSM software and Adobe Photoshop.
Supplementary Material
Acknowledgments
We thank Rami Reshef and Ulrike Kutay for the kind gift of reagents, Rachel Kaminsky for advice on confocal microscopy, Rina Rosenzweig and Dasha Krutauz for advice on proteasome methods, and Michael Elbaum and Asaf Rotem for helpful discussions. Mass spectrometry analysis was performed at the Smoler Proteomics Center, Technion. This work was supported by grants from the Israel Science Foundation to M.H.G. (890015) and to A.H. (813/05).
Abbreviations used:
- BAPTA
1,2-bis(o-aminophenoxy)ethane-N,N,N’,N’-tetra acetic acid
- CP
core particle
- FG
phenylalanine–glycine
- NLS
nuclear localization signal
- NPC
nuclear pore complex
- RP
regulatory particle
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E10-07-0595) on February 2, 2011.
REFERENCES
- Adori C, Low P, Moszkovkin G, Bagdy G, Laszlo L, Kovacs GG. Subcellular distribution of components of the ubiquitin-proteasome system in nondiseased human and rat brain. J Histochem Cytochem. 2006;54:263–267. doi: 10.1369/jhc.5B6752.2005. [DOI] [PubMed] [Google Scholar]
- Andersen KM, Madsen L, Prag S, Johnsen AH, Semple CA, Hendil KB, Hartmann-Petersen R. Thioredoxin Txnl1/TRP32 is a redox-active cofactor of the 26 S proteasome. J Biol Chem. 2009;284:15246–15254. doi: 10.1074/jbc.M900016200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arcangeletti C, Sutterlin R, Aebi U, De Conto F, Missorini S, Chezzi C, Scherrer K. Visualization of prosomes (MCP-proteasomes), intermediate filament and actin networks by “instantaneous fixation” preserving the cytoskeleton. J Struct Biol. 1997;119:35–58. doi: 10.1006/jsbi.1997.3871. [DOI] [PubMed] [Google Scholar]
- Bajorek M, Finley D, Glickman MH. Proteasome disassembly and downregulation is correlated with viability during stationary phase. Curr Biol. 2003;13:1140–1144. doi: 10.1016/s0960-9822(03)00417-2. [DOI] [PubMed] [Google Scholar]
- Cabrera R, Sha Z, Vadakkan TJ, Otero J, Kriegenburg F, Hartmann-Petersen R, Dickinson ME, Chang E. Proteasome nuclear import mediated by Arc3 can influence efficient DNA damage repair and mitosis in Schizosaccharomyces pombe. Mol Biol Cell. 2010;21:3125–3136. doi: 10.1091/mbc.E10-06-0506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Effantin G, Rosenzweig R, Glickman MH, Steven AC. Electron microscopic evidence in support of alpha-solenoid models of proteasomal subunits Rpn1 and Rpn2. J Mol Biol. 2009;386:1204–1211. doi: 10.1016/j.jmb.2009.01.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagotto F, Gluck U, Gumbiner BM. Nuclear localization signal-independent and importin/karyopherin-independent nuclear import of beta-catenin. Curr Biol. 1998;8:181–190. doi: 10.1016/s0960-9822(98)70082-x. [DOI] [PubMed] [Google Scholar]
- Fahrenkrog B, Koser J, Aebi U. The nuclear pore complex: a jack of all trades? Trends Biochem Sci. 2004;29:175–182. doi: 10.1016/j.tibs.2004.02.006. [DOI] [PubMed] [Google Scholar]
- Finlay DR, Forbes DJ. Reconstitution of biochemically altered nuclear pores: transport can be eliminated and restored. Cell. 1990;60:17–29. doi: 10.1016/0092-8674(90)90712-n. [DOI] [PubMed] [Google Scholar]
- Finley D. Recognition and processing of ubiquitin-protein conjugates by the proteasome. Annu Rev Biochem. 2009;78:477–513. doi: 10.1146/annurev.biochem.78.081507.101607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forbes DJ, Kirschner MW, Newport JW. Spontaneous formation of nucleus-like structures around bacteriophage DNA microinjected into Xenopus eggs. Cell. 1983;34:13–23. doi: 10.1016/0092-8674(83)90132-0. [DOI] [PubMed] [Google Scholar]
- Fried H, Kutay U. Nucleocytoplasmic transport: taking an inventory. Cell Mol Life Sci. 2003;60:1659–1688. doi: 10.1007/s00018-003-3070-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funakoshi M, Tomko RJ Jr, Kobayashi H, Hochstrasser M. Multiple assembly chaperones govern biogenesis of the proteasome regulatory particle base. Cell. 2009;137:887–899. doi: 10.1016/j.cell.2009.04.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girao H, Pereira P, Taylor A, Shang F. Subcellular redistribution of components of the ubiquitin-proteasome pathway during lens differentiation and maturation. Invest Ophthalmol Visual Sci. 2005;46:1386–1392. doi: 10.1167/iovs.04-0563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glickman M, Coux O. Purification and characterization of proteasomes from Saccharomyces cerevisiae. Curr Protoc Protein Sci. 2001;21.5.1 doi: 10.1002/0471140864.ps2105s24. [DOI] [PubMed] [Google Scholar]
- Glickman MH. Getting in and out of the proteasome. Semin Cell Dev Biol. 2000;11:149–158. doi: 10.1006/scdb.2000.0161. [DOI] [PubMed] [Google Scholar]
- Glickman MH, Ciechanover A. The ubiquitin-proteasome proteolytic pathway: destruction for the sake of construction. Physiol Rev. 2002;82:373–428. doi: 10.1152/physrev.00027.2001. [DOI] [PubMed] [Google Scholar]
- Glickman MH, Rubin DM, Fried VA, Finley D. The regulatory particle of the Saccharomyces cerevisiae proteasome. Mol Cell Biol. 1998;18:3149–3162. doi: 10.1128/mcb.18.6.3149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godon C, Lagniel G, Lee J, Buhler JM, Kieffer S, Perrot M, Boucherie H, Toledano MB, Labarre J. The H2O2 stimulon in Saccharomyces cerevisiae. J Biol Chem. 1998;273:22480–22489. doi: 10.1074/jbc.273.35.22480. [DOI] [PubMed] [Google Scholar]
- Gordon C. The intracellular localization of the proteasome. Curr Top Microbiol Immunol. 2002;268:175–184. doi: 10.1007/978-3-642-59414-4_7. [DOI] [PubMed] [Google Scholar]
- Gorlich D, Pante N, Kutay U, Aebi U, Bischoff FR. Identification of different roles for RanGDP and RanGTP in nuclear protein import. EMBO J. 1996;15:5584–5594. [PMC free article] [PubMed] [Google Scholar]
- Harel A, Chan RC, Lachish-Zalait A, Zimmerman E, Elbaum M, Forbes DJ. Importin beta negatively regulates nuclear membrane fusion and nuclear pore complex assembly. Mol Biol Cell. 2003a;14:4387–4396. doi: 10.1091/mbc.E03-05-0275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harel A, Forbes DJ. Importin beta: conducting a much larger cellular symphony. Mol Cell. 2004;16:319–330. doi: 10.1016/j.molcel.2004.10.026. [DOI] [PubMed] [Google Scholar]
- Harel A, Orjalo AV, Vincent T, Lachish-Zalait A, Vasu S, Shah S, Zimmerman E, Elbaum M, Forbes DJ. Removal of a single pore subcomplex results in vertebrate nuclei devoid of nuclear pores. Mol Cell. 2003b;11:853–864. doi: 10.1016/s1097-2765(03)00116-3. [DOI] [PubMed] [Google Scholar]
- Henderson BR, Fagotto F. The ins and outs of APC and beta-catenin nuclear transport. EMBO Rep. 2002;3:834–839. doi: 10.1093/embo-reports/kvf181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendil KB, Kriegenburg F, Tanaka K, Murata S, Lauridsen AM, Johnsen AH, Hartmann-Petersen R. The 20S proteasome as an assembly platform for the 19S regulatory complex. J Mol Biol. 2009;394:320–328. doi: 10.1016/j.jmb.2009.09.038. [DOI] [PubMed] [Google Scholar]
- Hendriksen J, Fagotto F, Van Der Velde H, van Schie M, Noordermeer J, Fornerod M. RanBP3 enhances nuclear export of active (beta)-catenin independently of CRM1. J Cell Biol. 2005;171:785–797. doi: 10.1083/jcb.200502141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetzer MW, Wente SR. Border control at the nucleus: biogenesis and organization of the nuclear membrane and pore complexes. Dev Cell. 2009;17:606–616. doi: 10.1016/j.devcel.2009.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano T, Mitchison TJ. Cell cycle control of higher-order chromatin assembly around naked DNA in vitro. J Cell Biol. 1991;115:1479–1489. doi: 10.1083/jcb.115.6.1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber J, Dickmanns A, Luhrmann R. The importin-beta binding domain of snurportin1 is responsible for the Ran- and energy-independent nuclear import of spliceosomal U snRNPs in vitro. J Cell Biol. 2002;156:467–479. doi: 10.1083/jcb.200108114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isono E, et al. The assembly pathway of the 19S regulatory particle of the yeast 26S proteasome. Mol Biol Cell. 2007;18:569–580. doi: 10.1091/mbc.E06-07-0635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaggi RD, Franco-Obregon A, Muhlhausser P, Thomas F, Kutay U, Ensslin K. Modulation of nuclear pore topology by transport modifiers. Biophys J. 2003;84:665–670. doi: 10.1016/S0006-3495(03)74886-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajava AV. What curves alpha-solenoids? Evidence for an alpha-helical toroid structure of Rpn1 and Rpn2 proteins of the 26 S proteasome. J Biol Chem. 2002;277:49791–49798. doi: 10.1074/jbc.M204982200. [DOI] [PubMed] [Google Scholar]
- Kaneko T, Hamazaki J, Iemura S, Sasaki K, Furuyama K, Natsume T, Tanaka K, Murata S. Assembly pathway of the mammalian proteasome base subcomplex is mediated by multiple specific chaperones. Cell. 2009;137:914–925. doi: 10.1016/j.cell.2009.05.008. [DOI] [PubMed] [Google Scholar]
- King RW, Peters JM, Tugendreich S, Rolfe M, Hieter P, Kirschner MW. A 20S complex containing CDC27 and CDC16 catalyzes the mitosis-specific conjugation of ubiquitin to cyclin B. Cell. 1995;81:279–288. doi: 10.1016/0092-8674(95)90338-0. [DOI] [PubMed] [Google Scholar]
- Klebe C, Bischoff FR, Ponstingl H, Wittinghofer A. Interaction of the nuclear GTP-binding protein Ran with its regulatory proteins RCC1 and RanGAP1. Biochemistry. 1995;34:639–647. doi: 10.1021/bi00002a031. [DOI] [PubMed] [Google Scholar]
- Kusmierczyk AR, Hochstrasser M. Some assembly required: dedicated chaperones in eukaryotic proteasome biogenesis. Biol Chem. 2008;389:1143–1151. doi: 10.1515/BC.2008.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutay U, Izaurralde E, Bischoff FR, Mattaj IW, Gorlich D. Dominant-negative mutants of importin-beta block multiple pathways of import and export through the nuclear pore complex. EMBO J. 1997;16:1153–1163. doi: 10.1093/emboj/16.6.1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann A, Janek K, Braun B, Kloetzel PM, Enenkel C. 20 S proteasomes are imported as precursor complexes into the nucleus of yeast. J Mol Biol. 2002;317:401–413. doi: 10.1006/jmbi.2002.5443. [DOI] [PubMed] [Google Scholar]
- Liu CW, Li X, Thompson D, Wooding K, Chang TL, Tang Z, Yu H, Thomas PJ, DeMartino GN. ATP binding and ATP hydrolysis play distinct roles in the function of 26S proteasome. Mol Cell. 2006;24:39–50. doi: 10.1016/j.molcel.2006.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohka MJ, Masui Y. Formation in vitro of sperm pronuclei and mitotic chromosomes induced by amphibian ooplasmic components. Science. 1983;220:719–721. doi: 10.1126/science.6601299. [DOI] [PubMed] [Google Scholar]
- Lyman SK, Guan T, Bednenko J, Wodrich H, Gerace L. Influence of cargo size on Ran and energy requirements for nuclear protein import. J Cell Biol. 2002;159:55–67. doi: 10.1083/jcb.200204163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macaulay C, Forbes DJ. Assembly of the nuclear pore: biochemically distinct steps revealed with NEM, GTP gamma S, and BAPTA. J Cell Biol. 1996;132:5–20. doi: 10.1083/jcb.132.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattsson K, Pokrovskaja K, Kiss C, Klein G, Szekely L. Proteins associated with the promyelocytic leukemia gene product (PML)-containing nuclear body move to the nucleolus upon inhibition of proteasome-dependent protein degradation. Proc Natl Acad Sci USA. 2001;98:1012–1017. doi: 10.1073/pnas.031566998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayr J, Wang HR, Nederlof P, Baumeister W. The import pathway of human and Thermoplasma 20S proteasomes into HeLa cell nuclei is different from that of classical NLS-bearing proteins. Biol Chem. 1999;380:1183–1192. doi: 10.1515/BC.1999.150. [DOI] [PubMed] [Google Scholar]
- Mitrousis G, Olia AS, Walker-Kopp N, Cingolani G. Molular basis for the recognition of snurportin 1 by importin beta. J Biol Chem. 2008;283:7877–7884. doi: 10.1074/jbc.M709093200. [DOI] [PubMed] [Google Scholar]
- Murata S, Yashiroda H, Tanaka K. Molular mechanisms of proteasome assembly. Nat Rev Mol Cell Biol. 2009;10:104–115. doi: 10.1038/nrm2630. [DOI] [PubMed] [Google Scholar]
- Murray AW, Desai AB, Salmon ED. Real time observation of anaphase in vitro. Proc Natl Acad Sci USA. 1996;93:12327–12332. doi: 10.1073/pnas.93.22.12327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray AW, Kirschner MW. Cyclin synthesis drives the early embryonic cell cycle. Nature. 1989;339:275–280. doi: 10.1038/339275a0. [DOI] [PubMed] [Google Scholar]
- Newport J, Kirschner M. A major developmental transition in early Xenopus embryos: I. characterization and timing of cellular changes at the midblastula stage. Cell. 1982;30:675–686. doi: 10.1016/0092-8674(82)90272-0. [DOI] [PubMed] [Google Scholar]
- Olink-Coux M, Arcangeletti C, Pinardi F, Minisini R, Huesca M, Chezzi C, Scherrer K. Cytolocation of prosome antigens on intermediate filament subnetworks of cytokeratin, vimentin and desmin type. J Cell Sci. 1994;107(Pt 3):353–366. [PubMed] [Google Scholar]
- Palmer A, Rivett AJ, Thomson S, Hendil KB, Butcher GW, Fuertes G, Knecht E. Subpopulations of proteasomes in rat liver nuclei, microsomes and cytosol. Biochem J. 1996;316(Pt 2):401–407. doi: 10.1042/bj3160401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pante N, Kann M. Nuclear pore complex is able to transport macromolecules with diameters of about 39 nm. Mol Biol Cell. 2002;13:425–434. doi: 10.1091/mbc.01-06-0308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S, Roelofs J, Kim W, Robert J, Schmidt M, Gygi SP, Finley D. Hexameric assembly of the proteasomal ATPases is templated through their C termini. Nature. 2009;459:866–870. doi: 10.1038/nature08065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pemberton LF, Paschal BM. Mechanisms of receptor-mediated nuclear import and nuclear export. Traffic. 2005;6:187–198. doi: 10.1111/j.1600-0854.2005.00270.x. [DOI] [PubMed] [Google Scholar]
- Peters JM, Cejka Z, Harris JR, Kleinschmidt JA, Baumeister W. Structural features of the 26 S proteasome complex. J Mol Biol. 1993;234:932–937. doi: 10.1006/jmbi.1993.1646. [DOI] [PubMed] [Google Scholar]
- Peters JM, Franke WW, Kleinschmidt JA. Distinct 19 S and 20 S subcomplexes of the 26 S proteasome and their distribution in the nucleus and the cytoplasm. J Biol Chem. 1994;269:7709–7718. [PubMed] [Google Scholar]
- Peters JM, Harris JR, Kleinschmidt JA. Ultrastructure of the approximately 26S complex containing the approximately 20S cylinder particle (multicatalytic proteinase/proteasome) Eur J Cell Biol. 1991;56:422–432. [PubMed] [Google Scholar]
- Pickart CM, Cohen RE. Proteasomes and their kin: proteases in the machine age. Nat Rev Mol Cell Biol. 2004;5:177–187. doi: 10.1038/nrm1336. [DOI] [PubMed] [Google Scholar]
- Rechsteiner M, Hoffman L, Dubiel W. The multicatalytic and 26 S proteases. J Biol Chem. 1993;268:6065–6068. [PubMed] [Google Scholar]
- Reits EA, Benham AM, Plougastel B, Neefjes J, Trowsdale J. Dynamics of proteasome distribution in living cells. EMBO J. 1997;16:6087–6094. doi: 10.1093/emboj/16.20.6087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribbeck K, Gorlich D. The permeability barrier of nuclear pore complexes appears to operate via hydrophobic exclusion. EMBO J. 2002;21:2664–2671. doi: 10.1093/emboj/21.11.2664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roelofs J, et al. Chaperone-mediated pathway of proteasome regulatory particle assembly. Nature. 2009;459:861–865. doi: 10.1038/nature08063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenzweig R, Osmulski PA, Gaczynska M, Glickman MH. The central unit within the 19S regulatory particle of the proteasome. Nat Struct Mol Biol. 2008;15:573–580. doi: 10.1038/nsmb.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotem A, Gruber R, Shorer H, Shaulov L, Klein E, Harel A. Importin beta regulates the seeding of chromatin with initiation sites for nuclear pore assembly. Mol Biol Cell. 2009;20:4031–4042. doi: 10.1091/mbc.E09-02-0150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryabova LV, Virtanen I, Olink-Coux M, Scherrer K, Vassetzky SG. Distribution of prosome proteins and their relationship with the cytoskeleton in oogenesis of Xenopus laevis. Mol Reprod Dev. 1994;37:195–203. doi: 10.1002/mrd.1080370210. [DOI] [PubMed] [Google Scholar]
- Shah S, Tugendreich S, Forbes D. Major binding sites for the nuclear import receptor are the internal nucleoporin Nup153 and the adjacent nuclear filament protein Tpr. J Cell Biol. 1998;141:31–49. doi: 10.1083/jcb.141.1.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart M. Molular mechanism of the nuclear protein import cycle. Nat Rev Mol Cell Biol. 2007;8:195–208. doi: 10.1038/nrm2114. [DOI] [PubMed] [Google Scholar]
- Strom AC, Weis K. Importin-beta-like nuclear transport receptors. Genome Biol. 2001;2 doi: 10.1186/gb-2001-2-6-reviews3008. reviews3008.1–3008.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonoki A, Kuranaga E, Tomioka T, Hamazaki J, Murata S, Tanaka K, Miura M. Genetic evidence linking age-dependent attenuation of the 26S proteasome with the aging process. Mol Cell Biol. 2009;29:1095–1106. doi: 10.1128/MCB.01227-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsvetkov P, Reuven N, Shaul Y. The nanny model for IDPs. Nat Chem Biol. 2009;5:778–781. doi: 10.1038/nchembio.233. [DOI] [PubMed] [Google Scholar]
- von Mikecz A, Chen M, Rockel T, Scharf A. The nuclear ubiquitin-proteasome system: visualization of proteasomes, protein aggregates, and proteolysis in the cell nucleus. Methods Mol Biol. 2008;463:191–202. doi: 10.1007/978-1-59745-406-3_14. [DOI] [PubMed] [Google Scholar]
- Walther TC, Pickersgill HS, Cordes VC, Goldberg MW, Allen TD, Mattaj IW, Fornerod M. The cytoplasmic filaments of the nuclear pore complex are dispensable for selective nuclear protein import. J Cell Biol. 2002;158:63–77. doi: 10.1083/jcb.200202088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang HR, Kania M, Baumeister W, Nederlof PM. Import of human and Thermoplasma 20S proteasomes into nuclei of HeLa cells requires functional NLS sequences. Eur J Cell Biol. 1997;73:105–113. [PubMed] [Google Scholar]
- Weis K. Regulating access to the genome: nucleocytoplasmic transport throughout the cell cycle. Cell. 2003;112:441–451. doi: 10.1016/s0092-8674(03)00082-5. [DOI] [PubMed] [Google Scholar]
- Wendler P, Lehmann A, Janek K, Baumgart S, Enenkel C. The bipartite nuclear localization sequence of Rpn2 is required for nuclear import of proteasomal base complexes via karyopherin alpha beta and proteasome functions. J Biol Chem. 2004;279:37751–37762. doi: 10.1074/jbc.M403551200. [DOI] [PubMed] [Google Scholar]
- Wilkinson CR, Wallace M, Morphew M, Perry P, Allshire R, Javerzat JP, McIntosh JR, Gordon C. Localization of the 26S proteasome during mitosis and meiosis in fission yeast. EMBO J. 1998;17:6465–6476. doi: 10.1093/emboj/17.22.6465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojcik C. Proteasome activator subunit PA28 alpha and related Ki antigen (PA28 gamma) are absent from the nuclear fraction purified by sucrose gradient centrifugation. Int J Biochem Cell Biol. 1999;31:273–276. doi: 10.1016/s1357-2725(98)00117-4. [DOI] [PubMed] [Google Scholar]
- Wojcik C, DeMartino GN. Intracellular localization of proteasomes. Int J Biochem Cell Biol. 2003;35:579–589. doi: 10.1016/s1357-2725(02)00380-1. [DOI] [PubMed] [Google Scholar]
- Yokoya F, Imamoto N, Tachibana T, Yoneda Y. Beta-catenin can be transported into the nucleus in a Ran-unassisted manner. Mol Biol Cell. 1999;10:1119–1131. doi: 10.1091/mbc.10.4.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zmijewski JW, Banerjee S, Abraham E. S-glutathionylation of the Rpn2 regulatory subunit inhibits 26 S proteasomal function. J Biol Chem. 2009;284:22213–22221. doi: 10.1074/jbc.M109.028902. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.