Abstract
We have used affinity chromatography to identify proteins that interact with Nap1, a protein previously shown to play a role in mitosis. Our studies demonstrate that a highly conserved protein called Sda1 binds to Nap1 both in vitro and in vivo. Loss of Sda1 function causes cells to arrest uniformly as unbudded cells that do not increase significantly in size. Cells arrested by loss of Sda1 function have a 1N DNA content, fail to produce the G1 cyclin Cln2, and remain responsive to mating pheromone, indicating that they arrest in G1 before Start. Expression of CLN2 from a heterologous promoter in temperature-sensitive sda1 cells induces bud emergence and polarization of the actin cytoskeleton, but does not induce cell division, indicating that the sda1 cell cycle arrest phenotype is not due simply to a failure to produce the G1 cyclins. The Sda1 protein is absent from cells arrested in G0 and is expressed before Start when cells reenter the cell cycle, further suggesting that Sda1 functions before Start. Taken together, these findings reveal that Sda1 plays a critical role in G1 events. In addition, these findings suggest that Nap1 is likely to function during G1. Consistent with this, we have found that Nap1 is required for viability in cells lacking the redundant G1 cyclins Cln1 and Cln2. In contrast to a previous study, we have found no evidence that Sda1 is required for the assembly or function of the actin cytoskeleton. Further characterization of Sda1 is likely to provide important clues to the poorly understood mechanisms that control passage through G1.
INTRODUCTION
Entry into the eukaryotic cell cycle occurs at a point in late G1 phase referred to as Start in yeast cells or the Restriction point in vertebrate cells (Hartwell et al., 1974; Pardee et al., 1978; Cross, 1995). Once cells pass through Start they become committed to a new round of cell division (Pringle and Hartwell, 1981). Entry into the cell cycle is highly regulated and occurs only when cells have reached a critical size and have received the appropriate external signals in the form of nutrients or growth factors (Johnston and Singer, 1990; Cross, 1995; Planas-Silva and Weinberg, 1997). At the molecular level, entry into the cell cycle is induced by the synthesis of G1 cyclins, which bind and activate cyclin-dependent kinases to initiate the events of cell division (Hadwiger et al., 1989; Richardson et al., 1989; Dirick et al., 1995).
Genetic studies in budding yeast have played an important role in identifying proteins that are required during G1 for entry into the cell cycle. Temperature-sensitive mutations that inactivate Cdc28, the major cyclin-dependent kinase in budding yeast, cause cells to arrest before passing through Start (Hartwell et al., 1974; Reed, 1980). Similarly, loss of function of the G1 cyclins, which activate Cdc28, results in an arrest before Start (Hadwiger et al., 1989; Richardson et al., 1989). Cells arrested in G1 by loss of function of Cdc28 or the G1 cyclins continue to increase in size, indicating that these proteins are not required for cell growth (Reed, 1980; Cross, 1990). Mutations in other genes cause cells to arrest before Start without increasing in size. These include CDC25, the guanine nucleotide exchange factor for Ras, and CDC35, which encodes adenylate cyclase (Dawes and Calvert, 1984; Broek et al., 1987). Cells arrested by loss of function of cdc25 or cdc35 undergo a cell cycle arrest that is similar to the arrest caused by nutrient deprivation, suggesting a defect in nutrient sensing (Werner-Washburne et al., 1993). Studies on these and other genes have demonstrated the existence of a cAMP-dependent signaling cascade used to link cell cycle progression to nutrient availability (Hubler et al., 1993; Baroni et al., 1994; Tokiwa et al., 1994; Thevelein and de Winde, 1999).
A number of studies have demonstrated that the G1 cyclin Cln3 plays a critical role in controlling entry into the cell cycle. For example, overexpression of Cln3, or mutations that cause stabilization of Cln3, lead to premature entry into the cell cycle, resulting in decreased cell size (Cross, 1988; Nash et al., 1988; Futcher, 1996). Conversely, loss of Cln3 function leads to a delay in cell cycle entry and an increase in cell size (Cross, 1988; Tyers et al., 1993). Cln3 appears to induce G1-specific expression of a large group of genes that includes additional G1 cyclins called Cln1 and Cln2 (Cross and Tinkelenberg, 1991; Tyers et al., 1993; Dirick et al., 1995; Stuart and Wittenberg, 1995). In the absence of Cln3, transcription of these G1-specific genes is delayed and occurs by a pathway that requires the function of the Bck2 protein (Di Como et al., 1995; Wijnen and Futcher, 1999). Transcription of G1-specific genes is regulated by two transcription factor complexes called SBF and MBF, composed of Swi4 and Swi6, or Mbp1 and Swi6, respectively (Nasmyth and Dirick, 1991; Ogas et al., 1991; Partridge et al., 1997). The pathways used to activate the transcription of SBF/MBF-dependent genes are poorly understood, as are the pathways that regulate the activity of Cln3 and Bck2.
Although a number of key regulators of G1 events have been identified, we still do not understand the molecular mechanisms that integrate cell size and external signals with entry into the cell cycle. A more complete understanding of the molecular mechanisms that control G1 events will require identification of additional proteins that are necessary for entry into the cell cycle. In this study, we have used a biochemical approach to identify a protein called Sda1 that is required for passage through Start in budding yeast. Because Sda1 is a highly conserved protein it may function to control G1 events in all eukaryotic cells.
MATERIALS AND METHODS
Yeast Strains
All yeast strains are in the W303 strain background (leu2-3,112 ura3-52 can1-100 ade2-1 his3-11 trp1-1), with the exception of the strain used for the two-hybrid screen (YD116: MATa ade2-101oc can1 gal4-542 gal80-538 his3-Δ200 leu2-3,112 lys2-801am trp1-901 ura3-52) (Shulewitz et al., 1999).
DK96: MATα,Δbar1, Δnap1::LEU2
DK186: MATa, Δbar1
DK209: MATa/MATα
ZZ6: MATa/MATα, SDA1/Δsda1::HIS3
ZZ13: MATa, Δsda1::HIS3, pZZ13 (CEN, SDA1, URA3)
ZZ28: MATa, Δbar1, sda1-2
ZZ35: MATa, Δbar1, sda1-2, Δnap1::LEU2
ZZ41: MATa, Δbar1, CLN2–3XHA::LEU2
ZZ42: MATa, Δbar1, sda1-2, CLN2–3XHA::LEU2
ZZ60: MATa, Δbar1, sda1-2, GAL1–3X HA-CLN2::HIS5
ZZ66: MATa, Δbar1, sda1-2, GAL1–3X HA-CLN3::HIS5
ZZ99: MATa, Δbar1, sda1-2, CLN2–3XHA::LEU2, pBA263V (CEN, GPD,TRP1)
ZZ100: MATa, Δbar1, sda1-2, CLN2–3XHA::LEU2, pBA1272 (CEN, GPD-SWI4, TRP1)
Biochemical Analysis
Nap1 affinity chromatography and identification of Nap1-binding proteins were performed as previously described (Altman and Kellogg, 1997). Coimmunoprecipitation experiments were performed as previously described (Altman and Kellogg, 1997). Briefly, immunoaffinity beads were made by binding 1 μg of affinity-purified antibody per microliter of protein A beads. A 25-ml culture of rapidly dividing DK186 (O.D600 of 0.6) was pelleted and immediately frozen on liquid nitrogen. The cells were broken open by bead beating for two 40-s intervals, separated by a 60-s incubation on ice, in the presence of 300 μl of acid-washed glass beads and 600 μl of IP buffer (50 mM K+-HEPES pH 7.6, 150 mM NaCl, 1 mM EGTA, 1 mM MgCl2, 0.1% Tween-20, 2 μg/ml leupeptin, 2 μg/ml pepstatin, 2 μg/ml chymostatin, 2 mM phenylmethylsulfonyl fluoride) at 4°C followed by centrifugation at 14,000 rpm for 10 min at 4°C. Supernatant (500 μl) was added to 15 μl of antibody-bound beads and rotated for 4 h at 4°C. The beads were washed four times in IP buffer containing 10% glycerol. Bound proteins were eluted by incubating the washed beads with 200 μl of IP buffer containing 1.0 M NaCl for 10 min at 20°C followed by a 15-s spin and removal of 150 μl of the supernatant. The elution step was repeated and the pooled elutions were trichloroacetic acid (TCA) precipitated, resuspended in 1× protein loading buffer (65 mM Tris-HCl, pH 6.8, 3% SDS, 5% β-mercaptoethanol, 10% glycerol), and separated on a SDS-polyacrylamide gel for Western blotting.
Yeast Two-Hybrid Analysis
Yeast two-hybrid analysis was performed as described (Shulewitz et al., 1999). Briefly, a Gal4 DNA-binding domain (Gal4-DBD) in-frame fusion to the SDA1 gene was generated by polymerase chain reaction (PCR) amplifying SDA1 by using the oligos GCGCGCGCGCATATGATGGGTAGAAGAAGTAGAGC (NdeI site underlined and the Met codon in bold) and GCGGGATCCCTAATACCCCTTCTTCTTTT (BamHI site underlined and the stop codon in bold). The PCR product was ligated into the NdeI and BamHI sites of pAS1 (2μ, TRP1) (Clonetech, Palo Alto, CA), yielding pAS1-SDA1 in which the Gal4-DBD-SDA1 fusion is expressed from the constitutive ADH1 promoter. Next, the yeast strain YD116 containing pAS1-SDA1 was transformed with a library of yeast cDNAs fused to the carboxyl terminus of the Gal4 transcriptional activation domain (Gal4-AD) driven from the ADH1 promoter on a LEU2-marked 2μ plasmid (purchased from the American Type Culture Collection, Vanassas, VA). Transformants were plated on -Trp-Leu-Ura+XGal media to select for cells that possessed the TRP1-marked pAS1-SDA1 and a LEU2-marked library plasmid, and that stimulated the expression of the URA3 and lacZ reporter genes. Candidate plasmids were recovered from yeast, amplified through Escherichia coli, and retested for activation of the URA3 and lacZ reporter genes by using the strain YD116 containing pAS1-SDA1. Specificity was confirmed by failure of the candidates to stimulate reporter expression in the absence of pAS1-SDA1. Partial nucleotide sequence of the candidate genes was obtained and then identified by comparison to the Saccharomyces cerevisiae Genome Database (Stanford University). We repeatedly identified NAP1 in this screen.
Generation of Temperature-sensitive sda1 Alleles
A deletion of the SDA1 gene was generated by a one-step gene disruption technique described previously (Guthrie and Fink, 1991). Briefly, the HIS3 gene was amplified from pRS423 by PCR with oligos possessing homology to regions 40 bp upstream of the SDA1 Start codon and 40 bp downstream of the SDA1 stop codon (oligos: AAGACGCACTGAATATACCAATCAAGGGCATCAACAATGGG -TAGAAGAAGCGGCATCAGAGCAGATTG and GTGTATATGT-ATGTATATATTAATATGGCTACGATGTCGTCTAATACCCCG -TGCGGTATTTCACACCG). The PCR product was transformed into the diploid strain DK209 and a transformant carrying a single SDA1 deletion was identified by selection on -His media and PCR analysis to create strain ZZ6.
To generate temperature sensitive sda1 alleles, a haploid strain dependent upon SDA1 carried on a CEN plasmid marked with the URA3 gene was generated. A DNA fragment containing the SDA1 open reading frame and 1000 bp upstream from the Start codon and 1000 bp downstream of the stop codon was amplified by PCR and cloned into the SacI and BamHI sites of YCplac33 (URA3) (oligos: GCGGAGCTCGAAAGAGAACGAATCTGGGC and GCGGGATCCGCATCTGGGGTTCTCTCAGC; SacI and BamHI sites underlined) to create pZZ13. This DNA fragment was also cloned into the SacI and BamHI sites of YCplac22 (TRP1) to create pZZ15. We then transformed the diploid yeast strain ZZ6 with pZZ13, induced sporulation, and selected a haploid that contained both the deletion of SDA1 and pZZ13 to generate strain ZZ13.
To mutagenize SDA1, the SDA1 coding sequence from nucleotides +190 to +1891 was amplified by PCR under mutagenic conditions as described (Muhlrad et al., 1992) (oligos: CGATGTAGGAAATGGATCTT and GCAATTTCACGGAAGGCAGC). A gapped-plasmid was created by digesting pZZ15 with AflII (cuts at +392) and BstEII (cuts at +1724). Next, the yeast strain ZZ13 was cotransformed with the mutagenized SDA1 PCR product and the gapped-pZZ15 plasmid. Transformants were selected on -Trp media, and cells that had lost the wild-type copy of SDA1 were selected by replica plating onto plates containing 5-fluoro-orotic acid. Temperature sensitive mutants were identified by screening for colonies that grew at 20°C but not at 37°C. To confirm that the mutagenized sda1-containing plasmids were responsible for the temperature-sensitive phenotype, the plasmids were recovered from yeast, amplified through E. coli, and then transformed back into the yeast strain ZZ13 for plasmid-shuffling and rescreening for temperature sensitivity (Guthrie and Fink, 1991).
The 10 temperature-sensitive sda1 mutant alleles were integrated into the genome at the SDA1 locus by a two-step gene replacement technique (Guthrie and Fink, 1991). Briefly, mutagenized sda1 was excised from the YCplac22 vector by using BamHI and SacI, and then cloned into the BamHI and SacI sites of the integrating vector YIplac211 (URA3). This construct was linearized with BglII to direct the integration of the sda1 temperature-sensitive allele to the chromosomal SDA1 locus, and was transformed into DK186, resulting in strains possessing a wild-type copy of SDA1 and a temperature-sensitive copy of sda1. The transformants were selected on −Ura plates and then streaked onto YPD to allow for recombination within the SDA1 coding regions, resulting in the looping out of a copy of SDA1 and the URA3 gene. Colonies that had lost the URA3 gene were selected on 5-fluoro-orotic acid-containing media, and then screened for temperature sensitivity to identify cells carrying the sda1 temperature-sensitive mutant allele.
Cell Cycle Arrests, Viability Assays, and Observation of Individual Cells
Cells were arrested in G0 by growth at 20°C for 6 d or at 30°C for 4 d in either liquid or solid YPD media. Strains were arrested in G1 with 1 μg/ml α factor for 3 h at 30°C or in G2/M with 30 μg/ml benomyl for 3 h at 30°C.
Viability assays were performed by plating 1 × 103 cells on 37°C prewarmed plates followed by incubation at the restrictive temperature (37°C) for varying amounts of time. At each time point, the plate was shifted from the restrictive to the permissive temperature (20°C). Colonies were counted after 4 d of growth at the permissive temperature and percentage of viability was determined by comparison with the number of colonies on a control plate incubated at the permissive temperature.
Individual cells were observed by plating onto thin solid media. A section of the media was excised and placed onto a glass slide, and then incubated at the permissive or restrictive temperature. At each time point the slide was quickly viewed under the microscope and the identical field of view was observed each time.
Antibody Production and Western Blotting
Antibodies that recognize Sda1 were raised by immunizing rabbits with a COOH-terminal fragment of Sda1, purified from bacteria as a glutathione S-transferase (GST) fusion protein. The COOH-terminal fragment was amplified by PCR and cloned into the BamHI and EcoRI sites of pGEX-1 to create pZZ8, which expresses the COOH-terminal fragment as a GST fusion (Amersham Pharmacia Biotech, Piscataway, NJ; oligos: GCGGGATCCGGAACTAAGCGATGACG and GCGGAATTCCTAATACCCCTTCTTCTTTTG; BamHI and EcoRI sites underlined and the stop codon in bold). An identical fragment of Sda1 was cloned into the BamHI and EcoRI sites of pMAL-c2 to create pZZ6, which expresses the COOH-terminal fragment as an MBP fusion (New England Biolabs, Beverly, MA). Sda1 antibodies were affinity purified from serum by using the purified Sda1-MBP fusion protein coupled to Affi-gel 10 (Bio-Rad Laboratories, Hercules, CA) as previously described (Kellogg and Alberts, 1992).
SDS-PAGE and Western blotting were carried out as previously described (Anderson et al., 1973; Harlow and Lane, 1988). For all Western blots, 1.6-ml samples of culture at O.D600 of 0.6 were taken at each of the indicated time points. The cells were then rapidly pelleted in a 1.8-ml screw-cap tube, the supernatant was removed, and the tube was frozen on liquid nitrogen. After all of the samples were collected, 150 μl of glass beads was added to each tube followed by 125 μl of 1× protein gel sample buffer containing 2 mM phenylmethylsulfonyl fluoride. The tubes were immediately placed in a Biospec Multibeater-8 and beaten at top speed for 2 min, centrifuged briefly, and immediately boiled in a heat block for 5 min. After centrifugation for 1 min, 15 μl of each sample was loaded onto a 10% SDS-polyacrylamide gel for Western blotting.
Immunofluorescence and Northern Blots
Actin staining was performed by fixing cells at room temperature in 37% formaldehyde at a final concentration of 3.7% for 1 h, washing twice in phosphate-buffered saline (PBS), and then rotating for 30 min in PBS containing 0.5% Tween. Afterward, the cells were washed twice in PBS and rotated in PBS containing rhodamine-phalloidin at a concentration of 0.3 μM in the dark for 1.5 h. After staining, the cells were washed three times in PBS and then resuspended in PBS containing one-tenth volume mounting solution (1 mg/ml phenylene-diamine, 90% glycerol, 0.1 M Tris-HCl, pH 8.8) before viewing by fluorescence microscopy.
Staining of mitotic spindles and DNA was carried out as previously described (Pringle et al., 1991). Northern blots were carried out as previously described (Kellogg and Murray, 1995).
Generation of HA-tagged Cln2 and Galactose-inducible Cln2 and Cln3
Galactose-driven 3XHA-tagged CLN2 and 3XHA-tagged CLN3 strains were generated by one-step replacement of the endogenous promoter with the GAL1 promoter followed by a triple HA tag fused in frame with either CLN2 or CLN3 coding regions as described (Longtine et al., 1998) (oligos for CLN2: ACTCTATAGCTGCCAATTCATTCGCTTACCACA-TCATAATGAATTCGAGCTCGTTTAAAC and TGATGACGAGTCC-CATACGGGGTCTTGGTTCAGCACTAGCGCACTGAGCAGCGTAA-TCTG; oligos for CLN3: CTCCTCTGCATTTCTTTTCTGACCC-ATAGCATTTCTTACAGAATTCGAGCTCGTTTAAAC and TTGCATTAGCGTATCTAATTATGGTATCCTTCAATATGGCGCACT-GAGCAGCGTAATCTG). For the experiment shown in Figure 9, strains were grown to saturation in YEP media containing 2% glycerol and 2% ethanol and were released from the nutrient arrest into YEP media containing 2% galactose. CLN2 at the endogenous locus was tagged with 3XHA at the COOH terminus by using pCLN2HA3 (gift of R. Deshaies, California Institute of Technology).
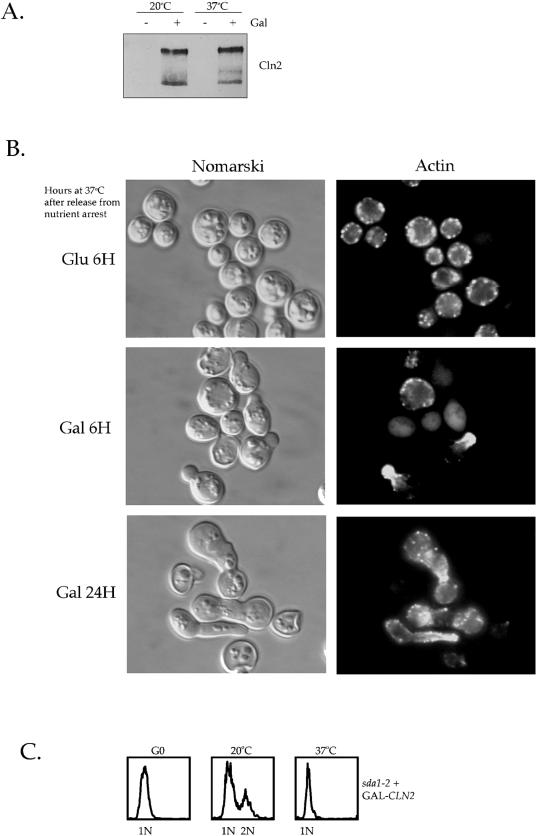
Figure 9.
Expression of CLN2 from a heterologous promoter induces bud formation and actin polarization in sda1-2 cells at the restrictive temperature. (A) G0-arrested sda1-2 cells containing GAL1–3XHA-CLN2 were released into fresh media containing glucose or galactose at the permissive or the restrictive temperatures for 8 h and levels of 3XHA-Cln2 were measured by Western blotting with anti-HA antibodies. (B) G0-arrested sda1-2 cells containing GAL1–3XHA-CLN2 were released into fresh media containing glucose or galactose at the restrictive temperature. Pictured are Nomarski images and actin staining in cells incubated for 6 h in glucose-containing media or for 6 and 24 h in galactose-containing media. (C) DNA content measured by fluorescence-activated cell sorter analysis of sda1-2 cells containing GAL1–3XHA-CLN2 in G0 or released into fresh media containing galactose at the permissive or restrictive temperatures for 8 h.
Flow Cytometry
Cells were prepared for flow cytometry by fixing in 70% ethanol for 30 min to overnight at room temperature. Cells were then washed twice in 50 mM Tris-HCl, pH 7.5, resuspended in 500 μl of 50 mM Tris-HCl, pH 7.5, 10 μg/ml DNase-free RNase, and incubated at 37°C for 1–4 h. The cells were pelleted, resuspended in 500 μl of 200 mM Tris-HCl, pH 7.5, 200 mM NaCL, 78 mM MgCl2, and sonicated for 10 s on “low.” After sonication, 1.5 μl of 10 mg/ml propidium iodide was added and the cells were stored overnight at 4°C or analyzed immediately.
RESULTS
Sda1 Interacts with Nap1
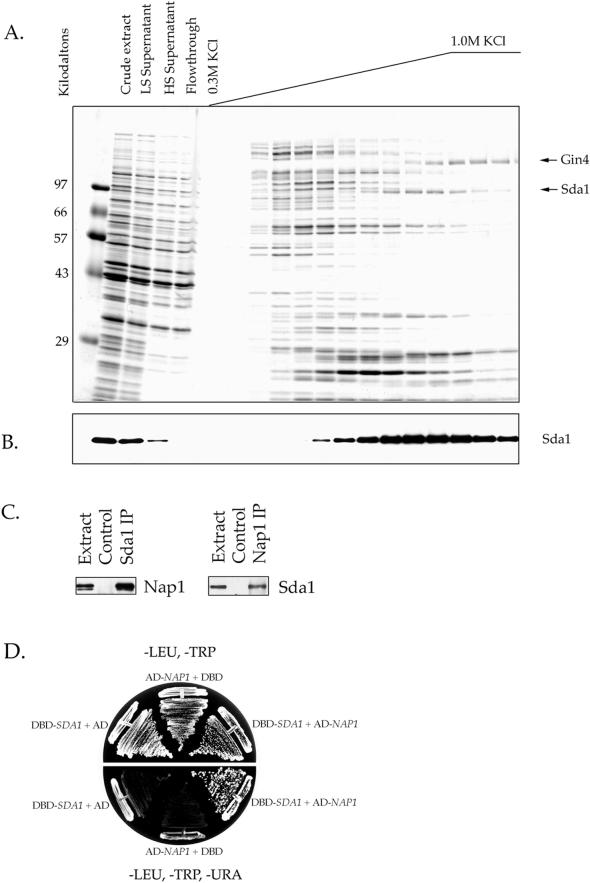
In previous work we found that Nap1 binds to Gin4 and Clb2, two proteins involved in the control of mitotic events (Kellogg et al., 1995; Altman and Kellogg, 1997). To learn more about the in vivo functions of Nap1, we have used affinity chromatography to identify additional Nap1-interacting proteins. For these experiments, a purified full-length Nap1 fusion protein was covalently coupled to a column matrix to create an affinity column (Altman and Kellogg, 1997). The column was loaded with a crude extract made from log phase yeast cells, washed extensively with buffer, and eluted with a gradient of increasing salt. The eluted proteins were then analyzed by SDS-PAGE (Figure 1A). The Gin4 protein kinase is one of the proteins that binds to the Nap1 affinity column (Figure 1A, arrow), and a variety of experiments have demonstrated that Nap1 and Gin4 function together in vivo (Altman and Kellogg, 1997). In this study, we have analyzed a 98-kDa protein that elutes from the Nap1 affinity column at ∼0.7 M KCl (Figure 1A, arrow). Analysis of this protein by mass spectrometry identified it as the open reading frame YGR245c, which has recently been given the name Sda1 (Buscemi et al., 2000). Western blot experiments with an anti-Sda1 antibody confirmed that Sda1 binds quantitatively to the Nap1 affinity column and elutes only under high salt conditions (Figure 1B). We also found that Nap1 binds to a Sda1-GST affinity column (our unpublished data), and that Sda1 and Nap1 coprecipitate (Figure 1C). Finally, we carried out a two-hybrid screen to find proteins that interact with Sda1 and repeatedly identified Nap1 (Figure 1D). Taken together, these results provide strong evidence that Sda1 interacts with Nap1 in vivo.
Figure 1.
Sda1 interacts with Nap1 in vitro and in vivo. (A) Affinity purification of Nap1-binding proteins. Crude extracts from rapidly growing yeast cells were loaded onto a Nap1 affinity column. After washing with buffer, the column was eluted with a KCl gradient. The samples from each fraction were TCA precipitated and loaded onto a 10% SDS-polyacrylamide gel. The gel is stained with Coomassie blue. The lane marked LS is a sample of the low-speed spin supernatant, whereas HS is the high speed supernatant. (B) The same fractions shown in A were probed by Western blotting with affinity-purified anti-Sda1 polyclonal antibody. The amount loaded onto the 10% SDS-polyacrylamide gel used for the Western blot in B was one-tenth the amount loaded in A. Note that the Sda1 protein is quantitatively depleted from the extract by the Nap1 affinity column. (C) Crude cell extracts were made from wild-type cells and used for Sda1 or Nap1 immunoprecipitations. The immunoprecipitations were washed in buffer containing 0.15 M NaCl. The Sda1 immunoprecipitation was probed with anti-Nap1 antibody to detect coprecipitated Nap1. The Nap1 immunoprecipitation was probed with anti-Sda1 antibody to detect coprecipitated Sda1. (D) Yeast two-hybrid analysis by using Sda1 cloned into the DBD vector pAS1 (marked with TRP1) as bait and Nap1 cloned into the transcriptional activating domain (AD) vector pACT (marked with LEU2) as prey. The interaction of Sda1 and Nap1 turns on the reporter genes URA3 and lacZ, which allows for growth on -URA media and the formation blue colonies on XGal media.
Sda1 Is Required for Passage through G1 from a G0 Arrest
Previous work has demonstrated that SDA1 is an essential gene (Buscemi et al., 2000). As a first step toward analyzing the function of Sda1 we generated a diploid strain that is heterozygous for a deletion of the SDA1 gene. Sporulation of this diploid yielded two viable spores and two inviable spores, as expected. Interestingly, examination of the inviable Δsda1 spores revealed that in every case they remained as single unbudded cells that failed to increase in size, suggesting that Sda1 has an essential role in entry into the cell cycle.
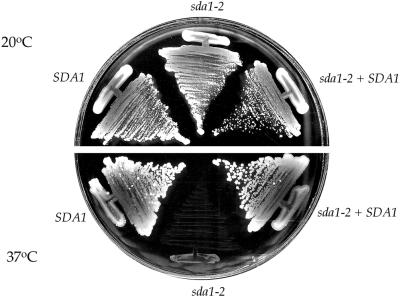
To further analyze the function of Sda1, we generated temperature-sensitive mutants by using error-prone PCR mutagenesis and plasmid shuffling techniques (see MATERIALS AND METHODS). We identified 10 temperature-sensitive sda1 mutant alleles that grow normally at the permissive temperature (20°C) but completely fail to grow at the restrictive temperature (37°C). The temperature-sensitive sda1 alleles were integrated at the SDA1 chromosomal locus, replacing the wild-type SDA1 gene. All of the temperature-sensitive sda1 alleles are recessive and can be rescued by wild-type SDA1 supplied on a low copy CEN plasmid or in a heterozygous diploid (Figure 2; our unpublished data). For the studies described below we show the results for the sda1-2 allele, although we have tested multiple alleles in each experiment and have obtained identical results.
Figure 2.
Isolation of temperature-sensitive sda1 mutants. The sda1–2 mutant forms colonies at the same rate as wild-type cells at the permissive temperature (20°C), but fails to grow at the restrictive temperature (37°C). The sda1-2 mutant can be rescued by SDA1 on a CEN plasmid.
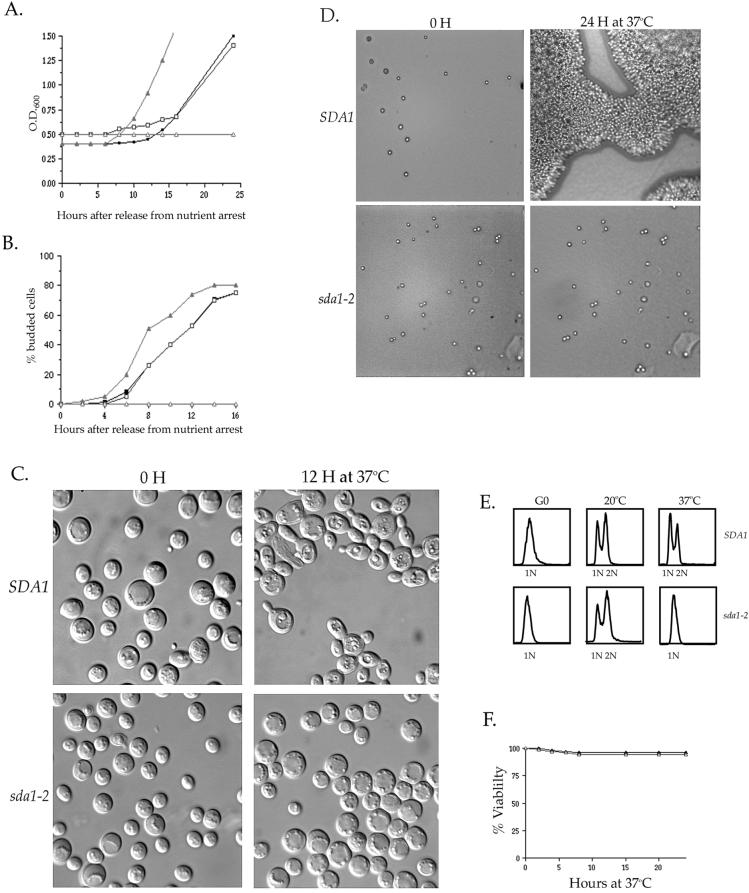
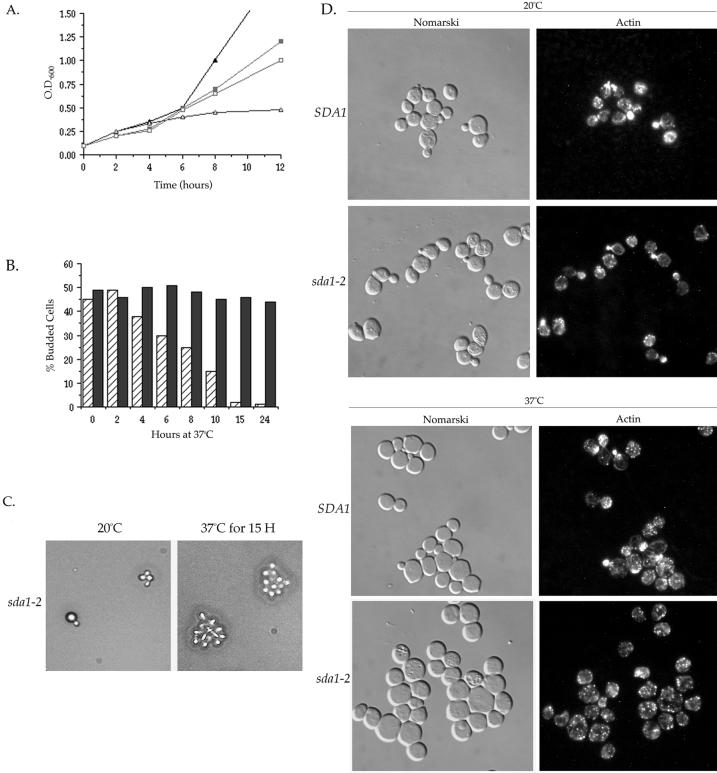
We used the temperature-sensitive sda1 alleles to determine the phenotype caused by inactivation of Sda1 function by two different approaches. Because haploid spores carrying a deletion of the SDA1 gene fail to enter the cell cycle, we hypothesized that Sda1 is required for entry into the cell cycle. Therefore, we first tested whether temperature-sensitive sda1 cells released from a G0 arrest are able to reenter the cell cycle at the restrictive temperature. We arrested wild-type and sda1-2 cells in G0 by nutrient depletion and then transferred them to fresh media at either the permissive or restrictive temperature. Cell proliferation was monitored by measuring the O.D.600 of the cultures over a period of 24 h (Figure 3A). We found that the sda1-2 cells proliferated at the permissive temperature in a manner nearly identical to the wild-type control strain. However, at the restrictive temperature the sda1-2 cells completely failed to proliferate. Bud emergence was also monitored during this experiment by determining the percentage of cells with buds at each time point (Figure 3B). At the permissive temperature, both wild-type and sda1-2 cells underwent bud emergence after 6 h. At the restrictive temperature, wild-type cells underwent bud emergence after 4 h; however, the sda1-2 cells completely failed to undergo bud emergence and >98% remained as unbudded cells (Figures 3, B and C). Interestingly, the sda1-2 cells do not significantly increase in size at the restrictive temperature. This result distinguishes sda1 mutants from cells lacking the function of Cdc28 or the G1 cyclins, which fail to pass through Start but continue to increase in size (Reed, 1980; Hadwiger et al., 1989; Richardson et al., 1989). To further confirm that the sda1-2 cells fail to reenter the cell cycle from a G0 arrest, we followed the behavior of individual nutrient-arrested cells plated onto fresh media at the restrictive temperature. Again, we found that >98% of the sda1-2 cells failed to initiate budding at the restrictive temperature (Figure 3D). Flow cytometry analysis demonstrated that sda1-2 cells do not undergo DNA replication when released from a G0 arrest into fresh media at the restrictive temperature (Figure 3E). The sda1-2 cells remain nearly 100% viable during the 24-h incubation at the restrictive temperature, as determined by a cell viability assay (Figure 3F).
Figure 3.
Sda1 is required for entry into the cell cycle from a G0 arrest. (A) Wild-type and sda1-2 cells were arrested in G0 by nutrient depletion and then released into fresh media at the permissive temperature (20°C) or the restrictive temperature (37°C). Cell proliferation was monitored by measuring the O.D600 of the culture over a period of 24 h. At the permissive temperature, wild-type (▪) and sda1-2 cells (□) proliferate at the same rates. At the restrictive temperature, wild-type cells (▴) proliferate, but the sda1-2 cells (▵) fail to proliferate. (B) The percentage of cells with buds was counted during an identical time course. At the permissive temperature, wild-type (▪) and sda1-2 cells (□) undergo budding at the same rates. At the restrictive temperature, wild-type cells (▴) bud, however the sda1-2 cells (▵) fail to bud. (C) Nomarski images of the G0-arrested wild-type and sda1-2 cells released into fresh media at the restrictive temperature. Pictured are the 0- and 12-h time points. (D) G0-arrested wild-type and sda1-2 cells were plated onto solid media at the restrictive temperature and the same field of view was observed under a microscope over a period of 24 h. Pictured are the 0- and 24-h time points. (E) DNA content measured by fluorescence-activated cell sorter analysis of wild-type and sda1-2 cells at G0 and after 8 h in fresh media at 20 and 37°C. (F) Measurement of the percentage of G0-arrested wild-type (▴) and sda1-2 cells (▵) that remain viable when released onto fresh YPD plates at the restrictive temperature. Percentage of viability was determined by dividing the number of colonies formed at each time point by the number of colonies at time 0 h.
The unbudded sda1-2 arrest phenotype suggested a possible defect in the actin cytoskeleton, because actin is required for bud emergence and bud growth (Pruyne and Bretscher, 2000). In addition, recent work has suggested that Sda1 is required for assembly or maintenance of cortical actin patches (Buscemi et al., 2000). Therefore, we used rhodamine-phalloidin to observe actin organization in the sda1-2 cells released from a G0 arrest at the restrictive temperature. Actin organization cannot be viewed in G0-arrested cells due to the presence of a thick cell wall that is resistant to enzymatic degradation and impermeable to phalloidin. Cells that have exited G0 reorganize their cell wall and become permeable to phalloidin. We could therefore use phalloidin staining to monitor both exit from G0 and the structure of the actin cytoskeleton. G0-arrested wild-type and sda1-2 cells were released into fresh media at the restrictive temperature and actin structures were observed over a period of 24 h. We found that wild-type cells became permeable to phalloidin 2 h before the sda1-2 cells; however, by 15 h 85% of both the wild-type and sda1-2 cells showed actin staining, indicating that both strains exited G0 (Figure 4A). The actin cytoskeleton in sda1-2 cells at the restrictive temperature consisted of randomly distributed actin patches and cables, similar to a cell in G1 before bud emergence. The actin structures in the sda1-2 cells remained the same throughout the 24 h time course.
Figure 4.
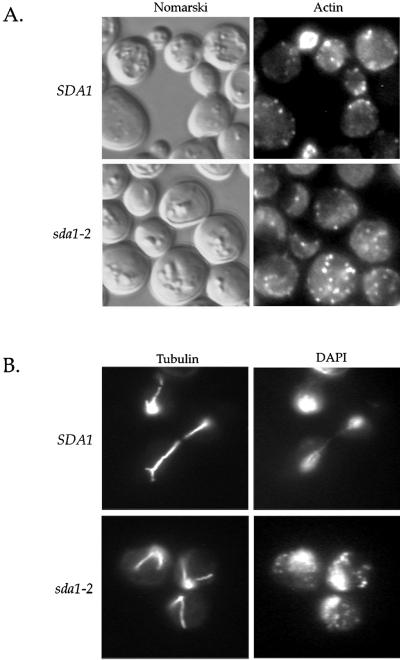
sda1-2 mutants arrest with a normal cortical actin cytoskeleton and a G1 microtubule array. Wild-type and sda1-2 cells were released from a G0 arrest into fresh media at the restrictive temperature for 12 h. (A) Nomarski images and actin staining by using rhodamine-phalloidin. (B) DNA and microtubule staining by using 4,6-diamidino-2-phenylindole and fluorescein isothiocyanate-labeled anti-tubulin antibodies.
We also performed DNA and tubulin staining on the wild-type and sda1-2 cells incubated at the restrictive temperature to observe nuclear morphology and microtubule organization (Figure 4B). After 15 h at the restrictive temperature the wild-type cells were in all stages of the cell cycle, whereas 95% of the sda1-2 cells arrested with a single nucleus and an interphase array of microtubules, indicative of a G1 arrest.
Rapidly Growing sda1-2 Mutant Cells Arrest in G1 at the Restrictive Temperature
Our results demonstrate that Sda1 is required for G0-arrested cells to reenter the cell cycle. Next, we wished to determine the phenotype caused by inactivation of Sda1 in rapidly dividing cells shifted from the permissive to the restrictive temperature. We found that all 10 of our temperature-sensitive sda1 alleles fail to undergo a rapid arrest when shifted to 37°C, as previously reported for an independently isolated sda1 temperature-sensitive allele (Buscemi et al., 2000). Upon the shift to the restrictive temperature, the sda1-2 cells continued to proliferate at a rate similar to wild-type for 6 h (Figure 5A). During this period of proliferation, the sda1-2 culture contained cells in all stages of the cell cycle, as indicated by the presence of cells with differently sized daughter buds. However, after 6 h at the restrictive temperature the sda1-2 cells began to proliferate more slowly until they arrested at ∼12 h (Figure 5A). The sda1-2 culture started to accumulate unbudded cells after 4 h and by 12 h the culture uniformly arrested as unbudded cells (Figure 5B).
Figure 5.
Rapidly growing sda1-2 cells arrest in G1 when shifted to the restrictive temperature. Wild-type and sda1-2 cells were grown to log phase and then either maintained at the permissive temperature (20°C) or shifted to the restrictive temperature (37°C). (A) Cell proliferation was monitored by measuring the O.D.600 of the culture over a period of 12 h. At the permissive temperature, wild-type (▪) and sda1-2 cells (□) proliferate at nearly the same rates. At the restrictive temperature, sda1-2 cells (▵) proliferate at the same rate as wild-type (▴) for the first 6 h, but then gradually slow down and cease growing at ∼10 h. (B) Wild-type (solid bars) and sda1–2 cells (hatched bars) were grown to log phase at the permissive temperature, shifted to the restrictive temperature, and the percentage of cells with buds was monitored over a period of 24 h. (C) Rapidly growing sda1-2 cells were plated onto solid media at the restrictive temperature (37°C) for 15 h to determine the number of cell divisions that sda1-2 cells undergo before arresting. (D) Nomarski images and actin staining of wild-type and sda1-2 cells grown to log phase at the permissive temperature, or 15 h after shifting rapidly growing cells from the permissive temperature to the restrictive temperature.
Because the sda1-2 arrest occurred over an extended period of time, we wished to determine the number of cell divisions the sda1-2 mutant underwent before arresting at the restrictive temperature. Rapidly growing sda1-2 cells were plated onto prewarmed media at the restrictive temperature and individual cells were observed over a 24-h period. We found that the sda1-2 cells underwent an average of three to four rounds of cell division at the restrictive temperature before arresting as a cluster of 8 to 16 cells after 15 h (Figure 5C). Use of a microneedle to disrupt the cluster of arrested sda1-2 cells revealed that they arrested as unbudded cells of a uniform size. The rapidly growing sda1-2 mutant cells shifted to the restrictive temperature remained viable for over 24 h, as was found for the previously reported temperature sensitive sda1-1 allele (Buscemi et al., 2000).
We next observed the actin cytoskeleton in rapidly growing sda1-2 shifted to the restrictive temperature. We found that sda1-2 cells possess normal cortical actin patches and cytoplasmic cables throughout the duration of the 24-h period (Figure 5D). Polarized actin was observed in budding sda1-2 cells until 15 h at the restrictive temperature, at which time the population of sda1-2 cells was uniformly arrested as unbudded cells containing actin patches and cables randomly distributed throughout the cell (Figure 5D). Note that a small fraction of both wild-type and sda1-2 cells failed to stain for actin at both the permissive and restrictive temperatures. We also performed DNA and tubulin staining on identical samples and found that the percentage of sda1-2 cells possessing an interphase array of microtubules increased during the incubation at the restrictive temperature until 95% of the cells arrested with an interphase array of microtubules and a single nucleus at the 15 h time point (our unpublished data).
The Sda1 Protein Is Only Present in Proliferating Cells and Is Synthesized before Start
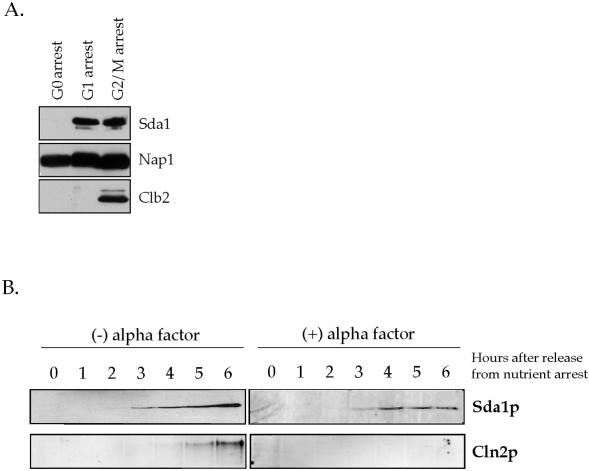
We next studied the behavior of the Sda1 protein during the cell cycle. Cells were arrested at different points in the cell cycle: either in G1 with α factor, in G2/M with the microtubule-destabilizing drug benomyl, or in G0 by nutrient depletion. The levels of the Sda1 protein at each cell cycle stage were then determined using an anti-Sda1 polyclonal antibody (Figure 6A). We also followed the behavior of the Nap1 protein, and as a cell cycle control we followed the mitotic cyclin Clb2. The Sda1 protein was present in G1 and G2/M arrested cells, but completely absent in G0 cells. The Nap1 protein was present during all stages of the cell cycle, whereas the Clb2 protein was only present in G2/M cells, as expected. We also followed the behavior of the Sda1 protein through a complete cell cycle after release from an α factor arrest, and observed no change in protein levels or noticeable posttranslational modifications of the Sda1 protein (our unpublished data).
Figure 6.
Sda1 is absent in G0 cells and appears before Start. (A) Rapidly growing wild-type cells were arrested in G0, G1, or mitosis and probed for the presence of Sda1, Nap1, and Clb2 by Western blotting. (B) G0-arrested wild-type cells containing a HA-tagged version of CLN2 were released into fresh media with or without 1 μg/ml α factor at 30°C and probed for the appearance of Sda1 and Cln2 by Western blotting.
Because Sda1 is present in dividing cells but not in G0-arrested cells, we wished to determine when the Sda1 protein first appears as cells exit G0 and reenter the cell cycle. Cells that enter the cell cycle and pass through Start express the G1 cyclins Cln1 and Cln2 (Wittenberg et al., 1990). Therefore, we determined when the Sda1 protein first appears relative to the appearance of the Cln2 protein by using a wild-type strain carrying a 3XHA-tagged version of the endogenous Cln2. G0-arrested cells were released into fresh media at 30°C and assayed for the appearance of Sda1 and Cln2 (Figure 6B). We found that the Sda1 protein was first detected after 3 h, whereas Cln2 does not appear until 5 h, suggesting that Sda1 is produced before the Start-induced expression of Cln2. To confirm that Sda1 appears before Start, we released G0-arrested cells into fresh media containing the mating pheromone α factor, which arrests cells before passage through Start. We found that Sda1 appeared 3 h after the release into fresh media containing α factor, further confirming that Sda1 is expressed before Start (Figure 6B). The Cln2 protein failed to appear in the presence of α factor, confirming that the cells arrested before passage through Start. We also found that the Sda1 protein appeared with normal kinetics in cdc28-4 cells released from a G0 arrest into fresh media at 37°C (our unpublished data). These experiments indicate that Sda1 is produced before passage through Start, and that Sda1 expression does not require a fully functional Cdc28 protein.
Newly Synthesized Mutant sda1 Protein Fails to Appear at the Restrictive Temperature
Our results demonstrate that cells lacking Sda1 function fail to pass through G1 when released from a G0 arrest. However, when rapidly dividing cells carrying a temperature sensitive allele of sda1 are shifted to the restrictive temperature they continue to divide for >6 h. Our studies on the behavior of the sda1-2 mutant protein provide an explanation for these differing results.
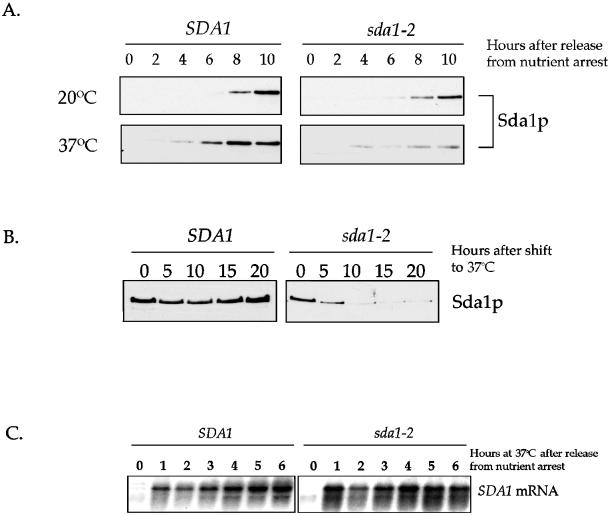
First, we released G0-arrested wild-type and sda1-2 cells into fresh media at the permissive or restrictive temperatures and then assayed Sda1 protein levels every 2 h (Figure 7A). Sda1 appeared after 6 h in both strains at the permissive temperature. At the restrictive temperature, wild-type Sda1 protein appeared after 4 h; however, the mutant sda1-2 protein was barely detectable throughout the time course. These results suggest that at the restrictive temperature newly made mutant sda1-1 protein is either unstable or is not synthesized at normal levels.
Figure 7.
Behavior of the Sda1 protein and mRNA at the restrictive temperature. (A) G0-arrested wild-type and sda1-2 cells were released into fresh media at the permissive (20°C) or restrictive temperature (37°C) and assayed for the appearance of Sda1 protein by Western blotting. (B) Rapidly growing wild-type and sda1-2 cells were shifted from the permissive temperature to the restrictive temperature and samples were taken every 5 h to assay for the presence of the Sda1 protein by Western blotting. At each time point, cells were diluted to O.D.600 of 0.6 to normalize total protein in each sample. (C) G0-arrested wild-type and sda1-2 cells were released into fresh media at the restrictive temperature (37°C) and monitored for the appearance of the SDA1 transcript by Northern blotting.
An additional experiment was performed to supply further evidence for this model. Rapidly dividing cells grown at the permissive temperature were shifted to the restrictive temperature and samples were taken every 5 h for 20 h. At each time point, the cultures were diluted to an O.D.600 of 0.6 to normalize the amount of total protein per sample, and the samples were probed for the Sda1 protein by Western blotting (Figure 7B). Wild-type Sda1 protein remained at constant levels; however, the mutant sda1-2 protein declined after 5 h and by 10 h was barely detectable. These results further suggest that newly made sda1-2 protein is either unstable or not synthesized, whereas preexisting sda1 protein made at the permissive temperature is stable but becomes depleted by continued cell division until cells eventually arrest in G1. We found this to be true for all 10 sda1 alleles.
We confirmed that the lack of Sda1 protein in the mutant strain is not due to a transcription defect by monitoring SDA1 message levels by Northern blot analysis (Figure 7C). The SDA1 mRNA appeared 1 h after release into fresh media from a G0 arrest at the restrictive temperature in both sda1-2 and wild-type strains, indicating that SDA1 is transcribed normally and that the basic transcriptional machinery is functional in the sda1-2 mutant.
The sda1–2 Mutant Fails to Produce the G1 Cyclins at the Restrictive Temperature
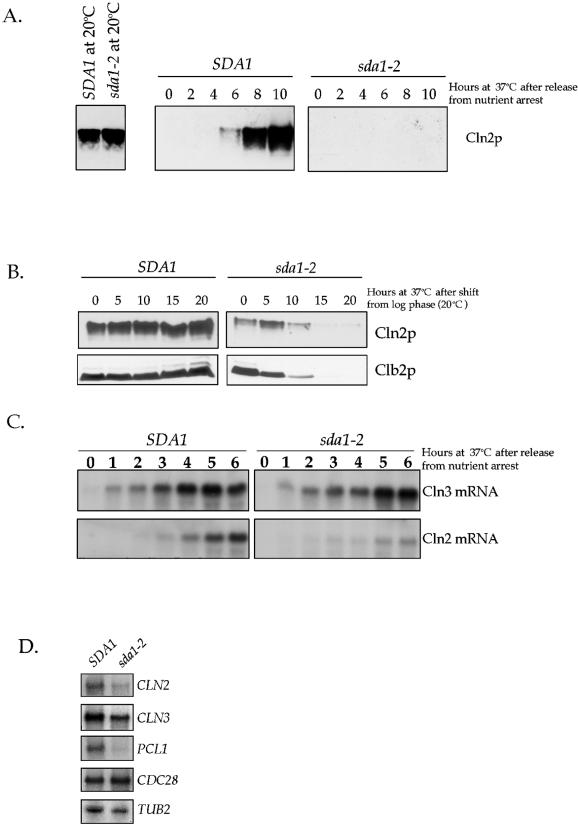
To learn more about the nature of the G1 arrest observed in the sda1-2 cells, we carried out experiments to determine which of the known molecular events that occur during G1 fail to occur in the absence of Sda1 activity. An important event in the passage through G1 is the synthesis of the G1 cyclins Cln1, Cln2, and Cln3. Cln3 is present at low levels during the entire cell cycle, whereas Cln1 and Cln2 appear specifically during G1 (Wittenberg et al., 1990; Tyers et al., 1992). Furthermore, Cln3 activity triggers transcription of CLN1 and CLN2, as well as other G1-specific genes, which leads to passage through Start (Cross and Tinkelenberg, 1991; Tyers et al., 1993; Stuart and Wittenberg, 1995; Spellman et al., 1998). Because sda1-2 cells arrest in G1, we wished to determine whether G1 cyclins are present in the arrested sda1-2 cells. For these experiments we generated a sda1-2 strain that contains a 3XHA-tagged version of the endogenous CLN2. G0-arrested wild-type and sda1-2 strains containing Cln2–3XHA were released into fresh media at the permissive or restrictive temperature and monitored for the appearance of Cln2–3XHA (Figure 8A). At the permissive temperature, Cln2–3XHA was present at the same level in both wild-type and sda1-2 cells. At the restrictive temperature, Cln2–3XHA appeared after 6 h in the wild-type cells, but completely failed to appear in the sda1-2 cells. We also attempted to determine whether the G1 cyclin Cln3 is made normally in sda1-2 cells. Although we were easily able to detect Cln3–3XHA at the permissive temperature, we were unable to consistently detect Cln3–3XHA at the restrictive temperature in either wild-type or sda1-2 cells (our unpublished data).
Figure 8.
Transcription of the G1 cyclin CLN2 is defective in sda1-2 cells, but Cln3 transcription is normal. (A) G0-arrested wild-type and sda1-2 cells containing an HA-tagged version of CLN2 were released into fresh media at the restrictive temperature, and Cln2 protein levels were assayed by Western blotting. The first panel is a control showing that Cln2 protein levels are the same in wild-type and sda1–2 cells grown at the permissive temperature. (B) Rapidly growing wild-type and sda1-2 cells carrying an HA-tagged version of Cln2 were shifted to the restrictive temperature and samples were taken every 5 h to assay for the presence of the Cln2 and Clb2 proteins by Western blotting with anti-HA or anti-Clb2 antibodies. At each time point, cells were diluted to O.D.600 of 0.6 to normalize total protein in each sample. (C) G0-arrested wild-type and sda1-2 cells were released into fresh media at the permissive (20°C) and restrictive (37°C) temperatures and CLN3 and CLN2 transcripts were monitored by Northern blotting. (D) G0-arrested wild-type and sda1-2 cells were released into fresh media at the restrictive temperature and the indicated transcripts were monitored by Northern blotting at 6 h.
To further confirm that temperature-sensitive sda1 cells fail to synthesize Cln2 at the restrictive temperature, we shifted rapidly growing wild-type and sda1-2 strains containing Cln2–3XHA from the permissive temperature to the restrictive temperature and then assayed Cln2–3XHA levels every 5 h for 20 h. As before, the cultures were diluted to an O.D.600 of 0.6 at each time point to normalize total protein levels (Figure 8B). In the wild-type cells, Cln2–3XHA levels remained constant; however, in the sda1-2 cells, Cln2–3XHA levels declined during the time course and after 15 h the cells arrested without detectable Cln2, indicative of a G1 arrest. In addition, the samples were probed for the mitotic cyclin Clb2 (Figure 8B). The levels of Clb2 in the sda1-2 cells decreased steadily during the time course and eventually disappeared, arguing against a delay in mitosis and further supporting a G1 arrest.
Next, we determined whether the defect in G1 cyclin expression is at the transcriptional level by monitoring the appearance of CLN2 and CLN3 mRNAs after sda1-2 cells were released from a G0 arrest at the restrictive temperature (Figure 8C). Wild-type and sda1-2 cells arrested in G0 were released into fresh media at the restrictive temperature, samples taken every hour for 6 h, and then probed for the presence of the CLN2 and CLN3 transcripts by Northern blotting. Interestingly, the CLN3 transcript appeared with the same kinetics in wild-type cells and sda1-2 cells. In contrast, the levels of the CLN2 transcript were greatly reduced in the sda1-2 mutant cells. We also assayed the appearance of the PCL1 transcript, another gene that is transcribed by the same G1-specific transcription factors as CLN1 and CLN2 (Figure 8D). As with CLN2, we found that the PCL1 transcript failed to appear as cells exited G0, whereas the levels of control transcripts (TUB2 and CDC28) were unaffected.
Expression of CLN2 Induces Budding in Arrested sda1-2 Cells
Because sda1-2 cells fail to transcribe CLN2 at the restrictive temperature, we asked whether the expression of CLN2 from a heterologous promoter could rescue the sda1-2 phenotype. For these experiments, a sda1-2 strain was constructed that contained a 3XHA-tagged version of the endogenous CLN2 gene under the control of the GAL1 promoter. We first determined that the Gal1-driven HA-tagged Cln2 protein was being made at equivalent amounts at the permissive and restrictive temperatures in cells released from a G0 arrest (Figure 9A). Next, we tested whether expression of GAL1–3XHA-CLN2 could permit sda1-2 cells to reenter the cell cycle from a G0 arrest at the restrictive temperature. G0-arrested sda1-2 GAL1–3XHA-CLN2 cells were released into fresh media containing either galactose or glucose at the restrictive temperature, and were monitored for bud emergence every 2 h. We found that expression of GAL1–3XHA-CLN2 induced polarization of the actin cytoskeleton and bud emergence in 44% of both wild-type and sda1-2 cells after 6 h (Figure 9B; our unpublished data). After 24 h, the majority of the sda1-2 GAL1–3XHA-CLN2 cells possessed abnormal, elongated buds with hyperpolarized actin. This phenotype was only observed when the sda1-2 GAL1–3XHA-CLN2 strain was grown in galactose-containing media. In addition, we obtained the same result if we arrested rapidly growing sda1-2 GAL1–3XHA-CLN2 cells at the restrictive temperature for 12 or 20 h and then added galactose (our unpublished data). Staining of DNA and microtubules revealed that the sda1-2 GAL1–3XHA-CLN2 cells incubated in galactose-containing media for 24 h at the restrictive temperature each contain a single nucleus localized to the bud neck, but no mitotic spindle (our unpublished data). Expression of GAL1–3XHA-CLN2 in wild-type cells caused a mild elongated bud phenotype, but did not affect cell proliferation.
We also determined whether the sda1-2 GAL1–3XHA-CLN2 cells are able to undergo cell division at the restrictive temperature by observing individual cells plated on galactose-containing solid media at the restrictive temperature. We found that the sda1-2 GAL1–3XHA-CLN2 cells did not undergo cell division and 75% of the cells arrested with large, elongated buds and the other 25% remained as unbudded cells (our unpublished data). In addition, flow cytometry analysis demonstrated that expression of Cln2 does not induce DNA replication in the sda1-2 cells (Figure 9C). We also tested whether GAL1–3XHA-CLN3 could induce sda1-2 cells to reenter the cell cycle. We found that galactose-induced expression of CLN3 in the sda1-2 GAL1-CLN3 cells did not induce bud emergence or polarized actin structures (our unpublished data). Galactose-induced expression of CLN3 in the sda1-2 GAL1-CLN3 cells was confirmed by Northern blotting (our unpublished data). These experiments demonstrate that the sda1-2 mutant can be induced to undergo bud emergence from a G0 arrest by the expression of CLN2, but not CLN3.
Transcription of the G1 cyclins is controlled by the transcription factor SBF, which is comprised of Swi4 and Swi6 (Nasmyth and Dirick, 1991; Ogas et al., 1991). Overexpression of Swi4 has been shown to drive cells through G1 by inducing the expression of the G1 cyclins and other G1 transcripts (Breeden and Mikesell, 1994; Dirick et al., 1995). Therefore, we tested whether the constitutive expression of SWI4 could rescue the sda1 mutant phenotype and induce bud emergence. A plasmid containing SWI4 controlled by the constitutive glycerol-3-phosphate dehydrogenase (GPD) promoter was transformed into the sda1-2 strain. We found that expression of SWI4 did not induce bud emergence or expression of Cln2 when G0-arrested sda1-2 cells were released into fresh media at the restrictive temperature (our unpublished data). At the permissive temperature, GPD-expression of SWI4 resulted in slightly longer buds and an increase in Cln2 protein levels compared with cells containing vector alone, confirming that the plasmid expressed Swi4.
sda1-2 Cells Arrested in G1 Respond to Mating Pheromone
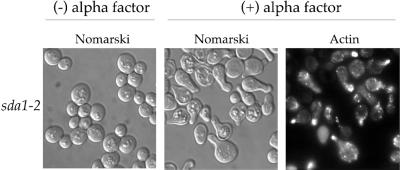
Once cells pass through Start they are unable to respond to mating pheromone until they complete cell division (Reid and Hartwell, 1977). We therefore used mating pheromone sensitivity as a means to determine whether sda1-2 cells arrest before or after Start. We first determined whether sda1-2 cells were able to respond to α factor at the restrictive temperature. To do this, we released G0-arrested sda1-2 cells into fresh media containing α factor at the restrictive temperature. After 6 h, polarization of the actin cytoskeleton and formation of mating projections occurred in 80% of both wild-type and sda1-2 cells (Figure 10; our unpublished data). We also found that polarization of the actin cytoskeleton and formation of mating projections persisted even after 24 h at the restrictive temperature in the presence of α factor (our unpublished data).
Figure 10.
sda1-2 cells are able to respond to α factor and polarize the actin cytoskeleton at the restrictive temperature. G0-arrested sda1-2 cells were released into fresh media with or without α factor at the restrictive temperature (37°C). Pictured are Nomarski images and actin staining taken 6 h after release into fresh media.
To further confirm that the arrested sda1-2 cells respond to α factor and to determine whether the cells arrest before or after Start, we released G0-arrested cells into fresh media at the restrictive temperature for 6 h, added α factor, and then immediately transferred the sda1-2 cells back to the permissive temperature and monitored cell cycle progression. We found that addition of α factor completely blocked cell cycle progression when the sda1-2 cells were transferred to the permissive temperature, indicating the sda1-2 cells arrest before Start. As a control, sda1-2 cells were transferred to the permissive temperature without α factor. These underwent bud emergence within 4 h and proliferated at a normal rate. All of the α factor-related experiments were repeated using rapidly dividing sda1-2 cells arrested in G1 by incubation at the restrictive temperature for 12 h, which yielded identical results. We were unable to perform the reciprocal experiment (e.g., arrest with α factor and release at the restrictive temperature) because cells arrested in G1 with α factor contain normal levels of functional sda1-2 protein, which is not rapidly inactivated when cells are shifted to the restrictive temperature (Figure 7). These results demonstrate that sda1-2 cells arrested in G1 are able to respond to α factor, suggesting that they have not passed through Start.
Sda1 Is Required for Increased Production of rRNA and Proteins in Cells Released from a G0 Arrest
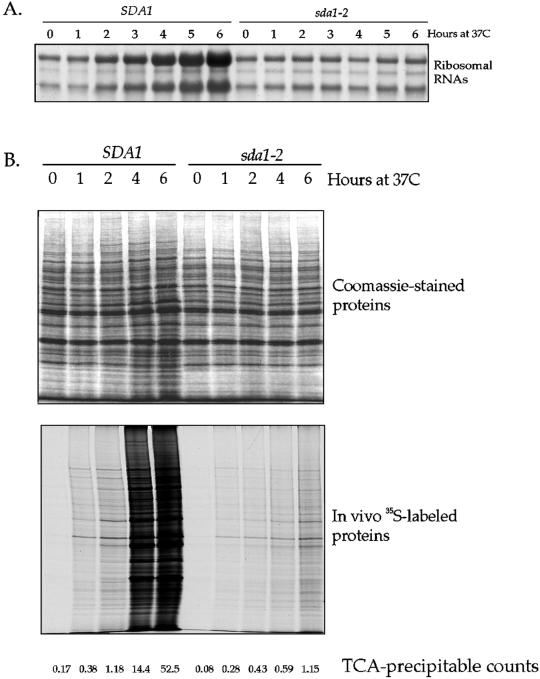
sda1-2 cells fail to increase significantly in size when released from a G0 arrest at the restrictive temperature. Synthesis of rRNAs and proteins are key events that must occur during G1 for cells to increase in size. We therefore assayed these two events in sda1-2 cells released from a G0 arrest at the restrictive temperature. In wild-type control cells, we found that levels of rRNAs began to increase 2 h after release from a G0 arrest, and 2 h before the initiation of bud emergence (Figures 11A and 3B). In contrast, the sda1-2 cells showed no increase in rRNA levels.
Figure 11.
sda1-2 cells released from a G0 arrest at the restrictive temperature fail to induce increased synthesis of rRNA and proteins. (A) Total RNA was isolated from wild-type or sda1-2 cells at the indicated times after release from a G0 arrest at the restrictive temperature. The RNA was then separated by electrophoresis, transferred to a nylon membrane, and stained with methylene blue. (B) The rate and pattern of protein synthesis were measured at each of the indicated time points after release from a G0 arrest by giving cells a 15-min pulse of 35S-labeled amino acids. The synthesized proteins were resolved by gel electrophoresis and were visualized by autoradiography. In addition, total TCA-precipitable counts were measured, and total proteins were visualized by Coomassie blue staining of a polyacrylamide gel.
To assay protein synthesis, we gave cells a 15-min pulse of 35S-labeled methionine and cysteine at 1-h intervals after release from a G0 arrest. Newly translated proteins that incorporated 35S were viewed by autoradiography, and the amount of total protein synthesized was assayed by counting TCA-precipitable protein (Figure 11B). We also separated total proteins by SDS-PAGE and stained with Coomassie blue. We found that neither wild-type or sda1 cells carry out detectable protein synthesis during the first 15 min after release from a G0 arrest. At 1 h, wild-type and sda1-2 cells synthesize approximately equal amounts of protein. Note that there is no Sda1 protein detectable in wild-type cells at this time point, making it unlikely that general protein synthesis requires Sda1. The amount of protein synthesized by wild-type cells increases dramatically as the cells enter the cell cycle and divide. In the sda1-2 cells, however, there is only a slight increase in the amount of protein synthesized, consistent with a slight increase in cell size that we observe. These data suggest that there is a low basal level of protein synthesis and turnover that occurs in cells lacking Sda1 function, even though the cells do not grow significantly or divide. The failure to produce increased levels of proteins in the sda1-2 mutant cells is not due to an increased rate of protein turnover, because we found that protein turnover rates are similar in the sda1-2 cells compared with wild type cells (our unpublished data). These results demonstrate that Sda1 is not required for protein synthesis, but is required for the increased synthesis of proteins and rRNA that accompanies cell growth and entry into the cell cycle.
Deletion of the NAP1 Gene Causes Synthetic Lethality in cln1cln2 Cells
We first identified Sda1 as a protein that interacts with Nap1, and previous work has demonstrated that Nap1 functions in a mitotic signaling network that includes the kinases Gin4 and Cla4 (Altman and Kellogg, 1997; Tjandra et al., 1998; Longtine et al., 2000). Interestingly, both Gin4 and Cla4 have been identified in screens for mutations that cause synthetic lethality in cells that lack the function of the G1 cyclins Cln1 and Cln2, suggesting that they carry out functions during G1 (Benton et al., 1993, 1997; Cvrckova and Nasmyth, 1993; Cvrckova et al., 1995). The finding that Sda1 plays a key role in passage through G1 suggests that Nap1 may also have a role in G1 events. We therefore tested whether loss of Nap1 function results in lethality in Δcln1Δcln2 cells. To do this, we mated Δnap1 cells to Δcln1Δcln2 cells, and then dissected tetrads and searched for spores that carry all three deletions. We dissected over 35 tetrads and were unable to recover the triple deletion, indicating that NAP1 is required for viability in Δcln1Δcln2 cells. We also tested for a genetic interaction between Nap1 and Sda1 by deleting or overexpressing Nap1 in the sda1 mutant background; however, we found that neither of these conditions affected the temperature sensitivity of the sda1 mutants.
DISCUSSION
G1 phase is a critical point in the cell cycle where cells assess conditions and decide whether to initiate a new round of cell division (Pardee, 1989; Murray and Hunt, 1993; Cross, 1995; Planas-Silva and Weinberg, 1997). In budding yeast, cells in G1 will not initiate a new round of cell division until they have reached a critical size and are in the presence sufficient nutrients. Similarly, vertebrate cells in G1 initiate cell division only when they have reached a critical size and have received the appropriate external signals from growth factors. Once these conditions are met, cells pass through a point late in G1 called Start (also referred to as the Restriction point). At the molecular level, passage through Start reflects production of the G1 cyclins, which activate cyclin-dependent kinase activity and thereby initiate the intricate series of events leading to cell division. The molecular mechanisms that operate during G1 to integrate cell size, external cues, and passage through Start are poorly understood. In this study, we demonstrate that the highly conserved Sda1 protein plays a key role in the mechanisms necessary for passage through Start in budding yeast.
Sda1 Is Required for Passage through G1
We have used several different approaches to demonstrate that Sda1 is required for passage through G1. First, we generated haploid spores that carry a complete deletion of the SDA1 gene and found that these fail to undergo budding or an increase in cell size. Second, we found that sda1 temperature-sensitive mutants released from a G0 arrest at the restrictive temperature completely fail to undergo budding, and rapidly growing sda1 temperature-sensitive mutants shifted to the restrictive temperature undergo a uniform G1 arrest. In each case, cells arrested by loss of Sda1 function do not increase significantly in size and contain a single nucleus with a 1N DNA content, an interphase microtubule array, and uniformly distributed cortical actin patches.
It is likely that the G1 arrest phenotype we have observed in temperature-sensitive sda1 mutants reflects a complete loss of Sda1 function, because Western blotting experiments demonstrate that the mutant sda1-2 protein fails to accumulate to significant levels at the restrictive temperature. In addition, all 10 sda1 mutant alleles result in an identical G1 arrest, and spores carrying a complete deletion of the SDA1 gene also produce a G1 arrest. Interestingly, rapidly growing sda1 mutants shifted to the restrictive temperature go through several divisions before arresting in G1. This appears to be due to the fact that preexisting sda1 mutant protein is stable and functional at the restrictive temperature, whereas newly made sda1 protein fails to accumulate, perhaps due to folding defects. Hence, the mutant phenotype is only observed when the preexisting Sda1 protein is depleted by several rounds of cell division. All 10 of our mutant alleles behave this way, suggesting that it is difficult to identify temperature-sensitive mutations that rapidly inactivate Sda1 function. This may explain why Sda1 was not identified in previous genetic screens for mutations that cause a rapid and specific cell cycle arrest. An alternative interpretation is that Sda1 is stringently required only upon emergence from a G0 arrest. To definitively demonstrate that Sda1 is required for passage through Start in every cell cycle, it will be necessary to identify conditional sda1 alleles that cause dividing cells to undergo a rapid G1 arrest.
Sda1 Is Required for Passage through Start
Previous work has demonstrated that production of the G1 cyclins is a key event that drives cells through Start (Hadwiger et al., 1989; Richardson et al., 1989). Budding yeast express three different G1 cyclins, referred to as Cln1, Cln2, and Cln3 (Cross, 1995). These cyclins are redundant, because expression of any one of the three is sufficient to induce passage through Start, although cells expressing only one G1 cyclin show a delay in exiting G1 and an increase in cell size (Hadwiger et al., 1989; Dirick et al., 1995). The Cln3 cyclin is the most distantly related of the three cyclins, and appears to function to induce transcription of Cln1 and Cln2 (Tyers et al., 1993; Dirick et al., 1995).
Loss of Sda1 function causes cells to arrest in G1 with 1N DNA, severely reduced levels of CLN2 mRNA, and undetectable levels of the Cln2 protein, suggesting that the cells arrest before Start. In addition, sda1 mutants arrest at a point where they are able to respond to mating pheromone. Taken together, these results argue strongly that sda1 mutants arrest before Start. Interestingly, sda1-2 cells exiting G0 at the restrictive temperature transcribe CLN3 mRNA normally, suggesting that the cells are able to initiate some of the events leading to passage through Start, but become blocked at a key step that requires Sda1.
Temperature-sensitive sda1 Mutants Are Not Rescued by Overexpression of G1 Cyclins
The finding that temperature-sensitive sda1 mutants fail to produce the G1 cyclin Cln2 suggested the possibility that the primary defect in sda1 mutants may be a failure to transcribe or translate G1 cyclins. To test this possibility, we determined whether expression of CLN2 or CLN3 from a heterologous promoter could rescue the sda1-2 mutant phenotype. Neither cyclin was able to rescue the proliferation defect; however, expression of CLN2 initiated polarization of the actin cytoskeleton and bud emergence. This finding suggests that loss of Sda1 function does not affect general transcription or translation, nor does it affect the ability of Cln2 to bind and activate Cdc28. In addition, these results distinguish the sda1 loss of function phenotype from the phenotype caused by loss of function of cdc28 or the G1 cyclins. Cells arrested in G1 by loss of function of cdc28 or the G1 cyclins arrest as unbudded cells that continue to grow, resulting in the formation of abnormally large cells (Reed, 1980; Cross, 1990). Loss of sda1 function, on the other hand, does not lead to a significant increase in cell size, supporting the idea that the primary defect in sda1 mutants is not in the formation or activation of Cln/Cdc28 complexes.
Sda1 Does Not Appear to Be Required for the Assembly or Function of the Actin Cytoskeleton
It has previously been reported that rapidly growing cells carrying a temperature-sensitive allele of SDA1 show a loss of actin staining after 16 h at the restrictive temperature (Buscemi et al., 2000). It was also reported that release of sda1 cells from an α factor arrest at the restrictive temperature results in 80% of the cells losing actin staining after 4 h. Finally, it was reported that 20% of sda1 cells arrest with large buds, separated nuclei, and interphase microtubule arrays, suggesting defects in cytokinesis.
In contrast, we have found no evidence that Sda1 is required for the assembly or function of the actin cytoskeleton. We found that sda1-2 mutants released from a G0 arrest at the restrictive temperature have normal actin staining, even after 24 h at the restrictive temperature. We also found that addition of α factor to cells arrested in G1 by loss of Sda1 function results in the formation of shmoos with normal actin staining, and expression of Cln2 results in polarization of the actin cytoskeleton and bud emergence. Because both shmoo formation and bud emergence are absolutely dependent upon the proper function of the actin cytoskeleton, these results argue strongly against a requirement for Sda1 in the assembly or function of the actin cytoskeleton. Finally, we found that shifting rapidly growing cells to the restrictive temperature results in >95% of the cells arresting without buds, inconsistent with a cytokinesis defect. It is important to note that in these experiments the cells arrest with barely detectable levels of the Sda1 protein, suggesting that the arrest phenotype reflects a complete loss of function. We tested all 10 of our sda1 alleles and found that all arrest with a normal actin cytoskeleton, arguing against the possibility that there are allele-specific differences in the actin phenotype of sda1 mutants. Finally, we tested whether the previously reported sda1-1 allele shows actin defects when shifted to the restrictive temperature from log phase or from a G0 arrest, and observed no defects, even after 15 h. At this point, we do not understand the discrepancy between our results and those previously reported for the sda1-1 allele.
Sda1 Binds to Nap1 In Vivo and In Vitro
We identified Sda1 as a protein that binds tightly to a Nap1 affinity column. In addition, we used coimmunoprecipitation and two-hybrid analysis to provide further evidence that Nap1 and Sda1 associate. Taken together, these experiments argue strongly that Sda1 and Nap1 interact in vivo. However, we do not yet know the functional significance of this interaction. A combination of genetics and biochemistry has shown that Nap1 plays an important role in a signaling network that functions during mitosis (Altman and Kellogg, 1997; Tjandra et al., 1998; Longtine et al., 2000). For example, Nap1 binds to the Clb2 mitotic cyclin and to the Gin4 kinase, and Nap1 is required in vivo for the Clb2/Cdc28-induced hyperphosphorylation of the Cla4 and Gin4 kinases. In addition, loss of Nap1 function causes prolonged mitotic delays in cells that are dependent upon the Clb2 mitotic cyclin for survival.
Our results argue that it is likely that Nap1 also carries out important functions during G1. In support of this, we found that Nap1 is required for viability in cells that have been made dependent upon CLN3 for survival by deletion of the genes for the redundant G1 cyclins CLN1 and CLN2. It is interesting to note that previous studies have shown that Gin4, Cla4, and members of the septin family are also required for viability in CLN3-dependent cells (Benton et al., 1993; Cvrckova and Nasmyth, 1993; Cvrckova et al., 1995; Benton et al., 1997). At this point we lack the information needed to understand these interesting and unusual genetic interactions. Perhaps Nap1, Cla4, Gin4, and the septins are components of signaling networks that mediate the functions or regulation of both G1 cyclins and mitotic cyclins. Previous work on mitogen-activated protein kinases has shown that the same proteins can function in signaling networks that control different processes (Madhani and Fink, 1998; Tan and Kim, 1999).
Sda1 and the Molecular Mechanisms That Control G1 Events
Why does loss of Sda1 function cause cells to arrest in G1? The arrest does not appear to be due to general defects in transcription, because we see that CLN3 and SDA1 are transcribed normally at the restrictive temperature, as are a number of genes fused to the GAL1 promoter (Figures 7C, 8C, and 9). In addition, we found that the HXT1 gene, which is expressed in response to glucose (Johnston, 1999), is transcribed normally in sda1-2 mutants at the restrictive temperature (our unpublished results). Similarly, the arrest does not appear to be due to general defects in protein synthesis, because we found that mRNAs expressed from the GAL1 promoter produce normal amounts of protein (Figure 9; our unpublished results), and the initial rate and pattern of protein synthesis after release from a nutrient arrest are nearly identical in wild-type and sda1-2 cells. It is interesting to note that the amount of Cln2 protein expressed from the Gal1 promotor is the same in wild-type and sda1-2 cells at the restrictive temperature, even though sda1-2 cells fail to show the large increase in protein synthesis observed for wild-type cells entering the cell cycle. This result seems to suggest that the protein synthesis machinery is functional in sda1-2 cells, but there is a failure in the initiation of increased synthesis of additional protein required for cell growth and division. The fact that sda1 mutants are able to respond to α factor further argues against defects in transcription or translation, because previous studies have demonstrated that transcription and translation are required for the response to α factor (Buehrer and Errede, 1997). It remains possible, however, that there are defects in the synthesis of specific proteins in sda1 mutants, or that there are subtle differences in the rate of protein synthesis. The fact that cells lacking Sda1 function remain viable for at least 24 h at the restrictive temperature argues that Sda1 does not play a role in a general metabolic function required for the viability of the cell.
To determine whether Sda1 may play a rate-limiting role in inducing entry into G1, we generated a strain that expresses Sda1 from the Gal1 promotor. We then tested whether overexpression of Sda1 advances the timing of bud emergence after release from a nutrient arrest, but observed no difference in the timing (our unpublished data). We also used flow cytometry to determine whether overexpression of Sda1 causes a decrease in cell size due to premature cell cycle entry, but observed no change in cell size (our unpublished data).
One possible model is that Sda1 functions in a signaling pathway that induces passage through Start by activating transcription of G1-specific genes. Previous work has shown that a large number of genes are specifically transcribed during G1 (Koch and Nasmyth, 1994; Spellman et al., 1998). All of these genes appear to be regulated by transcription complexes called SBF and MBF, and it appears that one of the primary functions of Cln3 is to activate transcription of SBF/MBF-dependent genes (Tyers et al., 1993; Dirick et al., 1995). We found that transcription of CLN3 is normal in sda1-2 cells, but transcription of CLN2 and PCL1, which are SBF/MBF-dependent genes, was greatly reduced. In addition, expression of CLN2 from the GAL1 promoter induces bud emergence in sda1-2 cells, but does not allow the cells to divide. Perhaps the most simple explanation for these results is that Sda1 is required for transcription of SBF/MBF-dependent genes. Transcription of CLN2 from a heterologous promoter would be expected to rescue the bud emergence defect because previous work has shown that Cln2 plays a key role in inducing actin polarization and bud emergence (Richardson et al., 1989; Lew and Reed, 1993). However, the cells would be unable to exit G1 because of a failure to transcribe other SBF/MBF-dependent genes. In this model, Sda1 could be functioning either directly in transcription, or in a signaling pathway that regulates transcription. The fact that sda1-2 cells exiting G0 fail to induce increased synthesis of proteins and rRNA perhaps suggests that Sda1 is part of a more general signaling pathway that controls initiation of a G1 program. To gain a better understanding of how Sda1 functions to control G1 events, it will be necessary to identify proteins that interact with Sda1. These experiments are currently in progress.
ACKNOWLEDGMENTS
We thank Brandt Schneider, Mike Tyers, Linda Breeden, Kristin Baetz, Andrew Murray, and members of the Kellogg lab for helpful discussions; and Kristin Baetz for supplying the SWI4 plasmids. We also thank Bill Sullivan for rhodamine phalloidin, Erik Samayoa for assistance in carrying out the two-hybrid screen, and Gina Costa and Gary Fathman for help with flow cytometry. This work was supported by the National Institutes of Health and the Pew Charitable Trusts
REFERENCES
- Altman R, Kellogg DR. Control of mitotic events by Nap1 and the Gin4 kinase. J Cell Biol. 1997;138:119–130. doi: 10.1083/jcb.138.1.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson CW, Baum PR, Gesteland RF. Processing of adenovirus 2-induced proteins. J Virol. 1973;12:241–252. doi: 10.1128/jvi.12.2.241-252.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baroni M, Monti P, Alberghina L. Repression of growth-regulated G1 cyclin expression by cyclic AMP in budding yeast. Nature. 1994;371:339–342. doi: 10.1038/371339a0. [DOI] [PubMed] [Google Scholar]
- Benton BK, Tinkelenberg AH, Gonzalez I, Cross FR. Cla4p, a Saccharomyces cerevisiae Cdc42p-activated kinase involved in cytokinesis, is activated at mitosis. Mol Cell Biol. 1997;17:5067–5076. doi: 10.1128/mcb.17.9.5067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benton BK, Tinkelenberg AH, Jean D, Plump SD, Cross FR. Genetic analysis of Cln/Cdc28 regulation of cell morphogenesis in budding yeast. EMBO J. 1993;12:5267–5275. doi: 10.1002/j.1460-2075.1993.tb06222.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breeden L, Mikesell G. Three independent forms of regulation affect expression of HO, CLN1 and CLN2 during the cell cycle of Saccharomyces cerevisiae. Genetics. 1994;138:1015–1024. doi: 10.1093/genetics/138.4.1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broek D, Toda T, Michaeli T, Levin L, Birchmeier C, Zoller M, Powers S, Wigler M. The S. cerevisiae CDC25 gene product regulates the RAS/adenylate cyclase pathway. Cell. 1987;48:789–799. doi: 10.1016/0092-8674(87)90076-6. [DOI] [PubMed] [Google Scholar]
- Buehrer BM, Errede B. Coordination of the mating and cell integrity mitogen-activated protein kinase pathways in Saccharomyces cerevisiae. Mol Cell Biol. 1997;17:6517–6525. doi: 10.1128/mcb.17.11.6517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buscemi G, Saracino F, Masnada D, Agostini Carbone M. The Saccharomyces cerevisiae SDA1 gene is required for actin cytoskeleton organization and cell cycle progression. J Cell Sci. 2000;113:1199–1211. doi: 10.1242/jcs.113.7.1199. [DOI] [PubMed] [Google Scholar]
- Cross FR. DAFI, a mutant gene affecting size control, pheromone arrest, and cell cycle kinetics of Saccharomyces cerevisiae. Mol Cell Biol. 1988;8:4675–4684. doi: 10.1128/mcb.8.11.4675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross F. Cell cycle arrest caused by CLN gene deficiency in Saccharomyces cerevisiae resembles START-I arrest and is independent of the mating-pheromone signaling pathway. Mol Cell Biol. 1990;10:6482–6490. doi: 10.1128/mcb.10.12.6482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross F. Starting the cell cycle: what's the point? Curr Opin Cell Biol. 1995;7:790–797. doi: 10.1016/0955-0674(95)80062-x. [DOI] [PubMed] [Google Scholar]
- Cross FR, Tinkelenberg AH. A potential positive feedback loop controlling CLN1 and CLN2 gene expression at the start of the yeast cell cycle. Cell. 1991;65:875–883. doi: 10.1016/0092-8674(91)90394-e. [DOI] [PubMed] [Google Scholar]
- Cvrckova F, De Virgilio C, Manser E, Pringle JR, Nasmyth K. Ste20-like protein kinases are required for normal localization of cell growth and for cytokinesis in budding yeast. Genes Dev. 1995;9:1817–1830. doi: 10.1101/gad.9.15.1817. [DOI] [PubMed] [Google Scholar]
- Cvrckova F, Nasmyth K. Yeast G1 cyclins CLN1 and CLN2 and a GAP-like protein have a role in bud formation. EMBO J. 1993;12:5277–5286. doi: 10.1002/j.1460-2075.1993.tb06223.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawes, I., and Calvert, G. (1984). Initiation of sporulation in Saccharomyces cerevesiae. Mutations causing derepressed sporulation and G1 arrest in the cell division cycle. J. Gen. Microbiol. 130, . [DOI] [PubMed]
- Di Como CJ, Chang H, Arndt KT. Activation of CLN1 and CLN2 G1 cyclin gene expression by BCK2. Mol Cell Biol. 1995;15:1835–1846. doi: 10.1128/mcb.15.4.1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dirick L, Bohm T, Nasmyth K. Roles and regulation of Cln-Cdc28 kinases at the start of the cell cycle of Saccharomyces cerevisiae. EMBO J. 1995;14:4803–4813. doi: 10.1002/j.1460-2075.1995.tb00162.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Futcher B. Cyclins and the wiring of the yeast cell cycle. Yeast. 1996;12:1635–1646. doi: 10.1002/(SICI)1097-0061(199612)12:16%3C1635::AID-YEA83%3E3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- Guthrie, C., and Fink, G. (1991). Guide to yeast genetics and molecular biology. 194, 273–372. [PubMed]
- Hadwiger JA, Wittenberg C, Richardson HE, de Barros-Lopes M, Reed SI. A novel family of cyclin homologs that control G1 in yeast. Proc Natl Acad Sci USA. 1989;86:6255–6259. doi: 10.1073/pnas.86.16.6255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harlow E, Lane D. Antibodies. A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1988. [Google Scholar]
- Hartwell LH, Culotti J, Pringle JR, Reid BJ. Genetic control of cell division cycle in yeast. Science. 1974;183:46–51. doi: 10.1126/science.183.4120.46. [DOI] [PubMed] [Google Scholar]
- Hubler L, Bradshaw-Rouse J, Heideman W. Connections between the Ras-cyclic AMP pathway and G1 cyclin expression in the budding yeast Saccharomyces cerevisiae. Mol Cell Biol. 1993;13:6274–6282. doi: 10.1128/mcb.13.10.6274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston M. Feasting, fasting, and fermenting. Trends Genet. 1999;15:29–33. doi: 10.1016/s0168-9525(98)01637-0. [DOI] [PubMed] [Google Scholar]
- Johnston G, Singer R. Regulation of proliferation by the budding yeast Saccharomyces cerevisiae. Biochem Cell Biol. 1990;68:427–435. doi: 10.1139/o90-060. [DOI] [PubMed] [Google Scholar]
- Kellogg DR, Alberts BM. Purification of a multiprotein complex containing centrosomal proteins from the Drosophila embryo by chromatography with low-affinity polyclonal antibodies. Mol Biol Cell. 1992;3:1–11. doi: 10.1091/mbc.3.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellogg DR, Kikuchi A, Fujii-Nakata T, Turck CW, Murray A. Members of the NAP/SET family of proteins interact specifically with B-type cyclins. J Cell Biol. 1995;130:661–673. doi: 10.1083/jcb.130.3.661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellogg DR, Murray AW. NAP1 acts with Clb2 to perform mitotic functions and suppress polar bud growth in budding yeast. J Cell Biol. 1995;130:675–685. doi: 10.1083/jcb.130.3.675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch C, Nasmyth K. Cell cycle regulated transcription in yeast. Curr Opin Cell Biol. 1994;6:451–459. doi: 10.1016/0955-0674(94)90039-6. [DOI] [PubMed] [Google Scholar]
- Lew DJ, Reed SI. Morphogenesis in the yeast cell cycle: regulation by Cdc28 and cyclins. J Cell Biol. 1993;120:1305–1320. doi: 10.1083/jcb.120.6.1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine MS, McKenzie A, DeMarini D, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Longtine MS, Theesfeld CL, McMillan JN, Weaver E, Pringle JR, Lew DJ. Septin-dependent assembly of a cell cycle-regulatory module in Saccharomyces cerevisiae. Mol Cell Biol. 2000;20:4049–4061. doi: 10.1128/mcb.20.11.4049-4061.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madhani HD, Fink GH. The riddle of MAP kinase signaling specificity. Trends Genet. 1998;14:151–155. doi: 10.1016/s0168-9525(98)01425-5. [DOI] [PubMed] [Google Scholar]
- Muhlrad D, Hunter R, Parker R. A rapid method for localized mutagenesis of yeast genes. Yeast. 1992;8:79–82. doi: 10.1002/yea.320080202. [DOI] [PubMed] [Google Scholar]
- Murray A, Hunt T. The Cell Cycle: An Introduction. New York: Oxford University Press; 1993. [Google Scholar]
- Nash R, Tokiwa G, Anand S, Erickson K, Futcher AB. The WHI1+ gene of Saccharomyces cerevisiae tethers cell division to cell size and is a cyclin homolog. EMBO J. 1988;7:4335–4346. doi: 10.1002/j.1460-2075.1988.tb03332.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasmyth K, Dirick L. The role of SWI4 and SWI6 in the activity of G1 cyclins in yeast. Cell. 1991;66:995–1013. doi: 10.1016/0092-8674(91)90444-4. [DOI] [PubMed] [Google Scholar]
- Ogas J, Andrews BJ, Herskowitz I. Transcriptional regulation of CLN1 and CLN2 and a putative new G1 cyclin (HCS26) by SWI4, a positive regulator of G1-specific transcription. Cell. 1991;66:1015–1026. doi: 10.1016/0092-8674(91)90445-5. [DOI] [PubMed] [Google Scholar]
- Pardee AB. G1 events and regulation of cell proliferation. Science. 1989;246:603–608. doi: 10.1126/science.2683075. [DOI] [PubMed] [Google Scholar]
- Pardee AB, Dunbrow R, Hamlin JL, Kletzein RF. Animal cell cycle. Annu Rev Biochem. 1978;47:715–750. doi: 10.1146/annurev.bi.47.070178.003435. [DOI] [PubMed] [Google Scholar]
- Partridge JF, Mikesell GE, Breeden LL. Cell cycle dependent transcription of CLN1 involves Swi4 binding to MCB-like elements. J Biol Chem. 1997;272:9071–9077. doi: 10.1074/jbc.272.14.9071. [DOI] [PubMed] [Google Scholar]
- Planas-Silva MD, Weinberg RA. The restriction point and control of cell proliferation. Curr Opin Cell Biol. 1997;9:768–772. doi: 10.1016/s0955-0674(97)80076-2. [DOI] [PubMed] [Google Scholar]
- Pringle JR, Adams AEM, Drubin DG, Haarer BK. Immunofluorescence methods for yeast. Methods Enzymol. 1991;194:565–602. doi: 10.1016/0076-6879(91)94043-c. [DOI] [PubMed] [Google Scholar]
- Pringle JR, Hartwell LH. The Saccharomyces cerevisiae cell cycle. In: Strathern JN, Jones EW, Broach JR, editors. The Molecular Biology of the Yeast Saccharomyces - Life Cycle and Inheritance. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1981. pp. 97–142. [Google Scholar]
- Pruyne D, Bretscher A. Polarization of cell growth in yeast II. J Cell Sci. 2000;113:571–585. doi: 10.1242/jcs.113.4.571. [DOI] [PubMed] [Google Scholar]
- Reed SI. The selection of S. cerevisiae mutants defective in the start event of cell division. Genetics. 1980;95:561–577. doi: 10.1093/genetics/95.3.561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid BH, Hartwell LH. Regulation of mating in the cell cycle of Saccharomyces cerevisiae. J Cell Biol. 1977;75:355–365. doi: 10.1083/jcb.75.2.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson HE, Wittenberg CW, Cross F, Reed SI. An essential G1 function for cyclin-like proteins in yeast. Cell. 1989;59:1127–1133. doi: 10.1016/0092-8674(89)90768-x. [DOI] [PubMed] [Google Scholar]
- Shulewitz MJ, Inouye CJ, Thorner J. Hsl7 localizes to a septin ring and serves as an adapter in regulatory pathway that relieves tyrosine phosphorylation of Cdc28 protein kinase in Saccharomyces cerevisiae. Mol Cell Biol. 1999;19:7123–7137. doi: 10.1128/mcb.19.10.7123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spellman PT, Sherlock G, Zhang MQ, Iyer VR, Anders K, Eisen MB, Brown PO, Botstein D, Futcher B. Comprehensive identification of cell cycle-regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Mol Biol Cell. 1998;9:3273–3297. doi: 10.1091/mbc.9.12.3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart D, Wittenberg C. CLN3, not positive feedback, determines the timing of CLN2 transcription in cyclin cells. Genes Dev. 1995;9:2780–2794. doi: 10.1101/gad.9.22.2780. [DOI] [PubMed] [Google Scholar]
- Tan PBO, Kim S. Signaling specificity the RTK/RAS/MAP kinase pathway in metazoans. Trends Genet. 1999;15:145–149. doi: 10.1016/s0168-9525(99)01694-7. [DOI] [PubMed] [Google Scholar]
- Thevelein JM, de Winde JH. Novel sensing mechanisms and targets for the cAMP-protein kinase A pathway in the yeast Saccharomyces cerevisiae. Mol Microbiol. 1999;33:904–918. doi: 10.1046/j.1365-2958.1999.01538.x. [DOI] [PubMed] [Google Scholar]
- Tjandra H, Compton J, Kellogg DR. Control of mitotic events by the Cdc42 GTPase, the Clb2 cyclin and a member of the PAK kinase family. Curr Biol. 1998;8:991–1000. doi: 10.1016/s0960-9822(07)00419-8. [DOI] [PubMed] [Google Scholar]
- Tokiwa G, Tyers M, Volpe T, Futcher B. Inhibition of G1 cyclin activity by the Ras/cAMP pathway in yeast. Nature. 1994;371:342–345. doi: 10.1038/371342a0. [DOI] [PubMed] [Google Scholar]
- Tyers M, Tokiwa G, Futcher B. Comparison of the Saccharomyces cerevisiae G1 cyclins: Cln3 may be an upstream activator of Cln1, Cln2 and other cyclins. EMBO J. 1993;12:1955–1968. doi: 10.1002/j.1460-2075.1993.tb05845.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyers M, Tokiwa G, Nash R, Futcher B. The Cln3-Cdc28 kinase complex of S. cerevisiae is regulated by proteolysis and phosphorylation. EMBO J. 1992;11:1773–1784. doi: 10.1002/j.1460-2075.1992.tb05229.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner-Washburne M, Braun E, Johnston G, Singer R. Stationary phase in the yeast Saccharomyces cerevisiae. Microbiol Rev. 1993;57:383–401. doi: 10.1128/mr.57.2.383-401.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijnen H, Futcher B. Genetic analysis of the shared role of CLN3 and BCK2 at the G(1)-S transition in Saccharomyces cerevisiae. Genetics. 1999;153:1131–1143. doi: 10.1093/genetics/153.3.1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittenberg C, Sugimoto K, Reed SI. G1-specific cyclins of S. cerevisiae: cell cycle periodicity, regulation by mating pheromone, and association with the p34CDC28 protein kinase. Cell. 1990;62:225–237. doi: 10.1016/0092-8674(90)90361-h. [DOI] [PubMed] [Google Scholar]