Abstract
Aims
To develop and evaluate two tumor-specific nanoprobes by functionalization of a PEG-immobilized nanoparticle with arginine-glycine-aspartic acid (RGD) or chlorotoxin (CTX) ligand that targets αvβ3 integrin and MMP-2 receptors, respectively.
Materials and Methods
The nanoprobes were made of iron oxide cores, biocompatible polymer coating, and surface-conjugated RGD or CTX peptide. The tumor-targeting specificity of the nanoprobes was evaluated both in vitro and in vivo.
Results and Discussion
Both nanoprobes were highly dispersive and exhibited excellent long-term stability in cell culture media. The RGD-conjugated nanoprobe displayed a strong initial accumulation near neovasculatures in tumors followed by quick clearance. Conversely, the CTX-enabled nanoprobe exhibited sustained accumulation throughout the tumor.
Conclusion
These findings revealed the influence of the targeting ligands on the intratumoral distribution of the ligand-enabled nanoprobes. With flexible surface chemistry, our nanoparticle platform can be used in a modular fashion to conjugate biomolecules for intended applications.
Keywords: Nanotechnology, nanoparticle, magnetic resonance imaging, cancer, targeting, chlorotoxin, RGD peptide, bioconjugation, stability, glioma
Introduction
The rapidly expanding nanotechnology is offering new approaches to diagnose and treat debilitating and deadly diseases, such as cancer [1–3]. Among these promising approaches, magnetic nanoparticle-based molecular imaging agents have drawn considerable attention in recent years [2, 4, 5]. These so-called “nanoprobes” are comprised of nanoscale magnetic cores that can be detected via magnetic resonance (MR) imaging and surface-immobilized targeting ligands that have high affinity to cancer biomarkers. These nanoprobes can facilitate visualization of cancer-related molecule expression, revealing biological processes occurring within the tumor, and thus can serve as powerful tools for ultra-sensitive and non-invasive detection, diagnosis and evaluation of cancer [3, 6, 7]. Despite significant progress made on the synthesis of various nanoscale magnetic cores, it remains a challenge to produce stable, biocompatible and target-specific nanoprobes for potential clinical applications.
In our previous study, we have developed a nanoparticle platform comprised of a superparamagnetic iron oxide core that was surface-modified with poly(ethylene glycol) (PEG) molecules bearing amine functional groups for further bioconjugation. This system was evaluated for stability under biological environments and demonstrated to be suitable for further development for in vivo use [8]. The aim of this study is to develop targeting nanoparticle systems capitalizing on this base nanoparticle to enable selective imaging of neoplastic tissue in vivo and to investigate the difference in targeting mechanism between ligands for different biomolecular targets. Critical to the development of these systems is the modification of the base nanoparticle with targeting ligands and optimization of its pharmacokinetic properties to ensure that the functionalized nanoparticle system remains stable under biological conditions and demonstrates intended functionality. Although having a stable base nanoparticle as a foundation is a necessary step towards the development of targeted nanoparticle systems, introduction of additional molecules on the surface of the base nanoparticle would most likely alter the physicochemical properties of the nanoparticle, such as hydrodynamic size and surface charge that regulate its in vitro and in vivo behavior and fate, and, further, there is no guarantee that the biomolecule conjugated on the nanoparticle surface would retain its functionality [9, 10]. Thus, a successfully functionalized base nanoparticle would demonstrate: (1) the base nanoparticle is suitable for conjugation of biomolecules for intended applications, and (2) the conjugation chemistry is appropriate for the tested ligand. In this study, we target at two biomarkers that are overexpressed in brain tumors, namely, αvβ3 integrin and matrix metalloproteinase-2 (MMP-2). These two biomarkers play a significant role in brain tumor cell invasion and angiogenesis [11], representing two major hallmark events in the progression of solid tumors [12–15]. However, while MMP-2 expression is limited to cancer cells, αvβ3 integrins are overexpressed on several invasive brain cancer varieties and nearly all neovascular endothelial cells [16]. To target the αvβ3 integrin biomarker, we chose cyclic arginine-glycine-aspartic acid (RGD) peptide as targeting ligand. RGD peptide has high affinity to αvβ3 integrin [17, 18], and has been used as targeting ligand of different nanomaterials for tumor imaging [19–24]. We chose chlorotoxin (CTX), a 36 amino acid peptide from scorpion venom, to target MMP-2 [25]. CTX has high affinity to tumors of the neuroectodermal origin, and has promising therapeutic potential due to its ability to inhibit cancer cell invasion [26–28]. By using two targeting agents in this study, we intended 1) to show that our base nanoparticle is versatile for conjugation of different targeting ligands and thus can be used to develop new nanoparticle systems for diagnosis and treatment of a variety of cancer types; 2) to show that each targeting ligand would retain their bioactivity after conjugation and enhance the specificity of the base nanoparticle to its targeted tumor cells; 3) to provide insight to the targeting mechanisms of different ligands used to target cancer cells.
The developed nanoprobes consist of an iron oxide core, biocompatible polymer coating, and near-infrared (NIR) fluorophores, and surface-conjugated targeting agent (either RGD or CTX peptide). We examined the hydrodynamic size, surface charge and colloidal stability in biological media. We evaluated in vitro target specificity of these nanoprobes via MRI, iron quantification assay and fluorescent microscopy. We further evaluated the tumor-specific accumulation of these nanoparticles in a xenograft mouse tumor model by MRI and histology, and examined the difference on targeting mechanism between these two nanoparticle systems.
Materials and Methods
Materials
All general chemicals, unless otherwise specified, were purchased from Sigma-Aldrich (St. Louis, MO). Chlorotoxin (MW: 4005, pI: 8.5) were purchased from Alomone Labs (Jerusalem, Israel). RGD peptide cyclo(Arg-Gly-Asp-D-Tyr-Cys) (c(RGDyC)) (MW: 594.65, pI: 5.9) and cyclo(Arg-Gly-Asp-D-Tyr-Lys) (c(RGDyK)) (MW: 619.68, pI: 10.4) were purchased from Peptide International (Louisville, KY). Dulbecco’s phosphate buffered saline (PBS), Dulbecco’s modified eagle’s medium (DMEM, with and without phenol red), fetal bovine serum (FBS), TrypLE, AF647 succinimidyl ester (AF647-NHS), wheat germ agglumin-Alexa Fluor 555 conjugates (WGA-AF555), ProLong Gold antifade solution containing DAPI (4',6-diamidino-2-phenylindole), donkey anti-hamster IgG Alexa Fluor 488 (AF488) conjugates, goat anti-rat IgG Alexa Fluor (AF555) conjugates were purchased from Invitrogen (Carlsbad, CA). Hamster anti-mouse CD61 IgG and rat anti-mouse CD31 IgG were purchased from BD Biosciences (San Jose, CA). Cassettes and O.C.T solution were acquired from Sakura Finetek (Torrance, CA). Acetone and hexane were purchased from EMD (San Diego, CA). N-Succinimidyl iodoacetate (SIA), N-succinimidyl 3-(2-pyridyldithio)propionate (SPDP), 2-iminothiolane (Traut’s reagent) and tris(2-carboxyethyl) phosphine hydrochloride (TCEP·HCl) were purchased from Molecular Bioscience (Boulder, CO). Sephacryl S-200 resin and empty PD-10 columns were purchased from GE Healthcare (Piscataway, NJ). The Micro BCA assay kit was purchased from Pierce Biotechnology (Rockford, IL). Amicon centrifugal filters (3 mL capacity, 5k molecular weight cut-off) were purchased from Millipore (Billerica, MA). U87-MG, MCF-7, and 9L cell lines were purchased from ATCC (Manassas, VA). All cell lines were maintained and propagated according to the ATCC’s protocol.
Xenograft Mice Preparation
All animal studies were conducted in accordance with University of Washington Institutional Animal Care and Use Committee (IACUC) approved protocols. Subcutaneous xenografts were established in athymic (nu/nu) mice (using U87-MG cell line. The xenografts were established by injection of 5 million of U87-MG suspended in serum free media and matrigel at a 1:1 ratio.
Synthesis of NP-PEG-NH2
12-nm core diameter, oleic acid coated magnetite nanoparticles (NP-OA) were synthesized via thermal decomposition of iron oleate complex [29]. Amine-functionalized, PEGylated nanoparticles were synthesized according to an established procedure [8]. The nanoparticles were stored in 0.1 M sodium bicarbonate buffer (pH 8.5).
Conjugation of RGD Peptide
RGD peptide was conjugated to NP-PEG-NH2 through iodoacetyl cross-linking chemistry. NP-PEG-NH2 nanoparticles were first functionalized with iodoacetyl groups. Specifically, 0.5 mg of AF647 succinimidyl ester was dissolved in 50 µL of anhydrous DMSO, and the solution was then added to 1 ml of NP-PEG-NH2 (5 mg Fe/mL) suspended in 0.1 M sodium bicarbonate (pH 8.5). The reaction mixture was under gentle shaking for 1 hr, and 5 mg of SIA dissolved in 100 µL of anhydrous DMSO was added. The resulting mixture was allowed to react for 2 hrs. Excessive AF647 and SIA were removed from the suspension through gel chromatography using Sephacryl S-200 column equilibrated with 0.1 M sodium bicarbonate, 5 mM EDTA buffer (pH 8.0). 0.5 mg of RGD peptide was then added to the SIA modified nanoparticle solution, and the mixture was allowed to react for overnight at 4°C. The resultant NP-RGD was passed through a Sephacryl S-200 column equilibrated with PBS to remove unreacted RGD peptide.
Conjugation of CTX
CTX was conjugated to NP-PEG-NH2 in a similar procedure as RGD conjugation. 1 mg of CTX was dissolved in 0.5 mL of 0.1 M sodium bicarbonate, 5 mM EDTA buffer (pH 8.0). To this solution, 11.2 µL of Traut’s reagent solution (10 mg/mL) was added. The reaction was allowed to proceed for 1 hr to produce thiolated CTX. Nanoparticles functionalized with AF647 and iodoacetyl groups were mixed with thiolated CTX and the reaction was allowed to react for overnight at 4°C. The resultant NP-CTX was passed through a Sephacryl S-200 column equilibrated with PBS to remove unreacted CTX peptide.
Nanoparticle Characterization
TEM samples were observed on a Phillips CM100 TEM (Philips, Eindhoven, The Netherlands) operating at 100 KV. FTIR spectra (resolution: 4 cm−1) were acquired using a Nicolet 5DXB spectrometer (Thermo Scientific, Boston, MA). 2 mg of nanoparticle powder was mixed with KBr (Sigma-Aldrich, St. Louis, MO) and milled, and the mixture was pressed into a pellet for analysis. The hydrodynamic size of nanoparticles was obtained via dynamic light scattering (DLS) using a Malvern Nano Series ZS particle size analyzer (Worcestershire, UK). Zeta potential measurements were performed by diluting the nanoparticle dispersion with 10 mM NaCl solution and then titrating to different pH values (pH = 3, 5, 7, 9 and 11) by addition of 0.1 N HCl or NaOH. For size and zeta potential measurements, the iron concentration of nanoparticle samples was 200 µg/mL. For long-term stability study, nanoparticles were sealed in sterilized DLS cuvettes and kept at 4°C for a desired time period. Size measurements were performed regularly up to a month. Inductively coupled plasma atomic emission spectroscopy (ICP-AES) was used to measure nanoparticle concentrations in various solutions at each stage of nanoparticle experiments including bioconjugations, in vitro cell uptake experiments and animal studies.
Quantification of Nanoprobe Components
The number of reactive amine groups per nanoparticle was quantified via SPDP assay [8, 30]. The number of RGD or CTX ligands per nanoparticle was determined by BCA assay. Briefly, micro BCA assay working reagent solution was prepared according to the vendor’s protocol. 0.6 mL of nanoparticle suspension (2 mg Fe/mL) were added to 0.6 mL of working reagent solution, and the mixture was incubated at 60°C for 2 hrs. The supernatant of reaction mixtures was isolated with Amicon centrifugal filter by centrifugation at 5,000 rpm for 10 min, The supernatants were loaded on 96-well plates and their UV-vis absorptions at 562 nm were determined. Concentrations of peptides on NPs were quantified by comparing free peptide standards. To exclude iron interference, the absorbance values of the NP-peptide conjugates were subtracted by the values of NP-PEG-NH2. In the case of RGD peptide, NP-RGD contains immobilized c(RGDyC) with thioether linkages that does to react with the BCA reagent, while the thiol group on the free c(RGDyC) reacts with the BCA reagent. Thus, c(RGDyK) was chosen as a standard, since the lysine group on c(RGDyK) does not react with the BCA reagent.
Quantification of Intracellular Iron Content
All cell lines were cultured in 12-well plates and grow to 80% of confluence before experiment. Cells were washed twice with PBS, and nanoparticles in growth media (50 µg Fe/mL) were then added. Cells incubated without nanoparticles were used as control for baseline intracellular iron content. After 2 h incubation at 37°C, 5% CO2, the cells were washed three times with PBS and lysed with 400 µL of 50 mM NaOH solution. Intracellular iron content was determined by the colorimetric ferrozine-based assay and determining cell counts by protein quantification [31]. The net cellular uptake of nanoprobes (expressed as weight of iron per cell) was derived by subtracting the baseline intracellular iron content from cells that were not treated with nanoprobes. All experiments were performed in triplicates.
Fluorescent Microscopy
All cell lines were cultured in 6-well plates. MCF-7 and U87-MG cells were seeded at 0.8 million per well two day before the assay, while 9L cells were seeded at 0.6 million per well one day before the assay. Cells were washed twice with PBS, and nanoprobes in growth media (50 µg Fe/mL) were then added. Cells incubated without nanoparticles were used as control for baseline intracellular iron content. After 2 h incubation at 37°C, 5% CO2, the cells were washed three times with PBS, detached from the well plates by TrypLE, and reseeded on glass cover-slips. After attachment, the cells were fixed with 4% formaldehyde solution for 1 h, and treated with WGA-AF555 for 30 min for membrane stain. The slides were mounted using ProLong Gold antifade solution containing DAPI and imaged using a LSM 510 Meta confocal fluorescence microscope (Carl Zeiss, Peabody, MA).
In vivo MRI Experiment
Nanoprobes were administered via tail-vein injections (n = 3 for each type of nanoprobes). Mice were anesthetized with 1 to 2.5% isoflurane (Abbott Laboratories, Abbott Park, IL) before they were placed in the imaging chamber. MR images were acquired before nanoprobe injection and at various time points post-injection on a 4.7T Bruker magnet (Bruker Medical Systems, Karlsruhe, Germany) equipped with Varian Inova spectrometer (Varian, Inc., Palo Alto, CA). A 6 cm volume coil and spin-echo imaging sequence were used to acquire spin-spin relaxation time (T2) weight images of the animals. A multi-spin-echo multi-slice imaging sequence was used to determine T2 values in tumor and normal tissues using the following imaging parameters: TR = 2 s, TE = 13.6, 30 and 60 ms, field of view of 60 × 30 mm2, number of averages of 2, matrix size of 256 × 128, slice number of 10, slice thickness of 1 mm, and gap of 0.5 mm. The T2 map was generated by NIH ImageJ (Bethesda, MD) based on the equation SI = A×exp(−TE/T2) + B, where SI is the signal intensity, TE is the echo time, A is the amplitude, and B is the offset. R2 maps were generated by taking the reciprocal of T2 map, and were colorized via Matlab program (The MathWorks, Natick, MA). The average proton relaxivity R2 (i.e. 1/T2) of whole tumor region (throughout different slices) were also calculated, and R2 changes were derived by subtracting pre-injection R2 values from post-injection R2 values.
Histological Examination of Tumors
Tumors were dissected, embedded in Tissue Tek O.C.T solution, and frozen in liquid nitrogen. Six-micrometer thin frozen slices were fixed in ice-cold methanol and washed with PBS. Slices were stained with primary hamster anti-mouse CD61 antibody followed by AF488-labeled anti-hamster secondary antibody. Endothelial cells were stained using rat anti-mouse CD31 antibody followed by AF555-labeled anti-rat secondary antibody. For Prussian blue / Nuclear fast red staining, standard clinical laboratory protocols were followed. Microscopic images of tissue were acquired using an E600 upright microscope (Nikon) equipped with a CCD color camera.
Statistical analysis
The data were expressed as mean ± SEM (standard error of the mean). We have prescreened all the original data to ensure that they followed normal distribution. Unpaired t-test was used to determine the significance of nanoprobe accumulation measured by Ferrozine assay, while paired t-test was used to determine the significance of R2 change in in vivo MRI experiment. We considered a P value of <0.05 as statistically significant.
Results
Conjugation of nanoparticles with CTX or RGD peptides and physicochemical properties of nanoprobes
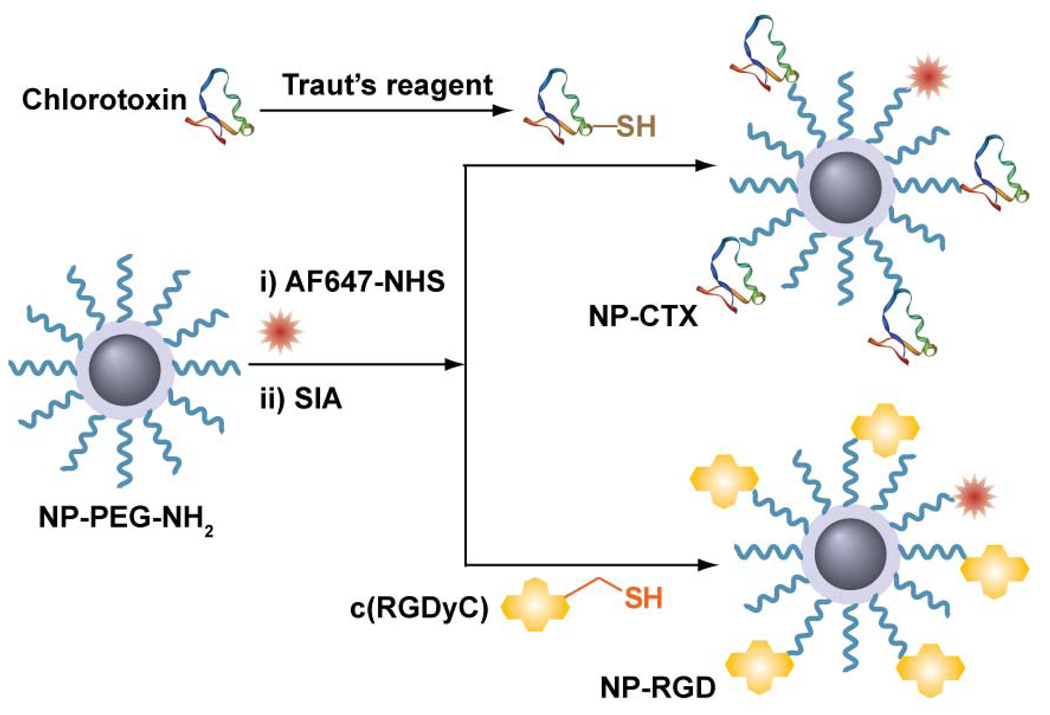
The PEG-coated, amine-functionalized base nanoparticles (NP-PEG-NH2) were synthesized as reported previously [8]. The hydrophilic PEG serves as exterior coating to render the nanoparticle biocompatible and stable in biological media. PEG also reduces protein adsorption and non-specific macrophage uptake, ultimately achieving prolonged serum half-life in vivo [9, 32]. Amine groups at the free termini of PEG chains allow for further conjugation of bioactive molecules or ligands. In this study, the targeting ligands, RGD and CTX peptides, were conjugated to the nanoparticle via thioether linkage (Figure 1). For conjugation CTX to NP-PEG-NH2, an extra thiol group was introduced on CTX molecule by reacting with 2-iminothiolane (Traut’s reagent). For RGD conjugation, a variant of RGD peptides, c(RGDyC) sequence was chosen because the cyclic backbone RGD has previously demonstrated to be better resistant to proteolysis and has high affinity to the αvβ3 integrin receptors [33].
Figure 1. The chemical scheme of nanoprobes synthesis.
Chemical scheme for conjugation of RGD (NP-RGD) or CTX (NP-CTX) to base nanoparticles (NP-PEG-NH2).
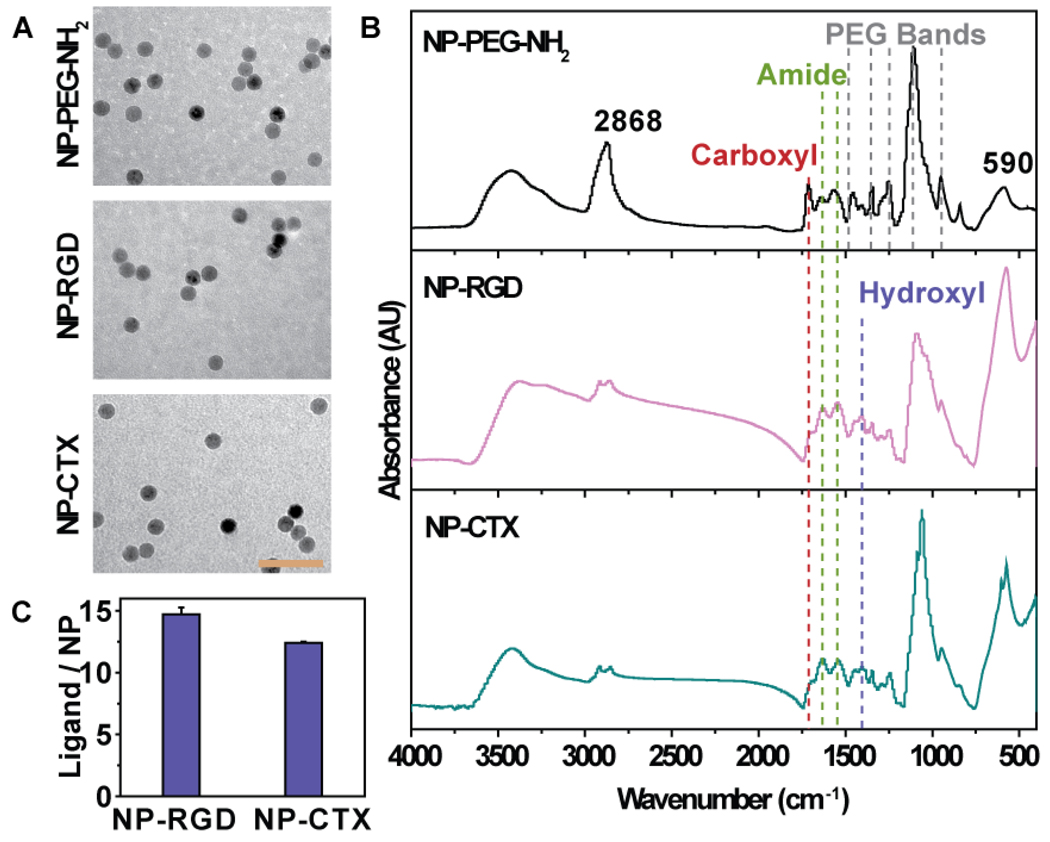
The size and morphology of the synthesized nanoprobes were characterized by TEM. Figure 2a shows that NP-PEG-NH2 and the nanoprobes conjugated with RGD (NP-RGD) or CTX (NP-CTX) were spherical and well dispersed, with a core size of ~12 nm, indicating that all the nanoprobes maintained their morphology after the conjugation of either RGD or CTX and no inter-particle cross-linking occurred during the conjugation process. The number of reactive amine groups per nanoparticle was found to be around 70–80 by quantifying released pyridine-2-thione following reaction with SPDP [8, 30].
Figure 2. Physicochemical properties of nanoparticles.
(A) TEM images of NP-PEG-NH2, NP-RGD and NP- CTX (scale bar: 60 nm). (B) FTIR spectra of NP-PEG-NH2, NP-RGD and NP-CTX. (c) Quantification of immobilized ligand by BCA assay.
Successful conjugation of RGD or CTX on NP-PEG-NH2 was confirmed by Fourier transform infrared spectroscopy (FTIR) (Figure 2B). The IR spectra of NP-PEG-NH2 contained multiple bands at 1458, 1346, 1244, 1112 and 949 cm−1, corresponding to the different vibrational modes of PEG’s C-O-C bonds [34]. The peaks at 1642 and 1559 cm−1 could be assigned to either primary amine groups or mono-substituted amide. On the spectrum of NP-RGD, the increased relative intensity of amide peaks at 1634 and 1542 cm−1 was observed, and the relative intensity of carboxyl peak at 1710 cm−1 was decreased. Furthermore, peak at ~1400 cm−1, representing the tyrosine hydroxyl groups of RGD peptide, could be seen on the spectrum. This suggests that the amide-rich RGD peptide was successfully conjugated to the base nanoparticle. The spectrum of NP-CTX followed the similar trend as the NP-RGD, displaying the increased intensity of amide peaks and dwarfed carboxyl peak, as well as a newly emerged hydroxyl peak. The number of immobilized ligands on each nanoprobe was determined via BCA protein quantification assay. The results showed that there were 14.9 RGD peptide on each NP-RGD nanoprobe, and 12.4 CTX on each NP-CTX nanoprobe (Figure 2C).
The hydrodynamic size of NP-PEG-NH2 in cell culture media was determined to be 38 nm by dynamic light scattering (DLS) (Figure 3A; and Table 1). The hydrodynamic size increased to 44 nm and 46 nm after being conjugated with RGD peptide and CTX, respectively. The zeta potential of NP-PEG-NH2 was −14 mV at pH 7, and remained at similar values after the conjugation of either RGD or CTX (Figure 3B). Both nanoprobes displayed excellent long-term colloidal stability in cell culture media, i.e., DMEM with 10% of FBS, as they maintained their hydrodynamic size for at least a month (Figure 3C). No flocculation or apparent size change was observed even after several months of storage. The major physiochemical properties of these nanoprobes are summarized in Table 1.
Figure 3. Colloidal properties of nanoprobes.
(A) Hydrodynamic size of nanoprobes in cell culture media. (B) Zeta potential of nanoprobes as a function of pH value. (C) Hydrodynamic sizes of nanoprobes in cell culture media as a function of storage time, showing long-term stability of the nanoprobes. Cell culture media was composed of 90% DMEM and 10% FBS.
Table 1.
Physicochemical properties of nanoprobes.
| Probe Name | Hydrodynamic size* (nm) |
Polydispersity index (PDI*) |
Zeta potential at pH 7 (mV) |
Number of ligand per nanoprobe |
|---|---|---|---|---|
| NP-PEG NH2 |
38 | 0.135 | −14 | 0 |
| NP-RGD | 44 | 0.147 | −16 | 14.9 |
| NP-CTX | 46 | 0.169 | −20 | 12.4 |
Measured in cell culture media (90% DMEM and 10% FBS).
In vitro evaluations of cell targeting specificity of nanoprobes
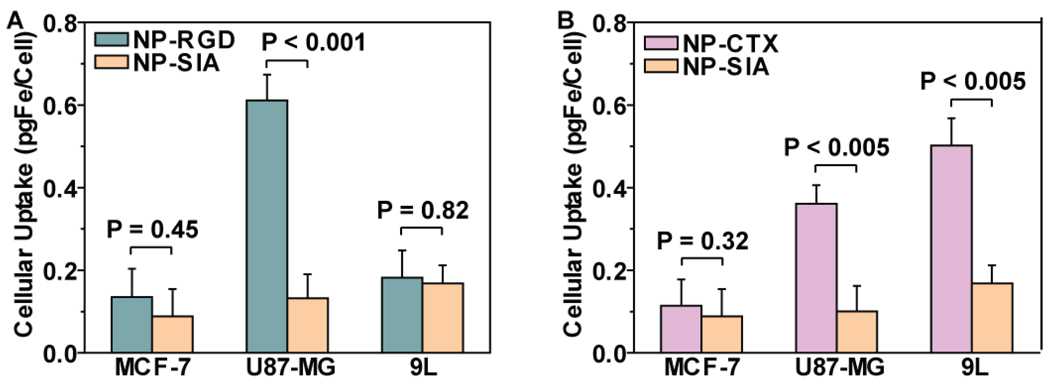
The specificity of the NP-CTX, and NP-RGD nanoprobes for tumor cells were assessed in vitro. Here we chose glioma cell lines as our model cells because they represent a common type of brain cancer with overall poor prognosis [35], and most of glioma cell lines overexpress MMP-2 while some of them also overexpress αvβ3 integrin [19, 28, 36, 37]. We tested our nanoprobes on human glioblastoma (U87-MG) and rat glioma (9L) cell lines. The former expresses both αvβ3 integrin and MMP-2 receptors, while the latter expresses MMP-2 only. We used human breast adenocarcinoma (MCF-7) cell line as negative control which does not express either receptor [37]. We have previously demonstrated that CTX conjugated on iron oxide nanoparticles promotes multivalent binding of nanoparticles to targeted receptors, which significantly increase the binding of nanoparticles to target cells compared to free CTX [28]. This effect has also been observed for RGD conjugated on iron oxide nanoparticle surfaces [20]. Thus the nanoprobes developed in this study, with such multivalent binding effect, are likely to interfere with their binding specificity through competitive binding inhibition assays. Therefore competitive binding assays may not be suitable for this nanoparticle system, and instead, negative control cells were used here to demonstrate receptor specific targeting. Iodoacetyl-conjugated nanoparticles (NP-SIA) that contain no targeting ligands were chosen as our control nanoparticles. NP-SIA is an intermediate product right before CTX conjugation.
To determine the amount of nanoparticles uptake by cells, the iron content of cells were determined by a ferrozine-based colorimetric assay after cells were cultured with nanoprobes. For U87-MG cell line, the cellular uptake of RGD peptide-conjugated nanoprobes (NP-RGD) was 4.6 times (P < 0.001) of that of NP-SIA. The difference in cellular uptake between NP-RGD and NP-SIA by MCF-7 and 9L cell lines was not as significant: 1.5 times (P = 0.45) for MCF-7, and 1.1 times (P = 0.82) for 9L (Figure 4A). These results showed that the NP-RGD nanoprobes were preferentially taken up by the U87-MG cells, which is the only cell line expressing αvβ3 integrins among the three tested. For CTX-conjugated nanoprobes (NP-CTX), 3.0 times (P < 0.005) and 3.6 times (P < 0.005) of uptake over those by control nanoprobes were observed in MMP-2 positive U87-MG and 9L cell lines, respectively. No significant difference in cellular uptake (P = 0.32) between NP-CTX and control nanoprobes for the MMP-2 negative MCF-7 cell line (Figure 4B).
Figure 4. In vitro assessment of cancer cell targeting specificity of nanoprobes.
Cellular uptake of (A) NP-RGD and (B) NP-CTX by MCF-7, U87-MG and 9L cells, with NPSIA as control nanoprobe. Results are presented as mean ± SEM.
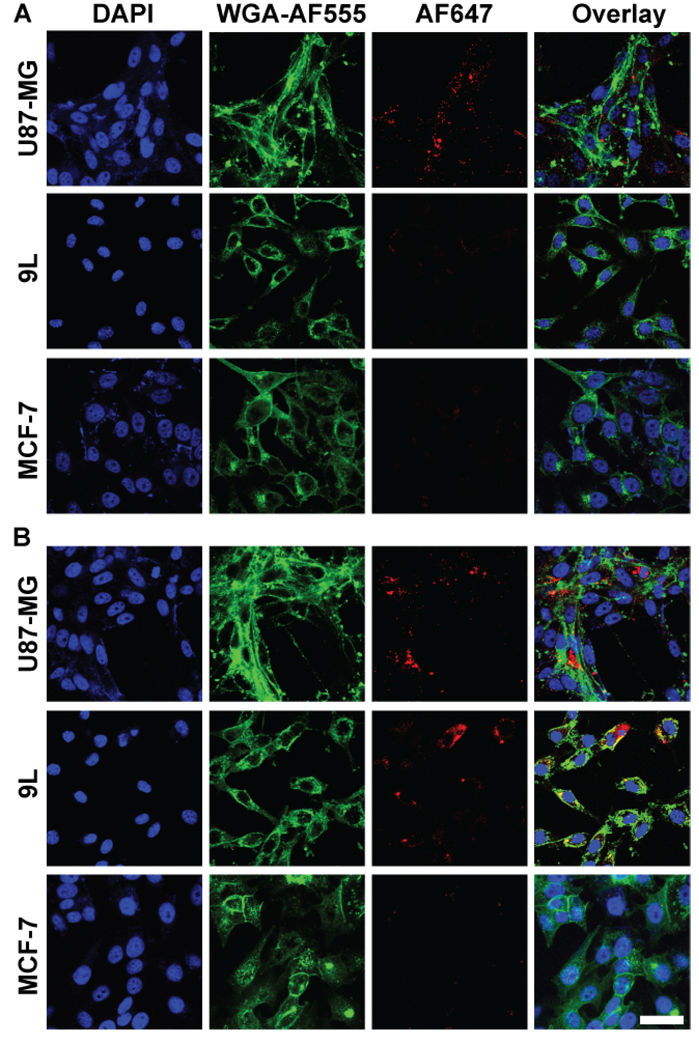
The preferential uptake of nanoprobes by target cells was further confirmed by fluorescence imaging, for which a near-infrared fluorophore, AF647-NHS succinimidyl ester, was conjugated onto the amine groups of nanoprobes. Cells were incubated with fluorescence-enabled nanoprobes for 2 hours before imaging. U87-MG cells treated with the NP-RGD can be clearly visualized in AF647 channel, while for 9L and MCF-7 cell lines, no AF647 signal was observed (Figure 5A). NP-CTX probe treatment of both U87-MG and 9L showed strong signal in AF647 channel, indicating strong uptake of NP-CTX by the cell lines expressing MMP-2 (Figure 5B). Conversely, little signal of NP-CTX can be found on the MCF-7 cell line.
Figure 5. Preferential uptake of nanoprobes by target cells assessed by confocal microscopy.
Images of U87-MG, 9L and MCF-7 cells incubated with (A) NP-RGD and (B) NP-CTX nanoprobes, where nuclei were stained with DAPI (blue, first column), cell membranes with WGA-AF555 (green, second column) and nanoprobes with AF647 (red, third column). Also shown are the overlay images (fourth column) (Scale bar: 40 µm).
In vivo tumor-specific accumulation and intratumoral distribution of nanoparticles
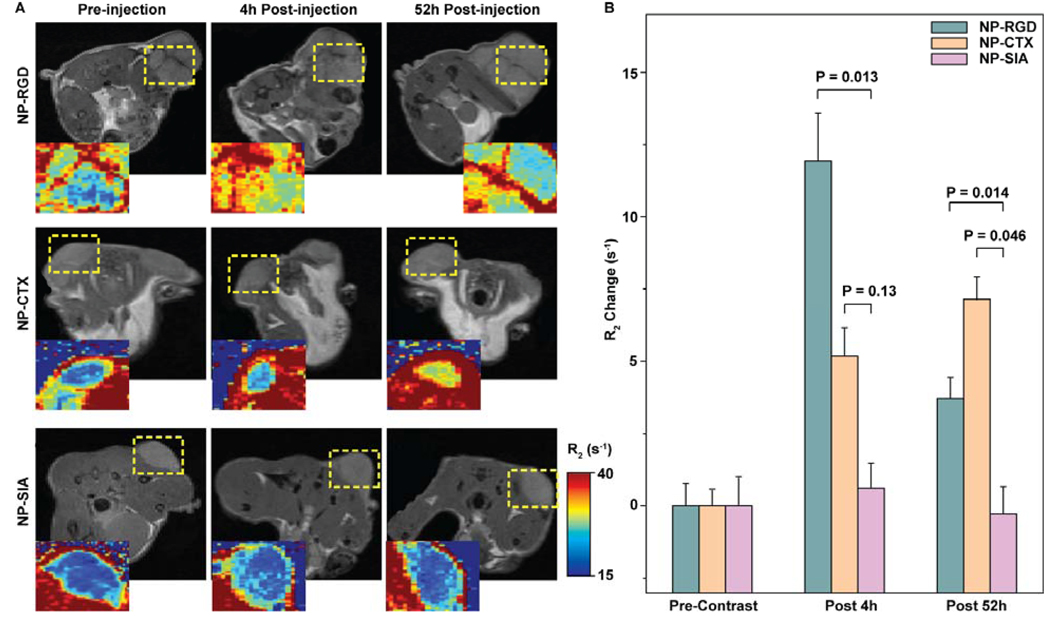
The tumor-specific targeting and contrast enhancement by the developed nanoprobes were assessed in nude mice bearing flank xenograft U87-MG tumors. The goal of our in vivo study was to investigate temporal and spatial behavior of NP-RGD and NP-CTX in mice bearing tumors that expressed receptors for both RGD and CTX ligands. Our in vitro experiments have identified that only U87-MG tumor cells expressed receptors for both RGD and CTX ligands. Thus we limited our in vivo study only to U87-MG tumors. The nanoprobes were administered intravascularly via tail vein injection. The MR images were acquired at 4 hrs and 52 hrs post injection. The targeting specificity towards U87-MG tumors was evaluated by comparing the relaxivity enhancement of tumor regions of mice receiving targeting nanoprobes and non-targeting nanoprobes. As shown in Figure 6A, the significant increase of R2 relaxivity in tumor regions (yellow dash-line circled areas) was visualized in colorized R2 maps of the mice injected with targeting nanoprobes NP-RGD (first row) or NP-CTX (second row). A slight increase in R2 within the tumor regions was observed at 4 hrs post injection for mice receiving non-targeting nanoprobes (NP-SIA), but the R2 fell back to the pre-injection level 52 hrs post injection (third row). Figure 6B shows quantitative results of R2. A contrast enhancement of 11.939 ± 2.746 s−1 (mean ± SEM) in terms of R2 was observed in the tumors 4 hrs post inject of NP-RGD, which is significant higher than in the tumors of the mice receiving NP-SIA (as control) (0.617 ± 1.447 s−1, P = 0.013). However, this contrast enhancement decreased to 3.712 ± 1.152 s−1 52 hours post NP-RGD injection, although it is still significant higher than that of NP-SIA (−0.267 ± 0.842 s−1, P = 0.014). For mice receiving NP-CTX nanoprobes, the contrast enhancement in tumors increased to 5.181 ± 1.567 s−1 (P = 0.13) 4 hrs post injection and progressively to 7.152 ± 1.311 s−1 (P = 0.046) 52 hrs post injection.
Figure 6. In vivo MRI assessment of tumor-targeting nanoprobes in xenograft U87-MG tumor mouse model.
(A) Representative T2-weighted MR images of mice injected with NP-RGD (first row), NP-CTX (second row) and NP-SIA (control, third row) nanoprobes, acquired before nanoprobe injection (first column), and 4 hrs (second column) and 52 hrs post injection. Axial cross sections of the lower body were analyzed. Expanded (2×) and colorized R2 maps of tumor region were shown next to the anatomical images. R2 (s−1) from low (blue) to high (red) were visually represented in colors generated from the gradient bar at right. The expended regions were enclosed with yellow dash-lines. (B) R2 changes in tumor regions quantified by calculating the difference in R2 before and after nanoprobe injection. Results are presented as mean ± SEM.
To further confirm the preferential accumulation of nanoprobes in tumors and to investigate their intra-tumoral distribution, the mice receiving nanoprobe injections were sacrificed after MR imaging, and histological analyses were performed on excised tumor tissues. CD61 and CD31 antibodies were used to stain β3 integrin, and PECAM-1 (Platelet Endothelial Cell Adhesion Molecule 1) receptors, respectively. β3 integrin are upregulated in U87-MG tumor cells and immature blood vessels, while PECAM-1 is strictly associated with endothelial cells. Thus, CD31 staining was used confirm blood vessel location, and the co-localization of CD31 and CD61 staining was used to confirm presence of neovasculature in the tumor. Immunofluorescence images showed that U87-GM tumors have developed heterogeneous and complex neovasculature (Figure 7A). The overlay images showed that the limited accumulation of NP-RGD in red in the tumor predominantly colocalizes with cells expressing CD61 in blue and CD31 in green, and are not far from blood vessels. Conversely, NP-CTX are dispersed within tumor and predominantly appeared not associated with blood vessels. Results from Prussian blue stain (Figure 7B) confirm the finding of the immunofluorescence staining, which showed that NP-RGD appeared near the vasculatures (labeled by arrows), while NP-CTX nanoprobes were distributed throughout the tumor, and a noticeable significant population of them were away from vasculatures.
Figure 7. Histological examination of xenograft U87-MG mouse tumors 52 hours post administration of AF647-labeled NP-CTX or NP-RGD.
(A) Immunofluorescence images of tumor sections. Antibodies were used to stain CD31 markers associated with mouse vasculature (first column, green), and CD61 markers up-regulated on U87-MG cells and mouse tumor neovasculature (second column, blue); the fluorescence signal of AF647 associated with nanoprobes are shown in red (third column). The merged images were shown in the fourth column (Scale bar: 40 µm). (B) Prussian blue staining images (counterstained with nucleus fast red) of tumors sections excised from mice injected with NP-RGD (left) or NP-CTX (right). Arrows in the images mark the location of blood vessels. The inset shows a cross-section of blood vessel at high magnification (Scale bar: 40 µm, inset: 10 µm).
Discussions
In the present study, we reported the development and evaluation of two iron oxide nanoparticlebased nanoprobes that bear targeting ligands for biomarkers of αvβ3 integrins or MMP-2 overexpressed on cancer cells.
Our results indicated these nanoprobes have superior colloidal stability in media containing high concentration of salts, amino acids, and proteins. This can be attributed to the presence of dense and stable hydrophilic coating on the nanoparticle surface. Stable silicon-oxygen-silicon (siloxane) linkages between succinic anhydride silane (SAS) molecules display good chemical stability against solvent leaching while highly hydrophilic PEG chains provide a steric repulsion force that prevents nanoprobe aggregation. Furthermore, these nanoprobes possess a moderate negative zeta potential that minimizes the ionic interaction between the nanoparticle surface and macromolecules in the cell media [38]. Unlike highly cationic nanoparticles, negative surface charge of nanoprobes does not cause severe disruption on the integrity of cell membrane.
We demonstrated the targeting efficacy of both NP-RGD and NP-CTX in cell lines that have different levels of αvβ3 integrins and MMP-2 expressions. Both NP-RGD and NP-CTX showed elevated cellular uptakes in receptor-positive cell lines as compared to those in receptor-negative cell lines. And in receptor-positive cell lines, targeting nanoprobes (NP-RGD and NP-CTX) showed much higher cellular uptake than non-targeting nanoparticles (NP-SIA).
Our in vivo MRI results demonstrated that the nanoprobes provided substantial contrast enhancement in the U87-MG xenograft mouse model by preferential accumulation in tumors. However, the temporal difference in particle accumulation in tumors between NP-RGD and NP-CTX was observed. While the NP-RGD exhibited a strong initial accumulation in tumors, the NP-CTX showed a more progressive and sustained accumulation. Histological analysis further revealed the spatial difference in intratumoral nanoprobe distribution between NP-RGD and NP-CTX. NP-RGD mainly accumulated near the blood vessels, while NP-CTX exhibited a highly diffusive distribution throughout the tumor. These differences in spatial and temporal nanoprobe accumulation can be attributed to the difference in molecular targets exhibited by RGD and CTX.
The tumor consists of neoplastic tumor cells and stromal cells that include neovascular endothelial cells [39]. The αvβ3 integrins are overexpressed on U87-MG tumor cells, as well as endothelial cells of angiogenic tumor vessels [12, 13]. Based on the nature of its targets, the RGD peptide is a dual-targeting ligand that binds to both neovascular endothelial cells and tumor cells, while CTX interacts with tumor cells only through its affinity to MMP-2. After NP-RGD was injected into the bloodstream, a significant amount of NP-RGD would be quickly associated with the neovascular endothelial cells while few others manage to reach the tumor cells, resulting in strong initial accumulation of the nanoprobe in the tumor. The decrease of contrast enhancement in later time points is consistent with previous reports [36, 40], and might be attributed to the dissociation and clearance of the vasculature-bound NP-RGD as a result of the relatively weak affinity of RGD peptides to the αvβ3 integrin expressed on endothelial cells (EC50 around micromolar range [20]). The results also justified that although NP-RGD showed a strong targeting effect in vitro, its contrast enhancement in vivo diminished over time in vivo. On the other hand, CTX is likely extravasated across the endothelium through Annexin A2 binding and then interact with MMP-2 receptors that are overexpressed on U87-MG tumor cells [37, 41–43]. Because the extravasation of NP-CTX nanoprobes across the endothelium is a receptor dependent process, it is likely to contribute to the gradual accumulation in the tumor. The increased contrast enhancement up to 52 hrs can be attributed to continuous nanoprobe accumulation. These results reveal how the nature of the molecular targets of the ligand affects the accumulation and retention of the ligand-enabled nanoprobe within the tumor.
Conclusions
We have developed two tumor-targeting nanoprobes that bear RGD and CTX peptides, respectively. The nanoprobes are highly dispersive and exhibit excellent long-term stability in the cell culture media. Colorimetric iron quantification assay and confocal fluorescent microscopy confirmed that nanoprobes conjugated with targeting ligands selectively bind to tumor cells overexpressing αvβ3 integrin or MMP-2 receptors. Results of in vivo MRI and histology showed that the both nanoprobes accumulated preferentially in tumors, and revealed the effect of the difference in molecular targets between ligands on nanoprobe accumulation and distribution in tumors.
Future perspective
This study demonstrated that with flexible surface chemistry, our base nanoparticle can be conjugated with biomolecules in a modular fashion to develop various nanoparticle-based diagnostic and therapeutic agents. Our study provided helpful information on selection of targeting ligands for development of new nanoparticle systems for intended applications.
Executive Summary
Two nanoprobes were developed with molecular targets of αvβ3 integrin or MMP-2 receptors, respectively.
The nanoprobes are highly stable in cell culture media and exhibits long-term stability.
The achieved high stability of the nanoprobes can be attributed to the dense PEG coating on the nanoprobe.
This study demonstrated that the highly stable base nanoparticle developed in our previous study could be conjugated with biomolecules in a modular fashion for intended applications.
Colorimetric iron quantification assay and confocal fluorescent microscopy confirmed that nanoprobes conjugated with targeting ligands selectively bind to cells that overexpress αvβ3 integrins or MMP-2.
MR imaging demonstrated that the both nanoprobes accumulated preferentially in tumors.
Targeted delivery of magnetic particle-based nanoprobes significantly enhances MR imaging contrast compared to non-targeted delivery.
The in vivo study revealed that the nanoprobes bearing different ligands accumulate differently in tumors both temporally and spatially. This provides insights in selecting targeting ligands in development of new nanoparticle systems for biomedical applications.
Acknowledgments
This work is supported in part by NIH grants (R01CA119408, R01EB006043, and R01CA134213). C.F. would like to acknowledge the support of the NCI/NSF IGERT fellowship from the University of Washington Center for Nanotechnology (UW-CNT). O.V. and F.K would like to acknowledge the support of the NIH training grant (T32CA138312).
Footnotes
The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed. No writing assistance was utilized in the production of this manuscript. We would like to thank Mr. Haobo Chen for laboratory assistance and Mr. Daniel Chen for MRI data processing.
References
- 1.Peer D, Karp JM, Hong S, Farokhzad OC, Margalit R, Langer R. Nanocarriers as an emerging platform for cancer therapy. Nature Nanotechnol. 2007;2:751–760. doi: 10.1038/nnano.2007.387. [DOI] [PubMed] [Google Scholar]
- 2.Sun C, Lee JSH, Zhang M. Magnetic nanoparticles in MR imaging and drug delivery. Adv Drug Deliv Rev. 2008;60:1252. doi: 10.1016/j.addr.2008.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Weissleder R, Pittet MJ. Imaging in the era of molecular oncology. Nature. 2008;452:580–589. doi: 10.1038/nature06917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cai WB, Chen XY. Nanoplatforms for targeted molecular imaging in living subjects. Small. 2007;3:1840–1854. doi: 10.1002/smll.200700351. [DOI] [PubMed] [Google Scholar]
- 5.Fang C, Zhang M. Multifunctional magnetic nanoparticles for medical imaging applications. J Mater Chem. 2009;19:6258–6266. doi: 10.1039/b902182e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Weissleder R. Molecular imaging in cancer. Science. 2006;312:1168–1171. doi: 10.1126/science.1125949. [DOI] [PubMed] [Google Scholar]
- 7.Waldman AD, Jackson A, Price SJ, Clark CA, Booth TC, Auer DP, et al. Quantitative imaging biomarkers in neuro-oncology. Nat Rev Clin Oncol. 2009;6:445–454. doi: 10.1038/nrclinonc.2009.92. [DOI] [PubMed] [Google Scholar]
- 8.Fang C, Bhattarai N, Sun C, Zhang M. Functionalized nanoparticles with long-term stability in biological media. Small. 2009;5:1637–1641. doi: 10.1002/smll.200801647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nel AE, Mädler L, Velegol D, Xia T, Hoek EM, Somasundaran P, et al. Understanding biophysicochemical interactions at the nano-bio interface. Nat Mater. 2009 doi: 10.1038/nmat2442. [DOI] [PubMed] [Google Scholar]
- 10.Lewinski N, Colvin V, Drezek R. Cytotoxicity of nanoparticles. Small. 2008;4:26–49. doi: 10.1002/smll.200700595. [DOI] [PubMed] [Google Scholar]
- 11.Lorusso G, Ruegg C. The tumor microenvironment and its contribution to tumor evolution toward metastasis. Histochem Cell Biol. 2008;130:1091–1103. doi: 10.1007/s00418-008-0530-8. [DOI] [PubMed] [Google Scholar]
- 12.Brooks PC, Clark RAF, Cheresh DA. Requirement Of Vascular Integrin Alpha(V)Beta(3) For Angiogenesis. Science. 1994;264:569–571. doi: 10.1126/science.7512751. [DOI] [PubMed] [Google Scholar]
- 13.Hood JD, Cheresh DA. Role of integrins in cell invasion and migration. Nat Rev Cancer. 2002;2:91. doi: 10.1038/nrc727. + [DOI] [PubMed] [Google Scholar]
- 14.Overall CM, Kleifeld O. Tumour microenvironment - opinion: validating matrix metalloproteinases as drug targets and anti-targets for cancer therapy. Nat Rev Cancer. 2006;6:227–239. doi: 10.1038/nrc1821. [DOI] [PubMed] [Google Scholar]
- 15.Stetler-Stevenson WG. Progelatinase A activation during tumor cell invasion. Invasion Metastasis. 1994;14:259–268. [PubMed] [Google Scholar]
- 16.Friedlander M, Theesfeld CL, Sugita M, Fruttiger M, Thomas MA, Chang S, et al. Involvement of integrins alpha(v)beta(3) and alpha(v)beta(5) in ocular neovascular diseases. Proc Natl Acad Sci USA. 1996;93:9764–9769. doi: 10.1073/pnas.93.18.9764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gurrath M, Muller G, Kessler H, Aumailley M, Timpl R. Conformation activity studies of rationally designed potent antiadhesive RGD peptides. Eur J Biochem. 1992;210:911–921. doi: 10.1111/j.1432-1033.1992.tb17495.x. [DOI] [PubMed] [Google Scholar]
- 18.Cai W, Chen X. Multimodality molecular imaging of tumor angiogenesis. J Nucl Med. 2008;49 Suppl 2:113S–128S. doi: 10.2967/jnumed.107.045922. [DOI] [PubMed] [Google Scholar]
- 19.Cai W, Chen X. Preparation of peptide-conjugated quantum dots for tumor vasculature-targeted imaging. Nat Protoc. 2008;3:89–96. doi: 10.1038/nprot.2007.478. [DOI] [PubMed] [Google Scholar]
- 20.Montet X, Funovics M, Montet-Abou K, Weissleder R, Josephson L. Multivalent effects of RGD peptides obtained by nanoparticle display. J Med Chem. 2006;49:6087–6093. doi: 10.1021/jm060515m. [DOI] [PubMed] [Google Scholar]
- 21.De La Zerda A, Zavaleta C, Keren S, Vaithilingam S, Bodapati S, Liu Z, et al. Carbon nanotubes as photoacoustic molecular imaging agents in living mice. Nature Nanotech. 2008;3:557–562. doi: 10.1038/nnano.2008.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen H, Wang L, Yeh J, Wu X, Cao Z, Wang YA, et al. Reducing non-specific binding and uptake of nanoparticles and improving cell targeting with an antifouling PEO-b-PgammaMPS copolymer coating. biomaterials. 2010 doi: 10.1016/j.biomaterials.2010.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen W, Jarzyna PA, van Tilborg GA, Nguyen VA, Cormode DP, Klink A, et al. RGD peptide functionalized and reconstituted high-density lipoprotein nanoparticles as a versatile and multimodal tumor targeting molecular imaging probe. FASEB J. 2010 doi: 10.1096/fj.09-139865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Danhier F, Vroman B, Lecouturier N, Crokart N, Pourcelle V, Freichels H, et al. Targeting of tumor endothelium by RGD-grafted PLGA-nanoparticles loaded with paclitaxel. J Control Release. 2009;140:166–173. doi: 10.1016/j.jconrel.2009.08.011. [DOI] [PubMed] [Google Scholar]
- 25.Deshane J, Garner CC, Sontheimer H. Chlorotoxin inhibits glioma cell invasion via matrix metalloproteinase-2. J Biol Chem. 2003;278:4135–4144. doi: 10.1074/jbc.M205662200. [DOI] [PubMed] [Google Scholar]
- 26.Veiseh O, Sun C, Gunn J, Kohler N, Gabikian P, Lee D, et al. Optical and MRI multifunctional nanoprobe for targeting gliomas. Nano Lett. 2005;5:1003–1008. doi: 10.1021/nl0502569. [DOI] [PubMed] [Google Scholar]
- 27.McFerrin MB, Sontheimer H. A role for ion channels in glioma cell invasion. Neuron Glia Biol. 2006;2:39–49. doi: 10.1017/S17440925X06000044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Veiseh O, Gunn JW, Kievit FM, Sun C, Fang C, Lee JS, et al. Inhibition of tumor-cell invasion with chlorotoxin-bound superparamagnetic nanoparticles. Small. 2009;5:256–264. doi: 10.1002/smll.200800646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Park J, An K, Hwang Y, Park J-G, Noh H-J, Kim J-Y, et al. Ultra-large-scale syntheses of monodisperse nanocrystals. Nat Mater. 2004;3:891–895. doi: 10.1038/nmat1251. [DOI] [PubMed] [Google Scholar]
- 30.Schellenberger EA, Sosnovik D, Weissleder R, Josephson L. Magneto/optical annexin V, a multimodal protein. Bioconjugate Chem. 2004;15:1062–1067. doi: 10.1021/bc049905i. [DOI] [PubMed] [Google Scholar]
- 31.Riemer J, Hoepken HH, Czerwinska H, Robinson SR, Dringen R. Colorimetric ferrozine-based assay for the quantitation of iron in cultured cells. Anal Biochem. 2004;331:370–375. doi: 10.1016/j.ab.2004.03.049. [DOI] [PubMed] [Google Scholar]
- 32.Sun C, Veiseh O, Gunn J, Fang C, Hansen S, Lee D, et al. In vivo MRI detection of gliomas by chlorotoxin-conjugated superparamagnetic nanoprobes. Small. 2008;4:372–379. doi: 10.1002/smll.200700784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Meyer A, Auernheimer J, Modlinger A, Kessler H. Targeting RGD recognizing integrins: drug development, biomaterial research, tumor imaging and targeting. Curr Pharm Des. 2006;12:2723–2747. doi: 10.2174/138161206777947740. [DOI] [PubMed] [Google Scholar]
- 34.Pouchert CJ, Aldrich Chemical C. The Aldrich library of FT-IR spectra. Milwaukee, Wis.: Aldrich Chemical Co.; 1985. [Google Scholar]
- 35.Wrensch M, Minn Y, Chew T, Bondy M, Berger MS. Epidemiology of primary brain tumors: current concepts and review of the literature. J Neurooncol. 2002;4:278–299. doi: 10.1093/neuonc/4.4.278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Montet X, Montet-Abou K, Reynolds F, Weissleder R, Josephson L. Nanoparticle imaging of integrins on tumor cells. Neoplasia. 2006;8:214–222. doi: 10.1593/neo.05769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Veiseh M, Gabikian P, Bahrami SB, Veiseh O, Zhang M, Hackman RC, et al. Tumor paint: a chlorotoxin:Cy5.5 bioconjugate for intraoperative visualization of cancer foci. Cancer Res. 2007;67:6882–6888. doi: 10.1158/0008-5472.CAN-06-3948. [DOI] [PubMed] [Google Scholar]
- 38.Petri-Fink A, Steitz B, Finka A, Salaklang J, Hofmann H. Effect of cell media on polymer coated superparamagnetic iron oxide nanoparticles (SPIONs): Colloidal stability, cytotoxicity, and cellular uptake studies. Eur J Pharm Biopharm. 2008;68:129–137. doi: 10.1016/j.ejpb.2007.02.024. [DOI] [PubMed] [Google Scholar]
- 39.Albini A, Sporn MB. The tumour microenvironment as a target for chemoprevention. Nat Rev Cancer. 2007;7:139–147. doi: 10.1038/nrc2067. [DOI] [PubMed] [Google Scholar]
- 40.Bibby DC, Talmadge JE, Dalal MK, Kurz SG, Chytil KM, Barry SE, et al. Pharmacokinetics and biodistribution of RGD-targeted doxorubicin-loaded nanoparticles in tumor-bearing mice. International journal of pharmaceutics. 2005;293:281–290. doi: 10.1016/j.ijpharm.2004.12.021. [DOI] [PubMed] [Google Scholar]
- 41.Jacoby DB, Dyskin E, Yalcin M, Kesavan K, Dahlberg W, Ratliff J, et al. Potent pleiotropic anti-angiogenic effects of TM601, a synthetic chlorotoxin peptide. Anticancer Res. 30:39–46. [PubMed] [Google Scholar]
- 42.Kesavan K, Ratliff J, Johnson EW, Dahlberg W, Asara JM, Misra P, et al. Annexin A2 is a molecular target for TM601, a peptide with tumor-targeting and anti-angiogenic effects. J Biol Chem. 285:4366–4374. doi: 10.1074/jbc.M109.066092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sontheimer H. An unexpected role for ion channels in brain tumor metastasis. Exp Biol Med (Maywood) 2008;233:779–791. doi: 10.3181/0711-MR-308. [DOI] [PMC free article] [PubMed] [Google Scholar]